Targeting of Aberrant mRNAs to Cytoplasmic Processing Bodies (original) (raw)
. Author manuscript; available in PMC: 2007 Jun 16.
SUMMARY
In eukaryotes, a specialized pathway of mRNA degradation termed nonsense-mediated decay (NMD) functions in mRNA quality control by recognizing and degrading mRNAs with aberrant termination codons. We demonstrate that NMD in yeast targets premature termination codon (PTC)-containing mRNA to P-bodies. Upf1p is sufficient for targeting mRNAs to P-bodies, whereas Upf2p and Upf3p act, at least in part, downstream of P-body targeting to trigger decapping. The ATPase activity of Upf1p is required for NMD after the targeting of mRNAs to P-bodies. Moreover, Upf1p can target normal mRNAs to P-bodies but not promote their degradation. These observations lead us to propose a new model for NMD wherein two successive steps are used to distinguish normal and aberrant mRNAs.
INTRODUCTION
Eukaryotic cells contain quality-control mechanisms that recognize and degrade aberrant mRNAs. One such quality-control system is referred to as nonsense-mediated decay (NMD). NMD degrades a variety of substrates with alterations in the normal spatial relationship between the termination codon and other RNA features (Hilleren and Parker, 1999; Baker and Parker, 2004; Maquat, 2004). The conserved core of the NMD machinery are the Upf1, Upf2, and Upf3 proteins, which are required for NMD in yeast, Drosophila, C. elegans, and mammalian cells (Culbertson and Leeds, 2003; Maquat, 2004). To trigger mRNA decay, NMD accelerates the rates of deadenylation, decapping, and in some cases endonucleolytic cleavage (Stevens et al., 2002; Mitchell and Tollervey, 2003; Lejeune et al., 2003; Chen and Shyu, 2003; Takahashi et al., 2003; Cao and Parker, 2003; Gatfield and Izaurralde, 2004). In yeast, NMD primarily leads to deadenylation-independent decapping (Muhlrad and Parker, 1994; Cao and Parker, 2003).
Recent studies have identified cytoplasmic foci in both yeast and mammals, referred to as P-bodies, that include the decapping enzymes (Dcp1p/Dcp2p), activators of decapping (Dhh1p, Pat1p, Lsm1–7p), and the 5′ to 3′ exonuclease Xrn1p and generally lack translation factors (van Dijk et al., 2002; Lykke-Andersen, 2002; Ingelfinger et al., 2002; Sheth and Parker, 2003; Cougot et al., 2004). P-bodies are dynamic structures and represent pools of nontranslating mRNPs (Sheth and Parker, 2003; Cougot et al., 2004; Kshirsagar and Parker, 2004; Ferraiuolo et al., 2005; Kedersha et al., 2005; Teixeira et al., 2005; Andrei et al., 2005). P-bodies have roles in mRNA decapping and 5′ to 3′ decay (Sheth and Parker, 2003; Cougot et al., 2004), in translational control (Coller and Parker, 2005), and in mRNA storage (Brengues et al., 2005). These roles suggest that cytoplasmic mRNAs shuttle in and out of P-bodies, and that P-body size and number will be a function of the rates of entry into P-bodies, decay within P-bodies, the aggregation rate of individual P-body mRNPs, and the rate of mRNAs exiting the P-body structure. An important and unresolved issue is whether the larger aggregates have compositional and/or functional differences from the individual mRNPs that are in the process of P-body assembly.
Three observations raise the possibility that NMD might involve targeting of PTC-containing mRNAs to P-bodies. First, previous work has shown that recognition of an mRNA as ‘‘nonsense containing’’ leads to its translational repression (Muhlrad and Parker, 1999). Second, Dcp1p, Dcp2p, and Xrn1p, which catalyze the degradative phase of NMD, are concentrated in P-bodies (Sheth and Parker, 2003). Third, SMG5, SMG7, and UPF1, proteins involved in NMD in higher eukaryotes, are localized to P-bodies in mammalian cells under certain conditions (Unterholzner and Izaurralde, 2004; Fukuhara et al., 2005).
Herein, we present evidence that NMD in yeast involves formation of an mRNP that accumulates within P-bodies. Moreover, we demonstrate that Upf1p is sufficient for targeting mRNAs to P-bodies with Upf2p and Upf3p acting later to trigger decapping of NMD substrates. Our results suggest that Upf1p also targets normal mRNAs to P-bodies and that normal and NMD substrates may be distinguished by a two-step sequential process, which would be expected to increase the accuracy of the NMD selection process.
RESULTS
Upf1p, Upf2p, and Upf3p Can Localize to P-Bodies
Since Upf1p, Upf2p, and Upf3p are required for NMD, the presence of these proteins in P-bodies would argue for the occurrence of NMD in P-bodies. To address this issue, a C-terminal GFP fusion of each Upf protein was constructed and the cellular localization of the chimeras determined in wild-type cells. Northern analysis of the CYH2 pre-mRNA, which is a substrate for NMD (He et al., 1993), indicated that the tagged Upf1p, Upf2p, and Upf3p are fully functional (data not shown). Consistent with prior work (Atkin et al., 1995; Shirley et al., 1998), GFP-tagged Upf1p, Upf2p, and Upf3p were uniformly distributed in the cytoplasm (Figure 1A, top panel). However, if Upf proteins target mRNA to P-bodies but are released rapidly after the degradation of the mRNA, they might only accumulate in P-bodies if degradation of the mRNA within P-bodies were blocked.
Figure 1. Upf Proteins Are Localized to P-Bodies.
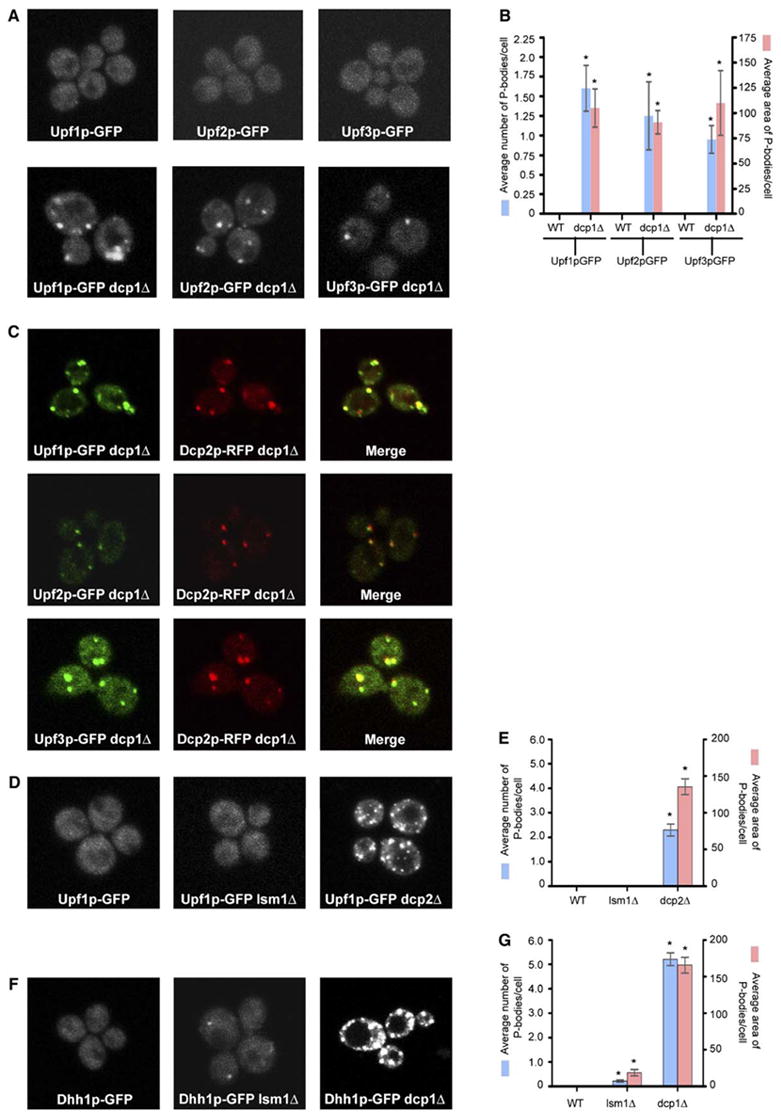
(A) Localization of Upf1p-GFP (left), Upf2p-GFP (middle), and Upf3p-GFP (right) in wt strain (top) and in dcp1Δ strain (bottom).
