E2A proteins promote development of lymphoid-primed multipotent progenitors (original) (raw)
. Author manuscript; available in PMC: 2009 Aug 1.
SUMMARY
The first lymphoid restricted progeny of HSCs are lymphoid-primed multipotent progenitors (LMPPs), which have little erythro-myeloid potential but retain lymphoid and granulocyte/macrophage differentiation capacity. Despite recent advances in the identification of LMPPs the transcription factors essential for their generation remain to be identified. Here we demonstrate that the E2A transcription factors are required for proper development of LMPPs. Within HSCs and LMPPs, E2A proteins prime expression of a subset of lymphoid-associated genes and prevent expression of genes that are not normally prevalent in these cells, including HSC-associated and non-lymphoid genes. E2A proteins also restrict proliferation of HSCs, MPPs, and LMPPs and antagonize differentiation of LMPPs toward the myeloid fate. Our results reveal that E2A proteins play a critical role in supporting lymphoid specification from HSCs and that the reduced generation of LMPPs underlies the severe lymphocyte deficiencies observed in E2A-deficient mice.
INTRODUCTION
Lymphocyte development from hematopoietic stem cells (HSCs) is accompanied by a loss of self-renewal capacity and progressive restriction of developmental potential. The pathway to the lymphocyte fate involves the generation of multipotent progenitors (MPPs) and lymphoid primed-multipotent progenitors (LMPPs) that have lymphoid and granulocyte/macrophage progenitor (GMP) but little or no megakaryocyte/erythrocyte progenitor (MEP) potential, respectively (Adolfsson et al., 2005; Lai et al., 2005). However, the precise stage where MEP potential is lost remains controversial (Forsberg et al., 2006). LMPPs are precursors to common lymphoid progenitors (CLPs), which have lost GMP potential but retain multi-lineage lymphoid differentiation capacity when placed in an appropriate microenvironment (Kondo et al., 1997). LMPPs also differentiate into early T-lymphocyte progenitors (ETPs), which undergo T-lymphocyte lineage specification upon activation of Notch receptors by Notch ligands in the thymus (Allman et al., 2003; Sambandam et al., 2005). While the molecular mechanisms for generation of committed B- or T-lymphocytes are beginning to be revealed, a major unanswered question is how the multipotent progeny of HSCs initiate differentiation toward lymphoid fate.
HSCs, MPPs and LMPPs are Lineage−Sca-1+c-_kit_high (LSK) cells that can be distinguished by increasing expression of the transmembrane receptor _fms_-like tyrosine kinase 3 (Flt3) (Adolfsson et al., 2001; Adolfsson et al., 2005). Up-regulation of Flt3 is associated with decreased megakaryocyte (Mk)/erythrocyte (E)-associated gene expression. Lymphoid-associated gene expression is initiated in LMPPs indicating priming of the lymphoid gene program (Igarashi et al., 2002; Mansson et al., 2007). A small number of transcriptional regulators have been implicated in the development or function of LMPPs. The zinc-finger transcription factors encoded by the Ikzf1 (Ikaros) gene are dispensable for the generation of LMPPs but required for high expression of Flt3 and subsequent lymphocyte differentiation (Yoshida et al., 2006). The Ets-related transcription factor PU.1, encoded by Sfpi1, is increased in a subset of LMPPs and Flt3high LSKs are absent in the liver of _Sfpi1_−/− embryos and bone marrow (BM) of adult mice with a conditional deletion of _Sfpi_1 (Arinobu et al., 2007; Dakic et al., 2005; Iwasaki et al., 2005). However, it remains to be determined whether PU.1 is required for development of functional LMPPs or, like Ikaros, is required specifically for Flt3 expression (Dakic et al., 2005; Iwasaki et al., 2005). Aside from these two factors, little is known about the transcriptional regulators that specify differentiation toward LMPPs in multipotent progenitors (Nutt and Kee, 2007).
Lymphoid priming involves the induction of lymphoid genes including Rag1, Dntt and Igh-6, which are known targets of the basic helix-loop-helix (bHLH) transcription factors E12 and E47, encoded by Tcfe2a(E2A) (Kee and Murre, 1998, 2001; Mansson et al., 2007). The E-box binding transcription factors (E-proteins) function in cell fate specification and differentiation in diverse cell types in both invertebrates and vertebrates (Massari and Murre, 2000). The E2A proteins have been characterized extensively as regulators of B- and T-lymphocyte development and are thought to function in specification of these lineages from lymphoid-restricted multipotent progenitors (Nutt and Kee, 2007; Welner et al., 2008). E2A proteins specify the B-lymphocyte fate by regulating the essential transcription factor encoded by the early B-cell factor 1 (Ebf1) gene, in collaboration with PU.1 and interleukin (IL)-7 receptor activated Stat5 (Dias et al., 2005; Kee and Murre, 1998; Kikuchi et al., 2005; Medina et al., 2004; Roessler et al., 2007; Seet et al., 2004). While no B-lymphocytes are generated in _E2A_−/− mice, T-lymphocytes develop, albeit in reduced numbers (Bain et al., 1997a; Bain et al., 1994; Zhuang et al., 1994). Recent in vitro studies suggest that E2A proteins regulate expression of the essential T-lymphocyte specification factor Notch1 and collaborate with Notch1 to activate target genes such as Hes1 (Ikawa et al., 2006). However, the requirements for E2A in vivo and in vitro differ since E2A is indispensable for T-lymphopoiesis in vitro on OP9 stromal cells expressing the Notch ligand Delta-like 1 but T cells develop in vivo with a partial arrest at the DN1 to DN2 transition (i.e. after the initial requirement for Notch1) (Bain et al., 1997a; Ikawa et al., 2006; Kee et al., 2002). Therefore, the role of E2A proteins in early T-lymphocyte development in vivo remains to be fully understood.
Multipotent hematopoietic progenitor cell lines can be generated from _E2A_−/− BM suggesting a role for E2A proteins in preventing expansion of multipotent cells and promoting differentiation (Ikawa et al., 2004). However, aside from their known roles in the regulation of B- and T-lymphocyte genes, a function for E2A proteins in earlier stages of hematopoiesis has not been reported. We show here that E2A proteins promote development of LMPPs and are required to establish an appropriate LMPP transcriptome. Importantly, E2A appears dispensible for extinction of the MEP fate en route to LMPPs but restricts proliferation and further myeloid differentiation in LMPPs. Our results reveal a dose-dependent requirement for E2A proteins at the earliest stages of lymphoid specification and indicate that reduced thymopoiesis in _E2A_−/− mice initiates from a failure to produce sufficient numbers of functional BM-derived LMPPs.
