Acquisition of a Functional T Cell Receptor during T Lymphocyte Development Is Enforced by HEB and E2A Transcription Factors (original) (raw)
. Author manuscript; available in PMC: 2008 Jan 10.
SUMMARY
The T cell receptor (TCR) is required for positive selection and the subsequent transition from the CD4+CD8+ double-positive (DP) to the CD4+ or CD8+ single-positive (SP) stage of αβ T cell development. The molecular mechanism that maintains DP fate prior to the acquisition of a functional TCR is not clear. We have shown here that the structurally and functionally related transcription factors HEB and E2A work together to maintain DP fate and to control the DP to SP transition. Simultaneous deletion of HEB and E2A in DP thymocytes was sufficient for DP to SP transition independent of TCR. Loss of HEB and E2A allowed DP cells to bypass the requirement for TCR-mediated positive selection, downregulate DP-associated genes, and upregulate SP-specific genes. These results identify HEB and E2A as the gatekeepers that maintain cells at the DP stage of development until a functional αβ TCR is produced.
INTRODUCTION
Production of a diverse, self-tolerant compartment of αβ T cells is dependent on precise coordination of antigen-receptor recombination, differentiation, and selection events during development in the thymus. The primary determinant of progression through αβ T cell development is the expression of a functional T cell receptor (TCR). The genes encoding the two components of the TCR, TCRα and TCRβ, undergo recombination in a lineage- and stage-specific manner (Goldrath and Bevan, 1999). First, thymocytes rearrange their TCRβ chain during the CD4+CD8+ double-negative (DN) stage of development. Cells that undergo in-frame rearrangement to assemble a functional TCRβ will express a pre-TCR, composed of TCRβ and pre-Tα, and will then progress to the CD4−CD8− double-positive (DP) stage. It is during the DP stage when thymocytes undergo TCRα rearrangement to produce a mature αβTCR. DP cells expressing a functional TCR, capable of recognition of antigen in the context of major histocompatibility complex (MHC) molecules, will receive a positive-selection signal and differentiate to the CD4+ or CD8+ single-positive (SP) stage. DP thymocytes that fail to produce a functional TCR cannot become SP cells and will die by neglect. Thymocytes expressing a functional TCR also undergo an additional selection process, termed negative selection, to eliminate autoreactive cells (von Boehmer and Kisielow, 2006). TCR expression and selection are obligatory events for the development of SP cells that will then emigrate from the thymus to establish the peripheral CD4 helper and CD8 cytotoxic T cell compartments.
The transition from DP to SP stage, directed by TCR-mediated positive selection, involves the activity of E protein transcription factors HEB and E2A encoded by the genes Tcf12 and Tcfe2a, respectively (Murre, 2005). E proteins, containing a basic-helix-loop-helix (bHLH) domain, function as dimers to recognize and bind E box sites (CANNTG), and HEB-E2A heterodimers have been shown to be the major form of E proteins functioning during T cell development (Barndt et al., 2000; Sawada and Littman, 1993). Although HEB and E2A are required and upregulated during the early stages of T cell development, E2A activity is subsequently downregulated as thymocytes progress through DP and SP stages (Bain et al., 1997; Barndt et al., 1999; Engel et al., 2001; Pan et al., 2002; Taghon et al., 2006). In particular, signaling by the pre-TCR in DN thymocytes has been shown to result in a downregulation of E2A activity for entry to DP stage, and a similar mechanism has been proposed downstream of TCR signaling for the DP to SP transition (Engel et al., 2001; Murre, 2005). TCR signaling during a positive-selection event has been suggested to reduce E2A activity by inducing expression of the E protein inhibitor Id3 (Bain et al., 2001). In addition, E2A-deficient mice demonstrate an increase in maturation from the DP to SP stage, whereas Id3-deficient mice demonstrate a decrease in DP-to-SP maturation (Bain et al., 1999; Rivera et al., 2000). These findings suggest that the downregulation of E protein activity upon positive selection is critical for proceeding past the TCR checkpoint to the SP stage of development.
Two major issues remain to be addressed. First, TCR signaling in late DP stage triggers multiple downstream events in addition to downregulation of E2A. Loss of E2A alone is clearly not sufficient to initiate the transition from DP to SP. It is not known whether the removal of both E2A and HEB provides the only switch or one of many parallel regulatory events leading to SP development. Second, although downregulation of E2A can facilitate the DP to SP transition, the exact role for E proteins during the DP stage prior to TCR expression is not known. Here we examined E protein function by simultaneous removal of both HEB and E2A at the DP stage. We found that the premature loss of HEB and E2A triggers development of CD8+ T cells even in the absence of a TCR. Our findings identify a function for HEB and E2A in maintaining DP fate and enforcing TCR-mediated positive selection. Loss of HEB and E2A activity is not only necessary but also sufficient for development to the SP stage.
RESULTS
T Cell-Specific Deletion of HEB and E2A in DP Thymocytes
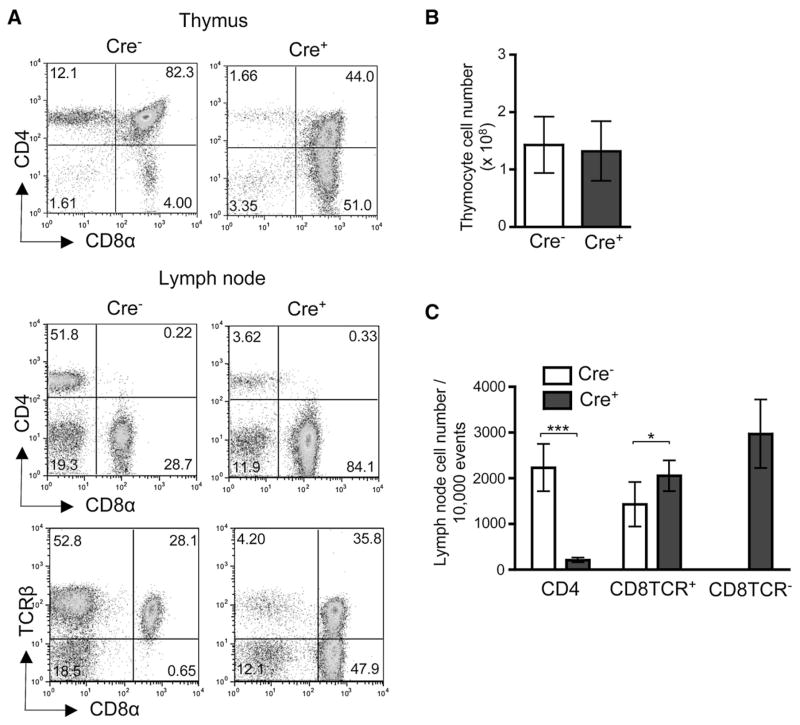
To further investigate the roles for E proteins in positive selection during DP to SP development, we have created a mouse model for T lineage-specific deletion of both HEB and E2A in DP thymocytes. We have crossed mice carrying HEB (Wojciechowski et al., 2007) and E2A (Pan et al., 2002) conditional alleles to CD4Cre transgenic mice (Wolfer et al., 2001), and these mice will hereafter be referred to as _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice. Upon initial characterization of _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice, we observed that the CD4 and CD8 populations were markedly altered in the thymus and periphery (Figure 1). Although the overall thymic cellularity remained unchanged (Figure 1B), there was a severe reduction of CD4SP and an increase of CD8SP cells in _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice compared to _Tcf12_f/f_Tcfe2a_f/fCD4Cre− control mice (Figure 1A). This reduction of CD4+ and increase of CD8+ cells was also observed in the periphery (Figures 1A and 1C). Even more striking, there was an abundant population of CD8+ cells lacking surface TCR expression in the periphery (Figures 1A and 1C). Deletion analysis demonstrated that both HEB and E2A were efficiently deleted in DP and subsequent stages of development (Figure S1 available online). However, to determine whether the CD8TCR− phenotype was dependent on deletion of all four alleles, we analyzed mice retaining either one HEB or E2A wild-type allele (Figure S2). Peripheral CD8TCR− cells were not detected in _Tcf12_f/+_Tcfe2a_f/f CD4Cre+ or _Tcf12_f/f_Tcfe2a_f/+CD4Cre+ mice, demonstrating that complete deletion of HEB and E2A was required for generation of this population.
