The MYCN oncogene is a direct target of miR-34a (original) (raw)
. Author manuscript; available in PMC: 2008 Oct 7.
Published in final edited form as: Oncogene. 2008 May 26;27(39):5204–5213. doi: 10.1038/onc.2008.154
Abstract
Loss of 1p36 heterozygosity commonly occurs with MYCN amplification in neuroblastoma tumors, and both are associated with an aggressive phenotype. Database searches identified 5 microRNAs that map to the commonly deleted region of 1p36 and we hypothesized that the loss of one or more of these microRNAs contributes to the malignant phenotype of _MYCN_-amplified tumors. By bioinformatic analysis, we identified that 3 out of the 5 microRNAs target MYCN and of these miR-34a caused the most significant suppression of cell growth through increased apoptosis and decreased DNA synthesis in neuroblastoma cell lines with _MYCN_-amplification. Quantitative RT-PCR showed that neuroblastoma tumors with 1p36 loss expressed lower level of miR-34a than those with normal copies of 1p36. Furthermore we demonstrated that MYCN is a direct target of miR-34a. Finally, using a series of mRNA expression profiling experiments, we identified other potential direct targets of miR-34a, and pathway analysis demonstrated that miR-34a suppresses cell cycle genes and induces several neural related genes. This study demonstrates one important regulatory role of miR-34a in cell growth and MYCN suppression in neuroblastoma.
Introduction
Neuroblastoma (NB) is the most frequent extra-cranial childhood solid tumor. Prognosis of this disease varies from spontaneous regression to aggressive progression, depending on several known clinical parameters and genetic aberrations. Among these, MYCN amplification, found in 20-30% of NB tumors, was the first and is the only molecular marker currently used in clinic to stratify patients for therapy (Brodeur, 2003). The MYCN oncogene encodes a transcription factor belonging to the MYC gene family, and is primarily expressed during normal developing embryos (Zimmerman et al., 1986). MYCN is thought to play a critical role in the tumorigenesis of NB (Weiss et al., 1997), as well as in normal brain and other neural development (Zindy et al., 2006). Patients with MYCN amplification and older than 1 year have <30% chance of survival despite aggressive multimodal therapies including surgery, chemotherapy, radiation therapy and stem cell transplant. Elevated expression levels of the MYCN gene subsequent to amplification is thought to be critical for the aggressive behavior of _MYCN_-amplified NBs (Kohl et al., 1984), but the mechanism of its oncogenic effect has not been determined.
Interestingly, loss of heterozygosity (LOH) of 1p36 (25-30% of NBs) is commonly found in NBs with MYCN amplification and is associated with poor outcome (Attiyeh et al., 2005; Guo et al., 1999; White et al., 2005). Since the loss of 1p36 was first reported in NB by Brodeur et al. (Brodeur et al., 1977), studies have identified the deletion of this region in many other human malignancies such as brain tumor (Hashimoto et al., 1995), melanoma (Poetsch et al., 1998), leukemia (Mori et al., 1998), breast cancer (Bieche et al., 1998), and cervix cancer (Cheung et al., 2005), suggesting the presence of tumor suppressor gene(s) in this region. Chromosomal transfer experiments support such a hypothesis, as NB cell lines transfected with 1p DNA showed neuronal differentiation and suppression of tumorigenicity (Bader et al., 1991). Studies have been performed to delineate the smallest region of overlap (SRO) on 1p36 in NBs, and have identified a consensus region of loss between 1p36.2 and 1pter (Chen et al., 2004; Fong et al., 1989; Hogarty et al., 2000; Maris et al., 2001; Mosse et al., 2005; White et al., 1995; White et al., 2005). Although many genes including CHD5 (Bagchi et al., 2007; Thompson et al., 2003), CAMTA1 (Barbashina et al., 2005; Henrich et al., 2006), HKR3 (Maris et al., 1997), Apo-3 (Eggert et al., 2002), ENO1 (Ejeskar et al., 2005), EXTL1 (Mathysen et al., 2004), DFF45 (Abel et al., 2004), p73 (Kong et al., 1999; Ozaki et al., 2005), and PITSLRE (Lahti et al., 1994) mapped to this consensus region have been implicated to have anti-tumorigenic effects, the identity of the single-causal tumor suppressor gene(s) on 1p36 remains elusive due to the similar effects of these genes on the behavior of NB cells.
While most conventional genes encode proteins to carry out their biological functions, a recently discovered class of genes transcribing small non-coding RNAs, namely microRNAs, was found to play important regulatory roles in normal development and physiology in plants and animals (Bartel and Bartel, 2003; Bartel, 2004). Mature microRNAs are 20-22 nucleotide molecules that can regulate gene expression through RNA interference effecter complex (RISC) mediated mRNA degradation and translational suppression via complimentary pairing predominantly to the 3’-untranslated region (3’-UTR) of their targeted messenger RNAs (Ambros, 2004; Bartel, 2004). An increasing number of studies have demonstrated a perturbation of the normal expression patterns of microRNAs in many human cancers (Calin et al., 2002; Cimmino et al., 2005; Costinean et al., 2006; Lu et al., 2005). Recently, a study by Welch et al. demonstrated that a microRNA located in 1p36, miR-34a, induced apoptosis in NB cells, suggesting its important role in regulating cell growth and death (Welch et al., 2007). Several studies further demonstrated that p53 directly regulates the members of miR-34 family including miR-34a, and miR-34 family members can mediate some of the p53 functions (Chang et al., 2007; He et al., 2007; Raver-Shapira et al., 2007).
In this study, we identified that among the 5 microRNAs mapping to 1p36 three are predicted to target MYCN, and miR-34a had the most growth suppressing effect on the _MYCN_-amplified NB cells among these three microRNAs. In addition, we provide evidence that miR-34a can directly regulate the MYCN protein level by targeting its 3’-UTR. Furthermore, global gene expression profiles demonstrated suppression of cell cycle genes and induction of neural genes in NB cells mediated by miR-34a. Therefore, our study reveals an important regulatory role of miR-34a in modulating MYCN activity in NB tumors.
