HP1 Binding to Chromatin Methylated at H3K9 Is Enhanced by Auxiliary Factors (original) (raw)
Abstract
A large portion of the eukaryotic genome is packaged into transcriptionally silent heterochromatin. Several factors that play important roles during the establishment and maintenance of this condensed form have been identified. Methylation of lysine 9 within histone H3 and the subsequent binding of the chromodomain protein heterochromatin protein 1 (HP1) are thought to initiate heterochromatin formation in vivo and to propagate a heterochromatic state lasting through several cell divisions. For the present study we analyzed the binding of HP1 to methylated chromatin in a fully reconstituted system. In contrast to its strong binding to methylated peptides, HP1 binds only weakly to methylated chromatin. However, the addition of recombinant SU(VAR) protein, such as ACF1 or SU(VAR)3-9, facilitates HP1 binding to chromatin methylated at lysine 9 within the H3 N terminus (H3K9). We propose that HP1 has multiple target sites that contribute to its recognition of chromatin, only one of them being methylated at H3K9. These findings have implications for the mechanisms of recognition of specific chromatin modifications in vivo.
Chromatin within the eukaryotic nucleus can be cytologically divided into active euchromatin and silent heterochromatin (19, 32, 56). Genetic analysis of position effect variegation in Drosophila melanogaster identified the methylation of lysine 9 within the H3 N terminus (H3K9) as a crucial factor for heterochromatin formation (60, 61, 68). The main histone methyl transferase (HMTase) responsible for this mark is SU(VAR)3-9 (60). This modification can be found at pericentric heterochromatin in virtually all higher eukaryotes and is currently viewed as a hallmark of silenced chromatin (13, 29, 56). Methylation at H3K9 (H3K9Me) is essential for the binding of heterochromatin protein 1 (HP1), a major constituent of heterochromatin (5, 40). HP1 homologues can be found in almost all eukaryotes ranging from Schizosaccharomyces pombe (18, 39, 43) to mammals and higher plants (26, 58, 62). Higher eukaryotes have at least three different isoforms of HP1 (HP1α, HP1β, and HP1γ in mammals and HP1a, HP1b, and HP1c in Drosophila) (47, 63), which differ in their subnuclear localization. HP1α/a and HP1β/b are primarily found within centromeric heterochromatin, whereas HP1γ/c is enriched at euchromatic sites (27, 48, 63). All HP1 molecules share a conserved architecture consisting of a chromo domain (CD), a flexible hinge region, and a chromo shadow domain (CSD) (2, 38). Genetic complementation assays (54) as well as structural data (67) showed that both globular domains (CD and CSD) are required for proper targeting of HP1. This is confirmed by experiments showing that a chimeric protein containing the CD of polycomb and the CSD of HP1 is targeted not only to heterochromatin but also to binding sites of the endogenous polycomb protein (53).
The CD of HP1 interacts specifically with a peptide resembling the N terminus of H3 that is di- or trimethylated at K9 (5, 34, 35, 40, 52). The interaction surface is highly conserved among different HP1 isoforms, and a mutation that abolishes binding results in a loss of function allele of HP1α/a (5, 40, 53). More recently, the CD of HP1 has also been shown to interact specifically with isoform 1.4 of the H1 linker histone when it is methylated at K26 (12). Although the CD of HP1 and its ability to recognize methylated histones are necessary for heterochromatin binding in vivo, they are not sufficient to support chromatin binding in vitro. Pulldown experiments using bacterially expressed glutathione _S_-transferase (GST)-HP1 showed that the CD alone could not efficiently pull down native soluble oligonucleosomes from chicken nucleated erythrocytes (46). The binding of recombinant HP1 to mononucleosomes required the presence of the full-length protein, suggesting that individual domains are not able to maintain a stable binding to a nucleosome (74). Moreover, in contrast to what is seen in vivo, the interaction of HP1 with chromatin is independent of the histone tails (46, 74) and could be mediated by an interaction between HP1 and the core region of H3 (51). This type of binding is contradictory to most data obtained in vivo that point out H3K9Me as a major factor in targeting. However, alternate ways of binding of HP1 to chromatin in vivo have been suggested as well. Treatment of nuclei with RNAses leads to a release of bound HP1 in mammalian tissue culture cells (45, 49). The putative RNA binding activity of HP1 could be assigned to a conserved region within the hinge domain, which binds RNA in vitro (49). The involvement of an RNA component in the targeting of HP1 to heterochromatin is further strengthened by the observation that mutations in components of the RNA interference (RNAi) machinery prevent SWI6/HP1-mediated formation of heterochromatin in fission yeast (30). The hinge region of HP1α has also been shown to directly interact with the linker histone H1 (51) and with native H1 containing chromatin (46). In addition, HP1α was shown to interact with the histone variant H2A.Z when incorporated into a nucleosomal array (21).
Besides the CD and the hinge domain, the CSD of HP1 is also crucial for targeting of HP1 to its site of action. The CSD mediates dimerization of HP1 and its binding to small peptide regions that can be recognized by key residues at the surface of an HP1 dimer (8). A consensus sequence that interacts with the CSD of HP1a has been identified by using a phage display method to enrich for peptides that have a high affinity to the CSD (64). This small motif can be found in many proteins, several of which have been shown to interact with the CSD and are thought to target HP1 to specific promoters (42, 65) in order to establish a silenced chromatin domain. The CSD of HP1α is also required to mediate its interaction with the large subunit of CAF1 (50). This interaction provided a link to the replication machinery and led to a model in which HP1 is targeted to replication foci by its interaction with CAF-1 and subsequently “handed over” to methylated chromatin (50).
Our goal was to reconstitute highly H3K9-methylated chromatin in vitro and to study the binding of recombinant Drosophila HP1a to methylated and nonmethylated chromatin. We found that HP1a binds to a unmodified chromatin array only weakly even though more than 85% of all H3 molecules within the reconstituted array were methylated at K9. The addition of auxiliary factors such as ACF1 or SU(VAR)3-9, which interact with the CSD of HP1a, facilitated its binding to methylated chromatin. Mutations inhibiting the interaction between HP1a and these factors abolished the binding, suggesting a bimodal binding of HP1 to methylated chromatin.
MATERIALS AND METHODS
Plasmids and cloning.
J. C. Eissenberg kindly provided HP1a in expression vector pET11a. Site-directed mutagenesis of full-length HP1a was performed using the QuickChange kit (Stratagene). To generate HP1 (V26M) we used primers HP1V26MNcoIfwd (5′-GAGGAGGAGTACGCCATGGAAAAGATCATCG-3′) and HP1V26MNcoIrew (5′-CGATGATCTTTTCCATGGCGTACTCCTCCTC-3′), and to generate HP1 (W200A) we used primers HP1aW200ABstNI (5′-CGAAGAGCGCCTATCCGCGTACTCTGATAATGAAG-3′) and HP1aW200ABstNIrev (5′-CTTCATTATCAGAGTACGCGGATAGGCGCTCTTCG-3′). HP1a (amino acids [aa] 2 to 206) was subcloned into XmaI and XhoI sites of pGEX4T-1 (Amersham) using primers pgexHP1aNtXmaI5 (5′-GTAGACCCGGGTGGCAAGAAAATCG-3′) and pgexHP1aCtXhoI3 (5′-TCTCACTCGAGTTAATCTTCATTATC-3′). SU(VAR)3-9 constructs were previously described in reference 20.
Antibodies and immunoblotting.
The HP1 (C1A9) mouse monoclonal antibody (36) and the HP1 rabbit polyclonal antibody (58) were kind gifts from S. C. R. Elgin. Dilutions for Western blots were 1:200 for C1A9 and 1:1,500 for polyclonal HP1. For all quantifications the HP1 polyclonal antibody was used. The FLAG antibody (Sigma) was used at a concentration of 1:2,000. SU(VAR)3-9 rat monoclonal antibody (SU3D9) was generated by E. Kremmer against purified His-tagged SU(VAR)3-9 Δ213. The supernatant was used at a concentration of 1:5. Proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore), probed with the indicated antibodies, detected with fluorescently labeled secondary antibodies, and quantified with an Odyssey system (Li-Cor). For quantification the background method was set to median with a border of 1 and a Top/Bottom segment. In Fig. 1C, the secondary antibody was conjugated to horseradish peroxidase (Amersham), and the detection was performed with chemiluminescence (Amersham).
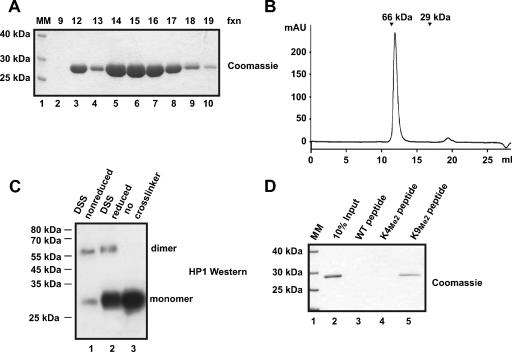
FIG. 1.
