Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome (original) (raw)
Abstract
Isogenic settings are routine in model organisms, yet remain elusive for genetic experiments on human cells. We describe the use of designed zinc finger nucleases (ZFNs) for efficient transgenesis without drug selection into the PPP1R12C gene, a “safe harbor” locus known as AAVS1. ZFNs enable targeted transgenesis at a frequency of up to 15% following transient transfection of both transformed and primary human cells, including fibroblasts and hES cells. When added to this locus, transgenes such as expression cassettes for shRNAs, small-molecule-responsive cDNA expression cassettes, and reporter constructs, exhibit consistent expression and sustained function over 50 cell generations. By avoiding random integration and drug selection, this method allows bona fide isogenic settings for high-throughput functional genomics, proteomics, and regulatory DNA analysis in essentially any transformed human cell type and in primary cells.
Transgenesis of human cells is used in functional genomics, proteomics, protein structure-function studies, and cell-based drug discovery, and is routinely accomplished by random integration combined with drug selection. Expression of a randomly integrated transgene is unpredictable and tends to be unstable over time due to epigenetic effects. Further, random integration often yields multiple integrants per cell, and this can result in the disruption or activation of host cell genes. Such unintended side effects produce a non-isogenic experimental setting, and this can confound data analysis and interpretation.
Work from the Soriano laboratory (Zambrowicz et al. 1997) has established Rosa26 as the standard locus for transgenesis via gene targeting (Thomas et al. 1986) in mouse embryonic stem cells. In the present study, we set out to develop a complementary approach for human somatic cell genetics, a research system where classical gene targeting remains challenging due to the relatively low frequency of correctly targeted events (e.g., Ruis et al. 2008). We turned to genome editing with designed zinc finger nucleases (ZFNs) (bottom of Fig. 1A; for review, see Carroll 2008). This technique emerged from pioneering work by the Jasin, Pabo, Klug, and Carroll laboratories (Choo and Klug 1994; Rebar and Pabo 1994; Rouet et al. 1994; Bibikova et al. 2001), and represents a generally applicable method for the efficient alteration of the genome in a targeted fashion. While applied most recently to drive multilocus gene disruption in mammalian tissue culture cells (Liu et al. 2010) and to make an allelic series of histone variant genes in mouse cells (Goldberg et al. 2010), ZFNs have also been used to drive targeted gene addition to investigator-specified endogenous loci in human cells (Lombardo et al. 2007; Moehle et al. 2007; Hockemeyer et al. 2009). ZFNs used for the first editing experiments (Bibikova et al. 2001) relied on three-finger zinc finger proteins and conventional endonuclease domains; the recent development of methods for engineering of ZFNs with composite recognition sites of up to 36 bp and carrying high-fidelity endonuclease domains has enabled highly specific genome editing in both transformed and primary human cells (Miller et al. 2007; Perez et al. 2008; Hockemeyer et al. 2009).
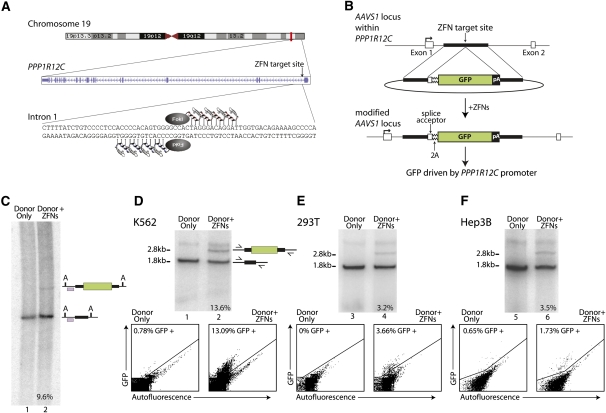
Figure 1.
ZFN-driven ORF addition to the PPP1R12C gene (also known as the AAVS1 locus) in various transformed cell types: (A) Schematic of the human PPP1R12C gene (http://www.genome.ucsc.edu), with the exon/intron structure and the ZFN target site indicated. (B) Schematic of donor construct and of the AAVS1 locus following GFP marker ORF addition. The first two exons of the PPP1R12C gene are shown as open boxes. Also annotated are the locations of the splice acceptor site, the 2A ribosome stuttering signal, and a polyadenylation signal (pA). (C) Southern blotting confirms efficient ZFN-dependent ORF addition to the AAVS1 locus in K562 cells. The positions of wild-type and transgenic chromatids are indicated to the right of the gel; the percentage of transgenic chromatids in this cell pool is indicated below lane 2. The PhosphorImager traces used for the quantitation are shown in Supplemental Figure 1. The probe used for Southern blotting, which corresponds to positions chr19:55,628,340–55,628,753 (GRCh37/hg19) is indicated as a purple-filled box; “A” indicates recognition sites for AccI that genomic DNA was cut with for this Southern. (D) Efficient ZFN-driven GFP ORF addition to AAVS1 in K562 cells. Results of a semiquantitative, body-labeled PCR-based assay (see Methods) on cells transfected with the indicated constructs are shown. Primers are located outside of the homology arms and are indicated on the schematic to the right of the gel. The positions of wild-type and transgenic chromatids are indicated to the right of the gel. The frequency of genome-edited chromatids is indicated below each lane. In this assay, when applied to this locus, weak nonspecific incorporation during early PCR cycles produces a band that appears in all samples and migrates above the one generated by the transgenic chromatid. The data below the autoradiograph represent analysis of the frequency of GFP-positive cells by FACS in the same cells genotyped above. (E) As in D, except HEK293T cells were used. (F) As in D, except Hep3B cells were used.
Therefore, we reasoned that ZFNs could be used to enable an isogenic setting for human cell transgenesis if a suitable genomic location could be found. Selection of such a locus to act as a “safe harbor” for ZFN-driven gene addition was guided by the observation that integration of adeno-associated virus (AAV) into the human genome disrupts the PPP1R12C gene on chromosome 19 (the locus is commonly referred to as AAVS1) (Kotin et al. 1992; Tan et al. 2001). AAV infection is not associated with a known pathophysiology (Smith et al. 2008); both hES (Smith et al. 2008; Hockemeyer et al. 2009) and hiPS (Hockemeyer et al. 2009) cells with a disruption of PPP1R12C retain pluripotency. Further, this gene is transcribed in all primary human cells studied (http://www.biogps.gnf.org) as well as in commonly used transformed cell lines, i.e., HEK293, K562, HeLa, DU-145, and Hep3B (RC DeKelver, data not shown). Thus, the AAVS1 locus fits two criteria for a transgene “safe harbor”: (1) no known adverse effect on the cell resulting from its disruption, and (2) transcriptional competence across cell types to maintain expression from an inserted gene cassette(s).