(B) Histogram represents average number of P-bodies/cell (left Y axis) and average area of P-bodies/cell in pixels^2 (right Y axis) for Upf1p-GFP, Upf2p-GFP, and Upf3p-GFP. * indicates p < 0.05 for wt versus dcp1Δ strain.
(C) Panel on the left (vertical) shows localization of GFP-tagged Upf proteins; middle panel (vertical) shows localization of RFP-tagged Dcp2p; and panel on the right (vertical) shows the merge generated using Adobe Photoshop. Top row shows localization of Upf1p, middle panel shows the localization of Upf2p, and bottom panel shows localization of Upf3p.
(D) shows localization of Upf1p-GFP in wt strain (left), lsm1Δ strain (middle), and dcp2Δ strain (right).
(E) Histogram represents average number of P-bodies/cell (left Y axis) and average area of P-bodies/cell in pixels^2 (right Y axis) for Upf1p-GFP. * indicates p < 0.005 for wt versus dcp2Δ strain.
(F) Localization of Dhh1p-GFP in wt strain (left), lsm1Δ strain (middle), and dcp1Δ strain (right).
(G) Histogram represents average number of P-bodies/cell (left Y axis) and average area of P-bodies/cell in pixels^2 (right Y axis) for Dhh1p-GFP. * indicates p < 0.05 for wt versus lsm1Δ and dcp1Δ strain.
Data are represented as mean of three experiments ± SD.
To test this model, we localized the Upf proteins in strains blocked at the catalytic steps of decay. In dcp1Δ, dcp2Δ, and xrn1Δ strains, Upf1p, Upf2p, and Upf3p were present in cytoplasmic foci (Figure 1A, bottom panel and data not shown), which in the dcp1Δ strain colocalized with Dcp2p-RFP (Figure 1C). Quantification of the images (see Experimental Procedures) indicated an increase in both the number and area of Upf-GFP signal within P-bodies (Figure 1B). These foci, and all other microscopic data presented in this manuscript, unless explicitly stated, were seen in greater than 80% of cells examined. This observation suggests that the Upf proteins cycle through P-bodies and accumulate in P-bodies when decapping is blocked.
Since Dcp1p is involved in the decay of both normal and nonsense-containing mRNAs (Beelman et al., 1996), the accumulation of Upf proteins in P-bodies in a dcp1Δ strain could be due to the accumulation of normal and/or nonsense-containing mRNAs in these foci. To determine if the accumulation of Upf proteins in P-bodies was specific to NMD, we examined the distribution of Upf1p-GFP in a lsm1Δ strain, which is only defective in the decapping of only normal mRNAs (Boeck et al., 1998). In the lsm1Δ strain, Upf1p-GFP is not concentrated in P-bodies (Figure 1D, middle panel) as compared to a dcp1Δ strain (Figure 1A, bottom panel) or a dcp2Δ strain (Figures 1D, right panel, and 1E). Similar results were also obtained for Upf2p-GFP and Upf3p-GFP (data not shown). In contrast, Dhh1p, an activator of decapping involved in the 5′ to 3′ decay of normal mRNA (Coller et al., 2001), accumulated in P-bodies in lsm1Δ, dcp1Δ, and dcp2Δ strains (Figures 1F, middle panel and left panel and 1G and data not shown). This result indicates that the presence of Upf proteins in P-bodies is due to a defect in NMD and implies that NMD targets PTC-containing mRNAs to P-bodies.
A Reporter mRNA Harboring a Nonsense Mutation Localizes to P-Bodies
To directly test if NMD targets mRNAs to P-bodies, we examined the subcellular distribution of PGK1 mRNAs with early (codon 22) or late (codon 225) nonsense codons. We examined reporter mRNAs with early and late nonsense codons since they differ in their rates of decapping, have different rate-limiting steps in decay, and therefore may respond differently in the NMD process (Cao and Parker, 2003). These reporter mRNAs contained 16 U1A binding sites in the 3′ UTR of the mRNA, which did not prevent NMD (Figure S1), and a U1A-GFP fusion protein was coexpressed, which allowed visualization of transcript localization (Bertrand et al., 1998; Brodsky and Silver, 2000; Teixeira et al., 2005). When expressed by itself, the U1A-GFP fusion protein showed no accumulation in P-bodies (Figures 2A, top panel, and 2B). As compared to the wild-type (wt) PGK1 mRNA, both the C22 and C225 mRNAs showed an increase in the number of P-bodies and their overall area, as detected by the tethered U1A-GFP (Figure 2A, right panel compare panel v (wt) to panel ix (C22) and panel xiii (C225) and Figure 2B). Furthermore, these mRNA-containing foci colocalized with RFP-tagged Dcp2p (Figure S2). These results show that PTC-containing mRNAs accumulate in P-bodies.
Figure 2. Nonsense-Containing Full-Length Reporter mRNA Localizes to P-Bodies.
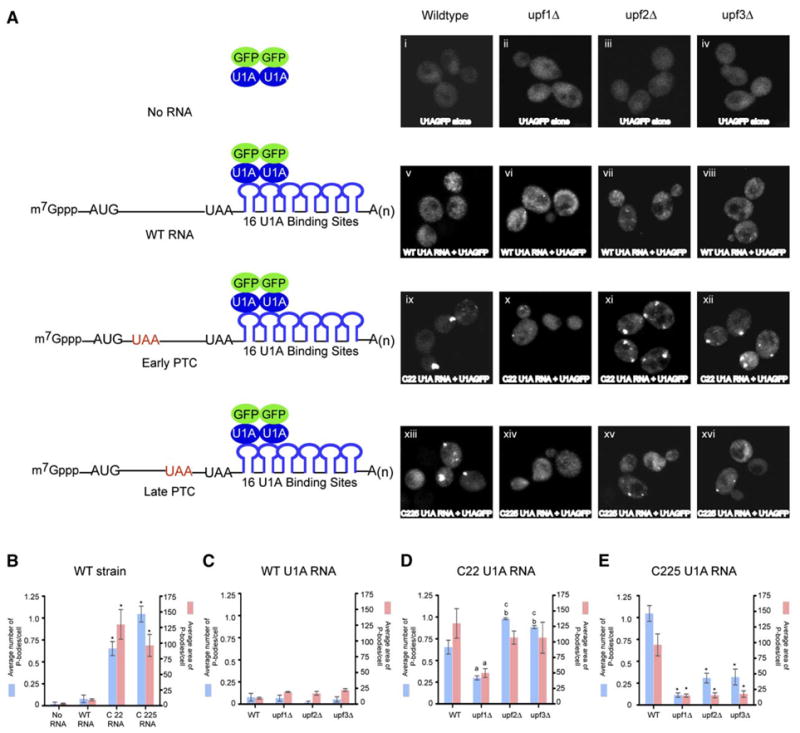
(A) First panel from left to right: Plasmid expressing U1A-GFP alone in wt (i), upf1Δ (ii), upf2Δ (iii), and upf3Δ (iv) strains.
Second panel from left to right: Wild-type PGK1 reporter mRNA with U1A binding sites and plasmid expressing U1A-GFP in wt (v), upf1Δ (vi), upf2Δ (vii), and upf3Δ (viii) strains.
Third panel from left to right: PGK1 mRNA with a premature stop codon at position 22 with U1A binding sites and plasmid expressing U1A-GFP in wt (ix), upf1Δ (x), upf2Δ (xi), and upf3Δ (xii) strains.
Fourth panel from left to right: PGK1 mRNA with a premature stop codon at position 225 with U1A binding sites and plasmid expressing U1A GFP in wt (xiii), upf1Δ (xiv), upf2Δ (xv), and upf3Δ (xvi) strains.
All the mRNAs lack the pG tract to trap the decay intermediate. The schematic diagram of the reporter RNA and its interaction with the U1A-GFP fusion protein is shown on the left.
(B) Cells containing no RNA, wt U1A RNA, C22 U1A RNA, and C225 U1A RNA mRNAs in a wt strain. * indicates p < 0.05 for wt U1A mRNA versus C22 and C225 U1A mRNAs.
(C) Cells containing wt U1A reporter mRNA in wt, upf1Δ, upf2Δ, and upf3Δ strains.