Bacterially expressed HP1 dimerizes and binds H3 peptides methylated at lysine 9. (A) Untagged recombinant HP1 was purified over four successive columns. A Coomassie-stained SDS-12% polyacrylamide (PAA) gel of 5 μl of fractions 9 to 19 from the last column, a MonoQ, is depicted. fxn, fractions. (B) Purified recombinant HP1 was loaded onto a gel filtration column (Superdex 200), and the elution profile (_A_280) of HP1 is shown. Molecular mass (MM) standards (bovine serum albumin [66 kDa] and carbonic anhydrase [29 kDa]) are labeled with arrows. (C) In vitro cross-linking of HP1 using DTSSP (DSS). Recombinant HP1 before (lane 3) or after (lanes 1 and 2) cross-linking was subjected to SDS-12% PAGE, transferred to a PVDF membrane, and detected with HP1 (C1A9) antibody. The DTSSP cross-linking can be partially reversed by reductive cleavage of the disulfide-containing cross-linking molecule (lane 2). The cross-linking revealed dimeric HP1. (D) Recombinant HP1 was assayed for binding to H3 peptides containing the first 19 amino acids of H3 immobilized onto Sulfolink Sepharose. The substrates were unmodified peptide (lane 3), peptide dimethylated at K4 (K4Me2; lane 4), and peptide dimethylated at K9 (K9Me2; lane 5). Bound HP1 was visualized by Coomassie staining.
Expression and purification of recombinant Drosophila HP1 and SU(VAR)3-9.
Bacterially expressed HP1 and point mutants were purified according to the method detailed in reference 73 and dialyzed against BC100 (25 mM HEPES [pH 7.6], 100 mM NaCl, 1 mM MgCl2, 0.5 mM EGTA, 0.1 mM EDTA, 10% [vol/vol] glycerol, 1 mM dithiothreitol [DTT], and 0.2 mM phenylmethylsulfonyl fluoride [PMSF]). All HP1s were quantified by Bradford (Bio-Rad), and Coomassie-stained proteins were quantified using the ImageMaster 1D Elite version 3.01 software package (Amersham), with bovine serum albumin (BSA) as a standard. SU(VAR)3-9 wild type (WT) and Δ213 were expressed and purified as described in reference 20.
H1 purification.
H1 was purified from Drosophila embryos (0 to 12 h) according to the method of Croston et al. (11a), and its identity was verified by mass spectrometry. For incorporation into chromatin, H1 was added after chromatin assembly, when the chromatin was linked to paramagnetic beads. H1 incubation with chromatin or DNA was performed for 1 h at 26°C. Washing steps were the same as those described below for HP1.
Histone purification and nucleosome assembly by salt dialysis.
Recombinant Drosophila histones were expressed and purified from Escherichia coli BL21(DE3)pLys and reconstituted into octamers as described previously (44). Nucleosomes were reconstituted by salt dialysis overnight at 4°C using NaCl concentrations of 2 M to 0.1 M (44). The dialysis buffer contained 10 mM Tris-HCl (pH 8), 1 mM EDTA, 0.1 M NaCl, 0.05% (vol/vol) NP-40, and 1 mM β-mecaptoethanol. Two micrograms of nucleosome particles was digested with 45 Boehringer units of micrococcal nuclease (MNase; Sigma), and reactions were stopped at 20, 60, and 120 s with 0.2 volumes of stop buffer (4% sodium dodecyl sulfate [SDS] and 10 mM EDTA). The reaction mixtures were digested with proteinase K (Genaxxon), and the DNA was separated on a 1.3% agarose gel.
Gel filtration.
Recombinant HP1 (128 μM/145 μg) was loaded onto a Superdex 200 column (HR 10/30; Amersham Pharmacia). The column was run isocratically with 0.2 ml/min in BC200 buffer at 1.4 column volumes, and 0.5-ml fractions were collected.
Cross-linking assay.
Bacterially expressed HP1 (0.17 μM) was cross-linked using 250 μM DTSSP [3,3′-dithiobis(sulfosuccinimydal propionate)] (Pierce) and incubated on ice for 2 h in BC100 buffer without DTT. The reaction was stopped by adding 100 mM Tris-HCl (pH 7.6) and boiled in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer with or without β-mercaptoethanol. After separation by SDS electrophoresis, the proteins were transferred to a PVDF membrane (Millipore) and incubated with α-HP1 (C1A9).
Peptide pulldown.
Various H3 peptides (aa 1 to 19) were coupled to Thiolink beads (Bio-Rad) and resuspended as 1:1 slurry in BC100. Twenty microliters of this slurry was incubated with 1 μg of recombinant HP1 in a total volume of 200 μl BC100. The incubation was performed on a rotating wheel at 4°C for 2 h. After washing three times with BC100 (containing 0.05% [vol/vol] NP-40) for 10 min each, the bound protein was eluted with 30 μl acidic elution buffer (100 mM Glycine [pH 2.5], 500 mM NaCl) for 20 min at 4°C. The eluted proteins were analyzed by SDS-12% PAGE and Coomassie stained. One microgram of biotinylated H3 peptides, unmodified from aa 1 to 21 (WT) and trimethylated at K9 aa 1 to 21 (Upstate), were mixed with 2 μg HP1 (WT or mutant proteins) and incubated for 1 h at 4°C. Then 10 μl of a 1:1 slurry of paramagnetic beads (Dynal) (preblocked in BSA) was added and incubated for 1 h at 26°C in BC100 (containing 0.05% [vol/vol] NP-40). The paramagnetic beads were concentrated on a magnetic concentrator (Dynal) and washed once with BC100 plus 0.05% (vol/vol) NP-40 and twice with BC200 plus 0.05% (vol/vol) NP-40. Bound proteins were separated on an SDS-12% PAGE gel and Coomassie stained.
H3K9-methylated octamer.
One hundred twenty micrograms of recombinant octamer was incubated in the presence of 9 μg of active recombinant Drosophila SU(VAR)3-9 (20) to retrieve 60 μg of a 70 to 80% H3K9 di- and trimethylated octamer. The reaction mixture was incubated at 30°C for 90 min in the presence of 40 μM _S_-adenosylmethionine (New England BioLabs) as methyl donor and 40 mM NaCl. After incubation concentrations were adjusted to 100 mM NaCl, and 0.2 mM PMSF and 2 mM DTT were added. To a 1-ml total volume, 80 μl (1:1 slurry) of Biorex70 beads (Bio-Rad) was added. The reaction mixture was rotated at 4°C for 4 h and washed five times with TEN200 (200 mM NaCl, 10 mM Tris, 1 mM EDTA, 0.2 mM PMSF, 1 mM DTT) and five times with TEN400. The methylated octamer was eluted with TEN2500, and 4 μl (2 μg) was analyzed by SDS-15% PAGE and stained with Coomassie blue.
MALDI-TOF analysis.
The Coomassie stained band corresponding to H3 was excised and subjected to chemical modification by treating with propionic anhydride to convert free amino groups to propionic amides of lysine residues as described (66). H3 was digested over night with 100 ng of sequencing-grade trypsin (Promega) in a total volume of 40 μl according to manufacturers protocol. In order to purify the methylated peptides from contaminating salts or acryl amide the peptide solution was passed over a pipette tip containing SCX material (ZipTip, Millipore) and eluted as previously described (20). The matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) spectra were acquired and analyzed according to the method described in reference 7. Quantification was performed as previously described in reference 28.
Chromatin assembly extract.
S150 chromatin assembly extract was prepared from 0- to 90-min Drosophila embryos according to the method described in reference 6.
Chromatin assembly on immobilized DNA and micrococcal nuclease digestion.
The assembly reactions for immobilized DNA were performed according to reference 57. In short, 2 μg DNA was immobilized to 0.8 mg paramagnetic streptavidin beads (Dynal) and, after extensive washing, blocked for 30 min at 4°C with BSA (1 μg/μl) in EX50 (10 mM HEPES [pH 7.6], 50 mM NaCl, 1.5 mM MgCl2, 0.5 mM EGTA, 10% [vol/vol] glycerol, 0.2 mM PMSF, 1 mM DTT) containing 0.05% (vol/vol) NP-40 before assembly. Unmodified or H3K9Me histone octamers (2 μg) were mixed with the assembly extract at time point zero together with an ATP regenerating system (3 mM ATP, 30 mM creatine phosphate, 10 μg creatine kinase/ml, 3 mM MgCl2, and 1 mM DTT). HP1 was then added at a concentration of 2 μg (8.3 ng/μl), and the reaction mixture was left to rotate for 6 h at 26°C. MNase digestions was performed as described in reference 57, with 30 Boehringer units of MNase (Sigma). For MNase digestion of chromatin assembled onto circular DNA, 150 Boerhringer units were used. A 123-bp ladder (Invitrogen) was used as a size marker.
Immobilization of salt-assembled chromatin and HP1 binding assays.
