Proteomics Analysis Reveals Stable Multiprotein Complexes in Both Fission and Budding Yeasts Containing Myb-Related Cdc5p/Cef1p, Novel Pre-mRNA Splicing Factors, and snRNAs (original) (raw)
Abstract
Schizosaccharomyces pombe Cdc5p and its Saccharomyces cerevisiae ortholog, Cef1p, are essential Myb-related proteins implicated in pre-mRNA splicing and contained within large multiprotein complexes. Here we describe the tandem affinity purification (TAP) of Cdc5p- and Cef1p-associated complexes. Using transmission electron microscopy, we show that the purified Cdc5p complex is a discrete structure. The components of the S. pombe Cdc5p/S. cerevisiae Cef1p complexes (termed Cwfs or Cwcs, respectively) were identified using direct analysis of large protein complex (DALPC) mass spectrometry (A. J. Link et al., Nat. Biotechnol. 17:676-682, 1999). At least 26 proteins were detected in the Cdc5p/Cef1p complexes. Comparison of the polypeptides identified by S. pombe Cdc5p purification with those identified by S. cerevisiae Cef1p purification indicates that these two yeast complexes are nearly identical in composition. The majority of S. pombe Cwf proteins and S. cerevisiae Cwc proteins are known pre-mRNA splicing factors including core Sm and U2 and U5 snRNP components. In addition, the complex contains the U2, U5, and U6 snRNAs. Previously uncharacterized proteins were also identified, and we provide evidence that several of these novel factors are involved in pre-mRNA splicing. Our data represent the first comprehensive analysis of CDC5-associated proteins in yeasts, describe a discrete highly conserved complex containing novel pre-mRNA splicing factors, and demonstrate the power of DALPC for identification of components in multiprotein complexes.
The spliceosome is a dynamic multiprotein/RNA complex that catalyzes the excision of introns from pre-mRNA. pre-mRNA splicing requires five snRNA molecules (U1, U2, U4, U5, and U6) that are closely associated with conserved protein components. These snRNA/protein combinations are referred to as small nuclear ribonucleoprotein particles (snRNPs). snRNPs interact with each other and mRNAs in a sequential order during the splicing reaction. The current model for snRNP assembly into an active spliceosome was developed from in vitro splicing experiments (reviewed in references 36, 46, and 59). In addition to the five core snRNP complexes, pre-mRNA splicing requires many other protein factors. Learning the identity of these proteins and defining their roles will further our understanding of how cells regulate the important cellular process of pre-mRNA splicing.
The Schizosaccharomyces pombe cdc5+ gene was identified in the first screen for fission yeast mutants defective for cell cycle progression (50). At the restrictive temperature, cells harboring the temperature-sensitive _cdc5_-120 mutation become arrested in the G2 phase of the cycle. The cloning and initial characterization of cdc5+ showed that its function is essential for viability and that its predicted protein product shares significant homology with the DNA binding domain of the vertebrate proto-oncoprotein c-Myb (52). Sequence similarity to a family of known DNA binding proteins led us to initially suggest that S. pombe Cdc5p might be required for entry into mitosis via regulation of transcription (52).
Since the initial characterization of S. pombe cdc5+ , Cdc5p-related proteins have been isolated from Saccharomyces cerevisiae (called Cef1) (51), Aradbidopsis thaliana (31), Drosophila melanogaster (51), Caenorhabditis elegans (51), Xenopus laevis (61), and Homo sapiens (hCDC5) (8, 27, 51). Substantial evidence has accumulated to suggest that these Myb-related proteins play an essential role in RNA processing rather than transcription (3, 10, 69). Genetic depletion of S. pombe Cdc5p or _S. cerevisia_e Cef1p causes accumulation of unspliced mRNAs in vivo (9, 43, 69). Inactivation of Cef1p by antibody interference or immunodepletion of hCDC5 inhibits splicing in vitro (3, 69). Also, Cdc5p, Cef1p, and hCDC5 are associated with multiprotein complexes that contain known splicing factors (3, 43, 69).
As a first step toward understanding the function of S. pombe Cdc5p, a multiprotein complex was purified by immunochromatography of Cdc5p-hemagglutinin (HA), and the identities of 10 Cwf (complexed with Cdc5p) proteins were reported (43). Most of the Cdc5p complex members identified have homologs that have been implicated in the process of pre-mRNA splicing (43). S. cerevisiae Cef1p also resides in a large protein complex identified through immunoaffinity purification of the splicing factor Prp19p (62, 69). This complex, termed the Nineteen complex (Ntc), appears to contain fewer proteins than the S. pombe Cwf complex. The S. cerevisiae Ntc complex includes Prp19p, Cef1p, Snt309p (Ntc25p), Isy1p (Ntc30p), Ntc20p, and at least six other polypeptides that have not been identified (11, 12, 69). All identified Ntc components are pre-mRNA splicing factors. Like its yeast counterparts, human CDC5 copurifies in a large multiprotein complex (3). However, homologs of only four identified S. pombe Cwf proteins (Cwf1p, Cwf7p, Cwf8p, and Cwf9p) and one identified S. cerevisiae Ntc component (Prp19p) have thus far been identified among the 30 copurifying proteins. Further, S. pombe homologs of Ntc components other than Prp19p and Cef1p were not identified in the analysis of Cdc5p-associated proteins. Similarly, few S. cerevisiae homologs of the S. pombe Cwf components correspond to identified Ntc members. It is not clear, then, whether multiple Cdc5p-containing complexes exist in different organisms, or whether the full complement of Cdc5p-associated proteins is known for any organism.
In order to comprehensively identify proteins stably associated with S. pombe Cdc5p and S. cerevisiae Cef1p and to compare the composition of the S. pombe and S. cerevisiae complexes, we used tandem affinity purification (TAP) (55, 65) to rapidly purify the yeast complexes. Negative staining and transmission electron microscopy of the S. pombe Cdc5p-TAP complex revealed distinct particles of similar sizes and recurring shapes, confirming the presence of a bona fide complex. The complex was found to contain the U2, U5, and U6 snRNAs in addition to proteins. To identify the polypeptide composition of the complexes in S. pombe and S. cerevisiae, we used direct-analysis-of-large-protein-complexes (DALPC) tandem mass spectrometry (39). We found that the S. pombe Cwf-and the S. cerevisiae Cwc (complexed with Cef1p)-TAP complexes from the two yeasts are nearly identical in composition. To validate our findings, we chose a subset of the known and unknown proteins in the complexes and tested for their interaction with Cdc5p/Cef1p by conventional methods. Involvement in pre-mRNA splicing was also assessed for some proteins of uncharacterized function. Our data confirm the association of Cdc5p/Cef1p with snRNPs, providing further compelling evidence that these proteins function in pre-mRNA processing. Further, our data reveal a previously undefined spliceosomal complex, perhaps related to the active spliceosome, involved in pre-mRNA splicing, and they illustrate the power of DALPC for the identification of multiprotein complex components.
MATERIALS AND METHODS
Strains and media.
S. pombe strains used in this study (Table 1) were grown in yeast extract medium or EMM minimal medium with appropriate supplements (44). S. cerevisiae strains (Table 1) were grown either in synthetic minimal medium with the appropriate nutritional supplements or in YPD (29). Transformations were performed by the lithium acetate method (25, 34).
TABLE 1.
Strain list
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| S. pombe | ||
| KGY246 | ade6-M210 ura4-D18 leu1-32 _h_− | Lab stock |
| KGY1994 | cwf10::cwf10-TAP::Kanr _ade6-M210 ura4-D18 leu1-32 h_− | This study |
| KGY2182 | u2a::u2a-TAP::Kanr _ade6-M210 ura4-D18 leu1-32 h_− | This study |
| KGY2183 | lsm8::lsm8-TAP::Kanr _ade6-M210 ura4-D18 leu1-32 h_− | This study |
| KGY2509 | cdc5::cdc5-TAP::Kanr_ade6-M210 ura4-D18 leu1-32 h_− | This study |
| KGY2056 | cwf5::cwf5-myc13::Kanr_ade6-M210 ura4-D18 leu1-32 h_− | This study |
| KGY2874 | cwf7::cwf7-HA:Kanr_ade6-M210 ura4-D18 leu1-32 h_− | This study |
| KGY3176 | cwf11::_cwf11_-HA:Kanr_ade6-M210 ura4-D18 leu1-32 h_− | This study |
| KGY3177 | cwf12::cwf12-HA:Kanr_ade6-M210 ura4-D18 leu1-32 h_− | This study |
| KGY2001 | cwf13::_cwf13-HA:_Kan_r ade6-M210 ura4-D18 leu1-32 h_− | This study |
| KGY2873 | cwf7::ura4+/cwf7+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 | This study |
| KGY3368 | cwf11::ura4+/cwf11+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 | This study |
| KGY2002 | cwf17::cwf17-HA:Kanr_ade6_-M210 _ura4-D18 leu1-32 h_− | This study |
| KGY3835 | prp4-l cdc5::cdc5-TAP::Kanr | This study |
| KGY3837 | prp1-1 cdc5::cdc5-TAP::Kanr | This study |
| KGY3838 | prp2-1 cdc5::cdc5-TAP::Kanr | This study |
| KGY3839 | prp12-1 cdc5::cdc5-TAP::Kanr | This study |
| S. cerevisiae | ||
| KGY823 | YPH98 MATaura3-52 lys2-801 ade2-101 leu2_-Δ_1 trp1_-Δ_1 | Lab stock |
| KGY1614 | _MAT_α _cef1-13 ade2-101 ade3_Δ ura3-52 lys2-801 trp1_-Δ_63 his3_-Δ_200 leu2_-Δ_1 | 51 |
| KGY2000 | MAT_α/MATa CWC2/CWC2::Kanr_ura3DO/ura3DO his3D1/his3D1 leuDO/leuDO met15DO/MET15 lysDO/lysDO | Research Genetics |
| KGY1989 | MATaCWC2::CWC2-TAP::Kanr_ura3-52 lys2-801 ade2-101 leu2_-Δ_1 trp1_-Δ_1_ | This study |
| KGY2080 | _MAT_α prp19-1 YDR163W::Kanr/pRS416PRP19 | This study |
| KGY2004 | MATaYCR063W::YCR063W-myc::Kanr_ura3-52 lys2-801 ade2-101 leu2_-Δ_1 trp1_-Δ_1_ | This study |
| KGY2006 | MATaYDR163W::YDR163W-myc::Kanr_ura3-52 lys2-801 ade2-101 leu2_-Δ_1 trp1_-Δ_1_ | This study |
| KGY2007 | MATaYPL064C::YPL064C-myc::Kanr_ura3-52 lys2-801 ade2-101 leu2_-Δ_1 trp1_-Δ_1_ | This study |
| KGY2100 | MAT_α CWC2::Kanr_ura3DO::CUP1-ha-CWC2 his3D1 leu2DO met15DO lysDO | This study |
| KGY2517 | MATaCEF1::CEF1-TAP::Kanr_ura3-52 lys2-801 ade2-101 leu2_-Δ_1 trp1_-Δ_1_ | This study |
| KGY2519 | MATaPRP19::PRP19-TAP::Kanr_ura3-52 lys2-801 ade2-101 leu2_-Δ_1 trp1_-Δ_1_ | This study |
| KGY2943 | MATaCWC1::CWC1-TAP::Kanr_ura3-52 lys2-801 ade2-101 leu2_-Δ_1 trp1_-Δ_1_ | This study |
| KGY2944 | MATaSNT309::SNT309-TAP::Kanr_ura3-52 lys2-801 ade2-101 leu2_-Δ_1 trp1_-Δ_1_ | This study |
In vivo tagging.
