Autoimmune diseases: insights from genome-wide association studies (original) (raw)
Abstract
Autoimmune diseases occur when an individual's own immune system attacks and destroys his or her healthy cells and tissues. Although it is clear that environmental stimuli can predispose someone to develop autoimmune diseases, twin- and family-based studies have shown that genetic factors also play an important role in modifying disease risk. Because many of these diseases are relatively common (prevalence in European-derived populations: 0.01–1%) and exhibit a complex mode of inheritance, many DNA sequence variants with modest effect on disease risk contribute to the genetic burden. Recently, the completion of the HapMap project, together with the development of new genotyping technologies, has given human geneticists the tools necessary to comprehensively, and in an unbiased manner, search our genome for DNA polymorphisms associated with many autoimmune diseases. Here we review recent progress made in the identification of genetic risk factors for celiac disease, Crohn's disease, multiple sclerosis, rheumatoid arthritis, systemic lupus erythematosus and type-1 diabetes using genome-wide association studies (GWAS). Strikingly, GWAS have increased the number of genetic risk variants associated with these autoimmune diseases from 15 before 2006 to 68 now. We summarize what this new genetic landscape teaches us in terms of the pathogenesis of these diseases, and highlight some of the outstanding challenges ahead. Finally, we open a discussion on ways to best maximize the impact of these genetic discoveries where it matters the most, that is for autoimmune disease patients.
INTRODUCTION
Our immune system protects us from infections and cancers by recognizing and neutralizing pathogens or abnormal cells. However, when this system goes awry and mounts hyperactive immune response against the organism's own healthy cells and tissues, consequences can be severe. Such attacks against ‘self’ normal antigens, resulting in inflammation and tissue damage, can lead to one of many forms of autoimmune diseases. In some cases, the immune response is specific to a particular cell type [e.g. pancreatic β cells in type-1 diabetes (T1D) or oligodendrocytes in multiple sclerosis (MS)], but it can also target a broader range of cell types and tissues [e.g. nuclear antigens in systemic lupus erythematosus (SLE)]. Although it is not always certain what triggers the initial immune attacks, it is apparent that both environmental and genetic factors play a role. Not surprisingly for autoimmune diseases, given that it contains multiple genes involved in immune functions such as antigen presentation, the major histocompatibility complex (MHC) region harbors several genetic variants that either protect or predispose individuals from developing celiac disease (CeD), MS, rheumatoid arthritis (RA), SLE and T1D (1). However, it is also clear that other DNA polymorphisms, unlinked to the MHC region, influence disease risk.
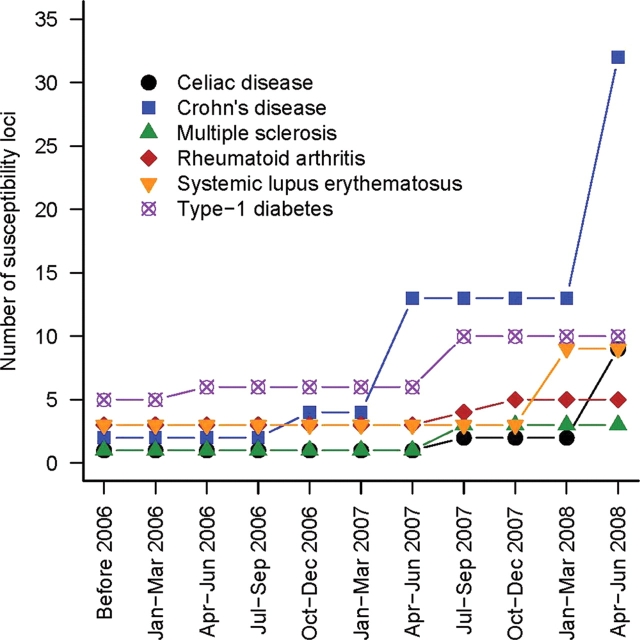
This Review does not aim at summarizing our current knowledge of the pathogenesis of some of the most common autoimmune diseases; these have been reviewed exquisitely elsewhere (2–7). Rather, we focus here on how genome-wide association studies (GWAS) have changed the genetic landscape of CeD, Crohn's disease (CD), MS, RA, SLE and T1D research in less than 3 years (Fig. 1). We report the lessons learned from these successful GWAS experiments, emphasize the exciting themes emerging from these genetic discoveries and highlight some remaining outstanding questions. Finally, we discuss ways to utilize this new genetic information to impact the quality of life of patients suffering from autoimmune diseases.
