Targeted Genome Modification in Mice Using Zinc-Finger Nucleases (original) (raw)
Abstract
Homologous recombination-based gene targeting using Mus musculus embryonic stem cells has greatly impacted biomedical research. This study presents a powerful new technology for more efficient and less time-consuming gene targeting in mice using embryonic injection of zinc-finger nucleases (ZFNs), which generate site-specific double strand breaks, leading to insertions or deletions via DNA repair by the nonhomologous end joining pathway. Three individual genes, multidrug resistant 1a (Mdr1a), jagged 1 (Jag1), and notch homolog 3 (Notch3), were targeted in FVB/N and C57BL/6 mice. Injection of ZFNs resulted in a range of specific gene deletions, from several nucleotides to >1000 bp in length, among 20–75% of live births. Modified alleles were efficiently transmitted through the germline, and animals homozygous for targeted modifications were obtained in as little as 4 months. In addition, the technology can be adapted to any genetic background, eliminating the need for generations of backcrossing to achieve congenic animals. We also validated the functional disruption of Mdr1a and demonstrated that the ZFN-mediated modifications lead to true knockouts. We conclude that ZFN technology is an efficient and convenient alternative to conventional gene targeting and will greatly facilitate the rapid creation of mouse models and functional genomics research.
CONVENTIONAL gene targeting technology in mice relies on homologous recombination in embryonic stem (ES) cells to target specific gene sequences, most commonly to disrupt gene function (Doetschman et al. 1987; Kuehn et al. 1987; Thomas and Capecchi 1987). Advantages of gene targeting in ES cells are selective target sequence modification, the ability to insert or delete genetic information, and the stability of the targeted mutations through subsequent generations. There are also potential limitations, including limited rates of germline transmission and strain limitations due to lack of conventional ES cell lines (Ledermann 2000; Mishina and Sakimura 2007). Moving the targeted allele from one strain to another requires 10 generations of backcrosses that take 2–3 years. A minimum of 1 year is necessary for backcrossing if speed congenics is applied (Markel et al. 1997).
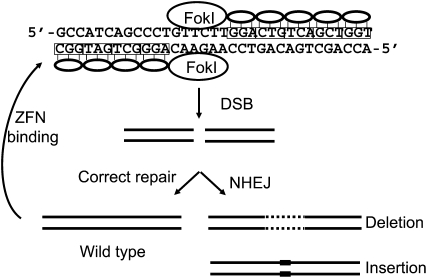
Zinc-finger nucleases (ZFNs) are fusions of specific DNA-binding zinc finger proteins (ZFPs) and a nuclease domain, such as the DNA cleavage domain of a type II endonuclease, _Fok_I (Kim et al. 1996; Smith et al. 1999; Bibikova et al. 2001). A pair of ZFPs provide target specificity, and their nuclease domains dimerize to cleave the DNA, generating double strand breaks (DSBs) (Mani et al. 2005), which are detrimental to the cell if left unrepaired (Rich et al. 2000). The cell uses two main pathways to repair DSBs: high-fidelity homologous recombination and error-prone nonhomologous end joining (NHEJ) (Lieber 1999; Pardo et al. 2009; Huertas 2010). ZFN-mediated gene disruption results from deletions or insertions frequently introduced by NHEJ. Figure 1 illustrates the cellular events following the injection of a pair of ZFNs targeting the mouse Mdr1a (also known as Abcb1a) gene.
Figure 1.—
The ZFN targeting mechanism. ZFN pairs bind to the target site, and _Fok_I endonuclease domain dimerizes and makes a double strand break between the binding sites. If a DSB is repaired so that the wild-type sequence is restored, ZFNs can bind and cleave again. Otherwise, nonhomologous end joining (NHEJ) introduces deletions or insertions, which change the spacing between the binding sites so that ZFNs might still bind but dimerization or cleavage cannot occur. Insertions or deletions potentially disrupt the gene function.
ZFNs have been successfully applied to generate genome modifications in plants (Shukla et al. 2009; Townsend et al. 2009), fruit flies (Bibikova et al. 2002), Caenorhabditis elegans (Morton et al. 2006), cultured mammalian cells (Porteus and Baltimore 2003; Santiago et al. 2008), zebrafish (Doyon et al. 2008; Meng et al. 2008), and most recently in rats (Geurts et al. 2009; Mashimo et al. 2010). The technology is especially valuable for rats because rat ES cell lines have only become available recently (Buehr et al. 2008; Li et al. 2008), and successful homologous recombination-mediated genome modification has not been reported. Previously, ENU mutagenesis (Zan et al. 2003) or transposons (Kitada et al. 2007) were the two main methods for generating gene knockout rats, both of which are random approaches and require labor-intensive and time-consuming screens to obtain the desired gene disruptions.
Although ES cell-based knockout technology is widely used in mice, ZFN technology offers three advantages: (i) high efficiency; (ii) drastically reduced timeline, similar to that of creating a transgene (Gordon et al. 1980); and (iii) the freedom to apply the technology in various genetic backgrounds. In addition, no exogenous sequences need to be introduced because selection is not necessary.
