Association of PPP2CA polymorphisms with SLE susceptibility in multiple ethnic groups (original) (raw)
. Author manuscript; available in PMC: 2012 Sep 1.
Published in final edited form as: Arthritis Rheum. 2011 Sep;63(9):2755–2763. doi: 10.1002/art.30452
Abstract
Objective
T cells from patients with SLE express increased amounts of PP2Ac which contribute to decreased production of IL-2. Because IL-2 is important in the regulation of several aspects of the immune response, it has been proposed that PP2Ac contributes to the expression of SLE. This study was designed to determine whether genetic variants of PPP2AC are linked to the expression of SLE and specific clinical manifestations and account for the increased expression of PP2Ac.
Methods
We conducted a trans-ethnic study consisting of 8,695 SLE cases and 7,308 controls from four different ancestries. Eighteen single-nucleotide polymorphisms (SNPs) across the PPP2CA were genotyped using an Illumina custom array. PPP2CA expression in SLE and control T cells was analyzed by real-time PCR.
Results
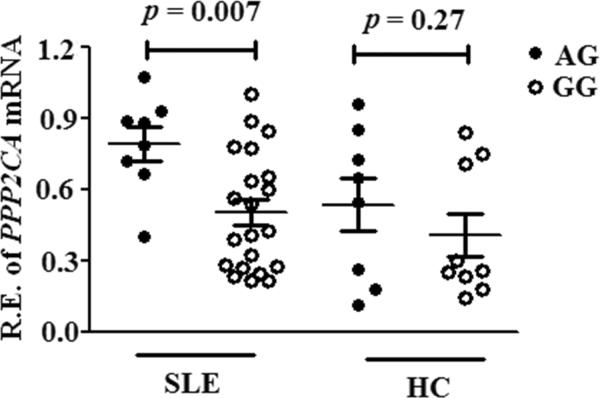
A 32-kb haplotype comprised of multiple SNPs of PPP2CA showed significant association with SLE in Hispanic Americans (HA), European Americans (EA) and Asians but not in African-Americans (AA). Conditional analyses revealed that SNP rs7704116 in intron 1 showed consistently strong association with SLE across Asian, EA and HA populations (_pmeta_=3.8×10−7, OR=1.3[1.14–1.31]). In EA, the largest ethnic dataset, the risk A allele of rs7704116 was associated with the presence of renal disease, anti-dsDNA and anti-RNP antibodies. PPP2CA expression was approximately 2-fold higher in SLE patients carrying the rs7704116 AG genotype than those carrying GG genotype (p = 0.008).
Conclusion
Our data provide the first evidence for an association between PPP2CA polymorphisms and elevated PP2Ac transcript levels in T cells, which implicates a new molecular pathway for SLE susceptibility in EA, HA and Asians.
Introduction
PP2A, a major serine/threonine phosphatase, negatively regulates NF-κB, MAPK and WNT signal transduction pathways that are involved in many cellular processes including cell cycle progression, DNA replication, gene transcription/translation and cell differentiation (1–3). PP2A consists of three subunits: a scaffold-type structural subunit (A), a regulatory subunit (B) and a catalytic subunit (C) (PP2Ac), which has serine/threonine phosphatase activity (4;5). There are two isoforms of the PP2Ac subunit. The α isoform (PP2Acα), encoded by PPP2CA (OMIM 176915), shares 97% sequence identity with the β isoform PP2Acβ and is about 10 times more abundant, probably because of an approximately 10-fold more active gene promoter (1).
T cells from patients with systemic lupus erythematosus (SLE [OMIM 152700]) exhibit abnormally elevated mRNA, protein, and enzymatic activity of PP2Ac, which results in decreased amounts of IL-2 via dephosphorylation (and thus inactivation) of cAMP response element-binding protein (CREB) (7;8). Silencing or inhibition of PP2Ac results in normalization of IL-2 production (8). Decreased IL-2 production is present in both human SLE patients and lupus-prone mice and has been considered to contribute to aberrant immunoregulation, including decreased regulatory T cell (Treg) function, deficient cytotoxic responses and activation-induced T cell death (9–12).
In the past two decades, multiple genes that predispose to SLE susceptibility have been identified and confirmed. In particular, advances in high throughput technology have enabled the genotyping of hundreds of thousands of single nucleotide polymorphisms (SN Ps) in a single individual, which facilitates the mapping of complex disease loci throughout the genome (2). Six recent genome-wide association studies (GWAS) of SLE performed in European and Asian populations have uncovered more than 30 common SLE risk loci that achieve genome-wide significance (p < 5×10−8) (14–20). Given the design of these studies and stringent statistical criteria used in GWAS, it is likely that many susceptibility genes remain hidden within the statistical “noise” inherent to GWAS (21;22); thus, the candidate gene strategy remains a useful approach to uncover genetic susceptibility to SLE (3). Here, we conducted a trans-ethnic case-control study in a large sample size containing 8,695 cases and 7,308 controls from HA, EA, Asian, and AA populations to examine the association of PPP2CA variants with SLE. We report that indeed PP2ACA variants are associated with SLE across ethnic and more importantly, a risk allele leads to increased production of PP2Ac.
