Genetic Coding Variant in GPR65 Alters Lysosomal pH and Links Lysosomal Dysfunction with Colitis Risk (original) (raw)
. Author manuscript; available in PMC: 2017 Jun 21.
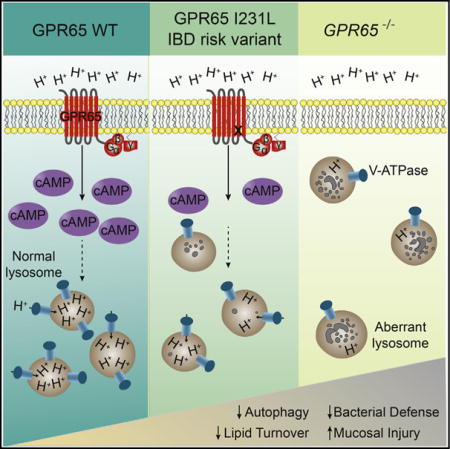
SUMMARY
Although numerous polymorphisms have been associated with inflammatory bowel disease (IBD), identifying the function of these genetic factors has proved challenging. Here we identified a role for nine genes in IBD susceptibility loci in antibacterial autophagy and characterized a role for one of these genes, GPR65, in maintaining lysosome function. Mice lacking Gpr65, a proton-sensing G protein-coupled receptor, showed increased susceptibly to bacteria-induced colitis. Epithelial cells and macrophages lacking GPR65 exhibited impaired clearance of intracellular bacteria and accumulation of aberrant lysosomes. Similarly, IBD patient cells and epithelial cells expressing an IBD-associated missense variant, GPR65 I231L, displayed aberrant lysosomal pH resulting in lysosomal dysfunction, impaired bacterial restriction, and altered lipid droplet formation. The GPR65 I231L polymorphism was sufficient to confer decreased GPR65 signaling. Collectively, these data establish a role for GPR65 in IBD susceptibility and identify lysosomal dysfunction as a potentially causative element in IBD pathogenesis with effects on cellular homeostasis and defense.
Graphical Abstract
INTRODUCTION
Gene mapping efforts have the potential to unlock profound insights into disease pathogenesis because each genetic association individually carries a biological link to disease. This approach is relevant to all genetic diseases as well as cancer (Reuter et al., 2015) and offers the potential to identify targets for development of novel therapeutic interventions. However, the swift progress of genome-wide association studies (GWASs) in many disease areas has exposed limitations in translating genetic loci to pathogenic insights, and it is thus becoming clear that the primary challenge for human genetics is no longer discovering genetic associations, but in the identification of how the identified genes and corresponding alleles exert their influence on the biology of health and disease (Altshuler et al., 2008).
Inflammatory bowel disease (IBD), including Crohn’s disease and ulcerative colitis, is a complex disease involving inflammation of the gastrointestinal tract. It is triggered by both genetic and environmental factors. There has been enormous progress in IBD genetics with the discovery of common genetic variants in more than 150 regions of the human genome that increase the risk and underlie the biology of IBD (Jostins et al., 2012). These common genetic variants have helped reveal key cell types controlling intestinal homeostasis and pathways such as antibacterial defense and autophagy as playing important roles in disease (Adolph et al., 2013; Cadwell et al., 2008; Cooney et al., 2010; Geremia et al., 2011; Lassen et al., 2014; Murthy et al., 2014). However, for the vast majority of these genetic associations, the specific implicated gene and causal variants have not been identified, limiting the near-term insights into pathogenesis and the longer-term ability to convert these associations into actionable therapeutic hypotheses.