Results
Biological effects of microRNAs on _MYCN_-amplified Cell Lines
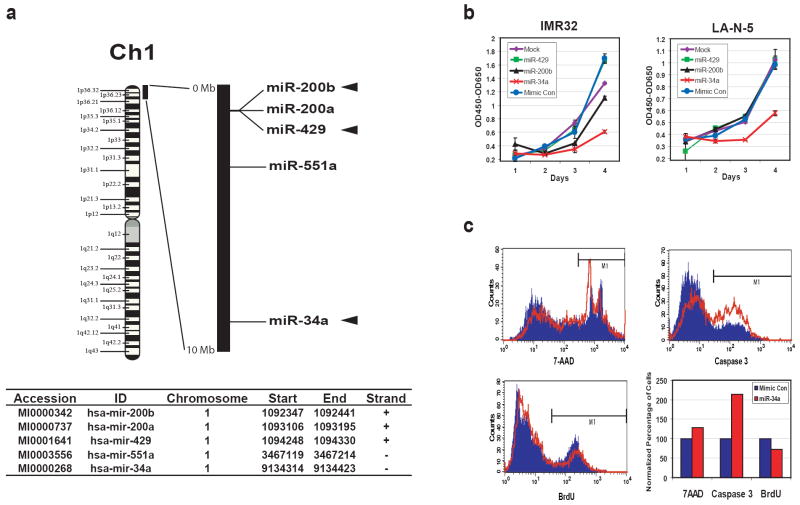
Due to the frequent association of 1p36 loss in neuroblastoma (NB) tumors with MYCN amplification, we investigated if microRNAs mapping to this region may affect growth of NB cells with MYCN amplification. We first identified that 5 microRNAs map within the first 10Mb on chromosome 1 short arm (1p36.22 to 1pter), which is commonly deleted in NB (Chen et al., 2004; Fong et al., 1989; Hogarty et al., 2000; Maris et al., 2001; Mosse et al., 2005; White et al., 1995; White et al., 2005), using the Sanger microRNA registry (http://microrna.sanger.ac.uk/sequences/index.shtml, Release 10.0) (Figure 1a). Of note, three of these (miR-200b, 429 and 34a) target the MYCN gene as predicted by computational analysis on the Sanger microRNA registry miRBase website. We thus investigated the effect on growth for these 3 microRNAs on IMR32 and LA-N-5 cell lines (both contain _MYCN_-amplification). We found that exogenous miR-34a resulted in a significant reduction in cell growth compared to the mimic control for both cell lines, whereas miR-200b and miR-429 had little or no effect (Figure 1b). To further characterize miR-34a-mediated inhibition of cell growth, we first measured the cell death using 7-AAD staining (uptake in dead cells) at 48 hours post transfection (Figure 1c upper left). IMR32 cells transfected with miR-34a had a 29% increase in cell death compared to those transfected with mimic control microRNA. Then, we investigated if the increased cell death was due to apoptosis using an antibody specific to the active form of caspase 3 in a flow cytometry analysis. The apoptosis rate doubled (213%) in miR-34a transfected cells in comparison to the mimic control (Figure 1c upper right). Finally, we measured the DNA synthesis using a BrdU incorporation assay (Figure 1c lower left) and demonstrated a modest reduction of BrdU incorporation in miR-34a transfected cells by 27% compared with the mimic control. A summary of the percentage change for cell death, apoptosis, and DNA synthesis of a representative experiment is shown in the lower right panel of Figure 1c. In addition, a cell cycle analysis on IMR32 cells transfected with miR-34a also demonstrated increased apoptosis and suppressed percentage of cells in the S and G2/M phases (Supplemental Figure 1). Therefore, exogenous miR-34a suppresses NB cell growth through both increasing apoptosis and decreasing DNA synthesis.
Figure 1.
miR-34a inhibits the growth of neuroblastoma cells through apoptosis and suppressed DNA synthesis. (a) microRNAs in the 1p36 region. Five microRNAs were mapped between 0-10Mb corresponding to 1p36.22-1pter, which is commonly deleted in NB tumors, using the Sanger miRNA Registry (http://microrna.sanger.ac.uk/sequences/index.shtml, Release 10.0). The relative positions of them are depicted on the black bar which represents 0-10Mb of chromosome 1. Among these 5 microRNAs, three (miR-200b, 429 and 34a; marked with arrowheads) are predicted to target MYCN genes by the miRBase Targets available on the same website. Start and End mark the genomic coordinates for the stem-loop pre-microRNA sequences of these microRNAs (Human Genome Build 36.2). (b) miR-34a inhibits the growth of neuroblastoma cells. _MYCN_-amplified NB cell lines, IMR32 and LA-N-5, were transfected with synthetic miR-34a, miR-200b, miR-429, and mimic control, the growth was monitored using WST-1 assays for 4 days. The optical densities (OD) at 450nm corrected by OD650nm were plotted over time after background subtraction of the readings from the media-only wells. miR-34a clearly suppressed the growth of both IMR32 and LA-N-5, but miR-200b and -429 had little or no effect. (c) A representative flow cytometry experiment shows miR-34a inducing apoptosis and suppression of DNA synthesis. Flow cytometry was used to study cell death (7-AAD staining), apoptosis (caspase 3 staining) and DNA synthesis (BrdU staining) in IMR32 cells at 48 hours after transfection. Blue: mimic control transfection; Red: miR-34a transfection. The bar graph summarizes the results of flow cytometry. Cell death and apoptosis were increased by 29% and 113% respectively, whereas DNA synthesis was suppressed by 27% in the miR-34a transfected cells.
Neuroblastoma tumors with 1p36 deletion express less miR-34a
In order to examine if 1p36 deletion results in any change in the expression of miR-34a in primary NB tumors, we performed Taqman® real-time RT-PCR on a set of stage 4 primary human NB tumors, of which 8 were _MYCN_-amplified with 1p36 LOH and 8 were not amplified and did not demonstrate LOH of this region. Consistent with a previous study (Welch et al., 2007), the level of miR-34a expression is lower in tumors with 1p36 deletion than in those with normal copy number (_p_=0.038) (Figure 2). Therefore, we demonstrated that genomic 1p36 deletion has an impact on the miR-34a expression in NB tumors, and the association of low miR-34a expression with this deletion implies a possible role of this microRNA in NB tumors with MYCN amplification.
Figure 2.

A box plot of miR-34a expression levels in 16 primary NB tumors of 1p36 normal (n=8) and 1p36 deletion (n=8). The expression levels of miR-34a was measured by real-time Taqman® RT-PCR assays, and represented as normalized log2 ratios between miR-34a and an internal control, RNU6B. The result shows that NB tumors with 1p36 deletion express less miR-34a than 1p36 normal tumors (p =0.038).