Here, we created the first genome-engineered mice using ZFN technology. Three genes were disrupted in two different backgrounds: Mdr1a, Jag1, and _Notch_3 in the FVB/N strain and Jag1 also in the C57BL/6 strain. All founders tested transmitted the genetic modifications through the germline.
MATERIALS AND METHODS
In vitro preparation of ZFN mRNAs:
The ZFN expression plasmids were obtained from Sigma's CompoZr product line. Each plasmid was linearized at the _Xba_I site, which is located at the 3′ end of the _Fok_I ORF. 5′ capped and 3′ poly(A)-tailed message RNA was prepared using either MessageMax T7 capped transcription kit and poly(A) polymerase tailing kit (Epicentre Biotechnology, Madison, WI) or mMessage (mMachine) T7 kit and poly(A) tailing kit (Ambion, Austin, TX). The poly(A) tailing reaction was precipitated with an equal volume of 5 m NH4OAc and then dissolved in injection buffer (1 mm Tris–HCl, pH 7.4, 0.25 mm EDTA). mRNA concentration was estimated using a NanoDrop 2000 Spectrometer (Thermo Scientific, Wilmington, DE).
ZFN validation in cultured cells:
National Institutes of Health (NIH) 3T3 cells were grown in DMEM with 10% FBS and antibiotics at 37° with 5% CO2. ZFN mRNAs were paired at 1:1 ratio and transfected into the NIH 3T3 cells to confirm ZFN activity using a Nucleofector (Lonza, Basel, Switzerland), following the manufacturer's 96-well shuttle protocol for 3T3 cells. Twenty-four hours after transfection, culturing medium was removed, and cells were incubated with 15 μl of trypsin per well for 5 min at 37°. The cell suspension was then transferred to 100 μl of QuickExtract (Epicentre) and incubated at 68° for 10 min and 98° for 3 min. The extracted DNA was then used as template in a PCR reaction to amplify 350- to 650-bp amplicons around the target site with the following primer pairs: Mdr1a Cel-I F, ctgtttcttgacaaaacaacactaggctc; Mdr1a Cel-I R, gggtcatgggaaagagtttaaaatc; Jag1 Cel-I F, cttcggggcacttgtcttag; Jag1 Cel-I R, gcgggactgatactccttga; Notch3 Cel-I F, tttaaagtgggcgtttctgg; and Notch3 Cel-I R, ggcagaggtacttgtccacc.
Each 50-μl PCR reaction contained 1 μl of template, 5 μl of buffer II, 5 μl of 10 μm each primer, 0.5 μl of AccuPrime Taq polymerase high fidelity (Invitrogen, Carlsbad, CA), and 38.5 μl of water. The following PCR program was used: 95°, 5 min, 35 cycles of 95°, 30 sec, 60°, 30 sec, and 68°, 45 sec, and then 68°, 5 min. Three microliter of the above PCR reaction was mixed with 7 μl of 1× buffer II and incubated under the following program: 95°, 10 min, 95° to 85°, at −2°/s, 85° to 25° at −0.1°/s.
One microliter each of nuclease S (Cel-I) and enhancer (Transgenomic, Omaha, NE) were added to digest the above reaction at 42° for 20 min. The mixture is resolved on a 10% polyacrylamide TBE gel (Bio-Rad, Hercules, CA).
Microinjection and mouse husbandry:
FVB/NTac and C57BL/6NTac mice were housed in static cages and maintained on a 14 hr/10 hr light/dark cycle with ad libitum access to food and water. Three- to 4-week-old females were injected with PMS (5 IU/mouse) 48 hr before hCG (5 IU/mouse) injection. One-cell fertilized eggs were harvested 10–12 hr after hCG injection for microinjection. ZFN mRNA was injected at 2 ng/μl. Injected eggs were transferred to pseudopregnant females [Swiss Webster (SW) females from Taconic Labs mated with vasectomized SW males] at 0.5 days post coitum (dpc).
Founder identification using mutation detection assay:
Toe clips were incubated in 100–200 μl of QuickExtract (Epicentre Biotechnology) at 50° for 30 min, 65° for 10 min, and 98° for 3 min. PCR and mutation detection assay were done under the same conditions as in ZFN validation in cultured cells using the same sets of primers.
TA cloning and sequencing:
To identify the modifications in founders, the extracted DNA was amplified with Sigma's JumpStart Taq ReadyMix PCR kit. Each PCR reaction contained 25 μl of 2× ReadyMix, 5 μl of primers, 1 μl of template, and 19 μl of water. The same PCR program was used as in ZFN validation in cultured cells. Each PCR reaction was cloned using TOPO TA cloning kit (Invitrogen) following the manufacture's instructions.
At least eight colonies were picked from each transformation, PCR amplified with T3 and T7 primers, and sequenced with either T3 or T7 primer. Sequencing was done at Elim Biopharmaceuticals (Hayward, CA).
PCR for detecting large deletions:
To detect larger deletions, which removed the original Cel-I priming sites, another set of distal primers were used for each of the targets: Mdr1a 800F, catgctgtgaagcagatacc; Mdr1a 800R, ctgaaaactgaatgagacatttgc; Jag1 600F, ggtgggaactggaagtagca; Jag1 600R, ggagtctctctcccgctctt; Notch3 800F, tctcaacaaacccacaacca; and Notch3 800R, gtcgtctgcaagagcaagtg.
