Type I interferon signaling is required for activation of the inflammasome during Francisella infection (original) (raw)
Abstract
Francisella tularensis is a pathogenic bacterium whose virulence is linked to its ability to replicate within the host cell cytosol. Entry into the macrophage cytosol activates a host-protective multimolecular complex called the inflammasome to release the proinflammatory cytokines interleukin (IL)-1β and -18 and trigger caspase-1–dependent cell death. In this study, we show that cytosolic F. tularensis subspecies novicida (F. novicida) induces a type I interferon (IFN) response that is essential for caspase-1 activation, inflammasome-mediated cell death, and release of IL-1β and -18. Extensive type I IFN–dependent cell death resulting in macrophage depletion occurs in vivo during F. novicida infection. Type I IFN is also necessary for inflammasome activation in response to cytosolic Listeria monocytogenes but not vacuole-localized Salmonella enterica serovar Typhimurium or extracellular adenosine triphosphate. These results show the specific connection between type I IFN signaling and inflammasome activation, which are two sequential events triggered by the recognition of cytosolic bacteria. To our knowledge, this is the first example of the positive regulation of inflammasome activation. This connection underscores the importance of the cytosolic recognition of pathogens and highlights how multiple innate immunity pathways interact before commitment to critical host responses.
The innate immune system is the first line of defense against microbes. Innate immune receptors called pattern recognition receptors (PRRs) recognize conserved microbial molecules. PRRs like Toll-like receptors (TLRs) at the plasma membrane detect extracellular ligands, whereas TLRs in phagosomes/endosomes and cytosolic PRRs detect intracellular infections (1). Most TLRs signal through the adaptor MyD88 to activate NF-κB. Some TLRs can signal through the adaptor TRIF to activate the transcription factor IRF-3, leading to the production of type I IFN, including IFN-β (2). IFN-β plays a crucial role in defense against viruses but is also induced in response to several bacterial infections (3).
The cytosolic PRRs Nod1/Nod2 activate NF-κB by signaling through the RIP2 adaptor after recognition of peptidoglycan fragments (4–6). Viral RNA is recognized in the cytosol by RIG-I and MDA5, which signal through the adaptor MAVS (7). Both viral RNA and bacterial DNA in the cytosol lead to an IRF-3–dependent induction of IFN-β, although, in the latter case, the PRR is unknown (8). In addition, cytosolic PRRs in the NALP family can activate a multimolecular complex called the inflammasome (9, 10). The cytosolic pathogens Francisella tularensis and Listeria monocytogenes, the vacuole-restricted pathogen Salmonella enterica serovar Typhimurium, and ATP induce inflammasome activation (11–13). These stimuli lead to the activation of caspase-1 through the adaptors ASC or Ipaf. In turn, in macrophages, caspase-1 activation leads to the release of two potent proinflammatory cytokines, IL-1β and -18, and eventually leads to cell death (10).
Inflammasome activation is critical for immune defense against many pathogens, including F. tularensis (10, 12). F. tularensis is a gram-negative bacterium and the causative agent of tularemia. Infections often occur through arthropod bites. The bacteria may spread from the skin to systemic sites, leading to colonization of the spleen and other organs (14). In the host, F. tularensis enters macrophages, escapes the phagosome/vacuole, and replicates within the cytosol before being found in autophagosome-like vacuoles (15). Mutants lacking the transcription factor MglA or genes within the Francisella pathogenicity island (FPI) are unable to escape the vacuole and to replicate within the host (12, 16, 17). Recently, we and others have shown that vacuolar escape/cytosolic localization is necessary to trigger inflammasome activation (12, 18).
Cytosolic localization of F. tularensis subspecies novicida (F. novicida) is required for its virulence but is also essential for its recognition by the host inflammasome. Thus, the macrophage cytosol is a critical host–pathogen interface during F. novicida infection. To analyze the events that occur at this site, we compared the macrophage responses to F. novicida strains that can or cannot reach that site. We report that a type I IFN response was associated with the recognition of cytosolic F. novicida. This was independent of signaling from TLRs, RIG-I, MDA5, Nod1/2, and inflammasome adaptors but dependent on IRF-3 signaling. Type I IFN signaling was necessary for activation of the inflammasome during infection with F. novicida as well as another cytosolic pathogen, L. monocytogenes, but not after infection with the vacuole-localized bacterium S. typhimurium or in response to extracellular ATP. Type I IFN secreted in vivo during F. novicida infection is required for extensive cell death and depletion of macrophages in the spleen. We propose that in response to cytosolic bacteria, type I IFN acts as a checkpoint to control inflammasome activation.