RESULTS
A dose dependent requirement for E2A in development of CLPs and ETPs
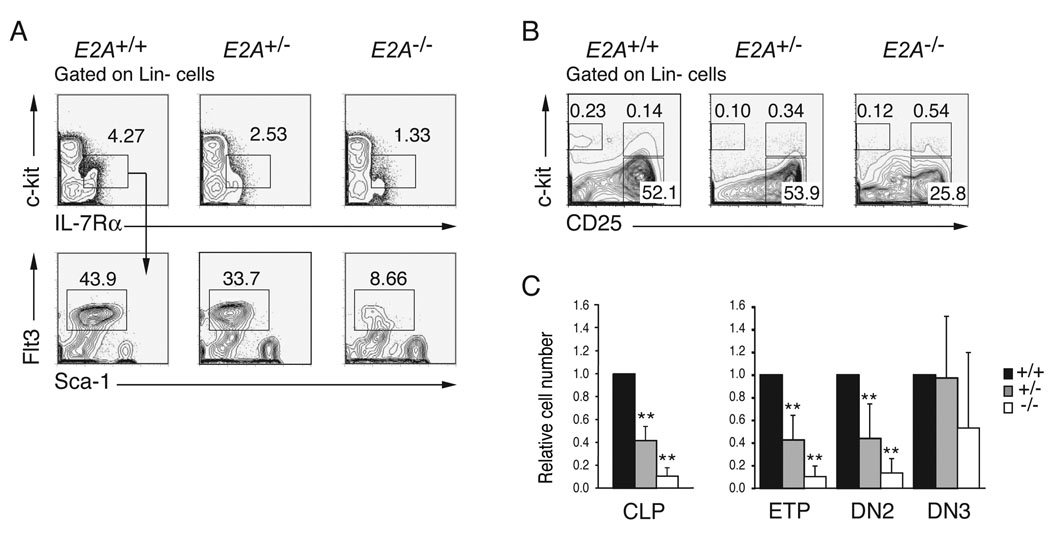
E2A proteins are required for B cell development in part because they promote expression of EBF1 (Kee and Murre, 1998; Seet et al., 2004). This observation contributed to the prevailing view that E2A proteins play a critical role at that CLP to pro-B lymphocyte transition, where EBF1 first becomes essential (Pongubala et al., 2008; Welner et al., 2008). A similar view has developed regarding the requirements for E2A in T cell development where these proteins are thought to be essential for commitment to the T lymphocyte lineage, despite recent evidence implicating E2A in the regulation of Notch1, which is required already by the ETP stage (Bain et al., 1997a; Ikawa et al., 2006; Kee et al., 2002; Rothenberg, 2007). However, it has not be determined whether E2A proteins function at earlier stages of lympho-hematopoiesis. Therefore, we undertook a rigorous quantitative analysis of lymphocyte progenitor populations in E2A+/+, E2A+/− and _E2A_−/− mice by flow cytometry. Consistent with a previous report (Borghesi et al., 2005), we found a significant reduction in the percent and total number of CLPs in _E2A_−/− as compared to E2A+/+ BM (10.8-fold, p<0.001). Interestingly however, we also found a reduced number of E2A+/− CLPs (2.5-fold, p<0.001) indicating a dose depend requirement for E2A at this stage of lymphopoiesis (Figure 1A and 1C). Analysis of Lin- thymocytes revealed a dose dependent requirement for E2A in development of ETPs and DN2 thymocytes (Figure 1B and 1C). However, the number of E2A+/− DN3 thymocytes is indistinguishable from wild-type (WT) while _E2A_−/− DN3 cells remain reduced, although the number was variable (Figure 1B and 1C). The reduced number of ETPs in E2A+/− and _E2A_−/− mice was not revealed in previous studies owing to contamination of the conventional DN1 subset with non-T lineage cells, which are not affected by deletion E2A (Allman et al., 2003; Bain et al., 1997a; Kee et al., 2002). Therefore, we conclude that there is a dose-dependent requirement for E2A during development of CLPs, ETPs and DN2 thymocytes.
Figure 1. A dose dependent requirement for E2A in the generation of CLPs, ETPs and DN2 cells.
(A) Lin− BM cells from E2A+/+, E2A+/− and _E2A_−/− mice were analyzed for surface expression of c-kit and IL-7Rα (upper panels) and the _c-kit_lowIL-7Rα+ subpopulation was analyzed for expression of Sca-1 and Flt3 (lower panels) by flow cytometry. CLPs are Lin−c-_kit_lowIL-7Rα+Sca-1+Flt3+. (B) Surface expression of c-kit and CD25 on Lin− thymocytes; numbers represent the percentage of cells in each gate. (C) Relative number of CLPs in BM and ETPs, DN2 and DN3 cells in the thymus of adult E2A+/− (grey) and _E2A_−/− (white) mice, as compared to wild type mice (black; set to 1). A minimum of 5 mice were analyzed in each group; bars represent the mean ± SD; ** p<0.01.
A dose-dependent requirement for E2A in development of LMPPs
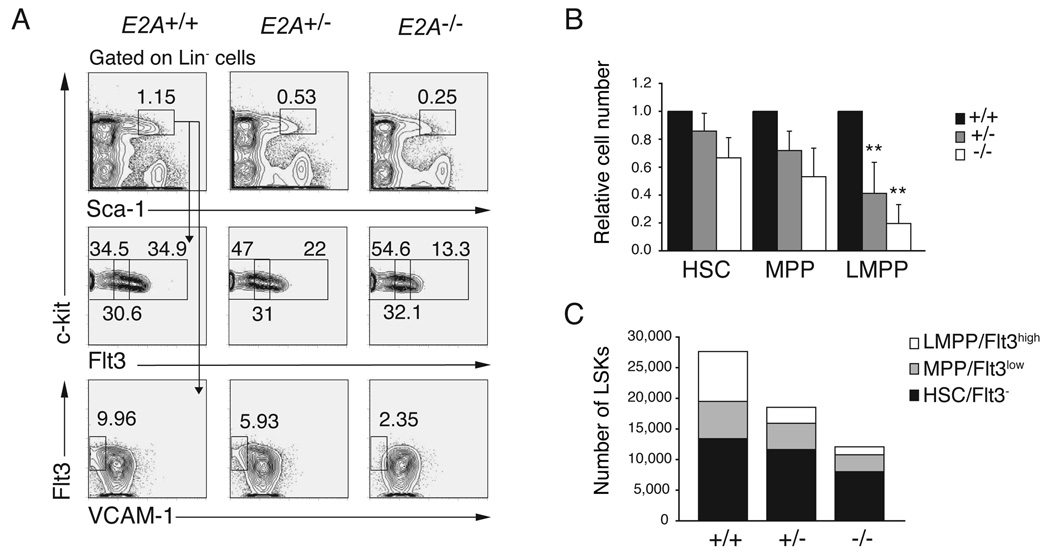
E2A proteins are widely expressed in BM multipotent hematopoietic progenitors (Figure S1) (Zhuang et al., 2004), and our data indicate that E2A may be required upstream of CLPs and ETPs. Therefore, we undertook a quantitative analysis of BM LSK cells, including HSCs, MPPs and LMPPs in E2A+/+, E2A+/− and _E2A_−/− mice. We found that the frequency and total number of LSK cells is decreased in E2A+/− and _E2A_−/− compared to E2A+/+ BM (Figure 2A and 2C). The number of HSCs and MPPs was not significantly different between each of these strains although a consistent mild decrease was observed (Figure 2A and 2C). In contrast, the number of LMPPs was significantly reduced in both E2A+/− (3.1-fold, _p_=0.008) and _E2A_−/− (6.3-fold, _p_=0.002) BM compared to E2A+/+ BM (Figure 2A, 2B and 2C). The decreased frequency of LMPPs does not appear to be the consequence of a requirement for E2A in transcription of the Flt3 gene since there is no compensatory increase in the number of Flt3− or Flt3low LSKs (Figure 2C). In addition, we have found no evidence that E2A can induce Flt3 mRNA in MPPs under conditions where E2A can induce other potential target genes (see Figure 4C). We also found a reduced frequency of VCAM-1−LSKs in E2A+/− and _E2A_−/− BM (Figure 2A), further supporting the conclusion that LMPPs are reduced in E2A+/− and _E2A_−/− mice (Lai and Kondo, 2006; Lai et al., 2005). In contrast, erythro-myeloid progenitors were not decreased, but rather, they are slightly increased in E2A+/− and _E2A_−/− mice (Figure S2). Taken together, our data indicate that E2A proteins play a dose-dependent role in development of LMPPs, which represent the earliest lymphoid-primed progeny of HSCs.
Figure 2. A dose dependent requirement for E2A in the generation of LMPPs.
(A) Lin− BM cells from E2A+/+, E2A+/− and _E2A_−/− mice were analyzed for surface expression of c-kit and Sca-1 (upper panels). The LSK subset was analyzed for expression of c-kit and Flt3 (middle panels) or Flt3 and VCAM-1 (lower panels). Numbers represent the percent of cells in the indicated gate. LMPPs are LSK Flt3high and LSK VCAM-1−. (B) Number of HSCs, MPPs, LMPPs and CLPs in E2A+/− (grey) and _E2A_−/− (white) mice relative to WT (black; set to 1). A minimum of 6 mice were analyzed in each group; bars represent the mean ± SD; ** p<0.01. (C) The total number of HSCs (black), MPPs (grey) and LMPPs (white) cells in E2A+/+, E2A+/− and _E2A_−/− mice is shown and the sum of these populations is the total number of LSK cells.
