Distribution of Surface Protein Variants among Hyperinvasive Meningococci: Implications for Vaccine Design (original) (raw)
Abstract
The bacterium Neisseria meningitidis is a major cause of meningitis and septicemia worldwide. Outer membrane proteins (OMPs) are candidates in the search for comprehensive meningococcal vaccines; however, the formulation of OMP vaccines is complicated by antigenic diversity, which is generated by high levels of genetic reassortment and strong positive selection in the meningococcal antigen genes. The genetic and antigenic diversity of three OMPs (FetA, PorA, and PorB) among a global collection of meningococcal isolates representative of the major hyperinvasive clonal complexes was determined. There was evidence for antigenic structuring among the three OMPs that could not be explained purely by descent. These observations violated the predictions of the clonal and epidemic clonal models of population structure but were in concordance with models of strain structure which propose that host immunity selects for nonoverlapping antigen combinations. The patterns of antigenic variant combinations suggested that an OMP-based vaccine with as few as six PorA and five FetA variant sequences could generate homologous immune responses against all 78 isolates examined.
Neisseria meningitidis, a common commensal of the human nasopharynx, continues to be a major cause of bacterial meningitis and septicemia worldwide. Unencapsulated meningococci are essentially avirulent, and only 5 of the 13 chemically and immunologically distinct meningococcal capsular polysaccharides, which define meningococcal serogroup, are frequently associated with invasive disease (43). The development of comprehensive capsule-based vaccines has, however, been constrained by the poor immunogenicity of one of the major disease-associated capsules, corresponding to serogroup B (48), and by concerns over the immunological identity of this antigen with host sialic acids (10, 11). A universal protein antigen that is a suitable basis for a vaccine effective against all disease-associated meningococci is yet to be described (16).
Many attempts have been made to develop vaccines based on the subcapsular antigens, especially the outer membrane proteins (OMPs), of this gram-negative bacterium (11). Most meningococcal OMPs are highly diverse (34, 37, 39), and the levels of potentially cross-protective immune responses to heterologous strains have been disappointing (36), although OMP-containing outer membrane vesicle (OMV) vaccines have been effective against the particular epidemic strain from which they were made (3, 31). Various OMPS have been proposed as vaccine components, including the PorA, PorB, FetA, TbpB, and NspA proteins; of these, the porin proteins and FetA are attractive, as they are present in OMVs and elicit immune responses in humans (4, 16, 45). From the perspective of vaccine development, the PorA and FetA proteins have the additional advantage that their major immunogenic epitopes are easily defined, as they correspond to contiguous peptide sequences located in putative surface-exposed loops of these proteins (20, 37, 41, 42). Various vaccine formulations comprising mixtures of OMP variants have been proposed, including the expression of multiple PorA variants by genetically engineered meningococci (6, 25). The choice of variants to be included in these multicomponent vaccines is complicated by the frequent occurrence of antigenic variants in all of the major surface antigens described to date (21, 27, 34, 37, 39, 44).
In addition to antigenic diversity, meningococcal populations are genetically highly diverse; many genotypes have been identified by examination of housekeeping genes that are subject to selection for conservation of metabolic function (9, 17, 19). These genotypes have been identified as electrophoretic types by multilocus enzyme electrophoresis (7) and more recently as sequence types (STs) by multilocus sequence typing (MLST) (19, 40). Population studies that exploit multilocus enzyme electrophoresis and MLST have identified equivalent groups of related genotypes that are referred to as clonal complexes (40); these are now named after a predominant, or central, ST, e.g., the ST-1 complex, and include all isolates that share identical alleles with this ST at four or more MLST loci. Isolate collections corresponding to populations of asymptomatically carried meningococci show the greatest genetic diversity, while most disease-associated meningococci belong to a limited number of clonal complexes known as hyperinvasive lineages (8, 17, 19).
To inform the choice of OMV variants to be included in possible meningococcal vaccines, we undertook a survey of the variation of three major OMPs that have been proposed as potential vaccine candidates, PorA, PorB, and FetA. The gene sequences encoding these three proteins were characterized in a collection of 78 meningococci representing the seven hyperinvasive lineages associated with most endemic and epidemic meningococcal disease of the latter half of the twentieth century (19). Our phylogenetic analyses of concatenated antigen gene sequences clustered isolates into groups that were largely congruent with the clonal complexes identified by analysis of housekeeping genes, although the topologies of the phylogenies differed. Further, there was evidence for both the persistence of particular combinations of antigen variants during decades of global spread and the presence of identical antigen combinations in otherwise unrelated isolates. This antigenic structuring greatly simplified the number of components required for a vaccine that potentially provided cross-protection against all seven hyperinvasive lineages.
MATERIALS AND METHODS
Growth of meningococci and DNA preparation.
A total of 78 meningococcal isolates, representative of the major hyperinvasive lineages, were chosen for this analysis (Table 1). These isolates were previously characterized by MLST (19) and included 37 serogroup A meningococci (14 ST-1 complex, 11 ST-4 complex, and 12 ST-5 complex), 10 isolates from the ST-11 complex (8 serogroup C and 2 serogroup B), 8 isolates from the ST-8 complex (5 serogroup B and 3 serogroup C), 10 ST-32 complex organisms (9 serogroup B and 1 serogroup C), and 13 isolates from the ST-41/44 complex (all serogroup B). Isolates were propagated on heated blood agar plates in an atmosphere of 5% CO2 for 8 to 16 h. Genomic DNA was prepared by using an Isoquick nucleic acid extraction kit (Orca Research Inc.).
TABLE 1.