miR-34a directly targets the MYCN gene
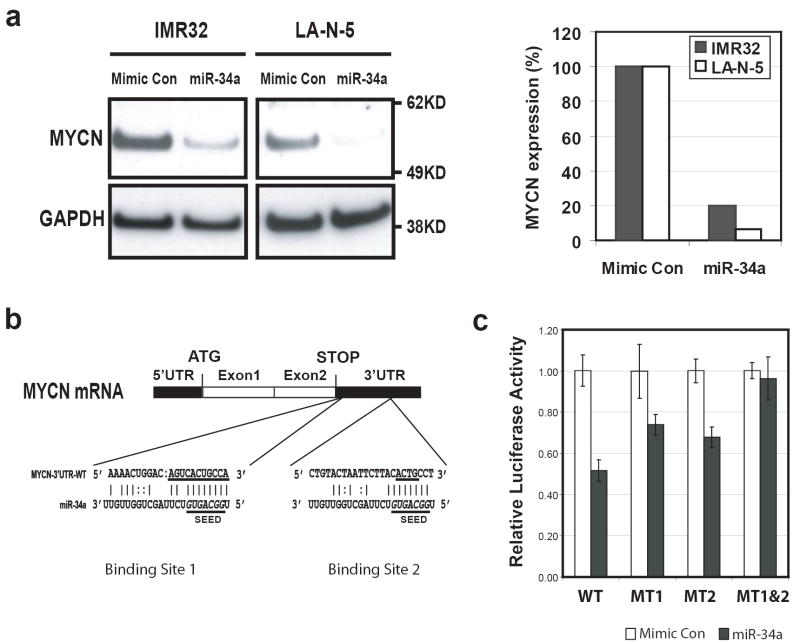
We next investigated if miR-34a directly targeted the MYCN gene. We first performed a Western analysis on the total protein extracts from IMR32 and LA-N-5 cells transfected with miR-34a 48 hours after transfection (Figure 3a, left panel). Quantification of the MYCN immuno-bands on the Western blot demonstrated that miR-34a caused an 80% or 95% reduction of MYCN protein in both IMR32 and LA-N-5 cells respectively after normalization by GAPDH (Figure 3a, right panel).
Figure 3.
MYCN is a direct target of miR-34a. (a) Left panel: a Western blot shows that miR-34a suppressed the expression of MYCN protein at 48 hours after transfection in both IMR32 and LA-N-5 cells. Right panel: quantification of MYCN protein showed a suppression of 80% and 95% by miR-34a in IMR32 and LA-N-5 cells respectively after normalization using the levels of GAPDH protein. (b) Sequence alignment of miR-34a with the two binding sites on the MYCN 3’-UTR. The underlined sequences on MYCN 3’-UTR were mutated to make luciferase reporter constructs containing a single miR-34a binding mutation (pMIR-MYCN-MT1 or MT2) or a double mutation (pMIR-MYCN-MT1&2). The underlined italicized letters (GGCAGUG) in miR-34a denote the seed sequence of miR-34a (position 2-8 at the 5’ end) (Lim et al., 2005). (c) Luciferase activity assays demonstrated that miR-34a directly suppresses MYCN by targeting the MYCN 3’-UTR. Luciferase reporter constructs containing full length wild-type (WT) and mutant (MT1, MT2, and MT1&2) 3’-UTR of MYCN were introduced into SK-N-AS cells with microRNAs, and luciferase activity was measured at 24 hours after transfection. Mutation of either miR-34a binding sites (MT1 or MT2) resulted in a reduction of suppression of luciferase activities, whereas double mutation totally abolished the suppression by miR-34a.
We found by bioinformatic analysis and search of the Sanger microRNA registry that the MYCN 3’-UTR contains two target sequences for the miR-34a at positions 10 and 567 (Figure 3b) (http://microrna.sanger.ac.uk/cgi-bin/targets/v5/detail_view.pl?transcript_id=ENST00000281043). In order to test if miR-34a directly targets MYCN gene, we cloned the entire wild type 3’-untranslated region (3’-UTR) of the MYCN gene into a luciferase reporter vector. Due to the endogenous expression of miR-34a in IMR32 and LA-N-5 cells, we transfected the resulting reporter construct (pMIR-MYCN-WT) into SK-N-AS cell, a neuroblastoma cell line that does not express miR-34a (data not shown), along with miR-34a or a mimic control microRNA. The luciferase activity assays at 24 hours post transfection demonstrated that miR-34a suppressed luciferase reporter activity by 50% (Figure 3c). To demonstrate the specificity of miR-34a directly targeting the MYCN gene, we generated mutation reporter constructs of each of the two predicted miR-34a binding sites on the MYCN 3’-UTR (pMIR-MYCN-MT1 and MT2) and a double mutation construct of both sites (pMIR-MYCN-MT1&2) (Figure 3b), and examined if these mutations would eliminate the suppression of the luciferase reporter activity. Figure 3c demonstrated that a mutation on either one of the predicted miR-34a binding sites attenuated the suppression of miR-34a on the luciferase activity, whereas mutations on both sites abolished the suppression of luciferase reporter by miR-34a. These experiments demonstrate that miR-34a directly targets the MYCN gene through its 3’-UTR, and both binding sites of miR-34a on MYCN 3’-UTR are required for the suppressive activity of miR-34a on MYCN.
Pathway analysis of mRNA global gene expression alterations in IMR32 cells transfected with miR-34a
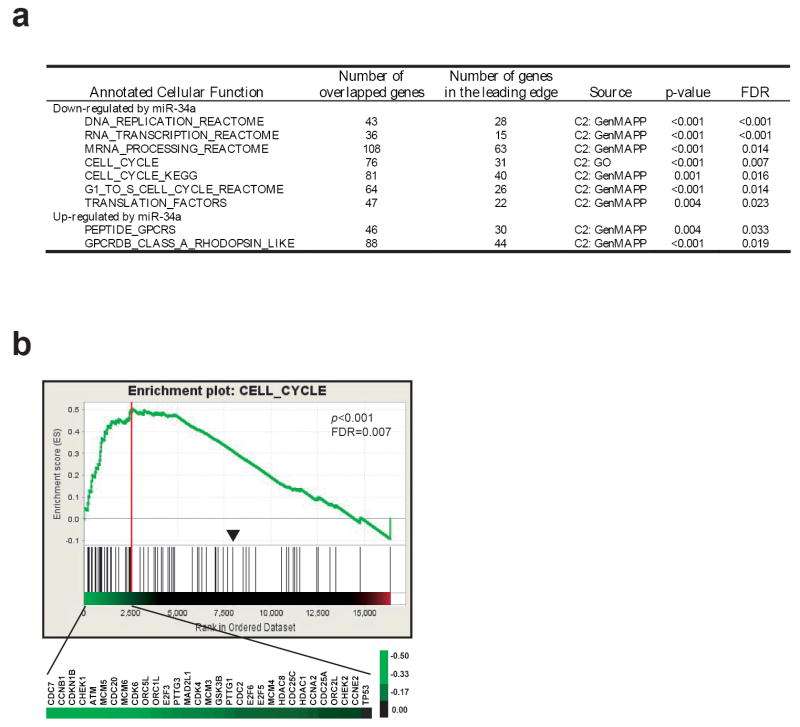
We next investigated the pathways regulated by miR-34a. Because microRNAs had been demonstrated to regulate messenger RNA levels (Ambros, 2004; Bartel, 2004; Lim et al., 2005), we performed global gene expression profiling on miR-34a transfected cells at 48 hours using Affymetrix U133 plus 2.0 chips. We used Gene Set Enrichment Analysis (GSEA), which is a computational method that determines whether a priorly defined set of genes or pathways shows statistically significant enrichment in the most altered genes (Subramanian et al., 2005). We found that miR-34a significantly altered 9 annotated cellular functions (p<0.01and FDR<0.05) including 7 associated with the down-regulated genes and 2 with the up-regulated genes by miR-34a (Figure 4a). Interestingly, the down-regulated genes have functions in cell proliferation (DNA replication, RNA transcription and mRNA processing, cell cycle, and translation), whereas the up-regulated genes belonged to G-protein coupled receptors which are involved in neuronal signaling (Figure 4a and Supplemental Table 1). Figure 4b shows an example of GSEA enrichment plot for one of the 9 function sets (CELL_CYCLE). Therefore, changes of global gene expression profile are consistent with what we observed at the cellular level, and the global gene expression profiling demonstrated that miR-34a inhibits growth and induces a neuronal phenotype in IMR32 cells.