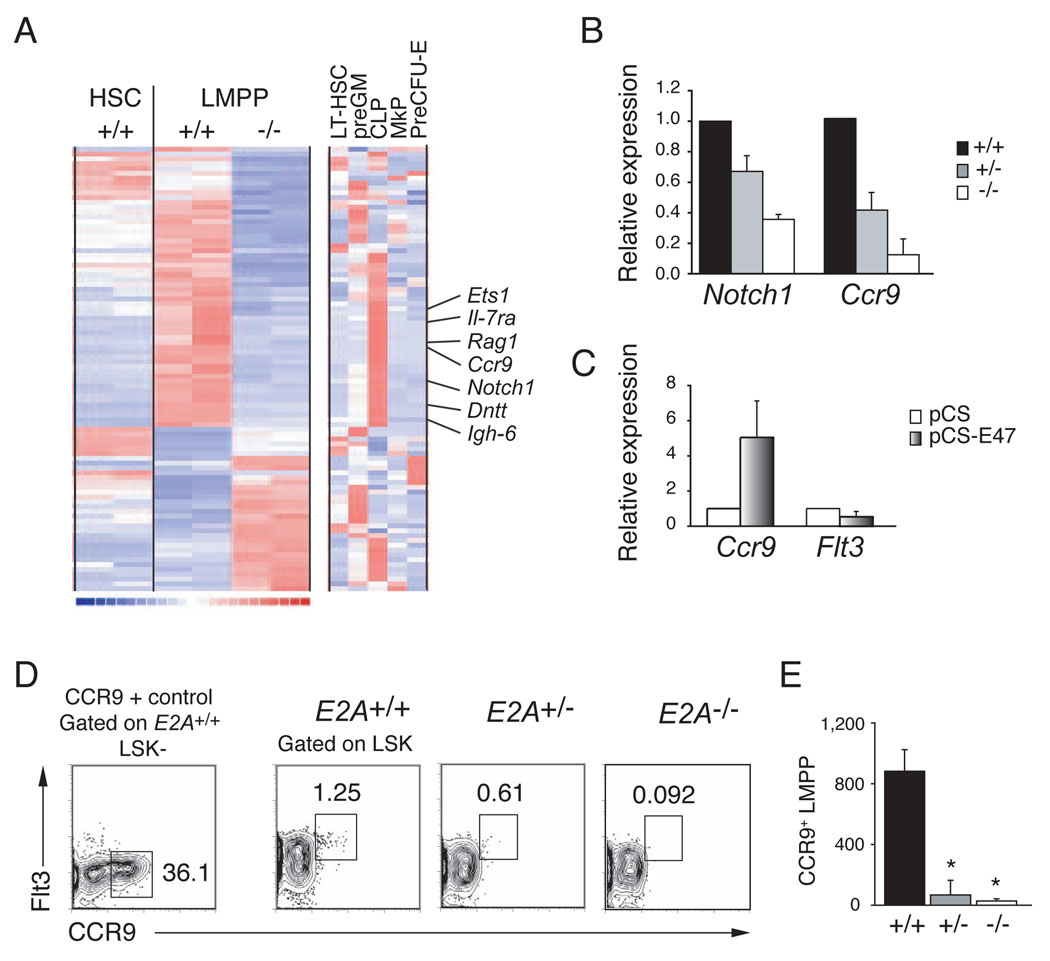
Figure 4. Analysis of the E2A+/+ and _E2A_−/− LMPP transcriptome.
(A) Clustering of genes that are differentially expressed in replicate samples (one per column) of E2A+/+ and _E2A_−/− LMPPs (Affymetrix MOE430 2.0 arrays). Expression of these genes in E2A+/+ HSCs is shown for comparison. The clustering includes all genes with expression levels >50 in at least one of the four LMPP arrays and differing by > 2- fold (using a lower 90% confidence bound of fold change). Each row corresponds to one unique identifier and a subset of CLP-associated genes is indicated (see Figure S3 for complete list of genes). The right-most clustering shows the lineage association of these differentially expressed genes in LT-HSC, preGM, CLP, MkP and preCFU-E (as defined in (Pronk et al., 2007b). Red indicates high, blue low, and white intermediate expression levels. (B) Relative expression of Notch1 and Ccr9 mRNA in purified E2A+/− and _E2A_−/− LMPPs, as compared to E2A+/+ progenitors (normalized to Hprt and set to 1), determined by QPCR. (C) QPCR analysis of Ccr9 and Flt3 mRNA in _E2A_−/− fetal liver multipotent progenitors, 36 hours after transduction with control or E47 producing retrovirus. (D) E2A+/+, E2A+/− and _E2A_−/− LSKs analyzed for surface expression of Flt3 and CCR9. CCR9 expression in E2A+/+ LK−S cells is shown for comparison. (E) Absolute number of CCR9+ LMPPs in the BM (femurs and tibias) of E2A+/+, E2A+/− and _E2A_−/− mice. Bars represent the mean ± SD; * p<0.05.
E2A proteins antagonize proliferation of HSCs, MPPs and LMPPs
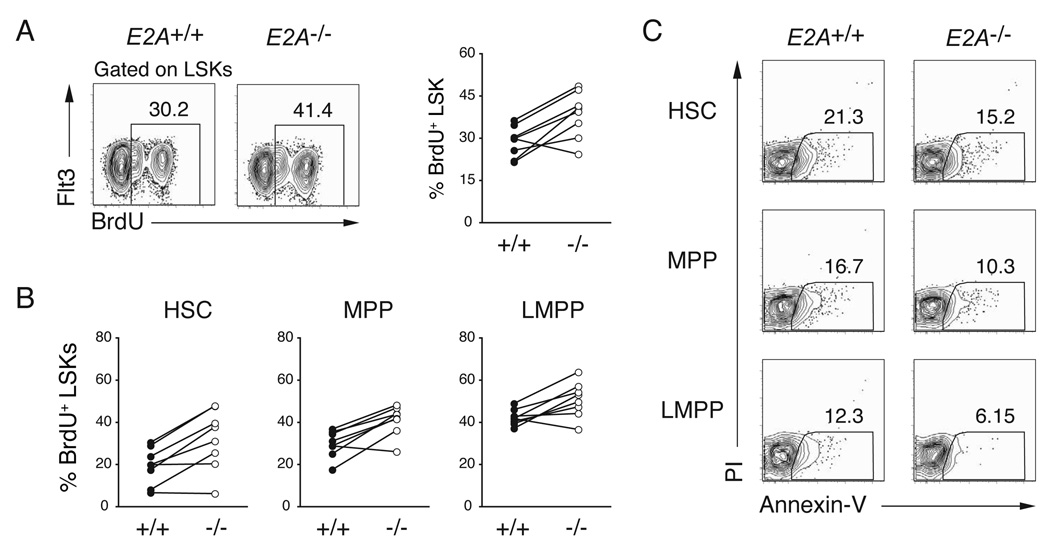
E proteins are necessary for pro-B lymphocyte proliferation (Kee, 2005; Seet et al., 2004), although high levels of E2A can restrain proliferation (Engel and Murre, 2004). Therefore, we considered the possibility that LMPPs are reduced in number in E2A+/− and _E2A_−/− mice because these cells, or their precursors, require E2A for expansion. To test this possibility we examined BrdU incorporation in E2A+/+ and _E2A_−/− BM LSK sub-populations after 24 hours of in vivo labelling. Surprisingly, we found that _E2A_−/− LSKs incorporated more BrdU than E2A+/+ LSKs (Figure 3A). Importantly, all _E2A_−/− LSK subsets incorporated more BrdU than the corresponding E2A+/+ population (Figure 3B). Therefore, the reduced number of LMPPs in E2A+/− and _E2A_−/− BM is not a consequence of reduced proliferation of LMPPs or their precursors. Indeed, E2A proteins appear to limit proliferation of these multipotent cells. Importantly, after in vitro culture, _E2A_−/− and E2A+/+ LSKs showed a similar, or slightly reduced, frequency of apoptotic cells as measured by annexin V binding indicating that the lack of E2A is not leading to increased cell death (Figure 3C). These data lead us to suggest that _E2A_−/− LSK cells preferentially adopt a fate other than LMPP.
