MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1-miRNA circuits (original) (raw)
Abstract
Mechanisms controlling resolution of acute inflammation are of wide interest. Here, we investigated microRNAs (miRNAs) in self-limited acute inflammatory exudates and their regulation by resolvin D1 (RvD1). Using real-time PCR analysis, we found in resolving exudates that miR-21, miR-146b, miR-208a, miR-203, miR-142, miR-302d, and miR-219 were selectively regulated (P<0.05) in self-limited murine peritonitis. RvD1 (300 ng/mouse or 15 μg kg−1) reduced zymosan-elicited neutrophil infiltration into the peritoneum 25–50% and shortened the resolution interval (_R_i) by ∼4 h. In peritonitis at 12 h, RvD1 up-regulated miR-21, miR-146b, and miR-219 and down-regulated miR-208a in vivo. In human macrophages overexpressing recombinant RvD1 receptors ALX/FPR2 or GPR32, these same miRNAs were significantly regulated (P<0.05) by RvD1 at concentrations as low as 10 nM, recapitulating the in vivo circuit. In addition, RvD1-miRNAs identified herein target cytokines and proteins involved in the immune system, e.g., miR-146b targeted NF-κB signaling, and miR-219 targeted 5-lipoxygenase and reduced leukotriene production. RvD1 also reduced nuclear translocation of NF-κB and SMAD and down-regulated phospho-IκB. Taken together, these results indicate that resolvin-regulated specific miRNAs target genes involved in resolution and establish a novel resolution circuit involving RvD1 receptor-dependent regulation of specific miRNAs.—Recchiuti, A., Krishnamoorthy, S., Fredman, G., Chiang, N., and Serhan, C. N. MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1-miRNA circuits.
Keywords: GPCR, resolution indices, lipid mediators, 5-lipoxygenase, cytokines, transcription factors
Resolution of acute inflammation is an essential physiological process required for homeostasis (ref. 1; for a review, see ref. 2). Results from this laboratory demonstrated for the first time that resolution of self-limited acute inflammation is an active programmed response involving a new genus of endogenous proresolving chemical mediators that are biosynthesized locally to control leukocyte trafficking and restore homeostasis by actively regulating proinflammatory mediators and efferocytosis (reviewed in ref. 3). Earlier, we introduced resolution indices that take into account temporal analysis of cellular components and chemical mediators in exudates (4) and permit the quantitative definition of the specific actions of endogenous proresolving mediators and pharmacologic agents within resolution (5–7). It is now evident that failure of resolution can contribute to ongoing pathogenesis of many classic and nonclassical inflammatory diseases (ref. 8; recently reviewed in ref. 9). Several epidemiological studies as well as basic investigations with transgenic models have uncovered links between inflammation, cancer (10), and atherosclerosis (11). Given the high incidence of these in humans and many other inflammation-related scenarios, appreciation of the molecular events in timely resolution are paramount, as harnessing endogenous chemical mediators of resolution may offer new therapeutic approaches.
The specialized proresolving lipid mediators (SPMs) include lipoxins (LXs), resolvins (Rvs), protectin, and maresins. These are endogenously biosynthesized from essential fatty acids during specific intervals within self-limited acute inflammation (reviewed in ref. 12). Each family of local mediators carries cell-type-specific actions that control key components in the host inflammatory response to limit collateral tissue damage and orchestrate resolution (3, 12). Among these, LXs are biosynthesized from arachidonic acid by transcellular mechanisms during cell-cell interactions via lipoxygenases (3). Aspirin treatment initiates the biosynthesis of carbon 15 epimers of LXs (i.e., 15-epi-LXs) via acetylation of cyclooxygenase-2 (3). 15-epi-LXA4 is produced in vivo in humans taking aspirin (13) and has been proved recently to mediate local anti-inflammatory actions of low-dose aspirin in healthy subjects (14). The resolvins are enzymatically produced potent local acting mediators that are biosynthesized from ω-3 essential fatty acids (i.e., EPA and DHA). For example, the D-series resolvin RvD1 is biosynthesized from DHA. Its complete stereochemistry is established as 7_S_,8_R_,17_S_-trihydroxy-4_Z_,9_E_,11_E_,13_Z_,15_E_,19_Z_-docosahexaenoic acid, and it is produced in vivo by exudates during resolution (15, 16). RvD1 exerts potent counterregulatory actions with human and murine polymorphonuclear neutrophil (PMN); stimulates nonphlogistic macrophage (MΦ) phagocytosis, termed efferocytosis (15–21); regulates cytokine and chemokines (19); and blocks nuclear factor (NF)-κB activation (18). Two G-protein-coupled receptors (GPCRs) were identified that mediate RvD1 actions expressed on human PMN and monocytes/MΦs, denoted ALX/FPR2 and GPR32 (18).
Resolution of acute inflammation is a tightly regulated process controlled by many soluble (e.g., cytokines, chemokines, lipid autacoids), as well as cell-associated receptors and growth factors (20, 22–24). An emerging line of investigation indicates that many processes, at cellular, organ, and systems level, are finely tuned by microRNAs (miRNAs) (25). miRNAs are small (22–24 nt) noncoding RNA sequences that act primarily as translational repressors of gene transcripts by interacting with their 3′ untranslated regions (3′UTR) (ref. 26; reviewed in ref. 27). miRNAs are involved in many physiological and pathological processes, including several cancers (25) and, more recently, in the immune response (28). Hence, whether miRNAs are regulated and play a functional role in the acute inflammatory response and its resolution is of considerable interest. Since RvD1 and other SPMs regulate cytokine production (19), NF-κB, and proinflammatory eicosanoids (e.g., certain prostaglandins and leukotrienes; refs. 2, 3, 18), and since miRNAs are involved in regulating these molecules (28), we postulated that miRNAs may be involved in the molecular mechanisms by which SPMs control inflammation and promote resolution.
Here, we identified a miRNA signature of resolution within resolving self-limited inflammatory exudates. The proresolving mediator RvD1 regulated resolution indices and controlled specific miRNA expression in exudates in vivo that were also activated via recombinant receptors ALX/FPR2 and GPR32 regulating miR-146b, miR-208a, and miR-219 (ex vivo). These RvD1-regulated miRNAs control target genes with roles in the human immune system and are the first identified miRNAs in resolvin resolution circuits.
MATERIALS AND METHODS
Materials
The following materials were obtained: Histopaque 1077 and zymosan A (from Saccharomyces cerevisiae; Sigma Aldrich, St. Louis, MO, USA); recombinant human granulocyte-MΦ colony-stimulating factor (GM-CSF; R&D Systems, Minneapolis, MN, USA); JetPEI MΦ transfection reagent (PolyPlus Transfection, Illkirch, France). Fluorochrome-conjugated antibodies (Abs) for flow cytometry were rat anti-mouse CD11b (Mac-1 α chain, clone M1/70) and CD16/32-blocking Ab (clone 2.462) from BD Bioscience (San Jose, CA, USA); Gr-1 (Ly-6G; clone RB6–8C5), F4/80 (clone BM8), and isotype-matched IgG controls were purchased from eBioscience (San Diego, CA, USA). Expression plasmids for ALX/FPR2 (UniProt Knowledgebase ID P25090), GPR32 (O75388) and control empty plasmid (pcDNA3) were obtained as in ref. 29. miRNA expression vectors for human miR-146b (miRBase ID MI0003129), miR-208a (MI0000555), mir-219 (MI0000296), and control pCMV-miR empty vector were obtained from Origene (Rockville, MD, USA). Reagents for cell culture were purchased from Lonza (Walkersville, MD, USA) and Life Technologies (Carlsbad, CA, USA). Murine IL-10 ELISA kit was purchased from R&D Systems. Resolvin D1 (7_S_,8_R_,17_S_-trihydroxy-4_Z_,9_E_,11_E_,13_Z_,15_E_,19_Z_-docosahexaenoic acid, as designated in ref. 16) was obtained from Cayman Chemical (Ann Arbor, MI, USA) and converted to its carboxy-methyl ester (ME) as in refs. 16, 18. Before each experiment, concentration and integrity were assessed by characteristic physical criteria, UV absorbance (in MeOH) and liquid chromatograhy–tandem mass spectrometry (LC-MS/MS). Compounds were taken to dryness with a gentle N2 stream, immediately dissolved in the appropriate vehicle, and kept in the dark on ice before the experiments. All other reagents were from Sigma Aldrich.