Figure 4.
(a) Summary of GSEA analysis. GSEA analysis was performed on the ranked genes according to the ratios of transcripts from mimic control and miR-34a transfected IMR32 cells at 48 hours. Nine gene sets with a _p_-value of <0.01 and a false discovery rate (FDR) of < 0.05 were considered significant. Of the 9 gene sets, 7 associate with the down-regulated genes by miR-34a, and 2 set associate with up-regulated genes. (b) One of the GSEA plots indicates significant enrichment of cell cycle genes in the transcripts suppressed by miR-34a (p < 0.001 and FDR = 0.007). Affymetrix U133 Plus 2 data for the duplicated experiments of IMR32 cells transfected with miR-34a and mimic control at 48h transfection were combined, and multiple probe sets for the same genes were averaged. Genes were ordered in their ranked ratios, and GSEA were performed using the GSEA tool at http://www.broad.mit.edu/gsea. Plot (the green curve) shows the running sum of enrichment score (ES) for ranked genes comparing with the cell_cycle gene set. Red vertical line specifies the maximum ES score. Heat map shows the log2 ratio (miR-34a vs. control) of leading edge subset genes in their ranked order which contribute to the significance. A scale of the log2 ratios is shown on the right. Black lines indicate gene hits in the cell_cycle gene set; and the black arrowhead indicates where the log2 ratio=0.
Identification of other direct targets of miR-34a
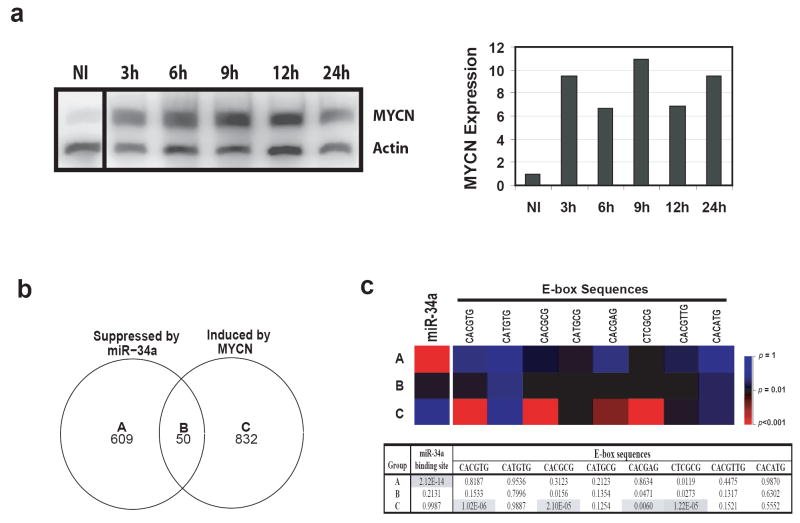
In order to delineate the direct targets of miR-34a from those genes controlled by MYCN, we induced MYCN expression in another NB cell line, MYCN-3, using a previously published inducible expression construct (Slack et al., 2005) and performed gene expression profiling at 12 hours after induction. Western analysis confirmed induction of MYCN protein expression (Figure 5a). The numbers of differentially expressed genes mediated by miR-34a or MYCN are shown in a Venn diagram (Figure 5b). Genes in group A (suppressed by miR-34a only) are the potential direct targets for miR-34a, since they are not up-regulated by MYCN induction, whereas genes in group B are potential indirect targets mediated by changes of the MYCN level (Figure 5b). In order to validate this approach, we examined if these is enrichment of miR-34a target sequences in the 3’-UTR of the genes in these 3 gene groups. A searched for sequence complementary to the miR-34a seed sequence (binding site) in 3’-UTRs in all groups showed that the group A genes are significantly enriched for the complimentary sequence of the miR-34a seed (_p_=2.12×10-14) (Figure 5c). On the contrary, all other groups did not show any significant enrichment (Figure 5c). Of the 609 genes in group A, 146 have at least one miR-34a binding site in their 3’-UTRs, and 140 of them are known genes (Supplemental Table 2). When we searched the functional annotation for these genes in a Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://www.david.niaid.nih.gov) (Dennis et al., 2003), and used a cutoff of p<0.01 for functional annotation enrichment, we found that they have diverse functions from primary metabolism to alternative splicing (Supplemental Table 3). Interestingly, one gene ontology term of phosphorylation consists of 23 genes including EPB41, CDK6, PDGFRA, PDGFRB, MAP2K1, KIT, etc., which are important molecules for cell signaling and proliferation. Down regulation of these genes by miR-34a is consistent with the observation of miR-34a induced growth inhibition in NB cells.
Figure 5.
Search for other direct targeted genes of miR-34a besides MYCN. (a) Identification of MYCN direct targets by induction of MYCN in NB cells. In order to delineate the genes directly targeted by miR-34a, but not through MYCN, we used a MYCN induction system to identify MYCN direct targets. MYCN-3 cells containing a tetracycline controlled MYCN expression construct were induced by tetracycline, and total protein was collected at 3, 6, 9, 12, and 24 hours. Left panel shows a Western blot for MYCN induction. Right panel: quantification of the Western blot demonstrates that MYCN protein is induced at least 6 fold after induction. MYCN expression is normalized by the non-induced (NI). (b) Venn diagram of the altered genes by miR-34a or MYCN induction. In order to identify the genes directly regulated by miR-34a but not through MYCN, gene expression profiling was performed on IMR32 cells transfected with miR-34a and its mimic control at 48 hours; or on MYCN-3 cells with MYCN induction at 12 hours compared to the non-induced (NI). Genes with a ratio of 33% difference was considered as changed. The numbers indicate the gene counts in the corresponding categories. (c) Enrichment analysis of miR-34a binding and E-box sequences shows the validity of our approach to identify the direct targets of miR-34a. Upper panel: a heat map of _p_-values for the enrichment of miR-34a binding sequence or E-box sequences in different categories. _P_-values were calculated for the enrichment of the complimentary sequences of miR-34a seed in 3’-UTRs, or the E-boxes (MYC binding motifs) in the 2.5 Kb promoter regions for gene groups A-C. A color scale of the _p_-values is shown on its right. Lower panel: a table of _p_-values of the enrichment analysis. Shaded cells indicate p<0.01.