Figure 1. T Cell-Specific Deletion of HEB and E2A Generates Peripheral CD8TCR− Cells.
(A) Representative staining of indicated tissues from 2-month-old _Tcf12_f/f_Tcfe2a_f/fCD4Cre− control and _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice for CD4, CD8α, and TCRβ. Percentages in each quadrant are displayed.
(B) Cell number in the thymus of 2–3-month-old mice (n = 9).
(C) Cell numbers per 10,000 events collected from lymph node (LN) of 2–7-month-old mice (_Tcf12_f/f_Tcfe2a_f/fCD4Cre− n = 7, _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ n = 8). ***p < 0.001 and *p = 0.012, Student’s t test, two-tailed. Graphed results in (B) and (C) are means with error bars representing SD.
Characterization of Peripheral CD8TCR− Cells
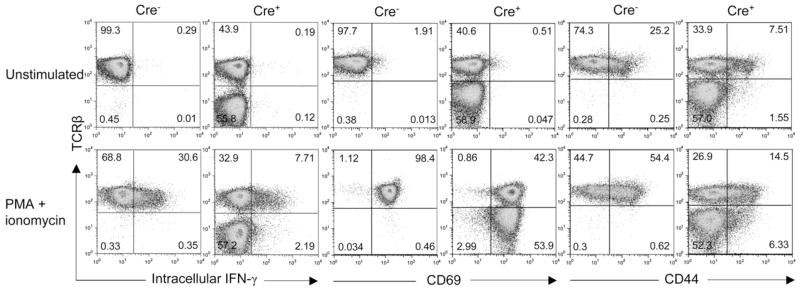
Further analysis of the surface phenotype of the peripheral CD8TCR− cells showed that they were CD8α+CD8β+ CD62L+CD44loCD69−TCRγδ−, consistent with a mature, resting αβ T cell phenotype (Figure S3 and data not shown). To test whether CD8TCR− cells displayed similar functionality to that of conventional CD8+ T cells, we analyzed their response to stimulation. CD8TCR− cells were able to produce IFN-γ, albeit at reduced amounts, and upregulate expression of activation markers CD44 and CD69 upon PMA and ionomycin stimulation (Figure 2). However, a defect in homeostatic proliferation was observed in the CD8TCR− cells when transferred to lymphopenic, _Rag2_−/− recipients (Figure S4). Because these cells resembled CD8+ T cells in terms of phenotype and function, but exhibited defective expansion in the periphery, they were probably generated by a constant output from the thymus.
Figure 2. HEB and E2A Double-Deficient CD8TCR+ and CD8TCR+ Cells Produce IFN-γ and Upregulate Activation Markers upon Stimulation.
In vitro culture of LN cells isolated from _Tcf12_f/f_Tcfe2a_f/fCD4Cre− and _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice with or without (unstimulated) PMA and ionomycin for 6 hr. Cells were analyzed by FACS analysis for intracellular IFN-γ and surface CD69 and CD44 expression. Plots are gated on CD8+ cells, and percentages in each quadrant are displayed. Data are representative of three independent experiments.
CD8TCR− Cells Are T Cells Developing in the Absence of a Functional TCR
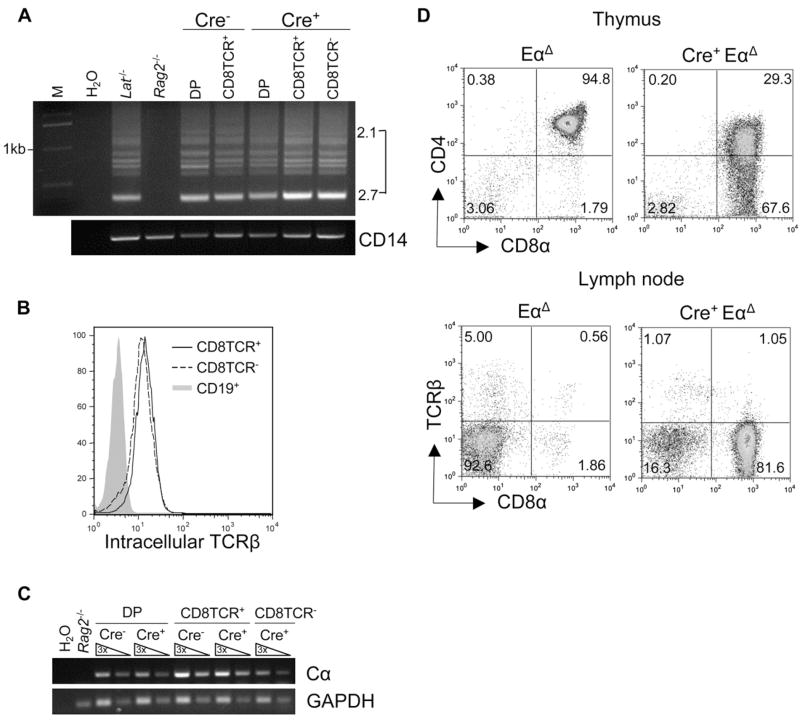
To begin investigating how and when CD8TCR− cells diverged from conventional CD8+ T cell development, we first examined their TCR rearrangement status. CD8TCR− cells had undergone TCRβ V to DJ recombination and expressed intracellular TCRβ chain at a comparable amount to their CD8TCR+ counterparts (Figures 3A and 3B). In addition, TCRα transcript expression was detected in the CD8TCR− cells (Figure 3C). We next performed sequencing analysis of TCRα V to J rearrangement by amplifying cDNA from _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ CD8TCR+ and CD8TCR− peripheral cells with Vα8- specific and Cα-specific (constant region) primers. Sequencing revealed that TCRα rearrangements were present in the CD8TCR− population, and they were nonfunctional, explaining why these cells lacked a functional surface TCR (Table 1). To then determine whether the development of CD8TCR− cells could occur independently of TCRα rearrangement, we crossed our _Tcf12_f/f_Tcfe2a_f/f CD4Cre+ mice onto a _Tcr_α enhancer-deficient background (EαΔ) (Sleckman et al., 1997). EαΔ mice exhibit a severe block in TCRα recombination and accumulation of cells at the DP stage (Sleckman et al., 1997). The presence of CD8TCR− cells in _Tcf12_f/f_Tcfe2a_f/fCD4Cre+EαΔ mice showed that development of this population did not require TCRα rearrangement (Figure 3D).