RESULTS AND DISCUSSION
Cytosolic localization of F. novicida induces a type I IFN response in an IRF-3–dependent TLR-independent manner
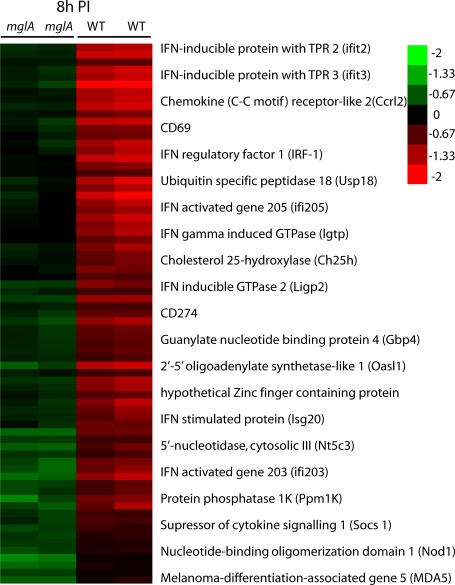
To gain insight into the host response to cytosolic F. novicida, we used microarrays to compare the transcriptome of mouse BM–derived macrophages (BMMs) infected for 8 h with WT F. novicida or a vacuole-restricted mutant, mglA. The transcriptional response associated with the presence of F. novicida in the host cytosol was statistically associated with 68 up-regulated genes (Fig. 1, Fig. S1, and Table S1; available at http://www.jem.org/cgi/content/full/jem.20062665/DC1). A striking feature of this list is the number of genes induced by type I IFN, suggesting that the detection of F. novicida in the host cytosol is associated with a type I IFN signature.
Figure 1.
WT cytosolic F. novicida induces a type I IFN transcriptional response in BMMs, whereas the vacuole-restricted mglA mutant does not. At 8 h postinfection (PI), 68 genes were statistically differentially regulated between mglA and WT _F. novicida_–infected BMMs. The arrays corresponding to the biological replicates and the 21 genes with the higher changes between mglA and WT infected macrophages are shown.
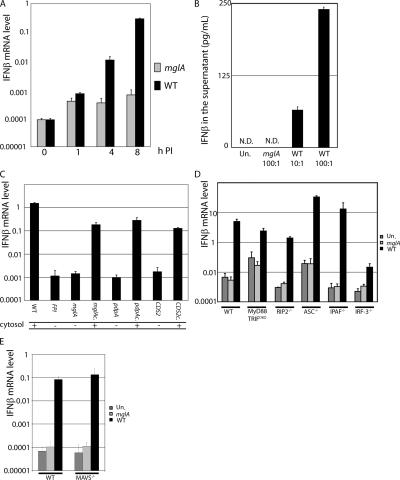
To confirm our transcriptional analysis, we quantified IFN-β transcript levels from infected macrophages. There was a 103-fold increase of IFN-β mRNA in macrophages infected for 8 h with WT bacteria, whereas macrophages infected with the mglA strain showed only a mild up-regulation of IFN-β transcript (Fig. 2 A). These results are consistent with our microarray data. Importantly, the two strains induced similar levels of TNF-α (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20062665/DC1), suggesting that the type I IFN transcription profile is specific for cytosolic F. novicida and not the result of a general difference in the responsiveness of macrophages to the two strains. Accordingly, IFN-β was present in the supernatant of macrophages infected with WT F. novicida in a dose-dependent manner but was undetectable in the supernatant of macrophages infected with the mglA strain (Fig. 2 B). F. novicida escape from the vacuole is dependent on FPI genes, which are transcriptionally regulated by MglA (12, 16). We measured the IFN-β mRNA levels in macrophages infected with F. novicida strains lacking the entire FPI or specific genes (pdpA and cds2) within the FPI. All of the vacuolar mutants failed to induce IFN-β transcript (Fig. 2 C). We conclude that the presence of F. novicida in the cytosol is necessary to induce IFN-β mRNA and secretion of the cytokine.
Figure 2.
F. novicida in the host cytosol induces IFN-β secretion in a TLR-independent IRF-3–dependent manner. IFN-β mRNA levels were determined by quantitative RT-PCR in uninfected BMMs (Un) or at various times PI (A) and 7 (C), 9 (D), or 8 (E) h PI with either mglA or WT F. novicida. IFN-β levels were determined by ELISA in the supernatant of BMMs infected at the indicated MOI for 9 h (ND, nondetectable; B). Various vacuole-restricted mutants do not induce IFN-β mRNA, whereas their complemented counterparts (c.) do. Cytosolic localization is shown (C). BMMs from WT or from IRF-3−/−, ASC−/−, MyD88/TRIFDKO, Ipaf−/−, RIP2−/− (D), MAVS−/− mice, or WT littermates (E) were analyzed for their IFN-β mRNA levels. Error bars represent SEM.