Salt-assembled unmodified or H3K9Me chromatin (1 μg) was immobilized onto 0.4-mg paramagnetic streptavidin beads (Dynal) in TEN100 buffer containing 0.05% (vol/vol) NP-40 and 250 ng/μl BSA. After 2 h of rotation at 4°C, the chromatin on paramagnetic beads was concentrated on a magnetic concentrator (Dynal) and washed once with EX100 buffer containing 0.05% (vol/vol) NP-40. Chromatin was immediately resuspended in a total volume of 80 μl containing 60 μl EX100 plus 0.05% (vol/vol) NP-40, BSA (100 ng/μl), ATP regenerating system (3 mM ATP, 30 mM creatine phosphate, 10 μg creatine kinase/ml, 3 mM MgCl2, and 1 mM DTT) and 2 μg (25 ng/μl) HP1. Purified SU(VAR3-9) WT and Δ213 were added in a total concentration of 100 ng in the presence of 1 μM Chaetocin dissolved in dimethyl sulfoxide or an equal volume of dimethyl sulfoxide only. FLAG-eluted ACF at a total concentration of 50 ng was added in the presence or absence of ATP. Drosophila assembly extract was added at a concentration of 100 μg in the presence of ATP or nonhydrolyzable ATP-γ-S analog.
Chromatin washes and HP1 detection.
Assembled chromatin was concentrated on a magnetic concentrator (Dynal), and the supernatant was removed. The chromatin beads were washed once with 100 μl EX100 (10 mM HEPES [pH 7.6], 100 mM NaCl, 1.5 mM MgCl2, 0.5 mM EGTA, 10% [vol/vol] glycerol, 0.2 mM PMSF, 1 mM DTT) containing 0.05% (vol/vol) NP-40 and twice with the same buffer containing 200 mM NaCl. The bound proteins were eluted with 10 μl SDS loading dye and separated by SDS-15% PAGE. The proteins on the gel that were smaller than 20 kDa, including the histones, were subjected to Coomassie staining, whereas the rest of the gel was transferred to a PVDF membrane (Millipore). Blots were probed with HP1 polyclonal rabbit antibody and incubated with fluorescently labeled secondary antibodies and visualized using the Odyssey system (Li-Cor) as described above.
ACF and ACF1 pulldowns.
ACF1-FLAG and imitation switch (ISWI) were expressed in Sf9 cells as described previously (15) The ACF complex was generated by coexpression of ACF1-FLAG with untagged ISWI. Sf9 cells were suspended in BC500 containing 0.05% (vol/vol) NP-40, 1 mM DTT, 0.2 mM PMSF, and protease inhibitors. The cells incubated on ice were sonicated two times for 15 s at 50% amplitude and centrifuged at maximum speed on a tabletop centrifuge for 30 min. A total of 500 ng of the expressed proteins was immobilized on M2 anti-FLAG agarose beads, washed with BC500 and BC1000 containing 0.05% (vol/vol) NP-40, and blocked with 1-μg/μl BSA. Recombinant HP1 or HP1 mutants were added at a concentration of 1 μg in a total volume of 200 μl BC100 containing 0.05% (vol/vol) NP-40, 100 μg BSA, and 5 μg ethidium bromide. After 30 min of incubation at room temperature, the beads were washed two times in the same buffer without ethidium bromide and BSA, containing 100 mM NaCl, and four times in a buffer containing 200 mM NaCl. The bound proteins were eluted with SDS sample buffer, analyzed by SDS-PAGE, and transferred onto a PVDF membrane (Millipore).
GST pulldowns.
GST pulldowns were performed as described in reference 20. ACF1 constructs were translated in vitro according to the method described in reference 31.
RESULTS
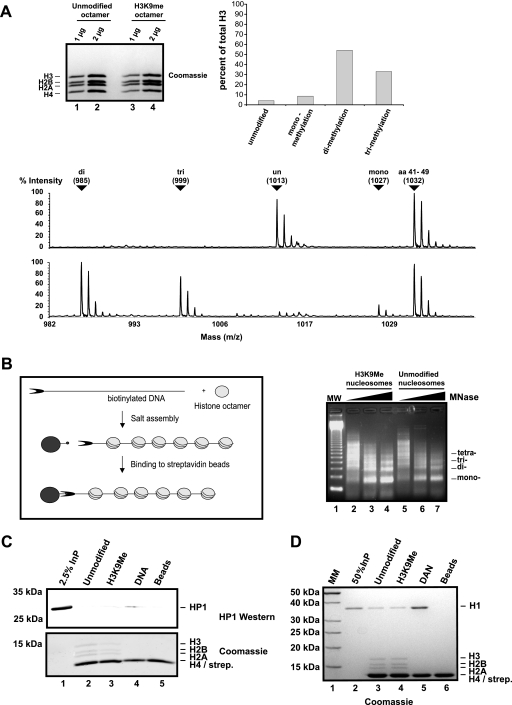
In order to generate chromatin fibers that contain HP1, we have expressed Drosophila HP1a (HP1) in bacteria and purified it to homogeneity over four consecutive columns (74) (Fig. 1A). Throughout the present article we will refer to this recombinant HP1a protein as HP1 unless stated otherwise. The purified HP1 dimerizes and interacts specifically with peptides that resemble the H3 N terminus dimethylated at K9 (Fig. 1). For the chromatin binding studies, we assembled recombinant Drosophila histones (16) onto DNA fragments containing 11 repeats of the 5S nucleosome positioning sequence using salt dialysis (9). The level of assembly was tested by micrococcal nuclease digestion (Fig. 2B, right panel). The DNA fragments were asymmetrically labeled with biotin and immobilized using streptavidin-coupled paramagnetic beads. Fully assembled arrays were coupled and used for binding assays after washing with a buffer containing 100 mM salt. The addition of the highly purified HP1 dimer at a 4:1 molar ratio of HP1/nucleosome to the immobilized chromatin fiber resulted in only weak binding (Fig. 2C, lane 2) of HP1. This is consistent with previous observations that report binding of HP1 only at HP1-to-nucleosome ratios of more than 500:1 (74). In contrast to HP1, the linker histone H1 binds very efficiently to chromatin fibers even at a molar ratio of 2:1 (Fig. 2D, lane 3). From these experiments we concluded that HP1 requires high-affinity docking sites in order to bind with a recognizable strength to chromosomal arrays.
FIG. 2.
Generation of H3K9-methylated chromatin. (A) To the left is a Coomassie blue-stained SDS-15% PAA gel of reconstituted unmodified (lanes 1 and 2) and H3K9-methylated (lanes 3 and 4) octamers. Displayed in the lower panel is a MALDI-TOF analysis of H3 peptide 9-17 from unmodified H3 (top) and H3 methylated at K9 (bottom). As an internal standard we show H3 peptide 41-49. Unmodified and mono-, di-, and trimethylated peptides with corresponding mass are labeled with arrows. The masses of unmodified and monomethylated peptide 9-17 are higher than those of di- and trimethylated peptide because the free N-terminal amines are propionylated. The quantification of the MALDI-TOF analysis is shown in the upper right panel. (B) Scheme of our chromatin reconstitution protocol. The DNA used for chromatin reconstitution is a linearized biotinylated fragment containing 12 repeats of the 5S nucleosome positioning sequence (69). A micrococcal digestion pattern of salt-reconstituted chromatin with unmodified or in vitro methylated histones is shown on the right. MW, molecular weight. (C) HP1 was assayed for binding to unmodified chromatin (lane 2), H3K9Me chromatin (lane 3), DNA immobilized on paramagnetic beads (lane 4), and beads alone (lane 5). Bound HP1 was separated by SDS-15% PAGE and visualized with an HP1 polyclonal antibody. The bottom panel shows the corresponding histones stained with Coomassie blue. Boiling of the streptavidin-coated beads resulted in the release of a strongly stained band with an apparent molecular weight similar to that of H4, which is therefore labeled H4/streptavidin (strep.). InP, input. (D) The same assay as described for panel C was performed with histone H1, also visualized by Coomassie blue. MM, molecular mass.
One of the best-characterized binding sites for HP1 in vivo is an H3 molecule that is methylated at K9 (40, 52). The enzyme responsible for creating the site in a living cell is the histone methyltransferase SU(VAR)3-9, which interacts with HP1 and has been suggested to create an autoregulatory loop that helps in maintaining the methylated state of heterochromatin (60). We wanted to generate a high-affinity binding site for HP1 by reconstituting chromatin using in vitro-methylated recombinant histones. To do this we used recombinant SU(VAR)3-9 (20) to methylate a mixture of four recombinant expressed core histones that were reconstituted into octamers (44). Subsequently, the recombinant SU(VAR)3-9 as well as the cofactors _S_-adenosylmethionine and _S_-adenosylhomocysteine were separated from the histone octamer using a cation exchange resin (70). Only histone preparations that contained no detectable SU(VAR)3-9 protein (as measured by Western blotting) were used for subsequent experiments. The purified histones were analyzed by mass spectrometry, which showed that more than 85% of all H3 molecules were methylated at K9, with more than 80% carrying two or three methyl groups (Fig. 2A, right panel and MALDI-TOF spectrum). No other lysine in H3, H2A, H2B, or H4 was found to be methylated, and no SU(VAR)3-9 was detectable in the purified histones (data not shown). The highly methylated histone octamers were then used to assemble chromatin fibers as described above. Micrococcal nuclease digestion showed that the methylated chromatin has a similar spacing and sensitivity toward the nuclease (Fig. 2B, right panel, compare lanes 2 to 4 and 5 to 7). However, despite the high content of methylated H3, recombinant HP1 showed only a weak binding that was independent of histone methylation and was even weaker than its affinity to free DNA (Fig. 2C). From these experiments we concluded that HP1 either binds to methylated histones before assembly of chromatin or it requires additional factors for the binding to its substrate.