Strains expressing epitope tagged versions of Cdc5p, Cwf5p, Cwf7p, Cwf8p, Cwf10p, Lsm8p (SPCC1840.10), SPBC646.02 (now termed Cwf11p), SPBC32F12.05 (now termed Cwf12p), SPCC188.11 (now termed Cwf13p), SPBC1289.11 (now termed Cwf17p), SPBC1861.08 (now termed U2Ap), Cef1p, Prp19p, Snt309p, Prp46p/Cwc1p, YDL209C (now termed Cwc2p), YCR063W (now termed Cwc14p), and YDR163W (now termed Cwc15p) were constructed as described previously (4, 40, 65). Each open reading frame (ORF) was tagged at its endogenous locus at its 3′ end with the _myc13-_Kanr, _HA3-_Kanr, or _TAP-_Kanr cassette. Appropriate tagging was confirmed by immunoblotting and PCR. All tagged strains were viable at temperatures ranging from 25 to 36°C.
S. pombe gene disruptions.
cwf5+, cwf7+, and cwf11+ were deleted by using DNA fragments containing the ura4+ gene flanked by 80 bp both upstream and downstream of the ORFs (4). Ura+ transformants were checked for gene deletion by PCR. Heterozygous diploids were sporulated at 25°C, and tetrads were dissected at both 25 and 32°C to determine whether the genes were essential for viability.
Immunoprecipitations, immunoblotting, and sucrose gradients.
Protein lysates were prepared in NP-40 buffer as detailed previously (26). Immunoprecipitations with anti-HA or anti-myc were performed as described previously (43). Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 6 to 20% gradient gels or on Novex NuPAGE 4 to 12% bis-Tris gels (Invitrogen, Carlsbad, Calif.). For immunoblotting, proteins were transferred by electroblotting to a polyvinylidene difluoride membrane (Immobilon P; Millipore Corp., Bedford, Mass.). Affinity-purified anti-Cef1p serum VU136#9 was used at a 1:300 dilution. The anti-Cdc5p serum JAM and anti-HA (12CA5) and anti-myc (9E10) antibodies were used as described previously (43). Antibodies were detected by using horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse secondary antibodies (0.8 mg/ml; Jackson Immunoresearch Laboratories, West Grove, Pa.) at a dilution of 1:50,000. Immunoblots were visualized by using ECL reagents (Amersham Pharmacia Biotech).
A 200-μl volume corresponding to 20% of the isolated Cdc5-TAP complex was layered onto a 10 to 30% sucrose gradient and centrifuged at 28,000 rpm for 18 h in an SW50 Ti rotor. Fractions from these gradients were collected, mixed with sample buffer, and resolved by SDS-PAGE. Parallel gradients were run; these contained thyroglobulin (19S) and catalase (11.35S) (HWM Standards; Pharmacia) or 2 mg of lysate from S. cerevisiae, which was subsequently probed with antibodies recognizing FAS as a 40S marker (a gift from S. J. Wakil).
Purification of Cwf/Cwc complexes by TAP.
To identify components of the Cwf and Cwc complexes, 8-liter cultures of cdc5-TAP, CEF1-TAP, PRP19-TAP, SNT309-TAP, CWC2-TAP, or PRP46/CWC1-TAP strains were grown to log phase and the tagged proteins were isolated as described previously (65). For silver staining, one-half of the eluate from the purification was precipitated with trichloroacetic acid, resuspended in lithium dodecyl sulfate (LDS)-PAGE buffer, and resolved on a 4 to 12% NuPAGE gel by using morpholinepropanesulfonic acid (MOPS) buffer (Novex). Silver staining was carried out by using the Plus One kit (Amersham Pharmacia Biotech) as recommended by the manufacturer.
Analysis of TAP complexes by DALPC-tandem mass spectrometry.
TAP complexes were precipitated with trichloroacetic acid and resuspended in 100 mM ammonium bicarbonate-5% acetonitrile. Purified proteins were reduced and alkylated with dithiothreitol and iodoacetamide, followed by digestion with modified sequencing-grade trypsin (Promega). Tryptic peptides were desalted using a reversed-phase (RP) cartridge (Michrom Bioresources, Auburn, Calif.), lyophilized, and resolubilized in 0.5% acetic acid. The entire peptide mixture was loaded onto a 75-μm (internal diameter) strong cation-exchange column (Partisphere SCX; Whatman) equilibrated in 0.5% acetic acid-2% acetonitrile. Iterative peptide fractions were displaced by using increasing salt step gradients of 250 mM ammonium acetate, 2% acetonitrile, and 0.5% acetic acid (0, 10, 20, 30, 40, 60, 80, and 100% 500 mM KCl and 1 M NaCl) at a flow rate of 1 μl/min. Each fraction (5 of 6 μl) was loaded onto a 75-μm (internal diameter) RP-high-pressure liquid chromatography (HPLC) column (Poros R2; Perceptive Biosystems) equilibrated in 0.5% acetic acid. Peptides were eluted by using a linear gradient of 0 to 40% acetonitrile over 60 min followed by 40 to 60% over 10 min at a flow rate of 0.3 μl/min. Eluting peptides were analyzed by electrospray ionization tandem mass spectrometry using an ion trap mass spectrometer (LCQ Deca; ThermoFinnigan) (24). All tandem spectra were searched against either the S. cerevisiae ORF database (SGD, Stanford University) or a merged S. pombe protein database (the Sanger Centre's pombe protein database combined with the S. pombe protein subset of the National Center for Biotechnology Information protein database) using the SEQUEST algorithm (20). Data processing of the SEQUEST output files into a list of proteins present in the TAP complex was performed as described previously (39).
RNA and Northern blotting.
To determine if cwf7+ was required for pre-mRNA splicing, the heterozygous diploid cwf7::ura4+ _/cwf7+ leu1_-_32/leu1_-_32 ura4+/ura4+ ade6-M210/ade6-M216 h+/h_− was grown to mid-log phase in minimal medium supplemented with leucine. The culture was then washed several times in minimal medium lacking nitrogen but containing leucine. Following complete sporulation of the culture and the breakdown of asci, spores were collected and reinoculated into minimal medium supplemented with adenine and leucine at 32°C. Under these conditions, only cwf7::ura4+ spores will germinate and grow.
Total RNA was prepared from cells by extraction with hot acidic phenol as described previously (16). To detect mRNAs, total RNA was resolved on formaldehyde-agarose gels and then capillary blotted to a Duralon-UV membrane (Stratagene) and UV cross-linked (UV Stratalinker 1800; Stratagene). For S. pombe, TFIID and HIS3 RNAs were detected by using 32P-labeled oligonucleotides complementary to both intronic and exonic sequences under conditions described previously (41, 53). For snRNAs, samples were resolved on a 6% Tris-borate-EDTA-urea gel (Invitrogen), transferred to a Duralon-UV membrane, and detected by using 32P-labeled oligonucleotides complementary to S. pombe U1 (SPU1), U2 (U2B), U4 (SPU4), U5 (YU5), and the exon of U6 (U6E). Blots were exposed to PhosphorImager screens and visualized by using MD IMAGE QUANT, version 3.3 (Molecular Dynamics). Reverse transcription-PCR (RT-PCR) was performed with a OneStep RT-PCR kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's directions. Five hundred nanograms of RNA was used for each reaction. Oligonucleotides specific to the third and fourth exons of cdc2+ were used to detect either spliced or unspliced transcript. The 5′ primer was 5′-AACTGGGGCCACTAGTTTAG-3′, and the 3′ primer was 5′-CACTAATGCGATGGGCAGGG-3′.
Negative-stain electron microscopy.
A 5-μl drop of a Cdc5p-TAP at a 1/5 dilution was adsorbed to a freshly glow-discharged, carbon-stabilized, Formvar-coated 400-mesh copper grid for 2 min. The sample was blotted, rinsed twice in droplets of elution buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM imidazole, 2 mM EGTA, 10 mM β-mercaptoethanol), and stained for 30 s with 0.75% uranyl formate; then it was thoroughly dried prior to being viewed under a JEOL 1200 EX electron microscope.
RESULTS
Purification of Cwf complex components by TAP tag purification.