We describe here a method for using ZFNs to achieve rapid, efficient transgenesis into the AAVS1 locus in many commonly used human transformed cell types (K562, HeLa, HEK293, U2OS, and others) in human fibroblasts and in hES cells. The gene addition process occurs following simple treatment of the cells with the ZFN/donor-carrying plasmid and produces a pool of cells harboring the donor-specified novel DNA at the ZFN-specified location. Both promoterless (i.e., reliant on the native PPP1R12C gene promoter) and promoter-containing inserts placed into the AAVS1 locus exhibit consistent levels of expression over extended passaging in culture.
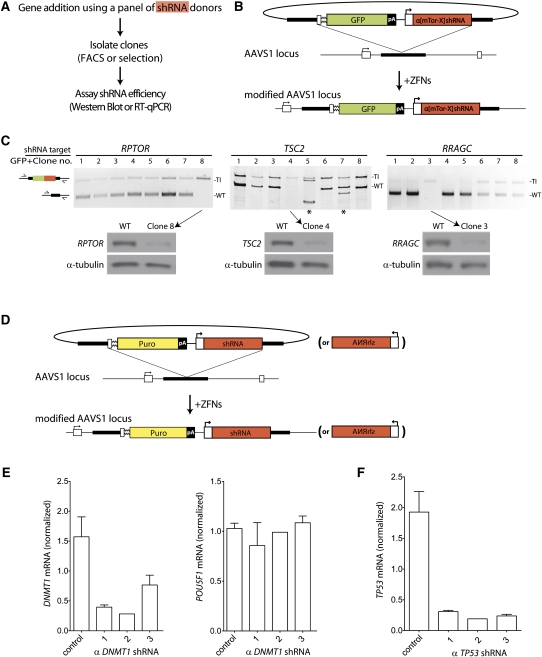
We describe three applications of this approach by way of illustrating some of the applications of isogenic transgenesis in human cells: (1) We use these ZFNs to construct, in a single step, an hES cell line transheterogygous at the AAVS1 locus for four distinct genetic entities that together allow inducible gene expression; (2) we construct a panel of isogenic U2OS cell lines, each carrying at _AAVS1_—i.e., in the same chromatin environment—a different reporter responsive to activation by liganded glucocorticoid receptor; (3) we generate a panel of isogenic HEK293 cell lines, each carrying at AAVS1 a distinct shRNA expression cassette directed against a component of the mTOR pathway, and each exhibiting long-term knockdown of the protein targeted by the small hairpin RNA (shRNA); we perform the same experiment—but now using shRNAs directed against TP53 or _DNMT1_—in hES cells, and observe comparably robust target gene mRNA knockdown by the shRNA.
Results
ZFN-driven open reading frame (ORF) addition to the AAVS1 locus: Use in distinct transformed and primary cell types, and expression stability over time
The PPP1R12C gene is transcribed in all cell types where this issue has been studied, and this allows the use of promoterless donor constructs. We designed a panel of ZFNs against nonrepetitive stretches of its intron 1 using an archive of prevalidated two-finger modules and screened this panel for endogenous gene disruption (RC DeKelver and JM Rock, data not shown). The most active ZFN pair introduces a double-strand break (DSB) ∼1800 bp downstream from the transcription start site of the PPP1R12C gene (Fig. 1A). The composite ZFN recognition site is unique in the human genome and is flanked by an extended stretch of single-copy genomic DNA suitable for donor construction.
To trap expression driven by the native PPP1R12C promoter, the donor construct used (Fig. 1B) contained two 800-bp stretches of sequence homologous to the region flanking the ZFN site interrupted by a promoterless green fluorescent protein (GFP) ORF and a polyadenylation signal. Since exon 1 contains the translational start site of AAVS1, the donor included a splice acceptor site, followed by the 2A ribosome stuttering signal (Fang et al. 2005) upstream of the GFP ORF (Fig. 1B). Addition of this cassette to intron 1 of the PPP1R12C gene yields a single transcript driven by the native promoter (Fig. 1B), translation of which produces the polypeptide encoded by exon 1 of PPP1R12C and, separately, GFP.
Genotyping of K562 cells 48 h following transient transfection with plasmid DNAs encoding the ZFN and donor DNA construct demonstrated that ∼10% of all AAVS1 chromatids in the cell population had acquired the donor-specified ORF cassette as gauged by Southern blotting (Fig. 1C; Supplemental Fig. 1) and a semiquantitative body-labeled PCR assay using primers that anneal to the chromosome outside of the region of homology with the donor (Fig. 1D; see Methods for experimental details). Sequencing of the transgenic chromatid from randomly chosen single-cell-derived clones carrying donor-specified transgenes at AAVS1 showed that transgenesis occurred in a homology-dependent fashion (e.g., Supplemental Fig. 2). We cannot exclude the possibility that in a fraction of cases, gene addition occurs via a process that uses both homology and end-joining (Richardson and Jasin 2000), but we have not observed such an outcome using the AAVS1 ZFNs and plasmid donors delivered to transformed cells using nucleofection. In agreement with our earlier data on the use of these and other ZFNs engineered to recognize extended DNA stretches and carrying high-fidelity FokI domains (Miller et al. 2007; Hockemeyer et al. 2009), a nucleus-wide survey of DSB frequency and measurement of donor plasmid random integration rates revealed high specificity of ZFN-driven gene addition (Supplemental Figs. 3, 8; Supplemental Discussion).
Control- and ZFN-treated cells were maintained in culture for 1 mo (∼35 population doublings) in the absence of selection and then analyzed by FACS. Consistent with the genotyping data, 13% GFP-positive cells were observed in the ZFN- and donor-treated cell pool (Fig. 1D, bottom right). Less than 1% of the cells treated with the donor plasmid alone expressed GFP (Fig. 1D, bottom left), and no chromatids transgenic for GFP at AAVS1 were detected in that sample by Southern blotting or by PCR (lanes 1 in Fig. 1, C and D, respectively). This showed that ZFN-driven addition of a promoterless GFP ORF to the human AAVS1 locus yields ∼10% GFP-positive K562 cells without selection for the desired event.