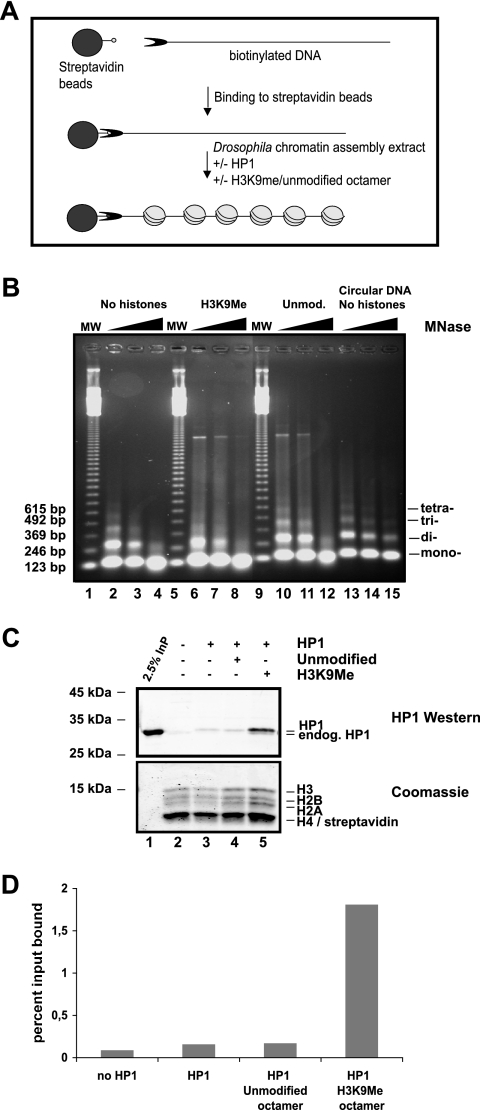
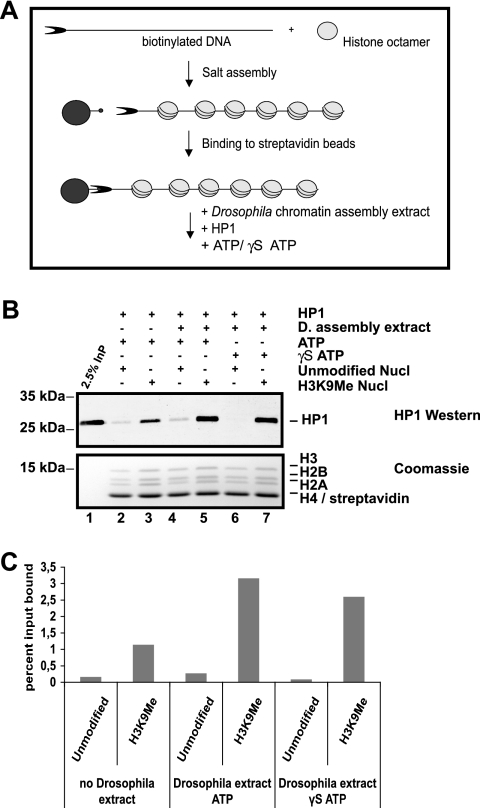
In addition to its interaction with the methylated H3 tail, HP1 has also been shown to interact with core residues of H3 and H1 (51), which are buried within chromatin, suggesting that HP1 may bind to H3 before assembly. In order to test this hypothesis, we had to use a different assembly method, as we reasoned that the HP1 binding would not sustain the high salt concentration during the salt assembly reaction. Therefore, we used a S150 chromatin assembly extract from early Drosophila embryos (6) that allowed us to assemble chromatin at lower salt concentrations (less than 100 mM) (Fig. 3A). However, even though recombinant HP1 was added at the same time as the assembly extract, we could detect only a weak association of HP1 with the assembled chromatin (Fig. 3C, lane 3). As we have previously shown that histones from early Drosophila embryos contain less than 5% H3K9 methylation (7), we added either unmodified or in vitro-methylated histones to the extract before the assembly reaction (Fig. 3C, lanes 4 and 5). The addition of exogenous histones led to a slight decrease in sensitivity towards MNase (Fig. 3B, compare lanes 2, 3, and 4 with lanes 6, 7, 8, 10, 11, and 12). However, we could not observe any difference in nucleosomal repeat length when supplementing the S150 with either unmodified or H3K9Me octamers. MS analysis of the chromatin after assembly showed that it contained K9-methylated chromatin only when the in vitro-methylated histones were added (data not shown), indicating that the exogenously added histones are incorporated by the assembly extract. Under these conditions, HP1 bound to chromatin arrays where methylated octamers were added before the assembly reaction but only weakly interacted with chromatin to which unmodified histones were added (Fig. 3C, compare lanes 4 and 5). A quantification of this experiment is shown in Fig. 3D. There is 10 times more HP1 bound to H3K9Me chromatin than to unmodified chromatin. As we assembled chromatin using a heterogeneous extract we could not directly conclude from these experiments whether HP1 bound to the methylated H3 before the assembly or whether the binding was enhanced by the action of accessory factors. To distinguish between these two possibilities, we assembled chromatin from unmodified or methylated histones by salt dialysis and added S150 extract together with recombinant HP1 after the assembly reaction. It turned out that the S150 extract was able to facilitate HP1 binding to methylated chromatin even at concentrations that were not sufficient to assemble nucleosomes in vitro (Fig. 4). As chromatin assembly is ATP dependent, we wondered whether the loading process required ATP hydrolysis. The addition of ATP stimulated binding of HP1 to the nucleosomal array irrespective of its methylation state (Fig. 4B, compare lanes 4 and 6). The stimulation of HP1 binding to the methylated chromatin, however, was not dependent on ATP hydrolysis (Fig. 4B, compare lanes 5 and 7). A quantification of this HP1 binding experiment is shown in Fig. 4C. In the presence of an assembly extract HP1 is bound more than 11 times better to H3K9Me chromatin compared to unmodified chromatin. From these results we reckoned that the assembly extract does indeed contain factors that facilitated HP1 binding to the methylated H3 tail and that can assist HP1 binding to assembled chromatin. The presence of ATP in the reaction moderately stimulates the affinity of HP1 to chromatin but does not increase the specific binding to methylated chromatin.
FIG. 3.
Reconstitution of methylated chromatin using an S150 Drosophila assembly extract and HP1 binding. (A) Scheme of the assay. (B) Micrococcal digestion pattern of chromatin assembly reactions as described for panel C, without HP1 added. MNase digestions were stopped after 30, 60, and 300 s. Assembly of circular DNA was used as a control. MW indicates lanes containing the 123-bp ladder as size marker. Unmod., unmodified. (C) Chromatin was reconstituted on a 2-μg linearized biotinylated fragment containing 12 repeats of the 5S nucleosome positioning sequence bound to paramagnetic beads in the presence or absence of 2 μg of HP1 for 6 h at 26°C. Before assembly, 2 μg of unmodified (lane 4) or H3K9Me (lane 5) histone octamers was supplemented to the extract. The paramagnetic beads were washed, and proteins remaining on the beads were separated on an SDS-15% polyacrylamide gel. HP1 was visualized with an HP1 polyclonal antibody. The corresponding histones were stained by Coomassie blue. Endogenous (endog.) HP1 from the assembly extract and recombinantly added HP1 are labeled. InP, input. (D) The graph corresponds to quantification of bound HP1 from panel C. Recombinant and endogenous HP1 are included in the quantification. The y axis displays the percentage of input bound. The graph is representative of three or more different experiments.
FIG. 4.
HP1 is bound to salt-assembled chromatin in the presence of Drosophila assembly extract. (A) Scheme of the assay. (B) Salt-assembled unmodified or H3K9Me chromatin attached to paramagnetic beads was incubated for 1 h at 26°C with HP1, plus and minus Drosophila (D.) assembly extract. The reactions were carried out in the presence of ATP or nonhydrolyzable ATP-γ-S analog (γS ATP). The assembly extract added was less than 5% of what is needed for the assembly reaction in Fig. 3. HP1 was detected with HP1 polyclonal antibody, and corresponding histones were detected with Coomassie blue. Nucl, nucleosome. (C) The graph corresponds to quantification of bound HP1 as shown in Fig. 3B. The y axis displays the percentage of input bound. The graph is representative of at least four individual experiments.