The juxtaposition of the polymicrobial community with gut tissue creates an environment in which host and/or cell sensing of microbes is critical to homeostasis. Understanding how gut microbes, including potentially pathogenic microbes, are sensed by gut tissues to coordinate an appropriate inflammatory response is a critical question in IBD (Deretic et al., 2013). Recent work has characterized the role of microbes and their bacterial products in altering gut dynamics (Kishino et al., 2013; Macia et al., 2015; Maslowski et al., 2009; Venkatesh et al., 2014). In turn, host genetics impinging on host response pathways, including intracellular bacterial defense, autophagy, metabolite receptor expression, and the ability to respond to cellular stresses, influences IBD susceptibility (Kaser et al., 2011; Knights et al., 2013). Autophagy (macroautophagy) is a prosurvival intracellular clearance system that directs cytoplasmic cargo to lysosomes for proteolytic degradation (Gomes and Dikic, 2014; Levine et al., 2011). Emerging data highlight the critical role of autophagy in host defense against a range of bacteria, viruses, and parasites (Deretic et al., 2015). Autophagy is critical for maintaining intestinal homeostasis in multiple cell types. To date, the IBD-associated genes ATG16L1 and IRGM have each been implicated in antibacterial autophagy (Lassen et al., 2014; McCarroll et al., 2008; Murthy et al., 2014; Singh et al., 2006). It is currently unclear whether there are additional IBD-associated genes that function in autophagy or intracellular bacterial defense.
In this study, we used a functional genomic approach to identify 30 IBD-associated genes in host bacterial defense and pinpointed the role for GPR65 in IBD pathogenesis. In vivo, loss of GPR65 increased susceptibility to colitis. At the cellular level, we showed that GPR65 maintained lysosomal function, thus preserving autophagy and pathogen defense. Expression of an IBD-associated missense variant of GPR65 (rs3742704), which encodes an isoleucine-to-leucine substitution at amino acid 231 (I231L), in cell lines and in patient lymphoblasts resulted in impaired lysosomal acidification and disrupted lysosomal function. Genetic rescue of this aberrant lysosomal activity in GPR65 I231L-expressing cells restored intracellular bacterial defense. Together these data highlight a critical role for lysosomal function in intestinal homeostasis and suggest a framework for placing disease-associated genes into functional pathways.
RESULTS
Functional Genomics Identifies a Role for IBD Risk Genes in Pathogen Defense
To begin assigning function to genes in IBD risk loci, we selected two known IBD-relevant pathways: intracellular bacterial replication and antibacterial autophagy. We targeted genes in each IBD risk locus by using short interfering RNAs (siRNAs) and monitored changes in intracellular replication of a bioluminescent Salmonella enterica and serovar Typhimurium after infection of immortalized cervical HeLa cells via a high-throughput assay (Figure 1A; Shaw et al., 2013). A second siRNA screen was used to identify genes involved in antibacterial autophagy by monitoring changes in colocalization of S. Typhimurium and the autophagy marker LC3 (MAP1LC3A, microtubule-associated proteins 1 light chain 3A) in HeLa cells stably expressing green fluorescent protein- LC3 (GFP-LC3) (Figure 1A; Lassen et al., 2014; Murthy et al., 2014). siRNAs against ATG16L1, a core autophagy protein, served as a positive control for both screens (Figures S1A and S1B). The two screens were performed using siRNA pools containing three siRNAs targeting each gene and found that knockdown of 9.7% of the genes increased intracellular replication and knockdown of 10.5% of the genes reduced LC3 colocalization (Figure 1B and Tables S1 and S2). These data reinforce the functional importance of antibacterial autophagy in IBD susceptibility.
Figure 1. Functional Genomic Analysis Identifies Genes Involved in Autophagy-Dependent Intracellular Pathogen Defense.