Murine peritonitis, flow cytometry, and resolution indices
Male FVB mice (6–8 wk old; Charles River Laboratories, Wilmington, MA, USA) were anesthetized with isoflurane (Aerrane; Baxter, Deerfield, IL, USA) and administered zymosan A (1 mg/mouse) in 1 ml of sterile saline alone or with RvD1-ME (300 ng/mouse or ∼15 μg/kg) by intraperitoneal (i.p.) injection. At selected time intervals, mice were euthanized with an overdose of isoflurane, peritoneal exudates were collected by lavaging with Ca2+/Mg2+-free PBS−/− (5 ml), and leukocytes were counted under a light microscope. All procedures were conducted in accordance with protocols approved by the Harvard Medical School Standing Committee on Animals guidelines for animal care (Protocol 02570). Differential cell numbers were used to determine resolution indices as in ref. 4, i.e., _T_max, time point of maximum PMN infiltration (Ψmax); _T_50, time necessary to achieve 50% reduction in PMN number (Ψ50) from Ψmax; resolution interval (Ri = _T_50 − _T_max); time interval between _T_max and _T_50.
miRNA nomenclature
The miRNAs investigated in the current study are reported in accordance with the miRBase registry gene annotation and nomenclature (http://www.mirbase.org; ref. 30) or the National Center for Biotechnology Information (NCBI; Bethesda, MD, USA) database.
miRNA array and quantitative real-time PCR
miRNA fractions from mouse and human cells were isolated (High Pure miRNA Isolation kit; Roche Applied Science, Indianapolis, IN, USA). All reagents for miRNA arrays were from SA Biosciences (MAM-3100 containing 376 miRNAs; SA Biosciences, Frederick, MD, USA). Reactions were carried out with an ABI 7900HT thermal cycler (Applied Biosystems, Foster City, CA, USA). Results were analyzed using the 2−Δ_Ct_ relative quantification method (31). miRNAs that displayed threshold cycles (Ct) >35 were excluded from the analysis. For each target miRNA, Δ_Ct_ was calculated by subtracting their Ct values from the average Ct value of small noncoding RNA (snoRNA251, snoRNA202, snoRNA142, Rnu6) used as reference housekeeping (HK) miRNA. Real-time PCR analyses of selected miRNAs were carried out using specific primer pairs (miScript; Qiagen, Germantown, MD, USA). For analysis of human miRNAs, small nucleolar RNA U1 (RNAU1A; NCBI accession no. NR_004421) was selected as reference miRNA; for in vivo experiments, RNAU1A and small Cajal body-specific RNA 17 (SCARNA17, NR_003003) were used as HK. Data analyses were carried out with StatMiner software (Integromics, Philadelphia, PA, USA).
Computational analysis of miRNA target genes
Computational prediction of putative interaction of selected candidate miRNA to 3′UTR of mRNAs was conducted with miRecords (http://mirecords.biolead.org; ref. 32). mRNA targets predicted by ≥3 algorithms were chosen for further analyses.
Putative mRNA targets were uploaded into Ingenuity Pathway Analysis software (IPA; http://www.ingenuity.com) to generate connection maps based on their biological functions and interaction (33). Statistical significance for the inferred association between target mRNAs and biological pathways/processes was assessed by both P value and score, i.e., a measure of the likelihood that the association between a set of genes and a given pathway is not due to random chance. Genes associated in IPA to inflammation and immune response with the highest relative score and the lowest P values (P<0.02) were reported and taken into consideration for further analysis.
Human MΦ isolation, transfection, and eicosanoids
Human MΦs were derived from peripheral blood monocytes isolated from deidentified healthy volunteers (Brigham and Women's Hospital protocol no. 88-02642) as described by Krishnamoorthy et al. (18). miRNA fractions were extracted, and expression levels of miRNAs (see text and figure legends) were assessed by real-time PCR with specific primers as described above. Human MΦs (2×106/10-cm dish) transfected with either miR-219 or empty vector were incubated with zymosan (5×106 particles/1×106 MΦs) for 2 h at 37°C in PBS+/+. Following incubation, 2 vol of methanol and deuterated internal standards (0.5 ng d4-PGE2, d4-LTB4, and d8-5S-HETE) were added. Cell suspensions were extracted by solid-phase extraction using a C-18 column (6 ml; Waters, Milford, MA, USA). The methyl formate fractions were analyzed for LTB4 and PGE2 levels by LC-MS/MS using an HPLC system (LCZO; Shimadzu, Kyoto, Japan) with a linear ion trap quadrupole mass spectrometer (3200 QTRAP; Applied Biosystems) equipped with an Eclipse Plus C18 column (4.6×50 mm×1.8 μm; Agilent Technologies, Palo Alto, CA, USA). Data acquisition was carried out in negative ionization mode. The enhanced product ion (EPI) mode was performed at a flow rate of 400 μl/min with a mobile phase composed of methanol/water/acetic acid at a ratio of 60:40:0.01 (v/v/v) and ramped to 80:20:0.01 over 7.5 min and to 95:5:0.01 in the next 4.5 min. For multiple reaction monitoring (MRM), the mobile phase consisted of methanol/water/acetic acid at 60:40:0.01, ramped to 90:10:0.01 after 10 min and to 100:0:0.01 after 12 min. Ion pair 335.2/195.1 was used to monitor LTB4 and 351.2/189.1 for PGE2; the levels were quantitated using calibration curves for each compound, and recoveries were calculated using the deuterated internal standards added prior to extraction (34).
Analysis of transcription factors
Human monocytes (2–5×106 cells/incubation) from healthy subjects were incubated with RvD1 (10.0 nM) or vehicle alone for 90 min at 37°C in PBS+/+. Nuclear protein fractions were isolated with the Nuclear Extraction kit (Signosis, Sunnyvale, CA, USA), and array analyses of transcription factors were carried out following manufacturer's instructions. Transcription factor heat map was generated by assigning fold changes to color intensities in a gradient from red to black to green. Evaluation of NF-κB pathway activation was carried out with whole-cell protein lysates from human monocytes (see above) using PathScan sandwich ELISA kits (Cell Signaling Technology, Beverly, MA, USA).
miRNA overexpression and target identification
Expression vectors containing the candidate hsa-miR-146b, miR-208a, and miR-219 or mock empty vector (all from Origene, Rockville, MD, USA) were transfected (each at 5 μg/2.5×106 cells) in primary human MΦs as described by Krishnamoorthy et al. (18). Total RNA was isolated, reverse transcribed to cDNA (Omniscript kit; Qiagen) and used to determine mRNA levels of potential targets involved in immune response and inflammation using the RT2 Profiler PCR Array (PAHS-O52E; SA Biosciences) and human inflammation TaqMan low-density array (4378722; Applied Biosystems). Data were acquired and analyzed using the 2−Δ_Ct_ method (31).
Western blots and ELISA
Proteins (50 μg) from human MΦs transfected with miR-containing or empty vectors were separated using SDS-PAGE and electroblotted to nitrocellulose membranes (Bio-Rad, Hercules, CA, USA). Membranes were probed with rabbit anti-human 5-lipoxygenase (5-LO; AB3796, 1:1,000; Millipore, Billerica, MA, USA) or goat anti-actin (1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA), followed by the HRP-conjugated secondary antibody. Immunoreactive signals were detected by incubating with a chemiluminescence system (Pierce, Rockford, IL, USA) and radiography. Semiquantitative densitrometric analysis was carried out using ImageJ software (U.S. National Institutes of Health, Bethesda, MD, USA). Supernatants from murine peritonitis at indicated time points were analyzed for IL-10 (ELISA, R&D Systems). IL-10 values obtained were then normalized to total protein in exudate supernatant, measured using Bradford's method.
Statistics
For statistical analysis of in vitro and in vivo experiments, unpaired or paired Student's t tests were used as appropriate, with values of P < 0.05 considered to be significant.