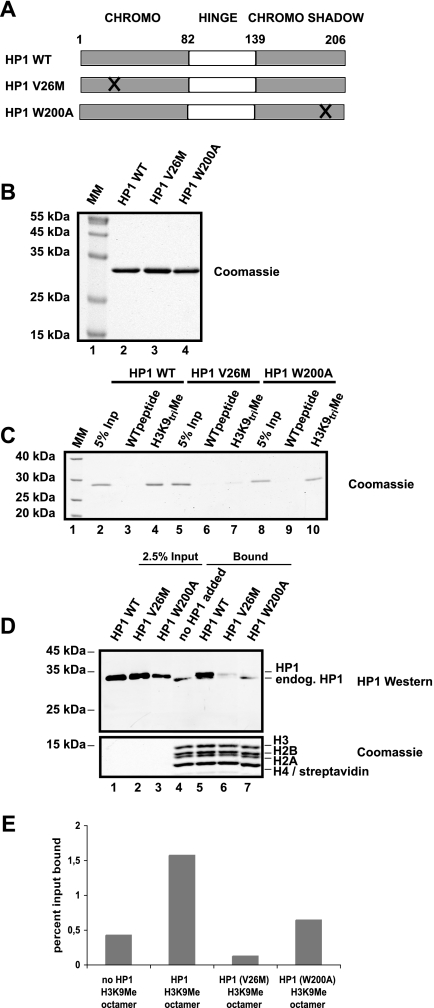
As discussed above, HP1 has three domains, all of which are involved in HP1 function. The CD binds histone H3 methylated at K9, the hinge domain is important for DNA and RNA binding, and the CSD carries a protein-protein interaction domain. In order to get more insight into the nature of HP1 binding to methylated chromatin, we expressed and purified mutant HP1 proteins (Fig. 5A and B) and added them to a chromatin assembly reaction as shown in Fig. 3A. As has been reported before (35), a V26M mutation within the CD of HP1 prevented binding to a peptide containing methylated K9 (Fig. 5C, lane 7). This mutation also resulted in a reduction of HP1 binding to the methylated chromatin (Fig. 5D, lane 6). A point mutation of W to A at position 200 in the CSD of HP1 that has been shown to selectively interfere with the interaction between HP1 and associated proteins (8) also resulted in a loss of HP1 binding to methylated chromatin (Fig. 5D, lane 7), despite its ability to interact with the methylated peptide (Fig. 5C, lane 10). A quantification of this experiment is shown in Fig. 5E. These results pointed towards a protein-protein interaction rather than an HP1 DNA or HP1 RNA interaction playing a key role in the loading of HP1 to heterochromatin.
FIG. 5.
Expression of HP1 mutant proteins and binding of these to H3K9Me chromatin during assembly. (A) Scheme of HP1 mutants generated. (B) Coomassie blue-stained 15% SDS-polyacrylamide gel of the purified HP1 proteins. MM, molecular mass. (C) Peptide pulldown of the recombinant HP1 WT and mutants using H3 peptide aa 1 to 21, unmodified (WT) versus trimethylated at K9 (H3K9triMe). Bound HP1s were visualized by use of Coomassie blue. InP, input. (D) Drosophila assembly reaction with 2 μg H3K9Me octamer as described for Fig. 3A. In lanes 1 to 3, 2.5% HP1 input was used. Lanes 4 to 7 correspond to proteins bound after 6 h of incubation. HP1 was detected with HP1 polyclonal antibody, and the corresponding histones were stained with Coomassie blue. Bound exogenous and endogenous (endog.) HP1 present in the Drosophila assembly extract are labeled. (E) The graph corresponds to quantification of bound HP1 from panel D. Recombinant and endogenous HP1 are included in the quantification. The y axis displays percentage of input bound. This quantification is representative of at least three different experiments.
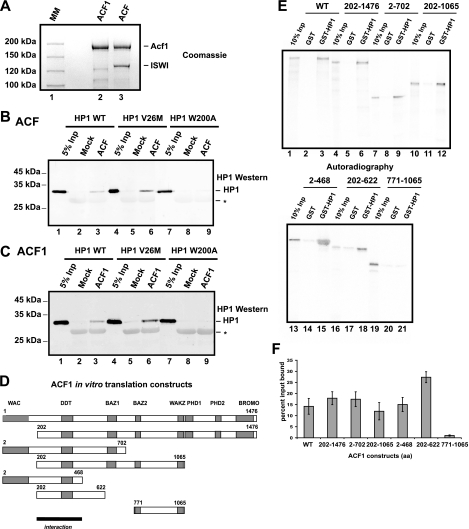
Several chromatin-associated factors have been suggested to play a role in heterochromatin formation and its function (55, 59). The chromatin-remodeling factor ACF consisting of the ISWI ATPase and the regulatory ACF1 protein is very abundant in early Drosophila embryos. A mutation in the gene ACF1 of Drosophila has been identified as a suppressor of position effect variegation, which places ACF1 in the same genetic pathway as HP1 (24). Mammalian ACF1 has been shown to colocalize with HP1β in NIH 3T3 cells and is suggested to have a role in replication of heterochromatin (11). As we have observed a strong impairment of chromatin binding of HP1 mutants that carry a mutation within the CSD, we first investigated whether recombinant ACF was able to interact directly with HP1 and whether this interaction was mediated by the CSD. We purified an ACF complex using a baculoviral system expressing a FLAG-tagged ACF1 protein together with untagged ISWI (Fig. 6A), and the immobilized complex was then incubated with various HP1 mutant proteins. We could detect binding of HP1 to the reconstituted ACF complex (Fig. 6B) as well as to the isolated ACF1 subunit (Fig. 6C). Consistent with previous findings showing that most heterotypic protein-protein interactions with HP1 are mediated by the CSD (67), the ACF1-HP1 interaction was also mediated by this domain, as the point mutation within the CSD motif impaired the interaction (Fig. 6B and C, compare lanes 3 and 6 with lane 9). The fact that the isolated ACF1 subunit is sufficient for the HP1 binding may explain the specific effect of an ACF1 mutation on heterochromatin formation (24). In order to map the interaction domain within ACF1 that is responsible for the HP1 interaction, we performed GST pulldown experiments using GST-HP1 and in vitro-translated ACF1 fragments (Fig. 6D). In these experiments we could detect binding of all fragments containing amino acids 202 to 468 (Fig. 6E and F).
FIG. 6.
HP1 interacts with the ACF complex and ACF1. (A) Coomassie-stained SDS-8% PAA gel of FLAG affinity-purified recombinant ACF1 and ACF complex from Sf9 cells coinfected with FLAG-ACF1 in the presence or absence of untagged ISWI. MM, molecular mass. (B) HP1 pulldown with FLAG beads incubated with mock Sf9 extract or extract containing FLAG-ACF1 and untagged ISWI. After extensive washing, the protein remaining on the beads was separated by SDS-12% PAGE, imunoblotted, and detected with HP1 antibody. Asterisks indicate FLAG antibody light chains. Inp, input. (C) Western blot of HP1 pulldown using FLAG beads incubated with mock Sf9 extract or extract containing FLAG-ACF1. Asterisks indicate FLAG antibody light chains. (D) ACF1 constructs used for in vitro translation. (E) GST and GST-HP1 pulldown with in vitro translated ACF1 constructs. (F) Quantification of the binding affinities of the various ACF constructs. Error bars represent standard deviations from three independent pulldown experiments.
The region responsible for ACF1 binding to HP1 contains the evolutionarily conserved DDT motif (14), suggesting that this motif most likely represents an HP1 interaction domain. For Drosophila ACF1, the region containing the DDT motif has been shown to be required for ISWI interaction (16, 25).
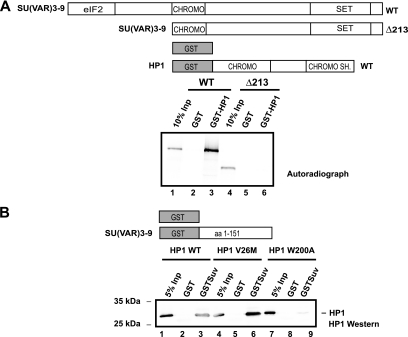
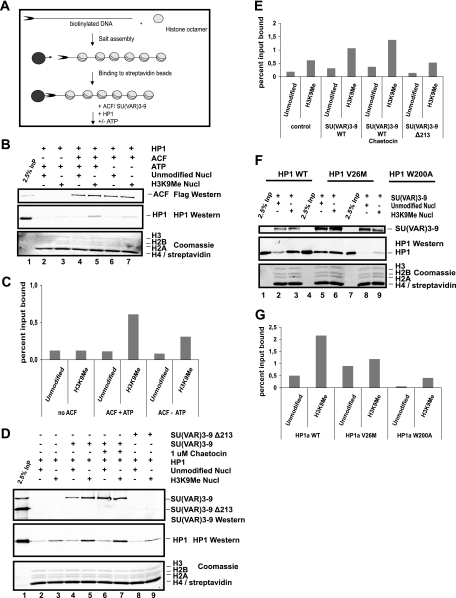
Another prominent factor that is known to interact with HP1 in vivo and which plays an important role in heterochromatin formation is the histone methyltransferase SU(VAR)3-9 (1, 60). We performed protein-protein interaction assays using different SU(VAR)3-9 or HP1 mutants in order to biochemically map the interaction regions for each protein. The N terminus of SU(VAR)3-9 was necessary for its association with HP1 (Fig. 7A), while the CSD of HP1 was required for SU(VAR)3-9 binding (Fig. 7B). This further demonstrated the importance of the HP1 CSD for protein-protein interaction (8, 41, 72). The in vivo target loci of HP1 and SU(VAR)3-9 have been mapped in Drosophila Kc cells (27). HP1 and SU(VAR)3-9 colocalized at multiple sites, suggesting a possible targeting of HP1 by SU(VAR)3-9, but the fact that HP1 can also be found at other chromatin sites supports the idea that SU(VAR)3-9 binding is not the sole way of stabilizing HP1 binding to chromatin. We therefore tested whether the known SU(VAR) proteins ACF1 and SU(VAR)3-9 could facilitate HP1 binding to methylated chromatin in vitro. In order to do this, we used salt-assembled chromatin that contained either methylated or nonmethylated histones and added recombinant HP1 together with recombinant SU(VAR)3-9 or ACF (Fig. 8A). Consistent with the model that HP1 requires multiple binding sites for efficient chromatin binding, we observed an increased association of HP1 with methylated chromatin when ACF complex (Fig. 8B, lane 5, and 8C [quantification]) or SU(VAR)3-9 (Fig. 8D, lane 5, and 8E [quantification]) was present. This was not due to an intrinsically higher affinity of the auxiliary factors to methylated chromatin, as both bound efficiently to the chromatin fiber independently from its modification state (Fig. 8B, compare lanes 4 and 5, and 8D, compare lanes 4 and 5). We also found that neither ATP nor HMTase activity was required for preferential binding of HP1 to methylated chromatin (Fig. 8B and 8D, compare lanes 5 and 7).