Figure 3. E2A proteins restrict HSC, MPP and LMPP proliferation.
(A) BrdU-incorporation in BM LSKs as determined 24 hours of in vivo exposure to BrdU. LSKs were analyzed for surface expression of Flt3 and intracellular BrdU; numbers indicate the percent of BrdU+ LSKs. The graph summarizes results of 5 independent experiments. (B) Percent of HSCs, MPPs, and LMPPs that were BrdU+ in E2A+/+ and _E2A_−/− mice, respectively. (C) Representative FACS analysis for Annexin-V and PI staining on cultured HSC, MPP or LMPPs cells.
E2A proteins are required for proper lymphoid gene expression in LMPPs
To further address the requirements for E2A in development of multipotent lymphoid progenitors we examined the transcriptome of purified E2A+/+ and _E2A_−/− LMPPs. Differentially expressed genes were further characterized based on their expression in E2A+/+ HSCs, as well as in LT-HSC, preGM, CLPs, MkP and preCFU-E (Pronk et al., 2007a). Comparing overall gene expression in E2A+/+ and _E2A_−/− LMPPs, the Pearson correlation was 0.99 as compared to 0.94 when either LMPP sample was compared to E2A+/+ HSCs. Therefore, the few Flt3high LSKs in _E2A_−/− mice are LMPPs and any alterations in gene expression are unlikely to result from differences in cell composition. A total of 47 genes failed to be appropriately expressed (i.e. decreased by > 2-fold) in _E2A_−/− LMPPs indicating that these genes are directly, or indirectly, regulated by E2A (Figure 4A). Interestingly, 26 of these genes are more highly express in CLPs than in other progenitor populations indicating that E2A proteins play a major role in establishing the lymphoid-associated gene expression signature. Among these genes were multiple known E2A targets, including Rag1, _Il-7r_α, Dntt, Igh-6, and Notch1 (Figure 4A). Interestingly, some of these lymphoid-associated genes were found to be expressed in HSCs and dependent on E2A indicating that they are being primed already in the HSC population, in an E2A-dependent manner (Table1). These genes are candidates for genes that influence lymphoid specification (Georgopoulos, 2002).
Table 1.
E2A-dependent lymphoid gene expression in HSCs.
| probe set | Gene Name | E2A+/+ HSC (mean)* | _E2A_−/− HSC (mean)* | Fold decrease in _E2A_−/− HSC (95% confidence interval) |
|---|---|---|---|---|
| 1449757_x_at | Dntt | 1370.74 | 135.61 | 10.11 (8.92–11.66) |
| 1450545_a_at | Dntt | 302.53 | 37.24 | 8.12 (6.43–10.8) |
| 1427351_s_at | Igh-6 | 662.31 | 112.67 | 5.88 (5.37–6.46) |
| 1427329_a_at | Igh-6 | 262.57 | 50.12 | 5.24 (4.92–5.6) |
| 1422851_at | Hmga2 | 291.41 | 79.5 | 3.67 (2.99–4.74) |
| 1420805_at | Mylc2pl | 87.59 | 25.35 | 3.45 (3.26–3.69) |
| 1455570_x_at | Cnn3 | 132.12 | 40.25 | 3.28 (2.87–3.81) |
| 1436759_x_at | Cnn3 | 139.58 | 42.86 | 3.26 (2.9–3.68) |
| 1449310_at | Ptger2 | 51.64 | 16.57 | 3.12 (2.47–3.86) |
| 1436836_x_at | Cnn3 | 186.31 | 62.16 | 3 (2.62–3.43) |
| 1450780_s_at | Hmga2 | 218.78 | 73.1 | 2.99 (2.49–3.73) |
| 1426725_s_at | Ets1 | 143.95 | 72.09 | 2 (1.62–2.46) |
| 1419872_at | Csf1r | 229.91 | 116.53 | 1.97 (1.73–2.22) |
| 1418634_at | Notch1 | 608.16 | 311.76 | 1.95 (1.82–2.1) |
| Fold increase in _E2A_−/− HSC (95% confidence interval) | ||||
| 1434465_x_at | Vldlr | 369.8 | 757.21 | 2.05 (1.66–2.44) |
| 1439814_at | Atp8b4 | 199.22 | 415.49 | 2.09 (1.82–2.44) |
| 1435893_at | Vldlr | 127.66 | 275.66 | 2.16 (1.66–3.08) |
| 1434278_at | Mtm1 | 229.19 | 505.02 | 2.21.96–2.52) |
| 1417900_a_at | Vldlr | 92.11 | 210.58 | 2.29 (1.95–2.75) |
| 1457139_at | Auts2 | 41.51 | 133.81 | 3.22 (2.71–3.94) |
In addition to the set of known E2A target genes, multiple genes were decreased in _E2A_−/− LMPPs that have not been shown previously to be E2A-dependent (Figure 4A and Figure S3). One of these genes is Ccr9, which encodes the receptor for the chemokine CCL25, and we confirmed that Ccr9 and Notch1 mRNA are reduced in E2A+/− and _E2A_−/− LMPPs by QPCR (Figure 4B). Importantly, Ccr9, but not Flt3 mRNA, could be induced by E47 in _E2A_−/− progenitors cultured in vitro (Figure 4C). CCR9 is expressed on a small population of LMPPs that possess efficient in vivo thymus repopulating capacity (Benz and Bleul, 2005; Schwarz et al., 2007; Scimone et al., 2006), and this subpopulation is severely decreased in both E2A+/− and _E2A_−/− mice (13-fold, _p_=0.021 and 32-fold, _p_=0.014, respectively) (Figure 4D and 4E). Therefore, the loss of Ccr9 expressing LMPPs is likely to contribute to the reduced number of ETPs observed in E2A+/− and _E2A_−/− mice.
E2A does not regulate expression of key lymphoid transcription factors in LMPPs
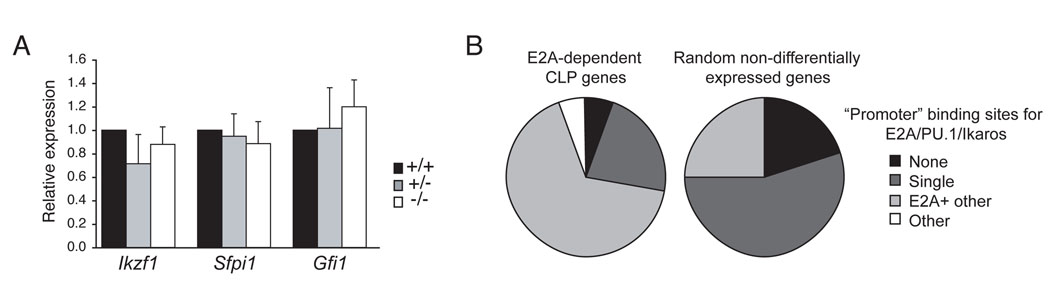
The microarray analysis of E2A+/+ and _E2A_−/− LMPPs revealed a central role for E2A in lymphoid priming. Importantly however, E2A proteins are not required for expression of the key lymphoid transcription factors Ikaros, PU.1, and Gfi1, which which are essential for lymphocyte development beyond the LMPP stage (Figure 5A) (Hock et al., 2004; Medina et al., 2004; Yoshida et al., 2006; Zeng et al., 2004). Nonetheless, known targets of PU.1 and Ikaros, including Dntt and _Il-7r_α, are not expressed appropriately (Figure 4A). An analysis of the genomic DNA surrounding the transcription start sites of CLP-associated E2A-dependent genes revealed that all but two of these genes (10%) have conserved (between human and mouse) potential binding sites for E2A (Figure 5B). Moreover, more than 60% of these genes had potential binding sites for E2A, Ikaros and/or PU.1 (Figure 5B). In contrast, within a set of randomly selected genes that were expressed equivalently in E2A+/+ and _E2A_−/− LMPPs, 40% lacked potential E2A binding sites and only 25% had sites for E2A, Ikaros and/or PU.1. Taken together, these data indicate that E2A is required for Ikaros and PU.1 to promote expression of at least a subset of their target genes in LMPPs.