PATIENTS AND METHODS
Study subjects
Study subjects consisting of 8,695 cases and 7,308 controls were from the collaborative Large Lupus Association Study 2 (LLAS2) that included four populations: Hispanic-Americans (HA) (1,508 cases and 812 controls), European-Americans (EA) (3,980 cases and 3,546 controls), Asians (1,273 cases and 1,269 controls), and African-Americans (AA) (1,934 cases and 1,681 controls) (Table 1). All study participants have given written informed consent that was approved by the Institutional Review Boards at each respective institution. Ethnicity was self-reported and verified by principal component and admixture proportion calculations. Patients met at least four of the 11 revised 1997 American College of Rheumatology criteria for SLE (4). Clinical data for the cases was obtained from medical records that were reviewed at each institution. We chose to examine the 11 ACR criteria and the presence of autoantibodies (anti-dsDNA, anti-Sm, anti-RNP, anti-SSA/Ro or anti-SSB/La) for association analyses. SLE onset occurs during adolescence in 15–20% of patients and early age of disease onset has been associated with a worse prognosis (5); thus, in our analyses we used the age of 20 years old to define early and late diagnosed cases (26).
Table 1.
Characteristics of studied patients and controls
| Cases | Controls | ||||
|---|---|---|---|---|---|
| Ethnicity | n | Male/Female | Age* | n | Male/Female |
| HA | 1508 | 118/1390 | 29.6±11.8 | 812 | 72/740 |
| EA | 3980 | 345/3635 | 33.6±13.6 | 3546 | 929/2617 |
| Asian | 1273 | 102/1171 | 26.4±10.8 | 1269 | 153/1116 |
| AA | 1934 | 592/1342 | 33.9±12.4 | 1681 | 137/1544 |
| Total | 8695 | 1157/7358 | 30.9±12.8 | 7308 | 1291/6017 |
SNP Markers and Genotyping
This genotyping project was part of the Lupus Large Association Study 2 (LLAS2) that involved multiple investigators and more than 32,000 SNPs. Genotyping was carried out using a customized Illumina Infinium II platform at the Lupus Genetics Studies Unit of the Oklahoma Medical Research Foundation. We initially genotyped 22 common SNPs spanning PPP2CA in four independent, ethnically divergent populations of SLE patients and controls containing 16,003 participants. After application of quality controls, a panel of 18 SNPs spanning a 32kb region from the PPP2CA promoter to 3.2kb downstream were genotyped in this study, including 5 EA haplotype tagging SNPs (rs10491322, rs2302599, rs2868600, rs7704116 and rs2292283), 9 SNPs with potential regulatory functions (rs2072494, rs7705270, rs4302636, rs254048, rs7704116, rs2300071, rs254051, rs2284317, and rs292283), and other common SNPs with known frequencies in non-European ethnic groups (minor allele frequency > 0.1).
Real-time PCR
Primary T cells were purified from the peripheral blood of 29 SLE patients and 17 healthy controls using a T-rosette purification kit (StemCell Technologies), and were 98% CD3+ by flow cytometry. RNA was extracted from 3×106 T cells using AllPrep RNA/DNA/Protein mini kit (Qiagen). Real-time PCR was performed in triplicate for every sample with a Light-Cycler 480 system by adding SYBR Green (Roche) to the reaction mixture. Primers used were: PPP2AC, F, 5'-TCCGAGTCCCAGGTCAAGAG-3', R, 5'-GCTACAAGCAGTGTAACTGTTTCA-3'; and GAPDH, F, 5'-CAACTACATGGTTTACATGTTCC-3', R, 5'-GGACTGTGGTCATGAGTCCT-3'. The average cycle threshold (CT) values of each reaction derived from the target gene, determined with LightCycler 480 system software, were normalized to GAPDH levels. ΔCt (CT of target - CT of GAPDH) was used to calculate relative mRNA expression by the ΔΔ_Ct_ relative quantification method.