FIG. 7.
HP1 interacts with SU(VAR)3-9. (A) SU(VAR)3-9 constructs used for in vitro translation and GST constructs are shown at the top. The GST pulldown is shown at the bottom. SU(VAR)3-9 was detected by autoradiography. CHROMO SH., chromo shadow. (B) The upper panel shows GST constructs used for the pulldown experiment. The lower panel represents an HP1 Western blot of GST pulldown with recombinant HP1 WT (lanes 1 to 3), HP1 (V26M) (lanes 4 to 6), and HP1 (W200A) (lanes 7 to 9). HP1 was detected with HP1 polyclonal antibody.
FIG. 8.
SU(VAR) 3-9 and ACF facilitate HP1 binding to H3K9Me chromatin. (A) Scheme of the assay. (B) Salt-assembled unmodified or H3K9Me chromatin bound to paramagnetic beads was incubated with HP1 in the presence or absence of ACF and ATP for 1 h at 26°C. After washing, the proteins remaining on the paramagnetic beads were separated by SDS-15% PAGE. ACF1 was detected with FLAG antibody, and HP1 was detected with HP1 polyclonal antibody (upper panels). The corresponding histones were detected with Coomassie blue (bottom panel). Lane 1 corresponds to 50% ACF input (InP) and 2.5% HP1 input. Nucl, nucleosome. (C) The graph displays bound HP1 as a percentage of input as shown in panel B. The y axis corresponds to percent input bound. The graph is representative of at least two individual experiments. (D) SU(VAR)3-9 couples HP1 to chromatin. HP1 was incubated with unmodified or H3K9Me chromatin in the presence of recombinant SU(VAR)3-9 WT (lanes 4 to 7) or Δ213 (lanes 8 and 9). The SU(VAR)3-9-specific HMTase inhibitor Chaetocin was added to a concentration of 1 μM (lanes 6 and 7). The remaining SU(VAR)3-9 and HP1 on paramagnetic beads was detected by Western analysis, and the histones were detected with Coomassie blue. Lane 1 corresponds to 100% SU(VAR)3-9 input and 2.5% HP1 input. (E) Quantification of HP1 bound as shown in panel D. The graph is representative of at least three individual experiments and displays percent input bound. (F) SU(VAR)3-9 was added in the presence of HP1 WT (lanes 1 to 3), HP1 (V26M) (lanes 4 to 6), and HP1 (W200A) (lanes 7 to 9) to unmodified and H3K9Me chromatin. Bound SU(VAR)3-9 and HP1 were detected by Western analysis, and histones were detected with Coomassie blue. (G) The graph corresponds to total HP1 binding as shown in panel F. The y axis displays percent input bound. The graph is representative of at least two individual experiments.
This evidence suggests that only the protein-protein interaction served as a second binding site within chromatin and stabilized the interaction of HP1 and the methylated chromatin. This finding is further strengthened by the observation that a mutation in the CSD of HP1 (W200A) that no longer interacts with SU(VAR)3-9 has a reduced binding affinity towards methylated chromatin (Fig. 8F, compare lanes 3 and 9).
DISCUSSION
It is of critical importance to understand how epigenetic information is stored and maintained. The finding that a combinatorial aspect of histone modification can contribute to this epigenetic information processing therefore represents a very attractive model (37, 71). However, it has not been shown so far how epigenetic marks in the form of specific histone modifications can be “read” within a defined chromatin context and how the factors that can bind specifically to these marks access them. We reconstituted HP1 containing chromatin fibers in the test tube using a highly purified reconstituted system containing unmodified histones with H3 molecules that are methylated at K9, recombinant HP1 and two known SU(VAR) proteins, ACF and SU(VAR)3-9. Both factors bind chromatin in a methylation-independent manner and can interact simultaneously with HP1 via the CSD of HP1. These findings suggest a bimodal interaction of HP1 with chromatin, in which a single binding site is not sufficient to stably anchor HP1 to chromatin. It is important to mention that the binding assay we are using throughout the study is not an equilibrium binding assay. As HP1 could bind to methylated chromatin with high affinity but with rapid association and dissociation rates, we actually detected the rate of dissociation of HP1 from chromatin. The auxiliary factors may increase the average time HP1 resides at methylated chromatin, which may be essential for heterochromatin formation.
These findings are in accordance with observations in vivo showing that HP1 can be released from its binding sites either by a peptide resembling the H3 N terminus that is methylated at K9 (5, 49), by treatment with RNase (45), or by a peptide that mediates the interaction with an associated factor (4). Our data provide a biochemical explanation for these seemingly contradictory observations.
HP1 is considered a major component of constitutive heterochromatin (56). Despite an enormous wealth of data regarding the localization of HP1 in vivo, the molecular details of how HP1 recognizes pericentric heterochromatin have been sparse. Our data show that even though HP1 is able to bind histones that are methylated at K9 when they are incorporated into chromatin, the affinity is rather weak and presumably not sufficient to maintain a heterochromatic structure in vivo or in vitro. This is in perfect agreement with biophysical studies that have measured a dissociation constant for H3K9-methylated peptides between 2 μM (nuclear magnetic resonance) (52) and 100 μM (isothermal titration calorimetry) (34), which is rather high compared to other protein-protein or protein-DNA interactions. Considering the picomolar constant for dissociation of a histone tail from the DNA (33) and the fact that histone tails can be UV cross-linked to DNA in vivo and in vitro (3), it is difficult to envision efficient binding of HP1 to the methylated tail only. The weak interaction and the corresponding low occupancy time at a given binding site is presumably also the reason for the dynamic nature of HP1 in the nuclei of eukaryotic cells (10, 23).
Our data also suggest that an efficient binding of HP1 to chromatin can be achieved only when several binding sites are present within the chromatin substrate. It had previously been reported that the general affinity of HP1 to mononucleosomes is rather low and independent of the histone tails (74). In contrast to this, native chromosomal fibers can be purified with reasonably efficiency using immobilized HP1 molecules (46). In our fully defined reconstituted system we also detected only weak binding of HP1 to chromatin that was moderately stimulated when the H3 tail is fully methylated at K9. This binding of HP1 to methylated chromatin was stimulated by the addition of factors that were able to bind HP1 and chromatin at the same time, thereby enhancing the affinity of HP1 to chromatin. A mutational analysis of the ability of the factors to load HP1 to methylated chromatin showed that the physical interaction with HP1 is required for their activity to assist HP1 binding. It may be that the native fibers still contain such additional factors and therefore increase the affinity of HP1 for heterochromatin.
HP1 has been shown to interact with several factors via its CSD (64), and the binding site is reconstituted by both HP1 molecules within the HP1 dimer (67). This interaction domain is required for heterochromatin localization in vivo as mutant HP1 proteins that can no longer form this domain also do not associate stably with centromeric heterochromatin or telomeric regions (22, 67). We also observe this failure of a HP1 mutant in the CSD to bind K9-methylated chromatin when assisted by a Drosophila S-150 chromatin assembly extract. This observation led us to the conclusion that HP1 has to interact with a factor present in this extract to bind methylated chromatin.