Each 50-μl PCR contained: 1 μl of template, 5 μl of 10× buffer II, 5 μl of 10 μm of each 800F/R primer, 0.5 μl of AccuPrime Taq polymerase high fidelity (Invitrogen), and 38.5 μl of water. The following program was used: 95°, 5 min, 35 cycles of 95°, 30 sec, 60°, 30 sec, and 68°, 3 min, and then 68°, 5 min. The samples were resolved on a 1% agarose gel. Distinct bands with lower molecular weight than the wild type (WT) were sequenced.
RNA preparation from tissues and RT–PCR:
_Mdr1a_−/− or Mdr1a+/+ littermates were sacrificed for tissue harvest at 5–9 weeks of age. Large intestine, kidney, and liver tissues were dissected and immediately used or stored in RNAlater solution (Ambion) at −20°. Total RNA was prepared using GenElute Mammalian Total RNA Miniprep kit (Sigma) following manufacturer's instructions. To eliminate any DNA contamination, the RNA was treated with DNAseI (New England Biolabs, Ipswich, MA) before being loaded onto the purification columns.
Mdr1a RT–PCR analysis was carried out with 1 μl of total RNA, primers RT-F (5′-GCCGATAAAAGAGCCATGTTTG) and RT-R (5′- GATAAGGAGAAAAGCTGCACC), using SuperScript III one-step RT–PCR system with platinum Taq high fidelity kit (Invitrogen). Reverse transcription and subsequent PCR were carried out with 1 cycle of 55° for 30 min and 94° for 2 min for cDNA synthesis and 40 cycles of 94° for 15 sec, 56° for 30 sec, and 68° for 1 min for amplification. The PCR product was loaded in a 1.2% agarose gel and visualized with ethidium bromide. Nested PCR used primers RT-F2 (5′- CTGGAGGAAGAAATGACCACG) and RT-R2 (5′-GATAGCTTTCTTTATCCCCAGCC).
Western blot analysis:
Mice were killed and the large intestine was immediately harvested and flushed with ice-cold PBS buffer, snap frozen on dry ice, and stored at −80°. For protein preparation, tissue pieces equivalent to ∼200 μl were shaved off the frozen samples and placed into an ice-cold microcentrifuge tube. Four hundred microliters of ice-cold PBS with 4× protease inhibitors was added, and the sample was dounce homogenized. The homogenate was pelleted at 20,000 × g for 5 min at 4°, and the supernatant (S1) was removed. The pellet, after being resuspended in 400 μl of ice-cold PBS with 4× protease inhibitors, was centrifugated at 4000 × g for 5 min at 4°. The supernatant (S2) was removed, and the pellet was resuspended in 500 μl lysis buffer (composition) (Gerlach et al. 1987), dounce homogenized, incubated on ice for 40 min with intermittent vortexing for 15 sec per interval, and finally pelleted at 20,000 × g for 20 min at 4°. The supernatant (S3) was collected, and the pellet was resuspended again in 250 μl of lysis buffer, dounce homogenized, spun at 4000 × g for 5 min at 4°, and the supernatant (S4) was kept. The S3 and S4 fractions were diluted 1:1 with 2× Laemmli buffer (Sigma) and incubated at 37° for 5–10 min. Lysates (15 μl, 10 μl, or 5 μl) were separated on a 4–20% Mini-PROTEAN TGX precast gel (BioRad) and transferred to nitrocellulose membrane using a semi-dry transblot (BioRad) at 25 V for 1 hr. The transfer buffer contained standard tris-glycine salts, 18% MeOH, and 0.25% SDS. Mouse anti-Mdr1a antibody C219 (Covance, Princeton, NJ) at 1:100 and mouse anti-actin antibody at 1:1000 (Sigma) were incubated together with the blot overnight in 5% milk/TBST, rocking at 4°, rinsed briefly in TBST, and the HRP-conjugated goat anti mouse secondary antibody (Jackson ImmunoResearch Labs, West Grove, PA) was incubated for 1 hr in 1% milk/TBST following a quick rinse with TBST, followed by 2 × 50 ml washes of 1% milk/TBST for 10 min. HRP was detected using the SuperSignal West Pico substrate (Thermo) and a ChemiDocXRS+ imaging system (Bio-Rad).
RESULTS
ZFN injection resulted in high-efficiency knockout at the Mdr1a locus:
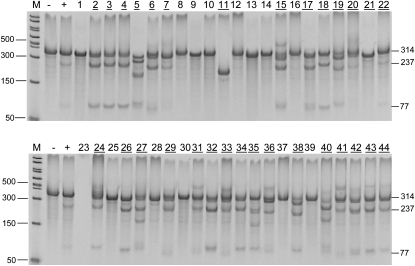
Validated Mdr1a ZFN mRNA (supporting information, Figure S1, File S1, and materials and methods) was microinjected into fertilized FVB/N eggs, which were transferred into pseudopregnant females. Pups born from the injected embryos were tested using a DNA mismatch endonuclease (Cel-I) assay (see materials and methods) for modifications at the target site. Thirty of the 44 live births contained deletions or insertions. Figure 2 shows the founders among wild-type littermates.