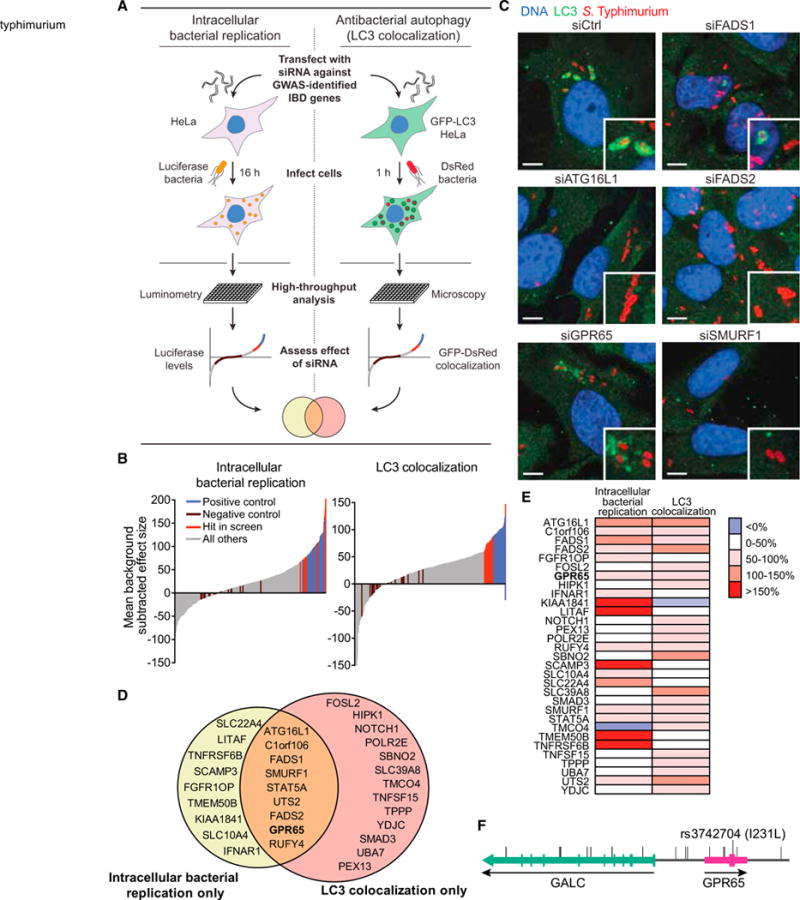
(A) Schematic of high-throughput genetic screens. HeLa or HeLa-GFP-LC3 cells were transfected with an siRNA library and infected with bioluminescent S. Typhimurium (intracellular bacterial replication) or DsRed-labeled S. Typhimurium (LC3 colocalization).
(B) Mean effect size of siRNAs in intracellular bacterial replication assay (left) and LC3 colocalization assay (right). n = 2 independent experiments.
(C) Representative confocal micrographs of selected siRNA-treated HeLa cells infected for 1 hr with DsRed-labeled S. Typhimurium. Scale bars represent 7.5 μm.
(D) Validated genes that scored in LC3 colocalization and intracellular bacterial replication assays using single siRNAs for a given gene.
(E) Heatmap illustration of effect size scores of confirmed hits.
(F) Schematic of GPR65 locus with identified IBD risk SNPs from the IBD fine-mapping project.
See also Figure S1 and Tables S1, S2, and S3–S5.
Genes that exhibited phenotypes in either primary assay screen were retested with three individual siRNAs per gene and endogenous LC3 staining. Genes were then filtered based on performance of individual siRNAs in both high-throughput and manual assays, as well as mRNA expression level in HeLa cells and correlation of activity with knockdown efficiency (Tables S3 and S4). This filtering resulted in a list of 30 genes, 9 of which were confirmed to have positive effects on both bacterial replication and LC3-bacteria colocalization, including our positive control, ATG16L1 (Figures 1C–1E and S1C). Most of the identified genes have not been previously suggested to alter pathogen defense or autophagy; however, some genes that scored in these screens were supported by published findings. RUFY4 (RUN and FYVE domain containing 4) has recently been shown to positively regulate autophagy in response to the cytokine IL-4 (Terawaki et al., 2015). SCAMP3 (secretory carrier membrane protein 3), a gene that scored only in the intracellular bacterial replication screen, has been shown to affect intracellular localization of S. Typhimurium, a phenotype that can have consequences for bacterial replication in host cells (Mota et al., 2009). Furthermore, the E3 ligase gene SMURF1 and the peroxisomal biogenesis gene PEX13 are implicated in selective autophagy of viruses and mitochondria, suggesting that these genes might function broadly in selective types of autophagy (Orvedahl et al., 2011).
Of the nine genes identified as positive regulators of pathogen defense in both screens, GPR65 (G protein coupled receptor 65) was selected for follow-up analysis because the association of IBD to the GPR65 locus implicates a set of 17 individual variants (Figure 1F and Table S5), including the missense mutation I231L (Jostins et al., 2012). The I231L variant has a frequency of 19.67% in the population. Additionally, protein expression and human transcriptome data indicate that GPR65 mRNA and protein expression is highest in immune cell compartments, including whole blood and spleen, as well as the intestinal tissue, suggesting that it plays a role in intestinal homeostasis (Kyaw et al., 1998; Melé et al., 2015). Although GPR65 is known to function as a H+-sensing G protein-coupled receptor (GPCR) that responds to acidic pH, the role of GPR65 in intestinal inflammation is currently unknown (Wang et al., 2004). Nonetheless, luminal changes in pH have been associated with inflammation and IBD (Nugent et al., 2001). Taken together, these data suggest a molecular link between GPR65 function and disease.