Using a similar strategy, we examined if there is overrepresentation of E-box sequences, MYCN binding sites, in a 2.5 kb promoter region of all gene groups. We found there is a significant enrichment for E-box sequences in group C, but not in A or B (Figure 5c) (p<0.01). A list of genes containing E-box sequences is provided in the Supplemental Table 4. Therefore, these sequence enrichment analyses validate our approach to identify the direct targets of miR-34a, and the genes in group A represent potential direct targets of miR-34a which requires further confirmatory studies.
Discussion
MicroRNAs have been demonstrated to play important roles in development and maintenance of normal cellular function, and alteration in expression of microRNAs can result in human cancers (Calin et al., 2002; Costinean et al., 2006; He et al., 2005; Iorio et al., 2005). Due to the frequent association of 1p36 loss in _MYCN_-amplified NB tumors, we examined the functional role of microRNAs mapping to the commonly deleted region within the first 10 Mb of chromosome 1 (1p36.22-1pter). Five known microRNAs were mapped to this region, and 3 of them (miR-200b, 429, and 34a) are predicted to target the MYCN gene. Introduction of these 3 microRNAs into NB cells with MYCN amplification showed that miR-34a, but not miR-200b or 429, can significantly suppress cell growth. We demonstrated that miR-34a directly inhibits MYCN protein by targeting the 3’-UTR of the MYCN gene. Gene expression profiling confirmed that miR-34a can suppress cell cycle genes and induces a neural phenotype. These results resemble the effect of transferring 1p DNA material into NB cell lines, which also resulted in neuronal differentiation and apoptosis (Bader et al., 1991).
MYCN amplification is the only molecular marker currently used in clinic to predict poor outcome for NB patients. The oncogenic function of MYCN and its homolog MYC has been recognized in several human malignancies including NB, Burkitts lymphoma, colon cancer, breast cancer, lung cancer, etc. (Nesbit et al., 1999). The MYC gene has been demonstrated to play important roles in both cell proliferation and apoptosis (Evan et al., 1992; Schwab and Bishop, 1988). Here, we showed that miR-34a-mediated inhibition of NB growth as a result of induction of apoptosis and modest suppression of DNA synthesis. Similar findings have been reported when MYCN is suppressed in _MYCN_-amplified cell lines (Kang et al., 2006; Nara et al., 2007; Woo et al., 2007). Therefore, some of the inhibitory effect miR-34a on cell growth shown in this study is likely mediated by the suppression of MYCN protein. In addition, E2F3 has been shown to be another direct target of miR-34a (Welch et al., 2007), and E2F family members can directly regulate MYCN transcriptionally (Strieder and Lutz, 2003). Thus, miR-34a can modulate MYCN expression at multiple levels. Although this study clearly demonstrated that overexpression of miR-34a can efficiently reduce the MYCN protein level, however, to what extent that MYCN protein level is regulated by the endogenous miR-34a is unclear at this point. Data from this study, as well as others showed that miR-34a is expressed in _MYCN_-amplified tumor but at a lower level (Welch et al., 2007). These results, together with the observation that MYCN amplification is almost always associated with the 1p36 loss (Attiyeh et al., 2005; Maris et al., 2000), raise the possibility that haploinsufficiency of miR-34a may contribute to the development of this type of neuroblastoma, similar to CHD5 and CDC42 in the literature (Bagchi et al., 2007; Valentijn et al., 2005). Recently it has been discovered that p53 directly controls the expression level of members of miR-34 family, and miR-34 family members can mediate some of the function of p53 (Chang et al., 2007; He et al., 2007; Raver-Shapira et al., 2007). Intriguingly, despite of 50% of all human cancers harboring mutations inactivating p53, TP53 mutation is rare in neuroblastoma (Hosoi et al., 1994; Vogan et al., 1993). Therefore we speculate that reduction of miR-34a expression from loss of 1p36 has a similar consequence to p53 deficiency, which results in increased cell growth and suppression of apoptosis as seen in _MYCN_-amplified NBs.
Our study has provided evidence that MYCN is a direct target of miR-34a which is contrary to a previous study by Welch et al. (Welch et al., 2007). These discrepant results may be due to the differences in the 3’-UTRs of the MYCN gene used in the two studies. We used a native full-length MYCN 3’-UTR (~1.3kb) containing 2 miR-34a binding sequences, whereas an oligonucleotide or a shorter version of MYCN 3’-UTR (625bp) containing only one of the miR-34a binding sites (binding site 1 in Figure 3b) were used in the other study. We further demonstrated that a double mutation of the miR-34a binding sites in the MYCN 3’-UTR completely abolished the suppression of luciferase activity by miR-34a. Therefore, both miR-34a binding sites are required for an efficient suppression of MYCN by miR-34a. Additionally, we have shown that miR-34a inhibited MYCN protein expression by >80% using Western blotting in two _MYCN_-amplified cell lines.
In conclusion, we have demonstrated an important role of miR-34a; a small non-coding microRNA in 1p36 region exerting potential tumor suppressor effects through direct modulation of MYCN protein levels as well as through the suppression of multiple other genes and pathways not directly related to MYCN.
Materials and Methods
Cell Culture, cell growth, cell viability, apoptosis, BrdU incorporation assay
IMR32 and LA-N-5, both with _MYCN_-amplification, were cultured in EMEM media (Quality Biological, Gaithersburg, MD, USA). SK-N-AS and MYCN-3, a NB cell line with an inducible MYCN gene construct (Slack et al., 2005) were maintained in RPMI-1640 media. All media were supplemented with 10% FBS (Hyclone, Logan, UT, USA), 1% Glutamine and 1% P/S (Quality Biological, Gaithersburg, MD, USA), and cells were cultured at 37°C.
Synthetic microRNAs (160 fmoles) were transfected into 1×106 IMR32 or LA-N-5 cells using an Amaxa Nucleofector kit according to the manufacture’s instruction (Amaxa Biosystems, Cologne, Germany). All synthetic miRIDIAN™ microRNAs were purchased from Dharmacon Technologies (Lafayette, CO, USA). Cell growth was monitored using the WST-1 assays (Roche Diagnostic GMbH, Mannheim, Germany) for 96 hours post transfection. Cell viability, apoptosis and BrdU incorporation were measured 48 hours after transfection using flow cytometry. Cell viability was monitored by staining cells with 7-AAD (BD Biosciences, San Jose, CA, USA), as described elsewhere (Wei et al., 2005). Apoptosis was measured by immunostaining cells using an antibody specific to the active form of caspase 3 (BD Biosciences, San Jose, CA, USA). DNA synthesis was measured by BrdU incorporation using a commercially available BrdU immunostaining kit (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s instructions.