We next investigated the possible roles of PRRs in the induction of IFN-β in response to cytosolic F. novicida. Macrophages deficient for the TLR adaptors MyD88 and TRIF are unable to transduce TLR signals (2). The IFN-β transcript level was only slightly lower in MyD88/TRIF double KO (DKO) macrophages as compared with WT cells (Fig. 2 D), indicating that the IFN-β response to F. novicida infection is mainly TLR independent. The cytosolic PRRs Nod1/2 signal through RIP2, whereas RIG-I and MDA5 signal through MAVS. Bacterial molecules in the host cytosol can lead to signaling through the inflammasome adaptors ASC and Ipaf (10). WT, RIP2-, ASC-, Ipaf-, and MAVS-deficient macrophages infected with WT F. novicida induced similar levels of IFN-β transcript (Fig. 2, D and E). This result suggests that the Nod1/2-, RIG-I–, and MDA5-dependent pathways and the inflammasome were not involved in IFN-β secretion in response to cytosolic F. novicida and that a still unknown PRR is involved in detecting F. novicida in the cytosol. IRF-3 is a major regulator of type I IFN. IRF-3−/− macrophages were severely impaired in their ability to up-regulate IFN-β transcript in response to F. novicida. Interestingly, this IRF-3–dependent IFN-β is similar to the one triggered by the cytosolic gram-positive bacteria L. monocytogenes but is different from the one observed upon infection with the cytosolic parasite Trypanosoma cruzi, which induces IFN-β in a TLR-dependent manner (19, 20). It is intriguing to speculate that the host is sensing cytosolic bacterial DNA and signaling through the IRF-3–dependent pathway to induce IFN-β in response to these intracellular bacteria (8, 21).
Type I IFN signaling is necessary for the cell death of macrophages infected with F. novicida
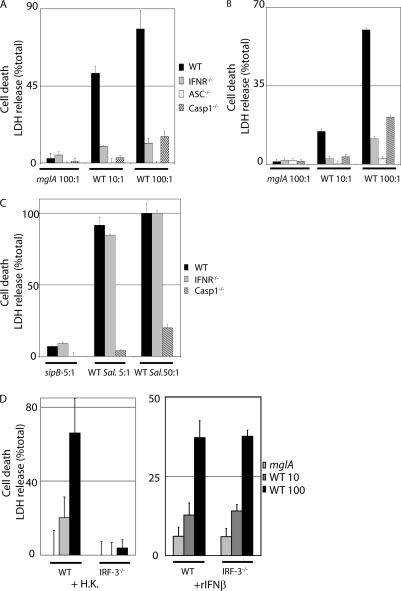
Cytosolic F. novicida leads to inflammasome activation, which results in host cell death. Because both inflammasome-mediated cell death and IFN-β production in _F. novicida_–infected macrophages requires cytosolic localization of the bacteria, we tested whether the two processes are linked. IFN-β signals through the type I IFN receptor (IFNR). To investigate the role of type I IFN in F. novicida infection, we used macrophages deficient for the IFNR. As shown previously (12), cell death in activated WT macrophages was dependent on ASC, caspase-1, and cytosolic bacteria because the vacuolar strain mglA did not induce cell death (Fig. 3 A). Strikingly, we found that cell death of activated IFNR−/− macrophages infected with WT F. novicida was completely abolished (Fig. 3 A). Similar to preactivated macrophages, cell death in unactivated macrophages was dependent on the IFNR and the ASC–caspase-1 axis (Fig. 3 B). The absence of cell death in IFNR−/− macrophages was not caused by a lack of bacterial replication because replication was similar or slightly higher in IFNR−/− macrophages compared with WT macrophages (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20062665/DC1). Collectively, our results demonstrate that the role of type I IFN in inflammasome-mediated cell death is independent of the activation state of the macrophages and is not related to bacterial replication.
Figure 3.
Type I IFN induction and signaling is required for _F. novicida_–mediated but not for _S. typhimurium_–mediated cell death. Cell death of WT, IFNR−/−, ASC−/−, and caspase-1 (casp1)−/− BMMs was assayed by lactate dehydrogenase (LDH) release. BMMs either unactivated (B) or preactivated with heat-killed F. novicida (A, C, and D [left]) or pretreated with recombinant IFN-β (D, right) were infected for 8 (A), 12.5 (B), 3 (C), or 6 h (D) with F. novicida (A, B, and D) or S. typhimurium (C) strains at the indicated MOI. In agreement with previous data (30), cell death required the S. typhimurium gene sipB. Error bars represent SEM.
We next investigated whether IFNR−/− macrophages have a general defect in inflammasome-dependent cell death by infecting WT and IFNR−/− macrophages with S. typhimurium, which also triggers caspase-1–dependent cell death. In contrast to _F. novicida_–induced cell death, _S. typhimurium_–mediated cell death was independent of type I IFN signaling because _S. typhimurium_–infected IFNR−/− macrophages were killed to the same extent as WT macrophages (Fig. 3 C). This result indicates that IFNR−/− macrophages do not have a general defect in inflammasome-mediated cell death but that type I IFN signaling is specifically required for _F. novicida_–mediated inflammasome-dependent cell death.
We have shown that IFN-β is secreted in response to cytosolic F. novicida and that signaling through the IFNR is required for host cell death. To examine the role of IFN-β in cell death, we infected WT macrophages with F. novicida in the presence of an IFN-β–neutralizing antibody. This resulted in a small but reproducible reduction of cell death (unpublished data), indicating that IFN-β is at least partially responsible for _F. novicida_–induced cell death. IRF-3−/− macrophages are deficient for the induction of IFN-β in response to F. novicida infection (Fig. 2 D). Accordingly, IRF-3−/− macrophages are resistant to _F. novicida_–mediated cell death, but the death of infected IRF-3−/− cells is restored upon the addition of recombinant IFN-β (Fig. 3 D). This result shows that type I IFN secreted in response to F. novicida is required for inflammasome-mediated cell death.