RESULTS
Mapping miRNA-dependent resolution circuits: temporal analysis of miRNAs in inflammation resolution
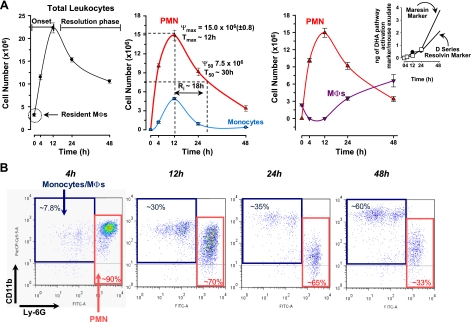
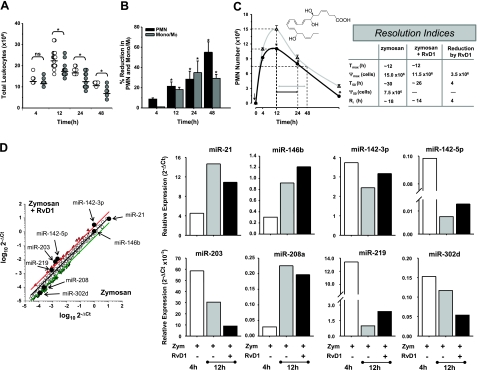
To identify miRNAs in self-resolving exudates, we used a systems approach with self-limited acute inflammatory exudates from murine peritonitis (Fig. 1). Zymosan A particles from S. cerevisiae, a Toll-like receptor 2 and 4 ligand, were administered by i.p. injection, and peritoneal exudates were collected to monitor temporal changes in the leukocyte composition. Rapidly after zymosan challenge, leukocytes started infiltrating the peritoneal cavity during the onset phase of acute inflammation (4 h), reaching a maximum of ∼22.0 ± 1.1 × 106 total cells at ∼12 h, which was followed by a slower decline in PMN numbers at 24–48 h, characteristic of the resolution phase (Fig. 2A, left and middle panels). Differential analysis with flow cytometry confirmed that the majority of extravasated leukocytes at 4 and 12 h were Ly-6G_high_/CD11b+ PMN (Fig. 2A, middle panel; B), which represented ∼90 and 70% of total leukocytes (10.0±0.3×106 cells at 4 h and 14.9± 0.8×106 cells at 12 h). PMN number declined during the resolution phase to 9.2 ± 0.8 × 106 and 3.3 ± 0.5 × 106 cells at 24 and 48 h after initiation of peritonitis. Conversely, resident peritoneal MΦs (i.e., F4/80+ cells), which represent the main leukocyte population in naive mice peritonea (35), were not detected in exudates at 4 h (Fig. 2A, right panel). Monocytes (Ly-6G_low/CD11b_high cells) gradually increased at 12 h to 5.6 ± 0.3 × 106 and differentiated into MΦs, as reflected by an increase in size, granularity, and CD11b surface expression (Fig. 2B). At 48 h after zymosan injection, ∼60% of exudate leukocytes were elicited MΦs (Fig. 2A, right panel; B). These results were used to calculate resolution indices, as introduced by Bannenberg et al. (4), and were characteristic of self-limited acute inflammation and resolution, i.e., Ψmax = 14.9 ± 0.8 × 106; _T_max ∼ 12 h; Ψ50 ∼ 7.5 × 106; _T_50 ∼ 30 h; Ri ∼ 18 h (see Materials and Methods).
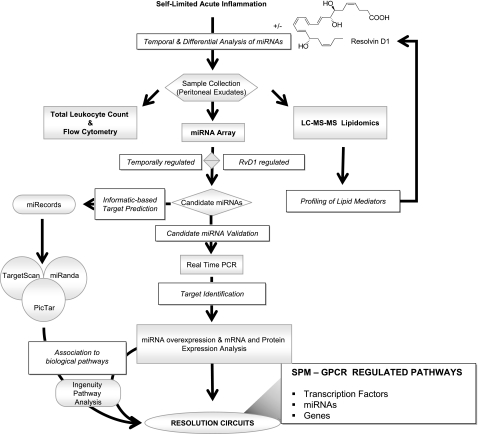
Figure 1.
Strategy for identifying RvD1-regulated miRNAs involved in resolution. Scheme of temporal and differential miRNA expression and analyses of resolving exudates.
Figure 2.
Strategy for identifying RvD1-regulated miRNAs involved in resolution. A) Self-limited acute inflammation (i.e., murine peritonitis) was used to obtain exudates (see Fig. 1). For temporal analysis, lavages were collected at 4, 12, 24, and 48 h after zymosan A injection. For resolution indices of self-limited peritonitis, peritoneal lavages were collected from zymosan A (1 mg/mouse, i.p.)-challenged mice (_n_=3–16 mice/time point). Total leukocytes (left panel), PMN (triangles) and monocytes (circles) (middle panel), and elicited MΦs (nablas; right panel) were determined (see Materials and Methods). Results are means ± se. Dotted lines indicate time points; PMN number values were used to determine resolution indices as in ref. 4; i.e., _T_max, time of maximum PMN infiltration (Ψmax); _T_50, time to achieve 50% reduction in PMN number (Ψ50) from Ψmax; _R_i, resolution interval (_T_50 − _T_max; time interval between _T_max and _T_50). Right panel, inset shows the rate of accumulation of 17- and 14-hydroxy-docosahexaenoic acid (HDHA) markers of D-series resolvin (solid circles) and maresin (open squares) pathway activation, respectively, in resolving exudates replotted here for comparison (50). B) Representative flow cytometry dot plots of Ly-6G and CD11b expression in exudate cells.
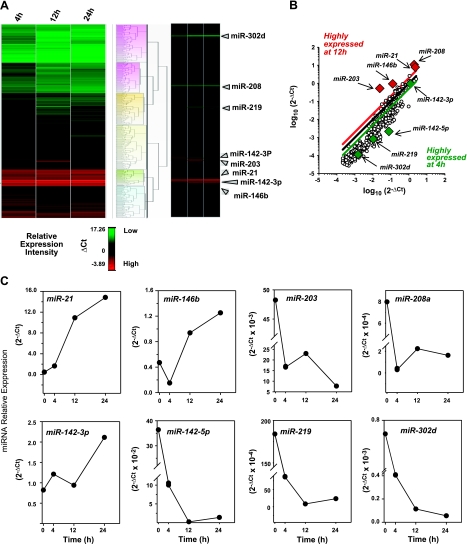
With the resolution phase characterized, we next investigated temporal changes in miRNA expression in self-resolving exudates collected 4, 12, and 24 h after zymosan challenge, which correspond to the onset, maximum, and resolution phase of the acute inflammatory response. Of the 376 mouse miRNA molecules examined using a miRNA real-time PCR array system, ∼95% were amplified with Ct values < 35 cycles by quantitative PCR (not shown) and were suitable for relative expression analysis. Hierarchical clustering (**Fig. 3_A_** and Supplemental Fig. S1) grouped these miRNAs into distinct clusters based on their relative abundance at the different time intervals taken within the inflammatory response. In particular, among the few miRNAs highly expressed in the exudates at 12 h, miR-21, miR-146b, miR-208a, and miR-203 displayed >2.5-fold change when compared to 4 h. Conversely, a large group of miRNAs showed lower expression (<0.4-fold change) between 12-h and 4-h time points (Fig. 3B, C). Among these, we focused on miR-142–5p and miR-142–3p, miR-219, and miR-302d because they showed consistent down-regulation at 12 h compared to 4 h in this system (Fig. 3B, C). Therefore, we next determined the expression levels of miR-21, miR-146b, miR-208a, miR-203, miR-142–3p, miR-142–5p, miR-219, and miR-302d along the full time course of acute peritonitis using real-time PCR.
Figure 3.
miRNA array from resolving exudates. A) Peritoneal leukocytes were collected at 4, 12, and 24 h after zymosan A (1 mg/mouse, i.p.) injection, and miRNA fractions were isolated. Heat map cluster represents relative expression of ∼300 target mouse miRNAs from resolving exudates (n_=3 mice/time point) determined with real-time PCR array after normalization with housekeeping small RNAs (see Materials and Methods). Dendrogram was generated to cluster miRNAs according to their relative expression with the Manhattan distance and Ward's method using StatMiner Software. High-resolution full-size heat map is included in Supplemental Fig. S1. Relative expression intensities are indicated in a green-red color code based on Δ_Ct values. B) Scatterplot of relative expression of ∼300 miRNAs analyzed in exudate cells at 4 and 12 h peritonitis. Red and green lines in the scatterplot indicate 2.5- and 0.4-fold changes in expression between 12 and 4 h, which were used for defining candidate miRNA (red and green diamonds). Black line represents no changes. C) Time course of miRNA relative expression in resolving exudates. miRNAs were selected among those that displayed fold changes >2.5 or <0.4 at 12 h of zymosan-induced peritonitis compared to 4 h. Relative expression was assessed with real-time PCR array (see Materials and Methods). Results are expressed as means of 3 mice.
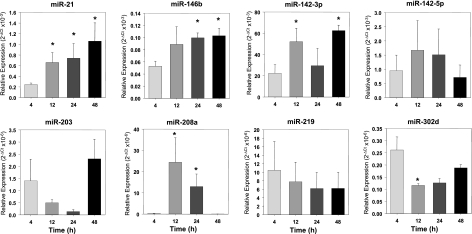
Consistent with results from the arrays, both miR-21 and miR-208a were significantly up-regulated at the peak of PMN infiltration compared to the onset (Fig. 4; _P_=0.037 and _P_=0.048 vs. 4 h, respectively). While miR-21 remained up-regulated at 24 and 48 h (i.e., early and late resolution), miR-208a expression gradually declined at 24 and 48 h. Also, miR-146b was up-regulated at 24 and 48 h (P<0.05 vs. 4 h). It is noteworthy that miR-302d was highly expressed in early exudates and rapidly decreased between _T_max and 48 h. In comparison, results for miR-142–3p, miR-142–5p, and miR-203 did not match the profiles obtained from the miRNA array. In particular, miR-203 showed a trend toward reduction at _T_max and _T_50 and an increase at 48 h, while miR-142–3p displayed an opposite trend compared to PCR array, and miR-142–5p did not change from 4 to 48 h (Fig. 4). We observed a gradual decrease for miR-219, which did not attain statistical significance, likely because of high variability in expression levels obtained for 4-h exudates. Supplemental Table S1 lists genes reported earlier as molecular targets of these miRNAs (see reference in Supplemental Table S1). Together, these findings indicate that specific miRNAs are temporally regulated during acute inflammation and its natural self-limited resolution.