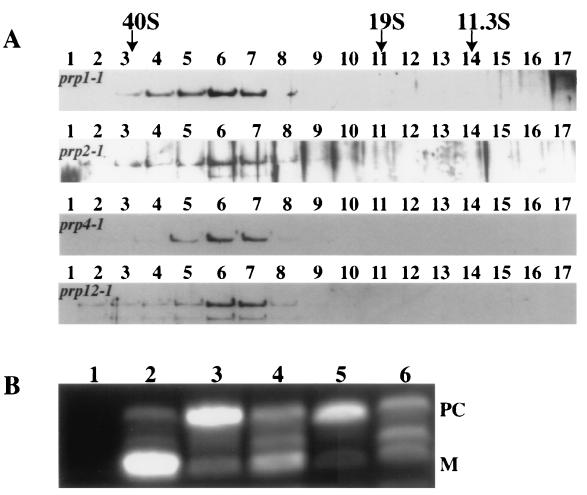
To determine the composition of the S. pombe and S. cerevisiae Cdc5p/Cef1p complexes and to address whether there are multiple Cdc5p/Cef1p-containing complexes in these organisms, we chose to TAP tag, purify, and identify proteins associated with six previously described components of the Cwf or Ntc complexes. To this end, the S. pombe cdc5+ and the S. cerevisiae CEF1, PRP19, SNT309, PRP46/CWC1, and CWC2 endogenous loci were engineered to encode proteins with a C-terminal TAP tag epitope. These six proteins were targeted for the following reasons. First, identification of S. pombe Cdc5p and S. cerevisiae Cef1p-associated proteins would allow us to compare complex composition between yeasts. Second, proteins associated with S. cerevisiae Prp19p and Snt309p, previously identified Ntc components that associate directly with each other, were of interest because they would indicate differences between the S. cerevisiae Ntc and S. pombe Cwf complexes. Finally, S. cerevisiae Prp46p/Cwc1p- and Cwc2p-associated proteins were investigated because both Prp46p/Cwc1p and Cwc2p are associated with S. cerevisiae Cef1p (M. D. Ohi and K. L. Gould, unpublished data) but have not been identified as Ntc components, again allowing us to probe for potential differences between the S. cerevisiae Cwc and S. cerevisiae Ntc complexes. In all cases, strains producing the epitope-tagged proteins exhibited growth rates and morphologies identical to those of the parental strains. The two-step immunoglobulin G (IgG)-Sepharose and calmodulin resin affinity steps were carried out as described previously (65) on 8-liter cultures of each strain. To confirm the presence of a complex in the S. pombe Cdc5p-TAP and to determine what fraction of Cdc5p was found associated with the complex, we analyzed the initial lysate and the final eluate of the purification by sucrose gradient centrifugation (Fig. 1A). All Cdc5p in both the lysate and the final eluate migrated in a discrete peak between the 19S and 40S markers (Fig. 1A). The protein composition of each fraction from the Cdc5-TAP gradient was examined by silver stain analysis. As predicted, multiple proteins migrated in a discrete peak between the 19S and 40S markers but not in other regions of the gradient (Fig. 1B).
FIG. 1.
Characterization of the TAP complexes. (A) Protein lysates from the Cdc5p-TAP-producing strain (upper panel) or the Cdc5p-TAP complex following its purification (lower panel) were resolved on a 10 to 30% sucrose gradient, and fractions were collected from the bottom (fraction 1). These were resolved by SDS-PAGE and immunoblotted with anti-Cdc5p serum. The migrations of FAS (40S), thyroglobulin (19S), and catalase (11.3S) collected from parallel gradients are indicated. (B) Silver-stained gel of fractions of the 10 to 30% sucrose gradient resolving the Cdc5p-TAP complex. Asterisks indicate fractions containing multiple peptides. Brackets labeled with the letter K indicate trace amounts of unavoidable human keratin contamination. (C) Silver-stained gels of a portion of each TAP complex. The protein compositions of mock purifications from wild-type S. pombe (KGY246) and wild-type S. cerevisiae (YPH09) were also examined by silver staining. (D) Electron microscopic analysis of the Cdc5p-TAP complex negatively stained with 0.75% uranyl formate. Shown is a gallery of selected particles representing different views of the complex. (E) Cdc5p-TAP associates with U2, U5, and U6 snRNAs. RNA was isolated from Cdc5p-TAP and from an anti-snRNA cap (antitrimethylguanosine [m3G]) immunoprecipitation from wild-type cells. Blots were probed with 32P-labeled oligonucleotides complementary to the S. pombe U1, U2, U4, U5, and U6 snRNAs. In the cases of the U2, U5, and U6 snRNAs for the TAP samples, exposures were 1/24 of the others. (F) Prp19p-TAP associates with U2, U5, and U6 snRNAs. RNA was isolated from Prp19p-TAP and from an anti-snRNA cap (anti-m3G) immunoprecipitation from wild-type cells. Blots were probed with 32P-labeled oligonucleotides complementary to the S. cerevisiae U1, U2, U4, U5, and U6 snRNAs. In the cases of the U2, U5, and U6 snRNAs for the TAP samples, exposures were 1/12 of the others.
The protein composition of each TAP complex was also examined directly by silver staining. In each case, multiple polypeptides were detected and the patterns of polypeptides from different S. cerevisiae purifications were very similar (Fig. 1C). Transmission electron microscopy was used to visualize the structure of the purified S. pombe Cdc5-TAP complex (Fig. 1D). Staining of the complex with uranyl formate revealed distinct particles of uniform size and a reproducible but complex asymmetrical shape (Fig. 1D). These different images most likely reflect the ability of the S. pombe Cdc5p-TAP complex to adsorb to the grid surface in many different orientations.
We previously detected snRNAs associated with the S. pombe Cdc5p-HA complex (43), whereas the S. cerevisiae Ntc complex has been reported not to associate with snRNAs (14, 64). To address this potential difference between the S. pombe Cwf and the S. cerevisiae Cwc and Ntc complexes, we analyzed the snRNA composition of S. pombe Cdc5p-TAP and S. cerevisiae Prp19p-TAP. snRNAs were not detected in an Arp2p-TAP (data not shown), demonstrating that the TAP protocol does not indiscriminately purify snRNAs. The U2, U5, and U6 snRNAs, but not the U1 or U4 snRNAs, were detected in both S. pombe Cdc5p-TAP (Fig. 1E) and S. cerevisiae Prp19-TAP (Fig. 1F).
The association of snRNAs with the Cwf and Cwc complexes led us to examine if Cdc5p complex stability is RNA dependent. To this end the Cdc5p-TAP complex was treated with RNase A and then analyzed by sucrose gradient sedimentation. The RNase A-treated Cdc5p-TAP complex comigrated with a mock-treated Cdc5p-TAP complex and contained the U2, U5, and U6 snRNAs in levels equal to those in the mock-treated sample (data not shown). These data suggest that the snRNAs in the Cwf complex are protected from RNase digestion.
Multidimensional microcapillary HPLC fractionation of the trypsin-digested TAP complexes coupled with electrospray ionization tandem mass spectrometry was then used to identify the polypeptide composition of the complexes (39). Proteins present in a mock TAP performed on a strain lacking tagged proteins (data not shown) were subtracted as nonspecific contaminants. Twenty-six proteins that copurified with at least four of the six targeted proteins were identified (Table 2). The 10 S. pombe Cwf proteins previously found to associate with Cdc5p (43) (Table 2, proteins 1 to 11) were again found in the S. pombe Cdc5p purification, and, with the exception of Cwf7p, their S. cerevisiae homologs were identified in each of the S. cerevisiae TAP tag purifications. S. cerevisiae lacks an obvious homolog of Cwf7p. Similarly, homologs of all previously identified S. cerevisiae Ntc complex members (Table 2, proteins 12 to 14) were found associated with S. pombe Cdc5p, with the exception of Snt309p and Ntc20p, which are without obvious S. pombe counterparts. The similarities between the S. pombe Cdc5p and S. cerevisiae Cef1p components suggest that the Cwf complex and the Cwc complex are nearly identical in composition. In addition to the previously identified Cwf and Ntc components, 13 other proteins were identified (Table 2, proteins 15 to 27). Of these, 10 are recognized pre-mRNA splicing factors, including known core Sm proteins and U2 and U5 snRNP components (Table 2, proteins 15 to 24). For both S. pombe and S. cerevisiae, three conserved components are uncharacterized proteins and have been named Cwf14 to Cwf16 in S. pombe and Cwc14 to Cwc16 in S. cerevisiae (Table 2, proteins 25 to 27).
TABLE 2.