Figure 1.
There is an increase in the rate of discovery of risk loci for many autoimmune diseases following the emergence of GWAS in 2006. We group in the ‘Before 2006’ category all loci identified before 2006 or loci identified after 2006 using candidate-gene association approaches as opposed to GWAS.
LESSONS LEARNED
To date, results from 18 GWAS aimed at identifying autoimmune genetic risk factors have been published (Table 1). This resulted in the impressive addition of 53 new common DNA sequence variants that influence risk of developing CeD, CD, MS, RA, SLE or T1D. All these studies had in common some key features to maximize their chance of success. First, the number of DNA markers genotyped was large (>100 000 single nucleotide polymorphisms or SNPs) in order to capture most of the common genetic variation in the human genome. Second, genotyping was performed in a large number of clinically well-defined patients and matched controls. Third, very stringent quality-control criteria were used to process genotype and phenotype data. And fourth, replication was done in equally large and well-designed cohorts using different genotyping platforms. These guidelines have now become common practice in the daily design and analysis of GWAS (8).
Table 1.
Summary of the new loci associated with autoimmune diseases and identified by genome-wide association studies (see http://www.genome.gov/26525384 for an updated list)
| Disease | Sibling relative risk ratio (λs) | GWAS no. of cases/controls | Replication no. of cases/controls | Chromosome | Genesa | Other associated autoimmune disease | References |
|---|---|---|---|---|---|---|---|
| Celiac disease | ∼30 | 778/1422 | 991/1489 | 4q27 | IL2–IL21 | MS, RA, T1D | (28) |
| Same study | 1643/3406 | 1q31 | RGS1 | T1D | (29) | ||
| 2q11–12 | IL18RAP (eQTL) | T1D | |||||
| 3p21 | CCR3 | ||||||
| 3q25–26 | IL12A | ||||||
| 3q28 | LPP | ||||||
| 6q25 | TAGAP | ||||||
| 12q24 | SH2B3 (R262W) | ||||||
| Crohn's disease | ∼20–35 | 547/548 | 401/433; 883b | 1p31 | IL23R (R381Q) | UC, psoriasis, AS | (10) |
| 735/368 | 498/1,032; 380b | 2q37 | ATG16L1 (T216A) | (11,13) | |||
| 547/928 | 1,266/559; 428b | 5p13 | PTGER4 (eQTL) | UC | (12) | ||
| 946/977 | 353/207, 530b | 10q21 | ZNF365 | GD, T1D | (13) | ||
| 1748/2938 | 1182/2024 | 3p21 | MST1 (R689C) | GD, RA, SLE, T1D | (9,14,44) | ||
| 3230/4829 (meta-analysis) | 2325/1809; 1339b | 5q33 | IRGM (20 kb deletion) | psoriasis | (30) | ||
| 10q24 | NKX2-3 | UC | |||||
| 18p11 | PTPN2 | ||||||
| 1p13 | PTPN22 (R620W) | ||||||
| 1q23 | ITLN1 | ||||||
| 1q24 | IL12B | ||||||
| 1q32 | CDKAL1 | ||||||
| 5q33 | CCR6 | ||||||
| 6p22 | JAK2 | ||||||
| 6p21 | C11orf30 | ||||||
| 6q21 | LRRK2, MUC19 | ||||||
| 6q27 | C13orf31 (V254I) | ||||||
| 7p12 | ORMDL3 (eQTL) | ||||||
| 8q24 | STAT3 | ||||||
| 9p24 | ICOSLG | ||||||
| 10p11 | |||||||
| 11q13 | |||||||
| 12q12 | |||||||
| 13q14 | |||||||
| 17q21 | |||||||
| 17q21 | |||||||
| 19p13 | |||||||
| 21q21 | |||||||
| 21q22 | |||||||
| Multiple | ∼20–40 | 0/2,431; 931a | 2322/2987; 609b | 5p13 | IL7RA (T244I) | (31) | |
| sclerosis | 10p15 | IL2RA | GD, T1D | ||||
| Rheumatoid | ∼5–10 | 1493/1831 | 1053/1858 | 9q34 | TRAF1-C5 | (35) | |
| arthritis | 397/1211 | 2283/3258 | 6q23 | TNFAIP3 | (9,34) | ||
| Systemic lupus | ∼30 | 720/2337 | 1846/1825 | 1q25 | (45) | ||
| erythematosus | 279/515 | 1757/1540 | 3p14 | PXK | (37) | ||
| 1311/3340 | 793/857 | 11p15 | KIAA1542 | (36) | |||