The addition of recombinant IFN-β to WT macrophages rendered them more susceptible to _F. novicida_–mediated cell death (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20062665/DC1), indicating that type I IFN is not only required for but can augment _F. novicida_–induced macrophage death. Cell death was not observed upon the addition of recombinant IFN-β to uninfected or _mglA_-infected macrophages, demonstrating that type I IFN is necessary but not sufficient to trigger cell death. Our results demonstrate that _F. novicida_–mediated cell death requires two sequential steps that are dependent on the recognition of F. novicida in the cytosol. The initial recognition step leads to IFN-β secretion, and the second step triggers inflammasome-dependent cell death in an IFNR-dependent manner and independently of TLR signaling (Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20062665/DC1.
Type I IFN signaling is necessary for activation of the inflammasome during F. novicida and L. monocytogenes infection but not upon treatment with ATP
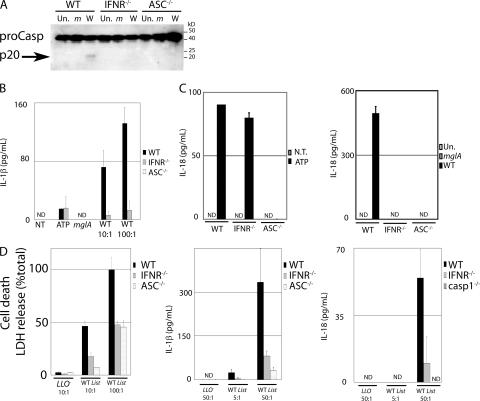
Type I IFN has diverse functions, including pro- and antiapoptotic actions. We show here that type I IFN is necessary for inflammasome-mediated cell death during F. novicida infection. However, it is unknown whether only the cell death is affected or whether all of the inflammasome-mediated activities are type I IFN dependent during F. novicida infection. Thus, we decided to determine whether the type I IFN signal was acting upstream or downstream of the inflammasome. Inflammasome activation leads to the proteolytic activation of procaspase-1, generating the processed p20 subunit, as well as the release of the proinflammatory cytokines IL-1β and -18. The p20 subunit was detected in WT macrophages upon infection with WT F. novicida but not in IFNR−/− or ASC−/− macrophages (Fig. 4 A). This suggests that type I IFN is acting upstream or at the level of the inflammasome. Consistent with the lack of caspase-1 processing, _F. novicida_–mediated release of IL-1β (Fig. 4 B) and -18 (Fig. 4 C) was severely impaired in IFNR−/− macrophages. This was not caused by the absence or reduction in the levels of caspase-1 or ASC in IFNR−/− macrophages because the amounts of these proteins were similar in WT and IFNR−/− macrophages (Fig. 4 A and Fig. S6, available at http://www.jem.org/cgi/content/full/jem.20062665/DC1). Collectively, these results show that type I IFN signaling is required for caspase-1 processing, cell death, and IL-1β and -18 release in response to F. novicida infection.
Figure 4.
Type I IFN signaling is necessary for activation of the inflammasome during F. novicida and L. monocytogenes infections but not upon ATP treatment. (A) WT, IFNR−/−, and ASC−/− BMMs uninfected (Un) or infected with mglA (m) or WT F. novicida (W) were lysed at 9 h PI. Caspase-1 processing was visualized by detection of the p20 subunit only in WT macrophages infected with WT F. novicida. (B and C) IL-1β (B) and -18 (C) were quantified by ELISA in the supernatant of preactivated BMMs. Similar levels were detected in WT and IFNR−/− BMMs treated for 3 h with 5 mM ATP (left), whereas high levels were only detected in WT macrophages upon infection with F. novicida for 5 h (right). (D) Cell death (left) was strongly reduced in IFNR−/− compared with WT BMMs at 6 h PI with L. monocytogenes. Similarly, IL-1β (middle) and -18 (right) levels were lower in activated BMMs infected for 2.5 h with L. monocytogenes at the indicated MOI.
To determine whether type I IFN plays a role in regulating inflammasome activation in response to other activators, we treated macrophages with ATP or infected them with L. monocytogenes (13, 22). Treatment of WT and IFNR−/− macrophages with ATP led to the release of similar levels of IL-1β (Fig. 4 B) and -18 (Fig. 4 C). Thus, as seen with _S. typhimurium_–induced cell death (Fig. 3 C), the inflammasome is activated normally in IFNR−/− macrophages in response to extracellular ATP. IFNR−/− macrophages were twice as resistant to _L. monocytogenes_–mediated cell death as WT macrophages, and both IL-1β and -18 release was highly impaired in IFNR−/− macrophages (Fig. 4 D). This result is consistent with previous results (23). Together, these results show that activation of the inflammasome during infection with cytosolic bacteria (F. novicida and L. monocytogenes) requires type I IFN signaling, whereas stimuli localized to other areas of the cell (S. typhimurium in the vacuole and extracellular ATP) activate the inflammasome independently of type I IFN.