Cwc and Cwf proteins identified by DALPC mass spectrometry
| No. | S. cerevisiae Cwc protein (accession no.a) | S. pombe homolog (accession no.a) | Mol wt (kDa) (S. cerevisiae) | Essential?b | Function or motif | No. of hitsc | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cdc5 | Cef1 | Prp19 | Snt309 | Cwc2 | Cwc1 | ||||||
| 1 | Cef1p (Q03654) | Cdc5p (P39964) | 68 | Yes | Required for splicing; Myb repeats | 35 | 11 | 13 | 14 | 16 | 19 |
| 2 | Prp46p/Cwc1p (Q12417) | Prp5p/Cwf1p (AAG01399) | 51 | Yes | WD-40 repeats | 22 | 20 | 13 | 16 | 14 | 20 |
| 3 | Cwc2p (CAA98787) | Cwf2p (P87126) | 41 | Yes | RRMd domains | 11 | 5 | 10 | 1 | 3 | 3 |
| 4 | Syf1p (Q04048) | Cwf3p (Q9P7R9) | 100 | Yes | Required for splicing; TPR repeats | 34 | 18 | 24 | 11 | 1 | 18 |
| 5 | C1f1p (Q12309) | Cwf4p (P87312) | 82 | Yes | Required for splicing; TPR repeats | 28 | 16 | 18 | 9 | 7 | 19 |
| 6 | Ecm2p (P38241) | Cwf5p (O59800) | 41 | Nots | Splicing factor; RRM domain | 15 | 8 | 11 | 5 | 8 | 1 |
| 7 | Prp8p (P33334) | Cwf6p (O10752) | 279 | Yes | Splicing factor, S-phase progression, U5 snRNP protein; polyproline domains | 92 | 38 | 67 | 18 | 7 | 16 |
| 8 | —c | Cwf7p (Q9USV3) | 21 | Yes | Required for splicing | 15 | — | — | — | — | — |
| 9 | Prp19p (P32523) | Cwf8p (O14011) | 51 | Yes | Splicing factor, U box, WD-40 repeats | 24 | 17 | 22 | 18 | 15 | 17 |
| 10 | SmD2p (Q06217) | Cwf9p (O14036) | 13 | Yes | Core Sm protein | 5 | 3 | 3 | 1 | 2 | 3 |
| 11 | Snu114p (P36048) | Cwf10p (SPBC215.12) | 114 | Yes | U5 snRNP component; GTP-binding domain | 38 | 24 | 34 | 12 | 11 | 9 |
| 12 | Isy1p/Ntc30p (P21374) | Cwf12p (O74370) | 28 | No | Auxiliary splicing factor, 2-hybrid interaction with Syf1p, Syf3p, and Cef1p | 9 | 2 | 6 | 3 | 7 | 5 |
| 13 | Snt309p/Ntc25p (Q06091) | — | 21 | Nots | Splicing factor | 9 | 7 | 4 | 13 | 12 | |
| 14 | Ntc20p (P38302) | — | 16 | No | Auxiliary splicing factor | 4 | 3 | 3 | 4 | 1 | |
| 15 | Lea1p (Q08963) | U2Ap (Q9USX8) | 27 | Nots | U2A protein, member of U2 snRNP | 10 | 6 | 5 | 4 | 5 | 4 |
| 16 | Msl1p (P40567) | (O13649) | 13 | No | U2 snRNP component | 1 | 2 | 3 | 2 | 1 | 1 |
| 17 | Brr2p (P32639) | (Q9UT24) | 246 | Yes | Required for splicing, U5 snRNP; DEAD/DEAH RNA helicase | 15 | 17 | 42 | 6 | 1 | 3 |
| 18 | Slu7p (Q02775) | (Q9Y7Y2) | 45 | Yes | 3′ splice choice in 2nd step in splicing | 2 | 1 | 1 | 1 | 2 | N |
| 19 | Syf2p (P53277) | (O59733) | 25 | No | 2-hybrid interaction with Syf1, Syf3, and Cef1p | 7 | 2 | 6 | 1 | 4 | 1 |
| 20 | Prp17p/Cdc40p (P40968) | Prp17p (O43071) | 52 | Nots | Splicing, G2/M progression; WD-40 repeats | 26 | 5 | 7 | 3 | 1 | N |
| 21 | Prp45p (P28004) | Cwf13p (Q09882) | 43 | Yes | SNW domain, polyproline domain, SH2-like domain | 13 | 16 | 13 | 12 | 18 | 18 |
| 22 | Smx3p (P54999) | Smf1p (Q59734) | 9.7 | Yes | Core Sm protein | 2 | 2 | 2 | 2 | 1 | N |
| 23 | SmB1p (P40018) | (Q10163) | 22 | Yes | Core Sm protein | 1 | 5 | 4 | 3 | 6 | 2 |
| 24 | SmD3p (P43321) | Smd3p (Q9UUC6) | 11 | Yes | Core Sm protein | 4 | 2 | 3 | 3 | 3 | N |
| 25 | Cwc14p (P25337) | Cwf14p (O74772) | 18 | No | Hypothetical protein; potential NLSf putative Zn finger | 4 | 2 | 1 | 1 | 1 | 1 |
| 26 | Cwc15p (Q03772) | Cwf15p (O74817) | 20 | No | Hypothetical protein | 6 | 3 | 4 | 3 | 5 | N |
| 27 | Cwc16p (P28320) | Cwf16p (Q9P7C5) | 32 | Yes | Hypothetical protein | 5 | 5 | 8 | N | 6 | 1 |
Proteins unique to the S. pombe Cdc5p purification are listed in Table 3. A homolog of a human U5 snRNP member (SPC1289.11) was detected, as were four hypothetical proteins. These have been named Cwf11p and Cwf17p to Cwf20p. None of these proteins have obvious S. cerevisiae homologs.
TABLE 3.
Cdc5p-TAP specific proteins identified by DALPC mass spectrometry
| Cwf protein | SwissProt accession no. | Mol wt (kDa) | Essential? | Putative orthologs in other species (accession no.) |
|---|---|---|---|---|
| Cwf11p | O94508 | 147 | No | Hypothetical protein; H. sapiens (O60306), D. melanogaster (Q9VGG7), A. thaliana (Q9ZVJ8), C. elegans (Q9U1Q7) |
| Cwf17p | O94620 | 37.5 | Hypothetical protein; H. sapiens U5 novel WD-40 protein (O95320), D. melanogaster (Q9VPL0), A. thaliana (O22826), C. elegans (O002097) | |
| Cwf18p | Q9UU80 | 16.7 | Hypothetical protein | |
| Cwf19p | Q09909 | 74.4 | Hypothetical protein; D. melanogaster (Q9VXT5), A. thaliana (Q9C7K4), C. elegans (AAK18872) | |
| Cwf20p | Q9USK4 | 32.0 | Hypothetical protein |
In addition, 18 proteins that copurified with at least 3 of the 6 targeted proteins were identified (Table 4). Of these, 11 are recognized pre-mRNA splicing factors, including core Sm proteins and U2 snRNP components (Table 4, proteins 28 to 39). For both S. pombe and S. cerevisiae, seven conserved components are uncharacterized proteins and have been named Cwf21p to Cwf27p in S. pombe and Cwc21p to Cwc27p in S. cerevisiae (Table 4, proteins 40 to 46).
TABLE 4.
Potential Cwc and Cwf proteins identified by DALPC mass spectrometry
| No. | S. cerevisiae Cwc protein (accession no.a) | S. pombe homolog (accession no.a) | Mol wt (kDa) (S. cerevisiae) | Essential?b | Function or motif | No. of hitsc | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cdc5 | Cef1 | Prp19 | Snt309 | Cwc2 | Cwc1 | ||||||
| 28 | SmD1p (Q02260) | SmD1p (O42661) | 13 | Yes | Core Sm protein | 4 | 1 | 4 | N | 2 | N |
| 29 | Smx2p (P40204) | Smg1 (O74966) | 8 | Yes | Core Sm protein | 4 | 2 | N | 1 | N | N |
| 31 | Ist3p (P40565) | (O94290) | 21 | Yes | U2 snRNP-associated protein | 1 | 1 | 4 | N | 1 | N |
| 32 | Prp9p (P19736) | Sap61p (O59706) | 63 | Yes | U2 snRNP component | 2 | 2 | 5 | N | N | N |
| 33 | Prp11p (Q07350) | SAP62p (Q9P7L8) | 30 | Yes | U2 snRNP component | 3 | 1 | 3 | N | N | N |
| 34 | Prp21p (P32524) | SAP114p (O13900) | 33 | Yes | U2 snRNP component | 1 | 2 | 2 | N | N | N |
| 35 | Hsh155p (P49955) | Prp10p (Q10178) | 110 | Yes | U2 snRNP component | 4 | 1 | 8 | N | N | N |
| 36 | Cus1p (Q02554) | Sap145p (Q9UU13) | 50 | Yes | U2 snRNP component | 2 | N | 4 | 1 | N | N |
| 37 | Rse1p (Q04693) | Prp12p (Q9UTT2) | 154 | Yes | U2 snRNP component | 5 | 1 | 1 | N | N | N |
| 38 | Spp2p (Q02521) | (O43031) | 21 | Yes | Pre-mRNA splicing factor required for final stages of spliceosome maturation | 1 | N | 5 | N | 4 | N |
| 39 | Prp22p (P24384) | Prp22p (O42643) | 130 | Yes | DEAD/DEAH RNA helicase | 16 | 2 | 8 | N | N | N |
| 40 | Cwc21p (Q03375) | Cwf21p (O14161) | 16 | No | Hypothetical RNA binding protein | 4 | 2 | 2 | N | 3 | N |
| 41 | Cwc22p (P53333) | Cwf22p (P78752) | 67 | Yes | Hypothetical protein; H. sapiens (Q9HCG8), D. melanogaster (Q9VJ87), A. thaliana (Q9SAG7), C. elegans (Q17336) | 6 | 2 | 9 | 2 | N | N |
| 42 | Cwc23p (P52868) | Cwf23p (Q9P7C6) | 41 | Yes | Hypothetical protein | N | 2 | 4 | N | N | 2 |
| 43 | Cwc24p (P53769) | Cwf24p (Q9P6R8) | 30 | Yes | Hypothetical protein | 1 | N | 4 | N | 2 | N |
| 44 | Cwc25p (P53854) | Cwf25p (Q9Y805) | 20 | Yes | Hypothetical protein | N | 1 | 3 | N | 3 | N |
| 45 | Cwc26p (P46947) | Cwf26p (O94417) | 31 | No | Hypothetical protein | N | 1 | 2 | N | 2 | N |
| 46 | Cwc27p (Q02770) | Cwf27p (O42941) | 36 | No | Putative peptidyl-prolyl _cis_-trans isomerase | 6 | 1 | 4 | N | N | N |
Validation of TAP/DALPC mass spectrometry results.