The same ZFN-donor combination was then used in HEK293 and Hep3B cells. In both cases, ORF addition frequencies as gauged by genotyping (Fig. 1E,F, top) and phenotyping (bottom) were ∼3% in the absence of selection and after 1 mo of passaging in culture. We next tested these ZFNs in a larger panel of transformed cells along with a donor DNA plasmid with homology arms flanking a 50-bp heterologous stretch with a novel RFLP (this arrangement allows for accurate measurement of ZFN-driven addition frequency in a nonradioactive assay) (Supplemental Fig. 4). With this approach, editing efficiency comparable to that seen in K562 cells was observed in HCT116 and U2OS cells (Supplemental Fig. 4, lanes 3,11), and greater than 1% transgenic chromatids were observed in A549, DU145, HeLa, HepG2, IMR90, and LNCap cells. Differences between cell lines in gene-addition frequency could potentially result from those in ZFN expression levels, in the epigenetic state of the AAVS1 locus, and in cell-type-specific bias for DSB repair pathways. To determine whether these ZFNs could drive gene addition in primary human cells, we generated integration-defective lentiviruses carrying the ZFNs and the donor (Lombardo et al. 2007); infection of hTERT-immortalized human fibroblasts (Rubio et al. 2002) resulted in ∼3% ORF addition to the AAVS1 locus as gauged both by genotyping and phenotyping (Supplemental Fig. 5, A and B, respectively).
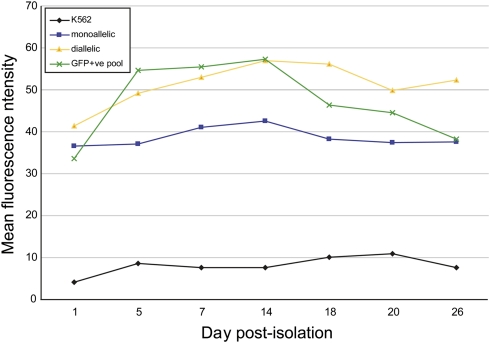
In the experiment shown in Figure 1 and Supplemental Figure 5, pools of ZFN-edited K562, HEK293, Hep3b cells, and fibroblasts remained marker positive after 1 mo of continuous passaging in culture. To more accurately measure the expression stability over time of a transgene resident in the AAVS1 locus, we used FACS to isolate GFP-positive K562 cells after transfection with the ZFN-donor combination shown in Figure 1B. Approximately 80% of the chromatids in that pool were found to be transgenic for the GFP ORF (Supplemental Fig. 7B; Supplemental Discussion). Limiting dilution without additional sorting for GFP expression generated a panel of single-cell-derived clonal lines exclusively carrying GFP in a monoallelic or diallelic state (RC DeKelver and EA Moehle, data not shown). This analysis showed that the ∼20% of nontransgenic chromatids in the GFP-positive pool (Supplemental Fig. 7B) derive from cells with a monoallelic GFP transgene at AAVS1. Representative control K562 cells, the GFP-positive FACS-enriched cell pool, and two single-cell clonal lines, monoallelic and diallelic for a GFP insertion at AAVS1, were grown for 50 cell doublings and assayed for GFP expression level biweekly. This analysis (Fig. 2) revealed: (1) no loss of mean fluorescence intensity over the course of the experiment; (2) consistently higher mean fluorescence intensity of cells diallelic for the GFP cassette at AAVS1 than monoallelic (cf. square- and diamond-annotated lines in Fig. 2). These data showed that ZFN-driven GFP-ORF addition to the AAVS1 gene locus results in stable long-term expression of the introduced transgene in transformed cells.
Figure 2.
Stability of GFP expression in K562 cells when driven by the endogenous PPP1R12C promoter. Shown is the mean fluorescence intensity of a GFP-positive cell pool (green X's), a clone derived by limiting dilution that is monoallelic at AAVS1 for the GFP ORF (blue squares), diallelic (yellow triangles), and negative control cells (black diamonds) measured over 25 d—∼30 cell doublings—of growth in nonselective medium. After 25 more days of passaging, the MFI remained essentially unchanged (VM Choi and EA Moehle, data not shown).
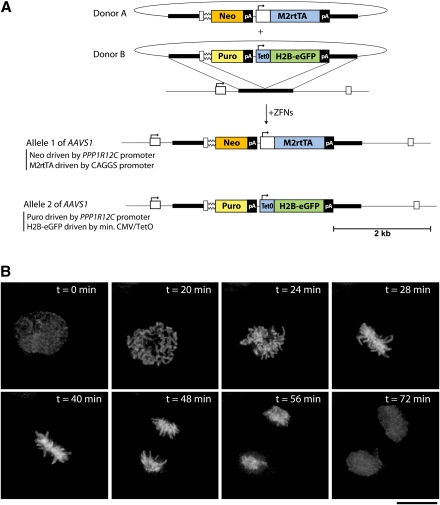
To demonstrate gene addition to the “safe harbor” in genetically unmodified primary cells, we turned to human embryonic stem cells (hESCs). DNA delivery to hESCs using electroporation is inefficient (<5%), and we included selectable markers in donor constructs solely to enrich for edited cells. In this experiment (Fig. 3) we made use of the fact that AAVS1 is autosomal; hence, a euploid cell provides two distinct cytogenetic locations for transgenes. Human embryonic stem cells (ESCs) were simultaneously treated with ZFNs and two distinct donors (Fig. 3A), each carrying a distinct promoterless selectable marker followed by an expression cassette for the tet reverse transactivator (Donor A) or histone H2B fused to eGFP (Donor B). Single-cell-derived clones resistant to both selection agents were genotyped and found to be _trans_-heterozygous for the two donor-specified cassettes at the AAVS1 locus (D Hockemeyer, data not shown). Following doxycyline treatment, uniform intranuclear fluorescence was observed. The cells retained normal hESC morphology and divided normally (Fig. 3B; Supplemental movie M1), with the H2B-eGFP fusion protein revealing condensed chromosomes during M phase. In these hESCs, therefore, the AAVS1 locus supported long-term function by four distinct expression constructs: two promoterless resistance markers driven by the PPP1R12C promoter, and two separate promoter-transcription units.