Loss of Gpr65 Increases Bacteria-Induced Colitis Susceptibility
To determine whether GPR65 plays an important role in intestinal homeostasis in vivo and to test host defense against pathogenic bacteria, we used Citrobacter rodentium, a murine pathogen that results in colonic lesions similar to the clinical enteropathogenic Escherichia coli strains associated with Crohn’s disease (Longman et al., 2014; Nell et al., 2010; Sasaki et al., 2007). Recent reports on Atg16l1 hypomorphic mice suggest that impaired autophagy can protect against C. rodentium infection (Marchiando et al., 2013). To determine whether _Gpr65_−/− mice were resistant to infectious colitis, we infected wild-type (WT) and _Gpr65_−/− mice with C. rodentium. Untreated _Gpr65_−/− mice were healthy with no signs of spontaneous colitis or inflammation (Figure 2A). At 14 days after infection, _Gpr65_−/− mice infected with C. rodentium showed more severe inflammation by histopathology (Figures 2A and 2B). Moreover, _Gpr65_−/− mice had significantly higher levels of C. rodentium in the cecum, colon, and feces, a phenotype consistent with decreased pathogen clearance (Figure 2C). Immunohistochemistry of infected colon tissue revealed enhanced infiltration of T cells (detected as CD3+) in _Gpr65_−/− mice although no differences in macrophage infiltration was observed (Figures 2D and S2). These data suggested that GPR65 was required for effective pathogen clearance and modulated susceptibility to infectious colitis in vivo. Additionally, these results were consistent with GPR65 having functions beyond autophagy.
Figure 2. _Gpr65_−/− Mice Are More Susceptible to Bacterial-Induced Colitis.
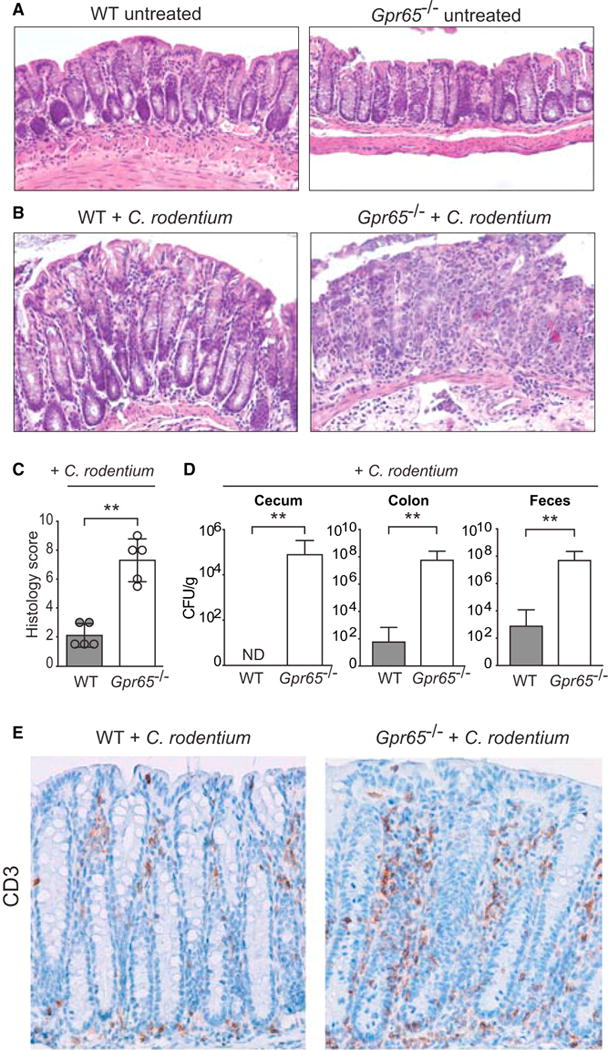
(A) Representative H&E-stained sections of distal colon tissue are shown from untreated WT and _Gpr65_−/− mice (20× magnification) (n = 4 mice per genotype).