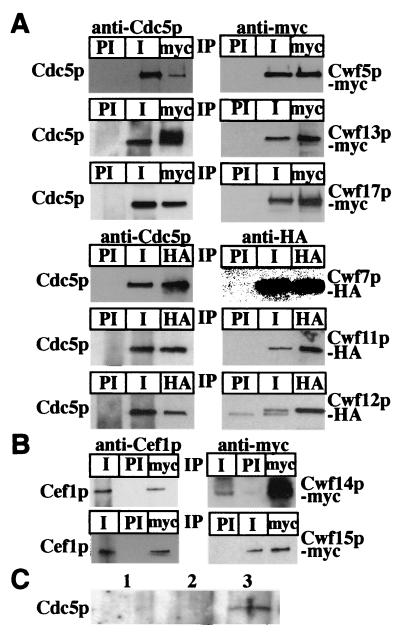
To validate the specificity of the TAP/DALPC mass spectrometry approach, we sought to confirm the interaction of a subset of the identified proteins with either S. pombe Cdc5p or S. cerevisiae Cef1p by coimmunoprecipitation. To this end, the S. pombe cwf5+, cwf7+, cwf11+, cwf12+, cwf13+, and cwf17+ loci were modified to encode proteins with either 13 copies of the myc or three copies of the HA epitope tag at their C termini. All epitope-tagged strains exhibited wild-type growth rates and morphologies (data not shown). S. pombe Cwf7p and Cwf11p are hypothetical proteins with no obvious S. cerevisiae homologs and no ascribed functions, while the S. cerevisiae homologs of Cwf5p, Cwf12p, and Cwf13p are known splicing factors (11, 18, 77). S. pombe Cwf17p is related to a human U5 snRNP protein (1) that does not have an obvious S. cerevisiae homolog. S. pombe Cwf5p-myc, Cwf7p-HA, Cwf11p-myc, Cwf12p-HA, Cwf13p-HA, and Cwf17p-myc were immunoprecipitated with either anti-myc or anti-HA antibodies and then immunoblotted with anti-Cdc5p antibodies. In each case, Cdc5p coimmunoprecipitated with the myc- or HA-tagged protein (Fig. 2A, left panels). In reciprocal experiments, anti-Cdc5p antibodies, but not preimmune sera, precipitated each of the myc- or HA-tagged proteins (Fig. 2A, right panels).
FIG. 2.
Validation of TAP/DALPC results. (A) S. pombe Cwf5p-myc, Cwf7p-HA, Cwf11p-myc, Cwf12p-HA, Cwf13p-HA, and Cwf17p-myc associate with Cdc5p in vivo. An anti-Cdc5 (left panel) and an anti-myc (right panel) immunoblot of immunoprecipitates (IP) from cwf5-myc, cwf7-HA, cwf11-myc, cwf12-HA, cwf13-HA, and cwf17-myc strains are shown. Immunoprecipitations were performed with preimmune sera (PI), anti-Cdc5p immune sera (I), anti-myc antibodies (myc), or anti-HA antibodies. (B) S. cerevisiae Cwc14p-myc and Cwc15p-myc associate with Cef1p in vivo. An anti-Cef1p (left panel) and an anti-myc (right panel) immunoblot of immunoprecipitates from cwc14-myc and cwc15-myc strains are shown. Immunoprecipitations were performed with preimmune sera (PI), anti-Cef1p immune sera (I), or anti-myc antibodies (myc). (C) S. pombe Lsm8p-TAP and Cdc5p coimmunoprecipitate. Shown is an anti-Cdc5p immunobolot of IgG pulldowns from wild-type (lane 1), arp2-TAP (lane 2), and lsm8-TAP (lane 3) strains.
In S. cerevisiae, the CWC14 and CWC15 loci were tagged with sequences encoding 13 copies of the myc epitope. Cwc14p and Cwc15p are hypothetical proteins of unknown function. Cwc14p-myc and Cwc15p-myc could be coimmunoprecipitated with Cef1p by using either the anti-myc antibody or the anti-Cef1p antibody (Fig. 2B). The results of our coimmunoprecipitation analyses confirm our DALPC results and indicate that at least portions of these proteins are associated with Cdc5p/Cef1p.
In yeast and human cells, seven Sm-like proteins, Lsm2p to Lsm8p, have been shown to associate with U6 snRNA (2, 42, 58). None of these were detected by DALPC analysis of our purifications. Therefore, we sought to confirm the interaction of Lsm proteins with S. pombe Cdc5p by coimmunoprecipitation. To this end, the S. pombe lsm8+ locus was modified to encode a protein with a C-terminal TAP tag epitope. Cdc5p coimmunoprecipitated with the TAP-tagged Lsm8p protein but not with Arp2-TAP (Fig. 2C, lanes 2 and 3). Therefore, Cdc5p does associate with Lsm proteins in vivo, as would be expected from a member of a complex containing the U6 snRNA.
Novel Cwfs/Cwcs function in pre-mRNA processing.
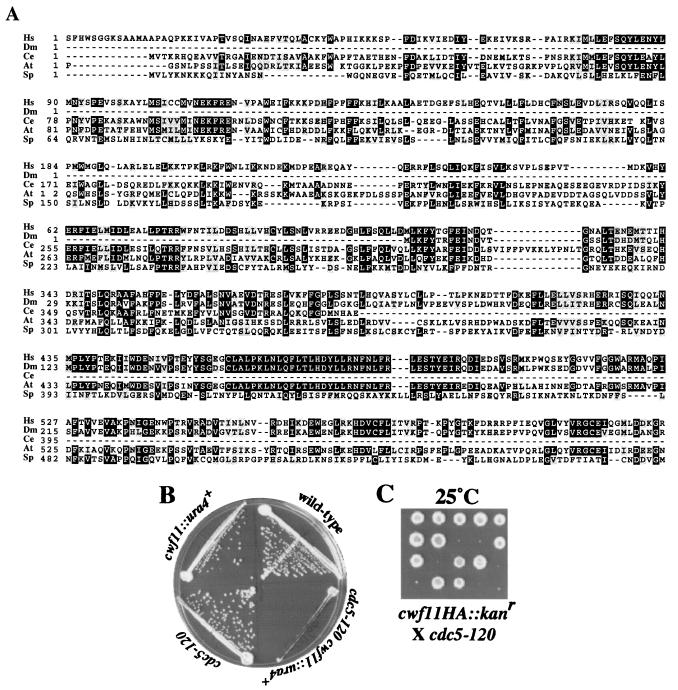
Because most S. pombe and S. cerevisiae Cwf/Cwc complex members are known pre-mRNA splicing factors, it seemed probable that hypothetical proteins identified in our purifications would also play a role in pre-mRNA splicing. To test this prediction, we examined two S. pombe proteins, Cwf11p and Cwf7p, which have no obvious S. cerevisiae homologs but have putative orthologs in higher eukaryotes. For Cwf11p, there are highly related proteins in humans (O60306), D. melanogaster (Q9VGG7), C. elegans (Q9U1Q7), and A. thaliana (Q9ZVJ8) (Fig. 3A). We first asked whether cwf11+ is an essential gene. It is not, since cwf11::ura4+ cells grew with wild-type kinetics at all temperatures tested (25 to 36°C) (Fig. 3B and data not shown). Because pre-mRNA splicing is an essential process, this result indicates that Cwf11p is not essential for pre-mRNA splicing. However, we determined that the absence of Cwf11p reduced the restrictive temperature of _cdc5_-120. _cdc5_-120 cwf11::ura4+ cells barely grew at 25°C (Fig. 3B). In addition, the _cwf11-HA cdc5_-120 strain was barely able to form colonies at 25°C (Fig. 3C). These negative genetic interactions, along with the physical interaction between the two proteins, are consistent with an ancillary role for S. pombe Cwf11p in pre-mRNA splicing.
FIG. 3.
Characterization of S. pombe Cwf11p. (A) The ClustalW 1.6 program (22, 67) was used to align Cwf11-related proteins from H. sapiens (Hs) (O060306), D. melanogaster (Dm) (Q9VGG7), C. elegans (Ce) (Q9U1Q7), A. thaliana (At) (Q9ZVJ8), and S. pombe (Sp) (O94508). Residues found to be identical or similar to those of human CWF11 are highlighted on a solid background or shaded, respectively. (B) _cdc5_-120 and cwf11::ura4+ cells show a reduced restrictive temperature. Wild-type, _cdc5_-120, cwf11::ura4, and _cdc5_-120 cwf11::ura4 mutant cells were streaked onto agar medium at 25°C. (C) Tetrads grown at 25°C from the cwf11HA/cwf11+ _cdc5_-120/cdc5+ diploid. Only very small colonies grew on plates containing G418, and these were temperature sensitive, indicating that they were double mutants.
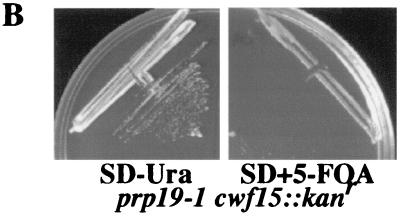
Another uncharacterized protein identified in our analysis was S. pombe and S. cerevisiae Cwf15p and Cwc15p, respectively. Cwf15p/Cwc15p homologs exist in humans (Q9UI29), D. melanogaster (Q9V3B6), C. elegans (O45766), and A. thaliana (Q9LK52) (Fig. 4A). Although this protein is not essential in S. cerevisiae, we determined that cwc15::kanr is synthetically lethal with _prp19_-1 (12) (Fig. 4B). This negative genetic interaction suggests a role for S. cerevisiae Cwc15p in pre-mRNA splicing.
FIG. 4.
Characterization of S. cerevisiae Cwc15p. (A) The ClustalW 1.6 program (22, 67) was used to align Cwc15-related proteins from H. sapiens (Hs) (Q9UI29), D. melanogaster (Dm) (Q9V3B6), C. elegans (Ce) (O45766), A. thaliana (At) (Q9LK52), S. pombe (Sp) (Cwf15p) (O74817), and S. cerevisiae (Sc) (Cwc15p) (Q03772). Residues found to be identical or similar to those of human CWF15 are highlighted on a solid background or shaded, respectively. (B) _prp19_-1 and cwc15::kanr strains are synthetically lethal. A CEN _URA3_-marked plasmid carrying the PRP19 gene was introduced into a MATa/α _prp19_-1/PRP19 cwc15::kanr heterozygote prior to sporulation. A _prp19_-1 cwc15::kanr double-mutant strain expressing PRP19 from a _URA3_-based plasmid was isolated, struck to synthetic complete medium containing dextrose (SD) without Ura (SD − Ura) (left panel) or SD with Ura plus 5-fluoroorotic acid (SD + 5-FOA) (right panel), and incubated for 3 days at 25°C.