Figure 5. Expression of key lymphoid transcription factors in the absence of E2A.
(A) QPCR for Ikzf1, Sfpi1, and Gfi1 mRNA in purified E2A+/+, E2A+/− and _E2A_−/− LMPPs. (B) Distribution of conserved potential E2A, Ikaros and PU.1 binding sites in the promoters (−7.5 kb to +2.5 kb) of E2A-dependent lymphoid-associated genes or a set of randomly selected LMPP genes whose expression is not E2A-dependent. Binding sites were identified using rVISTA through the ECR Browser.
E2A proteins restrict the GMP but not the MEP potential of LMPPs
While E2A is required for expression of multiple LMPP-associated genes we found that there is a set of genes that is expressed at higher levels in _E2A_−/− compared to E2A+/+ LMPPs, including genes expressed in HSCs, preGM, MkP and pre-CFU-E (Figure 4A and S3). Interestingly, a subset of these genes is also expressed at higher levels in _E2A_−/− HSCs indicating that these alterations in gene expression initiate within the HSC compartment and may influence lymphoid specification (Table 1). These data also show that the few _E2A_−/− LMPPs that develop have a significantly altered transcriptome and fail to appropriately extinguish a set of non-LMPP-associated genes.
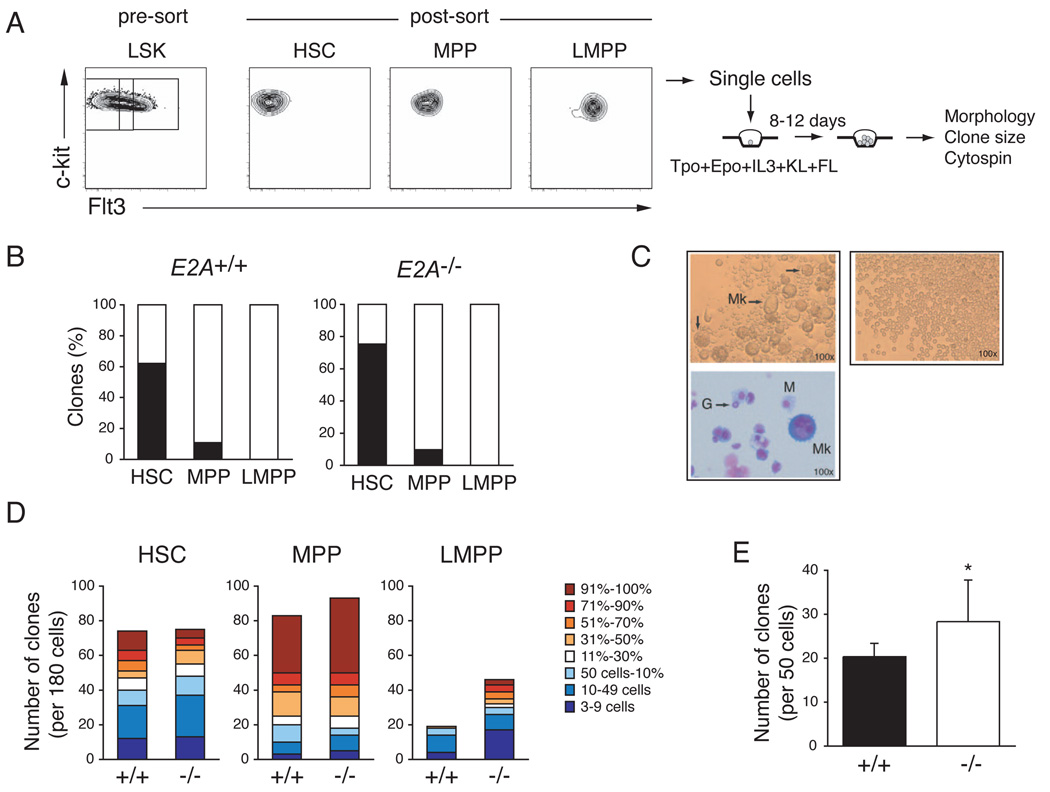
Our data indicate that in the absence of E2A there is a progressive loss of lymphoid progenitor potential that initiates within the HSC or MPP compartment and becomes significant by the time of emergence of LMPPs. The transition from HSC to LMPP is associated with a progressive loss of MEP developmental potential (Adolfsson et al., 2005; Mansson et al., 2007). Therefore, we questioned whether the loss of E2A influences the erythro-myeloid potential of MPPs or LMPPs. To test this possibility we examined the ability of single E2A+/+ and _E2A_−/− HSCs, MPPs and LMPPs to give rise to megakaryocytes (Mk) in vitro under conditions that efficiently support Mk and other myeloid lineage development (Figure 6A). As reported previously (Adolfsson et al., 2005; Arinobu et al., 2007), we found that WT HSCs, MPPs and LMPPs have a high cloning efficiency but that LMPPs do not develop into Mk under these conditions (Figure 6B and 6C). Importantly, sorted _E2A_−/− HSCs, MPPs, and LMPPs showed a similar restriction of Mk potential (Figure 6B). However, _E2A_−/− LMPPs had a higher clongenic potential than their WT counterparts and individual _E2A_−/− clones were significantly larger than WT (Figure 6D). _E2A_−/− MPP colonies were also, on average, larger than E2A+/+ MPP derived colonies (Figure 6D). Using a methycellulose based assay we also observed that Flt3+ LSKs from _E2A_−/− mice have an increased clonogenic potential compared to E2A+/+ cells (Figure 6D). Wright-Giemsa staining of individual clones, under either culture condition, revealed that WT and _E2A_−/− LMPPs gave rise only to granulocytes and/or macrophages (Figure 6C). We note that we also detected an increase in atypical granulocytes in _E2A_−/− LMPP clones, which appear to be cells undergoing apoptosis suggesting that loss of E2A may influence the rate of granulocyte differentiation, or the lifespan of granulocytes, under these conditions (Figure S4). Taken together, our data support a model in which E2A proteins promote differentiation toward the lymphoid fate and in the absence of E2A MPPs and LMPPs preferentially adopt the GMP fate. This model is consistent with previous studies showing that E2A proteins can induce lymphoid lineage conversion in a macrophage cell line and that E2A represses macrophage development from MPPs in vitro (Bhalla et al., 2007; Kee and Murre, 1998).
Figure 6. _E2A_−/− LMPPs restrict the MEP fate but show increased myeloid clonogenic potential.
(A) HSCs, MPPs, and LMPPs were isolated from the BM LSK population by cell sorting and single cells were seeded in wells of Terasaki plates in conditions supporting multilineage myeloid differentiation. (B) Frequency of E2A+/+ and _E2A_−/− clones giving Mk in vitro (black). One of 4 representative experiments is shown. (C) Representative examples of colonies positive (left) or negative (right) for the presence of megakaryocytes (Mk). Wright-Giemsa staining of the Mk-containing clone (bottom); G = granulocyte; M = macrophage. (D) Number of clones generated from 180 single cell cultures of E2A+/+ or _E2A_−/− HSCs, MPPs, or LMPPs. Color-coding from blue to red represents the size of colonies from smallest (3–9 cells) to largest (90–100% of well). One of 4 representative experiments is shown. (E) Frequency of clonable E2A+/+ (black) and _E2A_−/− (white) LSKFlt3+ progenitors determined at day 7 of culture in methylcellulose (mean of 3 replicate wells ± SD; * p<0.05; freq., frequency). One of 3 representative experiments is shown.