Figure 3.
Gene addition to the AAVS1 locus in hESCs using ZFNs. (A) Schematic overview depicting the editing strategy for both alleles of the PPP1R12C gene. Donor plasmids used to target the locus are shown above; gene elements are represented as in Figure 1B. (Puro) Puromycin resistance gene, (CAGGS) constitutively active CAGGS promoter, (M2rtTA) tetracycline reverse transactivator. (B) Still images from a time lapse movie (see Supplemental movie M1) imaging H2B-eGFP in a representative BGO1 cell targeted with the AAVS1 donor plasmids as shown in A. Scale bar, 10 μm.
Taken together, these data showed that (1) ZFNs can be used to add transgenes to the AAVS1 locus at a high frequency in a broad range of transformed and primary cell types; (2) the added transgene, when driven by the native PPP1R12C promoter, is expressed at a stable level over an extended time period in cell culture.
An isogenic panel of U2OS cells carrying glucocorticoid receptor response element reporter constructs
Next, we set out to determine whether the classical assay in gene promoter structure-function studies—transient transfection with reporter constructs—that has been used to study, among other things, allosteric effects of DNA on function by nuclear hormone receptors (NHRs) (Meijsing et al. 2009) can be performed at an isogenic chromosomal location. In this case, integration at the same genomic locus could allow a direct comparison of reporters in a defined genomic context with predetermined differences in the cis elements that orchestrate transcriptional activation by the glucocorticoid receptor (GR), an extensively studied member of the NHR superfamily that forms part of a complex response circuit to corticosteroids (Hager et al. 2009). Using this approach, the functional consequences of deleting or changing the exact sequence of a GR binding site (GBS) and the presence or absence of cis elements that functionally interact with the GBS can be assayed. Furthermore, recruitment of GR to such integrated reporters can be determined by chromatin immunoprecipitation (ChIP), an assay that is challenging to perform in transient transfection experiments.
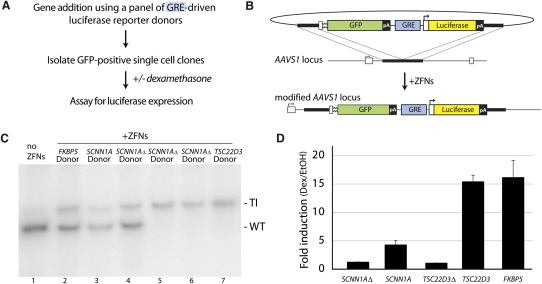
A panel of donor constructs was assembled (Fig. 4A), each harboring a distinct GR response element (GRE) of ∼1000 bp derived from endogenous GR target genes (Gerber et al. 2009) upstream of a basal promoter (FKBP5 and TSC22D3, alias GILZ) or harboring its own endogenous promoter (SCNN1A, alias ENAC), driving the expression of a luciferase reporter gene. In all cases, the donor retains the promoterless GFP cassette, such that if site-specific gene addition is successful, GFP expression, which is driven by the native PPP1R12C promoter, would be indicative of the presence of the reporter construct at the same locus.
Figure 4.
Use of ZFNs to generate a panel of U2OS cells carrying glucocorticoid receptor reporter constructs at AAVS1, and their functional characterization. (A) Outline of experiment. (B) Schematic of donor design and of the AAVS1 locus following GFP marker/GRE luciferase reporter addition. Gene elements are represented in the same way as in Figure 1B. (C) Genotype at th_e AAVS1_ locus for clones carrying reporters with GREs derived from genes indicated above each lane. The position of the wild-type (WT) and transgenic (TI) chromatid is indicated to the right of the gel. The “_SCNN1A_Δ” and “ _TSC22D3_Δ ” donors have the GR binding site deleted by site directed mutagenesis. Note that the transgenic chromatid is amplified less efficiently than the wild-type one due to a difference in size (see Supplemental Discussion). (D) The single-cell-derived clones genotyped in C were treated with vehicle (EtOH) or dexamethasone (dex); induction of the luciferase reporter was measured and is shown as fold activation by dex over treated with vehicle only.
U2OS cells expressing GR (Rogatsky et al. 1997) were transiently transfected with _AAVS1_-targeting ZFNs and each donor construct; GFP-positive cell pools for each donor were isolated, single-cell-derived clones produced without drug selection, and genotyped at the AAVS1 locus. In agreement with data on single-insert cassettes (Fig. 1), of nine randomly chosen single-cell-derived clones that expressed the luciferase reporter, all nine carried the desired donor-specified reporter cassette at the AAVS1 locus in either a monoallelic or diallelic configuration (Fig. 4C). Representative clones carrying each reporter were then treated with dexamethasone, a potent synthetic ligand for the GR. The reporters recapitulated (Fig. 4D) hormone-dependent activation of the endogenous target genes from which the regulatory sequence was derived (Gerber et al. 2009) and displayed distinct magnitudes of transcriptional up-regulation upon ligand treatment (Fig. 4D, cf. “_SCNN1A_” and “_FKBP5_” samples). Moreover, ligand-dependent activation was dependent on the presence of an intact GR binding site, as deleting the GR binding site of the SCNN1A GRE (Sayegh et al. 1999) ablated ligand-dependent transcriptional activation (Fig. 4D, sample “_SCNN1A_Δ”); the same observation was made with a line carrying the TSC22D3 promoter lacking GREs (Fig. 4D, sample “_TSC22D3_Δ”). Further, hormone-dependent recruitment of GR to the reporter constructs was observed by chromatin immunoprecipitation assays (Supplemental Fig. 6).
Taken together, the data show that the AAVS1 locus can be used to rapidly construct isogenic panels of cells harboring chromosomal reporters of steroid hormone receptor function that recruit the receptor and exhibit a biologically relevant response to hormone.
Isogenic panels of K562, HEK293, and hES cells carrying functional shRNA cassettes
Next, we set out to determine whether the AAVS1 locus would allow the long-term function of shRNA expression cassettes. In budding yeast, genome-wide reverse genetics experiments are conducted by systematic gene knockout in an isogenic background (Giaever et al. 2002). The recent development of several whole-genome collections of expression vectors for shRNAs (Paddison et al. 2004; Root et al. 2006; Berns et al. 2007) has, for the first time, allowed loss-of-function screens in human cells. These are currently performed in transient settings or by the random integration of the shRNA construct via lentivirus transgenesis.