porA, fetA, and porB allele sequences of 78 hyperinvasive meningococci
| Group and isolate | Yr, country | Phenotypea | ST | Allele | Epitopeb | PorB loops | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| porA | fetA | porB | PorA VR1 | PorA VR2 | FetA_VR | I | IV | V | VI | VII | VIII | ||||
| ST-1 complex/subgroup I/II | |||||||||||||||
| 6748 | 1971, Canada | A:4,21:P1.3,6 | 1 | 17 | 3 | 3-60 | 18-1 | 3 | F5-1 | 6 | 7 | 11 | 10 | 7 | 6 |
| 20 | 1963, Niger | A:4,21:P1.10 | 1 | 16 | 8 | 3-60 | 5-2 | 10 | F1-7 | 6 | 7 | 11 | 10 | 7 | 6 |
| 254 | 1966, Djibouti | A:4,21:P1.10 | 1 | 16 | 8 | 3-60 | 5-2 | 10 | F1-7 | 6 | 7 | 11 | 10 | 7 | 6 |
| 129 | 1964, West Germany | A:4,21:P1.10 | 1 | 16 | 57 | 3-60 | 5-2 | 10 | F3-6 | 6 | 7 | 11 | 10 | 7 | 6 |
| 371 | 1980, India | A:4,21:P1.10 | 1 | 16 | 30 | 3-60 | 5-2 | 10 | F5-1 | 6 | 7 | 11 | 10 | 7 | 6 |
| 139M | 1968, Philippines | A:—c | 1 | 16 | 3 | 3-60 | 5-2 | 10 | F5-1 | 6 | 7 | 11 | 10 | 7 | 6 |
| 120M | 1967, Pakistan | A:4,21:P1.10 | 1 | 16 | 56 | 3-60 | 5-2 | 10 | F5-1 | 6 | 7 | 11 | 10 | 7 | 6 |
| S5611 | 1977, Australia | A:— | 1 | 16 | 3 | 3-60 | 5-2 | 10 | F5-1 | 6 | 7 | 11 | 10 | 7 | 6 |
| 106 | 1967, Morocco | A:4,21:P1.10 | 1 | 16 | 3 | 3-60 | 5-2 | 10 | F5-1 | 6 | 7 | 11 | 10 | 7 | 6 |
| 393 | 1968, Greece | A:— | 1 | 16 | 3 | 3-59 | 5-2 | 10 | F5-1 | 11 | 7 | 11 | 10 | 7 | 6 |
| 322/85 | 1985, East Germany | A:4,21:P1.10 | 2 | 16 | 52 | 3-80 | 5-2 | 10 | F5-2 | 4 | 7 | 11 | 10 | 7 | 6 |
| 79128 | 1979, China | A:— | 3 | 54 | 10 | 3-60 | 7-1 | 10 | F5-5 | 6 | 7 | 11 | 10 | 7 | 6 |
| BZ133 | 1977, Netherlands | B:NT:— | 1 | 2 | 3 | 3-60 | 7 | 16 | F5-1 | 6 | 7 | 11 | 10 | 7 | 6 |
| 79126 | 1979, China | A:4:P1.7 | 3 | 22 | 10 | 3-01 | 7-3 | 10-5 | F5-5 | 4 | 7 | 11 | 9 | 5 | 5 |
| ST-4 complex/subgroup IV | |||||||||||||||
| A4/M1027 | 1937, United States | A:4,21:— | 4 | 16 | 45 | 3-26 | 5-2 | 10 | F1-5 | 4 | 6 | 11 | 10 | 7 | 6 |
| 26 | 1963, Niger | A:— | 4 | 45 | 5 | 3-26 | 7 | 13 | F1-5 | 4 | 6 | 11 | 10 | 7 | 6 |
| 243 | 1966, Cameroon | A:— | 4 | 45 | 5 | 3-26 | 7 | 13 | F1-5 | 4 | 6 | 11 | 10 | 7 | 6 |
| 2059001 | 1990, Mali | A:4,21:P1.7 | 4 | 45 | 5 | 3-46 | 7 | 13 | F1-5 | 4 | 6 | 11 | 10 | 7 | 10 |
| 10 | 1963, Burkina Faso | A:— | 4 | 21 | 5 | 3-26 | 7 | 13-1 | F1-5 | 4 | 6 | 11 | 10 | 7 | 6 |
| 255 | 1966, Burkina Faso | A:4,21:P1.7 | 4 | 24 | 5 | 3-26 | 7-5 | 13-1 | F1-5 | 4 | 6 | 11 | 10 | 7 | 6 |
| S3131 | 1973, Ghana | A:— | 4 | 21 | 5 | 3-26 | 7 | 13-1 | F1-5 | 4 | 6 | 11 | 10 | 7 | 6 |
| 690 | 1980, India | A:4,21:P1.7 | 4 | 21 | 5 | 3-26 | 7 | 13-1 | F1-5 | 4 | 6 | 11 | 10 | 7 | 6 |
| C751 | 1983, Gambia | A:— | 4 | 21 | 5 | 3-26 | 7 | 13-1 | F1-5 | 4 | 6 | 11 | 10 | 7 | 6 |
| 1014 | 1985, Sudan | A:— | 4 | 21 | 5 | 3-26 | 7 | 13-1 | F1-5 | 4 | 6 | 11 | 10 | 7 | 6 |
| D8 | 1990, Mali | A:— | 4 | 21 | 5 | 3-26 | 7 | 13-1 | F1-5 | 4 | 6 | 11 | 10 | 7 | 6 |
| ST-5 complex/subgroup III | |||||||||||||||
| IAL2229 | 1976, Brazil | A:— | .5 | 19 | 55 | 3-27 | 20 | 9 | F2-1 | 4 | 7 | 11 | 10 | 7 | 6 |
| 153 | 1966, China | A:4,21:P1.9 | 5 | 19 | 7 | 3-47 | 20 | 9 | F3-1 | 4 | 7 | 11 | 10 | 7 | 6 |
| 154 | 1966, China | A:4,21:P1.9 | 6 | 19 | 7 | 3-47 | 20 | 9 | F3-1 | 4 | 7 | 11 | 10 | 7 | 6 |
| 14/1455 | 1970, USSR | A:4,21:P1.20,9 | 5 | 19 | 7 | 3-47 | 20 | 9 | F3-1 | 4 | 7 | 11 | 10 | 7 | 6 |
| S4355 | 1974, Denmark | A | 5 | 20 | 7 | 3-27 | 5-1 | 9 | F3-1 | 4 | 7 | 11 | 10 | 7 | 6 |
| 7891 | 1975, Finland | A:4,21:P1.9 | 5 | 19 | 7 | 3-27 | 20 | 9 | F3-1 | 4 | 7 | 11 | 10 | 7 | 6 |
| F4698 | 1987, Saudi | A:— | 5 | 19 | 11 | 3-47 | 20 | 9 | F3-1 | 4 | 7 | 11 | 10 | 7 | 6 |
| H1964 | 1987, United Kingdom | A:— | 5 | 19 | 11 | 3-47 | 20 | 9 | F3-1 | 4 | 7 | 11 | 10 | 7 | 6 |
| F6124 | 1988, Chad | A:— | 5 | 19 | 11 | 3-47 | 20 | 9 | F3-1 | 4 | 7 | 11 | 10 | 7 | 6 |
| 92001 | 1992, China | A:— | 7 | 19 | 7 | 3-62 | 20 | 9 | F3-1 | 12 | 7 | 10 | 10 | 7 | 6 |
| 11-004 | 1984, China | A:— | 5 | 19 | 54 | 3-61 | 20 | 9 | F3-8 | 9 | 7 | 11 | 10 | 7 | 6 |
| 80049 | 1963, China | A:4:P1.10 | 5 | 16 | 5 | 3-56 | 5-2 | 10 | F1-5 | 4 | 7 | 15 | 9 | 7 | 6 |
| ST-8 complex/cluster A4 | |||||||||||||||
| BZ 10 | 1967, The Netherlands | B:2b:P1.2 | 8 | 7 | 4 | 2-20 | 5-1 | 2-2 | F3-9 | 1 | 8 | 2 | 2 | 1 | 1 |