Figure 3. CD8TCR− Cells Are T Cells Developing in the Absence of a Functional TCR.
(A) TCRβ V to DJ rearrangement analysis on DNA from _Tcf12_f/f_Tcfe2a_f/fCD4Cre− and _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ sorted thymus (DP) and LN (CD8TCR+, CD8TCR−) populations used Vβ8 5′ consensus and Jβ2.7 3′ primers. Rearrangement products involving Jβ2.1–Jβ2.7 are shown. _Lat_−/− (Zhang et al., 1999) and _Rag2_−/− total thymocyte DNA were used as positive and negative controls, respectively. CD14 was used as a loading control. Molecular weight marker is labeled (M).
(B) Intracellular TCRβ expression in specified populations from _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ LN.
(C) RT-PCR analysis for TCRα (Cα) expression in sorted populations from _Tcf12_f/f_Tcfe2a_f/fCD4Cre− and _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice. _Rag2_−/− total thymocyte cDNA was used as a negative control, and GAPDH was used as a loading control. Three-fold serial dilutions are as shown.
(D) Phenotype of thymus and LN cells from EαΔ and _Tcf12_f/f_Tcfe2a_f/fCD4Cre+Eα Δ mice. Percentages in each quadrant are displayed.
Table 1.
TCRα Rearrangements in CD8TCR− Cells Are Nonfunctional
| Jα | # of Clones | # in Frame | # out of Frame |
|---|---|---|---|
| CD8 TCR+ | |||
| 57 | 1 | 1 | 0 |
| 52 | 3 | 3 | 0 |
| 47 | 1 | 0 | 1 |
| 42 | 3 | 3 | 0 |
| Total | 8 | 7 | 1 |
| CD8 TCR− | |||
| 58 | 5 | 0 | 5 |
| 56 | 1 | 0 | 1 |
| 43 | 2 | 0 | 2 |
| Total | 8 | 0 | 8 |
The strategy used for the above sequencing of TCRα rearrangements also allowed for investigation of Jα segment usage. Rearrangement in the Jα locus proceeds in a proximal to distal (5′ to 3′ ) manner (Petrie et al., 1995; Thompson et al., 1990; Wang et al., 1998). Initial TCRα rearrangements use Jα segments at the 5′ end of the locus and can be followed by secondary rearrangements using more 3′ Jα segments. In our sequencing analysis, Jα usage in both CD8TCR+ and CD8TCR− populations from _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice appeared to be skewed to the 5′ end of the locus (Table 1). Analysis of Jα usage by PCR from genomic DNA of peripheral T cells also demonstrated this biased usage of the more 5′ Jα segments (Figure S5). Rearrangements using Jα22, the most 3′ Jα analyzed, were greatly reduced in _Tcf12_f/f _Tcfe2a_f/fCD4Cre+ CD8TCR+ and CD8TCR− T cells compared to _Tcf12_f/f_Tcfe2a_f/fCD4Cre− and heterozygous _Tcf12_f/+_Tcfe2a_f/+CD4Cre+ and _Tcf12_f/+_Tcfe2a_f/fCD4Cre+ controls, whereas rearrangements using the more 5′ Jα58 and Jα49 segments were equally if not more abundant compared to controls. These results were reminiscent of the RORγ-deficient mice that demonstrate a defect in 3′ Jα usage because of a survival defect limiting the lifespan of DP thymocytes (Guo et al., 2002; Kurebayashi et al., 2000; Sun et al., 2000). Because E2A has been shown to regulate the isoform RORγt in DP thymocytes (Xi et al., 2006), we next analyzed RORγt expression. As suspected, RORγt expression was decreased in _Tcf12_f/f _Tcfe2a_f/fCD4Cre+ DP cells (Figure 4A). In addition, _Tcf12_f/f _Tcfe2a_f/fCD4Cre+ DP thymocytes demonstrated reduced ex vivo survival when cultured in media alone compared to wild-type, _Tcf12_f/f_Tcfe2a_f/fCD4Cre−, and EαΔ controls (Figure 4B). Together, the Jα usage, decrease in RORγt expression, and reduced ex vivo survival suggested that _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ thymocytes also have a shortened time in the DP stage.
Figure 4. _Tcf12f/fTcfe2a_f/fCD4Cre+ DP Thymocytes Survive Poorly, but Differentiate to SP Cells fEfficiently in Culture.