DISCUSSION
We have identified a requirement for E2A proteins in supporting the development of LMPPs from HSCs. E2A proteins are essential for the initial priming of a subset of lymphoid-associated genes, some of which are detected already within HSCs. In the absence of E2A key lymphoid-promoting transcription factors such as PU.1, Ikaros and Gfi1 are expressed appropriately; however, E2A is necessary for these genes to activate some of their targets in LMPPs. Moreover, we found that E2A dependent lymphoid genes frequently have conserved potential binding sites for PU.1 and/or Ikaros indicating that these transcription factors may cooperatively regulate the lymphoid gene program. Nonetheless, the role of E2A extends beyond the induction of lymphoid genes since _E2A_−/− HSCs and LMPPs show aberrant expression of genes that are not normally expressed in these cells. We also found that E2A proteins restrict proliferation of HSCs, MPPs, and LMPPs and antagonize GMP differentiation from LMPPs in vitro. Our data indicate that the first essential function of E2A proteins in lymphocyte development is to support development of a lymphoid-primed population of cells that is capable of further restriction to specific lymphoid fates.
Our data reveal a severe loss of LMPPs in both E2A+/− and _E2A_−/− mice. However, unlike Ikaros and PU.1, which are also required for development of Flt3high LSKs, we found no evidence that E2A proteins directly regulate Flt3 (Medina et al., 2004; Yoshida et al., 2006). Flt3 mRNA was not differentially expressed in E2A+/+ and _E2A_−/− LMPPs and there was no compensatory increase in the number of Flt3− or Flt3low LSKs in _E2A_−/− mice as might be predicted if LMPPs lacking Flt3 are present. Moreover, ectopic expression of E2A in _E2A_−/− progenitors does not alter Flt3 mRNA even though Ccr9 mRNA is induced. We also found that LMPPs are reduced in _E2A_−/− mice using loss of VCAM-1 as a distinguishing feature of these cells (Lai and Kondo, 2007; Lai et al., 2005). Therefore, the loss of Flt3high LSKs in E2A+/− and _E2A_−/− mice reflects a decline in LMPP numbers. While E2A proteins do not regulate Flt3 mRNA it is possible that Flt3 signaling regulates E2A function. Flt3 could regulate E2A by phosphorylation or indirectly through Flt3-dependent down regulation of class II bHLH proteins such as SCL/TAL1 (Adolfsson et al., 2005; Neufeld et al., 2000).
E2A proteins clearly play a major role in the development of LMPPs. However, a small number of LMPPs develop in E2A+/− and _E2A_−/− mice and these cells are able to generate T-lymphocytes, but not B-lymphocytes, in vivo. A possible explanation for the development of these few LMPPs is that they rely on E-protein activity provided by the E-proteins HEB and E2-2. The E-proteins are functionally redundant and can cooperate with other transcription factors to activate E2A target genes, even when expressed at low levels (Seet et al., 2004; Zhuang et al., 1998). Nonetheless, a number of lymphoid genes fail to be appropriately expressed in _E2A_−/− LMPPs, including Rag1, _Il-7r_α, Dntt, Ets1, and Ccr9. It is unlikely that these particular genes regulate the development of LMPPs since LMPPs develop in mice that lack Rag1, Ccr9, _Il-7r_α or Ets1 (B. Kee, unpublished and (Schwarz et al., 2007). Regardless, our findings indicate that within LMPPs E2A is required for proper expression of a large subset of lymphoid genes. Interestingly, a subset of these genes can be detected in HSCs and are E2A-dependent indicating that the lymphoid gene program is beginning priming at a significantly earlier developmental stage than previously appreciated. This observation is consistent with other studies that have identified lineage-associated transcripts in HSCs (Manaia et al., 2000; Ye et al., 2003; Yoshida et al., 2008), and raise the possibility that this subset of E2A-dependent genes may play a role in specifying the lymphoid fate (Georgopoulos, 2002).
In addition to regulating lymphoid-associated genes, E2A is necessary to prevent expression of a set of genes not normally found in LMPPs. Interestingly, a subset of these genes is also increased in _E2A_−/− HSCs. This observation, in conjunction with the decline in MPP and LMPP numbers indicates that the function of E2A extends beyond lymphoid priming. Indeed, loss of E2A is associated with an increased proliferation of HSCs, MPPs and LMPPs as well as increased clongenic potential and GMP differentiation in LMPPs, effects that are unlikely to be attributed to a lack of lymphoid priming. The loss of LMPPs in E2A+/− and _E2A_−/− mice does not appear to be due to a failure of expansion or increased cell death. Rather, fewer cells are undergoing lymphoid specification and instead adopt the GMP fate. A role for E2A proteins in restricting GMP differentiation is consistent with a previous report demonstrating that E12 can induce lymphoid gene expression and repress myeloid traits in a macrophage cell line (Kee and Murre, 1998) and our recent finding that ectopic expression of E12 in _E2A_−/− MPPs promotes B lymphocyte and represses macrophage development in vitro (Bhalla et al., 2008). Therefore, E2A proteins, possibly in collaboration with other lymphoid transcription factors, promote lymphoid specification.
While E2A proteins promote lymphoid specification they are not required for restriction of Mk potential in LMPPs. Therefore, E2A proteins do not promote lymphomyeloid specification at the expense of erythro-myeloid specficiation, even though CMPs and MEPs are slightly increased in _E2A_−/− mice. This increase may be a result of the increased proliferation of _E2A_−/− HSCs which could lead to greater erythro-myeloid output over time. Interestingly however, E2A proteins are highly expressed in erythro-myeloid progenitors (see Figure S1), and can function as dimerizing partners for SCL and Lyl1, which play an essential role in Mk and E differentiation (Lecuyer and Hoang, 2004). Therefore, appropriate regulation of E2A protein homo- and hetero-dimer formation may play a major role in allowing differentiation of these two pathways downstream of HSCs. At the present time it is not know whether there is a single progenitor cell that makes a binary decision regarding the erythro-myeloid or lympho-myeloid fate, and recent studies suggest that these decisions may be temporally segregated (Arinobu et al., 2007). Therefore, considerable experimentation is still required to reveal the basic mechanisms dissociating these two branches of the hematopoietic system and the role of lymphoid promoting transcription factors in restricting MEP differentiation.
Our data also provide insight into the relationship between E2A and Notch1 and the regulation of T-lymphocyte specification. We show that E2A is required in vivo for optimal expression of Notch1 mRNA in LMPPs and HSCs. While the reduced level of Notch1 does not appear to prevent T-cell development, it may nonetheless decrease the “fitness” of these ETPs in vivo, as observed with Notch1+/− ETPs (Tan et al., 2005). The number of ETPs is reduced in E2A+/− and _E2A_−/− mice proportional to the decrease in LMPPs, taking into account the potential amplifying effect of the lack of CCR9 (Schwarz et al., 2007). Therefore, our data suggest that the primary cause of the reduced number of DN thymocytes in E2A+/− and _E2A_−/− mice is the failure to produce sufficient BM derived lymphoid-primed progenitors. However, the fact that _E2A_−/− thymocytes frequently fail to recover to WT levels by the DN3 stages suggests that there may be additional defects in these cells that prevent appropriate T cell development.