Statistical analysis
The statistical power of the association study was estimated by power calculator CaTS. Software Haploview was used to select tag SNPs, calculate LD values among pairs of SNPs and define haplotype blocks. Principal component analysis (PCA) and admixture proportions (using ADMIXMAP) for each ethnicity were computed using a total of 163 ancestry informative markers (AIMS) that passed statistical and laboratory quality controls among the 346 AIMs genotyped (27;28). Allele frequencies were calculated for each locus and tested for Hardy-Weinberg equilibrium (HWE) in controls. For each tested SNP, the quality of the genotype data was assessed by predetermined quality control inclusion criteria (MAF > 1%, SNP call rate > 90%, HWE (P > 0.001) among the controls). Case–control associations and Hardy–Weinberg proportions were calculated using PLINK. Allelic association tests were performed using Pearson's chi-square measures. Conditional haplotype-based likelihood ratio test was conducted by the software PLINK (29;30). Mantel-Haenszel analysis was performed to generate the trans-ethnic meta-analysis P value.
RESULTS
Identification of risk haplotype of PPP2CA for SLE in multiple ethnic groups
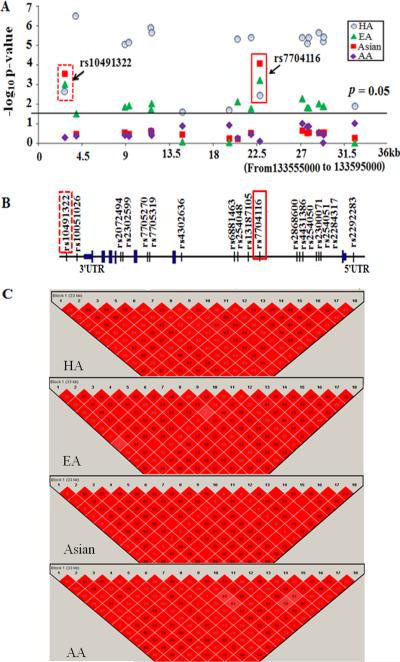
First, we constructed haplotypes using the genotype data from the tested 18 SNPs and analyzed the association of the formed haplotypes with SLE in each ethnic group. A single 32kb haplotype block was constructed in Asian, EA, HA and AA datasets that extended from the promoter of PPP2CA to downstream of the 3'UTR and which contained three common haplotypes (H1-H3) (frequency > 0.01; Figure 1C and Table 2). The H1 haplotype (GGCAAAACGAAAAAAAAG) that occurred with a higher frequency in SLE patients than controls (0.188 vs. 0.152 in HA, 0.084 vs. 0.07 in EA, 0.066 vs. 0.043 in Asian and 0.123 vs. 0.119 in AA, respectively) showed significant haplotypic association with SLE across Asian, EA and HA populations but not in AA (_p_meta = 0.03, OR=1.20 [1.11–1.29]) (Table 2). Of note, compared to the protective (H2) and the neutral (H3) haplotypes, the H1 haplotype uniquely carried both the risk `G' allele at rs10491322 and the risk `A' allele at rs7704116.
Figure 1.
PPP2CA gene structure with tested SNPs and haplotype blocks in HA, EA, Asian and AA patients. A. The −log10 base of allelic p value for each SNP. B. PPP2CA gene structure with tested SNPs. Two SNPs (rs7704116 and rs10491322) that showed consistently strong association with SLE across Asian, EA and HA are marked with frame. C. Haplotype blocks based on 18 tested SNPs in HA, EA, Asian and AA SLE patients.
Table 2.
Haplotype analysis of 18 SNPs in PPP2CA associated with SLE
Identification of risk alleles of PPP2CA in SLE in multiple ethnic groups
We next analyzed the allelic association of PPP2CA with SLE in different ethnic background populations in order to identify risk allele(s) that could capture the observed haplotypic association signal. We found that all 18 tested SNPs exhibited nominal levels of association to SLE at p < 0.05 in HA; 13 of 18 and 2 of 18 PPP2CA SNPs showed significant associations with SLE in EA and Asians, respectively, whereas none of the SNPs displayed a genetic association signal with SLE in AA (Figure 1A, Supplementary table 1). Notably, two SNPs (rs7704116 at intron1 and rs10491322 at the 3' downstream) showed consistently strong association with SLE across Asian, EA and HA, and were in almost complete pairwise linkage disequilibrium (LD) (r2 = 0.98–0.99) in these ethnic groups (Figure 1A, C). Of interest, SNP rs7704116 displayed the smallest p value both in Asians and EA (p = 8.1×10−5 in Asian, p = 5.8×10−4 in EA) and conferred the strongest odds ratio (OR = 1.6 [1.3–2.1]) in Asians.