(D) Cells containing C22 U1A reporter mRNA in wt, upf1Δ, upf2Δ, and upf3Δ strains. a indicates p < 0.05 for wt strain versus upf1Δ strain, b indicates p < 0.05 for wt strain versus upf2Δ and upf3Δ strains, and c indicates p < 0.0001 for upf1Δ versus upf2Δ and upf3Δ strains.
(E) Cells containing C225 U1A reporter mRNA in wt, upf1Δ, upf2Δ, and upf3Δ strains. * indicates p < 0.05 for wt strain versus upf1Δ, upf2Δ, and upf3Δ strains.
Data are represented as mean of three experiments ± SD. Total of 50 cells were analyzed for this quantification.
To determine if the accumulation of the C22 and C225 mRNAs in P-bodies was due to NMD, we examined how this localization was affected in upf1Δ, upf2Δ, and upf3Δ strains. In upf1Δ strain, the accumulation of both the C22 and C225 reporter mRNAs in P-bodies was reduced both in the number of P-bodies observed and their overall area (Figure 2A, x and xiv; Figures 2D and 2E). The continued accumulation of C22 mRNA in P-bodies in the upf1Δ strain is likely to be due to the short ORF on this transcript, and the inverse relationship seen between an mRNA being associated with ribosomes or accumulating in P-bodies (Brengues et al., 2005; Teixeira et al., 2005). It should be noted that the decrease in P-body accumulation in the upf1Δ strain was observed despite an increase in the levels of the C22 and C225 mRNAs in the upf1Δ strain due to an inhibition of NMD (Figure S1). This observation argues that Upf1p is required for the targeting of NMD substrates to P-bodies.
The C22 and C225 mRNAs behaved slightly differently in upf2Δ and upf3Δ strains. The C22 mRNA showed an approximately 2-fold increase in P-bodies both in number and area in upf2Δ and upf3Δ strains as compared to the upf1Δ strain (Figures 2A, xi and xii, and 2D). The C225 mRNA also accumulated more detectable P-bodies in upf2Δ and upf3Δ strains as compared to the upf1Δ strain, although the overall area of these P-bodies was not significantly different (Figure 2A, xv and xvi, and 2E). These results suggest that Upf1p is the critical protein for targeting mRNAs to P-bodies whereas Upf2p and Upf3p function, at least in part, after P-body targeting of the mRNA (see below). However, the reduction of the C225 mRNA in P-bodies in upf2Δ and upf3Δ strains as compared to wt strain suggests that Upf2p and Upf3p might also affect the efficiency with which the C225 mRNA is targeted to P-bodies (see Discussion).
Despite the differences in subcellular mRNA location, all three Upf proteins are required for the rapid decay of the C22 (early PTC) and C142 (intermediate PTC) transcripts (Figure S3 and data not shown). This suggests that localization of the PTC-containing mRNA to P-bodies by Upf1p is not sufficient for its degradation and that Upf2p and Upf3p are required to trigger decay of the mRNA.
Upf1p Acts upstream of Upf2p and Upf3p
The above observations suggest that a substep in NMD is the targeting of the mRNA to a specific mRNP complex, which can accumulate, and be detected, as P-bodies. This accumulation of mRNAs and proteins in P-bodies then becomes an assay for a substep in the NMD process. Thus, we examined whether each Upf protein acts upstream or downstream of P-body formation by using Dcp2p-GFP to monitor P-body formation in upf mutants. Both the number and overall area of Dcp2p-GFP in P-bodies increased in upf2Δ, upf3Δ, and upf2Δupf3Δ strains (Figure 3A, compare i to iii, iv, vii, and viii). In contrast, Dcp2p-GFP in the upf1Δ strain was distributed similarly to the wild-type strain (Figure 3A, compare i and ii, and viii). Moreover, GFP-tagged Dcp2p did not show increased accumulation in P-bodies in the upf1Δupf2Δ or upf1Δupf3Δ strain (Figure 3A, v, vi, and viii), indicating that the accumulation of P-bodies in upf2Δ and up3Δ strains is dependent on Upf1p. The changes in the P-bodies are not due to differences in Dcp2p-GFP abundance since the level of Dcp2p-GFP does not significantly differ in these strains (Figure S4). These observations argue that Upf1p functions upstream of Upf2p and Upf3p.
Figure 3. Analysis of Upf Mutants.
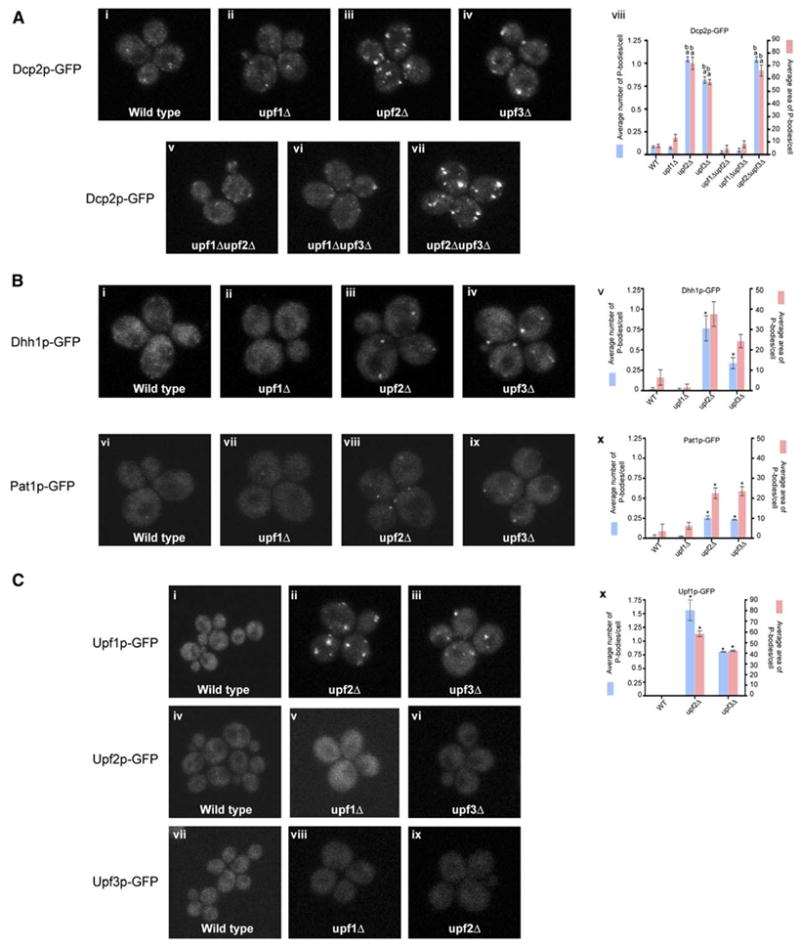
(A) Top panel (left to right): localization of Dcp2p-GFP in, wt (i), upf1Δ (ii), upf2Δ (iii), and upf3Δ (iv) strains.
Bottom panel (left to right): localization of Dcp2p-GFP in upf1Δupf2Δ (v), upf1Δupf3Δ (vi), and upf2Δupf3Δ (vii) strains.
(viii) Histogram represents average number of P-bodies/cell (left Y axis) and average area of P-bodies/cell in pixels^2 (right Y axis) for Dcp2p-GFP in wt, upf1Δ, upf2Δ, upf3Δ, upf1Δupf2Δ, upf1Δupf3Δ, and upf2Δupf3Δ strains. a indicates p < 0.0005 for wt strain versus upf2Δ, upf3Δ, and upf2Δupf3Δ strains (for wt strain versus upf1Δ, upf1Δ upf2Δ, and upf1Δupf3Δ strains the P values were high, indicating that they are very similar) and b indicates p < 0.0005 for upf1Δ strain versus upf2Δ, upf3Δ, and upf2Δupf3Δ strains.
(B) Top panel (left to right): localization of Dhh1p-GFP in, wt (i), upf1Δ (ii), upf2Δ (iii), and upf3Δ (iv) strains.
(v) Histogram represents average number of P-bodies/cell (left Y axis) and average area of P-bodies/cell in pixels^2 (right Y axis) for Dhh1p-GFP in wt, upf1Δ, upf2Δ, and upf3Δ strains. * indicates p < 0.05 for wt strain versus upf2Δ and upf3Δ strains.
Bottom panel (left to right): localization of Pat1p-GFP in wt (vi), upf1Δ (vii), upf2Δ (viii), and upf3Δ (ix) strains.