Figure 2.—
Identification of genetically engineered Mdr1a founders using the Cel-I mutation detection assay. Cleaved bands indicate a mutation is present at the target site (see materials and methods). Bands are marked with respective sizes in base pairs. M, PCR marker. One to 44 pups born from injected eggs. The numbers representing the mutant founder animals are underlined.
Larger deletions generated by ZFN activity:
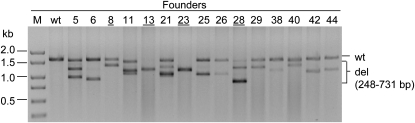
Some of the samples yielded no amplification product with the Cel-I primers. To detect potentially larger deletions that would have destroyed the priming sites used in Figure 2, a larger region spanning 800 bp on both sides of the cleavage site was PCR amplified. Figure 3 shows that 15 of the 44 pups indeed contain larger deletions, including 4 animals that were not identified as founders by the previous PCR assay. The PCR products for all founders were TA cloned and sequenced to reveal the exact sequences of modifications, and the deletions ranged between 3 and 731 bp in length as well as some small insertions (Table S1). Interestingly, three small deletions were each found in two or more founders: a 19-bp deletion in founders 7, 17, and 36, a 21-bp deletion in founders 17 and 20, and a 6-bp deletion in founders 34 and 44 (Figure S2). All three deletions are flanked by a 2-bp microhomology, which is predicted to create a common NHEJ junction (Lieber 1999).
Figure 3.—
Large deletions in Mdr1a founders. PCR products were amplified using primers located 800 bp upstream and downstream of the ZFN target site. Bands significantly smaller than the 1.6-kb wild-type band indicate large deletions in the target locus. Four founders that were not identified in Figure 2 are underlined.
High rate of germline transmission by Mdr1a founders:
Nine of the founders were chosen to backcross to the wild-type FVB/N mice to the F1 generation, all of which transmitted at least one mutant allele to their offspring. Seven founders transmitted more than two mutated alleles. Interestingly, in some cases, alleles that were not initially identified in the founders were also transmitted through the germline and discovered in the next generation, such as in founders 6, 8, 13, 21, and 44 (Table S2), most likely due to incomplete sequencing of the TA clones (see discussion).
Mdr1a expression by RT–PCR and Western:
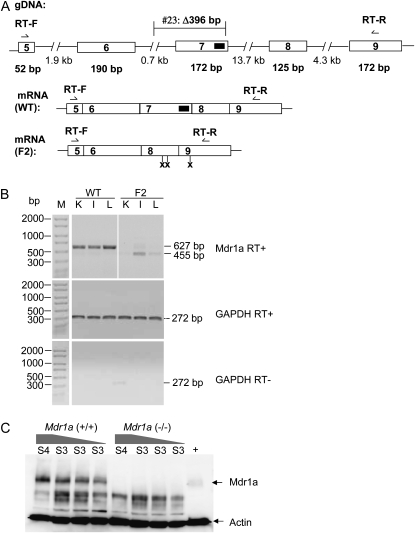
The Mdr1a protein is differentially expressed in tissues. Liver and large intestine predominantly express Mdr1a, and kidney expresses both Mdr1a and Mdr1b (Schinkel et al. 1994). To verify that a deletion in the Mdr1a gene abolishes its expression, we performed RT–PCR on total RNA from liver, kidney, and intestine of _Mdr1a_−/− mice established from founder 23, with a 396-bp deletion (Figure 4A), using a forward and a reverse primer located in exons 5 and 9, respectively. Samples from all the _Mdr1a_−/− tissues produced a smaller product with lower yield than those of corresponding wild-type samples, with a sequence correlating to exon 7 skipping and subsequent multiple premature stop codons in exon 8 in the mutant animals (Figure 4B). Furthermore, Western blotting with an anti-Mdr1a antibody showed absence of Mdr1a protein in the large intestine of _Mdr1a_−/− animals (Figure 4C), demonstrating that the 396-bp deletion leads to a true knockout.
Figure 4.—
Mdr1a expression in homozygous knockout animals. (A) A schematic of Mdr1a genomic and mRNA structures around the ZFN target site in exon 7, marked with a solid black rectangle. Exons are represented by open rectangles with respective numbers. The size of each exon in base pairs is labeled directly underneath it. Intron sequences are represented by broken bars with size in base pairs underneath. The position of the 396-bp deletion in founder 23 is labeled above intron 6 and exon 7. RT-F and RT-R are the primers used in RT–PCR, located in exons 5 and 9, respectively. (B) Mdr1a expression in tissues. For RT reactions, 40 ng of total RNA was used as template. Normalization of the input RNA was confirmed by GAPDH amplification with or without reverse transcriptase. M, PCR marker; WT, wild-type mouse; F2, _Mdr1a_−/− mouse; K, kidney; I, large intestine; L, liver. Amplicon sizes are marked on the right. (C) Western blot analysis with large intestine. +, positive control, lysate from the human Mdr1-overexpressing SK-N-FI cells (ATCC, Manassas, VA). S3 (15 μl, 10 μl, and 5 μl loaded in each of the three lanes) and S4 (15 μl loaded), the third and fourth supernatant fractions of large intestine membrane preparations (see materials and methods). Actin was used as a loading control. Mdr1a+/+, wild-type intestine; _Mdr1a_−/−, intestine from a homozygous knockout mouse derived from founder 23.