Given the consistent association of rs7704116 and rs10491322 with SLE observed in multiple ethnic groups, we carried out a meta-analysis of all ethnicities containing ~ 16,003 participants to maximize the number of samples for the association analysis. PMeta for rs7704116 was 3.8×10−7(OR=1.3[1.1–1.3]) and PMeta for rs10491322 was 3.0×10−4 (OR=1.2 [1.1–1.3]) without significant heterogeneity among populations (p = 0.82 and p = 0.58), again reinforcing the genetic association of PPP2CA with SLE (Table 3). By haplotype-based conditional test the genetic effect of rs10491322 disappeared after conditional on rs7704116 genotypes (Table 2). Thus, rs7704116 may be the sole polymorphism representing the genetic association for SLE in all the tested SNPs spanning from the promoter of PPP2CA to downstream of the 3'UTR.
Table 3.
Meta-analysis of the association of PPP2CA 2 SNPs with SLE in multiple ethnic groups
| SNP | Position | Ethnicity | MA | MAF | P | OR (95%CI) | Conditional on rs7704116 | |
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | |||||||
| rs7704116 | Intron 1 | HA | A | 0.19 | 0.15 | 0.003 | 1.3 [1.08–1.5] | |
| EA | A | 0.09 | 0.07 | 5.8×10−4 | 1.2 [1.1–1.4] | |||
| Asian | A | 0.07 | 0.04 | 8.1×10−5 | 1.6 [1.28–2.11] | - | ||
| AA | A | 0.13 | 0.13 | 0.68 | 1.0 [0.9–1.18] | |||
| Meta analysis | 3.8×10−7 | 1.3 [1.14–1.31] | ||||||
| rs10491322 | Downstream of 3' UTR | HA | G | 0.19 | 0.15 | 2.1×10−3 | 1.3 [1.1–1.52] | 0.28 |
| EA | G | 0.09 | 0.07 | 9.3×10−4 | 1.2 [1.09–1.38] | 0.29 | ||
| Asian | G | 0.07 | 0.04 | 3.0×10−4 | 1.6 [1.23–2.02] | 0.14 | ||
| AA | G | 0.12 | 0.12 | 0.50 | 1.1 [0.91–1.22] | 0.89 | ||
| Meta analysis | 3.8×10−4 | 1.2 [1.07–1.27] |
Association of PPP2CA with SLE clinical features
To determine whether rs7704116 was associated with particular clinical or serological manifestations of SLE, we first examined each subphenotype based on the presence of each of the 11 ACR classification criteria (malar rash, discoid rash, photosensitivity, oral ulcers, arthritis, serositis, renal disorder, neurologic disorder, hematologic disorder, immunologic disorder and antinuclear antibodies), specific autoantibodies (anti-dsDNA, anti-Sm, anti-RNP, anti-SSA/Ro and anti-SSB/La) and age of disease onset for association with rs7704116. In the SLE cases stratified by the presence (SLE+) vs. the absence (SLE−) analyses, we found that the risk A allele of rs7704116 displayed an association with the presence of renal disease (p = 0.03, OR = 1.2[1.0–1.5]), anti-dsDNA (p = 1×10−3, OR = 1.7[1.2–2.3]), and anti-RNP antibody (p = 6×10−4, OR = 1.4[1.1–1.7]; respectively) in the largest dataset of European-derived SLE patients (Table 4). When SLE subjects where compared to controls, in both EA and Asians the risk A allele yielded significant risk for the presence of renal disease and anti-DNA antibody (p = 6×10−4, OR = 1.3 [1.1–1.6] for renal disorder and p = 1×10−4, OR = 1.4[1.2–1.6] for anti-dsDNA antibody in EA; and p = 4×10−3, OR = 1.7[1.3–2.3] for renal disorder and p = 1×10−4, OR = 1.7[1.3–2.2] for anti-dsDNA antibody in Asians). After Bonferroni correction for multiple comparisons, the association with anti-DNA antibody (_p_Bonferroni = 0.0016 in EA and Asians, receptively) remained significant in both EA and Asians; however, the association with renal was observed in EA (_p_Bonferroni = 0.01) but not in Asians (_p_Bonferroni = 0.06). No association was detected between the presence of the risk allele and specific clinical manifestations in HA patients.
Table 4.