The data we have presented indicate that E2A proteins function in the transcriptional network that specifies the lympho-myeloid fate in multipotent progenitors and for lymphoid-priming within these cells and their precursors. These newly identified functions for E2A, together with its role in regulation of B- and T-lymphocyte specific gene expression, resembles the known functions of the Drosophila homologue of E2A, daughterless, in neurogenesis, where it is required to establish the identity of cells with potential to develop into neuronal lineages (the proneural field) and subsequently functions within the proneural field to activate lineage-specifying genes (Bertrand et al., 2002). Similar functions have been described for class II bHLH proteins, which require E-proteins to mediate transcription, in determination and differentiation of olfactory neurons in mammals (Cau et al., 2002). Therefore, the E-proteins may play universal functions in cell fate determination and differentiation.
EXPERIMENTAL PROCEDURES
Mice
Mice were housed at the University of Chicago Animal Resource Center and experiments were performed in accordance to the guidelines of the National Institute of Health and approved IACUC protocol. C57Bl/6 E47−/− mice (here referred to as _E2A_−/−) and genotyping protocols have been described previously (Bain et al., 1997b). Mice were analyzed between 6 and 12 weeks of age.
Flow cytometry and sorting
Cells were harvested from thymus or BM and stained with specific combinations of antibodies (see Supplemental experimental procedures Table I) purchased from BD Biosciences and eBiosciences, with the exception of CCR9 (R&D Systems). Lineage-antibodies were biotinylated (revealed with streptavidin-PECy5.5) and the remainders were conjugated to FITC, PE, PE-Cy7 or APC. Cells were analyzed using a FACS Canto and FlowJo (Tree Star) software or sorted using a FACS Aria. For sorting, Lineage+ cells were first depleted by Magnetic-Activated Cell Separation using streptavidin MicroBeads (Miltenyi Biotec) according to manufacturer’s instructions. Dead cells were stained with PI and excluded by electronic gating. Intracellular staining was performed as described previously using purified anti-hE47 monoclonal antibody (BD Biosciences) (Engel et al., 2001). Negative staining controls were used to distinguish negative and positive populations. LSK gates were determined using the strategy described by Kondo et al. (Kondo et al., 1997). Flt3 gates were determined using the strategy described by Adolfsson et al. (Adolfsson et al., 2005).
Determination of cell numbers in each progenitor population
We determined the absolute number of viable cells in the BM (2 femurs and 2 tibias) or thymus by Trypan blue exclusion. This number was multiplied by the percent of Lin− cells in the viable cell gate (PI−) and then by the percent of cells in each sub-gate thereafter, to determine the absolute number of progenitors in each subpopulation per mouse. For BM progenitors, this number was further normalized to 50×106 total BM cells, to minimize differences due to animal size. Relative cell numbers were calculated by dividing the number of cells in each subpopulation by the number of cells in that same subpopulation in WT mice (therefore, WT=1 for each cell population and in each set of mice analyzed). Statistical significance was assessed by the Student’s _t_-test.
In vivo BrdU-incorporation assay
Mice were injected intraperitoneally with 1 mg BrdU per 6g of body weight (BD Biosciences), 24 and 12 h prior to analysis of BM subpopulations by flow cytometry. BrdU staining was performed using the FITC BrdU Flow Kit (BD Biosciences) following manufacturer’s instructions.
In vitro apoptosis assay
Sorted HSC, MPP or LMPP were cultured in media lacking serum for 20h. Staining was performed using the Annexin-V-FITC Apoptosis Detection Kit (BD Biosciences) following manufacturer’s instructions.
Affymetrix Gene Expression and Data Analysis
RNA was extracted using the RNeasy Micro Kit (Qiagen), labeled, amplified, and hybridized to MOE430 2.0 Affymetrix gene expression arrays according to Affymetrix standard protocols. Probe level expression values were calculated with RMA (Irizarry et al., 2003). Further analysis was performed using dChip (www.dchip.org). Array data are accessible through the gene expression omnibus (GEO; GSE8407 and GSE7302).
Transcription factor binding sites conserved between human and mouse genomic sequences were identified in the promoters of E2A-dependent lymphoid-associated genes and a set of randomly selected genes using rVISTA (Dubchak and Ryaboy, 2006). The promoters were defined as the region −7.5 kb to +2.5 kb of the transcription start site as defined by gene annotation in the ECRBrowser (http://ecrbrowser.dcode.org/). The E2A-dependent lymphoid-associated genes examined were: Apcdd1, Arpp21, Blnk, Ccr9, Cnn3, Dntt, Ets1, Fgf13, Gsn, Hmga2, _Il7r_α, Klf3, Mylc2pl, Notch1, Ptger2, Rag1, Slc9a9, Slc27a2; and the randomly selected genes were: Appbp1, Cdk4, Cdk9, Cog2, Ctsc, Entpd5, Gdi2, Hnrpa2, Mrps14, Ndufb7, Rplp1, Rpl14, sdfr1, Smc6l1, Snrpa1, Snsx3, Timeless, Tpd52l2, Uba52, Ywhab.
Quantitative PCR
RNA was extracted using the RNeasy Micro Kit and cDNA was prepared using random primers and SuperScriptIII reverse transcriptase (Invitrogen), following manufacturer’s instructions. QPCR reactions were performed in triplicate, using iQ SYBR Green Supermix (Biorad) and detected by MyiQ Single Color Real-Time PCR System (Biorad). Most primers (Supplemental experimental procedures Table II) amplify multi-exon sequences; for the remainder, DNaseI (Qiagen) treatment and a non-RT negative control were included.
Retroviral Transduction
Sorted Lin−_c-kit_highCD27+ fetal liver progenitors were spinoculated (2h at 2,500rpm) in the presence of 5µg/ml polybrene, using pCS and pCS-E47-ER (Sayegh et al., 2003) retroviral supernatants produced in Plat-E cells. Transduced cells were cultured on OP9-DL1 stroma in the presence of _c-kit_-ligand, Flt3-ligand, IL7, and 1µM 4-hydroxytamoxifen for 36 hours.
In vitro evaluation of erythro-myeloid differentiation potential
Single BM HSCs, MPPs or LMPPs were seeded in Terasaki plates (180 cells per population in each experiment) and cultured as previously described (Mansson et al., 2007). Wells were scored using an inverted light microscope on day 9–12 of culture for clonal growth, clone size and frequency of Mk. Mk detection was confirmed by Wright-Giemsa staining of cytospin preparations. 50 BM LSKFlt3+ cells were seeded per well in 6-well plates, in 1.1 mL MethoCult® 03434 (StemCell Technologies) supplemented with 50 ng/mL Flt3-ligand and 10 ng/mL thrombopoietin (PeproTech) and clonal growth evaluated on day 7.