| B6116/77 | 1977, Iceland | B:— | 10 | 7 | 17 | 2-03 | 5-1 | 2-2 | F1-4 | 1 | 2 | 2 | 2 | 1 | 1 |
| BZ 163 | 1979, Netherlands | B:2b:P1.16 | 9 | 1 | 38 | 2-03 | 21 | 16 | F1-7 | 1 | 2 | 2 | 2 | 1 | 1 |
| G2136 | 1986, England | B:— | 8 | 46 | 20 | 2-03 | 5-2 | 10-1 | F3-6 | 1 | 2 | 2 | 2 | 1 | 1 |
| SB25 | 1990, South Africa | C:— | 8 | 51 | 4 | 2-03 | 18-1 | 3 | F3-9 | 1 | 2 | 2 | 2 | 1 | 1 |
| AK22 | 1992, Greece | B:— | 8 | 16 | 4 | 2-30 | 5-2 | 10 | F3-9 | 1 | 3 | 2 | 2 | 1 | 1 |
| 94/155 | 1994, New Zealand | C:— | 66 | 9 | 4 | 2-21 | 5 | 2 | F3-9 | 1 | 2 | 2 | 2 | 1 | 11 |
| 312 901 | 1996, England | C:— | 8 | 9 | 18 | 2-03 | 5 | 2 | F1-7 | 1 | 2 | 2 | 2 | 1 | 1 |
| ST-11 complex/ET-37 complex | |||||||||||||||
| 38VI | 1964, United States | B:—:P1.5,2 | 11 | 9 | 47 | 2-02 | 5 | 2 | F1-1 | 1 | 1 | 1 | 1 | 1 | 1 |
| NG P20 | 1969, Norway | B:2a:P1.2 | 11 | 9 | 9 | 2-19 | 5 | 2 | F1-1 | 1 | 1 | 1 | 1 | 1 | 1 |
| F1576 | 1984, Ghana | C:2a:P1.5,2 | 11 | 9 | 9 | 2-47 | 5 | 2 | F1-1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 500 | 1984, Italy | C:2a | 11 | 9 | 9 | 2-02 | 5 | 2 | F1-1 | 1 | 1 | 1 | 1 | 1 | 1 |
| MA-5756 | 1985, Spain | C:2a:P1.5 | 11 | 27 | 6 | 2-02 | 5 | 2-1 | F5-5 | 1 | 1 | 1 | 1 | 1 | 1 |
| M597 | 1988, Israel | C:2a:P1.5 | 11 | 27 | 6 | 2-02 | 5 | 2-1 | F5-5 | 1 | 1 | 1 | 1 | 1 | 1 |
| D1 | 1989, Mali | C:2a:P1.5 | 11 | 27 | 37 | 2-02 | 5 | 2-1 | F5-4 | 1 | 1 | 1 | 1 | 1 | 1 |
| 90/18311 | 1990, Scotland | C:NT:P1.5 | 11 | 27 | 6 | 2-31 | 5 | 2-1 | F5-5 | 1 | 1 | 5 | 14 | 16 | 1 |
| L93/4286 | 1993, England | C:— | 11 | 67 | 27 | 2-02 | 5-1 | 10-4 | F3-6 | 1 | 1 | 1 | 1 | 1 | 1 |
| BRAZ10 | 1976, Brazil | C:2a:P1.5 | 11 | 70 | 36 | 2-02 | 5-1 | 10-1 | F1-10 | 1 | 1 | 1 | 1 | 1 | 1 |
| ST-32 complex/ET-5 complex | |||||||||||||||
| 8680 | 1987, Chile | B:15:P1.3 | 32 | 41 | 15 | 3-24 | 7-2 | 3 | F3-1 | 7 | 7 | 10 | 12 | 11 | 6 |
| BZ 83 | 1984, The Netherlands | B:NT | 34 | 16 | 53 | 3-01 | 5-2 | 10 | F5-1 | 4 | 7 | 11 | 9 | 5 | 5 |
| 204/92 | 1992, Cuba | B:— | 33 | 4 | 26 | 3-08 | 19 | 15 | F5-1 | 4 | 5 | 7 | 9 | 5 | 5 |
| EG 329 | 1985, East Germany | B:15:P1.16 | 32 | 60 | 49 | 3-24 | 7-1 | 16 | F1-2 | 7 | 7 | 10 | 12 | 11 | 6 |
| NG 080 | 1981, Norway | B:15:P1.16 | 32 | 2 | 1 | 3-24 | 7 | 16 | F3-3 | 7 | 7 | 10 | 12 | 11 | 6 |
| BZ 169 | 1985, The Netherlands | B:NT:P1.16 | 32 | 14 | 1 | 3-14 | 5-2 | 16 | F3-3 | 3 | 7 | 6 | 7 | 6 | 8 |
| 44/76 | 1976, Norway | B:— | 32 | 2 | 1 | 3-24 | 7 | 16 | F3-3 | 7 | 7 | 10 | 12 | 11 | 6 |
| NG144/82 | 1982, Norway | B:15:P1.16 | 32 | 2 | 1 | 3-63 | 7 | 16 | F3-3 | 7 | 7 | 10 | 12 | 11 | 6 |
| 196/87 | 1987, Norway | C:15 | 32 | 61 | 1 | 3-24 | 7-2 | 16-12 | F3-3 | 7 | 7 | 10 | 12 | 11 | 6 |
| NG PB24 | 1985, Norway | B:NT:P1.16 | 32 | 57 | 1 | 3-24 | 7-2 | 16-7 | F3-3 | 7 | 7 | 10 | 12 | 11 | 6 |
| ST-41/44 complex/lineage 3 | |||||||||||||||
| 931905 | 1993, The Netherlands | B:— | 41 | 39 | 2 | 3-16 | 7-2 | 4 | F1-5 | 3 | 7 | 8 | 6 | 5 | 5 |
| 50/94 | 1994, Norway | B:— | 45 | 39 | 2 | 3-51 | 7-2 | 4 | F1-5 | 3 | 7 | 8 | 6 | 9 | 8 |
| 88/03415 | 1988, Scotland | B:— | 46 | 39 | 2 | 3-49 | 7-2 | 4 | F1-5 | 3 | 7 | 11 | 9 | 5 | 5 |
| 91/40 | 1991, New Zealand | B:4:P1.4 | 42 | 39 | 2 | 3-01 | 7-2 | 4 | F1-5 | 4 | 7 | 11 | 9 | 5 | 5 |
| AK50 | 1992, Greece | B:— | 41 | 39 | 16 | 3-52 | 7-2 | 4 | F1-5 | 4 | 7 | 11 | 9 | 15 | 5 |
| BZ198 | 1986, The Netherlands | B:NT | 41 | 39 | 2 | 3-01 | 7-2 | 4 | F1-5 | 4 | 7 | 11 | 9 | 5 | 5 |
| M-101/93 | 1993, Iceland | B:— | 41 | 39 | 2 | 3-01 | 7-2 | 4 | F1-5 | 4 | 7 | 11 | 9 | 5 | 5 |
| M40/94 | 1994, Chile | B:— | 41 | 39 | 2 | 3-53 | 7-2 | 4 | F1-5 | 4 | 7 | 12 | 11 | 9 | 8 |
| N45/96 | 1996, Norway | B:— | 41 | 39 | 16 | 3-01 | 7-2 | 4 | F1-5 | 4 | 7 | 11 | 9 | 5 | 5 |
| 400 | 1991, Austria | B:— | 40 | 52 | 2 | 3-36 | 7-2 | 13-2 | F1-5 | 3 | 7 | 12 | 11 | 9 | 8 |
| NG E30 | 1988, Norway | B:4:P1.16 | 44 | 1 | 46 | 3-45 | 21 | 16 | F1-7 | 4 | 5 | 7 | 9 | 5 | 5 |
| NG H15 | 1988, Norway | B:8:P1.15 | 43 | 65 | 2 | 3-54 | 19 | 15-2 | F1-5 | 3 | 7 | 16 | 6 | 5 | 5 |
| NG H36 | 1988, Norway | B:8:P1.2 | 47 | 18 | 41 | 3-16 | 5-1 | 2-2 | F1-7 | 3 | 7 | 8 | 6 | 5 | 5 |
Nucleotide sequence determination and gene nomenclature.