| 16p11 | ITGAM | ||||||
| Type-1 diabetes | ∼15 | 2029/1755 | 2471/4593; 2134b | 2q24 | IFIH1 (A946T) | GD, RA | (39) |
| 1963/2938 | 4000/5000; 2997b | 12q24 | SH2B3 (R262W) | CeD | (9,46) | ||
| 563/1,146; 483b | 364b; 549c | 12q13 | ERBB3 | CD, GD | (47,48) | ||
| 16p13 | CLEC16A | ||||||
| 18p11 | PTPN2 | ||||||
| 12q13 | ERBB3 | ||||||
| 16p13 | CLEC16A |
Generally, the effect size of the autoimmune disease risk variants identified through GWAS is small, increasing liability by ∼10–30%; this observation is also true for disease risk alleles associated with most non-autoimmune human complex diseases (9). When considering these published effect sizes, it is important to remember that a single SNP from each promising locus was often taken into the replication stage of the experimental design. It is, therefore, possible that many of these disease loci carry more than one functional disease variant, and that together multiple variants (common and rare) within each of the causal genes might have a larger effect on disease liability. Furthermore, in most cases, because we are not genotyping directly the causal marker(s) but we rely on linkage disequilibrium to test association to disease, we might underestimate effect sizes. Fully assessing effect size will require the identification and characterization of all causal markers at each of these disease loci.
Because the effect size of the DNA polymorphisms associated with complex human diseases is small, the major determinant in the identification of new genetic risk factors for autoimmune diseases by GWAS has been the number of patients and controls genotyped (Table 1). This is exemplified most convincingly by the progress made in the study of the genetics of CD. Four initial GWAS of CD scanned 547, 547, 946 and 1748 CD patients (9–14) and identified eight new risk loci for CD which, with the three previously reported associations (NOD2, IBD5 and TNFSF15) (15–18), brought the total of risk loci for CD to 11. In a large meta-analysis of three of these groups’ association results, Barrett et al. (19) reported the identification of an additional 21 new loci, bringing the total of CD-associated susceptibility loci to 32 (Table 1). This, and results from the analysis of other complex traits and diseases using GWAS (20–25), indicates that larger sample size results in more genetic discoveries. In theory, another factor that might influence the ease to find genetic risk factors for diseases is the heritability of these diseases. There is a hint that this is indeed the case for autoimmune diseases: when taking into account the number of individuals scanned by GWAS, more risk loci have been found for more heritable diseases (Table 1). For instance, analyzing 1890 RA patients and 778 CeD patients by GWAS identified two and eight new susceptibility loci for these diseases, respectively. CeD, with a sibling relative risk ratio (λs) of ∼30, is more heritable than RA (λs ∼5–10).
NEW GENETIC RISK FACTORS
The power of GWAS is to interrogate common genetic variation across the entire human genome in an unbiased manner. For autoimmune diseases, this has yielded very satisfying, but also surprising, discoveries, implicating both adaptive and innate immunity. Obviously, more work is required to confirm the causal gene(s) at each of these loci, but it is nevertheless informative to comment on some of these likely candidate functional genes.