Figure 4.
Real-time PCR analysis of miRNAs reveals their specific temporal regulation in resolving exudates. Kinetics of relative expression for indicated miRNAs was determined from exudates collected at different time intervals after zymosan A injection using real-time PCR after normalization (see Materials and Methods). Results are expressed as means ± se (_n_=3–6 mice) *P < 0.05 vs. 4 h.
Resolvin D1 controls acute inflammation, accelerates resolution, and regulates leukocyte miRNA expression
RvD1 is biosynthesized during resolution (15), regulates further PMN infiltration, and stimulates nonphlogistic MΦ phagocytic activity (16–19). Resolution indices were used here to define and pinpoint RvD1 actions in resolution. To this end, RvD1-methyl ester was administered by i.p. injection (300 ng/mouse) with zymosan A (1 mg/mouse), and leukocyte numbers and composition were assessed in the exudates. The carboxyl methyl ester was used here because it is more chemically stable, and mouse esterase activity is likely to rapidly convert RvD1 methyl ester to the bioactive mediator in vivo (36). Administration of RvD1 resulted in a significant decrease in leukocyte infiltration at 12 h, consistent with RvD1 defined actions (refs. 15, 16 and Fig. 5A). Differential analysis revealed reduction in PMN, monocyte, and elicited MΦ presence in the peritoneal exudates (Fig. 5B). Next, quantitative resolution indices were determined, providing an unbiased definition of RvD1 effect (Fig. 5C). Locally administered RvD1 significantly lowered Ψmax from 15.0 to 11.5 × 106 of extravasated PMN, and it accelerated resolution by shortening the _R_i ∼ 4 h from 18 to 14 h. Results from these experiments led us to carry out differential analyses to identify specific RvD1-regulated miRNAs that might favor anti-inflammatory and proresolving actions (Supplemental Fig. S2). miRNA array analysis showed that RvD1 increased levels of miR-146b, miR-142–3p, miR-142–5p, and miR-219 in exudate cells at _T_max (Fig. 5D). RvD1 slightly down-regulated miR-21, miR-203, miR-208, and miR-302d at this time point (Fig. 5D). Computational analysis was performed using miRecords software to search for mRNA targets of RvD1 regulated miRNAs (see Materials and Methods). Among these, we focused on those genes with functions in immune systems identified with the IPA program (Table 1).
Figure 5.
Resolvin D1 controls acute inflammation, accelerates resolution, and selectively regulates miRNA expression in exudates. A, B) Male FVB mice were administered zymosan alone (1 mg/mouse, i.p.; A, open circles) or zymosan plus RvD1 (300 ng/mouse, i.p.; A, shaded circles); exudates were collected at indicated time points to determine total leukocytes (A) and PMN and monocytes/MΦs (B) using flow cytometry, as described in Materials and Methods. C) Resolution indices. Solid circles and open triangles (left panel) indicate each of the parameters used to define the resolution indices (right panel) in zymosan (left panel, gray lines) or zymosan plus RvD1 (black lines) group, as described by Bannenberg et al. (4). Arrow denotes time of zymosan and RvD1 addition. Results are expressed as means ± se (_n_=3–16 mice). *P < 0.05 vs. zymosan. Inset: RvD1 structure. D) Scatterplot and temporal profile of miRNA expression from exudates collected 12 h after injection of zymosan alone or zymosan plus RvD1 (300 ng/mouse, i.p.). Red and green lines in scatterplot represent boundaries of 2.5- and 0.4-fold changes by RvD1 compared to zymosan-treated mice, which were used as cutoff values to define RvD1-regulated miRNA. Results are means of 3 mice.
Table 1.
Targets for RvD1-regulated miRNAs
| miRNA symbol | Fold change by RvD1, 12 h | miRecords IPA | ||
|---|---|---|---|---|
| Total putative target mRNA | Putative targets associated with inflammation and immune response | P value | ||
| miR-142-3p | +3.3 | 6,861 | IL-6, IL-23A, TGFBR1 | 1.4 × 10−3 |
| miR-142-5p | +5.1 | 10,018 | c-Fos, ATF2, SRF, CREB | 1.9 × 10−5 |
| miR-203 | −6.2 | 9,310 | STAT5, SOCS3, JAK1 | 3.2 × 10−4 |
| miR-219 | +2.2 | 1,507 | TNF-α, TNF-αR, IL-1, IL-1R accessory protein | 1.8 × 10−2 |
Resolvin D1 temporally regulates miRNA expression
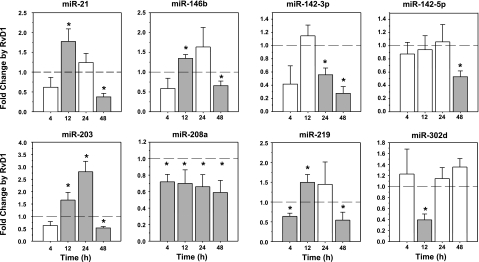
To validate array results and to further examine the time-dependent RvD1 regulation of identified miRNAs, real-time PCR analyses were conducted. RvD1 significantly up-regulated miR-146b (P<0.01) and miR-219 (_P_=0.02), while reducing miR-208 (_P_=0.01), and miR-302d (_P_=0.03) when compared to zymosan-treated mice at _T_max values, which were in agreement with array results (Fig. 6). Both miR-21 and miR-203 showed significant up-regulation by RvD1 at 12 h (_P_=0.02 and _P_=0.04, respectively), whereas no significant changes were observed for the 142 family at the peak of PMN infiltration at 12 h. The 142–3p was significantly decreased in resolution at 24 and 48 h, and 142–5p was reduced at 48 h (Fig. 6). Interestingly, mice that were administered RvD1 together with zymosan had significant reduction in expression levels of all miRNAs analyzed at 48 h, with the exception of miR-302d, which was not regulated in RvD1-treated mice at this time point. These findings suggest that RvD1 temporally controls specific sets of miRNAs in exudates in vivo.
Figure 6.
Resolvin D1 temporally regulates miRNA expression in resolving exudates: real-time PCR. miRNA fractions were extracted from peritoneal exudates at 4, 12, 24, and 48 h after injection of zymosan alone or zymosan plus RvD1 and quantified using real-time PCR after normalization (see Materials and Methods). Dotted lines indicate no changes in miRNA expression. Shaded bars indicate statistically significant expression levels. Results are expressed as means ± se (_n_=3–6 mice). *P < 0.05 vs. zymosan alone at each time point.
RvD1 controls miRNAs via receptor interaction.
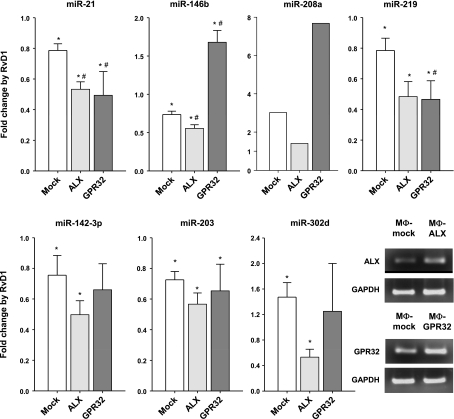
For translation to humans, we next investigated whether these regulatory actions of RvD1 on mouse miRNA expression were dependent on the specific RvD1 receptors ALX/FPR2 and GPR32 in isolated human cells (18). Since MΦs are key players in innate immunity and resolution, we chose to overexpress GPCRs and miRNAs in this cell type. As shown in Fig. 7, RvD1 (10.0 nM) down-regulated miR-21 (P<0.01), miR-146b (P<0.01), and miR-219 (_P_=0.04) by ∼25% in human MΦs, whereas it increased levels of miR-208a by ∼3-fold (cells from 5 separate donors gave consistent increases that ranged from 2- to 7-fold) and miR-302d by ∼1.5-fold (P<0.01). Of interest, miR-21 was significantly decreased in both ALX/FPR2- and GPR32-overexpressing MΦs compared to mock-transfected cells, whereas miR-146b was significantly up-regulated in MΦ-GPR32. Also, miR-302d and miR-219 down-regulation was evident in MΦ-ALX- as well as MΦ-GPR32-overexpressing cells. Although RvD1 treatment of mock-transfected cells resulted in a significant reduction (∼33%) of miR-142-3p and miR-203, they were not further regulated in ALX/FPR2- and/or GPR32 receptor-overexpressing MΦs. Some miRNAs (e.g., miR-21 and miR-146b) are differentially regulated by RvD1 in vivo when compared to the isolated in vitro MΦs overexpressing either ALX or GPR32. This could likely be a result of species difference, a lower signal-to-noise ratio in vivo, and the differences in cell types between in vivo and in vitro experiments. These results indicate that RvD1 regulates specific miRNA molecules via its interactions with specific RvD1 receptors.