In a pilot study (Supplemental Fig. 7), we used a donor construct carrying promoterless GFP upstream of a U6-promoter-driven shRNA expression cassette targeting the cell surface marker CD58 (known to be expressed in K562 cells). Cells were transfected with _AAVS1_-targeting ZFNs and this donor, GFP-positive cells (∼8% of total cells; EA Moehle, data not shown) isolated by FACS, and this cell pool was found to contain ∼80% chromatids transgenic at AAVS1 (Supplemental Fig. 7B, lane 3; see also Supplemental Discussion). To evaluate the efficacy of the inserted shRNA cassette, we compared control cells and AAVS1 ZFN/donor modified cells by FACS staining for CD58—the shRNA target molecule (Supplemental Fig. 7C). Cell-surface staining for CD58 was significantly reduced even after 30 cell population doublings (Supplemental Fig. 7C, last sample) and was comparable in magnitude to that seen 48 h post-transient transfection with the shRNA expression plasmid itself (Supplemental Fig. 7C, third sample); cells carrying GFP at the AAVS1 locus were indistinguishable in their CD58 levels from control cells (Supplemental Fig. 7C).
We next constructed a panel of donor plasmids (Fig. 5B), each harboring an expression cassette for a shRNA directed against three distinct genes in the mammalian target of rapamycin (mTOR) pathway (Sancak et al. 2008). Plasmids encoding the _AAVS1_-specific ZFNs and the individual donor DNA constructs were transfected into HEK293 cells, and GFP-positive cells isolated by FACS. Genotyping of each cell pool revealed significant single-step enrichment for cells carrying each shRNA at the AAVS1 locus (RC DeKelver and VM Choi, data not shown).
Figure 5.
Use of ZFNs to generate a panel of isogenic HEK293 cells and hESCs carrying distinct shRNA expression cassettes at the AAVS1 locus. (A) Experimental outline. (B) Schematic of donor construct for the HEK293 experiment. Gene elements are represented in the same way as in Figure 1B. shRNA cassettes are driven by a pol III promoter, which is annotated as a white box with an arrow, followed by a red box. (C) Genotypes of the AAVS1 locus (top) and protein expression (bottom) of a panel of single-cell derived HEK293 cell clones. The clones were obtained by FACS and genotyped using primers that lie outside the region of homology with the donor construct (schematic of PCR to the left of the autoradiograph). In all cases, the upper band corresponds to the transgenic, and the lower to the wild-type chromatid, respectively. In a small subset of cases (indicated by asterisks), the clone contains an additional allele of the AAVS1 locus, most likely the result of a DSB-induced deletion. The indicated clones were assayed by Western blot (bottom) for levels of proteins encoded by genes targeted by the indicated shRNAs. A Western blot for a loading control (α-tubulin) is shown at the bottom. (D) Schematic overview depicting the editing strategy for adding shRNA expression cassettes to the AAVS1 locus in hES cells. Annotations are as in Figure 1B. (E) Real-time PCR data (normalized to GAPDH mRNA) for DNMT1 (left) or POU5F1 (right) in pools of single-cell-derived hES clones carrying control shRNAs or three distinct shRNAs directed against DNMT1. See Supplemental Figure 8 for Southern blot clone genotyping data. (F) As in panel E, but with TP53 as the shRNA target. Note that in E and F, for shRNA construct no. 2 for each gene target, a single-cell-derived hES clone was analyzed in this experiment.
Western blotting of the pool of GFP-positive cells was performed to assess protein levels for the shRNA-targeted gene products. For two of the three shRNA targets studied (TSC2 and RPTOR) we observed lower target protein levels than control cells (Y Sancak and DM Sabatini, data not shown). This was not the case for either of the _RRAGC_-targeting shRNA constructs we tested, despite comparable transgenesis levels. We next isolated and genotyped a panel of GFP-positive single-cell-derived clones for each shRNA and identified clonal cell lines monoallelic and diallelic for gene addition at AAVS1 (Fig. 5C, top). In the case of all three gene targets, single-cell-derived clones carrying diallelic gene addition of the relevant shRNA cassette exhibited a significant knockdown of the target protein level (Fig. 5C, bottom). Of note, ∼8 wk of continued passaging had elapsed since the cells were initially treated with the ZFNs and the donor cassette. Since ZFN-driven gene addition occurs within 24–48 h of transient transfection, these data indicate that the AAVS1 locus provides a suitable genomic environment for long-term function by shRNA expression cassettes.
In earlier work (Hockemeyer et al. 2009) and in Figure 3 we have demonstrated the feasibility of ZFN-driven addition of transgenes carrying RNA pol II promoters to AAVS1 in hiPS and hES cells. We next asked whether the AAVS1 locus could function as a safe harbor for shRNA expression cassettes in hESCs. Single-cell-derived hESC clones carrying shRNAs directed against TP53, DNMT1, or control shRNAs were generated using ZFNs and donor constructs (Fig. 5D). In agreement with nucleus-wide data from transformed cells (Supplemental Fig. 3), Southern blotting showed that >90% of hESC clones that carry the desired transgene at AAVS1 lack additional random donor integrants (Supplemental Fig. 8; Supplemental Discussion). Clones were expanded, mRNA isolated, and expression levels of the genes targeted by the shRNAs measured (Fig. 5E,F). In the case of both genes, single-cell-derived clones carrying target-specific, but not control shRNA expression cassettes, exhibited stable long-term knockdown of mRNA levels (Fig. 5E,F); importantly, hESC clones carrying _DNMT1_-targeted shRNAs transcribed POU5F1, a key marker of pluripotency, to the same level as control cells (Fig. 5E, right).
Taken together, these data reveal the feasibility of using a ZFN-driven “safe harbor” gene addition process to rapidly, and in some cases without drug selection, obtain a panel of isogenic cells carrying investigator-specified shRNAs at a defined location, and exhibiting robust knockdown of the mRNA (Fig. 5E) or protein (Fig. 5C) encoded by the gene that the shRNA targets.