Type I IFN signaling is necessary for extensive cell death and macrophage depletion in the spleen of infected animals
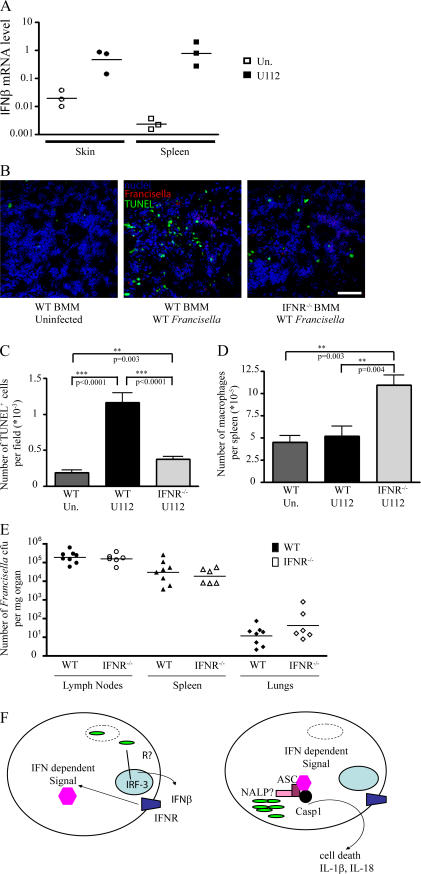
We have shown in vitro that macrophages infected with F. novicida secrete IFN-β. To determine whether this occurs in vivo, we injected WT mice with F. novicida subcutaneously. At 48 h postinfection (PI), we observed a large up-regulation of IFN-β mRNA in the skin and spleen (Fig. 5 A). Our in vitro model predicts that higher numbers of macrophages would die in infected WT mice as compared with IFNR−/− mice. To further validate our in vitro results, we performed TdT-mediated 2′-deoxyuridine 5′-triphosphate–biotin nick end labeling (TUNEL) staining on spleen sections. Extensive TUNEL labeling was observed in the spleens of infected WT mice, indicating that F. novicida infection triggered cell death in vivo (Fig. 5, B and C). In contrast, there were few TUNEL-positive cells in the spleens of infected IFNR−/− mice. Consistent with this result, FACS analysis revealed a significant decrease in the number of splenic macrophages in infected WT mice as compared with IFNR−/− mice (P = 0.004; Fig. 5 D). Collectively, these results show that in vivo, F. novicida infection triggers a type I IFN–dependent cell death that results in the depletion of macrophages, thereby validating our in vitro data.
Figure 5.
IFN-β is up-regulated in vivo upon infection with F. novicida, leading to IFNR-dependent cell death and depletion of BMMs in the spleen at 48 h PI. Each symbol represents the value of one individual mouse. Horizontal bars correspond to the geometric mean. (A) IFN-β mRNA levels in the skin and spleen of WT uninfected (open symbols) or infected (closed symbols) mice were determined by quantitative RT-PCR. (B) TUNEL staining (green) performed on a spleen section of uninfected (left), infected (middle), WT, or infected IFNR−/− mice (right) nuclei (blue), and F. novicida (red) stainings are shown. Representative images are shown. Bar, 100 μm. (C–E) Quantification of the number of TUNEL-positive cells (C), splenic macrophages (D), and F. novicida CFU in various organs from WT (closed symbols) and IFNR−/− (open symbols) mice (E) are presented. (F) Model for the inflammasome activation in response to F. novicida (see Conclusion and model: cytosolic sensing of F. novicida … for details). Error bars represent SEM.
The difference in the levels of TUNEL-positive cells in the spleens of WT and IFNR−/− mice could not be attributed to differences in bacterial colonization because both mouse backgrounds harbored similar levels of bacteria (Fig. 5 E). In contrast, ASC−/− and caspase-1−/− mice harbor significantly higher levels of bacteria than WT mice upon F. novicida infection (12). Type I IFN signaling induces neutropenia in vivo, an effect that is detrimental for host control of L. monocytogenes (24). Therefore, the levels of F. novicida in WT compared with IFNR−/− mice may be the sum of host detrimental neutropenia (25) and host protective activation of the inflammasome (12). Future work will address this hypothesis.
Conclusion and model: cytosolic sensing of F. novicida leads to two sequential signals integrated at the inflammasome
Our results show that cytosolic F. novicida induces type I IFN secretion and that signaling through the IFNR is necessary for activation of the inflammasome in infected macrophages. Extensive IFNR-dependent cell death and depletion of macrophages was also observed in vivo in a mouse model of tularemia. Secretion of IFN-β by infected cells could act as a paracrine danger signal to activate the neighboring cells and accelerate activation of the inflammasome when they become infected with F. novicida, thereby removing the bacterial replication niche before extensive replication occurs. Because IFN-β is a major cytokine secreted upon viral infection, it would be interesting to test whether inflammasome activation in virus-infected cells (26) is also IFNR dependent.