PCR amplification and nucleotide sequence determination of the meningococcal porA, porB, and fetA genes were as described previously (33, 37, 38). Nucleotide sequence data for forward and reverse strands were assembled with the STADEN software package (32), reformatted into Genetics Computer Group (GCG) format, and aligned to maintain maximum positional homology by using the SEQLAB program within the GCG software package (version 10.1: GCG, Madison, Wis.) (47). Using the Molecular Evolutionary Genetics Analysis software package (version 2.0) (18), pairwise comparisons were performed on each set of aligned sequences to identify distinct alleles; allele numbers were then assigned to each unique porA, porB, and fetA gene sequence on the basis of previously defined nomenclature systems (29, 37).
VR identification.
The identification of the variable regions (VRs) of PorA and FetA was straightforward. Nucleotide sequences encoding the defined PorA variable epitopes, VR1 and VR2 (41), were translated and identified by querying the PorA VR sequence database located at http://neisseria.org/nm/typing/pora/. The amino acid sequence variants determined for the FetA VR (42) (37) were also identified by database interrogation at http://neisseria.org/nm/typing/feta/.
PorB VR identification was more complicated, as PorB proteins often have discontinuous epitopes, where several of the eight PorB surface-exposed variable loops are involved in epitope formation (49). As loops II and III are essentially invariant in PorB, it was necessary to identify amino acid sequence variation in loops I and IV to VIII when determining PorB epitope diversity. Variation in PorB was therefore determined by pairwise comparisons of the aligned amino sequences corresponding to each surface loop, with each unique sequence at that loop assigned a number in order of discovery. All PorB variable-loop amino acid sequences are listed at http://neisseria.org/nm/typing/porb/. Results were reported by using a scheme slightly modified from that of Frasch et al. (12), with the format serogroup:serotype:subtype:FetA type (e.g., A:4,21:P1.5-2,10:F5-1).
Data manipulation and analysis.
Phylogenetic trees were constructed by using the maximum-likelihood (ML) method available in the PAUP* package (35). The general time reversible model of nucleotide substitution was used, with values for the nucleotide substitution matrix, the proportion of invariant sites, and the shape parameter (α) of a gamma distribution of rate variation among sites (with four categories) estimated during tree reconstruction (trees and parameter values are available on request). ML phylogenies were constructed by using concatenated DNA sequences for (i) the seven housekeeping gene fragments employed in MLST (19), giving a total sequence length of 3,284 bp, and (ii) the three antigen gene sequences, giving a total length of 4,209 bp.
RESULTS
Diversity of OMP genes and proteins.
The lengths of the nucleotide sequences when aligned for analysis were as follows: fetA, 2,031 bp; porA, 1,155 bp; and porB, 1,023 bp. Similar levels of sequence diversity were observed at the three loci (Table 2). Two distinct allele classes were present at the porB locus, porB2 and porB3, and the majority of differences among porB alleles were due to differences between these classes. Peptide sequence variation was identified in regions of the proteins implicated in immune responses in animals and humans. At the FetA VR, 16 unique peptide sequences were identified. There were 12 PorA VR1 and 18 PorA VR2 peptides sequences (26 unique VR1-VR2 combinations) and 26 unique combinations of the peptide sequences corresponding to loops I and IV to VIII of the PorB protein. The allele number designations, nucleotide sequences, and alignments are accessible at http://pubmlst.org/neisseria/links.htm.
TABLE 2.
Genetic and antigenic diversity among 78 hyperinvasive meningococci
| Locus | Nucleotide sequence length (bp) | No. of alleles | Proportion of segregating sites | Mean p distance | No. of peptide sequences | No. of VR combinationsa |
|---|---|---|---|---|---|---|
| porA | 1,086-1,149 | 33 | 0.14 | 0.041 | 33 | 26 |
| fetA | 1,977-2,019 | 33 | 0.19 | 0.051 | 31 | 16 |
| porBb | 927-1,020 | 31 | 0.36 | 0.131 | 28 | 26 |
| porB2 | 1,020 | 8 | 0.05 | 0.016 | 8 | 6 |
| porB3 | 927-939 | 23 | 0.10 | 0.031 | 20 | 20 |
Comparison of phylogenies obtained from antigen genes and housekeeping genes.