Figure 7.
Resolvin D1 regulates miRNA expression via recombinant receptors ALX/FPR2 and GPR32. Regulation of miRNA levels by RvD1 in human MΦs transfected with ALX/FPR2 or GPR32. Relative expression of indicated miRNAs from MΦs incubated with RvD1 (10.0 nM, 6 h, 37°C) compared to vehicle alone was determined using real-time PCR analysis. Results are expressed as means ± se (_n_=3–5 healthy subjects). Results for miR-208 are representative of 5 healthy subjects. Bottom right panel: receptor expression levels by PCR in ALX/FPR2- and GPR32-overexpressing MΦs from a representative healthy subject. *P < 0.05 vs. vehicle alone; *P < 0.05 vs. MΦ-mock transfected cells.
Identification of gene targets for RvD1-regulated miRNAs and RvD1-activated transcription factors
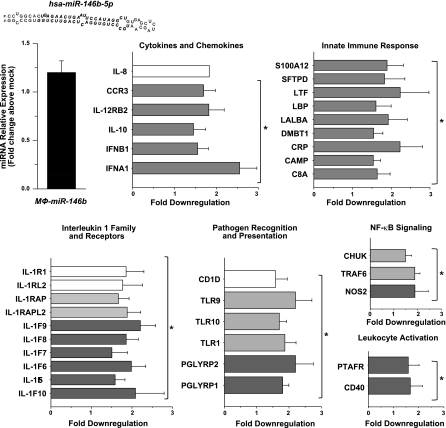
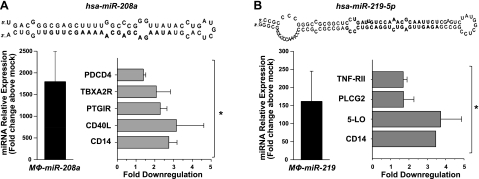
Next, we sought to identify target genes regulated by miR-146b, miR-208, and miR-219, since they were regulated by RvD1 both in vivo and in vitro in a receptor-dependent manner. To this end, human MΦs were transfected with expression vectors for human miR-146b, miR-208a, and miR-219. Up-regulation of each miRNA was confirmed using real-time PCR as early as 24 h after transfection (not shown) and remained stable up to 48 h (Figs. 8–11). Therefore, we selected 48 h as an end point to assess down-regulation of mRNA of genes involved in the inflammatory and immune response in MΦ-miR. As shown in Fig. 8, overexpression of miR-146b resulted in significant down-regulation of transcripts of several cytokines, chemokines, and their receptors, such as interleukin (IL)-8 and IL-10, IL-12 receptor β2, chemokine CC motif receptor 3 (CCR3), interferon (IFN) α1 and IFNβ1, and members of IL-1 family. Also, genes involved in the innate immune response and pathogen recognition, i.e., S100A12, LPS binding protein (LBP), C-reactive protein, complement component 8 α polypeptide (C8a), Toll-like receptor (TLR) 9 and 10, and peptidoglycan recognition protein (PGLYRP) 1 and 2, were significantly reduced in MΦ-miR-146b. In addition, we found reductions in the mRNAs for conserved helix-loop-helix ubiquitous kinase (CHUK), also known as IκB kinase (IKK) α, tumor necrosis factor receptor-associated factor 6 (TRAF6), nitric oxide synthase 2, platelet-activating factor receptor (PTAFR), and CD40 (Fig. 8). In miR-208a-overexpressing MΦs, reductions were obtained for mRNAs encoding for CD14, CD40 ligand (CD40L), prostacyclin I2 receptor (PTGIR), thromboxane A2 receptor (TBXA2R), and programmed cell death 4 (PDCD4; Fig. 9A), a tumor suppressor molecule that regulates NF-κB activation and decreases IL-10 production (37). This result was consistent with the existence of a putative seed region for miR-208a in the 3′ UTR of PDCD4, which was predicted using miRecords software. miR-219 overexpression gave significant reduction in CD14, TNF receptor II, phospholipase Cγ2, and arachidonate 5-lipoxygenase (5-LO; Figs. 9B and 12A), as well as leukotriene production (Fig. 12B).
Figure 8.
RvD1-regulated miRNAs target genes with specific roles in inflammation and resolution. Human MΦs were transfected with plasmids containing hsa-miR-146b, as described in Materials and Methods. Figure shows the expression level of indicated miRNA (top left) and down-regulated target mRNAs in MΦ-miR compared to MΦ-mock transfected cells determined using real-time PCR 48 h after transfection. Results are expressed as means ± se (_n_=3–4 healthy subjects). Result for IL-8 mRNA level is representative of 4 healthy subjects. *P < 0.05 vs. MΦ-mock transfected cells. C8A, complement component 8, polypeptide α; CAMP, cathelicidin antimicrobial peptide; CHUK, conserved helix-loop-helix ubiquitous kinase (IKK-α); CRP, C reactive protein; DMBT1, deleted in malignant brain tumors 1; IFN, interferon; IL, interleukin; IL1F, IL-1 family member; IL1R1, IL-1 receptor 1; IL1RAP, IL-1 receptor accessory protein; IL1RAPL2, IL-1 receptor accessory protein-like 2; IL1RL2, IL-1 receptor-like 2; LALBA, lactalbumin α; LPB, lipopolysaccharide binding protein; LTF, lactotransferrin; NOS2, nitric oxide synthase 2 (iNOS); PGLYRP, peptidoglycan recognition protein; PTAFR, platelet-activating factor receptor; S100A12, S100 calcium-binding protein A12; SFTPD, surfactant protein D; TLR, Toll-like receptor; TRAF, tumor necrosis factor receptor-associated factor.
Figure 9.
RvD1-regulated miRNAs target genes with specific roles in inflammation and resolution. Human MΦs were transfected with plasmids containing miR-208a (A) or miR-219 (B), as described in Materials and Methods. Each panel shows the expression levels of indicated miRNAs (left) and down-regulated target mRNAs in MΦ-miR compared to MΦ-mock transfected cells determined using real-time PCR 48 h after transfection. Results are expressed as means ± se (_n_=3–4 healthy subjects). *P < 0.05 vs. MΦ-mock transfected cells. 5-LO, arachidonate 5-lipoxygenase (ALOX5); CD40L, CD40 ligand; PDCD4, programmed cell death 4; PLCG2, phospholipase Cγ2; TBXA2R, thromboxane A2 receptor; TNF-RII, tumor necrosis factor-α receptor 2.
Figure 10.
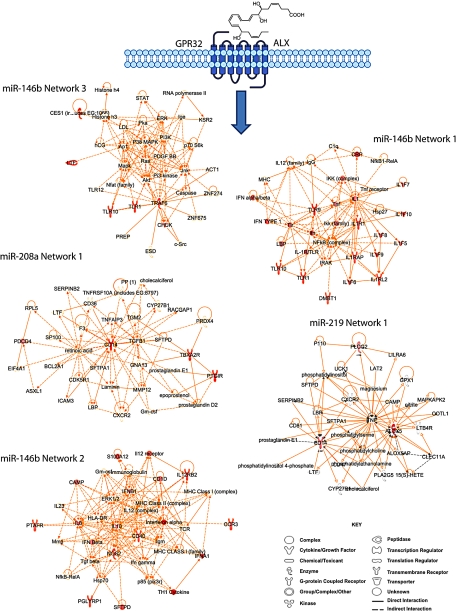
IPA networks connecting target genes of RvD1-regulated miRNAs. Genes that were significantly down-regulated in MΦ-miR are indicated in red within each network.
Figure 11.
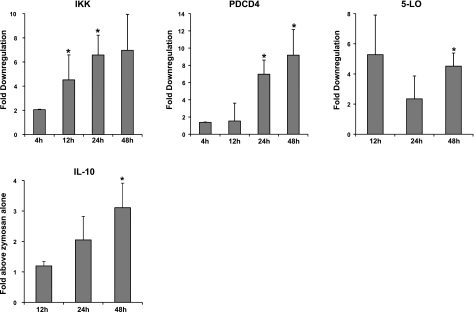
RvD1 regulates miRNA target genes in self-limited murine acute peritonitis. Messenger RNA fractions were isolated from peritoneal exudates 4, 12, 24, and 48 h after injection of zymosan with or without RvD1. Top panels: relative expression levels of IKK, PDCD4, and 5-LO measured using real-time PCR and normalized to GAPDH levels. Bottom panel: IL-10 protein levels determined by ELISA and normalized to total protein. Results are expressed as means ± se of fold change above zymosan alone (_n_=3–9 mice). *P < 0.05 vs. zymosan alone. 5-LO, arachidonate 5-lipoxygenase (ALOX5); IKK, IκB kinase [conserved helix-loop-helix ubiquitous kinase (CHUK)]; IL-10, interleukin 10; PDCD4, programmed cell death 4.