High-efficiency targeting and germline transmission in C57BL/6 strain:
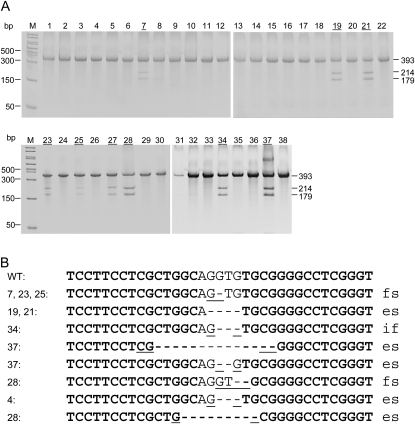
Next, we microinjected Jag1 ZFN mRNA into fertilized eggs from C57BL/6 strain and identified 24% founders among live births (Figure 5A). The Jag1 ZFNs precisely target the junction of intron 1 and exon 2; therefore, even small deletions can destroy the recognition site for splicing. Deletions among Jag1 founders range from 1 to 14 bp (Figure 5B). Five founders, 4, 19, 21, 28, and 37, carry deletions that mutated the conserved G residue at the end of the intron and will likely lead to exon 2 skipping and deletion of 102 amino acids from the protein. Except for founders 28 and 37, both with two mutant alleles, the rest of the founders only bear one mutated allele. Similar to some Mdr1a founders, some Jag1 founders carry the same deletions. Founders 7, 23, and 25 share the same 1-bp deletion. Founders 19 and 21 bear the same 4-bp deletion. Except for the mutant allele in founders 19 and 21, the rest of the deletions are flanked by 1- to 2-bp microhomology (Figure 5B, also see discussion). Founder 28 has a 2-bp deletion, both resulting in frameshift and premature stop codons shortly downstream. Founder 19 was backcrossed to wild-type C57BL/6 and achieved germline transmission in the first mating (three heterozygotes among eight F1 pups).
Figure 5.—
Identification and genotype of Jag1 founders. (A) _Jag_1 founders identified using the Cel-I mutation detection assay. M, PCR marker; 1–38, pups born from two injection sessions. The numbers of founders are underlined. The sizes in base pairs of uncut and cut bands are labeled on the right. (B) Genotype of the Jag1 founders. Target site sequences of wild type and founders are aligned. ZFN binding sites are in boldface type. A dash represents a deleted nucleotide. One to 4 bp of microhomology that was likely used by NHEJ is underlined. The frameshift (fs), exon skipping (es), or in-frame amino acid loss (if) resulting from each deletion is indicated to the right of each sequence.
Notch3 targeting in FVB/N mice:
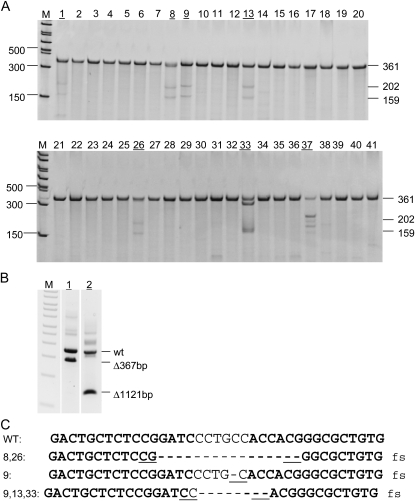
We targeted a third gene, Notch3, again in FVB/N and obtained 20% founder rate (Figure 6A). Founders 1 and 2 have large deletions, 367 bp and 1121 bp, respectively (Figure 6B). Number 9 is the only founder carrying two different mutated alleles, a 1-bp deletion, and an 8-bp deletion. Again, the same 8-bp deletion in founder 9 was also identified in founders 13 and 23, and founders 8 and 26 both carry an identical 16-bp deletion. All three deletions are flanked by a 2-bp microhomology (Figure 6C, also see discussion). All deletions are completely within exon 11, resulting in a frameshift that introduces premature translational stop codons within the exon.
Figure 6.—
Identification and genotype of Notch3 founders. M, PCR marker. (A) The Cel-I mutation detection assay was used to identify founders, whose numbers are underlined. (B) Large deletions were detected in founders 1 and 2. (C) Genotype of the Notch3 founders. ZFN binding sites are in boldface type. A dash represents a deleted nucleotide. One to 4 bp of microhomology that was likely used by NHEJ is underlined. All deletions result in frameshift (fs), which is labeled to the right of each sequence.
Potential off-target sites validation:
We identified 20 sites in the mouse genome that are most similar to the Mdr1a target site, all with 5-bp mismatches from the ZFN binding sequence, and top potential off-target sites for Jag1 and Notch3, all with at least 6-bp mismatches from their respective target sites (Table S3, Table S4, and Table S5). To validate specificity of the Mdr1a ZFNs, we tested the site in the Mdr1b gene, which is 88% identical to Mdr1a, in all 44 Mdr1a F0 pups using mutation detection assay. None of the 44 pups had an NHEJ event at the Mdr1b site (Figure S3). To further and more fully characterize the Mdr1a mutant animals, we tested all the predicted potential off-target sites in four founder animals and found no spurious mutations (Figure S4).