Discussion
Isogenic settings are standard in experimental systems such as budding yeast, Drosophila, and laboratory mice, where they can be achieved via well-established protocols (e.g., see Osborne et al. 2009). The work we describe adds transgenesis in human transformed and primary cells to the list of such systems. Gene addition occurs in a single step within 48 h of transient transfection, relies on plasmid DNA constructs, functions across a range of transformed human cell types commonly used in laboratory practice, in fibroblasts and hES cells (this work) and in induced pluripotent stem (iPS) cells (Hockemeyer et al. 2009). Constructs relying on the native PPP1R12C gene promoter, as well as carrying their own pol II and pol III promoters, function when placed into the AAVS1 locus over extended passaging in culture. The method we describe provides a complement to other techniques available for transgene placement into the same location in human cells, including two-step recombination-based cassette exchange (Sauer and Henderson 1988) and gene targeting with recombinant AAV vectors (Kohli et al. 2004). The frequency of the addition process observed with ZFNs often obviates the need for a drug selection step on the path to obtaining the cells of interest, which is required with other approaches. In further contrast, the use of ZFNs and donors carrying promoterless markers allows the rapid, single-step isolation of a cell pool or single-cell-derived clones that carry the marker-linked transgene at the desired location (e.g., Figs. 4, 5).
Earlier data on gene targeting (Smith et al. 2008) or ZFN-driven gene addition to AAVS1 (Hockemeyer et al. 2009) showed that hES and iPS cells disrupted at that locus retain pluripotency. Further, ZFN-edited human ES cells carrying transgenes at the AAVS1 locus retained a normal karyotype, expression of pluripotency markers, and remained pluripotent as well as wild type at a panel of putative off-target sites (Hockemeyer et al. 2009). In our experiments, K562, Hep3B, HEK293, U2OS, fibroblast, and hES cells carrying transgenes at AAVS1 proliferated indistinguishably from control cells; the hES cells shown growing and dividing normally in Supplemental movie 1, for instance, carry _PPP1R12C_-disrupting transgenes at both its alleles. This said, it remains formally possible that some pathway may be adversely affected by the lack of regulatory subunit 12C of protein phosphatase 1 (which is thought to regulate actomyosin-based contractility) (Mulder et al. 2004). If such a pathway is found, which could be attempted, for instance, by genome-wide expression analysis of edited cells or by knocking out the orthologous gene in mouse or in rat (Geurts et al. 2009), we have shown that single-cell-derived clones possessing one wild-type and one transgenic AAVS1 allele can be readily isolated (e.g., see Figs. 2, 4, 5) (in a typical experiment, 60%–70% of edited cells fall into that category). We are unaware of evidence that PPP1R12C is haploinsufficient.
We describe three applications for isogenic transgenesis into a “safe harbor.” We show that four distinct coding regions—two promoterless and two with their own promoters—can be introduced site specifically in allelic positions at the AAVS1 locus to generate an inducible expression system (Fig. 3). In the present work, we describe hES cells carrying a histone H2B-eGFP fusion in such an inducible setting. Studies by Grunstein and colleagues (Kayne et al. 1988) launched an ever-growing line of genetic investigation into histone structure and function; the system we describe allows the rapid construction of a panel of isogenic cells carrying, for instance, an allelic series of histone genes (see also Goldberg et al. 2010). More generally, high-throughput proteomic (e.g., structure-function) studies can be performed using this approach in a setting that is normalized across different cell lines for expression of the transgene and its integration site in the genome. The native PPP1R12C promoter is active in all cells where we and others have studied this issue; in our hands, K562 and HeLa cells carrying promoterless GFP transgenes at AAVS1 show lower mean fluorescence intensity than such cells carrying the same transgene driven by the PGK promoter (VM Choi and EA Moehle, data not shown). While weaker by this criterion, the native PPP1R12C promoter drives sufficient transcription to allow FACS or selection-based isolation of pools and clones of cells with correctly added promoterless markers (e.g., Figs. 2–5).
We also show that the AAVS1 locus can be used to carry a panel of reporter constructs for the study of transcriptional _cis_-regulatory elements (Fig. 4). In the wake of initial studies in the 1980's (Zaret and Yamamoto 1984), a significant body of evidence has been accumulated that connects targeted chromatin remodeling and modification to action by nuclear hormone receptors (Stallcup et al. 2003; Hager et al. 2009) and most transcription factors where this issue has been studied. The ability to place investigator-specified reporters into the AAVS1 locus offers an opportunity to study genome control on templates chromatinized via normal physiological pathways; preliminary studies (SH Meijsing, unpubl.) show the reporter transgenes exhibit kinetics of induction by hormone and epigenetic marks similar to those observed at native GR gene targets.
Finally, we demonstrate that the AAVS1 locus serves as a safe harbor for shRNA expression cassettes in transformed and hES cells (Fig. 5). Functional genomics using RNAi is an important research tool and is commonly performed in transient settings where RNAi hairpins or transient transfection approaches suffice. The data we show demonstrate the feasibility of rapidly obtaining single-cell-derived clones isogenic except for a construct expressing a shRNA hairpin against the gene of choice. Robust target gene knockdown is still observed 2 mo after transgenesis. In transformed cells, linear donors with short homology arms produced by PCR may be used (Orlando et al. 2010), or ZFNs and a donor construct can be combined onto the same plasmid without significant loss of efficiency (Supplemental Fig. 9), illustrating two simple paths to establishing genome-wide screening with shRNA constructs stably integrated into the AAVS1 locus.
These data, therefore, expand the toolbox of human somatic cell genetics to include transgenesis in isogenic settings.
Methods
Zinc finger nucleases and donor constructs
ZFNs against the AAVS1 locus (Hockemeyer et al. 2009) were used in their obligate heterodimer, high-fidelity FokI (Miller et al. 2007) form cloned into pVAX as a 2A fusion construct (Perez et al. 2008). Donor constructs for RFLP addition and gene addition were assembled exactly as described (Urnov et al. 2005; Moehle et al. 2007; see also below); shRNA expression constructs for the experiment shown in Figure 5A were obtained from Sigma-Aldrich.
Cell culture
Cell culture techniques for K562, HeLa, and HEK293 cells (Moehle et al. 2007), U2OS cells (Meijsing et al. 2009), hTERT fibroblasts (Lombardo et al. 2007), and hESCs (Hockemeyer et al. 2008) have been described previously. All other cell types were cultured as per ATCC guidelines.