Figure 12.
miR-219 regulates 5-LO protein levels and LTB4 generation. A) Western blot analyses show 5-LO protein levels in MΦ-vector and MΦ-miR-219, determined 72 h after transfection. Results are expressed as means ± se of relative intensities of immunoreactive bands of target proteins quantitated and normalized to actin, used as loading control (_n_=3 healthy subjects). *P < 0.05 vs. MΦ-vector. Inset: representative immunoblot radiograph visualized using chemiluminescence. B) Quantitation of LTB4 and PGE2 levels using LC-MS/MS of MΦ-vector and MΦ-miR-219 cells incubated with 5 × 106 zymosan particles/106 MΦs (2 h, 37°C). Representative chromatogram from n = 3 healthy subjects is shown. Inset: mean ± se percentage reduction in LTB4 levels in miR-219 transfected MΦs and MS/MS spectrum of LTB4 (_n_=3 healthy subjects). *P < 0.05 vs. MΦ-vector. C) RvD1 regulates activation of transcription factors in human monocytes. Peripheral blood monocytes (see Materials and Methods) were incubated with RvD1 (10 nM) or vehicle alone for 90 min at 37°C. Activation of indicated transcription factors was assessed using a competitive ELISA kit. Heat map depicts fold change in nuclear transcription factor levels by RvD1 compared to vehicle alone (heat map generated from mean of _n_=4 healthy subjects). See Supplemental Data for list of TF abbreviations. D) Left panel: percentage reduction by RvD1 in nuclear translocation of NF-κB and SMAD in human monocytes. Results are expressed as means ± se (_n_=4 healthy subjects). *P < 0.05 vs. vehicle alone). Right panel: RvD1 regulation of phospho-IκB levels in human monocytes. Peripheral blood monocytes were incubated with 10 nM RvD1 or vehicle for 15 min, followed by 1 ng TNF-α or vehicle for 90 min at 37°C. Phospho-IκB levels were measured. Results are from 4 healthy subjects.
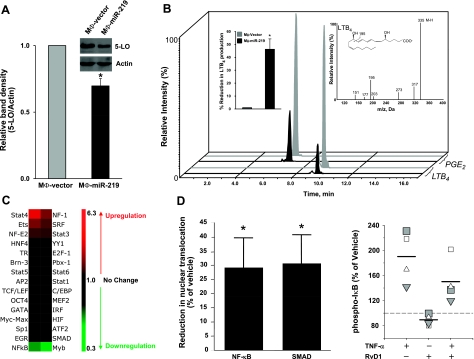
To further evaluate and validate these results, we assessed changes in 5-LO protein levels in MΦ-miR-219. Using Western blot analysis, we observed a 20–30% reduction in 5-LO in miR-219-overexpressing MΦs compared to mock-transfected cells (Fig. 12A), findings consistent with real-time PCR and leukotriene production (Figs. 9B and 12A, B). Moreover, using the IPA knowledge database, we constructed connection maps where these genes occupy nodal positions in the matrices connected by edges. Targets of RvD1-regulated miR-146b, miR-208a, and miR-219 clustered in 5 gene networks (Fig. 10). Biological functions and canonical pathways regulated by these genes included inflammatory response; antigen presentation; antimicrobial response; infectious, inflammatory, and immunological diseases; gene expression; lipid metabolism; and cell signaling (Supplemental Fig S3). To identify in vivo correlates for RvD1-miR-dependent gene regulation, we determined the levels of IKK, PDCD4, 5-LO, and IL-10 in self-limited acute murine peritonitis. Administration of RvD1 (300 ng, i.p.) significantly reduced the mRNA levels of IKK at 12 and 24 h (P<0.05), PDCD4 at 24 and 48 h (P<0.05), and 5-LO at 48 h (P<0.05) in peritoneal exudate cells (Fig. 11). Conversely, IL-10 protein levels were increased by RvD1 in comparison to zymosan alone (Fig. 11).
Since many of the target genes of RvD1-regulated miRNA networks were involved in transcription factor (TF) regulation, we screened a panel of TFs with human monocytes to gain insight into potential translation. In these experiments, GM-CSF, an inducer of monocyte differentiation, was not used in order to address the RvD1-dependent regulation of TFs. The heat map in Fig. 12C shows the screened TFs that are regulated by RvD1 (10 nM, 90 min, 37°C) in human monocytes. Of interest, RvD1 significantly reduced nuclear translocation of NF-κB and SMAD compared to vehicle (P<0.05; Fig. 12D, left panel). Since network analyses revealed that central positions in RvD1-miRNA circuits are occupied by molecules belonging to the TNF-α–NF-κB axis, and because RvD1 blocked NF-κB nuclear translocation, we investigated whether RvD1 counterregulated TNF-α-dependent regulation of phosphorylation of IκB. Indeed, RvD1 reduced TNF-α-induced phosphorylation of IκB, a critical step in NF-κB activation and nuclear translocation (Fig. 12D, right panel). Therefore, these results suggest that RvD1 regulates the NF-κB pathway in human monocytes.
DISCUSSION
In the present study, we identified specific endogenous miRNAs and resolvin-dependent miRNAs that were temporally regulated in a self-limited acute inflammatory response. These include miR-21, miR-146b, miR-208a, and miR-219, which represent a new class of innate and proresolving miRNAs. The potent proresolving mediator RvD1 regulated these miRNAs in a GPCR-dependent manner. In addition, we identified target genes of miRNAs regulated by RvD1-GPCR interactions that are components of this resolution axis. These findings indicate that RvD1 regulates miRNAs as an underlying mechanism to control the magnitude and duration of the inflammatory response, as well as promote resolution.
Post-translational control of target genes by miRNAs is a widely conserved and rapid mechanism of regulation of cellular pathways (Supplemental Fig. S4 and refs. 27, 38). Among RvD1-regulated miRs, miR-146b decreases protein levels of proinflammatory IL-8 and RANTES, which are well-appreciated potent chemoattractants that drive leukocytes in inflamed areas (20, 22, 23). RvD1, also regulated miR-21, which increases anti-inflammatory IL-10 in response to LPS (37), and can promote cell survival and proliferation by acting simultaneously on both proapoptotic and antiapoptotic genes, such as caspase-3, Bax, and TGF-β (Supplemental Fig. S4 and ref. 39).
miRNA-208a is known to be involved in cardiac function via regulation of thyroid-hormone-associated protein 1 and myostatin 2, two negative regulators of cardiomyocyte growth (40). In addition, miR-219 was recently identified as critical for normal oligodendrocyte differentiation and myelination (41). In the present report, we identified genes that are molecular targets of miR-146b, miR-208a, and miR-219, which display key roles in inflammation and the immune response (Figs. 8–11). Using the IPA knowledge database, we identified 5 networks that connect these genes. Network analysis enables unbiased inferences of molecular pathways and biological functions controlled by specific genes. In each network, genes occupy nodal positions and are connected by edges based on their physical and/or functional interactions previously identified. Also, since miRNAs target hundreds to thousands of putative genes, network analysis is a useful approach to evaluate miRNA actions within their multinodal contexts (27, 33, 38).
These analyses showed that central positions in RvD1-miRNA networks are occupied by NF-κB and associated genes. RvD1 counteracted TNF-α-induced phosphorylation of IκBα, an inhibitory protein of the NF-κB complex that undergoes proteasomal degradation on phosphorylation by IKK. Of interest along these lines, RvD1 inhibits NF-κB activation evoked by TNF-α via its GPCRs ALX/FPR2 and GPR32 (18), and dampens PMN infiltration in response to TNF-α acute inflammation in the murine dorsal air pouch (15). RvD1 also inhibited nuclear translocation-activation of NF-κB in human monocytes. Therefore, it is likely that the miRNA–NF-κB axis is a key component in the RvD1-GPCR downstream signaling pathways.