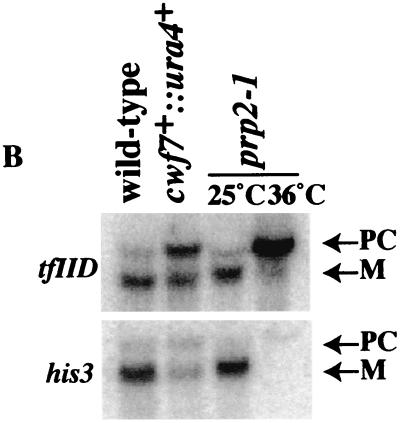
S. pombe Cwf7p was of particular interest because it was identified in our previous immunoaffinity purification of Cdc5p (43). Cwf7p homologs exist in a variety of organisms (humans [SwissProt no. O75934], D. melanogaster [Q9VAY6], and C. elegans [Q22417]) but not in S. cerevisiae. Alignment of Cwf7p family members shows that these proteins share significant sequence identity (Fig. 5A). As with cwf11+, we first asked whether cwf7+ is an essential gene. Following replacement of the cwf7+ ORF by the ura4+ gene in a diploid, we determined that it is essential for viability at all temperatures tested (25° to 36°C). We then performed a spore germination experiment (described in Materials and Methods) to collect cells lacking Cwf7p function to test for accumulation of pre-mRNAs by Northern blot analysis. In cwf7::ura4+ cells, there were elevated levels of both TF11D and HIS3 pre-mRNAs (Fig. 5B) as well as reductions in the mRNA levels. These data indicate that S. pombe cwf7+ is necessary for pre-mRNA splicing.
FIG. 5.
Characterization of S. pombe Cwf7p. (A) The ClustalW 1.6 program (22, 67) was used to align Cwf7p-related proteins from H. sapiens (Hs) (O75934), D. melanogaster (Dm) (Q9VAY6), C. elegans (Ce) (Q22417), and S. pombe (Sp) (Q9USV3). Residues found to be identical or similar to those of human CWF7 are highlighted on a solid background or shaded, respectively. (B) cwf7+ is required for pre-mRNA splicing. RNAs were prepared from cwf7::ura4+ spores after germination, _prp2_-1 cells grown at 25°C or shifted to 36°C for 2 h, and wild-type cells and were then hybridized to oligonucleotides complementary to the tfIId or his3 mRNAs. PC, precursor; M, mature.
The Cwf complex remains stable in pre-mRNA splicing mutants.
The detection of the U2, U5, and U6 snRNAs along with core Sm proteins, U2 snRNP components, and U5 snRNP components in the Cwf/Cwc complexes has led us to speculate that Cdc5p/Cef1p and associated proteins are equivalent to a mature form of the spliceosome. Therefore, we wished to determine if Cdc5p would remain in a high-molecular-weight complex if early steps in the pre-mRNA splicing reaction were blocked. To this end we created strains containing S. pombe Cdc5p-TAP in either a _prp1_-1, _prp2_-1, _prp4_-1, or _prp12_-1 background. S. pombe Prp1p is homologous to S. cerevisiae Prp6p (72, 73), which is required for the accumulation of the U5:U6/U4 tri-snRNP (23). S. pombe Prp2p and Prp12p are considered to be required for addition of the U2 snRNP to pre-mRNA (30, 72). S. pombe Prp4p, a serine/threonine kinase, is also necessary for early steps in pre-mRNA splicing (28, 54, 56). We examined the sedimentation characteristics of Cdc5p-TAP from lysates of these splicing mutants that had been shifted to a restrictive temperature for 3 h. Interestingly, all Cdc5p-TAP sedimented as it did from wild-type cells (Fig. 6A), although pre-mRNA splicing had been blocked (Fig. 6B), suggesting that the complex remains largely unchanged when early steps in pre-mRNA processing fail to occur.
FIG. 6.
Cdc5p remains in a large complex in pre-mRNA splicing mutants. (A) Lysates from _prp1_-1 cdc5-TAP, _prp2_-1 cdc5-TAP, _prp4_-1 cdc5-TAP, and _prp12_-1 cdc5-TAP strains after 3 h at 36°C, probed with anti-Cdc5p antibodies. Migration of FAS (40S), thyroglobulin (19S), and catalase (11.3S) collected from parallel gradients is indicated. (B) RT-PCR analysis of cdc2+ RNA isolated from wild-type (lane 2), _prp1_-1 cdc5-TAP (lane 3), _prp2_-1 cdc5-TAP (lane 4), _prp4_-1 cdc5-TAP (lane 5), and _prp12_-1 cdc5-TAP (lane 6) strains after 2 h at 36°C. Lane 1 represents a mock RT-PCR that lacks RNA but includes all other components of the reaction. PC, precursor; M, mature.
DISCUSSION
In S. pombe, S. cerevisiae, and human cells, Cdc5p, Cef1p, and hCDC5, respectively, have been implicated in the process of pre-mRNA splicing and are components of multiprotein complexes (3, 43, 47, 69). In this study we analyzed S. pombe Cdc5p- and S. cerevisiae Cef1p-associated proteins using TAP (55, 65) coupled with DALPC tandem mass spectrometry. This powerful approach revealed the existence of a stable complex containing at least 26 proteins. The majority of these proteins are known components of the U2 or U5 snRNPs or have otherwise been implicated in the process of pre-mRNA splicing, although several uncharacterized proteins were also identified. This work marks the first comprehensive analysis of CDC5-associated proteins in yeasts and provides evidence that the members of this complex must cooperate in some aspect of pre-mRNA processing.
We have confirmed the protein associations detected by DALPC by conventional methods. In all cases we have tested, at least a portion of each identified protein is able to coimmunoprecipitate with either Cdc5p or Cef1p. In addition, we have tested three of the uncharacterized proteins for a role in pre-mRNA splicing. We have found that the essential component (S. pombe Cwf7p) is required for pre-mRNA splicing, and genetic analysis of the nonessential components (S. pombe Cwf11p and S. cerevisiae Cwc15p) suggests that they positively contribute to Cdc5p/Cef1p function. These results suggest that all components identified in our purifications function in the process of pre-mRNA splicing.
One striking result of the DALPC analysis is the conservation between the S. pombe Cwf complex and the S. cerevisiae Cwc complex. These complexes contain similar numbers of components, with 24 proteins being conserved between species. Significantly, homologs of all Cwf proteins previously identified in S. pombe were found in each S. cerevisiae purification, and homologs of all S. cerevisiae Ntc components were identified in the S. pombe Cdc5p purification, with a few exceptions. The exceptions were those proteins without obvious counterparts in the other yeast (S. pombe Cwf7p, S. cerevisiae Snt309p [Ntc25p], and S. cerevisiae Ntc20p). Homologs of S. cerevisiae Snt309p and Ntc20p are also not obvious in higher eukaryotes, but there is a mammalian homolog of S. pombe Cwf7p, termed SPF27, that copurified with hCDC5 (3, 13).
The S. cerevisiae Ntc complex includes Prp19p, Cef1p (Ntc85p), Snt309p (Ntc25p), Isy1p (Ntc30p), Ntc20p (11, 12, 62, 69), and at least six other unidentified proteins (Ntc120p, Ntc90p, Ntc81p, Ntc77p, Ntc50p, and Ntc40p). A number of proteins identified here could represent these unknown polypeptides. For example, the 40-kDa S. cerevisiae Cwc2p is probably Ntc40p, S. cerevisiae Snu114p might represent Ntc120p, S. cerevisiae Syf1p might represent Ntc90p, S. cerevisiae Clf1p could correspond to Ntc81p, and S. cerevisiae Cwc1p/Prp46p may represent Ntc50p.
The identification of Ntc components within the Cdc5p-TAP purification strongly suggests that Ntc components are members of a larger protein complex. DALPC analysis of Prp19p-TAP- and Snt309p-TAP-associated proteins further strengthens this conclusion. Both of these S. cerevisiae Ntc members copurified with the same contingent of polypeptides as did S. pombe Cdc5p-TAP, S. cerevisiae Cef1p-TAP, S. cerevisiae Cwc1p-TAP, and S. cerevisiae Cwc2p-TAP. Like S. pombe Cdc5p-TAP, S. cerevisiae Prp19-TAP was also found to associate with the U2, U5, and U6 snRNAs. Therefore, Ntc proteins might represent a smaller unit of more stably associated proteins. It should be noted, however, that our DALPC analyses are not quantitative and do not provide information on the stoichiometry of any component within the larger complex we have examined. Therefore, it is possible that a smaller complex(s) does exist as a separate unit in cells, especially in S. cerevisiae, and joins with other proteins at certain times during the pre-mRNA splicing reaction. However, we have no data to support this interpretation from our studies in S. pombe. Under all conditions we have tested, Cdc5p exists in a large, discrete complex that contains numerous polypeptides and all three snRNAs (Fig. 1; Table 2)(43). An alternative explanation for the existence of a smaller unit in S. cerevisiae is that the larger complex might simply be less stable in the buffer conditions employed for its purification than its S. pombe counterpart. If so, by either breaking apart during cell lysis, sucrose gradient fractionation, or purification, several S. cerevisiae complex members could behave as if they were in smaller complexes.