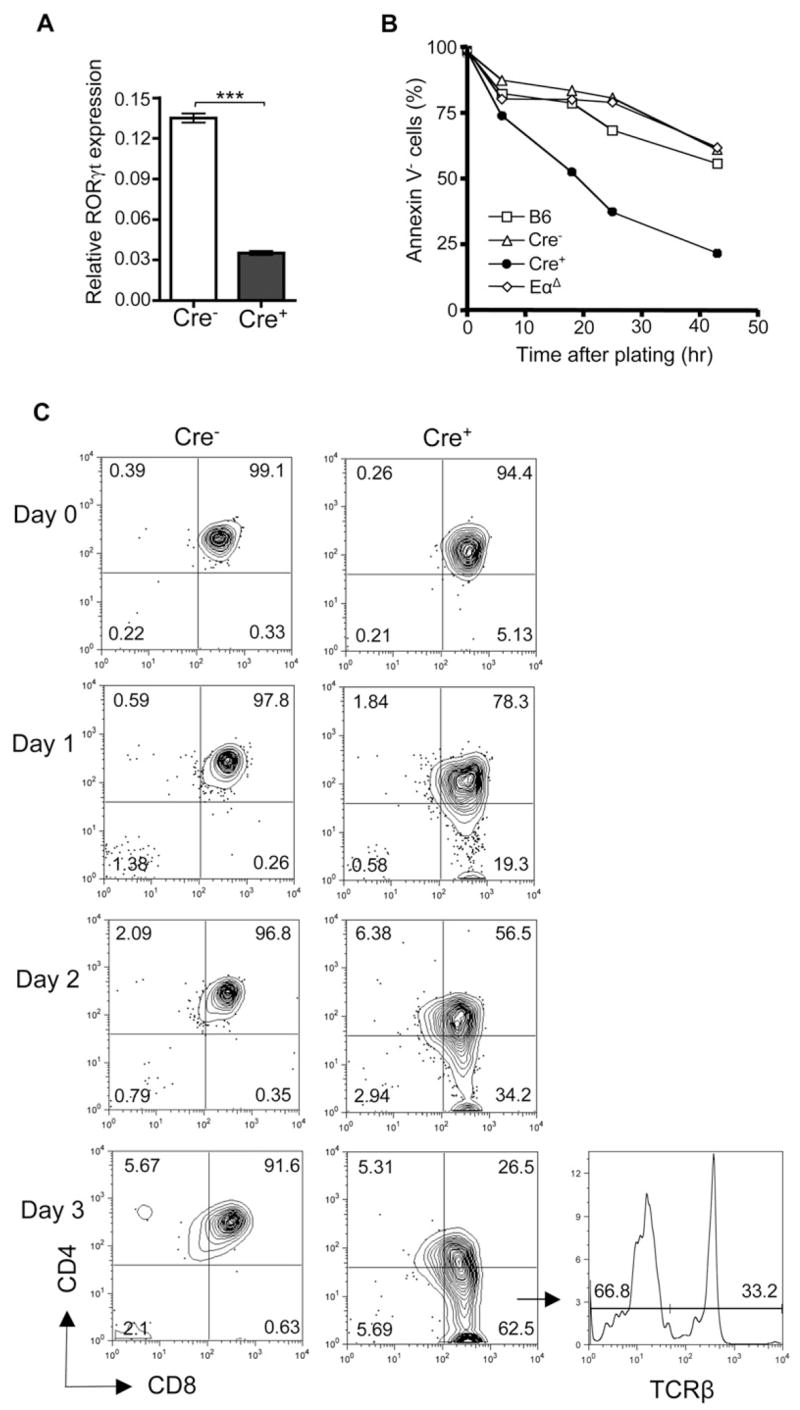
(A) Quantitative RT-PCR analysis of RORγt expression in sorted _Tcf12_f/f_Tcfe2a_f/fCD4Cre− and _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ DP cells. Samples were normalized to the expression of GAPDH. Data are from duplicates of two independent experiments (n = 4). ***p < 0.001, Student’s t test, two-tailed. Graphed results are means with error bars representing standard error of the mean (SEM).
(B and C) Ex vivo culture analysis of sorted DP thymocytes from _Tcf12_f/f_Tcfe2a_f/fCD4Cre− and _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice. (B) DP cells were plated in media alone and analyzed for Annexin V expression by FACS at 0, 6, 18, 25, and 43 hr after plating. Wild-type (B6) and Eα ΔDP cells were used as additional controls. Data are representative of two independent experiments. (C) DP cells were plated on a layer of total thymic stromal cells (day 0) and analyzed by FACS analysis for CD4, CD8, and TCRβ expression on day 1–3. Percentages in each quadrant are displayed. TCRβ expression within the _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ CD8SP gate is shown for day 3. Data are representative of three independent experiments.
Even though the data suggested that CD8TCR− cells developed from DP thymocytes, it still remained possible that these cells, being CD8+, could develop directly from the earlier CD8+ immature single-positive (ISP) stage. ISP cells represent a transitional stage from DN to DP development when cells first upregulate CD8 prior to CD4. To therefore verify DP stage as the developmental source of our CD8TCR− cells, we cultured DP thymocytes ex vivo on a thymic stromal layer. DP thymocytes from _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice not only gave rise to both CD8TCR+ and CD8TCR− SP cells, but also did so very efficiently compared to the _Tcf12_f/f_Tcfe2a_f/fCD4Cre− control cells (Figure 4C). _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ DP cells appeared to have been rescued from an enhanced cell-death phenotype (Figure 4B) because the loss of HEB and E2A also triggered maturation of DP cells to the SP stage. It is this latter event that led to the development of CD8TCR− cells.
We then wanted to analyze the rate of CD8SP cell production in vivo. _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ and wild-type (B6) mice were injected with BrdU for 4 and 24 hr pulse analysis. Percent labeling in DN, DP, and CD8SP populations did not demonstrate a dramatic difference between _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ and control mice at either time point (Figure S6). However, _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice did display an increase in BrdU+ CD8SP cell numbers at 24 hr. Because cell-cycle analysis by Hoechst staining indicated that _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ CD8SP cells were not proliferating (data not shown), the increase in BrdU+ CD8SP cell number could be due to more DP cells giving rise to CD8SP, an increased rate of DP to CD8SP differentiation, or a combination of both.