Computational analysis using miRecords software deduced that miR-208a could interact with the 3′ UTR of PDCD4, which we validated using real-time PCR analysis from miR-208a-overexpressing MΦs (Fig. 9A). PDCD4 is a translational repressor that binds to the eukaryotic initiation factors eIF4a and eiF4G, and inhibits protein synthesis of many cytokines, including IL-10 (42, 43). In MΦ PDCD4, expression is rapidly induced in response to TLR4 ligands, such as LPS through MyD88 and NF-κB. In addition, in MΦs, LPS also induces miR-21 expression, which, in turn, suppresses PDCD4 and subsequently enhances IL-10 (37). In accordance with these pathways, we found that RvD1 down-regulated PDCD4 and up-regulated IL-10 (Fig. 11) during zymosan-induced acute peritonitis, which may suggest the involvement of these pathways.
miRNA 219 down-regulates 5-LO, an essential enzyme involved in the biosynthesis of both leukotrienes and lipoxins (44), as well as D-series resolvins from DHA (15). Each of these autacoids has pivotal roles in inflammation and its resolution. miR-219 overexpression in human MΦs also resulted in reduction of LTB4 levels on exposure to zymosan (Fig. 12B). Leukotriene B4 causes leukocyte adhesion to postcapillary venules and is a potent chemoattractant for neutrophils and monocytes (45), whereas cysteinyl LTs mediate anaphylaxis and bronchoconstriction, and increase vascular permeability (45). The LXs and resolvins play specialized roles in resolution in that they reduce further PMN chemotaxis and infiltration, regulate cytokines, and promote proresolving actions of monocytes/MΦs (3, 4). Transcriptional regulation of the 5-LO gene, which is mainly expressed in leukocytes, is dependent on several cytokines (e.g., TGFβ), and growth factors (GM-CSF) (reviewed by Radmark et al.; ref. 46), consistent with the function of this enzyme within the immune system. In addition to this, we uncovered a mechanism of control of 5-LO expression via miRNAs, which may provide a more rapid regulation of this enzyme in cells. Interestingly, the endonuclease Dicer, which is critical for processing precursors pri-miRNA into mature hairpin miRNAs (27), also binds 5-LO via its C-terminal domain. Interaction of Dicer and 5-LO results in a mutual enhancement of their enzymatic activity, providing an additional link between 5-LO and the miRNA synthesis machinery (47).
Since RvD1 regulates cytokines (19) and miRNAs that target NF-κB pathway molecules, it was of interest in the present studies to determine whether RvD1 regulates additional transcription factors involved in cytokine gene expression. Along these lines, RvD1 inhibited nuclear translocation of SMAD, which are effectors of TGF-β receptor signaling, a pleiotropic cytokine with actions on cell proliferation and differentiation. The roles of TGF-β in the immune system and inflammation include inhibition of leukocyte differentiation, stimulation of wound healing, and fibrosis (22, 23). RvD1 has potent organoprotective (17) and antiangiogenesis effects in vivo (48). Hence, the inhibitory actions on TGF-β-SMAD pathway in monocytes (Fig. 12D) may link tissue and cell-specific regulations by RvD1.
LXA4 and RvE1 both stimulate nonphlogistic recruitment of monocytes during resolution and simultaneously stop further PMN recruitment (4, 5). RvD1 shares the ability to enhance efferocytosis by MΦs such as RvE1 and LXA4, but RvD1 (300 ng, i.p.) did not change the monocyte numbers in peritonitis in this study (Fig. 5). Hence, specific SPM differentially regulate leukocyte traffic. It is possible that the action of RvD1 in lowering PMN numbers at 24 and 48 h could also reflect enhanced nonphlogistic clearance of PMN by MΦs, as well as potential differences in RvD1 pharmacokinetics.
Targets of RvD1-regulated miRNAs uncovered herein include cytokines, chemokines, and their receptors that are associated with leukocyte trafficking. It is likely that these miRNAs contribute to RvD1 in vivo actions in reducing further PMN recruitment and stimulating nonphlogistic MΦ responses. Therefore, these miRNAs each represent novel mechanisms by which resolvins and, specifically, RvD1 regulate phagocyte trafficking, lowering Ψmax and shortening _R_i (Fig. 5). The actions of SPMs as local autacoids are demonstrable days and weeks following tissue exposure to, for example, lipoxins (49), at time intervals when the SPMs are inactivated and apparently no longer present. Hence, miRNAs identified herein are likely to participate in resolvin-mediated return of target tissues to homeostasis and may be involved in recurring bouts of local acute inflammation that may lead to chronicity. In summation, we identified here miRNAs miR-21, miR-146b, miR-142 family, miR-203, miR-208a, miR-219, and miR-302d that are temporally and differentially expressed in resolving exudates. RvD1, biosynthesized in resolution, regulates miR-21, miR-146b, miR-208a, and miR-219, and its actions in human MΦs proved to be dependent on ALX/FPR2 and GPR32 in the GPCR-recombinant cell system. Target genes of miR-146b, miR-208a, and miR-219 are involved in the immune system and form RvD1-dependent networks that govern inflammation and resolution. Together, these results establish a novel RvD1-GPCR-dependent miRNA axis in resolution circuits.
Supplementary Material
Supplemental Data
Acknowledgments
The authors thank Thad Vickery for LC-MS-MS lipidomics, Dr. Sungwhan F. Oh for generating the transcription factor heat map, Padmini S. Pillai for helping with animal experiments, and Mary H Small for assistance with manuscript preparation. The authors also acknowledge the Institute of Chemistry and Cell Biology (ICCB)–Longwood for the use of their real-time PCR equipment and IPA software license.
This work was supported by U.S. National Institutes of Health (NIH) grants R37GM38765, R01DK074448, and R01DE019938. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Science, the National Institute of Diabetes and Digestive and Kidney Diseases, or the NIH. C.N.S. is the inventor on patents assigned to Brigham and Women's Hospital and Partners HealthCare on the composition of matter, uses, and clinical development of anti-inflammatory and proresolving lipid mediators. These are licensed for clinical development. C.N.S. retains founder stock in Resolvyx Pharmaceuticals.
Footnotes
REFERENCES
- 1.Ryan G. B., Majno G. (1977) Acute inflammation. A review. Am. J. Pathol. 86, 183–276 [PMC free article] [PubMed] [Google Scholar]
- 2.Gilroy D. W., Lawrence T., Perretti M., Rossi A. G. (2004) Inflammatory resolution: new opportunities for drug discovery. Nat. Rev. Drug Discov. 3, 401–416 [DOI] [PubMed] [Google Scholar]
- 3.Serhan C. N. (2007) Resolution phase of inflammation: novel endogenous anti-inflammatory and proresolving lipid mediators and pathways. Annu. Rev. Immunol. 25, 101–137 [DOI] [PubMed] [Google Scholar]
- 4.Bannenberg G. L., Chiang N., Ariel A., Arita M., Tjonahen E., Gotlinger K. H., Hong S., Serhan C. N. (2005) Molecular circuits of resolution: formation and actions of resolvins and protectins. J. Immunol. 174, 4345–4355 [DOI] [PubMed] [Google Scholar]
- 5.Schwab J. M., Chiang N., Arita M., Serhan C. N. (2007) Resolvin E1 and protectin D1 activate inflammation-resolution programmes. Nature 447, 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haworth O., Cernadas M., Yang R., Serhan C. N., Levy B. D. (2008) Resolvin E1 regulates interleukin 23, interferon-gamma and lipoxin A4 to promote the resolution of allergic airway inflammation. Nat. Immunol. 9, 873–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Navarro-Xavier R. A., Newson J., Silveira V. L., Farrow S. N., Gilroy D. W., Bystrom J. (2010) A new strategy for the identification of novel molecules with targeted proresolution of inflammation properties. J. Immunol. 184, 1516–1525 [DOI] [PubMed] [Google Scholar]
- 8.Serhan C. N., Brain S. D., Buckley C. D., Gilroy D. W., Haslett C., O'Neill L. A., Perretti M., Rossi A. G., Wallace J. L. (2007) Resolution of inflammation: state of the art, definitions and terms. FASEB J. 21, 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nathan C., Ding A. (2010) Nonresolving inflammation. Cell 140, 871–882 [DOI] [PubMed] [Google Scholar]
- 10.Mantovani A., Allavena P., Sica A., Balkwill F. (2008) Cancer-related inflammation. Nature 454, 436–444 [DOI] [PubMed] [Google Scholar]
- 11.Randolph G. J. (2009) The fate of monocytes in atherosclerosis. J. Thromb. Haemost. 7(Suppl. 1), 28–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serhan C. N. (2009) Systems approach to inflammation resolution: identification of novel anti-inflammatory and pro-resolving mediators. J. Thromb. Haemost. 7(Suppl. 1), 44–48 [DOI] [PubMed] [Google Scholar]