Luciferase reporter constructs, site-directed mutagenesis, and luciferase reporter assay
We cloned the full-length 3’-UTR of the MYCN gene into the pMIR-REPORT™ miRNA expression vector (Ambion, Austin, TX, USA) between SacI and Mlu1 restriction sites using a directional RT-PCR cloning strategy (Forward primer: GCCGAGCTCCCACCAGCAGCACAACTATG; reverse primer: GCGACGCGTTAATACGACTCACTATAGGGAGGCGG). The resulting plasmid, pMIR-MYCN-WT, containing the wild-type 3’-UTR of the MYCN gene was sequenced to ensure accuracy.
To ablate the two miR-34a binding sites in the pMIR-MYCN-WT construct (Figure 3c), we used a QuikChange® XL site-direct mutagenesis kit (Stratagene, La Jolla, CA, USA) to generate a mutation on one of the two predicted miR-34a binding sites (pMIR-MYCN-MT1 and MT2) or on both sites (pMIR-MYCN-MT1&2) of the MYCN 3’-UTR according to the manufacturer’s instruction. All 3 mutation constructs were sequenced to ensure sequence accuracy.
For luciferase assays, we used Dharmafect 1 (Dharmacon Technologies, Lafayette, CO, USA) to co-transfect SK-N-AS cells with luciferase reporter construct plasmids, a β-galactosidase control plasmid and microRNAs (10nM) per well in 48-well cell culture plates according to the manufacture’s instruction. Luciferase activity was measured using a Dual light® Luciferase and β-Galactosidase Reporter Gene Assay System (Applied Biosystems, Foster City, CA, USA) at 24 hours after transfection. Luciferase activity was then normalized by the β-galactosidase activity for transfection in each well.
RNA extraction and TaqMan® real-time PCR
RNA extraction was described previously (Wei and Khan, 2002) with modification. In brief, cells or tissues were homogenized in TRIzol® solution and larger RNA (>200nt) was purified by RNeasy columns (Qiagen, Valencia, CA, USA). Ethanol (1.4 volume of the flow through) was added to the flow-through of the RNeasy columns to achieve a final concentration of 70%. Then, small size RNA (<200nt) containing micro RNAs (enriched small RNA) was recovered and purified using an Invitrogen PureLink miRNA isolation kit according to the manufacture’s instruction (Invitrogen, Carlsbad, CA, USA).
MicroRNA expression was measured using a Taqman® MicroRNA RT-PCR assay (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol, and microRNA expression levels were normalized against RNU6B expression. We used the Wilcoxon rank test to calculate the _p_-value.
Western blotting
Total proteins were extracted from the cells using RIPA buffer with 3% proteinase inhibitor cocktail (Sigma, St. Louis, MO, USA). Twenty micrograms of protein (5 μg for the MYCN induction experiment) was separated on a polyacrylamide gel under denaturing conditions and transferred to a nitrocellulose membrane (Invitrogen, Carlsbad, CA, USA). The membranes were blocked for 1 hour at room temperature, and then incubated overnight at 4°C in TBST containing 5 % BSA and following antibodies: MYCN (OP13, Calbiochem, San Diego, CA); GAPDH (MAB374 , Chemicon, Temecula, CA); actin (Sigma, St. Louis, MO, USA ). Membranes were washed 3 times in TBST, and incubated with a secondary antibody conjugated with horseradish peroxidase (Rockland Immunochemicals, Gilbertsville, PA, USA) in TBST and 0.5% BSA for 1 hour at room temperature. After two washes with TBS, bands were detected by chemiluminescence using a SuperSignal Chemiluminescence kit (Pierce, Rockford, IL, USA) on Biomax MR X-ray film (Kodak, Rochester, NY, USA). Intensity of the bands was determined using ImageQuant software (GE Healthcare, Piscataway, NJ, USA) in the volume mode.
Gene expression analysis
Gene expression profiling was performed using Affymetrix U133 Plus 2.0 arrays for miR-34a transfection experiments; or on Affymetrix U133 AB arrays for MYCN induction experiment, according to the manufacturer’s instructions (Affymetrix, Santa Clara, CA, USA). Gene expression data were normalized using DNA Chip Analyzer (dChip) in the PM-only model (Li and Wong, 2001). We used mean intensity ≥200 to remove probe sets of low quality. Genes suppressed by miR-34a were identified by comparing the miR-34a transfected IMR32 cells to the mimic control transfected cells at 48 hours post transfection. We used the average of two replicate experiments at this time point. Genes induced by MYCN were identified by comparing expression profiles of MYCN-3 cells at 12 hours after MYCN induction with the non-induced control MYCN-3 cells. A threshold of 33% deviation from a ratio =1 compared to each control was used to select for differentially expressed genes. Affymetrix probes were mapped to Ensembl gene identifiers using the Bioconductor biomaRt package (Durinck et al., 2005). Probes that mapped to none or more than one Ensembl gene identifier by Ensembl were removed from the analysis. Data from different probes that are mapped to the same Ensembl gene identifier were averaged.
Pathway analysis
In order to investigate the global effect of miR-34a on NB cells, we used Gene Set Enrichment Analysis (GSEA) at http://www.broad.mit.edu/gsea for the pathway analysis (Subramanian et al., 2005) on the gene expression profiling data of IMR32 cells transfected with miR-34a. Intensities of signals for every probe set from two duplicated experiments were averaged. A total of 35830 probe sets passed the quality filtering, and they represent 16782 known genes by the symbol identifier in the GSEA. The ratios of mimic control vs. miR-34a were calculated for every probe set, and the mean of ratios from all probe sets was used for genes with multiple probe sets. Genes were ranked using the log2 ratio, and a total of 404 gene sets from a collection of curated biological function databases (http://www.broad.mit.edu/gsea/msigdb/msigdb_index.html) including BioCarta (http://www.biocarta.com/), GenMAPP (http://www.genmapp.org/), and GO (http://www.geneontology.org/), were used in this analysis. After applied a minimal size of 25 overlapping genes per pathway, 80 gene sets remained. Because of the limited number of samples, we performed a gene set type permutation test. Gene sets with a _p_-value of <0.01 and a false discovery rate (FDR) of < 0.05 were considered significant.
Analysis of enrichment of miR-34a binding and E-box sequences
We first retrieved the 3’-UTR sequences of all genes represented on Affymetrix U133 chips as annotated by Ensembl using the biomaRt package (Durinck et al., 2005). When multiple 3’-UTRs were annotated to the same gene, the longest 3’-UTR was used. Next, the number of genes that have at least one perfect complementary match to the miR-34a microRNA seed sequence (GGCAGUG) and the number of genes without any complementary seed match were counted. We defined the seed sequence as 2-8 nucleotides from 5’-end of miR-34a (Lim et al., 2005). We repeated this process for the different gene groups displayed in the Venn diagram (Figure 5b). Finally, we computed _p_-values for overrepresentation of sequence complementary to miR-34a seed sequence using the counts in a hypergeometric test. To calculate the enrichment of the E-box motifs in the promoter of genes, we retrieved 2.5 kb sequences upstream of the annotated start site of the coding region for each gene, and used the same method to calculate the _p_-values. Sequences of the E-box motif were published in a previous study (Blackwell et al., 1993).