We propose the following model (Fig. 5 F) for activation of the inflammasome in response to cytosolic bacteria. First, the initial sensing of bacteria in the cytosol by an unidentified sensor leads to IRF-3–dependent IFN-β secretion (our results and reference 23) and IFNR-dependent signaling. Second, the recognition of cytosolic bacteria, probably by a PRR from the NALP family, leads to activation of the inflammasome if the first signal originating from the IFNR has been received. Activation of the inflammasome is a critical event leading to the release of proinflammatory cytokines and cell death. Both cell death and inflammation are processes that the host must tightly control to avoid extensive tissue damage. We propose that the IFNR signal controls the commitment to such a terminal pathway.
The nature of the type I IFN–dependent signaling events are unknown. There is no obvious correlation between the requirement for specific inflammasome components and IFNR signaling. Indeed, inflammasome activation triggered by both ATP and L. monocytogenes is dependent on NALP3 and ASC, but only in the latter case is IFNR signaling required. Among the 68 genes identified as being up-regulated in response to cytosolic F. novicida, three genes (ifi-203, -204, and -205) encode proteins predicted to have a pyrin domain capable of interacting with the pyrin domain of ASC (27). Activation of the inflammasome is thought to result from the formation of a multiprotein complex through homotypic domain interactions. The ifi-203, -204, and -205 genes are induced in an IFNR-dependent manner during F. novicida infection (unpublished data). Therefore, those proteins might represent the connection between the type I IFN signaling and the inflammasome by providing a required interacting partner for ASC, a key adaptor molecule of the F. novicida and L. monocytogenes inflammasome.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and growth conditions are listed in the supplemental materials and methods (available at http://www.jem.org/cgi/content/full/jem.20062665/DC1).
Mice and F. novicida.
infections. IFNRA1−/− (IFNR−/−) mice in the C57BL/6 background were obtained from the D. Portnoy laboratory (University of California, Berkeley, Berkeley, CA). Caspase-1 −/− (12) mice were bred at the Stanford University animal facility. Mice were kept under specific pathogen-free conditions in filter-top cages at Stanford University and provided with sterile water and food ad libitum. Experimental studies were in accordance with Institutional Animal Care and Use Committee guidelines. All mice were inoculated with 105 WT F. novicida subcutaneously in 50 μl PBS, and organs were taken at 48 h PI. Frozen sections of spleen were stained using the In Situ Cell Death Detection kit (TUNEL; Roche), anti–F. tularenis chicken polyclonal antibody, anti–chicken AlexaFluor594 antibody, and TO-PRO-3 iodide. Two mice per condition were used, and six spleen sections per mouse were stained. The number of TUNEL+ cells was scored in 10 fields per section. For FACS analysis, single-cell suspensions from spleens from four WT uninfected mice, eight WT infected mice, and six IFNR−/−-infected mice were prepared and stained with 7/4-FITC, F480-PE, Gr-1-Cy7PE, CD11b-Cy7APC, CD3ɛ-Pacblue, and CD19-Cy5.5APC. Macrophage numbers were determined as the number of Gr1Int, 7/4low, F480+, and CD11b+ cells. F. novicida colonization levels were determined by plating serial dilutions of homogenized organs from the same mice on Mueller agar plates and counting the CFU after incubation of the plates for 24 h at 37°C.
Macrophage infections.
BMMs were prepared from mice femurs, cultured, infected at the indicated multiplicity of infection (MOI), and assayed for cell death by lactate dehydrogenase release as previously described (28). Cell death was also assayed by crystal violet staining or direct visualization under the microscope with similar results. Femurs from IFNR−/−, ASC−/−, Ipaf−/−, IRF-3−/−, MyD88/TRIFDKO, and MAVS−/− mice were obtained from the laboratories of D. Portnoy, V. Dixit (Genentech, South San Francisco, CA), R. Medzhitov (Yale University, New Haven, CT) and Z.J. Chen (University of Texas Southwestern Medical Center, Dallas, TX). Where indicated, macrophages were preactivated overnight. Unless otherwise indicated, preactivation was performed by adding heat-killed F. novicida. Recombinant IFN-β fused at 1,000 U/ml was purchased from R&D Systems.
Western blotting.
BMMs infected at an MOI of 100:1 were lysed in 1% NP-40 lysis buffer (50 mM Tris buffer, pH 7.4, and 150 mM NaCl) supplemented with Complete protease inhibitor cocktail (Roche). Rabbit anticaspase-1 p10 (Santa Cruz Biotechnology, Inc.), rat anticaspase-1 p20 (11), mouse anti–α-tubulin (clone B-5-1-2; Sigma-Aldrich), and rat anti-ASC (11) antibodies were used.
Cytokine measurements.
At the indicated times PI before substantial cell death was detected, supernatant was collected and cleared by centrifugation. ELISA kits were obtained from R&D Systems, MBL International Corporation, or PBL Biomedical Laboratories.
RNA preparation and quantitative real-time RT-PCR.