DISCUSSION
We generated mice with modifications at three loci by direct injection of ZFN mRNA into the pronucleus of one-cell mouse embryos. ZFN technology offers a few obvious advantages when compared to conventional methods in producing knockout mice. By bypassing ES cells, ZFN technology enables the generation of homozygous mice with targeted modifications in a matter of months, with no need for selection. Highly efficient targeting (20–75%) allows one to identify founders by screening relatively small number of pups. Many founders carry more than one mutant allele in addition to the wild-type allele, implying that ZFNs remain active beyond one-cell stage. Every cell division doubles the number of the wild-type allele, which is the only allele cleavable by ZFNs. Deletions or insertions change the space between ZFN binding sites, preventing _Fok_I domains from dimerization. For those founders harboring up to five different alleles, ZFN-mediated cleavage likely did not happen before the first embryonic cell division. Thus, most founders are mosaics. All tested founders transmitted at least one mutant allele through the germline (Table S1).
Most Mdr1a founders transmitted more than one allele, as observed in rats as well (Geurts et al. 2009). Some alleles that were not identified in the founders were inherited in F1 generation (Table S2), which was likely caused by PCR bias and incomplete sampling of the TA clones. PCR reactions for detecting large deletions, which favor amplification of smaller products resulting from larger deletions, were used to TA clone, followed by sequencing to identify mutant alleles. We only sequenced 8–16 clones from each founder. Some of the small deletions, especially if they were also low representing, could be missed. Although all live births were tested with Cel-I assay (with a detection limit ∼1%), some of the negative pups may carry low-representing alleles that are still germline competent. It is also possible that toe or tail clips do not necessarily have the same genotype as germ cells, of which we observed only one confirmed example. Founder 23 did not have wild-type allele amplification in either toe or tail DNA. Yet when mated to wild-types, only 50% of its F1's were heterozygous. The other half was wild type. Thus wild-type allele was present in the germline but was not represented in the toe or tail samples we analyzed.
We examined the effect of modifications on gene expression in one of the _Mdr1a_−/− strains in further detail. The RT–PCR results demonstrate that the samples from the _Mdr1a_−/− founder 23 produce a transcript missing the 172-bp exon 7 that causes exon skipping during mRNA splicing and immediately creates multiple premature translational stop codons in the message (Figure 4B). Such mutations often lead to nonsense-mediated decay (NMD) of the mutant mRNA (Chang et al. 2007), and this is supported by an apparently reduced level of exon-skipping transcript, compared to that of the wild type, detected in RT–PCR analyses (Figure 4B), implying likely mRNA degradation provoked by NMD. In the _Mdr1a_−/− samples, there were faint bands at and above the size of the wild-type transcript, which are most likely PCR artifacts because amplification of those bands excised from the gel yielded mostly the exon-skipped product. The bands at the wild-type size in secondary rounds of PCR were mixtures that did not yield readable sequences (not shown). This conclusion is supported by Western blot analysis using an anti-Mdr1a antibody that detected abundant protein expression in the large intestine (which highly expresses Mdr1a but not Mdr1b) of wild-type littermates but no detectable Mdr1a protein in the same tissue of homozygous animals derived from founder 23. Thus, the _Mdr1a_−/− mice derived from founder 23 represent a functional knockout. Consistent with the theory of possible NMD, we obtained similar RT–PCR results on another animal, a compound homozygote from founder 11, harboring 417- and 533-bp deletions in respective alleles. A smaller amplicon corresponding to exon skipping was detected at a lower level than that of wild-type PCR product (not shown), as in the case of Mdr1a−/− from founder 23. This observation extends to the rat as well. A 19-bp deletion in the rat Mdr1a locus, greatly reduced the mRNA level, though sizewise it was similar to the wild-type and again, Western blots showed complete lack of Mdr1a expression in Mdr1−/− large intestine (I. D. Carbery and X. Cui, unpublished data).
The mouse Mdr1a gene has 28 exons, and the encoded protein is composed of two units of six transmembrane domains (TMs 1–6 and TMs 7–12), each unit with an ATP binding site and with a linker region in between the units (Mitzutani and Hattori 2005). All 12 TM domains as well as the two ATP-binding motifs are essential for Mdr1a function (Pippert and Umbenhauer 2001). The Mdr1a ZFNs target exon 7, which encodes TMs 3 and 4. On the basis of previous work in this field, any partial protein that might result from the described frameshift and nonsense mutations we observed (assuming such protein fragments could be stable) should not be functional (Pippert and Umbenhauer 2001). Among the mutant alleles, 41% cause exon skipping, 37% result in frameshift, and the rest carry in-frame deletions (Table S1). It is safe to conclude that the majority of the mutants obtained will be true knockouts.