Although cytokine signaling was thought to play a major role in the pathogenesis of many autoimmune diseases, there was no convincing evidence—with the exception of the association between a IL2RA SNP and T1D (26,27)—that common genetic variations at cytokine-related genes were genetic risk factors for these diseases before the GWAS era. There is now a plethora of such new associations, involving interleukin genes (or their receptors) with disease liability for CeD (28,29), CD (10,30), and MS (31–33), and tumor necrosis factor alpha (TNFα) signaling with RA (34,35) (Table 1). SLE being a primary disease affecting B cells, it is rewarding that two of the new SLE risk loci contain genes (BANK1 and BLK) that might be involved in B cell hyperactivation and production of autoantibodies (36,37) (Table 1). It now becomes essential to determine functionally how genetic variation within these genes modulates cytokine signaling or autoantibody synthesis, and how these modulations contribute to the etiology of autoimmune diseases (see below).
The innate immune system had previously been implicated in the development of CD. Two non-synonymous SNPs and one truncation variation in NOD2, which encodes a protein that mediates the interaction between microbes and the intestinal mucosa, were the first DNA sequence variants associated with CD risk (15,16). GWAS results have reinforced the importance of innate immunity in the pathogenesis of CD since two of the new CD disease loci include the autophagy genes ATG16L1 and IRGM, which likely defend the host against intestinal pathogens (13,14). Autophagy is a process by which cellular ‘debris’ are degraded by lysosomes, and can be used as an innate immune response to clear intracellular pathogens (38). The discovery of a non-synonymous SNP in the interferon-induced helicase IFIH1 gene suggested that innate immunity might also play a role in the development of T1D (39). This helicase can detect viral dsRNA; thus, it might be one of the answers to the reported correlation between viral infection and T1D susceptibility (40). In the coming years, it will be important to learn whether innate immunity is involved in the pathogenesis of other autoimmune diseases, and to resolve the extent to which adaptive and innate immunity interact together, and with the environment, to increase or decrease disease risk.
One of the most striking observations that can be made from for the list of DNA polymorphisms associated with risk of developing CeD, CD, MS, RA, SLE and T1D is that many of them cluster around the same loci (Fig. 2 and Table 1). For instance, SNPs around the protein tyrosine phosphatase PTPN22 gene have been convincingly associated with CD, RA, SLE, T1D as well as with Graves’ disease (GD), an autoimmune disease affecting the thyroid. In this particular example, we know that the same coding SNP (R602W) in PTPN22 is associated with CD, GD, RA, SLE and T1D, but that the specific allele that protects against CD (602W) actually predisposes to GD, RA, SLE and T1D (30,41). This clustering of genetic risk factors for many autoimmune diseases suggests that these diseases might share, at least partly, similar underlying causal mechanisms. It also offers a new strategy to identify additional genetic risk factors: if there are common disease mechanisms, combining GWAS results from different autoimmune diseases could provide sufficient statistical power to identify them.
Figure 2.
There is an overlap in the genetic risk loci for CeD, CD, MS, RA, SLE and T1D. This may suggest common mechanisms that lead to the development of autoimmune diseases. We did not include the MHC region in this network, but variants in this region are associated with all of these diseases.
REMAINING QUESTIONS
All the published GWAS for autoimmune studies have been performed in populations of European descent, often only in adults. It will be extremely informative to test whether these new risk alleles associate with disease susceptibility in other ethnic groups, in children, and whether they have stronger effect in men or women. These issues require additional work, but are otherwise relatively straightforward to address. More difficult is the identification and functional validation of the causal genes and alleles at each of these loci: currently, excellent candidates for actual causal sequence variants have been identified for only a small subset (<25%) of the loci listed in Table 1. Of tremendous importance if we are to understand how these sequence variants influence disease pathogenesis, this task will require very strong collaborations between human geneticists and experimental biologists. It will also require the establishment of bio-banks with large collection of relevant tissues and other bio-specimens to facilitate experiments in the appropriate live cell types or cellular environments. And then there is the study of other forms of genetic variation, rare or structural and their role in the etiology of complex autoimmune diseases. One great example is the recent discovery of a 20 kb deletion polymorphism that controls the expression of the autophagy IRGM gene and associates with CD (42). New technological development, such as next-generation sequencing technologies and improved genotyping platforms (43), should help answer the question about the importance of rare or structural variants in complex human diseases in the near future, although challenges remain on how to interpret data from these new technologies and how to leverage this information to establish causality of associated variants.