(x) Histogram represents average number of P-bodies/cell (left Y axis) and average area of P-bodies/cell in pixels^2 (right Y axis) for Pat1p-GFP in wt, upf1Δ, upf2Δ, and upf3Δ strains. * indicates p < 0.05 for wt strain versus upf2Δ and upf3Δ strains.
(C) Top panel: localization of Upf1p-GFP in wt (i), upf2Δ (ii), and upf3Δ (iii) strains. Middle panel: localization of Upf2p-GFP in wt (iv), upf1Δ (v), and upf3Δ (vi) strains. Bottom panel: Localization of Upf3p-GFP in wt (vii), upf1Δ (viii), and upf2Δ (ix) strains.
(x) Histogram represents average number of P-bodies/cell (left Y axis) and average area of P-bodies/cell in pixels^2 (right Y axis) for Upf1p-GFP in wt, upf2Δ, and upf3Δ strains. * indicates p < 0.005 for wt strain versus upf2Δ and upf3Δ strains.
Data are represented as mean of three experiments ± SD.
To identify other proteins that accumulate in P-bodies in upf2Δ and upf3Δ strains, we examined the localization of Dcp1p-GFP, Xrn1-GFP, Dhh1p-GFP, Pat1p-GFP, and Lsm1p-GFP in different upf mutants. As assessed by both average P-body number and area, Dcp1p, Xrn1p, Dhh1p, Pat1p, and Lsm1p accumulated in P-bodies in upf2Δ and upf3Δ strains (Figure 3B, iii, iv, v, viii, ix, and x and data not shown). This observation suggests two possibilities. First, normal mRNAs in the process of decay might accumulate in P-bodies in upf2Δ and upf3Δ strains. However, this possibility is unlikely since normal mRNAs do not accumulate in P-bodies in upf2Δ and upf3Δ strains (Figure 2C). More plausibly, when Upf1p targets an mRNA to assemble a P-body complex it may recruit the decapping enzyme as part of a larger complex consisting of the decapping enzymes (Dcp1p/Dcp2p), activators of decapping (Dhh1p, Pat1p, and Lsm1–7p), and the 5′ to 3′ exonuclease Xrn1p.
Interdependence of Upf Proteins
Examining the location of each of the Upf proteins in strains lacking one of the other two Upf proteins provided three observations. First, Upf1p-GFP accumulated in P-bodies in upf2Δ or upf3Δ and upf2Δupf3Δ strains (Figure 3C, ii, iii, and x and data not shown). This further supports the model that Upf1p functions upstream, and independently, of Upf2p and Upf3p. This result also implies that Upf2p and Upf3p are required for degradation of the mRNA after P-body targeting. A second observation was that Upf2p and Upf3p do not accumulate in P-bodies in an upf1Δ strain (Figure 3C, v and viii) consistent with Upf1p acting upstream of Upf2p and Upf3p. Third, in contrast to Upf1p, neither Upf2p nor Upf3p accumulated in foci in upf3Δ or upf2Δ strains, respectively (Figure 3C, vi and ix). This suggests that the stable accumulation of Upf2p and Upf3p in P-bodies is interdependent.
These results argue that Upf1p is required for targeting of PTC-containing mRNAs to P-bodies. In contrast, in strains lacking Upf2p and Upf3p, mRNAs and associated proteins accumulate in P-bodies, presumably due to the action of Upf1p, but are not degraded. This is also supported by the observation that the localization of a reporter mRNA harboring an early PTC to P-bodies is decreased in upf1Δ, whereas its levels increase in upf2Δ and upf3Δ strains (Figures 2A and 2D).
Upf1p Has a Central Role in Targeting mRNAs to P-Bodies
Upf1p is a 5′ to 3′ ATP-dependent RNA helicase and its ATPase activity is essential for NMD in an unknown manner (Weng et al., 1996). One possibility is that the ATPase activity of Upf1p may be required for the initial targeting of mRNAs to P-bodies. This model predicts that a strain expressing an ATPase-defective allele of Upf1p would not be able to target mRNAs to P-bodies and therefore P-bodies would decrease in size. Alternatively, the ATPase domain could be required for degradation of the mRNA substrate, after targeting to a P-body. In this case, a strain expressing an ATPase-defective allele of Upf1p would show an increase in P-bodies.
To test these predictions, we expressed wt Upf1p and DE572AA, an ATPase-defective upf1 allele (Weng et al., 1996) from a low-copy plasmid in an upf1Δ strain and examined P-body number and area. We observed that Dcp2p-GFP accumulated in P-bodies in cells expressing the DE572AA upf1 allele (Figure 4A, iii and iv) but not in strains containing either wt Upf1p or empty vector (Figure 4A, i, ii, and iv). This suggests that the ATP hydrolysis activity of Upf1p is not required for P-body targeting and instead may promote a rearrangement of the mRNP within P-bodies that can trigger mRNA degradation.
Figure 4. Analysis of the DE572AA upf1 Allele.
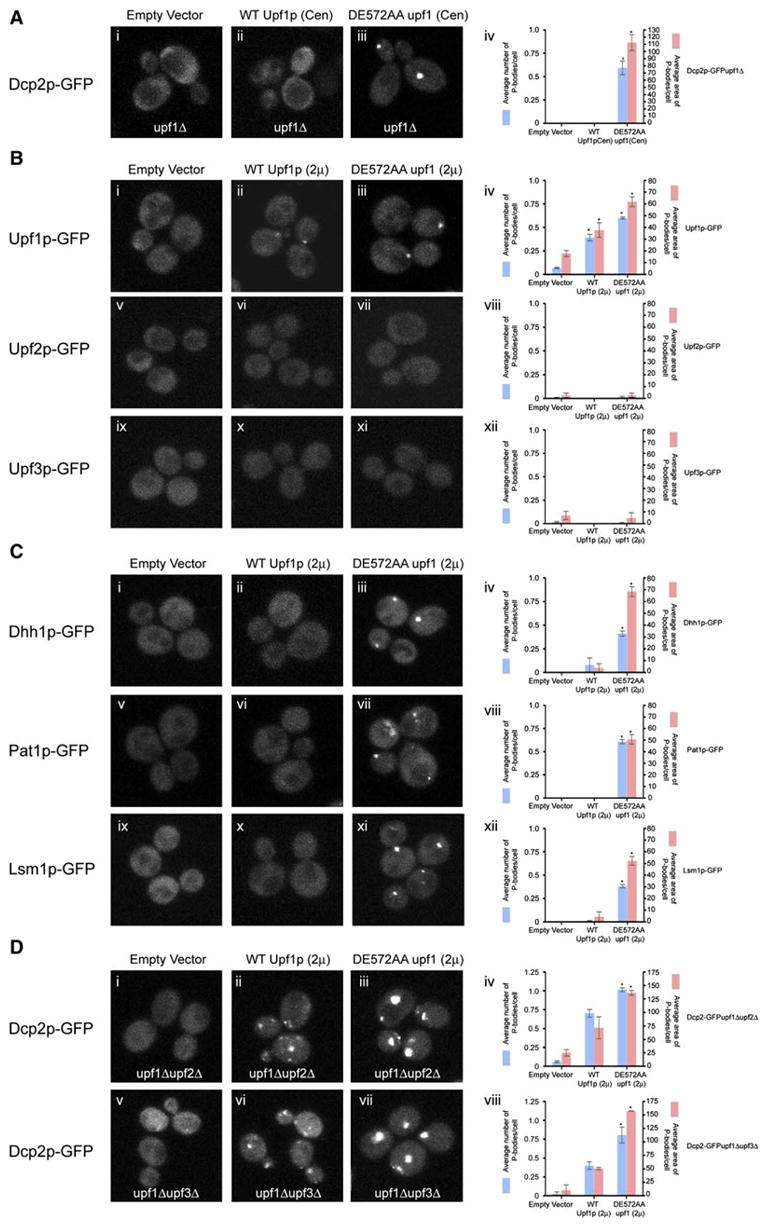
(A) upf1Δ strain transformed with low-copy plasmid expressing empty vector (pRP415) (i), wt Upf1p (pRP910) (ii), DE572AA upf1 allele (pRP912) (iii). All strains are transformed with plasmid containing Dcp2p-GFP as a marker for P-bodies.