Selected results from association analyses of rs7704116 in SLE phenotype subsets based on ACR clinical criteria
| Phenotype | Ethnicity | SLE(+) | SLE(−) | Controls | SLE(+) vs. SLE(−) | SLE(+) vs. Control | SLE(−) vs. Control | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | MAF | n | MAF | n | MAF | P | OR | P | OR | P | OR | ||
| Renal | EA | 1128 | 9.4% | 2154 | 7.6% | 3980 | 7.0% | 0.03 | 1.2(1.0–1.5) | 6×10−4 | 1.3(1.1–1.6) | 0.21 | 1.1(1.0–1.3) |
| Asian | 529 | 7.2% | 610 | 6.5% | 1269 | 4.4% | 0.53 | 1.1(0.8–1.5) | 4×10−3 | 1.7(1.3–2.3) | 0.11 | 1.1(0.8–1.2) | |
| HA | 651 | 17.9% | 750 | 16.7% | 812 | 15.4% | 0.47 | 1.1(0.8–1.3) | 0.13 | 1.2(1.0–1.4) | 0.35 | 1.1(0.9–1.4) | |
| Anti-dsDNA | EA | 1210 | 9.6% | 1514 | 7.1% | 3980 | 7.0% | 1×10−3 | 1.7(1.2–2.3) | 1×10−4 | 1.4(1.2–1.6) | 0.78 | 1.0(0.9–1.2) |
| Asian | 888 | 7.0% | 233 | 6.4% | 1269 | 4.4% | 0.69 | 1.1(0.7–1.6) | 1×10−4 | 1.7(1.3–2.2) | 0.09 | 1.2(0.8–1.5) | |
| HA | 731 | 17.6% | 571 | 16.7% | 812 | 15.4% | 0.71 | 1.0(0.9–1.3) | 0.16 | 1.1(1.0–1.4) | 0.35 | 1.1(0.9–1.4) | |
| Anti-RNP | EA | 268 | 11.9% | 1401 | 7.1% | 3980 | 7.0% | 6×10−4 | 1.4(1.1–1.7) | 1×10−4 | 1.7(1.3–2.3) | 0.78 | 1.0(0.8–1.2) |
| Asian | 261 | 6.1% | 574 | 6.9% | 1269 | 4.4% | 0.6 | 0.9(0.6–1.4) | 0.06 | 1.5(1.0–2.2) | 1×10−3 | 1.6(1.2–2.2) | |
| HA | 174 | 19.3% | 449 | 16.7% | 812 | 15.4% | 0.37 | 1.2(1.0–1.5) | 0.14 | 1.2(0.9–1.7) | 0.48 | 1.1(0.9–1.4) |
PPP2CA SNP rs7704116 affects PPP2CA expression level
The location of rs7704116 in intron 1 suggested its potential role in transcription regulation. We next carried out real-time PCR assays to determine whether the rs7704116 variant would impact on PPP2CA mRNA expression in T cells from 29 EA SLE patients (8 AG and 21 GG genotype for rs7704116) and 17 healthy controls (8 AG and 9 GG genotype for rs7704116). PPP2CA expression was approximately 2-fold higher in SLE patients carrying rs7704116 AG genotype (0.07 ± 0.03) than in those carrying GG genotype (0.04 ± 0.02; p = 0.008; Figure 2). No significant allelic-specific difference in PPP2CA expression was observed among healthy controls. These results support that T cells from SLE patients carrying th_e_ rs7704116 risk A allele of PPP2CA produce higher levels of PP2Ac mRNA which has been shown to suppress IL-2 production by dephosphorylating CREB (7).
Figure 2.
Association of PPP2CA genotypes with mRNA expression levels. Relative mRNA expression levels of PPP2CA in isolated T cells from SLE and healthy controls (HC) with the AG or GG genotype at rs7704116 (AG = 8 vs. GG = 21 in SLE and AG = 8 vs. GG = 9 in healthy controls) were determined by real-time PCR and normalized to GAPDH. Relative mRNA expression levels were log 10 transformed to normalize its distribution. The results represent the mean±SD of four independent experiments. None of the subjects were homozygous for the minor A allele.
DISCUSSION
To our knowledge, this is the first report of genetic association between PPP2CA with SLE in multiple ethnic groups. In our study, a novel susceptibility locus in PPP2CA (rs7704116) accounts for association signals with SLE observed in HA, EA and Asian but not in AA datasets (a total of 8,695 cases and 7,308 controls). Although six published GWAS in patients with SLE have identified and replicated more than 30 risk loci, PPP2CA was not included in any of the risk loci described thus far (14–20). A variety of factors including modest sample size, limited genome-wide coverage of the genotyping platforms, difference in demographics and disease subphenotypes, analytical strategies and environmental factors might influence the detection of association in published GWAS (14–20). Our candidate gene study has a robust sample size including four ethnically diverse populations, and dense marker coverage. Although the results of this study did not meet a strict genome-wide association significance level, the robust and consistent association signal of rs7704116 (_p_meta = 3.8×10−7, OR = 1.3 [1.14–1.31]) presented in multiple independent and ethnically diverse sample sets strongly implicates PPP2CA in the development of SLE.