Loss of HEB and E2A Initiates CD8 T Cell Development
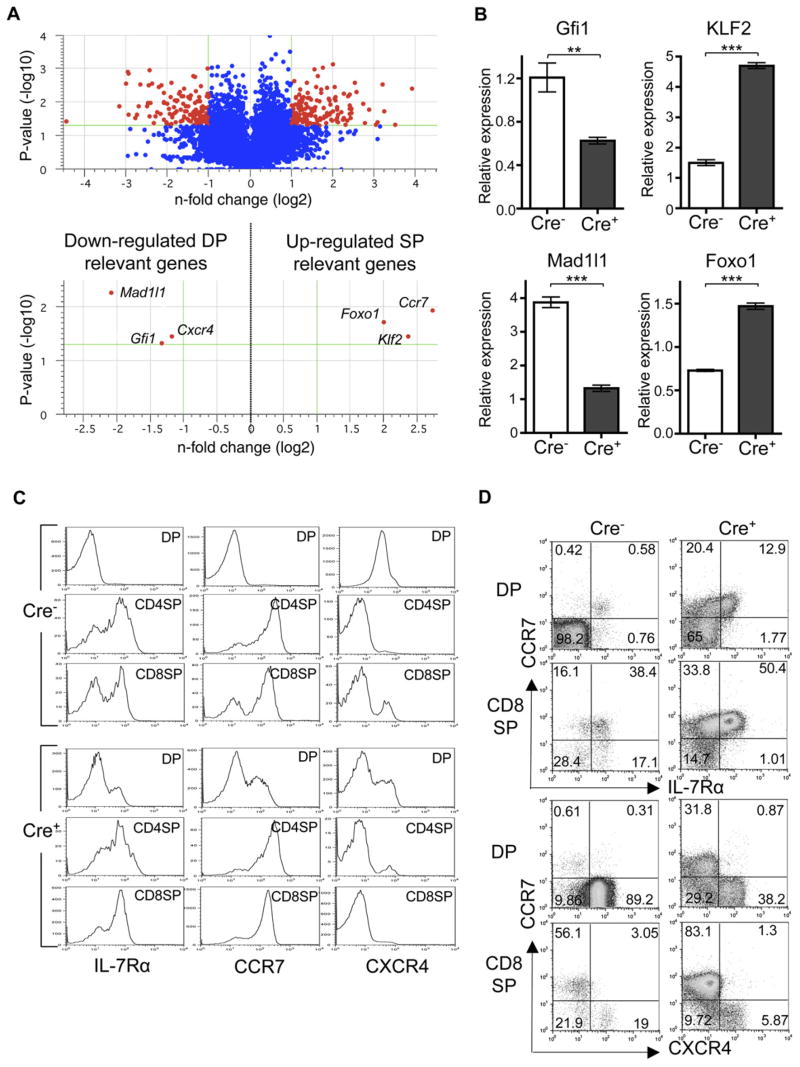
The above data suggested that HEB and E2A are required to maintain DP fate and prevent premature differentiation to SP stage. To get a comprehensive view of HEB- and E2A-mediated gene expression during the DP stage, we performed microarray analysis with sorted DP cells. The results yielded a group of 285 genes that were either upregulated or downregulated greater than 2-fold in the _Tcf12_f/f _Tcfe2a_f/fCD4Cre+ DP population compared to _Tcf12_f/f _Tcfe2a_f/fCD4Cre− control DP cells (Figure 5A and 3-fold change listed in Tables S1 and S2). Within these groups, an interesting trend emerged. Genes known to be highly expressed at DP stage were downregulated within the _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ DP population, whereas genes known to be upregulated at SP stage were already being upregulated. For example, Gfi1 and Mad1l1, which are two genes whose expression has been shown to drop dramatically upon differentiation to SP stage (Rudolph et al., 2001; Yucel et al., 2003), were both downregulated in _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ DP cells. In contrast, Foxo1, which is most highly expressed in positively selected DP cells and SP cells (Leenders et al., 2000), was upregulated in _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ DP thymocytes. Of these, Gfi1 is of particular interest because _Gfi1_−/− mice also demonstrate an enhanced development of CD8SP cells; however, CD8SP development in these mice is shown to occur in only a TCR-dependent manner (Yucel et al., 2003). Altered expression of Gfi1, Mad1l1, and Foxo1 in _Tcf12_f/f _Tcfe2a_f/fCD4Cre+ DP thymocytes was also confirmed by quantitative RT-PCR (Figure 5B).
Figure 5. Loss of HEB and E2A Initiates CD8 T Cell Maturation and Thymic Egress in the Absence of a TCR-Mediated Positive-Selection Signal.
(A) Volcano plot from microarray data comparing gene expression in _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ and _Tcf12_f/f_Tcfe2a_f/fCD4Cre− DP thymocytes. Changes in gene expression are shown as a ratio of _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ to _Tcf12_f/f_Tcfe2a_f/fCD4Cre− cells. Upper plot shows the 15,730 genes remaining after quality filtering, with the 285 genes with greater than 2-fold change and t test p value ≤ 0.05 in red. Lower plot highlights a few genes of interest.
(B) Quantitative RT-PCR analysis of Gfi1, Mad1l1, KLF2, and Foxo1 expression in sorted _Tcf12_f/f_Tcfe2a_f/fCD4Cre− and _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ DP cells. Samples were normalized to the expression of GAPDH. Data are from duplicates of two independent experiments (n = 4). ***p < 0.001 and **p = 0.0054, Student’s t test, two-tailed. Graphed results are means with error bars representing SEM.
(C and D) FACS analysis of IL-7Rα, CCR7, and CXCR4 expression in DP compared to SP stage in thymus from _Tcf12_f/f_Tcfe2a_f/fCD4Cre− and _Tcf12_f/f _Tcfe2a_f/fCD4Cre+ mice. Cells are pregated on CD4+CD8+ (DP), CD4+CD8− (CD4SP), and CD4−CD8+ (CD8SP) populations.
(C) Histograms display IL-7Rα, CCR7, and CXCR4 expression in designated populations from individual stainings.
(D) FACS plots demonstrate coordinated expression of CCR7 and IL-7Rα or CXCR4 in designated populations with percentages in each quadrant displayed. Data are representative of two independent experiments.
The DP microarray analysis also identified genes that are relevant to thymocyte migration. Klf2, encoding a transcription factor critical for activating expression of the sphingosine-1-phosphate receptor S1P1 during SP maturation to allow thymic egress of mature T cells (Carlson et al., 2006), was upregulated in our _Tcf12_f/f _Tcfe2a_f/fCD4Cre+ DP cells. KLF2 expression was also verified by quantitative RT-PCR analysis (Figure 5B). The chemokine receptor CXCR4 is expressed in early stages of T cell development for cortical retention within the thymus (Plotkin et al., 2003) and then downregulated upon positive selection. Concurrently, the chemokine receptor CCR7 is upregulated at this time to induce migration of selected SP thymocytes from the cortex to the medulla, where they will then undergo negative selection (Kurobe et al., 2006). We found that Cxcr4 and Ccr7 were downregulated and upregulated, respectively, in _Tcf12_f/f_Tcfe2a_f/f CD4Cre+ DP thymocytes. In accordance with these microarray results, the surface expressions of IL-7Rα and CCR7, which are both upregulated from DP to SP, were already upregulated in a fraction of _Tcf12_f/f_Tcfe2a_f/f CD4Cre+ DP cells at the level of that in SP stage cells (Figure 5C). In addition, surface expression of CXCR4 was predominantly downregulated in _Tcf12_f/f_Tcfe2a_f/f CD4Cre+ DP cells. To ensure that these were coordinated events in individual cells, we analyzed staining of CCR7 and IL-7Rα or CXCR4 together. _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ DP cells that had upregulated CCR7 expression had concurrently upregulated IL-7Rα and downregulated CXCR4 in a manner similar to that of the _Tcf12_f/f_Tcfe2a_f/f CD4Cre− SP cells (Figure 5D).