Supplementary Material
01
ACKNOWLEDGMENTS
We thank the members of the Flow Cytometry Core Facility at the University of Chicago for cell sorting, the Microarray facility at Lund University and Katia Georgopoulos for Ikaros primer sequences. We are grateful to members in the Kee Lab for helpful discussions. S.D. was supported by the Committee on Cancer Biology. This work was supported by the NIH/NCI (CA99978). The authors declare that they have no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adolfsson J, Borge OJ, Bryder D, Theilgaard-Monch K, Astrand-Grundstrom I, Sitnicka E, Sasaki Y, Jacobsen SE. Upregulation of Flt3 expression within the bone marrow Lin(−)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–669. doi: 10.1016/s1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. Identification of Flt3+lympho-myeloid stem cells lacking erythro-megakaryocytic potential: a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- Arinobu Y, Mizuno S, Chong Y, Sigematsu H, Iino T, Iwasaki H, Graf T, Mayfield R, Chan S, Kastner P, Akashi K. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and mylolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Bain G, Engel I, Robanus Maandag EC, te Riele HPJ, Voland JR, Sharp LL, Chun J, Huey B, Pinkel D, Murre C. E2A deficiency leads to abnormalities in alpha/beta T-cell development and to rapid development of T-cell lymphomas. Mol. Cell Biol. 1997a;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Robanus Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- Bain G, Robanus Maandag EC, te Riele HP, Feeney AJ, Sheehy A, Schlissel M, Shinton SA, Hardy RR, Murre C. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997b;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- Benz C, Bleul CC. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J Exp Med. 2005;202:21–31. doi: 10.1084/jem.20050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of the neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bhalla S, Spaulding C, Brumbaugh RL, Zagort DE, Massari ME, Murre C, Kee BL. Differential roles for the E2A activation domains in B lymphocytes and macrophages. J. Immunol. 2008;180:1694–1703. doi: 10.4049/jimmunol.180.3.1694. [DOI] [PubMed] [Google Scholar]
- Borghesi L, Aites J, Nelson S, Lefterov P, James P, Gerstein RM. E47 is required for V(D)J recombinase activity in common lymphoid progenitors. J. Exp. Med. 2005;202:1669–1677. doi: 10.1084/jem.20051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129:1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med. 2005;201:1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S, Silva HJ, Cumano A, Vieira P. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J. Exp. Med. 2005;201:971–979. doi: 10.1084/jem.20042393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubchak I, Ryaboy DV. VISTA family of computational tools for comparative analysis of DNA sequences and shole genomes. Methods Mol Biol. 2006;338:69–89. doi: 10.1385/1-59745-097-9:69. [DOI] [PubMed] [Google Scholar]
- Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J Exp Med. 2001;194:733–745. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I, Murre C. E2A proteins enforce a proliferation checkpoint in developing thymocytes. EMBO J. 2004;23:202–211. doi: 10.1038/sj.emboj.7600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg EC, Serwold T, Kogan S, Weissman IL, Passegue E. New evidence supporting megakaryocyte-erythrocyte potential of flt2/flt3+ multipotent hematopoietic progenitors. Cell. 2006;2 doi: 10.1016/j.cell.2006.06.037. 126. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol. 2002;2:162–174. doi: 10.1038/nri747. [DOI] [PubMed] [Google Scholar]
- Hock H, Hamblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y, Orkin SH. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 2002;17:117–130. doi: 10.1016/s1074-7613(02)00366-7. [DOI] [PubMed] [Google Scholar]
- Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J. Exp. Med. 2006;203:1329–1342. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa T, Kawamoto H, Wright LYT, Murre C. Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent. Immunity. 2004;20:349–360. doi: 10.1016/s1074-7613(04)00049-4. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno S, Arinobu Y, Geary K, Zhang P, Dayaram T, et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee BL. Id3 Induces Growth Arrest and Caspase-2-Dependent Apoptosis in B Lymphocyte Progenitors. J. Immunol. 2005;175:4518–4527. doi: 10.4049/jimmunol.175.7.4518. [DOI] [PubMed] [Google Scholar]
- Kee BL, Bain G, Murre C. IL7Ra and E47: independent pathways required for the development of multipotent lymphoid progenitors. EMBO J. 2002;20 doi: 10.1093/emboj/21.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee BL, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J. Exp. Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee BL, Murre C. Transcription factor regulation of B lineage commitment. Current Opin. Immunol. 2001;13:180–185. doi: 10.1016/s0952-7915(00)00202-8. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Lai AY, Hsu CL, Kondo M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J. Exp. Med. 2005;201:1197–1203. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Lai AY, Kondo M. Asymmetrical lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. J Exp Med. 2006;203:1867–1873. doi: 10.1084/jem.20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AY, Kondo M. Identifcation of a bone marrow precursor of the earliest thyocytes in adult mouse. Proc. Natl. Acad. Sci. 2007;104:6311–6316. doi: 10.1073/pnas.0609608104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AY, Lin SM, Kondo M. Heterogeneity of Flt3-expressing multipotent progenitors in mouse bone marrow. J. Immunol. 2005;175:5016–5023. doi: 10.4049/jimmunol.175.8.5016. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Hoang T. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp. Hematol. 2004;32:11–24. doi: 10.1016/j.exphem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Manaia A, Lemarchandel V, Klaine M, Max-Audit I, Romeo PH, Dieterlen-Lievre F, Godin I. Lmo2 and GATA-3 associated expression in intraembryonic hemogenic sites. Development. 2000;127:643–653. doi: 10.1242/dev.127.3.643. [DOI] [PubMed] [Google Scholar]
- Mansson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al-Hashmi S, Liuba K, Thoren L, Adolfsson J, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina KL, Pongubala JM, Reddy KL, Lancki DW, DeKoter RP, Kieslinger M, Grosschedl R, Singh H. Assembling a gene regulatory network for specification of the B cell fate. Dev. Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Neufeld B, Grosse-Wilde A, Hoffmeyer A, Jordan BW, Chen P, Dinev D, Ludwig S, Rapp UR. Serine/threonine kinases 3pK and MAPK-activated protein kinase 2 interact with the basic helix-loop-helix transcription factor E47 and repress its transcriptional activity. J. Biol. Chem. 2000;275:20239–20242. doi: 10.1074/jbc.C901040199. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Kee BL. The transcriptional regulation of B-cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- Pronk CJH, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CKF, Sigvardsson M, Weissman IL, Bryder D. Elucidation of the phenotypic, functional, and molecular topography of myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007a;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Pronk CJH, Rossi DJ, Månsson R, Attema JL, Norddahl GL, Chan CKF, Sigvardsson M, Weissman IL, Bryder D. Elucidation of the Phenotypic, Functional, and Molecular Topography of a Myeloerythroid Progenitor Cell Hierarchy. Cell Stem Cell. 2007b;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Roessler S, Gyory I, Imhof S, Spivakov M, Williams RR, Busslinger M, Fisher AG, Grosschedl R. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol. Cell Biol. 2007;27:579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV. Negotiation of the T lineage fate decision by transcription-faactor interplay and microenvironmental signals. Immunity. 2007;26:690–702. doi: 10.1016/j.immuni.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat. Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat. Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- Schwarz BA, Sambandam A, Maillard I, Harman BC, Love PE, Bhandoola A. Selective thymus settling regulated by cytokine and chemokine receptors. J. Immunol. 2007;178:2008–2017. doi: 10.4049/jimmunol.178.4.2008. [DOI] [PubMed] [Google Scholar]
- Scimone ML, Aifantis I, Apostolou I, von Boehmer H, von Andrian UH. A multistep adhesion cascade for lymphpoid progenitor homing to the thymus. Proc. Natl. Acad. Sci. 2006;103:7006–7011. doi: 10.1073/pnas.0602024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet CS, Brumbaugh RL, Kee BL. Early B-cell factor promotes B-lymphopoiesis with reduced interleukin-7-responsiveness in the absence of E2A. J. Exp. Med. 2004;199:1689–1700. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat. Immunol. 2005;6:671–679. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- Welner RS, Pelayo R, Kincade PW. Evolving views on the genealogy of B cells. Nat Rev Immunol. 2008;8:95–106. doi: 10.1038/nri2234. [DOI] [PubMed] [Google Scholar]
- Ye M, Iwasaki H, Laiosa CV, Stadtfeld M, Xie H, Heck S, Clausen B, Akashi K, Graf T. Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multlineage repopulation potential. Immunity. 2003;19:689–699. doi: 10.1016/s1074-7613(03)00299-1. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Zi X, Lawton LN, Williams CJ, Georgopoulos K. The role of the chromatin remodeler Mi-2(beta) in hematopoietic stem cell self-renewal and multilineage differentaition. Genes & Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, S.Y N, Zunig-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol. 2006;7:382–391. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Yucel R, Kosan C, Klein-Hitpass L, Moroy T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 2004;23:4116–4125. doi: 10.1038/sj.emboj.7600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Barndt RJ, Pan L, Kelley R, Dai M. Functional replacement of the mouse E2A gene with a human HEB cDNA. Mol. Cell. Biol. 1998;18:3340–3349. doi: 10.1128/mcb.18.6.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Jackson A, Pan L, Shen K, Dai M. Regulation of E2A gene expression in B-lymphocyte development. Mol. Immunol. 2004;40:1165–1177. doi: 10.1016/j.molimm.2003.11.031. [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01