BMMs were infected at an MOI of 100:1 or 10:1 for microarray analysis and quantitative RT-PCR, respectively. To isolate RNA, 1 ml TRIzol reagent (Invitrogen) was added to BMMs in a six-well plate or to 5–100 mg of the various mice organs. RNA was isolated using the RNeasy Mini kit (QIAGEN). Quantitative real-time RT-PCR was performed on a real-time detection system (iCycler; Bio-Rad Laboratories) using rTth enzyme (Applied Biosystems), SYBR green, and primers for β-actin, IFN-β, and TNF-α that were previously described (29). Gene-specific transcript levels were normalized to the amount of β-actin mRNA.
Microarray analysis.
Mouse exonic evidence-based oligonucleotide arrays consisting of 70-nucleotide-long probes were used. Detailed protocols are available in supplemental material. Two biological replicates were performed for each sample at each time point. The list of genes whose expression was statistically different at 8 h PI between WT F. novicida and _mglA_-infected BMMs was generated using the Significance Analysis for Microarrays program using a false discovery rate of 1.5%. All results are available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/).
Online supplemental material.
Table S1 provides a list of genes differentially regulated in BMMs infected with WT F. novicida or the vacuole-restricted mutant mglA. Table S2 provides the bacterial strains used in this study. Fig. S1 shows microarray analysis of BMM infection with WT F. novicida and the mglA strain. Fig. S2 shows TNF-α induction in BMMs upon mglA or WT F. novicida infection. Fig. S3 shows a replication of F. novicida in WT and IFNR−/− BMMs. Fig. S4 shows the cell death of _F. novicida_–infected BMMs upon activation with recombinant IFN-β. Fig. S5 shows the cell death of WT, RIP2−/−, MyD88/TRIFDKO, and MAVS−/− BMMs after F. novicida infection. Fig. S6 shows the protein levels of caspase-1 and ASC in WT and IFNR−/− BMMs. Supplemental materials and methods provides information about bacterial growth conditions, intracellular replication assay, microarray analysis, and statistical analysis. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20062665/DC1.
Supplemental Material
[Supplemental Material index]
Acknowledgments
We thank N. Meyer-Morse, D. Portnoy for the IFNR−/− mice, I. Brodsky, R. Medzhitov for the IRF-3−/− and MyD88/TRIFDKO femurs, V. Dixit for the RIP-2−/−, ASC−/−, and Ipaf−/− femurs, R. Vance, and Q. Sun and Z.J. Chen for the MAVS−/− BMMs. We also thank K. Elkins for the gift of bacterial strains, J. Cox, A. Sil, all PO1 members for stimulating discussions, and S. Falkow for critical reading.
T. Henry and D. Weiss were supported by fellowships from the European Molecular Biology Organization and the Giannini Family Foundation. This work was supported by grants RO1 AI063302 and AI-65359 from the National Institutes of Health (National Institute of Allergy and Infectious Diseases).
The authors have no conflicting financial interests.
A. Brotcke and D.S. Weiss contributed equally to this paper.
References
- 1.Janeway, C.A., Jr. 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54:1–13. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto, M., S. Sato, H. Hemmi, K. Hoshino, T. Kaisho, H. Sanjo, O. Takeuchi, M. Sugiyama, M. Okabe, K. Takeda, and S. Akira. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 301:640–643. [DOI] [PubMed] [Google Scholar]
- 3.Decker, T., M. Muller, and S. Stockinger. 2005. The yin and yang of type I interferon activity in bacterial infection. Nat. Rev. Immunol. 5:675–687. [DOI] [PubMed] [Google Scholar]
- 4.Girardin, S.E., I.G. Boneca, L.A. Carneiro, A. Antignac, M. Jehanno, J. Viala, K. Tedin, M.K. Taha, A. Labigne, U. Zahringer, et al. 2003. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 300:1584–1587. [DOI] [PubMed] [Google Scholar]
- 5.Girardin, S.E., I.G. Boneca, J. Viala, M. Chamaillard, A. Labigne, G. Thomas, D.J. Philpott, and P.J. Sansonetti. 2003. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278:8869–8872. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi, K., N. Inohara, L.D. Hernandez, J.E. Galan, G. Nunez, C.A. Janeway, R. Medzhitov, and R.A. Flavell. 2002. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 416:194–199. [DOI] [PubMed] [Google Scholar]
- 7.Sun, Q., L. Sun, H.H. Liu, X. Chen, R.B. Seth, J. Forman, and Z.J. Chen. 2006. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 24:633–642. [DOI] [PubMed] [Google Scholar]
- 8.Stetson, D.B., and R. Medzhitov. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 24:93–103. [DOI] [PubMed] [Google Scholar]
- 9.Martinon, F., K. Burns, and J. Tschopp. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell. 10:417–426. [DOI] [PubMed] [Google Scholar]