The conservation of Cdc5p/Cef1p complexes between yeasts raises the possibility that CDC5 complexes in higher eukaryotes also contain similar proteins. Human CDC5 has been shown to copurify with a core complex of six proteins, as well as a number of other polypeptides (3). Four of the mammalian core proteins correspond to proteins identified in the yeast complexes (S. pombe Cdc5p and S. cerevisiae Cef1p; S. pombe Cwf8p and S. cerevisiae Prp19p; S. pombe Cwf7p; and S. pombe Prp5p/Cwf1p and S. cerevisiae Prp46p/Cwc1p). However, homologs of the majority of hCDC5-copurifying proteins were not found in the yeast preparations. None of the hCDC5-copurifying proteins was tested for its interaction with hCDC5 by direct coimmunoprecipitation or other follow-up methods (3), and therefore, their reported association with hCDC5 might be misleading. On the other hand, our fairly comprehensive analysis of the yeast Cdc5p/Cef1p complexes represents the minimum complement of Cdc5p/Cef1p-associated proteins. Only proteins whose binding is not altered by the presence of Ca2+ and that remain stably associated through the two affinity purification steps can be identified by the TAP strategy. Thus, it is likely that still other proteins will be found to interact with those identified in this study.
The detection of the U2, U5, and U6 snRNAs along with core Sm proteins, U2 snRNP components, and U5 snRNP components in the Cwf/Cwc complexes has led us to speculate that Cdc5p/Cef1p and associated proteins are a part of, or equivalent to, the active, mature form of the spliceosome. Such a conclusion is consistent with in vitro studies in which hCDC5 was found associated with core Sm proteins throughout the pre-mRNA splicing reaction (10). It is possible that many of the novel proteins identified in our purifications will be found to be members of the U6 snRNP, which has yet to be well characterized in any organism. Structural analysis of individual snRNPs and core Sm-protein complexes have confirmed that snRNAs found in an assembled spliceosome would be buried in the center of a protein core (17, 32, 33, 45, 60, 68, 76). The stability of the Cwf complex in the presence of RNase A provides further evidence that that the Cwf/Cwc complex could represent the active spliceosome.
Although this report is the first to link all of the proteins listed in Table 2 into one stable unit, many genetic and physical interactions between S. cerevisiae Cwc components have been noted previously (6, 7, 11, 12, 19, 57, 69, 70, 74, 75, 77). Also, a number of S. cerevisiae Cwc components are known to interact with U2, U5, and/or U6 snRNAs either by photo-cross-linking or by coimmunoprecipitation experiments (18, 19, 43, 75). Cef1p, Prp19p, Snt309p, Isy1p/Ntc30p, and Ntc20p associate with U2, U5, and U6 snRNPs in vitro, concomitant with U4 snRNP dissociation (11, 12, 63, 69). Other S. cerevisiae Cwc components, such as Brr2p, Prp8p, Snu114p, Clf1p, and Slt11p/Ecm2p, are known to initiate and/or stabilize key RNA-RNA and protein-RNA transitions during pre-mRNA splicing (5, 15, 19, 21, 35, 37, 38, 48, 49, 59, 66, 71, 77, 78). Taken in combination, these data provide further support for the idea that the complex described here represents a portion of the active spliceosome. Indeed, it is possible that the Cdc5-TAP complex simply represents U2, U5, and U6 snRNPs containing proteins not previously recognized as U2, U5 and U6 snRNP components.
It was unexpected, however, that our attempts to prevent formation of the Cdc5p complex by blocking pre-mRNA splicing using a variety of pre-mRNA splicing mutants were not successful. In all cases tested, Cdc5p was detected only in a high-molecular-weight complex, indistinguishable from the complex present in wild-type cells. This could indicate that at steady state, most of the pre-mRNA splicing factors associated with Cdc5p are actively engaged in the splicing reaction and only transiently dissociate to perform the initial pre-mRNA recognition steps. Given the relative stability of the purified S. pombe Cdc5p-TAP complex, further analysis of its RNA composition and structure should help explain these observations and provide insight into its role in pre-mRNA processing.
Acknowledgments
We thank Peg Coughlin from Tim Mitchision's laboratory for the negative-stained EM pictures of the Cdc5p-TAP complex, Anna Feoktistova for valuable technical assistance, and Jim Patton for helpful comments on the manuscript.
This work was supported by National Institutes of Health grant GM47728 (to K.L.G.). M.D.O. was supported by National Cancer Institute grant T32 CA09592. A.J.L. was supported by the Vanderbilt-Ingram Cancer Support Grant (5P30CA68485), the HHMI, and an Ingram Family gift. K.L.G. is an Associate Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Achsel, T., K. Ahrens, H. Brahms, S. Teigelkamp, and R. Luhrmann. 1998. The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 18**:**6756-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achsel, T., H. Brahms, B. Kastner, A. Bachi, M. Wilm, and R. Luhrmann. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18**:**5789-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajuh, P., B. Kuster, K. Panov, J. C. Zomerdijk, M. Mann, and A. I. Lamond. 2000. Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J. 19**:**6569-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14**:**943-951. [DOI] [PubMed] [Google Scholar]
- 5.Beggs, J. D., S. Teigelkamp, and A. J. Newman. 1995. The role of PRP8 protein in nuclear pre-mRNA splicing in yeast. J. Cell Sci. Suppl. 19**:**101-105. [DOI] [PubMed] [Google Scholar]
- 6.Ben Yehuda, S., I. Dix, C. S. Russell, S. Levy, J. D. Beggs, and M. Kupiec. 1998. Identification and functional analysis of hPRP17, the human homologue of the PRP17/CDC40 yeast gene involved in splicing and cell cycle control. RNA 4**:**1304-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Yehuda, S., C. S. Russell, I. Dix, J. D. Beggs, and M. Kupiec. 2000. Extensive genetic interactions between PRP8 and PRP17/CDC40, two yeast genes involved in pre-mRNA splicing and cell cycle progression. Genetics 154**:**61-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein, H. S., and S. R. Coughlin. 1997. Pombe Cdc5-related protein. A putative human transcription factor implicated in mitogen-activated signaling. J. Biol. Chem. 272**:**5833-5837. [DOI] [PubMed] [Google Scholar]
- 9.Burns, C. G., and K. L. Gould. 1999. Connections between pre-mRNA processing and regulation of the eukaryotic cell cycle. Front. Horm. Res. 25**:**59-82. [DOI] [PubMed] [Google Scholar]
- 10.Burns, C. G., R. Ohi, A. R. Krainer, and K. L. Gould. 1999. Evidence that Myb-related CDC5 proteins are required for pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 96**:**13789-13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, C. H., W. Y. Tsai, H. R. Chen, C. H. Wang, and S. C. Cheng. 2001. Identification and characterization of two novel components of the Prp19p-associated complex, Ntc30p and Ntc20p. J. Biol. Chem. 276**:**488-494. [DOI] [PubMed] [Google Scholar]
- 12.Chen, H. R., S. P. Jan, T. Y. Tsao, Y. J. Sheu, J. Banroques, and S. C. Cheng. 1998. Snt309p, a component of the Prp19p-associated complex that interacts with Prp19p and associates with the spliceosome simultaneously with or immediately after dissociation of U4 in the same manner as Prp19p. Mol. Cell. Biol 18**:**2196-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, H. R., T. Y. Tsao, C. H. Chen, W. Y. Tsai, L. S. Her, M. M. Hsu, and S. C. Cheng. 1999. Snt309p modulates interactions of Prp19p with its associated components to stabilize the Prp19p-associated complex essential for pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 96**:**5406-5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng, S. C., W. Y. Tarn, T. Y. Tsao, and J. Abelson. 1993. PRP19: a novel spliceosomal component. Mol. Cell. Biol 13**:**1876-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung, S., M. R. McLean, and B. C. Rymond. 1999. Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA 5**:**1042-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collart, M. A., and S. Oliviero. 1993. Basic protocol: preparation of yeast RNA, p. 13.12.1-13.12.50. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. John Wiley & Sons, New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 17.Collins, B. M., S. J. Harrop, G. D. Kornfeld, I. W. Dawes, P. M. Curmi, and B. C. Mabbutt. 2001. Crystal structure of a heptameric Sm-like protein complex from Archaea: implications for the structure and evolution of snRNPs. J. Mol. Biol. 309**:**915-923. [DOI] [PubMed] [Google Scholar]
- 18.Dix, I., C. Russell, S. Yehuda, M. Kupiec, and J. Beggs. 1999. The identification and characterization of a novel splicing protein, Isy1p, of Saccharomyces cerevisiae. RNA 5**:**360-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dix, I., C. S. Russell, R. T. O'Keefe, A. J. Newman, and J. D. Beggs. 1998. Protein-RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA 4**:**1675-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eng, J. K., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5**:**976-989. [DOI] [PubMed] [Google Scholar]
- 21.Fabrizio, P., B. Laggerbauer, J. Lauber, W. S. Lane, and R. Luhrmann. 1997. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 16**:**4092-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng, D. F., and R. F. Doolittle. 1996. Progressive alignment of amino acid sequences and construction of phylogenetic trees from them. Methods Enzymol. 266**:**368-382. [DOI] [PubMed] [Google Scholar]
- 23.Galisson, F., and P. Legrain. 1993. The biochemical defects of prp4-1 and prp6-1 yeast splicing mutants reveal that the PRP6 protein is required for the accumulation of the [U4/U6.U5] tri-snRNP. Nucleic Acids Res. 21**:**1555-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatlin, C. L., G. R. Kleemann, L. G. Hays, A. J. Link, and J. R. Yates III. 1998. Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal. Biochem. 263**:**93-101. [DOI] [PubMed] [Google Scholar]
- 25.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11**:**355-360. [DOI] [PubMed] [Google Scholar]