Since SLE is a complex and heterogeneous disease with respect to organ involvement and autoantibody production, we sought to investigate whether specific PPP2CA variants contributed to phenotypic heterogeneity in SLE. rs7704116 showed an association with renal disease and specific autoantibodies to dsDNA in EA and Asians when lupus patients were compared to control subjects that was maintained only in the EA subjects when the patients were subset into those with and those without a specific clinical manifestation (Table 4). Several reasons may account for our failure to detect associations between the risk allele and clinical manifestations across ethnicities, such as insufficient statistical power or the appearance of each manifestation at different time points during the course of the disease. We were impressed with the absence of association between PP2Ac risk alleles and AA SLE patients. Although it could be argued that PP2Ac risk alleles contribute to less severe disease, the finding that they were associated with renal disease and anti-dsDNA antibody, among all clinical manifestations, in the other patient populations, negates this claim. In addition, previously we had noticed that both AA and other ethnic subgroups displayed decreased IL-2 production and increased PP2Ac levels, suggesting that the levels of PP2Ac expression in SLE T cells are determined by additional factors and not only by the identified herein risk allele.
When investigating the consequences of the risk allele on the expression of PPP2CA, we found that the expression of PPP2CA mRNA was increased approximately 2-fold in T cells of EA SLE patients carrying a single copy of the risk allele of rs7704116 compared to non-carriers. Abnormalities in the expression of PPP2CA have been linked to SLE regardless of disease activity and treatment and in a disease-specific manner, which resulted in decreased IL-2 production (31). IL-2 is essential for both the promotion and the suppression of the immune response, contributing to both the generation of effector and memory T cells and the maintenance of Tregs (32;33). Our data support the notion that PPP2CA polymorphism(s) that upregulate the expression of PPP2CA in SLE T cells, which in turn negatively regulates IL-2 production is a plausible molecular mechanism for how PPP2CA variants contribute to SLE susceptibility. The fact that in normal T cells the risk allele did not correlate with higher expression levels of PP2Ac suggests the presence of additional molecular modifiers in the regulation of expression of PP2Ac.
PPP2CA are composed of seven exons and six introns localized in chromosome 5q23–q31. SLE-associated rs7704116 is located in a region of intron 1. There is no known or obvious functional role for this region that explains why this SLE-associated SNP would affect PPP2CA expression. It is likely that there might be an unknown functional polymorphism within the PPP2CA common haplotype, which is in tight linkage disequilibrium with rs7704116. Thus, identification of the causal allele will ultimately require the resequencing of the PPP2CA common risk haplotype region in a large number of patients with SLE, together with functional studies. Bioinformatic searches showed rs7704116 might be located in two putative microRNA target sites of hsv2-miR-H3 and mmu-miR-709. Although the functions of these two microRNA have not been delineated, it is possible that the SLE-associated SNP may modify microRNA binding sites and thus interfere with microRNA mediated regulation.
Multiple PPP2CA SNPs showed stronger allelic association with SLE in HA subjects than in other ethnic groups. To exclude the possibility that spurious associations were the result of underlying population admixture in this ethnic group, we tested each SNP for association with SLE using logistic regression including the major principal components and admixture estimates from AIMs as covariates, respectively. Very similar results were observed using both models, which supported the stronger association of PPP2CA with SLE in HA (data not shown).
The risk haplotype of SLE containing rs7704116 spanned 32kb of PPP2CA. Our current data cannot exclude other PPP2CA genetic variants or rare SNP alleles that might also contribute to the development of SLE. Nevertheless, this study provides the first evidence for consistent association between PPP2CA and SLE across multi-ancestral SLE cohorts. Of importance is our finding that the PPP2CA risk allele is associated with the presence of renal disease and anti-dsDNA antibody and dictates higher levels of PP2Ac expression in SLE T cells. Lastly, our study indicates that GWAS may not detect all risk alleles and a gene-oriented approach such as the one presented here is of significant value, particularly with genes whose products have been amply demonstrated to contribute to the immunopathology of the disease.
Supplementary Material
1
ACKNOWLEDGMENTS
We thank the study participants and physicians who provided samples (Peter K. Gregersen, BIOLUPUS Network and GENLES Network). The members of BIOLUPUS Network are Sandra D'Alfonso in Italy; Bernard R. Lauwerys in Belgium; Emoke Endreffy and László Kovács in Hungary; Carlos Vasconcelos and Berta Martins da Silva in Portugal; Iñigo Rúa Figueroa in Spain. The members of GENLES Network are Hugo R. Scherbarth, Pilar C. Marino, Estela L. Motta, Susana Gamron, Cristina Drenkard, Emilia Menso, Alberto Allievi, Guillermo A. Tate, Jose L. Presas, Simon A. Palatnik, Marcelo Abdala, Mariela Bearzotti, Alejandro Alvarellos, Francisco Caeiro, Ana Bertoli, Sergio Paira, Susana Roverano, Cesar E. Graf, Estela Bertero, Cesar Caprarulo, Griselda Buchanan, Carolina Guillerón, Sebastian Grimaudo, Jorge Manni, Luis J. Catoggio, Enrique R. Soriano, Carlos D. Santos, Cristina Prigione, Fernando A. Ramos, Sandra M. Navarro, Guillermo A. Berbotto, Marisa Jorfen, Elisa J. Romero, Mercedes A. Garcia, Juan C Marcos, Ana I. Marcos, Carlos E. Perandones, Alicia Eimon and Cristina G. Battagliotti in Argentina; Eduardo Acevedo and Mariano Cucho in Perú; Ignacio García de la Torre, Mario Cardiel Ríos, José Francisco Moctezuma and Marco Maradiaga Ceceña in Mexico. We acknowledge support (CDL, AHW, JTZ) and computing resource provided by the Wake Forest University Health Sciences Center for Public Health Genomics.