These SP-like changes in chemokine-receptor expression at DP stage in _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice together with the decrease in RORγt expression (Figure 4A) and skewed Jα usage (Table 1 and Figure S5) were consistent with a shorter DP lifespan. To further investigate the dwell time of cells in the thymus, we chose to analyze HSA (CD24) expression. HSA is a useful marker for T cell maturation but is not functionally critical for thymocyte development (Nielsen et al., 1997). HSA is highly expressed on ISP cells, slightly lower on DP cells, begins to be further downregulated on SP cells, and is low or absent on mature peripheral T cells. The CD4SP and CD8SP thymocytes in _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice demonstrated a DP-like staining for HSA, suggesting that SP cells had not yet had time to begin downregulating HSA expression (Figure S7). However, the HSA expression in peripheral T cells from _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice was similar to that of the _Tcf12_f/f_Tcfe2a_f/fCD4Cre− control cells. This indicated that _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ T cells did down-regulate HSA by the time they reached the periphery. Together, the microarray and surface-expression analysis showed that DP cells were prematurely acquiring an SP phenotype upon deletion of HEB and E2A.
DISCUSSION
The TCR-independent maturation of _Tcf12_f/f_Tcfe2a_f/f CD4Cre+ DP cells exhibited various aspects of CD8SP cell development, including the silencing of DP-specific genes, maturation to a CD8 single-positive phenotype, and activation of factors critical for migration and thymic egress. These findings have demonstrated that HEB and E2A are critical at the DP stage to block further development until a proper TCR-mediated positive-selection signal is received. Premature withdrawal of HEB and E2A prior to this signal was sufficient to activate the developmental program for CD8 lineage, whether the cell had produced a functional TCR or not. We propose that HEB and E2A function as gatekeepers for a default pathway from DP to CD8SP stage.
The _Tcf12_f/f_Tcfe2a_f/fCD4Cre mouse model will now allow us to further elucidate the downstream transcriptional network directing this pivotal differentiation step from DP to SP. It appears that E proteins are regulating, perhaps directly, two sets of genes. HEB and E2A are required to both maintain DP gene expression and prevent SP gene activation. One candidate gene from our microarray data mentioned above is Gfi1, encoding a transcriptional repressor that may act with or downstream of E proteins to suppress SP genes at the DP stage. Gfi1 was also identified in a microarray study by Schwartz et al., (2006) as a gene suggested to be directly upregulated by E2A. In addition, Foxo1 was identified as an E2A target in this microarray analysis and was suggested to be directly repressed by E2A (Schwartz et al., 2006). This finding also concurs with our microarray data, indicating that Foxo1 may be an E2A-repressed gene at DP stage that is activated upon differentiation to SP stage. These targets and others identified suggest a network of transcription factors functioning downstream of E proteins to orchestrate proper DP to SP development.
Our observation that _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ T cells developed primarily into CD8 lineage but not CD4 suggests a role for E proteins in lineage choice. Depending on their TCR specificity, DP cells give rise to either MHC class II-restricted CD4 SP cells or class I-restricted CD8 SP cells. How this process of lineage choice occurs and is regulated has been under intensive investigation and ongoing debate (Kappes and He, 2006). Focus on transcriptional regulation of this event has led to some more recent advances, mainly the identification of a key transcription factor, Th-POK (also known as cKrox). Th-POK is both required and sufficient for commitment to the CD4 lineage (He et al., 2005; Sun et al., 2005). It has been suggested that CD8 development is a default pathway, and an additional or prolonged signal is required for activation of Th-POK to direct cells to the CD4 lineage (Aliahmad and Kaye, 2006; Kappes and He, 2006). The loss of HEB and E2A in our system initiated premature DP to CD8SP development. Although HSA staining of _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ thymocytes suggests an increased maturation rate, whether or not this DP to SP transition was accelerated once initiated remains to be determined. If the development to SP stage was accelerated upon deletion of HEB and E2A in our model, it would therefore be possible that the cells did not have enough time for CD4 lineage instruction. Alternatively, HEB and E2A may be required specifically for CD4 development or for suppression of CD8 development. Analysis of Th-POK expression in _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ versus _Tcf12_f/f_Tcfe2a_f/fCD4Cre− CD4SP cells demonstrated that HEB and E2A are not required for maintenance of Th-POK expression (data not shown). However, this finding does not rule out a role for E proteins upstream of initiation of Th-POK expression.
One consideration for why the loss of E protein activity favors CD8 over CD4 is the role of E proteins in regulating CD4 expression. Previous studies have suggested the CD4 enhancer as a target of E proteins during thymocyte development (Sawada and Littman, 1993). The presence of HEB- and E2A-deficient CD4 T cells in our _Tcf12_f/f _Tcfe2a_f/fCD4Cre+ mice, albeit at greatly reduced numbers, indicates that HEB and E2A are not required for CD4 expression in mature T cells. However, it still remains possible that loss of CD4 expression, or a CD4-lineage-specific gene, at DP stage upon deletion of HEB and E2A could contribute to the development of only CD8+ T cells.
If the loss of HEB and E2A triggers development of CD8+ T cells, why did we see this small population of peripheral CD4+ T cells in the _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice? These peripheral CD4+ T cells were HEB and E2A deficient, as demonstrated by deletion analysis. Because all of these CD4+ T cells were TCR+, they probably originated from a small number of MHC class II-restricted (CD4-specific) DP cells receiving a positive-selection signal prior to complete deletion of HEB and E2A. These CD4+ T cells were capable of survival and homeostatic proliferation upon transfer (data not shown), so it is expected that the peripheral CD4+ T cell population results from an accumulation of these rare events. It also remains possible that most of the CD4+ T cells in the _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice were negatively selected and those that survived were CD4+ T cells with the lowest binding affinity for MHC-peptide. A role for E proteins in negative selection is also currently being investigated.
Transcriptional regulation by E proteins may provide a means to coordinate DP to SP differentiation, selection, and CD4 versus CD8 lineage commitment. Our results have shown that HEB and E2A function as critical determinants of the DP to SP transition by enforcing the requirement for TCR-mediated positive selection. Our results also suggest that E proteins may serve as key regulators during CD4 versus CD8 lineage choice. Positive selection and lineage choice are suggested to be tightly linked processes; however, the regulation of each remains controversial. Future analysis of the _Tcf12_f/f_Tcfe2a_f/f CD4Cre+ mice in predefined selective backgrounds may help to distinguish the regulatory pathways driving each of these events.