How can we use this genetic information to help autoimmune patients now? It is essential to know whether genotyping these SNPs can help predict whether someone is likely to develop one of these diseases. Although this is an avenue of research that should be pursued, it is a formidable task that will necessitate very large prospective cohorts and long epidemiological studies. On a more contemporary time scale, it would be valuable to know if these SNPs associate more strongly with forms of each of these diseases that are characterized by severe complications. In such case, diagnosed patients could be genotyped and, depending on the genotype score, a decision be made to introduce earlier more aggressive preventive therapies. The emergence of genetic risk factors that fall within distinct biological pathways (e.g. ATG16L1 and IRGM in autophagy or PTPN2 and PTPN22 in T-cell signaling for CD) also suggests the possibility to classify disease subtypes using molecular diagnostics. In turn, this provides rationale for different therapies being targeted to different patients based on genetic information rather than by trial and error.
CONCLUSION
The advent of GWAS approaches has had an unprecedented impact on our knowledge of the genetics of many autoimmune diseases, bringing to light many unexpected candidate genes and biological pathways. And there is more to come given that the current risk loci do not fully account for the genetic contribution to diseases susceptibility [e.g. 32 risk loci for CD explain one-fifth of the CD heritability (19)], and that large GWAS meta-analyses are in progress. Appropriately in this Olympic year, autoimmune disease geneticists are now beginning to relay the torch to experimental immunologists and clinicians to discover new pathological mechanisms and appropriate treatment strategies, and hopefully to bring comfort to autoimmune disease patients in the nearest possible future.
Conflict of Interest statement. None declared.
FUNDING
J.D.R. is funded by grants from the National Institutes of Allergy and Infectious Diseases (AI065687; AI067152), from the National Institute of Diabetes and Digestive and Kidney Diseases (DK064869; DK062432) and the Crohn's and Colitis Foundation of America (SRA512).
REFERENCES
- 1.Fernando M.M., Stevens C.R., Walsh E.C., De Jager P.L., Goyette P., Plenge R.M., Vyse T.J., Rioux J.D. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 2008;4:e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oksenberg J.R., Baranzini S.E., Sawcer S., Hauser S.L. The genetics of multiple sclerosis: SNPs to pathways to pathogenesis. Nat. Rev. Genet. 2008;9:516–526. doi: 10.1038/nrg2395. [DOI] [PubMed] [Google Scholar]
- 3.van Heel D.A., Hunt K., Greco L., Wijmenga C. Genetics in coeliac disease. Best Pract. Res. Clin. Gastroenterol. 2005;19:323–339. doi: 10.1016/j.bpg.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Cho J.H. The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 5.Gregersen P.K. Pathways to gene identification in rheumatoid arthritis: PTPN22 and beyond. Immunol. Rev. 2005;204:74–86. doi: 10.1111/j.0105-2896.2005.00243.x. [DOI] [PubMed] [Google Scholar]
- 6.Wakeland E.K., Liu K., Graham R.R., Behrens T.W. Delineating the genetic basis of systemic lupus erythematosus. Immunity. 2001;15:397–408. doi: 10.1016/s1074-7613(01)00201-1. [DOI] [PubMed] [Google Scholar]
- 7.Ounissi-Benkalha H., Polychronakos C. The molecular genetics of type 1 diabetes: new genes and emerging mechanisms. Trends Mol. Med. 2008;14:268–275. doi: 10.1016/j.molmed.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Chanock S.J., Manolio T., Boehnke M., Boerwinkle E., Hunter D.J., Thomas G., Hirschhorn J.N., Abecasis G., Altshuler D., Bailey-Wilson J.E., et al. Replicating genotype–phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 9.