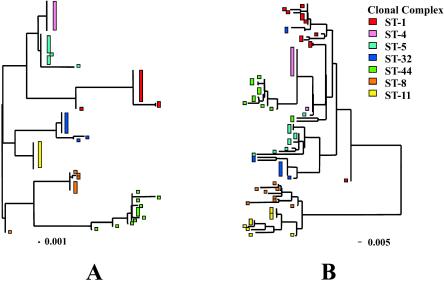
An ML tree constructed with concatenated housekeeping genes clustered the isolates by clonal complex (Fig. 1A), with the exception of the ST-8 complex isolate B6116/77, which possessed a pdhC allele that was divergent from those present among other ST-8 isolates. Similar clusters of isolates were observed in the ML tree constructed from the concatenated sequences of the three antigen genes, although the deeper branching patterns of the two trees were different (Fig. 1B). The isolates belonging to the ST-1, ST-4, and ST-5 complexes, which were mostly serogroup A, formed a clade in the housekeeping gene tree but not in the antigen gene tree. Conversely, the isolates belonging to the ST-8 and ST-11 complexes formed a clade in the antigen gene tree, as a result of these isolates possessing porB alleles belonging to the porB2 allele class, while the remaining isolates possessed porB alleles of the porB3 class. The clustering of two ST-32 complex isolates (204/92 and BZ83) with ST-1 isolates reflected identity or similarity of fetA, porA, and porB alleles with ST-1 complex organisms. The oldest isolate in the collection (A4/M1027, ST-4), isolated in 1937 in the United States, fell outside the otherwise closely related cluster of sequences formed by ST-4 complex meningococci, because it had a porA16 allele at the porA locus (Table 1), an allele predominant among ST-1 complex organisms. Isolate A4/M1027 clustered more closely with a single ST-5 complex isolate (80049), with which it shared an identical porA allele and a closely related fetA allele. Two ST-41/44 complex isolates (NG E30 and NG H36) did not cluster with the remaining isolates belonging to this complex as a result of divergent fetA and porA alleles.
FIG. 1.
Phylogenetic analysis of 78 hyperinvasive meningococci by using seven concatenated housekeeping gene sequences (3,284 bp) (A) and three concatenated antigen gene sequences (4,209 bp) (B). Each isolate is color coded according to clonal complex, as defined by MLST.
Distribution of FetA and PorA antigenic variants among clonal complexes.
There was similarity in patterns of variation identified in FetA and PorA protein sequences. Furthermore, variants of the major immunogenic regions of both FetA and PorA proteins were unevenly distributed among clonal complexes, with particular combinations of the three antigenic regions associated with particular clonal complexes (Table 1). The most common combination among the ST-1 complex isolates was P1.5-2,10;F5-1 (6 of 10 isolates), while ST-4 complex isolates were predominantly (6 of 10) P1.7,13-1:F1-5 and ST-5 complex isolates were predominantly (9 of 12) P1.20,9:F3-1. The majority (9 of 13) of ST-41/44 isolates were P1.7-2,4:F1-5. The ST-8 complex isolates contained a number of variants with no predominant VR sequences, although there was a prevalence of P1.5 sequence variants at PorA VR1, variants of P1.2 at PorA VR2, and the FetA VR F3-9. Variants of the P1.7,16:F3-3 combination dominated the ST-32 complex organisms. In two cases, isolates with unrelated STs exhibited identical PorA and FetA types: the ST-41/44 complex isolate NG E30 was identical to ST-8 complex isolate BZ163, both being P1.21,16:F1-7, although these isolates did not share identical porA and fetA alleles. One of the ST-32 isolates was identical to the majority antigenic type exhibited by ST-1 complex organisms, P1.5-2,10:F5-1, although again the allele sequences were not identical.
Distribution of PorB antigenic variants among clonal complexes.
Despite evidence of mosaic porB gene structure, conservation of PorB sequences was observed in all clonal complexes. The majority of ST-1 complex organisms (11 of 14) possessed PorB3 proteins that were identical in all surface loops and were phenotypically identified as serotype 4,21. The serotype 4,21 PorB3 protein identified among ST-4 complex organisms was distinguished from that identified in ST-1 complex and ST-5 complex meningococci by two amino acid changes in variable loop IV. Most ST-8 complex isolates (five of eight) had identical serotype 2b PorB2 amino acid sequences, with amino acid changes observed in either loop IV or loop VIII of the remaining three isolates, 94/155, BZ10, and AK22 (BZ10 still reacted with the serotype 2b monoclonal antibody despite this variation). The PorB protein was well conserved among ST-11 complex meningococci: 9 of 10 isolates possessed identical PorB2 variable loops and corresponded to serotype 2a. Seven of 10 ST-32 complex isolates examined had identical PorB3 proteins corresponding to serotype 15, while divergent sequences associated with reactivity with serotype 4 monoclonal antibodies were identified in isolates 204/92 and BZ83. The remaining isolate (BZ169) possessed a PorB3 amino acid sequence identical to that of a serotype 1 reference strain. Among ST-41/44 complex organisms, four isolates shared identical PorB3 amino acid sequences corresponding to serotype 4. Three isolates (88/03415, NG E30, and AK50) had variant PorB sequences but were still defined as serotype 4 as they possessed the loop VI amino acid sequence recognized by serotype 4 monoclonal antibodies (38). Three isolates (NG H15, NG H36, and N31905) had variant PorB3 sequences corresponding to serotype 8, while the remaining three ST-41/44 complex organisms possessed variant PorB3 proteins composed of novel combinations of variable loop sequences that were not typeable by serological methods.
Temporal and geographic distribution of antigen gene variants and combinations.
A number of antigen gene alleles exhibited wide temporal and geographical distributions. The porA16 allele was identified in 16 meningococci isolated in 15 countries from five continents over a period of 55 years; _porB3_-26 was identified in 10 isolates obtained from nine countries, including India, the United States, and several African countries, over a period of 53 years. The most widely distributed fetA allele in this collection was fetA5, which was present in 11 isolates originating from nine countries, mainly from Africa but including China and India, over 27 years. The VRs of these proteins also exhibited longevity and global distribution; the FetA VR variant F1-5 was present in 26 members of the isolate collection, including the oldest (A4/M1027, isolated in the United States in 1937) and the most recent isolate examined (N45/96, isolated in Norway in 1996).
DISCUSSION
It is well established that meningococcal OMPs are highly diverse in peptide sequence and that their genes are subject to strong positive selection in regions encoding those parts of the proteins exposed to immune attack (37, 39). The strength of positive selection recorded for the porB gene, for example, exceeds that reported for the human immunodeficiency virus type 1 envelope protein (39). These observations raise the possibility of poor efficacy and the rapid spread of escape variants following introduction of vaccines that include meningococcal OMPs as immunodominant constituents. There is, however, evidence for structure in the extent of antigenic diversity present in meningococcal populations, and, if understood and exploited, these limitations could simplify vaccine design and implementation.
Associations of particular serotypes (PorB variants) and serosubtypes (PorA variants) with clonal complexes have been identified in several studies (9, 33, 44), and variants observed in PorA VR1 and VR2 are structured into nonoverlapping combinations (14). These associations appear to be maintained despite diversifying selection and high rates of recombination which reassort genes among clonal complexes and generate mosaic genes encoding novel antigen variants (13). In addition, a study of antigenic variants of the OMP TbpB in the ST-5 complex indicated that while antigenic variants emerged during epidemic spread, they were less fit than the parent genotype and were lost during subsequent transmission (1). The present analysis has, for the first time, assessed the allele diversity present at three distinct OMP-encoding loci in the major hyperinvasive meningococcal lineages. The results confirmed the extensive diversity of these proteins, with between 28 and 33 peptide sequences observed for each protein, and provided evidence for the structuring of combinations of these variants.