EXPERIMENTAL PROCEDURES
Mice
Mice have been described previously (Pan et al., 2002; Sleckman et al., 1997; Wojciechowski et al., 2007; Wolfer et al., 2001; Zhang et al., 1999). All research with mice was performed in accordance with relevant guidelines, and protocols were approved by the Duke University Animal Care and Use Committee.
Cell Staining and Flow Cytometry
Intracellular staining was done by 2% paraformaldehyde fixation followed by permeabilizing in 0.5% saponin. Annexin V staining was done according to manufacturer’s protocol (BD PharMingen). FACS analysis was done with a FACSCalibur (BD Biosciences) or FACSVantage SE with DiVa option (BD Biosciences) and FlowJo software (Tree Star). FACS plots are pregated on 7-aminoactinomycin D (7AAD, Molecular Probes) negative lymphocytes. FACSVantage SE with DiVa option (BD Biosciences) was used for cell sorting.
In Vitro Lymphocyte Stimulation
Total lymph node (LN) cells isolated from _Tcf12_f/f_Tcfe2a_f/fCD4Cre− and _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ mice were cultured in RPMI (5% FBS) and 2 ng/mL IL-2, with or without 10 ng/mL phorbol 12-myristate 13-acetate (PMA) and 1 μg/mL ionomycin for 6 hr. Three micromolars of monensin was added for the last 4 hr to prevent release of cytokine.
TCRβ Recombination
The protocol has been described previously (Wojciechowski et al., 2007). The following primers were used: Vβ8 5′ consensus (5′-GCATG GGCTGAGGCTGATCCATTA-3′), Jβ2.7 3′ (5′-TGAGAGCTGTCTC CTACTATGGATT-3′). CD14 primers were used as a loading control (Lazorchak et al., 2006).
RT-PCR
For TCRα (Cα constant region) analysis, RNA extraction, DNase I treatment, and reverse transcription has been described previously (Lazorchak et al., 2006). For RORγt, Gfi1, Mad1l1, KLF2, and Foxo1 analysis, RNA was extracted with RNeasy QIAGEN kit with DNase I step following the manufacturer’s protocol. Quantitative real-time PCR analysis was performed with a Roche LightCycler and the Fast-Start DNA master SYBR green kit I (Roche) as per the manufacturer’s instructions. The following primers were used: RORγt primers (Xi et al., 2006) and GAPDH primers (Lazorchak et al., 2006); see the list in Table S3 for the other RT-PCR primers.
TCRα Sequencing
PCR amplification of cDNA from sorted CD8TCR+ and CD8TCR− _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ LN cells with primers specific for Vα8 and Cα yielded a product from any expressed TCRα transcripts when a Vα8 rearrangement was used. The PCR product was cloned into a PCR4 TOPO vector (Invitrogen) and sequenced. A total of eight clones were analyzed for each population. The Jα gene segment usage and V to J rearrangement frame status were determined from the sequences. The following primer sequences were used: Vα8 5′-CAGACAGAAGGCCTGGTCAC-3′, Cα 5′-TGGCGTTGGTCTCTTT GAAG-3′.
In Vitro DP Cultures
For survival assay, sorted CD4+CD8+ DP cells were plated in RPMI media, 10% FBS, 50 uM β-mercaptoethanol, L-glut/Pen/Strep, 25 mM HEPES. For thymic stromal culture, thymus was harvested from wild-type CD45.1 congenic mice, cut into approximately eight pieces, and digested for 30 min at 37°C with 1 mg/mL collagenase 1A (Sigma). A single-cell suspension was made and plated for 6 hr. Resulting adherent cell layer was washed to remove most suspension cells. CD45.2+ CD4+CD8+ DP cells were sorted and plated on thymic stromal layer the following day in IMDM media 5% FBS, 50 uM β-mercaptoethanol, NaPyr/L-glut/Pen/Strep. FACS plots are pregated on 7AAD−CD45.1−Gr-1−Mac-1−B220− lymphocytes.
Microarray Analysis
CD4+CD8+ DP cells were sorted from _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ and _Tcf12_f/f_Tcfe2a_f/fCD4Cre− thymus. The same CD4hiCD8hi gate was used for both _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ and _Tcf12_f/f_Tcfe2a_f/fCD4Cre− sorting to assure analysis of populations expressing similar CD4 and CD8 levels. Independent sorts from two mice per genotype were done. RNA was extracted with RNeasy QIAGEN kit with DNase I step following the manufacturer’s protocol. Array analysis was performed by the Duke Microarray Core Facility (http://microarray.genome.duke.edu/services/spotted-arrays/protocols). In brief, one round of amplification was performed, RNA samples were labeled with Cy3 or Cy5 dyes, and samples were hybridized to the Mouse Operon oligo set 4.0 Chip. Two comparisons were performed: _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ to a Universal Mouse Reference RNA (Strata-gene) and _Tcf12_f/f_Tcfe2a_f/fCD4Cre− to this same reference, each done in duplicate. Data filtering and statistical analysis were performed with GeneSpring software. _Tcf12_f/f_Tcfe2a_f/fCD4Cre+ versus _Tcf12_f/f_Tcfe2a_f/f CD4Cre− comparisons were done with averages of the duplicates, filtering out any genes with > 2 standard deviation (SD) within the groups. Genes that have neither Ensembl nor Unigene identification are excluded from Tables S1 and S2. The complete data set has been submitted to the GEO database (http://www.ncbi.nlm.nih.gov/geo;GSE9749).
Statistical Analysis
Statistical significance was assessed by the two-tailed Student’s t test.
Supplementary Material
Acknowledgments
We thank Michael Krangel, Motonari Kondo, and Jingquan Jia for discussions and reading of the manuscript; M. Krangel for providing Eα Δ mice, _Rag2_−/− mice, and TCRα primers for sequencing and recombination analysis; Meifang Dai for genotyping and i.v. injection assistance; the Duke Comprehensive Cancer Center Flow Cytometry facility and M. Kondo for cell sorting; and the Duke Microarray Core Facility for microarray analysis. This work was supported by funding from the National Institutes of Health to Y.Z.
Footnotes
Accession Numbers
The microarray accession number for the analysis reported in this paper is GSE9749.