Support for this work was obtained from the US National Institutes of Health grants: R01AR043814 (B.P.T), RO1 AI068787 (G.C.T.), R01AR043274 (K.L.M.), R01 AI063274 (P.M.G.), N01AR62277 (J.B.H.), R37AI024717 (J.B.H.), RO1AR042460 (J.B.H.), P01 AI083194 (J.B.H.), P20RR020143 (J.B.H.), PO1AR49084 (R.P.K.), R01AR33062 (R.P.K.), K08 AI083790 (T.B.N.), LRP AI071651 (T.B.N.), R01CA141700 (M.E.A.R.), RC1 AR058621 (M.E.A.R.) and UL1 RR024999 (T.B.N.), R01AR051545- 01A2 (A.M.S.), P30 AR053483 (J.A.J and J.M.G), AR 43727(M.A.P), UL1RR025005(M.A.P), K24 K24AR002138(R.R.G), P602AR30692(R.R.G), P01AR49084(R.R.G), UL1RR025741(R.R.G), P20 RR015577 (J.A.J), N01 AI50026 (J.A.J and J.M.G), R21 AI070304 (S.A.B.), P60 AR053308 (L.A.C.), M01 RR-00079 (L.A.C.), UL1-RR029882 (G.S.G. and D.L.K.), and P60-AR049459 (G.S.G. and D.L.K.).. This study was also supported by a grant from the Korea Healthcare Technology R&D Project, Ministry for Health and Welfare, Republic of Korea (A010252, A080588; S.C.B.), Korean R&D Program of MKE/KEIT (10035615; Y.W.S), the Merit Award from the US Department of Veterans Affairs (J.B.H. and G.S.G.), Lupus Research Institute (B.P.T., T.B.N., A.M.S.), The Alliance for Lupus Research (K.L.M.,T.B.N., L.A.C. and C.O.J.), the Arthritis National Research Foundation Eng Tan Scholar Award (T.B.N. and JZ), the Arthritis Foundation (A.M.S, P.M.G.), and the US Department of Defense PR094002 (J.B.H.). Additional funding awarded from the Swedish Research Council, Swedish Association Against Rheumatism and the King Gustaf Vth 80th Jubilee. Foundation and the Fundación Instituto de Salud Carlos III PS0900129 and the Consejería de Salud de Andalucía PI-0012 (M.E.A.R.), the Wellcome Trust (T.J.V.), Arthritis Research UK (T.J.V.), Arthritis Research UK project grant 17761 (T.J.V.), CTSA Grant Number I ULI RR025014-02 (A.M.S.) from the National Center for Research Resources (NCRR), Kirkland Scholar Award (L.A.C.). The work reported on in this publication has been in part financially supported by the ESF, in the framework of the Research Networking Programmers European Science Foundation – The Identification of Novel Genes and Biomarkers for Systemic Lupus Erythematosus (BIOLUOUS)-RNP-083
Reference List
- (1).Eichhorn PJ, Creyghton MP, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta. 2009;1795(1):1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- (2).Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353(Pt 3):417–39. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Roberts KG, Smith AM, McDougall F, Carpenter H, Horan M, Neviani P, et al. Essential requirement for PP2A inhibition by the oncogenic receptor c-KIT suggests PP2A reactivation as a strategy to treat c-KIT+ cancers. Cancer Res. 2010;70(13):5438–47. doi: 10.1158/0008-5472.CAN-09-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Purev E, Giordano A, Soprano DR, Soprano KJ. Interaction of PP2A catalytic subunit with Rb2/p130 is required for all-trans retinoic acid suppression of ovarian carcinoma cell growth. J Cell Physiol. 2006;206(2):495–502. doi: 10.1002/jcp.20490. [DOI] [PubMed] [Google Scholar]
- (5).Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139(3):468–84. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- (6).Janssens V, Longin S, Goris J. PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail) Trends Biochem Sci. 2008;33(3):113–21. doi: 10.1016/j.tibs.2007.12.004. [DOI] [PubMed] [Google Scholar]
- (7).Sunahori K, Juang YT, Tsokos GC. Methylation status of CpG islands flanking a cAMP response element motif on the protein phosphatase 2Ac alpha promoter determines CREB binding and activity. J Immunol. 2009;182(3):1500–8. doi: 10.4049/jimmunol.182.3.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Katsiari CG, Kyttaris VC, Juang YT, Tsokos GC. Protein phosphatase 2A is a negative regulator of IL-2 production in patients with systemic lupus erythematosus. J Clin Invest. 2005;115(11):3193–204. doi: 10.1172/JCI24895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Linker-Israeli M, Bakke AC, Kitridou RC, Gendler S, Gillis S, Horwitz DA. Defective production of interleukin 1 and interleukin 2 in patients with systemic lupus erythematosus (SLE) J Immunol. 1983;130(6):2651–5. [PubMed] [Google Scholar]