(B) Representative H&E-stained sections of distal colon tissue are shown from infected WT and _Gpr65_−/− mice (20× magnification) (n = 8 mice per genotype).
(C) Histological score for inflammation in colon tissues 14 days after C. rodentium infection. Data shown as mean ± SD; n = 5/group. **p < 0.01 (Mann-Whitney U test).
(D) WT and _Gpr65_−/− mice were orally infected with C. rodentium and bacterial numbers (CFU) were measured in the cecum, colon, feces, and liver 14 days after infection. Data are means + SEM (n = 10 [WT], 10 [_Gpr65_−/−]; 2 experiments). **p < 0.01 (Mann-Whitney U test).
(E) Immunohistochemistry image of CD3 staining on WT and _Gpr65_−/− colon tissue 14 days after infection with C. rodentium.
See also Figure S2.
Gpr65 Expression in Non-hematopoietic and Hematopoietic Cells Limits C. rodentium Infection
Gpr65 is highly expressed in various immune cell types as well as intestinal tissue. Both non-hematopoietic and hematopoietic cells have been shown to contribute to defense against C. rodentium (Song-Zhao et al., 2014; Vallance et al., 2002). To determine which cellular compartment was responsible for _Gpr65_-mediated protection, bone marrow (BM) cells from WT and _Gpr65_−/− mice were transferred into lethally irradiated WT and _Gpr65_−/− mice, respectively. At 8 weeks after transfer, mice were infected with C. rodentium for 11 days to analyze the immune response closer to the peak of infection. Consistent with our results in Figure 2, _Gpr65_−/− mice that received _Gpr65_−/− BM (_Gpr65_−/− → _Gpr65_−/− BM chimeric mice) exhibited higher levels of C. rodentium in the colon compared to WT → WT BM chimeric mice and decreased colon length, a marker of colitis (Figures 3A and 3B). Restoration of Gpr65 expression in the non-hematopoietic compartment (_Gpr65_−/− →WT) or in the hematopoietic compartment (WT → _Gpr65_−/−) alone was not sufficient to reduce C. rodentium bacterial loads to that of WT mice (Figure 3A). Data obtained using in vivo imaging of BM chimeric mice infected with a bioluminescent C. rodentium were consistent with bacterial CFU data (Figures S3A and S3B). Additionally, _Gpr65_−/− → WT BM chimeric mice displayed significantly shorter colon lengths (Figure 3B). Major differences in histopathology were not observed at this time point after infection (Figure 3C). Cytokine analysis demonstrated reduced expression of tumor necrosis factor α (TNF-α), interleukin 17 (IL-17), and interferon-γ (IFN-γ) in infected colons associated with loss of Gpr65 (Figures 3D and 3E). This reduction was most significant in _Gpr65_−/− → _Gpr65_−/− BM chimeric mice and was apparent at both the mRNA level and the protein level (Figures 3D and 3E). No difference was observed in IL-1β or IL-6 mRNA or protein expression in any BM chimeric mice (Figures 3D, 3E, S3C, and S3D). Additionally, levels of IL-23 mRNA were significantly reduced in mice lacking Gpr65 in the hematopoietic compartment (Figure S3C). Taken together, these data are consistent with a role for epithelial Gpr65 expression in limiting pathogen replication and a role for hematopoietic Gpr65 expression in the inflammatory response to intestinal pathogens.
Figure 3. Gpr65 Expression in Non-hematopoietic and Hematopoietic Cells Limits C. rodentium Infection.