(iv) Histogram represents average number of P-bodies/cell (left Y axis) and average area of P-bodies/cell in pixels^2 (right Y axis) for Dcp2p-GFP in upf1Δ strain containing empty vector, wt Upf1p, or DE572AA upf1 allele. * indicates p < 0.005 for vector versus DE572AA upf1 allele.
(B) Localization of Upf1p-GFP (top), Upf2p-GFP (middle), Upf3p-GFP (bottom) in wt strain overexpressing empty vector (pRP415) (i, v, ix), or wt Upf1p (pRP913) (ii, vi, x), or DE572AA upf1 allele (pRP915) (iii, vii, xi), respectively.
Histograms represent average number of P-bodies/cell (left Y axis) and average area of P-bodies/cell in pixels^2 (right Y axis) for:
(iv) Upf1-GFP strain containing empty vector, wt Upf1p, or DE572AA upf1 allele. * indicates p < 0.05 for vector versus wt Upf1p and DE572AA upf1 allele.
(viii) Upf2p-GFP strain containing vector, wt Upf1p, or DE572AA upf1 allele.
(xii) Upf3p-GFP strain containing vector, wt Upf1p, or DE572AA upf1 allele.
(C) Localization of Dhh1p-GFP (top), Pat1p-GFP (middle), Lsm1p-GFP (bottom) in wt strain overexpressing empty vector (i, v, ix), or wt Upf1p (ii, vi, x), or DE572AA upf1 allele (iii, vii, xi).
Histograms represent average number of P-bodies/cell (left Y axis) and average area of P-bodies/cell in pixels^2 (right Y axis) for:
(iv) Dhh1p-GFP strain containing empty vector, wt Upf1p, or DE572AA upf1 allele. * indicates p < 0.0001 for vector versus DE572AA upf1 allele. (viii) Pat1p-GFP strain containing empty vector, wt Upf1p, or DE572AA upf1 allele. * indicates p < 0.0001 for vector versus DE572AA upf1 allele. (xii) Lsm1p-GFP strain containing empty vector, wt Upf1p, or DE572AA upf1 allele. * indicates p < 0.0001 for vector versus DE572AA upf1 allele. (D) Localization of Dcp2p-GFP in upf1Δupf2Δ strain (top) or in upf1Δupf3Δ strain (bottom) overexpressing empty vector (i, v), or wt Upf1p (ii, vi), or DE572AA upf1 allele (iii, vii).
Histograms represent average number of P-bodies/cell (left Y axis) and average area of P-bodies/cell in pixels^2 (right Y axis) for:
(iv) Dcp2p-GFP in upf1Δupf2Δ strain containing empty vector, wt Upf1p, or DE572AA upf1 allele. * indicates p < 0.005 for vector versus DE572AA upf1 allele.
(viii) Dcp2p-GFP in upf1Δupf3Δ strain containing empty vector, wt Upf1p, or DE572AA upf1 allele. * indicates p < 0.05 for vector versus DE572AA upf1 allele.
Data are represented as mean of three experiments ± SD.
Additional evidence for the role of the ATP hydrolysis activity of Upf1p comes from overexpression of the DE572AA upf1 allele from a two-micron plasmid in wild-type strains expressing GFP-tagged Upf1p, Upf2p, Upf3p, Dhh1p, Pat1p, or Lsm1p. GFP-tagged Upf1p, Dhh1p, Pat1p, and Lsm1p localized to P-bodies in strains overexpressing the DE572AA upf1 allele (Figures 4B, iii and iv; 4C, iii, iv, vii, viii, xi and xii). Overexpressing wt Upf1p, or presence of empty vector, had no effect on the localization of all the proteins tested except for Upf1p where overexpression of wt Upf1p led to a slight increase in P-bodies (Figures 4B, i, ii and iv; 4C, i, ii, iv, v, vi, viii, ix, x, and xii). Moreover, the accumulation of P-bodies in strains overexpressing the DE572AA upf1 allele is independent of Upf2p and Upf3p, as shown by the accumulation of Dcp2p-GFP in upf2Δ and upf3Δ strains (Figure 4D, iii, iv, vii, and viii). In contrast to Upf1p, GFP-tagged Upf2p and Upf3p did not localize to P-bodies in strains overexpressing either wt Upf1p or the DE572AA upf1 allele (Figure 4B, vi, vii, viii, x, xi, and xii). This observation indicates that either the DE572AA upf1 allele may have lost its ability to recruit Upf2p or Upf3p or that Upf2p and Upf3p may associate with the mRNP after the ATP hydrolysis step.
Upf1p Can Target Normal mRNAs to P-Bodies
The accumulation of proteins in P-bodies in the DE572AA allele of Upf1p argues that some mRNAs are accumulating in P-bodies in this strain. Thus, we examined the localization of both normal and nonsense mRNAs in an upf1Δ strain expressing the DE572AA upf1 allele. The P-body accumulation of the PTC (C22)-containing PGK1 reporter mRNA slightly increased in the strain expressing the DE572AA upf1 allele as compared to a strain expressing wt Upf1p (Figures 5A, bottom panel compare left to right, and 5B, bottom panel). These results argue that ATP hydrolysis by Upf1p is not required for localization of nonsense mRNAs, or associated proteins, to a P-body.
Figure 5. ATPase Activity of Upf1p Is Not Required for Targeting of mRNAs to P-Bodies.
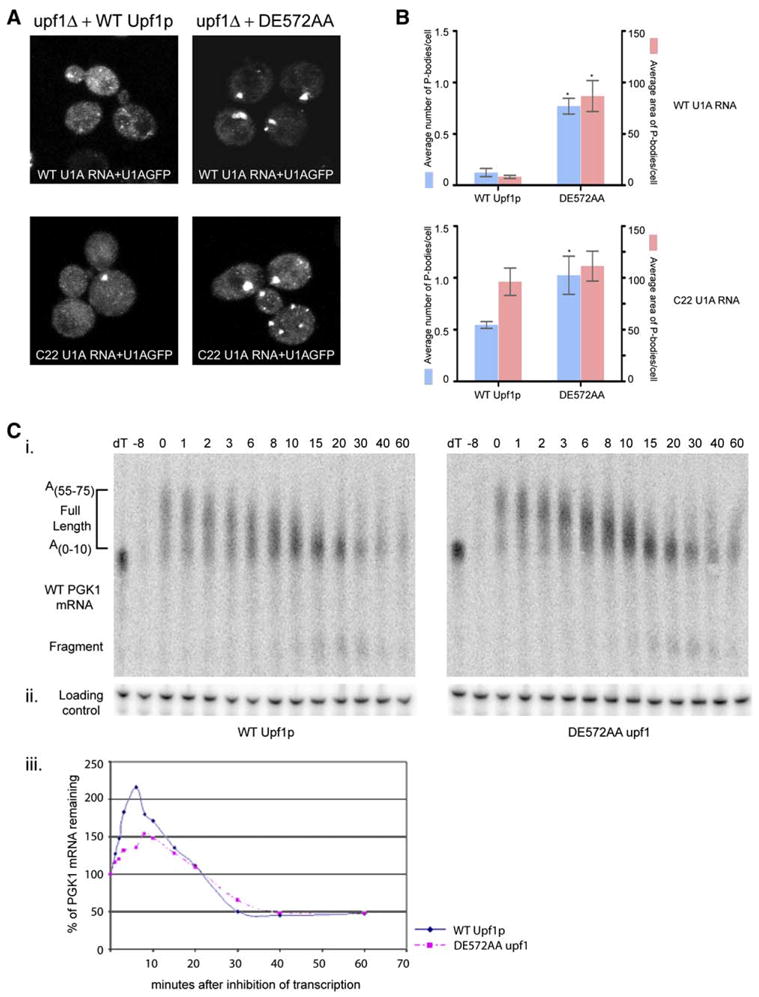
(A) Localizing wt U1A RNA (top) or C22 U1A RNA (bottom) in upf1Δ strain transformed with low copy plasmid expressing wt Upflp (left panel) or DE572AA upf1 allele (right panel).
(B) Histograms represent average number of P-bodies/cell (left Y axis) and average area of P-bodies/cell in pixels^2 (right Y axis) for:
Top panel: wt U1A RNA in upf1Δ strain expressing either wt Upflp or DE572AA upf1 allele. * indicates p < 0.05 for wt Upflp versus DE572AA upf1 allele. Bottom panel: C22 U1A RNA in upf1Δ strain expressing either wt Upflp or DE572AA upf1 allele. * indicates p < 0.05 for wt Upflp versus DE572AA upf1 allele.