Supplementary Material
3
Acknowledgments
We would like to thank Dr. Joon-Yong Chung and Xinyu Wen, MS for their excellent technical support. We also thank Drs. John Maris, Wendy London (Children’s Oncology Group, Philadelphia, PA), and Steven Qualman (Cooperative Human Tissue Network, Columbus, OH) for the NB tumor samples, and for invaluable discussion of this study with Dr. John Maris and his group.
This study is supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
References
- Abel F, Sjoberg RM, Krona C, Nilsson S, Martinsson T. Mutations in the N-terminal domain of DFF45 in a primary germ cell tumor and in neuroblastoma tumors. Int J Oncol. 2004;25:1297–302. [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Attiyeh EF, London WB, Mosse YP, Wang Q, Winter C, Khazi D, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–53. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- Bader SA, Fasching C, Brodeur GM, Stanbridge EJ. Dissociation of suppression of tumorigenicity and differentiation in vitro effected by transfer of single human chromosomes into human neuroblastoma cells. Cell Growth Differ. 1991;2:245–55. [PubMed] [Google Scholar]
- Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–75. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- Barbashina V, Salazar P, Holland EC, Rosenblum MK, Ladanyi M. Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene. Clin Cancer Res. 2005;11:1119–28. [PubMed] [Google Scholar]
- Bartel B, Bartel DP. MicroRNAs: at the root of plant development? Plant Physiol. 2003;132:709–17. doi: 10.1104/pp.103.023630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bieche I, Khodja A, Lidereau R. Deletion mapping in breast tumor cell lines points to two distinct tumor-suppressor genes in the 1p32-pter region, one of deleted regions (1p36.2) being located within the consensus region of LOH in neuroblastoma. Oncol Rep. 1998;5:267–72. doi: 10.3892/or.5.1.267. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Huang J, Ma A, Kretzner L, Alt FW, Eisenman RN, et al. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993;13:5216–24. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–16. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- Brodeur GM, Sekhon G, Goldstein MN. Chromosomal aberrations in human neuroblastomas. Cancer. 1977;40:2256–63. doi: 10.1002/1097-0142(197711)40:5<2256::aid-cncr2820400536>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 Broadly Influences Gene Expression and Promotes Apoptosis. Mol Cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QR, Bilke S, Wei JS, Whiteford CC, Cenacchi N, Krasnoselsky AL, et al. cDNA array-CGH profiling identifies genomic alterations specific to stage and MYCN-amplification in neuroblastoma. BMC Genomics. 2004;5:70. doi: 10.1186/1471-2164-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TH, Lo KW, Yim SF, Poon CS, Cheung AY, Chung TK, et al. Clinicopathologic significance of loss of heterozygosity on chromosome 1 in cervical cancer. Gynecol Oncol. 2005;96:510–5. doi: 10.1016/j.ygyno.2004.10.035. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–9. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–40. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- Eggert A, Grotzer MA, Zuzak TJ, Ikegaki N, Zhao H, Brodeur GM. Expression of Apo-3 and Apo-3L in primitive neuroectodermal tumours of the central and peripheral nervous system. Eur J Cancer. 2002;38:92–8. doi: 10.1016/s0959-8049(01)00347-1. [DOI] [PubMed] [Google Scholar]
- Ejeskar K, Krona C, Caren H, Zaibak F, Li L, Martinsson T, et al. Introduction of in vitro transcribed ENO1 mRNA into neuroblastoma cells induces cell death. BMC Cancer. 2005;5:161. doi: 10.1186/1471-2407-5-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–28. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Fong CT, Dracopoli NC, White PS, Merrill PT, Griffith RC, Housman DE, et al. Loss of heterozygosity for the short arm of chromosome 1 in human neuroblastomas: correlation with N-myc amplification. Proc Natl Acad Sci U S A. 1989;86:3753–7. doi: 10.1073/pnas.86.10.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, White PS, Weiss MJ, Hogarty MD, Thompson PM, Stram DO, et al. Allelic deletion at 11q23 is common in MYCN single copy neuroblastomas. Oncogene. 1999;18:4948–57. doi: 10.1038/sj.onc.1202887. [DOI] [PubMed] [Google Scholar]
- Hashimoto N, Ichikawa D, Arakawa Y, Date K, Ueda S, Nakagawa Y, et al. Frequent deletions of material from chromosome arm 1p in oligodendroglial tumors revealed by double-target fluorescence in situ hybridization and microsatellite analysis. Genes Chromosomes Cancer. 1995;14:295–300. doi: 10.1002/gcc.2870140408. [DOI] [PubMed] [Google Scholar]
- He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–80. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich KO, Fischer M, Mertens D, Benner A, Wiedemeyer R, Brors B, et al. Reduced expression of CAMTA1 correlates with adverse outcome in neuroblastoma patients. Clin Cancer Res. 2006;12:131–8. doi: 10.1158/1078-0432.CCR-05-1431. [DOI] [PubMed] [Google Scholar]
- Hogarty MD, Liu X, Guo C, Thompson PM, Weiss MJ, White PS, et al. Identification of a 1-megabase consensus region of deletion at 1p36.3 in primary neuroblastomas. Med Pediatr Oncol. 2000;35:512–5. doi: 10.1002/1096-911x(20001201)35:6<512::aid-mpo2>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Hosoi G, Hara J, Okamura T, Osugi Y, Ishihara S, Fukuzawa M, et al. Low frequency of the p53 gene mutations in neuroblastoma. Cancer. 1994;73:3087–93. doi: 10.1002/1097-0142(19940615)73:12<3087::aid-cncr2820731230>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Kang JH, Rychahou PG, Ishola TA, Qiao J, Evers BM, Chung DH. MYCN silencing induces differentiation and apoptosis in human neuroblastoma cells. Biochem Biophys Res Commun. 2006;351:192–7. doi: 10.1016/j.bbrc.2006.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl NE, Gee CE, Alt FW. Activated expression of the N-myc gene in human neuroblastomas and related tumors. Science. 1984;226:1335–7. doi: 10.1126/science.6505694. [DOI] [PubMed] [Google Scholar]