- 26.Gould, K. L., S. Moreno, D. J. Owen, S. Sazer, and P. Nurse. 1991. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 10**:**3297-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Groenen, P. M., G. Vanderlinden, K. Devriendt, J. P. Fryns, and W. J. Van de Ven. 1998. Rearrangement of the human CDC5L gene by a t(6;19)(p21;q13.1) in a patient with multicystic renal dysplasia. Genomics 49**:**218-229. [DOI] [PubMed] [Google Scholar]
- 28.Gross, T., M. Lutzelberger, H. Weigmann, A. Klingenhoff, S. Shenoy, and N. F. Kaufer. 1997. Functional analysis of the fission yeast Prp4 protein kinase involved in pre-mRNA splicing and isolation of a putative mammalian homologue. Nucleic Acids Res. 25**:**1028-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guthrie, C., and G. R. Fink (ed.). 1991. Methods in enzymology, vol. 194. Guide to yeast genetics and molecular biology. Academic Press, Inc., San Diego, Calif. [PubMed]
- 30.Habara, Y., S. Urushiyama, T. Shibuya, Y. Ohshima, and T. Tani. 2001. Mutation in the prp12+ gene encoding a homolog of SAP130/SF3b130 causes differential inhibition of pre-mRNA splicing and arrest of cell-cycle progression in Schizosaccharomyces pombe. RNA 7**:**671-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirayama, T., and K. Shinozaki. 1996. A cdc5+ homolog of a higher plant. Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93**:**13371-13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kambach, C., S. Walke, R. Young, J. M. Avis, E. de la Fortelle, V. A. Raker, R. Luhrmann, J. Li, and K. Nagai. 1999. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96**:**375-387. [DOI] [PubMed] [Google Scholar]
- 33.Kastner, B., M. Bach, and R. Luhrmann. 1990. Electron microscopy of small nuclear ribonucleoprotein (snRNP) particles U2 and U5: evidence for a common structure-determining principle in the major U snRNP family. Proc. Natl. Acad. Sci. USA 87**:**1710-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keeney, J. B., and J. D. Boeke. 1994. Efficient targeted integration at _leu1_-32 and _ura4_-294 in Schizosaccharomyces pombe. Genetics 136**:**849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, D. H., and J. J. Rossi. 1999. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA 5**:**959-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65**:**367-409. [DOI] [PubMed] [Google Scholar]
- 37.Lauber, J., P. Fabrizio, S. Teigelkamp, W. S. Lane, E. Hartmann, and R. Luhrmann. 1996. The HeLa 200 kDa U5 snRNP-specific protein and its homologue in Saccharomyces cerevisiae are members of the DEXH-box protein family of putative RNA helicases. EMBO J. 15**:**4001-4015. [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, J., and J. J. Rossi. 1996. Identification and characterization of yeast mutants that overcome an experimentally introduced block to splicing at the 3′ splice site. RNA 2**:**835-848. [PMC free article] [PubMed] [Google Scholar]
- 39.Link, A. J., J. Eng, D. M. Schieltz, E. Carmack, G. J. Mize, D. R. Morris, B. M. Garvik, and, J. R. Yates III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17**:**676-682. [DOI] [PubMed] [Google Scholar]
- 40.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14**:**953-961. [DOI] [PubMed] [Google Scholar]
- 41.Lundgren, K., S. Allan, S. Urushiyama, T. Tani, Y. Ohshima, D. Frendewey, and D. Beach. 1996. A connection between pre-mRNA splicing and the cell cycle in fission yeast: cdc28+ is allelic with prp8+ and encodes an RNA-dependent ATPase/helicase. Mol. Biol. Cell 7**:**1083-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayes, A. E., L. Verdone, P. Legrain, and J. D. Beggs. 1999. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 18**:**4321-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDonald, W. H., R. Ohi, N. Smelkova, D. Frendewey, and K. L. Gould. 1999. Myb-related fission yeast Cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol. 19**:**5352-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194**:**795-823. [DOI] [PubMed] [Google Scholar]
- 45.Mura, C., D. Cascio, M. R. Sawaya, and D. S. Eisenberg. 2001. The crystal structure of a heptameric archaeal Sm protein: implications for the eukaryotic snRNP core. Proc. Natl. Acad. Sci. USA 98**:**5532-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray, H. L., and K. A. Jarrell. 1999. Flipping the switch to an active spliceosome. Cell 96**:**599-602. [DOI] [PubMed] [Google Scholar]
- 47.Neubauer, G., A. Gottschalk, P. Fabrizio, B. Seraphin, R. Luhrmann, and M. Mann. 1997. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc. Natl. Acad. Sci. USA 94**:**385-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newman, A. J., and C. Norman. 1992. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68**:**743-754. [DOI] [PubMed] [Google Scholar]
- 49.Noble, S. M., and C. Guthrie. 1996. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics 143**:**67-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nurse, P., P. Thuriaux, and K. Nasmyth. 1976. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146**:**167-178. [DOI] [PubMed] [Google Scholar]
- 51.Ohi, R., A. Feoktistova, S. McCann, V. Valentine, A. T. Look, J. S. Lipsick, and K. L. Gould. 1998. Myb-related Schizosaccharomyces pombe Cdc5p is structurally and functionally conserved in eukaryotes. Mol. Cell. Biol. 18**:**4097-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohi, R., D. McCollum, B. Hirani, G. J. Den Haese, X. Zhang, J. D. Burke, K. Turner, and K. L. Gould. 1994. The Schizosaccharomyces pombe cdc5+ gene encodes an essential protein with homology to c-Myb. EMBO J. 13**:**471-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Potashkin, J., and D. Frendewey. 1989. Splicing of the U6 RNA precursor is impaired in fission yeast pre-mRNA splicing mutants. Nucleic Acids Res. 17**:**7821-7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Potashkin, J., D. Kim, M. Fons, T. Humphrey, and D. Frendewey. 1998. Cell-division-cycle defects associated with fission yeast pre-mRNA splicing mutants. Curr. Genet. 34**:**153-163. [DOI] [PubMed] [Google Scholar]
- 55.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17**:**1030-1032. [DOI] [PubMed] [Google Scholar]
- 56.Rosenberg, G. H., S. K. Alahari, and N. F. Kaufer. 1991. prp4 from Schizosaccharomyces pombe, a mutant deficient in pre-mRNA splicing isolated using genes containing artificial introns. Mol. Gen. Genet. 226**:**305-309. [DOI] [PubMed] [Google Scholar]
- 57.Russell, C. S., S. Ben-Yehuda, I. Dix, M. Kupiec, and J. D. Beggs. 2000. Functional analyses of interacting factors involved in both pre-mRNA splicing and cell cycle progression in Saccharomyces cerevisiae. RNA 6**:**1565-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salgado-Garrido, J., E. Bragado-Nilsson, S. Kandels-Lewis, and B. Seraphin. 1999. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 18**:**3451-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92**:**315-326. [DOI] [PubMed] [Google Scholar]
- 60.Stark, H., P. Dube, R. Luhrmann, and B. Kastner. 2001. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature 409**:**539-542. [DOI] [PubMed] [Google Scholar]
- 61.Stukenberg, P. T., K. D. Lustig, T. J. McGarry, R. W. King, J. Kuang, and M. W. Kirschner. 1997. Systematic identification of mitotic phosphoproteins. Curr. Biol. 7**:**338-348. [DOI] [PubMed] [Google Scholar]
- 62.Tarn, W. Y., C. H. Hsu, K. T. Huang, H. R. Chen, H. Y. Kao, K. R. Lee, and S. C. Cheng. 1994. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 13**:**2421-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tarn, W. Y., K. R. Lee, and S. C. Cheng. 1993. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc. Natl. Acad. Sci. USA 90**:**10821-10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tarn, W. Y., K. R. Lee, and S. C. Cheng. 1993. The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol. Cell. Biol. 13**:**1883-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tasto, J. J., R. H. Robert, H. Carnahan, W. H. McDonald, and K. L. Gould. 2001. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast 18**:**657-662. [DOI] [PubMed] [Google Scholar]
- 66.Teigelkamp, S., E. Whittaker, and J. D. Beggs. 1995. Interaction of the yeast splicing factor PRP8 with substrate RNA during both steps of splicing. Nucleic Acids Res. 23**:**320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22**:**4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toro, I., S. Thore, C. Mayer, J. Basquin, B. Seraphin, and D. Suck. 2001. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 20**:**2293-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsai, W. Y., Y. T. Chow, H. R. Chen, K. T. Huang, R. I. Hong, S. P. Jan, N. Y. Kuo, T. Y. Tsao, C. H. Chen, and S. C. Cheng. 1999. Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing. J. Biol. Chem. 274**:**9455-9462. [DOI] [PubMed] [Google Scholar]
- 70.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403**:**623-627. [DOI] [PubMed] [Google Scholar]
- 71.Umen, J. G., and C. Guthrie. 1995. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev. 9**:**855-868. [DOI] [PubMed] [Google Scholar]
- 72.Urushiyama, S., T. Tani, and Y. Ohshima. 1996. Isolation of novel pre-mRNA splicing mutants of Schizosaccharomyces pombe. Mol. Gen. Genet. 253**:**118-127. [DOI] [PubMed] [Google Scholar]
- 73.Urushiyama, S., T. Tani, and Y. Ohshima. 1997. The prp1+ gene required for pre-mRNA splicing in Schizosaccharomyces pombe encodes a protein that contains TPR motifs and is similar to Prp6p of budding yeast. Genetics 147**:**101-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Nues, R. W., and J. D. Beggs. 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157**:**1451-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vidal, V. P., L. Verdone, A. E. Mayes, and J. D. Beggs. 1999. Characterization of U6 snRNA-protein interactions. RNA 5**:**1470-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walke, S., E. Bragado-Nilsson, B. Seraphin, and K. Nagai. 2001. Stoichiometry of the Sm proteins in yeast spliceosomal snRNPs supports the heptamer ring model of the core domain. J. Mol. Biol. 308**:**49-58. [DOI] [PubMed] [Google Scholar]
- 77.Xu, D., and J. D. Friesen. 2001. Splicing factor slt11p and its involvement in formation of U2/U6 helix II in activation of the yeast spliceosome. Mol. Cell. Biol. 21**:**1011-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu, D., S. Nouraini, D. Field, S. J. Tang, and J. D. Friesen. 1996. An RNA-dependent ATPase associated with U2/U6 snRNAs in pre-mRNA splicing. Nature 381**:**709-713. [DOI] [PubMed] [Google Scholar]