The two candidate factors that we used in order to test their ability to assist HP1 in its binding to K9-methylated chromatin both show a SU(VAR) phenotype when mutated in Drosophila melanogaster (24, 68). One, ACF1, has been shown to be a major chromatin assembly factor in Drosophila and has an effect on position effect variegation (24). In human cells, ACF1 has also been shown to colocalize with HP1β (11). ACF1 has been shown to bind DNA via its WAC domain (25) and to interact with histone molecules via its PHD fingers (16). Those two domains are presumably responsible for its interaction with the chromatin template. It is intriguing that the region spanning the DDT motif, which we found crucial for interaction with HP1, does not seem to be involved in binding to the chromatin substrate and could therefore be used to recruit HP1 to chromatin. It is tempting to speculate that ACF1-HP1 interaction may increase the local concentration of HP1 within heterochromatin, where the binding could be stabilized by its interaction via the CD with chromatin methylated at K9.
The second factor that we have tested is the histone methyltransferase SU(VAR)3-9. SU(VAR)3-9 is responsible for methylating H3 at K9 (60), thereby generating a potential binding site for HP1 within heterochromatin. SU(VAR)3-9 interacts with HP1 via the N terminus, and it has been suggested that this interaction serves as an autoregulatory loop, helping to maintain the methylated state of heterochromatin (60). We observed a high affinity of full-length SU(VAR)3-9 for in vitro assembled chromatin irrespective of its methylation state. Similar to our observations for ACF, described above, we also see increased binding of HP1 after adding SU(VAR)3-9 that is independent of its ability to methylate H3. However, when the interaction between SU(VAR)3-9 and HP1 was impaired due to a mutation either in HP1 or in SU(VAR)3-9, no increase in binding of HP1 to methylated chromatin could be observed. This additional function of SU(VAR)3-9 in stabilizing HP1 binding could help explain the strong dose dependency the gene has, which is rather unusual for enzymatic activity. This may also be an explanation that some hypomorphic alleles of SU(VAR)3-9 can be isolated that show a Su(var) phenotype despite having normal HMTase activity (17).
The finding that HP1 binding is not only dependent on methylation at H3K9 but also requires additional auxiliary factors explains many in vivo observations that have been previously considered to be contradictory. It would also enable a cell to fine-tune its level of heterochromatin in response to external signals by modulating the different binding sites of HP1 within chromatin. We tested two known chromatin-associated factors for their ability to help increasing the affinity of HP1 to methylated chromatin. However, there may be additional factors and probably redundant mechanisms that can help loading HP1 to chromatin. It will be interesting to see whether the different targeting factors have different contributions to the localization of HP1 at different stages of the cell cycle, during different stages of development, or in different cell types. Our in vitro system for looking at HP1 binding to methylated chromatin will certainly be useful for identification of additional targeting factors in the future.
Acknowledgments
We acknowledge Sally Elgin for the HP1 antibodies and Joel Eisenberg for the HP1 plasmid, Jeff Hansen for the 208-12 plasmid, Elisabeth Kremmer for generation of the SU(VAR)3-9 antibody, Gernot Langst and Peter Becker for helpful discussions and advice, and Josephine Sutcliffe for critical reading of the manuscript.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (IM23/4-3).
Footnotes
▿
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Aagaard, L., G. Laible, P. Selenko, M. Schmid, R. Dorn, G. Schotta, S. Kuhfittig, A. Wolf, A. Lebersorger, P. B. Singh, G. Reuter, and T. Jenuwein. 1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 18**:**1923-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aasland, R., and A. F. Stewart. 1995. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 23**:**3168-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angelov, D., J. M. Vitolo, V. Mutskov, S. Dimitrov, and J. J. Hayes. 2001. Preferential interaction of the core histone tail domains with linker DNA. Proc. Natl. Acad. Sci. USA 98**:**6599-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badugu, R., M. M. Shareef, and R. Kellum. 2003. Novel Drosophila heterochromatin protein 1 (HP1)/origin recognition complex-associated protein (HOAP) repeat motif in HP1/HOAP interactions and chromocenter associations. J. Biol. Chem. 278**:**34491-34498. [DOI] [PubMed] [Google Scholar]
- 5.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410**:**120-124. [DOI] [PubMed] [Google Scholar]
- 6.Becker, P. B., and C. Wu. 1992. Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Mol. Cell. Biol. 12**:**2241-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonaldi, T., A. Imhof, and J. T. Regula. 2004. A combination of different mass spectroscopic techniques for the analysis of dynamic changes of histone modifications. Proteomics 4**:**1382-1396. [DOI] [PubMed] [Google Scholar]
- 8.Brasher, S. V., B. O. Smith, R. H. Fogh, D. Nietlispach, A. Thiru, P. R. Nielsen, R. W. Broadhurst, L. J. Ball, N. V. Murzina, and E. D. Laue. 2000. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 19**:**1587-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carruthers, L. M., C. Tse, K. P. Walker III, and J. C. Hansen. 1999. Assembly of defined nucleosomal and chromatin arrays from pure components. Methods Enzymol. 304**:**19-35. [DOI] [PubMed] [Google Scholar]
- 10.Cheutin, T., A. J. McNairn, T. Jenuwein, D. M. Gilbert, P. B. Singh, and T. Misteli. 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299**:**721-725. [DOI] [PubMed] [Google Scholar]
- 11.Collins, N., R. A. Poot, I. Kukimoto, C. Garcia-Jimenez, G. Dellaire, and P. D. Varga-Weisz. 2002. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat. Genet. 32**:**627-632. [DOI] [PubMed] [Google Scholar]
- 11a.Croston, G. E., L. M. Lira, and J. T. Kadonaga. 1991. A general method for purification of H1 histones that are active for repression of basal RNA polymerase II transcription. Protein Expr. Purif. 2**:**162-169. [DOI] [PubMed]
- 12.Daujat, S., U. Zeissler, T. Waldmann, N. Happel, and R. Schneider. 2005. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J. Biol. Chem. 280**:**38090-38095. [DOI] [PubMed] [Google Scholar]
- 13.Dillon, N., and R. Festenstein. 2002. Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 18**:**252-258. [DOI] [PubMed] [Google Scholar]
- 14.Doerks, T., R. Copley, and P. Bork. 2001. DDT—a novel domain in different transcription and chromosome remodeling factors. Trends Biochem. Sci. 26**:**145-146. [DOI] [PubMed] [Google Scholar]
- 15.Eberharter, A., S. Ferrari, G. Langst, T. Straub, A. Imhof, P. Varga-Weisz, M. Wilm, and P. B. Becker. 2001. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J. 20**:**3781-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberharter, A., I. Vetter, R. Ferreira, and P. B. Becker. 2004. ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. EMBO J. 23**:**4029-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebert, A., G. Schotta, S. Lein, S. Kubicek, V. Krauss, T. Jenuwein, and G. Reuter. 2004. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 18**:**2973-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ekwall, K., J. P. Javerzat, A. Lorentz, H. Schmidt, G. Cranston, and R. Allshire. 1995. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269**:**1429-1431. [DOI] [PubMed] [Google Scholar]
- 19.Elgin, S. C., and S. I. Grewal. 2003. Heterochromatin: silence is golden. Curr. Biol. 13**:**R895-R898. [DOI] [PubMed] [Google Scholar]
- 20.Eskeland, R., B. Czermin, J. Boeke, T. Bonaldi, J. T. Regula, and A. Imhof. 2004. The N-terminus of Drosophila SU(VAR)3-9 mediates dimerization and regulates its methyltransferase activity. Biochemistry 43**:**3740-3749. [DOI] [PubMed] [Google Scholar]
- 21.Fan, J. Y., D. Rangasamy, K. Luger, and D. J. Tremethick. 2004. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol. Cell 16**:**655-661. [DOI] [PubMed] [Google Scholar]
- 22.Fanti, L., G. Giovinazzo, M. Berloco, and S. Pimpinelli. 1998. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell 2**:**527-538. [DOI] [PubMed] [Google Scholar]
- 23.Festenstein, R., S. N. Pagakis, K. Hiragami, D. Lyon, A. Verreault, B. Sekkali, and D. Kioussis. 2003. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science 299**:**719-721. [DOI] [PubMed] [Google Scholar]
- 24.Fyodorov, D. V., M. D. Blower, G. H. Karpen, and J. T. Kadonaga. 2004. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 18**:**170-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fyodorov, D. V., and J. T. Kadonaga. 2002. Binding of Acf1 to DNA involves a WAC motif and is important for ACF-mediated chromatin assembly. Mol. Cell. Biol. 22**:**6344-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaudin, V., M. Libault, S. Pouteau, T. Juul, G. Zhao, D. Lefebvre, and O. Grandjean. 2001. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 128**:**4847-4858. [DOI] [PubMed] [Google Scholar]