- Kong XT, Valentine VA, Rowe ST, Valentine MB, Ragsdale ST, Jones BG, et al. Lack of homozygously inactivated p73 in single-copy MYCN primary neuroblastomas and neuroblastoma cell lines. Neoplasia. 1999;1:80–9. doi: 10.1038/sj.neo.7900010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti JM, Valentine M, Xiang J, Jones B, Amann J, Grenet J, et al. Alterations in the PITSLRE protein kinase gene complex on chromosome 1p36 in childhood neuroblastoma. Nat Genet. 1994;7:370–5. doi: 10.1038/ng0794-370. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Maris JM, Guo C, Blake D, White PS, Hogarty MD, Thompson PM, et al. Comprehensive analysis of chromosome 1p deletions in neuroblastoma. Med Pediatr Oncol. 2001;36:32–6. doi: 10.1002/1096-911X(20010101)36:1<32::AID-MPO1009>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Maris JM, Jensen J, Sulman EP, Beltinger CP, Allen C, Biegel JA, et al. Human Kruppel-related 3 (HKR3): a candidate for the 1p36 neuroblastoma tumour suppressor gene? Eur J Cancer. 1997;33:1991–6. doi: 10.1016/s0959-8049(97)00279-7. [DOI] [PubMed] [Google Scholar]
- Maris JM, Weiss MJ, Guo C, Gerbing RB, Stram DO, White PS, et al. Loss of heterozygosity at 1p36 independently predicts for disease progression but not decreased overall survival probability in neuroblastoma patients: a Children’s Cancer Group study. J Clin Oncol. 2000;18:1888–99. doi: 10.1200/JCO.2000.18.9.1888. [DOI] [PubMed] [Google Scholar]
- Mathysen D, Van Roy N, Van Hul W, Laureys G, Ambros P, Speleman F, et al. Molecular analysis of the putative tumour-suppressor gene EXTL1 in neuroblastoma patients and cell lines. Eur J Cancer. 2004;40:1255–61. doi: 10.1016/j.ejca.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Mori N, Morosetti R, Spira S, Lee S, Ben-Yehuda D, Schiller G, et al. Chromosome band 1p36 contains a putative tumor suppressor gene important in the evolution of chronic myelocytic leukemia. Blood. 1998;92:3405–9. [PubMed] [Google Scholar]
- Mosse YP, Greshock J, Margolin A, Naylor T, Cole K, Khazi D, et al. High-resolution detection and mapping of genomic DNA alterations in neuroblastoma. Genes Chromosomes Cancer. 2005;43:390–403. doi: 10.1002/gcc.20198. [DOI] [PubMed] [Google Scholar]
- Nara K, Kusafuka T, Yoneda A, Oue T, Sangkhathat S, Fukuzawa M. Silencing of MYCN by RNA interference induces growth inhibition, apoptotic activity and cell differentiation in a neuroblastoma cell line with MYCN amplification. Int J Oncol. 2007;30:1189–96. [PubMed] [Google Scholar]
- Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–16. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- Ozaki T, Hosoda M, Miyazaki K, Hayashi S, Watanabe K, Nakagawa T, et al. Functional implication of p73 protein stability in neuronal cell survival and death. Cancer Lett. 2005;228:29–35. doi: 10.1016/j.canlet.2004.12.050. [DOI] [PubMed] [Google Scholar]
- Poetsch M, Woenckhaus C, Dittberner T, Pambor M, Lorenz G, Herrmann FH. An increased frequency of numerical chromosomal abnormalities and 1p36 deletions in isolated cells from paraffin sections of malignant melanomas by means of interphase cytogenetics. Cancer Genet Cytogenet. 1998;104:146–52. doi: 10.1016/s0165-4608(97)00471-8. [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, et al. Transcriptional Activation of miR-34a Contributes to p53-Mediated Apoptosis. Mol Cell. 2007;26:731–43. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Schwab M, Bishop JM. Sustained expression of the human protooncogene MYCN rescues rat embryo cells from senescence. Proc Natl Acad Sci U S A. 1988;85:9585–9. doi: 10.1073/pnas.85.24.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack A, Chen Z, Tonelli R, Pule M, Hunt L, Pession A, et al. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci U S A. 2005;102:731–6. doi: 10.1073/pnas.0405495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strieder V, Lutz W. E2F proteins regulate MYCN expression in neuroblastomas. J Biol Chem. 2003;278:2983–9. doi: 10.1074/jbc.M207596200. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Gotoh T, Kok M, White PS, Brodeur GM. CHD5, a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene. 2003;22:1002–11. doi: 10.1038/sj.onc.1206211. [DOI] [PubMed] [Google Scholar]
- Valentijn LJ, Koppen A, van Asperen R, Root HA, Haneveld F, Versteeg R. Inhibition of a new differentiation pathway in neuroblastoma by copy number defects of N-myc, Cdc42, and nm23 genes. Cancer Res. 2005;65:3136–45. doi: 10.1158/0008-5472.CAN-04-2469. [DOI] [PubMed] [Google Scholar]
- Vogan K, Bernstein M, Leclerc JM, Brisson L, Brossard J, Brodeur GM, et al. Absence of p53 gene mutations in primary neuroblastomas. Cancer Res. 1993;53:5269–73. [PubMed] [Google Scholar]
- Wei JS, Khan J. Purification of Total RNA from Mammalian Cells and Tissues. In: Bowtell D, Sambrook J, editors. DNA Microarrays: A Molecular Cloning Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2002. pp. 110–119. [DOI] [PubMed] [Google Scholar]
- Wei JS, Whiteford CC, Cenacchi N, Son CG, Khan J. BBC3 mediates fenretinide-induced cell death in neuroblastoma. Oncogene. 2005;24:7976–83. doi: 10.1038/sj.onc.1208947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. Embo J. 1997;16:2985–95. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007 doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- White PS, Maris JM, Beltinger C, Sulman E, Marshall HN, Fujimori M, et al. A region of consistent deletion in neuroblastoma maps within human chromosome 1p36.2-36.3. Proc Natl Acad Sci U S A. 1995;92:5520–4. doi: 10.1073/pnas.92.12.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PS, Thompson PM, Gotoh T, Okawa ER, Igarashi J, Kok M, et al. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–94. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- Woo CW, Tan F, Cassano H, Lee J, Lee KC, Thiele CJ. Use of RNA interference to elucidate the effect of MYCN on cell cycle in neuroblastoma. Pediatr Blood Cancer. 2007 doi: 10.1002/pbc.21195. [DOI] [PubMed] [Google Scholar]
- Zimmerman KA, Yancopoulos GD, Collum RG, Smith RK, Kohl NE, Denis KA, et al. Differential expression of myc family genes during murine development. Nature. 1986;319:780–3. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- Zindy F, Knoepfler PS, Xie S, Sherr CJ, Eisenman RN, Roussel MF. N-Myc and the cyclin-dependent kinase inhibitors p18Ink4c and p27Kip1 coordinately regulate cerebellar development. Proc Natl Acad Sci U S A. 2006;103:11579–83. doi: 10.1073/pnas.0604727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3