References
- Aliahmad P, Kaye J. Commitment issues: Linking positive selection signals and lineage diversification in the thymus. Immunol Rev. 2006;209:253–273. doi: 10.1111/j.0105-2896.2006.00345.x. [DOI] [PubMed] [Google Scholar]
- Bain G, Engel I, Robanus Maandag EC, te Riele HP, Voland JR, Sharp LL, Chun J, Huey B, Pinkel D, Murre C. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Quong MW, Soloff RS, Hedrick SM, Murre C. Thymocyte maturation is regulated by the activity of the helix-loop-helix protein, E47. J Exp Med. 1999;190:1605–1616. doi: 10.1084/jem.190.11.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol. 2001;2:165–171. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- Barndt R, Dai MF, Zhuang Y. A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during alpha beta thymopoiesis. J Immunol. 1999;163:3331–3343. [PubMed] [Google Scholar]
- Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol Cell Biol. 2000;20:6677–6685. doi: 10.1128/mcb.20.18.6677-6685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, Jameson SC. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J Exp Med. 2001;194:733–745. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldrath AW, Bevan MJ. Selecting and maintaining a diverse T-cell repertoire. Nature. 1999;402:255–262. doi: 10.1038/46218. [DOI] [PubMed] [Google Scholar]
- Guo J, Hawwari A, Li H, Sun Z, Mahanta SK, Littman DR, Krangel MS, He YW. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat Immunol. 2002;3:469–476. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- Kappes DJ, He X. Role of the transcription factor Th-POK in CD4:CD8 lineage commitment. Immunol Rev. 2006;209:237–252. doi: 10.1111/j.0105-2896.2006.00344.x. [DOI] [PubMed] [Google Scholar]
- Kurebayashi S, Ueda E, Sakaue M, Patel DD, Medvedev A, Zhang F, Jetten AM. Retinoid-related orphan receptor gamma (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc Natl Acad Sci USA. 2000;97:10132–10137. doi: 10.1073/pnas.97.18.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurobe H, Liu C, Ueno T, Saito F, Ohigashi I, Seach N, Arakaki R, Hayashi Y, Kitagawa T, Lipp M, et al. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity. 2006;24:165–177. doi: 10.1016/j.immuni.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Lazorchak AS, Wojciechowski J, Dai M, Zhuang Y. E2A promotes the survival of precursor and mature B lymphocytes. J Immunol. 2006;177:2495–2504. doi: 10.4049/jimmunol.177.4.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenders H, Whiffield S, Benoist C, Mathis D. Role of the forkhead transcription family member, FKHR, in thymocyte differentiation. Eur J Immunol. 2000;30:2980–2990. doi: 10.1002/1521-4141(200010)30:10<2980::AID-IMMU2980>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- Nielsen PJ, Lorenz B, Muller AM, Wenger RH, Brombacher F, Simon M, von der Weid T, Langhorne WJ, Mossmann H, Kohler G. Altered erythrocytes and a leaky block in B-cell development in CD24/HSA-deficient mice. Blood. 1997;89:1058–1067. [PubMed] [Google Scholar]
- Pan L, Hanrahan J, Li J, Hale LP, Zhuang Y. An analysis of T cell intrinsic roles of E2A by conditional gene disruption in the thymus. J Immunol. 2002;168:3923–3932. doi: 10.4049/jimmunol.168.8.3923. [DOI] [PubMed] [Google Scholar]
- Petrie HT, Livak F, Burtrum D, Mazel S. T cell receptor gene recombination patterns and mechanisms: Cell death, rescue, and T cell production. J Exp Med. 1995;182:121–127. doi: 10.1084/jem.182.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin J, Prockop SE, Lepique A, Petrie HT. Critical role for CXCR4 signaling in progenitor localization and T cell differentiation in the postnatal thymus. J Immunol. 2003;171:4521–4527. doi: 10.4049/jimmunol.171.9.4521. [DOI] [PubMed] [Google Scholar]
- Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 2000;12:17–26. doi: 10.1016/s1074-7613(00)80155-7. [DOI] [PubMed] [Google Scholar]
- Rudolph B, Hueber AO, Evan GI. Expression of Mad1 in T cells leads to reduced thymic cellularity and impaired mitogen-induced proliferation. Oncogene. 2001;20:1164–1175. doi: 10.1038/sj.onc.1204196. [DOI] [PubMed] [Google Scholar]
- Sawada S, Littman DR. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol Cell Biol. 1993;13:5620–5628. doi: 10.1128/mcb.13.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci USA. 2006;103:9976–9981. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleckman BP, Bardon CG, Ferrini R, Davidson L, Alt FW. Function of the TCR alpha enhancer in alphabeta and gamma-delta T cells. Immunity. 1997;7:505–515. doi: 10.1016/s1074-7613(00)80372-6. [DOI] [PubMed] [Google Scholar]
- Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, Mebius RE, Littman DR. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Thompson SD, Pelkonen J, Hurwitz JL. First T cell receptor alpha gene rearrangements during T cell ontogeny skew to the 5′ region of the J alpha locus. J Immunol. 1990;145:2347–2352. [PubMed] [Google Scholar]
- von Boehmer H, Kisielow P. Negative selection of the T-cell repertoire: Where and when does it occur? Immunol Rev. 2006;209:284–289. doi: 10.1111/j.0105-2896.2006.00346.x. [DOI] [PubMed] [Google Scholar]
- Wang F, Huang CY, Kanagawa O. Rapid deletion of rearranged T cell antigen receptor (TCR) Valpha-Jalpha segment by secondary rearrangement in the thymus: Role of continuous rearrangement of TCR alpha chain gene and positive selection in the T cell repertoire formation. Proc Natl Acad Sci USA. 1998;95:11834–11839. doi: 10.1073/pnas.95.20.11834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol. 2007;178:5717–5726. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfer A, Bakker T, Wilson A, Nicolas M, Ioannidis V, Littman DR, Lee PP, Wilson CB, Held W, MacDonald HR, Radtke F. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat Immunol. 2001;2:235–241. doi: 10.1038/85294. [DOI] [PubMed] [Google Scholar]
- Xi H, Schwartz R, Engel I, Murre C, Kersh GJ. Interplay between RORgammat, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity. 2006;24:813–826. doi: 10.1016/j.immuni.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Yucel R, Karsunky H, Klein-Hitpass L, Moroy T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp Med. 2003;197:831–844. doi: 10.1084/jem.20021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Sommers CL, Burshtyn DN, Stebbins CC, DeJarnette JB, Trible RP, Grinberg A, Tsay HC, Jacobs HM, Kessler CM, et al. Essential role of LAT in T cell development. Immunity. 1999;10:323–332. doi: 10.1016/s1074-7613(00)80032-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.