- 13.Chiang N., Bermudez E. A., Ridker P. M., Hurwitz S., Serhan C. N. (2004) Aspirin triggers antiinflammatory 15-epi-lipoxin A4 and inhibits thromboxane in a randomized human trial. Proc. Natl. Acad. Sci. U. S. A. 101, 15178–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris T., Stables M., Hobbs A., de Souza P., Colville-Nash P., Warner T., Newson J., Bellingan G., Gilroy D. W. (2009) Effects of low-dose aspirin on acute inflammatory responses in humans. J. Immunol. 183, 2089–2096 [DOI] [PubMed] [Google Scholar]
- 15.Serhan C. N., Hong S., Gronert K., Colgan S. P., Devchand P. R., Mirick G., Moussignac R. L. (2002) Resolvins: a family of bioactive products of omega-3 fatty acid transformation circuits initiated by aspirin treatment that counter proinflammation signals. J. Exp. Med. 196, 1025–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y. P., Oh S. F., Uddin J., Yang R., Gotlinger K., Campbell E., Colgan S. P., Petasis N. A., Serhan C. N. (2007) Resolvin D1 and its aspirin-triggered 17R epimer. Stereochemical assignments, anti-inflammatory properties, and enzymatic inactivation. J. Biol. Chem. 282, 9323–9334 [DOI] [PubMed] [Google Scholar]
- 17.Kasuga K., Yang R., Porter T. F., Agrawal N., Petasis N. A., Irimia D., Toner M., Serhan C. N. (2008) Rapid appearance of resolvin precursors in inflammatory exudates: novel mechanisms in resolution. J. Immunol. 181, 8677–8687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishnamoorthy S., Recchiuti A., Chiang N., Yacoubian S., Lee C. H., Yang R., Petasis N. A., Serhan C. N. (2010) Resolvin D1 binds human phagocytes with evidence for proresolving receptors. Proc. Natl. Acad. Sci. U. S. A. 107, 1660–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merched A. J., Ko K., Gotlinger K. H., Serhan C. N., Chan L. (2008) Atherosclerosis: evidence for impairment of resolution of vascular inflammation governed by specific lipid mediators. FASEB J. 22, 3595–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossi A. G., Sawatzky D. A., eds (2008) The Resolution of Inflammation, Birkhäuser Verlag AG, Basel [Google Scholar]
- 21.Tabas I. (2010) Macrophage death and defective inflammation resolution in atherosclerosis. Nat. Rev. Immunol. 10, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majno G. (1982) Inflammation and infection: historic highlights. In Current Topics in Inflammation and Infection (Majno G., Cotran R. S., Kaufman N., eds) pp. 1–17, Williams & Wilkins, Baltimore: [PubMed] [Google Scholar]
- 23.Majno G., Cotran R. S., Kaufman N., eds (1982) Current Topics in Inflammation and Infection, Vol. 23, Williams & Wilkins, Baltimore [Google Scholar]
- 24.Shimizu T. (2009) Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu. Rev. Pharmacol. Toxicol. 49, 123–150 [DOI] [PubMed] [Google Scholar]
- 25.Iorio M. V., Croce C. M. (2009) MicroRNAs in cancer: small molecules with a huge impact. J. Clin. Oncol. 27, 5848–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee R. C., Feinbaum R. L., Ambros V. (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 [DOI] [PubMed] [Google Scholar]
- 27.Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheedy F. J., O'Neill L. A. (2008) Adding fuel to fire: microRNAs as a new class of mediators of inflammation. Ann. Rheum. Dis. 67(Suppl. 3), iii50–iii55 [DOI] [PubMed] [Google Scholar]
- 29.Arita M., Bianchini F., Aliberti J., Sher A., Chiang N., Hong S., Yang R., Petasis N. A., Serhan C. N. (2005) Stereochemical assignment, antiinflammatory properties, and receptor for the omega-3 lipid mediator resolvin E1. J. Exp. Med. 201, 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambros V., Bartel B., Bartel D. P., Burge C. B., Carrington J. C., Chen X., Dreyfuss G., Eddy S. R., Griffiths-Jones S., Marshall M., Matzke M., Ruvkun G., Tuschl T. (2003) A uniform system for microRNA annotation. RNA 9, 277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 32.Xiao F., Zuo Z., Cai G., Kang S., Gao X., Li T. (2009) miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 37, D105–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naga Prasad S. V., Duan Z. H., Gupta M. K., Surampudi V. S., Volinia S., Calin G. A., Liu C. G., Kotwal A., Moravec C. S., Starling R. C., Perez D. M., Sen S., Wu Q., Plow E. F., Croce C. M., Karnik S. (2009) Unique microRNA profile in end-stage heart failure indicates alterations in specific cardiovascular signaling networks. J. Biol. Chem. 284, 27487–27499 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Serhan C. N., Lu Y., Hong S., Yang R. (2007) Mediator lipidomics: search algorithms for eicosanoids, resolvins and protectins. Methods Enzymol. 432, 275–317 [DOI] [PubMed] [Google Scholar]
- 35.Winyard P. G., Willoughby D. A., eds (2003) Inflammation Protocols, Humana, Totowa, NJ, USA [Google Scholar]
- 36.Clish C. B., O'Brien J. A., Gronert K., Stahl G. L., Petasis N. A., Serhan C. N. (1999) Local and systemic delivery of a stable aspirin-triggered lipoxin prevents neutrophil recruitment in vivo. Proc. Natl. Acad. Sci. U. S. A. 96, 8247–8252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheedy F. J., Palsson-McDermott E., Hennessy E. J., Martin C., O'Leary J. J., Ruan Q., Johnson D. S., Chen Y., O'Neill L. A. (2009) Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 11, 141–147 [DOI] [PubMed] [Google Scholar]
- 38.Inui M., Martello G., Piccolo S. (2010) MicroRNA control of signal transduction. Nat. Rev. Mol. Cell. Biol. 11, 252–263 [DOI] [PubMed] [Google Scholar]
- 39.Papagiannakopoulos T., Shapiro A., Kosik K. S. (2008) MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 68, 8164–8172 [DOI] [PubMed] [Google Scholar]
- 40.Callis T. E., Pandya K., Seok H. Y., Tang R. H., Tatsuguchi M., Huang Z. P., Chen J. F., Deng Z., Gunn B., Shumate J., Willis M. S., Selzman C. H., Wang D. Z. (2009) MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest. 119, 2772–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dugas J. C., Cuellar T. L., Scholze A., Ason B., Ibrahim A., Emery B., Zamanian J. L., Foo L. C., McManus M. T., Barres B. A. (2010) Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron 65, 597–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang H. S., Jansen A. P., Komar A. A., Zheng X., Merrick W. C., Costes S., Lockett S. J., Sonenberg N., Colburn N. H. (2003) The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell. Biol. 23, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loh P. G., Yang H. S., Walsh M. A., Wang Q., Wang X., Cheng Z., Liu D., Song H. (2009) Structural basis for translational inhibition by the tumour suppressor Pdcd4. EMBO J. 28, 274–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samuelsson B., Dahlen S. E., Lindgren J. A., Rouzer C. A., Serhan C. N. (1987) Leukotrienes and lipoxins: structures, biosynthesis, and biological effects. Science 237, 1171–1176 [DOI] [PubMed] [Google Scholar]
- 45.Samuelsson B. (1983) Leukotrienes: mediators of immediate hypersensitivity reactions and inflammation. Science 220, 568–575 [DOI] [PubMed] [Google Scholar]
- 46.Radmark O., Werz O., Steinhilber D., Samuelsson B. (2007) 5-Lipoxygenase: regulation of expression and enzyme activity. Trends Biochem. Sci. 32, 332–341 [DOI] [PubMed] [Google Scholar]
- 47.Dincbas-Renqvist V., Pepin G., Rakonjac M., Plante I., Ouellet D. L., Hermansson A., Goulet I., Doucet J., Samuelsson B., Radmark O., Provost P. (2009) Human Dicer C-terminus functions as a 5-lipoxygenase binding domain. Biochim. Biophys. Acta 1789, 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Connor K. M., SanGiovanni J. P., Lofqvist C., Aderman C. M., Chen J., Higuchi A., Hong S., Pravda E. A., Majchrzak S., Carper D., Hellstrom A., Kang J. X., Chew E. Y., Salem N., Jr., Serhan C. N., Smith L. E. (2007) Increased dietary intake of omega-3-polyunsaturated fatty acids reduces pathological retinal angiogenesis. Nat. Med. 13, 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bannenberg G., Moussignac R. L., Gronert K., Devchand P. R., Schmidt B. A., Guilford W. J., Bauman J. G., Subramanyam B., Perez H. D., Parkinson J. F., Serhan C. N. (2004) Lipoxins and novel 15-epi-lipoxin analogs display potent anti-inflammatory actions after oral administration. Br. J. Pharmacol. 143, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serhan C. N., Yang R., Martinod K., Kasuga K., Pillai P. S., Porter T. F., Oh S. F., Spite M. (2009) Maresins: novel macrophage mediators with potent antiinflammatory and proresolving actions. J. Exp. Med. 206, 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data