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duerr R.H., Taylor K.D., Brant S.R., Rioux J.D., Silverberg M.S., Daly M.J., Steinhart A.H., Abraham C., Regueiro M., Griffiths A., et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hampe J., Franke A., Rosenstiel P., Till A., Teuber M., Huse K., Albrecht M., Mayr G., De La Vega F.M., Briggs J., et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 12.Libioulle C., Louis E., Hansoul S., Sandor C., Farnir F., Franchimont D., Vermeire S., Dewit O., de Vos M., Dixon A., et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rioux J.D., Xavier R.J., Taylor K.D., Silverberg M.S., Goyette P., Huett A., Green T., Kuballa P., Barmada M.M., Datta L.W., et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkes M., Barrett J.C., Prescott N.J., Tremelling M., Anderson C.A., Fisher S.A., Roberts R.G., Nimmo E.R., Cummings F.R., Soars D., et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat. Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugot J.P., Chamaillard M., Zouali H., Lesage S., Cezard J.P., Belaiche J., Almer S., Tysk C., O'Morain C.A., Gassull M., et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 16.Ogura Y., Bonen D.K., Inohara N., Nicolae D.L., Chen F.F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R.H., et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 17.Rioux J.D., Daly M.J., Silverberg M.S., Lindblad K., Steinhart H., Cohen Z., Delmonte T., Kocher K., Miller K., Guschwan S., et al. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat. Genet. 2001;29:223–228. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
- 18.Yamazaki K., McGovern D., Ragoussis J., Paolucci M., Butler H., Jewell D., Cardon L., Takazoe M., Tanaka T., Ichimori T., et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn's disease. Hum. Mol. Genet. 2005;14:3499–3506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- 19.Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudbjartsson D.F., Walters G.B., Thorleifsson G., Stefansson H., Halldorsson B.V., Zusmanovich P., Sulem P., Thorlacius S., Gylfason A., Steinberg S., et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 21.Lettre G., Jackson A.U., Gieger C., Schumacher F.R., Berndt S.I., Sanna S., Eyheramendy S., Voight B.F., Butler J.L., Guiducci C., et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weedon M.N., Lango H., Lindgren C.M., Wallace C., Evans D.M., Mangino M., Freathy R.M., Perry J.R., Stevens S., Hall A.S., et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer C.J., Sanna S., Jackson A.U., Scuteri A., Bonnycastle L.L., Clarke R., Heath S.C., Timpson N.J., Najjar S.S., Stringham H.M., et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N.P., Rieder M.J., Cooper G.M., Roos C., Voight B.F., Havulinna A.S., et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeggini E., Scott L.J., Saxena R., Voight B.F., Marchini J.L., Hu T., de Bakker P.I., Abecasis G.R., Almgren P., Andersen G., et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vella A., Cooper J.D., Lowe C.E., Walker N., Nutland S., Widmer B., Jones R., Ring S.M., McArdle W., Pembrey M.E., et al. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am. J. Hum. Genet. 2005;76:773–779. doi: 10.1086/429843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe C.E., Cooper J.D., Brusko T., Walker N.M., Smyth D.J., Bailey R., Bourget K., Plagnol V., Field S., Atkinson M., et al. Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat. Genet. 2007;39:1074–1082. doi: 10.1038/ng2102. [DOI] [PubMed] [Google Scholar]
- 28.van Heel D.A., Franke L., Hunt K.A., Gwilliam R., Zhernakova A., Inouye M., Wapenaar M.C., Barnardo M.C., Bethel G., Holmes G.K., et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat. Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt K.A., Zhernakova A., Turner G., Heap G.A., Franke L., Bruinenberg M., Romanos J., Dinesen L.C., Ryan A.W., Panesar D., et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., et al. Genome-wide association defines more than thirty distinct susceptibility loci for Crohn's disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hafler D.A., Compston A., Sawcer S., Lander E.S., Daly M.J., De Jager P.L., de Bakker P.I., Gabriel S.B., Mirel D.B., Ivinson A.J., et al. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 32.Gregory S.G., Schmidt S., Seth P., Oksenberg J.R., Hart J., Prokop A., Caillier S.J., Ban M., Goris A., Barcellos L.F., et al. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat. Genet. 2007;39:1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- 33.Lundmark F., Duvefelt K., Iacobaeus E., Kockum I., Wallstrom E., Khademi M., Oturai A., Ryder L.P., Saarela J., Harbo H.F., et al. Variation in interleukin 7 receptor alpha chain (IL7R) influences risk of multiple sclerosis. Nat. Genet. 2007;39:1108–1113. doi: 10.1038/ng2106. [DOI] [PubMed] [Google Scholar]