Although recombination has erased most deep phylogenetic signal in meningococcal populations (15), groups of related genotypes, which are likely to comprise meningococci that share a common ancestor, can be identified (19). MLST resolves these related genotypes as clonal complexes, defined by allelic differences rather than nucleotide sequence comparisons per se, to account for frequent recombination (19, 40). The ML trees for both housekeeping gene sequences and antigen gene sequences generated clades that were largely congruent with clonal complex for the representatives of the seven hyperinvasive lineages analyzed here. While congruence of phylogenies based on housekeeping genes with clonal complex designation was unremarkable, the distribution of antigen gene sequences illustrated in the phylogenetic analysis required further explanation.
Evolution by simple clonal processes cannot account for congruence of the combinations of antigen variants with clonal complex, as the meningococcus experiences frequent recombination (20, 46). The predictions of clonality are further violated by the presence of identical antigen gene sequences throughout the data set, regardless of clonal complex. The epidemic clone concept, in which nonclonal populations can be transiently dominated by particular genotypes that share a recent common ancestor (23), could be invoked to explain the observations. However, this would not explain the longevity of antigen combinations over decades of global spread or the two examples of the presence of identical antigen variant combinations in otherwise genetically unrelated isolates.
The epidemic clone population structure also provides no explanation for the nonoverlapping nature of the antigen combinations observed. In the majority of cases, the clonal complexes representative of particular hyperinvasive lineages have predominant antigen gene combinations that share no major antigen variants with other clonal complexes. There were some examples of sharing of identical antigen variants among combinations; for example, the ST-4 complex shared the P1.7 variant with the ST-32 complex and the F1-5 variant with ST-41/44 complex. However, neither of the latter clonal complexes have been recently reported in Africa, where all of the recent ST-4 isolates originated, so it is possible that these overlapping antigen variant complexes are not present simultaneously in the same transmission system. By contrast, although possibly related by descent, all serogroup A-associated clonal complexes that have recently caused disease in Africa (ST-1, ST-4, and ST-5) (24) have distinct, nonoverlapping antigen gene combinations.
Models of strain structuring on the basis of host immunity provide a theoretical framework that can accommodate the observed combinations of antigenic variants (13, 14). These models postulate that, within a given transmission system, pathogen strains sharing immunological variants are disadvantaged. In this respect, it was noteworthy that a number of clonal complexes contained antigenic variant combinations that were altered at multiple antigens. There was evidence for two globally circulating strains within the ST-11 complex, P1.5,2:F1-1 and P1.5,2-1:F5-5, and one of these variants was associated with two different capsules, with two isolates that were B:2a:P1.5,2:F1-1 and two that were C:2a:P1.5,2:F1-1. At least two ST-32 complex variants spread widely from the mid-1970s onwards, and these also displayed nonoverlapping combinations of OMP antigen variants. The “Norwegian” strain was typically B:15:P1.7,16;F3-3, while the “Spanish” strain was B:4:P1.19,15;F5-1. A further variant identified in The Netherlands and the United Kingdom had an OMP profile (4:P1.5-2,10:F5-1) with many ST-1 complex isolates. A final variant identified in Chile, while bearing some similarities with the Norwegian strain, also differed at many OMPs, being B:15:P1.7,3:F3-1. Many examples of these strains within the ST-32 complex have been described previously by use of serological and molecular techniques (5, 7, 46). Many of the exceptions to the nonoverlapping structure involved PorB and PorA VR1; there is some evidence that these antigens may be relatively less immunogenic than PorA VR2 (22, 28). In conclusion, it is possible that meningococcal strains or transmission variants may, at least in part, be defined by OMP variant combinations, especially those of FetA VR and PorA VR2.
Whatever the mechanism by which meningococcal antigen variants are structured, the observed structuring greatly simplifies vaccine design. PorA and FetA are attractive vaccine candidates, as they are known to be protective and to generate bactericidal responses in both humans and animal models of infection and protection (4, 26, 30). PorA has been a major constituent of strain-specific vaccines included in vaccine trials or in development (3, 6, 25, 31). While FetA has been included in vaccines and is protective, it is relatively less exploited, as it is not expressed well in vitro (28), although it is a major outer membrane component in the iron-limited conditions prevalent in vivo (2). The PorA VRs, especially VR2, and the FetA VR are relatively long surface-exposed peptides (VR2, 8 to 24 amino acids long; FetA VR, 20 to 42 amino acids long) that are easily defined. These data suggest that a combination of PorA and FetA variants in a vaccine could be particularly effective given the strong structuring of immunogenic variants of these two antigens. This survey of 78 isolates showed that as few as six PorA variants (P1.5-1,2-2, P1.5-2,16, P1.5,10, P1.7,13-1, P1.7-2,4, and P120,9) combined with five FetA variants (F1-5, F3-1, F5-1, F3-9, and F5-5) would potentially provide homologous protection against all 78 isolates. This combination of PorA and FetA variants would potentially protect against 95 (89%) of the 107 diverse meningococcal isolates used to develop MLST, from which the 78 representatives of the hyperinvasive lineages were derived (19).
The results of this survey are consistent with a limited repertoire of antigen variant combinations dominating the major meningococcal hyperinvasive lineages. The nonoverlapping nature of combinations observed was consistent with strain structure imposed by herd immunity. If this is the case, some low-frequency variants in the population may have a short-term selective advantage within a single host, although there is some evidence that such variants are likely to be less fit during epidemic spread (1). This framework further predicts that novel variants can emerge and spread from time to time if they are distinct at multiple loci encoding immunodominant antigen variants and that distinct genotypes can possess identical antigen types; both of these phenomena are present in this data set. Moreover, as bacteria are less likely to change at two distantly located genes simultaneously, a vaccine that targets at least two distinct variable antigens will exhibit improved efficacy over a vaccine with multiple variants of a single antigen.
The isolates employed in this analysis represent the major disease-associated lineages reported over the last 60 years, and, while they are sufficient to establish the general features of meningococcal antigenic diversity discussed above, continued surveillance of current endemic and epidemic meningococcal disease is likely to be required to identify the emergence of new hyperinvasive lineages or novel antigen combinations. Analysis of additional representatives from each hyperinvasive lineage, together with carrier population samples, is also necessary if these findings are to be exploited as the basis for rational vaccine development. Such studies provide the prospect of relatively simple meningococcal OMP-based vaccines that are effective against all hyperinvasive lineages, regardless of serogroup.
Acknowledgments
This work was supported by grants from the Wellcome Trust (to R.U. and M.C.J.M.), The Meningitis Research Foundation (to J.E.R.), and the BBSRC (to E.A.L.T.). M.C.J.M. is a Wellcome Trust Senior Research Fellow.