- 10.Mariathasan, S., and D.M. Monack. 2007. Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat. Rev. Immunol. 7:31–40. [DOI] [PubMed] [Google Scholar]
- 11.Mariathasan, S., K. Newton, D.M. Monack, D. Vucic, D.M. French, W.P. Lee, M. Roose-Girma, S. Erickson, and V.M. Dixit. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 430:213–218. [DOI] [PubMed] [Google Scholar]
- 12.Mariathasan, S., D.S. Weiss, V.M. Dixit, and D.M. Monack. 2005. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J. Exp. Med. 202:1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mariathasan, S., D.S. Weiss, K. Newton, J. McBride, K. O'Rourke, M. Roose-Girma, W.P. Lee, Y. Weinrauch, D.M. Monack, and V.M. Dixit. 2006. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 440:228–232. [DOI] [PubMed] [Google Scholar]
- 14.Francis, E. 1927. Tularemia. The Atlantic Medical Journal. 30:337–344. [Google Scholar]
- 15.Checroun, C., T.D. Wehrly, E.R. Fischer, S.F. Hayes, and J. Celli. 2006. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. USA. 103:14578–14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santic, M., M. Molmeret, K.E. Klose, S. Jones, and Y.A. Kwaik. 2005. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7:969–979. [DOI] [PubMed] [Google Scholar]
- 17.Baron, G.S., and F.E. Nano. 1998. MglA and MglB are required for the intramacrophage growth of _Francisella novicida._Mol. Microbiol. 29:247–259. [DOI] [PubMed] [Google Scholar]
- 18.Gavrilin, M.A., I.J. Bouakl, N.L. Knatz, M.D. Duncan, M.W. Hall, J.S. Gunn, and M.D. Wewers. 2006. Internalization and phagosome escape required for Francisella to induce human monocyte IL-1beta processing and release. Proc. Natl. Acad. Sci. USA. 103:141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaena de Avalos, S., I.J. Blader, M. Fisher, J.C. Boothroyd, and B.A. Burleigh. 2002. Immediate/early response to Trypanosoma cruzi infection involves minimal modulation of host cell transcription. J. Biol. Chem. 277:639–644. [DOI] [PubMed] [Google Scholar]
- 20.Koga, R., S. Hamano, H. Kuwata, K. Atarashi, M. Ogawa, H. Hisaeda, M. Yamamoto, S. Akira, K. Himeno, M. Matsumoto, and K. Takeda. 2006. TLR-dependent induction of IFN-beta mediates host defense against _Trypanosoma cruzi._J. Immunol. 177:7059–7066. [DOI] [PubMed] [Google Scholar]
- 21.Ishii, K.J., C. Coban, H. Kato, K. Takahashi, Y. Torii, F. Takeshita, H. Ludwig, G. Sutter, K. Suzuki, H. Hemmi, et al. 2006. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 7:40–48. [DOI] [PubMed] [Google Scholar]
- 22.Kanneganti, T.D., N. Ozoren, M. Body-Malapel, A. Amer, J.H. Park, L. Franchi, J. Whitfield, W. Barchet, M. Colonna, P. Vandenabeele, et al. 2006. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 440:233–236. [DOI] [PubMed] [Google Scholar]
- 23.Stockinger, S., T. Materna, D. Stoiber, L. Bayr, R. Steinborn, T. Kolbe, H. Unger, T. Chakraborty, D.E. Levy, M. Muller, and T. Decker. 2002. Production of type I IFN sensitizes macrophages to cell death induced by _Listeria monocytogenes._J. Immunol. 169:6522–6529. [DOI] [PubMed] [Google Scholar]
- 24.Navarini, A.A., M. Recher, K.S. Lang, P. Georgiev, S. Meury, A. Bergthaler, L. Flatz, J. Bille, R. Landmann, B. Odermatt, et al. 2006. Increased susceptibility to bacterial superinfection as a consequence of innate antiviral responses. Proc. Natl. Acad. Sci. USA. 103:15535–15539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sjostedt, A., J.W. Conlan, and R.J. North. 1994. Neutrophils are critical for host defense against primary infection with the facultative intracellular bacterium Francisella tularensis in mice and participate in defense against reinfection. Infect. Immun. 62:2779–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanneganti, T.D., M. Body-Malapel, A. Amer, J.H. Park, J. Whitfield, L. Franchi, Z.F. Taraporewala, D. Miller, J.T. Patton, N. Inohara, and G. Nunez. 2006. Critical role for cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J. Biol. Chem. 281:36560–36568. [DOI] [PubMed] [Google Scholar]
- 27.Albrecht, M., D. Choubey, and T. Lengauer. 2005. The HIN domain of IFI-200 proteins consists of two OB folds. Biochem. Biophys. Res. Commun. 327:679–687. [DOI] [PubMed] [Google Scholar]
- 28.Brotcke, A., D.S. Weiss, C.C. Kim, P. Chain, S. Malfatti, E. Garcia, and D.M. Monack. 2006. Identification of MglA-regulated genes reveals novel virulence factors in _Francisella tularensis._Infect. Immun. 74:6642–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auerbuch, V., D.G. Brockstedt, N. Meyer-Morse, M. O'Riordan, and D.A. Portnoy. 2004. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med. 200:527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hersh, D., D.M. Monack, M.R. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA. 96:2396–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material index]