Data are represented as mean of three experiments ± SD. Total of 50 cells were analyzed for this quantification.
(C) (i) Transcriptional pulse-chase of wt PGK1 mRNA in upf1Δ strain expressing wt Upflp (left panel) or DE572AA upf1 allele (right panel). Plasmid pRP469 was introduced into the strains by transformation and transcriptional pulse-chase performed as previously described (Decker and Parker, 1993).
(ii) shows the loading control using the oligo (oRP100) for the 7S RNA.
(iii) Graph showing the decay profile for wt PGK1 mRNA in upf1Δ strain expressing wt Upflp (blue) or DE572AA upf1 allele (pink).
Surprisingly, the expression of the DE572AA upf1 allele also led to clear accumulation of wt mRNAs in P-bodies as assessed both by the number of P-bodies containing this mRNA and their overall area (Figures 5A, top panel compare left to right, and 5B, top panel). This observation implies that Upf1p can also target normal mRNAs to P-bodies and that they accumulate in P-bodies either because ATP hydrolysis is required for their degradation or for their release back into translation. Transcriptional pulse-chase experiments, which allow measurement of the rates of both deadenylation and decapping of the mRNA (Cao and Parker, 2003), indicated that deadenylation, decapping, and overall decay rates of the wt PGK1 mRNA were not affected by the DE572AA upf1 allele (Figure 5C, i, ii, and iii). Moreover, we did not observe even a small pool of mRNAs which might be stuck in a nonproductive intermediate in the DE572AA allele and thereby be responsible for the increased P-bodies. This suggests that the ATP hydrolysis activity of Upf1p is likely to be required to release a pool of normal mRNAs that can be targeted to P-bodies by Upf1p back into the translating pool.
The observation that Upf1p leads to the accumulation of normal mRNAs in P-bodies suggests that Upf1p can repress the translation of normal mRNAs. This predicts that overexpression of Upf1p might generally affect translation. To test this possibility, we overexpressed either wt Upf1p or the DE572AA upf1 allele from the galactose promoter in wild-type strain and examined P-body formation and polysome profiles 2 hr after induction with galactose. Compared to vector alone (Figure 6A), overexpression of the DE572AA upf1 allele reduced polysomes (Figure 6C, compare the red polysome trace [sucrose] to the gray [galactose] polysome trace) and showed a clear accumulation of P-bodies (Figure 6C, compare ii to iii). In addition to a reduction in polysomes and an increase in P-body size, overexpression of the DE572AA upf1 allele also inhibited growth (Figure 6C, compare iv to v) whereas overexpression of wt Upf1 caused a minor, but clear, decrease in growth rate (Figure 6B, compare iv to v). These results are consistent with Upf1p repressing the translation of a pool of ‘‘normal’’ mRNAs and targeting those transcripts to P-bodies. Moreover, because the DE572AA allele is more severe than overexpression of the wt Upf1p, it implies that the DE572AA is inhibiting the recycling of ‘‘normal’’ mRNAs back into translation (see Discussion).
Figure 6. Overexpression of DE572AA upf1 Allele Causes Translational Repression, an Increase in P-Bodies, and Inhibition of Growth.
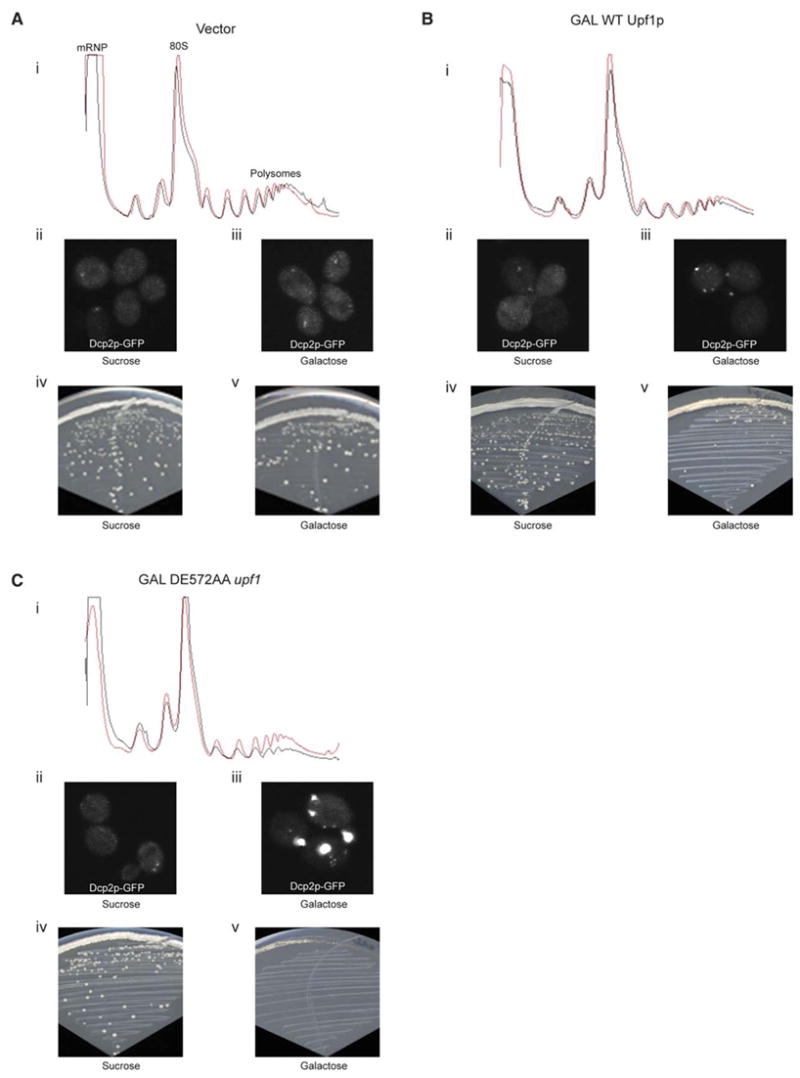
Polysome profiles (i, sucrose in red and galactose in gray), P-body accumulation (ii and iii), and growth assays (iv and v) in wt strain expressing empty vector (A), or overexpressing wt Upf1p (B), or overexpressing DE572AA upf1 allele (C). All the polysome profiles were obtained using UV monitor Uvicord SII, Pharmacia at 254 nm wavelength and 0.5AUFS sensitivity. P-bodies were visualized using Dcp2p-GFP.
DISCUSSION
NMD Involves Targeting of PTC-Containing Substrates to P-Bodies
Several observations indicate that the process of NMD involves the assembly of an mRNP, which targets the mRNA for decapping and to P-bodies. First, Upf1p, Upf2p, and Upf3p are localized to P-bodies when NMD is inhibited at the nucleolytic steps in degradation (Figure 1). Second, this accumulation of the Upf proteins in P-bodies is specific to the NMD pathway and does not occur when decapping of normal mRNAs is inhibited (Figure 1). Third, reporter mRNAs harboring PTCs are localized to P-bodies in an Upf1p-dependent manner (Figure 2). Fourth, when NMD is blocked in an upf2Δ or upf3Δ strain, or by an ATPase-defective upf1 allele, P-bodies increase in number and area (Figures 3 and 4). These observations argue that NMD in yeast occurs within complexes that can accumulate into P-bodies. Moreover, because the basic principles of NMD are broadly conserved, and SMG5, SMG7, and UPF1 proteins can accumulate in P-bodies in mammalian cells (Unterholzner and Izaurralde, 2004), it is anticipated that NMD will occur within P-bodies in other eukaryotes, including mammals. An unresolved issue is whether NMD requires assembly of aggregates into a larger structure to occur, or if decapping can occur within individual mRNPs that can eventually accumulate within P-bodies.