Concatenated DNA sequences were generated with a Perl script written by Keith Jolley, University of Oxford.
REFERENCES
- 1.Achtman, M., A. van der Ende, P. Zhu, I. S. Koroleva, B. Kusecek, G. Morelli, I. G. Schuurman, N. Brieske, K. Zurth, N. N. Kostyukova, and A. E. Platonov. 2001. Molecular epidemiology of serogroup a meningitis in Moscow, 1969 to 1997. Emerg. Infect. Dis. 7**:**420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beucher, M., and P. F. Sparling. 1995. Cloning, sequencing, and characterization of the gene encoding FrpB, a major iron-regulated, outer membrane protein of Neisseria gonorrhoeae. J. Bacteriol. 177**:**2041-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjune, G., E. A. Høiby, J. K. Grønnesby, O. Arnesen, J. H. Fredriksen, A. Halstensen, E. Holten, A. K. Lindbak, H. Nøkleby, E. Rosenqvist, L. K. Solberg, O. Closs, J. Eng, L. O. Frøholm, A. Lystad, L. S. Bakketeig, and B. Hareide. 1991. Effect of outer membrane vesicle vaccine against group B meningococcal disease in Norway. Lancet 338**:**1093-1096. [DOI] [PubMed] [Google Scholar]
- 4.Black, J. R., D. W. Dyer, M. K. Thompson, and P. F. Sparling. 1986. Human immune response to iron-repressible outer membrane proteins of Neisseria meningitidis. Infect. Immun. 54**:**710-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bygraves, J. A., R. Urwin, A. J. Fox, S. J. Gray, J. E. Russell, I. M. Feavers, and M. C. J. Maiden. 1999. Population genetic and evolutionary approaches to the analysis of Neisseria meningitidis isolates belonging to the ET-5 complex. J. Bacteriol. 181**:**5551-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartwright, K., R. Morris, H. Rümke, A. Fox, R. Borrow, N. Begg, P. Richmond, and J. Poolman. 1999. Immunogenicity and reactogenicity in UK infants of a novel meningococcal vesicle vaccine containing multiple class 1 (PorA) outer membrane proteins. Vaccine 17**:**2612-2619. [DOI] [PubMed] [Google Scholar]
- 7.Caugant, D. A., L. O. Frøholm, K. Bovre, E. Holten, C. E. Frasch, L. F. Mocca, W. D. Zollinger, and R. K. Selander. 1986. Intercontinental spread of a genetically distinctive complex of clones of Neisseria meningitidis causing epidemic disease. Proc. Natl. Acad. Sci. USA 83**:**4927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caugant, D. A., B. E. Kristiansen, L. O. Frøholm, K. Bovre, and R. K. Selander. 1988. Clonal diversity of Neisseria meningitidis from a population of asymptomatic carriers. Infect. Immun. 56**:**2060-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caugant, D. A., L. F. Mocca, C. E. Frasch, L. O. Frøholm, W. D. Zollinger, and R. K. Selander. 1987. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein pattern. J. Bacteriol. 169**:**2781-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finne, J., M. Leinonen, and P. H. Makela. 1983. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet ii**:**355-357. [DOI] [PubMed] [Google Scholar]
- 11.Frasch, C. E. 1995. Meningococcal vaccines: past, present, and future, p. 246-283. In K. Cartwright (ed.), Meningococcal disease. John Wiley & Sons, Chichester, England.
- 12.Frasch, C. E., W. D. Zollinger, and J. T. Poolman. 1985. Serotype antigens of Neisseria meningitidis and a proposed scheme for designation of serotypes. Rev. Infect. Dis. 7**:**504-510. [DOI] [PubMed] [Google Scholar]
- 13.Gupta, S., and M. C. J. Maiden. 2001. Exploring the evolution of diversity in pathogen populations. Trends Microbiol. 9**:**147-192. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, S., M. C. J. Maiden, I. M. Feavers, S. Nee, R. M. May, and R. M. Anderson. 1996. The maintenance of strain structure in populations of recombining infectious agents. Nat. Med. 2**:**437-442. [DOI] [PubMed] [Google Scholar]
- 15.Holmes, E. C., R. Urwin, and M. C. J. Maiden. 1999. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol. Biol. Evol. 16**:**741-749. [DOI] [PubMed] [Google Scholar]
- 16.Jodar, L., I. M. Feavers, D. Salisbury, and D. M. Granoff. 2002. Development of vaccines against meningococcal disease. Lancet 359**:**1499-1508. [DOI] [PubMed] [Google Scholar]
- 17.Jolley, K. A., J. Kalmusova, E. J. Feil, S. Gupta, M. Musilek, P. Kriz, and M. C. Maiden. 2002. Carried meningococci in the Czech Republic: a diverse recombining population. J. Clin. Microbiol. 40**:**3549-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17**:**1244-1245. [DOI] [PubMed] [Google Scholar]
- 19.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95**:**3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maiden, M. C. J., J. Suker, A. J. McKenna, J. A. Bygraves, and I. M. Feavers. 1991. Comparison of the class 1 outer membrane proteins of eight serological reference strains of Neisseria meningitidis. Mol. Microbiol. 5**:**727-736. [DOI] [PubMed] [Google Scholar]
- 21.Malorny, B., G. Morelli, B. Kusecek, J. Kolberg, and M. Achtman. 1998. Sequence diversity, predicted two-dimensional protein structure, and epitope mapping of neisserial Opa proteins. J. Bacteriol. 180**:**1323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin, S. L., R. Borrow, P. van der Ley, M. Dawson, A. J. Fox, and K. A. V. Cartwright. 2000. Effect of sequence variation in meningococcal PorA outer membrane protein on the effectiveness of a hexavalent PorA outer membrane vesicle vaccine. Vaccine 18**:**2476-2481. [DOI] [PubMed] [Google Scholar]
- 23.Maynard Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90**:**4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olyhoek, T., B. A. Crowe, and M. Achtman. 1987. Clonal population structure of Neisseria meningitidis serogroup A isolated from epidemics and pandemics between 1915 and 1983. Rev. Infect. Dis. 9**:**665-682. [DOI] [PubMed] [Google Scholar]
- 25.Peeters, C. C. A. M., H. C. Rümke, L. C. Sundermann, E. M. Rouppe van der Voort, J. Meulenbelt, M. Schuller, A. J. Kuipers, P. van der Ley, and J. T. Poolman. 1996. Phase I clinical trial with a hexavalent PorA containing meningococcal outer membrane vesicle vaccine. Vaccine 14**:**1009-1015. [DOI] [PubMed] [Google Scholar]