Genome editing in transformed cells and in fibroblasts
ZFN expression and donor constructs, assembled using standard recombinant DNA techniques (detailed protocols are available from the authors upon request) were introduced into transformed cells by nucleofection (Amaxa) exactly as described (Urnov et al. 2005; Moehle et al. 2007) and into hTERT fibroblasts using integration-defective lentivirus generated exactly as described (Lombardo et al. 2007). Genomic DNA was harvested using QuickExtract (Epicentre) for PCR-based assays and DNEasy (Qiagen) for Southern blotting performed exactly as described (Urnov et al. 2005; Moehle et al. 2007); for the Southern blot shown in Figure 1, genomic DNA was digested with AccI. Targeted integration frequency was measured using a body-labeled PCR assay as described (Moehle et al. 2007) with minor modifications. One hundred nanograms of genomic DNA was amplified (forward primer: CGGAACTCTGCCCTCTAACG; reverse primer, CTGGGATACCCCGAAGAGTG—both anneal to the chromosome outside the region of homology with the donor) in the presence of 5 μCi each of [α-32P]dCTP and [α-32P]dATP using Accuprime HiFi Taq polymerase (Invitrogen) and the following PCR settings: 95°C, 3 min; 24 cycles of 95°C, 30 sec; 62°C, 30 sec; 68°C, 4 min; 68°C, 7 min. The radiolabeled PCR and products were then purified using a column-based kit (Qiagen) and resolved on a 5% nondenaturing PAGE; the gel was dried and analyzed using a PhosphorImager (Molecular Dynamics). Cells were phenotyped for fluorescence using a Guava benchtop flow cytometer exactly as described (Moehle et al. 2007).
Genome editing in BGO1 hESCs using ZFNs
hESCs were cultured in Rho Kinase (ROCK)-inhibitor (Calbiochem; Y-27632) 24 h prior to electroporation. Cell were harvested using 0.25% trypsin/EDTA solution (Invitrogen) and 1 × 107 cells resuspended in phosphate buffered saline (PBS) were electroporated with a total of 40 μg of donor plasmids (designed and assembled by D Hockemeyer) and 5 μg of the AAVS1 ZFN encoding plasmid using a Gene Pulser Xcell System, Bio-Rad: 250 V, 500 μF, 0.4 cm cuvettes (Costa et al. 2007). Cells were subsequently plated on MEF feeder layers DR4 MEFs in hESC medium supplemented with ROCK-inhibitor for the first 24 h. Individual colonies were picked after puromycin selection (0.4 μg/mL) and neomycin selection (50 ng/mL) and expanded 10–14 d after electroporation. Southern blot analysis as described in Hockemeyer et al. (2009) was used to identify hESC clones with unique integrations of each donor plasmid in one of the AAVS1 alleles.
Live cell imaging
Targeted hESCs (BG01 [NIH Code: BG01; BresaGen, Inc.]) were cultured for the duration of imaging on mitomycin C inactivated mouse embryonic fibroblast (MEF) feeder layers and on glass-bottom dishes (MatTek Corporation) in CO2 independent medium (Gibco) supplemented with 15% fetal bovine serum (FBS) (Hyclone), 5% KnockOut Serum Replacement (Invitrogen), 1 mM glutamine (Invitrogen), 1% nonessential amino acids (Invitrogen), 0.1 mM β-mercaptoethanol (Sigma), and 4 ng/mL FGF2 (R&D systems). Cells were cultured and maintained in the presence of 2 μg/mL doxycycline for more than 2 wk prior to imaging to induce and maintain H2B-eGFP expression. Cells were imaged over a period of 20 h using a Detavision microscope (40× objective) and a HQ2 camera. Time lap intervals were 4 min, and seven 1.5 μm Z-stack images were taken for each time point with an exposure time of 0.25 sec and 10% transmittance.
Luciferase reporter assays and Western blotting
For reporter activity assays, single-cell derived clonal lines were seeded into 24-well plates in DMEM/5% FBS at ∼20,000 cells per well and treated the next day with either vehicle (ethanol) or 100 nM dexamethasone. After treatment for ∼13 h, cells were lysed in 100 μL per well of 1× lysis buffer (PharMingen) and assayed for luciferase activity, which was normalized for cell number. Western blotting was performed as described (Sancak et al. 2008).
Reverse transcription of total RNA and real-time PCR
RNA was isolated from hESCs, which were mechanically separated from feeder cells using TRIzol extraction and subsequent precipitation. Reverse transcription was performed on 1 μg of total RNA using oligo dT priming and thermoscript reverse transcriptase at 50°C (Invitrogen). Real-time PCR was performed in an ABI Prism 7900 (Applied Biosystems) with Platinum SYBR green pPCR SuperMIX-UDG with ROX (Invitrogen). Primers for the analysis of endogenous gene expressions were: hrtdnmt1_F ggttcagcaaaaccaatctatgatg; hrtdnmt1_R gccaagatttttgccattaacac; hrtoct4_F gctcgagaaggatgtggtcc; hrtoct4_R cgttgtgcatagtcgctgct; hrtp53_F gcccccagggaggagcacta; hrtp53_R gggagaggagctggtgttg (the latter two from Boley et al. 2000). Gene expression was normalized using GAPDH primers: hrtgapdh_F cagtcttctgggtggcagtga; hrtgapdh_R cgtggaaggactcatgacca.
Acknowledgments
We thank Judith Campisi for the gracious gift of hTERT fibroblasts, William Hahn for proposing the experiment that resulted in the data shown in Figure 5, Edward Weinstein for shRNA reagents, and Edda Einfeldt for technical assistance. We deeply appreciate suggestions provided by Aaron Klug, Jasper Rine, and Michael Holmes, and thank the three anonymous referees for comments on the manuscript. D.H. is a Merck Fellow of the Life Science Research Foundation. R.J. was supported by US National Institutes of Health grants R37-CA084198, RO1-CA087869, and RO1-HD045022 and by the Howard Hughes Medical Institute. R.J. is an adviser to Stemgen and a cofounder of Fate Therapeutics. S.H.M. and K.R.Y. received research support from NIH grants, K.R.Y. is a paid consultant with Merck and Company. Y.S. and D.M.S. were supported by grants from the NIH (R01 CA103866 and AI47389), Department of Defense (W81XWH-07-0448), and W.M. Keck Foundation. D.E.P., J.C.M., X.M., L.Z., E.J.R., P.D.G., and F.D.U. are full-time current, and R.D., V.M.C., E.A.M., I.R., S.M.G., H.C.S., K.J.P., and J.M.R. are past employees of Sangamo BioSciences, which has filed a patent application based on this work; X.C. and G.D.D. are full-time current employees of Sigma-Aldrich Biotechnology.