Upf1p Acts upstream of Upf2p and Upf3p to Promote P-Body mRNP Formation
Multiple lines of evidence suggest that Upf1p acts first in NMD and is required for targeting of PTC-containing mRNAs to a P-body mRNP whereas Upf2p and Upf3p act later. First, Upf1p-GFP and Dcp2p-GFP accumulate in P-bodies in upf2Δ or upf3Δ strains (Figure 3). Moreover, the accumulation of Dcp2p-GFP in P-bodies is dependent on Upf1p since P-body number and area decrease in upf1Δupf2Δ and upf1Δupf3Δ strains as compared to upf2Δ or upf3Δ strains (Figure 3). Second, Upf2p and Upf3p do not accumulate in P-bodies in an upf1Δ strain (Figure 3). Third, overexpression of the DE572AA upf1 allele causes accumulation of Dcp2p in P-bodies independent of Upf2p or Upf3p (Figure 4). Moreover, reporter mRNA localizations are also consistent with this model, as mRNAs harboring both early as well as late nonsense codons localize to P-bodies in an Upf1p-dependent manner and independent of Upf2p and Upf3p (Figure 2). Indeed, mRNAs with early nonsense codons also show a slight increase in their accumulation in P-bodies in upf2Δ and upf3Δ strains as compared to upf1Δ strain (Figure 2). These observations argue that Upf1p acts first, and independent of Upf2p and Upf3p, in formation of a P-body mRNP, while Upf2p and Upf3p function, at least in part, to subsequently trigger decapping and 5′ to 3′ decay of NMD substrates. Note that because Upf1p and Dcp2p accumulate in P-bodies in upf2Δ and upf3Δ strains, yet the mRNA is not subjected to NMD, these results imply that interaction of the mRNA substrate with Upf1p and Dcp2p, and its accumulation within P-bodies, is not sufficient to trigger mRNA decay.
Since the efficient targeting of a reporter mRNA harboring a late PTC to P-bodies requires all three Upf proteins (Figure 2), it suggests that Upf2p and Upf3p may also increase the efficiency of recognition of aberrant termination events. However, some phenotypes of the upf2Δ and upf3Δ strains could be due to the accumulation of Upf1p in P-bodies, thereby limiting Upf1p. Consistent with this possibility, it has been observed that some of the phenotypes of the upf2Δ and upf3Δ strains are partially suppressed by overexpression of Upf1p (Maderazo et al., 2000). Thus, whether Upf2p and Upf3p also have a direct role early in the NMD process remains to be clarified.
Another line of evidence for the role of Upf1 in the formation of the P-body complex comes from our studies of the DE572AA upf1 allele. Strains expressing the DE572AA upf1 allele accumulate P-body complexes, demonstrated by the accumulation of Upf1p, Dcp2p, Dhh1p, Pat1p, and Lsm1p (Figure 4). In addition, the expression of the DE572AA upf1 allele leads to an increase in accumulation of nonsense mRNAs in P-bodies (Figure 5). These observations imply that that the ATP hydrolysis function of Upf1p is not required for the initial recognition and targeting of mRNAs to P-bodies.
Recent results suggest that Upf1p also acts upstream and independently of Upf2p and Upf3p in mammalian cells. The key observation is that siRNA knockdown of Upf2p or Upf3p leads to the accumulation of a ‘‘SURF’’ complex consisting of eRF1, eRF3, Upf1, and SMG-1, a kinase that acts on Upf1 (Kashima et al., 2006). This observation, the direct effect of Upf1p on termination in vitro (Amrani et al., 2004), and Upf1p’s ability to target mRNAs to P-bodies in yeast argue that Upf1p acts first during recognition of aberrant translation termination events, independent of Upf2p and Upf3p. Upf1p may also have a second functional role, after ATP hydrolysis and its interaction with Upf2p and Upf3p, in triggering the actual decay of the PTC-containing transcript.
Upf1p Can Target Normal mRNAs to P-Bodies
Several lines of evidence argue that Upf1p can also target normal mRNAs to P-bodies. First, and most importantly, strains containing the DE572AA upf1 allele show an accumulation of the normal PGK1 mRNAs in P-bodies (Figure 5). Second, overexpression of the DE572AA upf1 allele leads to a small, but reproducible, reduction in polysomes and an increase in P-body size (Figure 6). Third, overexpression of DE572AA upf1 allele also inhibits growth, even in the absence of Upf2p and Upf3p (Figure 6 and data not shown). Because overexpression of wt Upf1p also has a minor and reproducible effect on growth rate and accumulation of P-bodies, the translational repression of normal mRNA is not unique to the DE572AA upf1 allele. Consistent with that view, several other ATPase defective alleles of Upf1p also lead to accumulation of P-bodies (Muhlrad and Parker, unpublished observation). This suggests that the stronger defect in the DE572AA upf1 allele is due to the ATPase mutant trapping mRNAs in P-bodies that would otherwise normally return to translation. Interestingly, Upf1p can affect the rate of decapping of normal mRNAs in xrn1Δ strains, indicating that under some conditions targeting of normal mRNAs to P-bodies might affect their decay rate (He and Jacobson, 2001). These observations suggest that Upf1p can target at least a subset of normal mRNAs to P-bodies. More importantly, these observations imply that NMD in yeast may require at least two phases of substrate distinction, one outside of P-bodies during translation termination event and another within the P-body following ATP hydrolysis.
A Model for the Process of Nonsense-Mediated Decay
The above experiments can be incorporated into an emerging model for NMD with the following features (Figure 7). First, there are two kinds of translation termination events: normal and aberrant, with aberrant terminations leading to the recruitment of Upf1p to the termination complex. The presence of Upf1p in the termination complex promotes the mRNA to stop translation and recruits Dcp1p, Dcp2p, Dhh1p, Pat1p, and Lsm1p-7p, thereby forming an mRNP capable of aggregation into a larger P-body. Because Pab1p is thought to compete with Upf1 for access to the termination complex (Amrani et al., 2004), this initial discrimination between normal and aberrant termination, on both normal and PTC-containing mRNAs, may simply be a result of competition between Pab1p and Upf1p for access to the termination complex. This suggests that, following deadenylation, a subset of normal mRNAs could be targeted to P-bodies by Upf1p. However, since most normal mRNAs are not affected by Upf1p, and decay rates of the normal PGK1 mRNA after deadenylation are not affected by Upf1p (Cao and Parker, 2003), this targeting of normal mRNAs to P-bodies does not generally appear to trigger their degradation, perhaps due to the absence of Upf2p and Upf3p.
Figure 7.
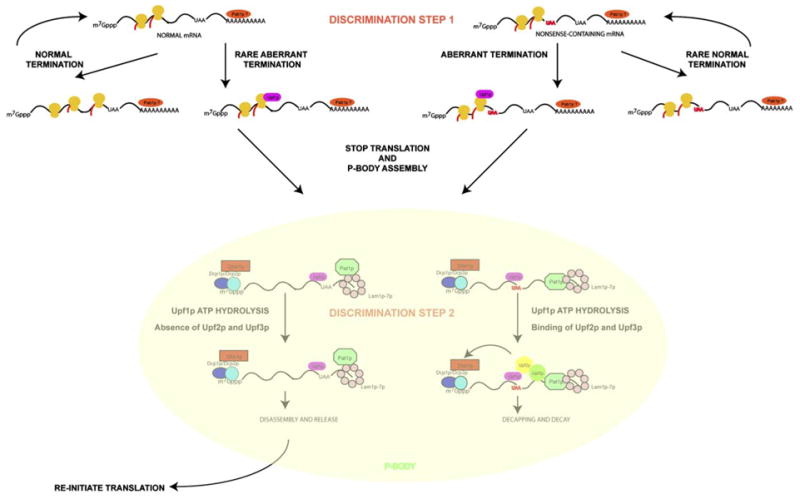
A Model for the Process of Nonsense-Mediated Decay
In a downstream step, Upf1p must hydrolyze ATP and recruit Upf2p and Upf3p to trigger mRNA degradation. In yeast, ATP hydrolysis appears to be required for Upf2p/Upf3p recruitment since in the DE572AA allele the nonsense mRNA and associated proteins accumulate in P-bodies but Upf2p and Upf3p do not (Figure 5). Whether there is an obligate order to ATP hydrolysis by Upf1p and its interaction with Upf2p/Upf3p is unclear since in mammalian cells, ATPase-defective alleles of Upf1p show increased coimmunoprecipitation with Upf2p and Upf3p, suggesting that Upf1p interaction with Upf2p and Upf3p can be independent of ATP hydrolysis (Kashima et al., 2006). Following ATP hydrolysis by Upf1p within the P-body mRNP, we hypothesize that normal mRNAs are unable to recruit Upf2p or Upf3p and therefore reinitiate translation. In contrast, PTC-containing mRNAs efficiently recruit Upf2p and Upf3p, allowing a second discrimination between normal and nonsense mRNAs and thereby increasing the fidelity of NMD. The interaction of Upf1p with Upf2p and Upf3p on the nonsense mRNAs leads to decapping and 5′ to 3′ degradation of the mRNA within the P-body. In mammalian cells and C. elegans, Upf2p and Upf3p may activate decay in part by phosphorylation of Upf1p (Page et al., 1999; Kashima et al., 2006). However, since phosphorylation of Upf1p is not known to be required for NMD in yeast, Upf2p and Upf3p might also activate decapping by other means.