Interestingly, large deletions were introduced in both targets, Mdr1a and Notch3 in the FVB/N strain but not in Jag1 in C57BL/6, suggesting a possible difference in DNA repair that may be related to the host genetic background. However, injection of Jag1 ZFNs into FVB/N embryos resulted in similar founder rate and deletion sizes (not shown) as in C57BL/6, indicating the difference in deletion size might not have resulted from variation in genetic background. The Mdr1a locus also has a higher percentage of large deletions than Notch3, although both were targeted in FVB/N. It is possible that the target site per se contributes at least partially to the determination of modifications. Table S6 contains data from all the injections in both FVB/N and C57BL/6, including number of eggs injected, number of pups born from each injection, and number of founders identified among live births. Due to procedural similarity between generation of a transgene and ZFN-mediated genome modifications, any background that is competent for traditional transgenesis should in theory be a good candidate to use for creating a ZFN-mediated knockout. We have not accumulated enough data to analyze differences on targeting efficiency or the types of modifications that can be caused by different mouse backgrounds. However, we and others have observed similar targeting rates in various rat strains, and the size of deletions seems to also be target dependent (SAGE Labs, unpublished data; Mashimo et al. 2010).
Another interesting observation was that for all three targets, some small deletions were identical in multiple founders (Figures 5 and 6 and Figure S2), assuming deletion occurs randomly during NHEJ. We considered the possibility that these deletions were merely PCR artifacts caused by GC-rich microhomology flanking some of the deletions. However, several of the small deletions transmitted germline (Table S2), proving that these small deletions are true targeting events. Our data support the notion that microhomology of 1–4 bp at the ends of DSBs promotes, but is not necessary for, NHEJ (Lieber 1999). We noticed that most of the deletions, regardless of whether identified in single or multiple founders, contain 1–4 bp microhomology at the deletion boundary (Figures 5 and 6 and Figure S2). In alleles such as that shared by founders 19 and 21 of Jag1, where microhomology is not present, we hypothesize that sequence-dependent DNA secondary structures might form around the target site that pause the resection of the ends by exonucleases before ligation (Huertas 2010), so that certain deletions resulted in multiple founders. _Mdr1a_−/− founder 11 contains an unusual allele with discontinuous deletions, a 417-bp deletion from −528 to −112, >100 bp upstream of the cleavage site and flanked by a 5-bp microhomology GACAA, and a 19-bp deletion at the cleavage site, −14 to +5 (Table S1). This complex allele was transmitted through the germline (Table S2). One explanation could be that two sequential ZFN cleavages occurred in the same chromatid. The repair of the first DSB was initiated as homologous recombination using the sister chromatid as template but was completed by NHEJ using the 5-bp microhomology, as observed previously (Richardson and Jasin 2000), leading to a 417-bp deletion upstream of the target site. The restored target site was cleaved again by ZFNs and repaired by NHEJ, resulting in a 19-bp deletion.
We identified sequences in the mouse genome that are most similar to the Mdr1a, Jag1, and Notch3 target sites and tested the potential off-target sites for the Mdr1a ZFNs. No modifications were detected at the Mdr1b site in any of the 44 live births, and of 80 other off-targets tested (20 sites in four independent founders), none harbored modifications, illustrating the specificity of the Mdr1a ZFNs (see Figure S3). Doing the best we could have done without performing costly whole genome sequencing, these data do not exclude that there are off-target sites that do not resemble the target site. Assuming hypothetical, unlinked off-target modifications will be diluted through breeding, an indirect way to detect potential off-target events could be to compare phenotypically early-generation to later-generation homozygotes. The lack of difference in phenotypes implies the absence of off-target events. To include wild-type littermates as controls in phenotyping assays is another way to reduce the possible interference of off-target modifications on phenotype. In the meantime, we do realize that the ultimate proof of absence or presence of off-target events has to come from whole-genome sequencing, which will hopefully be affordable in the near future with the continuous reduction in sequencing cost.
Altogether, we conclude that ZFN technology is a valuable alternative to conventional knockout technology for generating genome modifications in mice.
Acknowledgments
We thank Fyodor Urnov for helping interpret the puzzling allele in founder 11, Thom Saunders for suggestions on improving mRNA preparation for injection, Dave Briner for ZFN assembly, and Danhui Wang for her assistance in off-target search.