- 34.Plenge R.M., Cotsapas C., Davies L., Price A.L., de Bakker P.I., Maller J., Pe'er I., Burtt N.P., Blumenstiel B., DeFelice M., et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat. Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plenge R.M., Seielstad M., Padyukov L., Lee A.T., Remmers E.F., Ding B., Liew A., Khalili H., Chandrasekaran A., Davies L.R., et al. TRAF1-C5 as a risk locus for rheumatoid arthritis—a genomewide study. N. Engl. J. Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hom G., Graham R.R., Modrek B., Taylor K.E., Ortmann W., Garnier S., Lee A.T., Chung S.A., Ferreira R.C., Pant P.V., et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 37.Kozyrev S.V., Abelson A.K., Wojcik J., Zaghlool A., Linga Reddy M.V., Sanchez E., Gunnarsson I., Svenungsson E., Sturfelt G., Jonsen A., et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat. Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 38.Levine B., Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth D.J., Cooper J.D., Bailey R., Field S., Burren O., Smink L.J., Guja C., Ionescu-Tirgoviste C., Widmer B., Dunger D.B., et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat. Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 40.Knip M., Veijola R., Virtanen S.M., Hyoty H., Vaarala O., Akerblom H.K. Environmental triggers and determinants of type 1 diabetes. Diabetes. 2005;54(Suppl. 2):S125–S136. doi: 10.2337/diabetes.54.suppl_2.s125. [DOI] [PubMed] [Google Scholar]
- 41.Vang T., Congia M., Macis M.D., Musumeci L., Orru V., Zavattari P., Nika K., Tautz L., Tasken K., Cucca F., et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat. Genet. 2005;37:1317–1319. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 42.McCarroll S.A., Huett A.S., Kuballa P., Chilewski S.D., Landry A., Goyette P., Zody M.C., Hall J.L., Brant S.R., Cho J.H., et al. Deletion polymorphism upstream of IRGM is associated with altered IRGM expression and Crohn's disease. Nat. Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCarroll S.A., Kuruvilla F.G., Korn J.M., Cawley S., Nemesh J., Wysoker A., Shapero M.H., de Bakker P.I.W., Maller J.B., Kirby A., et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008 doi: 10.1038/ng.238. published online 6 September 2008, doi:10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 44.Goyette P., Lefebvre C.N.G.A., Brant S.R., Cho J.H., Duerr R.H., Silverberg M.S., Taylor K.D., Latiano A., Aumais G., et al. Gene-centric association mapping of chromosome 3p implicates MST1 in IBD pathogenesis. Mucosal. Immunol. 2008;1:131–138. doi: 10.1038/mi.2007.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harley J.B., Alarcon-Riquelme M.E., Criswell L.A., Jacob C.O., Kimberly R.P., Moser K.L., Tsao B.P., Vyse T.J., Langefeld C.D., Nath S.K., et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Todd J.A., Walker N.M., Cooper J.D., Smyth D.J., Downes K., Plagnol V., Bailey R., Nejentsev S., Field S.F., Payne F., et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hakonarson H., Grant S.F., Bradfield J.P., Marchand L., Kim C.E., Glessner J.T., Grabs R., Casalunovo T., Taback S.P., Frackelton E.C., et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 48.Hakonarson H., Qu H.Q., Bradfield J.P., Marchand L., Kim C.E., Glessner J.T., Grabs R., Casalunovo T., Taback S.P., Frackelton E.C., et al. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes. 2008;57:1143–1146. doi: 10.2337/db07-1305. [DOI] [PubMed] [Google Scholar]