Footnotes
References
- Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, et al. 2007. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell 12: 395–402 [DOI] [PubMed] [Google Scholar]
- Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S 2001. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol 21: 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boley SE, McManus TP, Maher VM, McCormick JJ 2000. Malignant transformation of human fibroblast cell strain MSU-1.1 by _N_-methyl-_N_-nitrosourea: Evidence of elimination of p53 by homologous recombination. Cancer Res 60: 4105–4111 [PubMed] [Google Scholar]
- Carroll D 2008. Progress and prospects: Zinc-finger nucleases as gene therapy agents. Gene Ther 15: 1463–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Y, Klug A 1994. Toward a code for the interactions of zinc fingers with DNA: Selection of randomized fingers displayed on phage. Proc Natl Acad Sci 91: 11163–11167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Dottori M, Sourris K, Jamshidi P, Hatzistavrou T, Davis R, Azzola L, Jackson S, Lim SM, Pera M, et al. 2007. A method for genetic modification of human embryonic stem cells using electroporation. Nat Protoc 2: 792–796 [DOI] [PubMed] [Google Scholar]
- Fang J, Qian JJ, Yi S, Harding TC, Tu GH, VanRoey M, Jooss K 2005. Stable antibody expression at therapeutic levels using the 2A peptide. Nat Biotechnol 23: 584–590 [DOI] [PubMed] [Google Scholar]
- Gerber AN, Masuno K, Diamond MI 2009. Discovery of selective glucocorticoid receptor modulators by multiplexed reporter screening. Proc Natl Acad Sci 106: 4929–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, et al. 2009. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325: 433 doi: 10.1126/science.1172447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli T 2009. Transcription dynamics. Mol Cell 35: 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R 2008. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell 3: 346–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. 2009. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayne PS, Kim UJ, Han M, Mullen JR, Yoshizaki F, Grunstein M 1988. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell 55: 27–39 [DOI] [PubMed] [Google Scholar]
- Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B 2004. Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses. Nucleic Acids Res 32: e3 doi: 10.1093/nar/gnh009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotin RM, Linden RM, Berns KI 1992. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J 11: 5071–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PQ, Chan EM, Cost GJ, Zhang L, Wang J, Miller JC, Guschin DY, Reik A, Holmes MC, Mott JE, et al. 2010. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol Bioeng 106: 97–105 [DOI] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. 2007. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol 25: 1298–1306 [DOI] [PubMed] [Google Scholar]
- Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR 2009. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324: 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. 2007. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25: 778–785 [DOI] [PubMed] [Google Scholar]
- Moehle EA, Rock JM, Lee YL, Jouvenot Y, Dekelver RC, Gregory PD, Urnov FD, Holmes MC 2007. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci 104: 3055–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder J, Ariaens A, van den Boomen D, Moolenaar WH 2004. p116Rip targets myosin phosphatase to the actin cytoskeleton and is essential for RhoA/ROCK-regulated neuritogenesis. Mol Biol Cell 15: 5516–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando S, Santiago Y, Dekelver RC, Freyvert Y, Boydston EA, Moehle EA, Choi VM, Gopalan SML, Li JF, Miller J, et al. 2010. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res (in press). doi: 10.1093/nar/gkq512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne EA, Dudoit S, Rine J 2009. The establishment of gene silencing at single-cell resolution. Nat Genet 41: 800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison P, Silva J, Conklin D, Schlabach M, Li M, Aruleba S, Balija V, O'Shaughnessy A, Gnoj L, Scobie K, et al. 2004. A resource for large-scale RNA-interference-based screens in mammals. Natured 428: 427–431 [DOI] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. 2008. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26: 808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebar EJ, Pabo CO 1994. Zinc finger phage: Affinity selection of fingers with new DNA-binding specificities. Science 263: 671–673 [DOI] [PubMed] [Google Scholar]
- Richardson C, Jasin M 2000. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol Cell Biol 20: 9068–9075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogatsky I, Trowbridge JM, Garabedian MJ 1997. Glucocorticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol Cell Biol 17: 3181–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM 2006. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods 3: 715–719 [DOI] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M 1994. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol 14: 8096–8106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MA, Kim SH, Campisi J 2002. Reversible manipulation of telomerase expression and telomere length. Implications for the ionizing radiation response and replicative senescence of human cells. J Biol Chem 277: 28609–28617 [DOI] [PubMed] [Google Scholar]
- Ruis BL, Fattah KR, Hendrickson EA 2008. The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol Cell Biol 28: 6182–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B, Henderson N 1988. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci 85: 5166–5170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh R, Auerbach SD, Li X, Loftus RW, Husted RF, Stokes JB, Thomas CP 1999. Glucocorticoid induction of epithelial sodium channel expression in lung and renal epithelia occurs via _trans_-activation of a hormone response element in the 5′-flanking region of the human epithelial sodium channel alpha subunit gene. J Biol Chem 274: 12431–12437 [DOI] [PubMed] [Google Scholar]
- Smith JR, Maguire S, Davis LA, Alexander M, Yang F, Chandran S, ffrench-Constant C, Pedersen RA 2008. Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells 26: 496–504 [DOI] [PubMed] [Google Scholar]
- Stallcup MR, Kim JH, Teyssier C, Lee YH, Ma H, Chen D 2003. The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J Steroid Biochem Mol Biol 85: 139–145 [DOI] [PubMed] [Google Scholar]
- Tan I, Ng CH, Lim L, Leung T 2001. Phosphorylation of a novel myosin binding subunit of protein phosphatase 1 reveals a conserved mechanism in the regulation of actin cytoskeleton. J Biol Chem 276: 21209–21216 [DOI] [PubMed] [Google Scholar]
- Thomas KR, Folger KR, Capecchi MR 1986. High frequency targeting of genes to specific sites in the mammalian genome. Cell 44: 419–428 [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC 2005. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651 [DOI] [PubMed] [Google Scholar]
- Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P 1997. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci 94: 3789–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Yamamoto KR 1984. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell 38: 29–38 [DOI] [PubMed] [Google Scholar]