References
- Bibikova, M., D. Carroll, D. J. Segal, J. K. Trautman, J. Smith et al., 2001. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 21 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova, M., M. Golic, K. G. Golic and D. Carroll, 2002. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr, M., S. Meek, K. Blair, J. Yang, J. Ure et al., 2008. Capture of authentic embryonic stem cells from rat blastocysts. Cell 135 1287–1298. [DOI] [PubMed] [Google Scholar]
- Chang, Y.-F., J. S. Imam and M. F. Wilkinson, 2007. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76 51–74. [DOI] [PubMed] [Google Scholar]
- Doetschman, T., R. G. Gregg, N. Maeda, M. L. Hooper, D. W. Melton et al., 1987. Targeted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature 330 576–578. [DOI] [PubMed] [Google Scholar]
- Doyon, Y., J. M. McCammon, J. C. Miller, F. Faraji, C. Ngo et al., 2008. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 26 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach, J. H., D. R. Bell, C. Karakousis, H. K. Slocum, N. Kartner, et al., 1987. P-glycoprotein in human sarcoma: evidence for multidrug resistance. J. Clin. Oncol. 5 1452–1460. [DOI] [PubMed] [Google Scholar]
- Geurts, A. M., G. J. Cost, Y. Freyvert, B. Zeitler, J.C. Miller et al., 2009. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, J. W., G. A. Scangos, D. J. Plotkin, J. A. Barbosa and F. H. Ruddle, 1980. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc. Natl. Acad. Sci. USA 77 7380–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas, P., 2010. DNA resection in eukaryotes: deciding how to fix the break. Nat. Struct. Mol. Biol. 17 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.-G., J. Cha and S. Chandrasegaran, 1996. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 93 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada, K., S. Ishishita, K. Tosaka, R. Takahashi, M. Ueda et al., 2007. Transposon-tagged mutagenesis in the rat. Nat. Methods 4 131–133. [DOI] [PubMed] [Google Scholar]
- Kuehn, M. R., A. Bradley, E. J. Robertson and M. J. Evans, 1987. A potential animal model for Lesch-Nyhan syndrome through introduction of HPRT mutations into mice. Nature 326 295–298. [DOI] [PubMed] [Google Scholar]
- Ledermann, B., 2000. Embryonic stem cells and gene targeting. Exp. Physiol. 85 603–613. [PubMed] [Google Scholar]
- Li, P., C. Tong, R. Mehrian-Shai, L. Jia, N. Wu et al., 2008. Germline competent embryonic stem cells derived from rat blastocysts. Cell 135 1299–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber, M. R., 1999. The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukabryotes. Genes Cells 4 77–85. [DOI] [PubMed] [Google Scholar]
- Mani, M., J. Smith, K. Kandavelou, J. M. Berg and S. Chandrasegaran, 2005. Binding of two zinc finger nuclease monomers to two specific sites is required for effective double-strand DNA cleavage. Biochem. Biophys. Res. Comm. 334 1191–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markel, P., P. Shu, C. Ebeling, G. A. Carlson, D. L. Nagle et al., 1997. Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nat. Genet. 17 280–284. [DOI] [PubMed] [Google Scholar]
- Mashimo, T., A. Takizawa, B. Voigt, K. Yoshimi, H. Hiai et al., 2010. Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PloS One 5 e8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X., M. B. Noyes, L. J. Zhu, N. D. Lawson and S. A. Wolfe, 2008. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 26 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina, M., and K. Sakimura, 2007. Conditional gene targeting on the pure C57BL/6 genetic background. Neurosci. Res. 58 105–112. [DOI] [PubMed] [Google Scholar]
- Mitzutani, T., and A. Hattori, 2005. New Horizon of MDR1 (P-glycoprotein) study. Drug Metab. Rev. 37 489–510. [DOI] [PubMed] [Google Scholar]
- Morton, J., M. W. Davis, E. M. Jorgensen and D Carroll, 2006. Induction and repair of zincfinger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc. Natl. Acad. Sci. USA 103 16370–16375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo, B., B. Gomez-Gonzalez and A. Aguilera, 2009. DNA double-strand break repair: how to fix a broken relationship. Cell. Mol. Life Sci. 66 1039–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pippert, T. R., and D. R. Umbenhauer, 2001. The subpopulation of CF-1 mice deficient in P-glycoprotein contains a murine retroviral insertion in the mdr1a gene. J. Biochem. Mol. Toxicol. 15 83–89. [DOI] [PubMed] [Google Scholar]
- Porteus, M. H., and D. Baltimore, 2003. Chimeric nucleases stimulate gene targeting in human cells. Science 300 763. [DOI] [PubMed] [Google Scholar]
- Rich, T., R. L. Allen and A. H. Wyllie, 2000. Defying death after DNA damage. Nature 407 777–783. [DOI] [PubMed] [Google Scholar]
- Richardson, C., and M. Jasin, 2000. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol. Cell. Biol. 20 9068–9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago, Y., E. Chan, P. Q. Liu, S. Orlando, L. Zhang et al., 2008. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 105 5809–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinkel, A. H., J. J. Smit, O. van Tellingen, J. H. Beijnen, E. Wagenaar et al., 1994. Disruption of the mouse mdr1a P-glycoprotein gene leads to a deficiency in the blood-brain barrier and to increased sensitivity to drugs. Cell 77 491–502. [DOI] [PubMed] [Google Scholar]
- Shukla, V. K., Y. Doyon, J. C. Miller, R. C. DeKelver, E. A. Moehle et al., 2009. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459 437–441. [DOI] [PubMed] [Google Scholar]
- Smith, J., J. M. Berg and S. Chandrasegaran, 1999. A detailed study of the substrate specificity of a chimeric restriction enzyme. Nucleic Acids Res. 27 674–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, K. R., and M. R. Capecchi, 1987. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51 503–512. [DOI] [PubMed] [Google Scholar]
- Townsend, J. A., D. A. Wright, R. J. Winfrey, F. Fu, M. L. Maeder et al., 2009. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459 442–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan, Y., J. D. Haag, K. S. Chen, L.A. Shepel, D. Wigington et al., 2003. Production of knockout rats using ENU mutagenesis and a yeast-based screening assay. Nat. Biotechnol. 21 645–651. [DOI] [PubMed] [Google Scholar]