- 26.Pettersson, A., B. Kuipers, M. Pelzer, E. Verhagen, R. H. Tiesjema, J. Tommassen, and J. T. Poolman. 1990. Monoclonal antibodies against the 70-kilodalton iron-regulated protein of Neisseria meningitidis are bactericidal and strain specific. Infect. Immun. 58**:**3036-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rokbi, B., M. Mignon, D. A. Caugant, and M. J. Quentin-Millet. 1997. Heterogeneity of tbpB, the transferrin-binding protein B gene, among serogroup B _Neisseria meningitidi_s strains of the ET-5 complex. Clin. Diagn. Lab. Immunol. 4**:**522-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenqvist, E., E. Arne Høiby, E. Wedege, K. Bryn, J. Kolberg, A. Klem, E. Rønnild, G. Bjune, and H. Nøkleby. 1995. Human antibody responses to meningococcal outer membrane antigens after three doses of the Norwegian group B meningococcal vaccine. Infect. Immun. 63**:**4642-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell, J. E., K. A. Jolley, I. M. Feavers, M. C. Maiden, and J. S. Suker. 2004. PorA variable regions of Neisseria meningitidis. Emerg. Infect. Dis. 10**:**674-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saukkonen, K., M. Leinonen, H. Abdillahi, and J. T. Poolman. 1989. Comparative evaluation of potential components for group B meningococcal vaccine by passive protection in the infant rat and in vitro bactericidal assay. Vaccine 7**:**325-328. [DOI] [PubMed] [Google Scholar]
- 31.Sierra, G. V. G., H. C. Campa, N. M. Varcacel, I. L. Garcia, P. L. Izquierdo, P. F. Sotolongo, G. V. Casanueva, C. O. Rico, C. R. Rodriguez, and M. H. Terry. 1991. Vaccine against group B Neisseria meningitidis: protection trial and mass vaccination results in Cuba. Nat. Inst. Public Health Ann. 14**:**195-207. [PubMed] [Google Scholar]
- 32.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5**:**233-241. [DOI] [PubMed] [Google Scholar]
- 33.Suker, J., I. M. Feavers, M. Achtman, G. Morelli, J.-F. Wang, and M. C. J. Maiden. 1994. The porA gene in serogroup A meningococci: evolutionary stability and mechanism of genetic variation. Mol. Microbiol. 12**:**253-265. [DOI] [PubMed] [Google Scholar]
- 34.Suker, J., I. M. Feavers, and M. C. J. Maiden. 1996. Monoclonal antibody recognition of members of the P1.10 variable region family: implications for serological typing and vaccine design. Microbiology 142**:**63-69. [DOI] [PubMed] [Google Scholar]
- 35.Swofford, D. 1998. PAUP*. Phylogenetic analysis using parsimony and other methods. Sinauer, Sunderland, Mass.
- 36.Tappero, J. W., R. Lagos, A. M. Ballesteros, B. Plikaytis, D. Williams, J. Dykes, L. L. Gheesling, G. M. Carlone, E. A. Hoiby, J. Holst, H. Nokleby, E. Rosenqvist, G. Sierra, C. Campa, F. Sotolongo, J. Vega, J. Garcia, P. Herrera, J. T. Poolman, and B. A. Perkins. 1999. Immunogenicity of 2 serogroup B outer-membrane protein meningococcal vaccines: a randomized controlled trial in Chile. JAMA 281**:**1520-1527. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, E. A., I. M. Feavers, and M. C. Maiden. 2003. Antigenic diversity of meningococcal enterobactin receptor FetA, a vaccine component. Microbiology 149**:**1849-1858. [DOI] [PubMed] [Google Scholar]
- 38.Urwin, R., I. M. Feavers, D. M. Jones, M. C. J. Maiden, and A. J. Fox. 1998. Molecular variation of meningococcal serotype 4 antigen genes. Epidemiol. Infect. 121**:**95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urwin, R., E. C. Holmes, A. J. Fox, J. P. Derrick, and M. C. Maiden. 2002. Phylogenetic evidence for frequent positive selection and recombination in the meningococcal surface antigen PorB. Mol. Biol. Evol. 19**:**1686-1694. [DOI] [PubMed] [Google Scholar]
- 40.Urwin, R., and M. C. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11**:**479-487. [DOI] [PubMed] [Google Scholar]
- 41.van der Ley, P., and J. T. Poolman. 1991. The class 1 outer membrane protein of Neisseria meningitidis: prediction of topology and construction of a multivalent vaccine strain, p. 295-300. In M. Achtman (ed.), Neisseria 1990. Walter de Gruyter, Berlin, Germany.
- 42.van der Ley, P., J. van der Biezen, R. Sutmuller, P. Hoogerhout, and J. T. Poolman. 1996. Sequence variability of FrpB, a major iron-regulated outer-membrane protein in the pathogenic neisseriae. Microbiology 142**:**3269-3274. [DOI] [PubMed] [Google Scholar]
- 43.Vedros, N. A. 1987. Development of meningococcal serogroups, p. 33-37. In N. A. Vedros (ed.), Evolution of meningococcal disease, vol. II. CRC Press Inc., Boca Raton, Fla. [Google Scholar]
- 44.Wang, J.-F., D. A. Caugant, G. Morelli, B. Koumaré, and M. Achtman. 1993. Antigenic and epidemiological properties of the ET-37 complex of Neisseria meningitidis. J. Infect. Dis. 167**:**1320-1329. [DOI] [PubMed] [Google Scholar]
- 45.Wedege, E., E. A. Høiby, E. Rosenqvist, and G. Bjune. 1998. Immune responses against major outer membrane antigens of Neisseria meningitidis in vaccinees and controls who contracted meningococcal disease during the Norwegian serogroup B protection trial. Infect. Immun. 66**:**3223-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wedege, E., J. Kolberg, A. Delvig, E. A. Høiby, E. Holten, E. Rosenqvist, and D. A. Caugant. 1995. Emergence of a new virulent clone within the electrophoretic type 5 complex of serogroup B meningococci in Norway. Clin. Diagn. Lab. Immunol. 2**:**314-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Womble, D. D. 2000. GCG: the Wisconsin package of sequence analysis programs. Methods Mol. Biol. 132**:**3-22. [DOI] [PubMed] [Google Scholar]
- 48.Wyle, F. A., M. S. Artenstein, B. L. Brandt, E. C. Tramont, D. L. Kasper, P. L. Altieri, S. L. Berman, and J. P. Lowenthal. 1972. Immunologic response of man to group B meningococcal polysaccharide vaccines. J. Infect. Dis. 126**:**514-521. [DOI] [PubMed] [Google Scholar]
- 49.Zapata, G. A., W. F. Vann, Y. Rubinstein, and C. E. Frasch. 1992. Identification of variable region differences in Neisseria meningitidis class 3 protein sequences among five group B serotypes. Mol. Microbiol. 6**:**3493-3499. [DOI] [PubMed] [Google Scholar]