An important issue is how Upf2p and Upf3p interact differently with normal and aberrant mRNAs. In yeast, mRNAs with PTCs might recruit Upf2p and Upf3p effectively due to their interactions with RNA binding proteins, such as Hrp1p, still bound to the coding region of the mRNA (Gonzalez et al., 2000). On mRNAs with a complete open reading frame, such proteins could have been dislodged by elongating ribosomes. In mammalian cells, a reasonable model is that the exon-junction complex, which is deposited by splicing, contains Upf2p and Upf3p and functions in the discrimination of normal and aberrant mRNAs in mammals (Maquat, 2004) and effectively delivers Upf2p and Upf3p to Upf1p. This provides a possible explanation for observations that NMD in mammalian cells can function, albeit less efficiently, independent of an EJC (Exon Junction Complex) (Zhang et al., 1998; Buhler et al., 2006). This suggests that the EJC may only affect NMD when Upf1p interaction with Upf2p and Upf3p is the limiting step in the process, thereby providing a possible explanation for why the EJC is only required for NMD in mammalian cells.
EXPERIMENTAL PROCEDURES
Yeast Strains
See Table S1.
Preparing Cells for Confocal Microscopy
The cells were grown to an O.D600 of 0.3 at 30°C in YEP or synthetic medium containing 2% dextrose as a carbon source, washed two times with complete minimal medium containing 2% dextrose, resuspended in the same medium used for washing, and rapidly observed. Observations were made using a Nikon PCM 2000 Confocal Microscope using a 100× objective with 2× or 3× zoom using Compix Software. All images are a compilation of 8–12 images in a stack.
Colocalization Experiments
GFP-tagged strains were transformed with a TRP1 cen plasmid containing Dcp2p-RFP (pRP1156). The transformants were grown to an O.D600 of 0.3 in synthetic media containing 2% dextrose lacking tryptophan. The cells were collected and observed as described above.
Method for Visualizing the Full-Length Reporter mRNA
Wild-type yeast cells were transformed with plasmid pRP1187 (expressing the U1A-GFP fusion protein), and either plasmid pPS 2037 (centromeric plasmid expressing the wt PGK1 mRNA with U1A binding sites), or plasmid pRP1295 (centromeric plasmid expressing PGK1 mRNA harboring an early nonsense mutation at position 22 and U1A binding sites), or plasmid pRP1296 (centromeric plasmid expressing the PGK1 mRNA with a nonsense codon at position 225 and U1A binding sites). The cells were grown, collected, and observed as described above. For colocalization, the same cells were transformed with plasmid pRP1156 (expressing Dcp2p-RFP on a cen plasmid with the TRP marker).
RNA Analysis
Transcriptional pulse-chases were done as described earlier (Decker and Parker, 1993).
Polysome Analysis
Analysis was performed as described in Coller and Parker (2005). Cells were grown in selective media containing 2% sucrose, harvested, and split into two cultures. They were resuspended in media containing either 2% Sucrose or 0.25% Sucrose and 2% Galactose. Cultures were incubated for 120 min and then harvested. The polysomes were analyzed using 15%–50% sucrose gradients.
P-Body Quantification
Randomly selected images from each experiment were analyzed using Image J (Abramoff et al., 2004). After image smoothing and background cellular fluorescence subtraction (established independently for each protein), automatic image thresholding was performed using the Otsu thresholding plugin (Otsu, 1979). The number and area of P-bodies was determined by using the ‘‘Analyze particles’’ function in Image J. Minimum pixel size and maximum pixel sizes were set and average number and area of P-bodies per cell were obtained. It should be noted that because of threshold subtractions, such quantifications are not necessarily absolute numbers or areas of P-bodies but provide a systematic and unbiased measure of relative P-body number and area within experiments. Total of 75 cells were analyzed for this quantification analysis unless otherwise indicated. P values were calculated using Two-tailed Student’s t test for both the number and area of P-bodies.
Supplementary Material
Supplementary Material
Acknowledgments
We thank all the members of the Parker Lab with special thanks to Dr. Carolyn Decker and Dr. Tracy Nissan for their helpful advice. We would also like to thank Manuj Bhandari, Jennifer Tenlen, and Anne Webb for assistance with the preparation of the manuscript. We are grateful to the confocal facility in the MCB department for its technical support. This work was supported by Howard Hughes Medical Institute and NIH (GM45443).
Footnotes
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3[prime]-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin AL, Altamura N, Leeds P, Culbertson MR. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell. 1995;6:611–625. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KE, Parker R. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr Opin Cell Biol. 2004;16:293–299. doi: 10.1016/j.ceb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Boeck R, Lapeyre B, Brown CE, Sachs AB. Capped mRNA degradation intermediates accumulate in the yeast spb8–2 mutant. Mol Cell Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky AS, Silver PA. Pre-mRNA processing factors are required for nuclear export. RNA. 2000;6:1737–1749. doi: 10.1017/s1355838200001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Steiner S, Mohn F, Paillusson A, Muhlemann O. EJC-independent degradation of nonsense immunoglobulin-[mu] mRNA depends on 3′UTR length. Nat Struct Mol Biol. 2006;13:462–464. doi: 10.1038/nsmb1081. [DOI] [PubMed] [Google Scholar]
- Cao D, Parker R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell. 2003;113:533–545. doi: 10.1016/s0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol. 2003;23:4805–4813. doi: 10.1128/MCB.23.14.4805-4813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson MR, Leeds PF. Looking at mRNA decay pathways through the window of molecular evolution. Curr Opin Genet Dev. 2003;13:207–214. doi: 10.1016/s0959-437x(03)00014-5. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14–3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- Gonzalez CI, Ruiz-Echevarria MJ, Vasudevan S, Henry MF, Peltz SW. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol Cell. 2000;5:489–499. doi: 10.1016/s1097-2765(00)80443-8. [DOI] [PubMed] [Google Scholar]
- He F, Jacobson A. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol Cell Biol. 2001;21:1515–1530. doi: 10.1128/MCB.21.5.1515-1530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc Natl Acad Sci USA. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, Parker R. Mechanisms of mRNA surveillance in eukaryotes. Annu Rev Genet. 1999;33:229–260. doi: 10.1146/annurev.genet.33.1.229. [DOI] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fitzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshirsagar M, Parker R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics. 2004;166:729–739. doi: 10.1534/genetics.166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maderazo AB, He F, Mangus DA, Jacobson A. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol Cell Biol. 2000;20:4591–4603. doi: 10.1128/mcb.20.13.4591-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′/5′ degradation. Mol Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. Recognition of yeast mRNAs as ‘‘nonsense containing’’ leads to both inhibition of mRNA translation and mRNA degradation: Implications for the control of mRNA decapping. Mol Biol Cell. 1999;10:3971–3978. doi: 10.1091/mbc.10.11.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N. Threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9:62–66. [Google Scholar]
- Page MF, Carr B, Anders KR, Grimson A, Anderson P. SMG-2 Is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley RL, Lelivelt MJ, Schenkman LR, Dahlseid JN, Culbertson MR. A factor required for nonsense-mediated mRNA decay in yeast is exported from the nucleus to the cytoplasm by a nuclear export signal sequence. J Cell Sci. 1998;111:3129–3143. doi: 10.1242/jcs.111.21.3129. [DOI] [PubMed] [Google Scholar]
- Stevens A, Wang Y, Bremer K, Zhang J, Hoepfner R, Antoniou M, Schoenberg DR, Maquat LE. Beta -Globin mRNA decay in erythroid cells: UG site-preferred endonucleolytic cleavage that is augmented by a premature termination codon. Proc Natl Acad Sci USA. 2002;99:12741–12746. doi: 10.1073/pnas.192442399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Araki Y, Sakuno T, Katada T. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′ -to-5′ mRNA decay in yeast. EMBO J. 2003;22:3951–3959. doi: 10.1093/emboj/cdg374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun X, Qian Y, Maquat LE. Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material