- 27.Greil, F., I. van der Kraan, J. Delrow, J. F. Smothers, E. de Wit, H. J. Bussemaker, R. van Driel, S. Henikoff, and B. van Steensel. 2003. Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes Dev. 17**:**2825-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greiner, D., T. Bonaldi, R. Eskeland, E. Roemer, and A. Imhof. 2005. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat. Chem. Biol. 1**:**143-145. [DOI] [PubMed] [Google Scholar]
- 29.Grewal, S. I., and J. C. Rice. 2004. Regulation of heterochromatin by histone methylation and small RNAs. Curr. Opin. Cell Biol. 16**:**230-238. [DOI] [PubMed] [Google Scholar]
- 30.Hall, I. M., G. D. Shankaranarayana, K. Noma, N. Ayoub, A. Cohen, and S. I. Grewal. 2002. Establishment and maintenance of a heterochromatin domain. Science 297**:**2232-2237. [DOI] [PubMed] [Google Scholar]
- 31.Hartlepp, K. F., C. Fernandez-Tornero, A. Eberharter, T. Grune, C. W. Muller, and P. B. Becker. 2005. The histone fold subunits of Drosophila CHRAC facilitate nucleosome sliding through dynamic DNA interactions. Mol. Cell. Biol. 25**:**9886-9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henikoff, S. 2000. Heterochromatin function in complex genomes. Biochim. Biophys. Acta 1470**:**O1-O8. [DOI] [PubMed] [Google Scholar]
- 33.Hong, L., G. P. Schroth, H. R. Matthews, P. Yau, and E. M. Bradbury. 1993. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J. Biol. Chem. 268**:**305-314. [PubMed] [Google Scholar]
- 34.Jacobs, S. A., and S. Khorasanizadeh. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295**:**2080-2083. [DOI] [PubMed] [Google Scholar]
- 35.Jacobs, S. A., S. D. Taverna, Y. Zhang, S. D. Briggs, J. Li, J. C. Eissenberg, C. D. Allis, and S. Khorasanizadeh. 2001. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 20**:**5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.James, T. C., J. C. Eissenberg, C. Craig, V. Dietrich, A. Hobson, and S. C. Elgin. 1989. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 50**:**170-180. [PubMed] [Google Scholar]
- 37.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293**:**1074-1080. [DOI] [PubMed] [Google Scholar]
- 38.Kellum, R. 2003. HP1 complexes and heterochromatin assembly. Curr. Top. Microbiol. Immunol. 274**:**53-77. [DOI] [PubMed] [Google Scholar]
- 39.Klar, A. J., and M. J. Bonaduce. 1991. swi6, a gene required for mating-type switching, prohibits meiotic recombination in the mat2-mat3 “cold spot” of fission yeast. Genetics 129**:**1033-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410**:**116-120. [DOI] [PubMed] [Google Scholar]
- 41.Lechner, M. S., D. C. Schultz, D. Negorev, G. G. Maul, and F. J. Rauscher III. 2005. The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem. Biophys. Res. Commun. 331**:**929-937. [DOI] [PubMed] [Google Scholar]
- 42.Li, Y., J. R. Danzer, P. Alvarez, A. S. Belmont, and L. L. Wallrath. 2003. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development 130**:**1817-1824. [DOI] [PubMed] [Google Scholar]
- 43.Lorentz, A., K. Ostermann, O. Fleck, and H. Schmidt. 1994. Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene 143**:**139-143. [DOI] [PubMed] [Google Scholar]
- 44.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304**:**3-19. [DOI] [PubMed] [Google Scholar]
- 45.Maison, C., D. Bailly, A. H. Peters, J. P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein, and G. Almouzni. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30**:**329-334. [DOI] [PubMed] [Google Scholar]
- 46.Meehan, R. R., C. F. Kao, and S. Pennings. 2003. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 22**:**3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minc, E., Y. Allory, H. J. Worman, J. C. Courvalin, and B. Buendia. 1999. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 108**:**220-234. [DOI] [PubMed] [Google Scholar]
- 48.Minc, E., J.-C. Courvalin, and B. Buendia. 2000. HP1γ associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet. Cell Genet. 90**:**279-284. [DOI] [PubMed] [Google Scholar]
- 49.Muchardt, C., M. Guilleme, J. S. Seeler, D. Trouche, A. Dejean, and M. Yaniv. 2002. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 3**:**975-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murzina, N., A. Verreault, E. Laue, and B. Stillman. 1999. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell 4**:**529-540. [DOI] [PubMed] [Google Scholar]
- 51.Nielsen, A. L., M. Oulad-Abdelghani, J. A. Ortiz, E. Remboutsika, P. Chambon, and R. Losson. 2001. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell 7**:**729-739. [DOI] [PubMed] [Google Scholar]
- 52.Nielsen, P. R., D. Nietlispach, H. R. Mott, J. Callaghan, A. Bannister, T. Kouzarides, A. G. Murzin, N. V. Murzina, and E. D. Laue. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416**:**103-107. [DOI] [PubMed] [Google Scholar]
- 53.Platero, J. S., T. Hartnett, and J. C. Eissenberg. 1995. Functional analysis of the chromo domain of HP1. EMBO J. 14**:**3977-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Powers, J. A., and J. C. Eissenberg. 1993. Overlapping domains of the heterochromatin-associated protein HP1 mediate nuclear localization and heterochromatin binding. J. Cell Biol. 120**:**291-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reuter, G., and P. Spierer. 1992. Position effect variegation and chromatin proteins. Bioessays 14**:**605-612. [DOI] [PubMed] [Google Scholar]
- 56.Richards, E. J., and S. C. Elgin. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108**:**489-500. [DOI] [PubMed] [Google Scholar]
- 57.Sandaltzopoulos, R., T. Blank, and P. B. Becker. 1994. Transcriptional repression by nucleosomes but not H1 in reconstituted preblastoderm Drosophila chromatin. EMBO J. 13**:**373-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saunders, W. S., C. Chue, M. Goebl, C. Craig, R. F. Clark, J. A. Powers, J. C. Eissenberg, S. C. Elgin, N. F. Rothfield, and W. C. Earnshaw. 1993. Molecular cloning of a human homologue of Drosophila heterochromatin protein HP1 using anti-centromere autoantibodies with anti-chromo specificity. J. Cell Sci. 104 (Pt. 2)**:**573-582. [DOI] [PubMed] [Google Scholar]
- 59.Schotta, G., A. Ebert, R. Dorn, and G. Reuter. 2003. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell Dev. Biol. 14**:**67-75. [DOI] [PubMed] [Google Scholar]
- 60.Schotta, G., A. Ebert, V. Krauss, A. Fischer, J. Hoffmann, S. Rea, T. Jenuwein, R. Dorn, and G. Reuter. 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 21**:**1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schotta, G., A. Ebert, and G. Reuter. 2003. SU(VAR)3-9 is a conserved key function in heterochromatic gene silencing. Genetica 117**:**149-158. [DOI] [PubMed] [Google Scholar]
- 62.Singh, P. B., J. R. Miller, J. Pearce, R. Kothary, R. D. Burton, R. Paro, T. C. James, and S. J. Gaunt. 1991. A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res. 19**:**789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smothers, J. F., and S. Henikoff. 2001. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol. Cell. Biol. 21**:**2555-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smothers, J. F., and S. Henikoff. 2000. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol. 10**:**27-30. [DOI] [PubMed] [Google Scholar]
- 65.Stewart, M. D., J. Li, and J. Wong. 2005. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol. Cell. Biol. 25**:**2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taipale, M., S. Rea, K. Richter, A. Vilar, P. Lichter, A. Imhof, and A. Akhtar. 2005. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol. Cell. Biol. 25**:**6798-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thiru, A., D. Nietlispach, H. R. Mott, M. Okuwaki, D. Lyon, P. R. Nielsen, M. Hirshberg, A. Verreault, N. V. Murzina, and E. D. Laue. 2004. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 23**:**489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tschiersch, B., A. Hofmann, V. Krauss, R. Dorn, G. Korge, and G. Reuter. 1994. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13**:**3822-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tse, C., and J. C. Hansen. 1997. Hybrid trypsinized nucleosomal arrays: identification of multiple functional roles of the H2A/H2B and H3/H4 N-termini in chromatin fiber compaction. Biochemistry 36**:**11381-11388. [DOI] [PubMed] [Google Scholar]
- 70.Wade, P. A., P. L. Jones, D. Vermaak, and A. P. Wolffe. 1999. Purification of a histone deacetylase complex from Xenopus laevis: preparation of substrates and assay procedures. Methods Enzymol. 304**:**715-725. [DOI] [PubMed] [Google Scholar]
- 71.Wang, Y., W. Fischle, W. Cheung, S. Jacobs, S. Khorasanizadeh, and C. D. Allis. 2004. Beyond the double helix: writing and reading the histone code. Novartis Found. Symp. 259**:**3-17, 17-21, 163-169. [PubMed] [Google Scholar]
- 72.Yamamoto, K., and M. Sonoda. 2003. Self-interaction of heterochromatin protein 1 is required for direct binding to histone methyltransferase, SUV39H1. Biochem. Biophys. Res. Commun. 301**:**287-292. [DOI] [PubMed] [Google Scholar]
- 73.Zhao, T., and J. C. Eissenberg. 1999. Phosphorylation of heterochromatin protein 1 by casein kinase II is required for efficient heterochromatin binding in Drosophila. J. Biol. Chem. 274**:**15095-15100. [DOI] [PubMed] [Google Scholar]
- 74.Zhao, T., T. Heyduk, C. D. Allis, and J. C. Eissenberg. 2000. Heterochromatin protein 1 binds to nucleosomes and DNA in vitro. J. Biol. Chem. 275**:**28332-28338. [DOI] [PubMed] [Google Scholar]