- (10).Theofilopoulos AN, Dixon FJ. Murine models of systemic lupus erythematosus. Adv Immunol. 1985;37:269–390. doi: 10.1016/s0065-2776(08)60342-9. [DOI] [PubMed] [Google Scholar]
- (11).Crispin JC, Kyttaris VC, Terhorst C, Tsokos GC. T cells as therapeutic targets in SLE. Nat Rev Rheumatol. 2010;6(6):317–25. doi: 10.1038/nrrheum.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Crispin JC, Liossis SN, Kis-Toth K, Lieberman LA, Kyttaris VC, Juang YT, et al. Pathogenesis of human systemic lupus erythematosus: recent advances. Trends Mol Med. 2010;16(2):47–57. doi: 10.1016/j.molmed.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6(12):683–92. doi: 10.1038/nrrheum.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358(9):900–9. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- (15).Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40(2):204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40(2):211–6. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- (17).Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40(9):1059–61. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1228–33. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1234–7. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- (20).Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6(2):e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kruglyak L. The road to genome-wide association studies. Nat Rev Genet. 2008;9(4):314–8. doi: 10.1038/nrg2316. [DOI] [PubMed] [Google Scholar]
- (22).Eleftherohorinou H, Wright V, Hoggart C, Hartikainen AL, Jarvelin MR, Balding D, et al. Pathway analysis of GWAS provides new insights into genetic susceptibility to 3 inflammatory diseases. PLoS One. 2009;4(11):e8068. doi: 10.1371/journal.pone.0008068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4(2):45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- (24).Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- (25).Brunner HI, Gladman DD, Ibanez D, Urowitz MD, Silverman ED. Difference in disease features between childhood-onset and adult-onset systemic lupus erythematosus. Arthritis Rheum. 2008;58(2):556–62. doi: 10.1002/art.23204. [DOI] [PubMed] [Google Scholar]
- (26).Jakab L, Laki J, Sallai K, Temesszentandrasi G, Pozsonyi T, Kalabay L, et al. Association between early onset and organ manifestations of systemic lupus erythematosus (SLE) and a down-regulating promoter polymorphism in the MBL2 gene. Clin Immunol. 2007;125(3):230–6. doi: 10.1016/j.clim.2007.08.020. [DOI] [PubMed] [Google Scholar]
- (27).Lessard CJ, Adrianto I, Kelly JA, Kaufman KM, Grundahl KM, Adler A, et al. Identification of a Systemic Lupus Erythematosus Susceptibility Locus at 11p13 between PDHX and CD44 in a Multiethnic Study. Am J Hum Genet. 2011;88(1):83–91. doi: 10.1016/j.ajhg.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Halder I, Shriver M, Thomas M, Fernandez JR, Frudakis T. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications. Hum Mutat. 2008;29(5):648–58. doi: 10.1002/humu.20695. [DOI] [PubMed] [Google Scholar]
- (29).Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Purcell S, Daly MJ, Sham PC. WHAP: haplotype-based association analysis. Bioinformatics. 2007;23(2):255–6. doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
- (31).Katsiari CG, Kyttaris VC, Juang YT, Tsokos GC. Protein phosphatase 2A is a negative regulator of IL-2 production in patients with systemic lupus erythematosus. J Clin Invest. 2005;115(11):3193–204. doi: 10.1172/JCI24895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Barron L, Dooms H, Hoyer KK, Kuswanto W, Hofmann J, O'Gorman WE, et al. Cutting edge: mechanisms of IL-2-dependent maintenance of functional regulatory T cells. J Immunol. 2010;185(11):6426–30. doi: 10.4049/jimmunol.0903940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hoyer KK, Dooms H, Barron L, Abbas AK. Interleukin-2 in the development and control of inflammatory disease. Immunol Rev. 2008;226:19–28. doi: 10.1111/j.1600-065X.2008.00697.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1