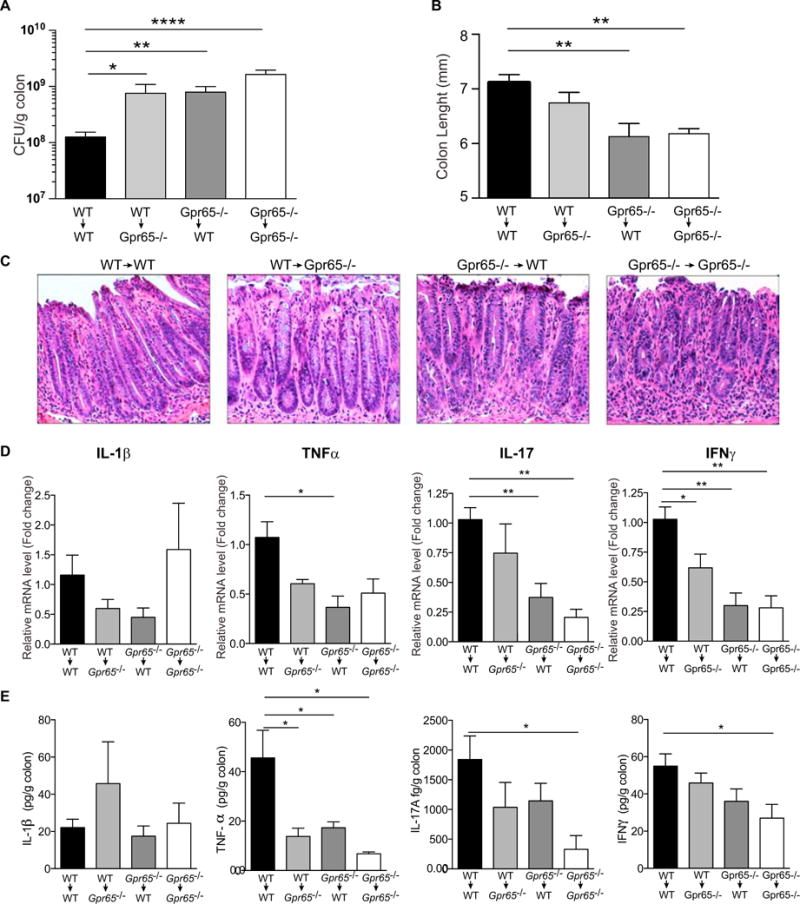
(A) Bone marrow chimeric mice were orally infected with C. rodentium and bacterial numbers (CFU) were measured in the colon at 11 days after infection. Data are means + SEM (n = 11 [WT → WT], n = 6 [WT → _Gpr65_−/−], n = 10 [_Gpr65_−/− → WT], n = 4 [_Gpr65_−/−/_Gpr65_−/−]). *p < 0.05, **p < 0.01, ****p < 0.0001 (unpaired t test).
(B) Colon length from bone marrow chimeric mice infected with C. rodentium for 11 days. Data are means + SEM. **p < 0.01 (unpaired t test).
(C) Representative H&E-stained sections of distal colon tissue are shown from infected bone marrow chimeric mice at 11 days after infection (20× magnification).
(D) Cytokine expression in _C. rodentium_-infected mice, as quantified by qRT-PCR. Relative mRNA levels of the indicated cytokine are shown. *p < 0.05; **p < 0.01 (unpaired t test).
(E) Secretion of cytokines from colon tissues 11 days after infection with C. rodentium. Data are means + SEM. *p < 0.05 (unpaired t test).
See also Figure S3.
Loss of GPR65 Alters Pathogen Clearance and Confers Intracellular Degradative Defects
To investigate the role of GPR65 in intracellular pathogen defense, we generated _GPR65_-null HeLa cell lines using the CRISPR/Cas9 system. To confirm that GPR65 ablation was specifically responsible for the observed phenotype and to control for any off-target effects of the CRISPR/Cas9 system, we stably re-expressed GPR65 or an empty vector control. We observed increased S. Typhimurium replication in _GPR65_-null cells compared to cells expressing GPR65, consistent with the knockdown results (Figure 4A). To determine whether GPR65 functions broadly in defense against other intracellular bacteria and in other cell types, we infected WT and _Gpr65_−/− primary bone-marrow-derived macrophages (BMDMs) with a strain of Listeria monocytogenes lacking the autophagy-evading protein ActA (L. monocytogenes ΔactA) (Rich et al., 2003). In agreement with our S. Typhimurium infection results, _Gpr65_−/− BMDMs showed significantly increased CFUs of intracellular L. monocytogenes, indicative of increased survival of the bacteria (Figure 4B). These data suggest a broad role for GPR65 in controlling intracellular microbial clearance.
Figure 4. Cells Lacking GPR65 Exhibit Impaired Antibacterial Autophagy and Accumulation of Aberrant Degradative Compartments.
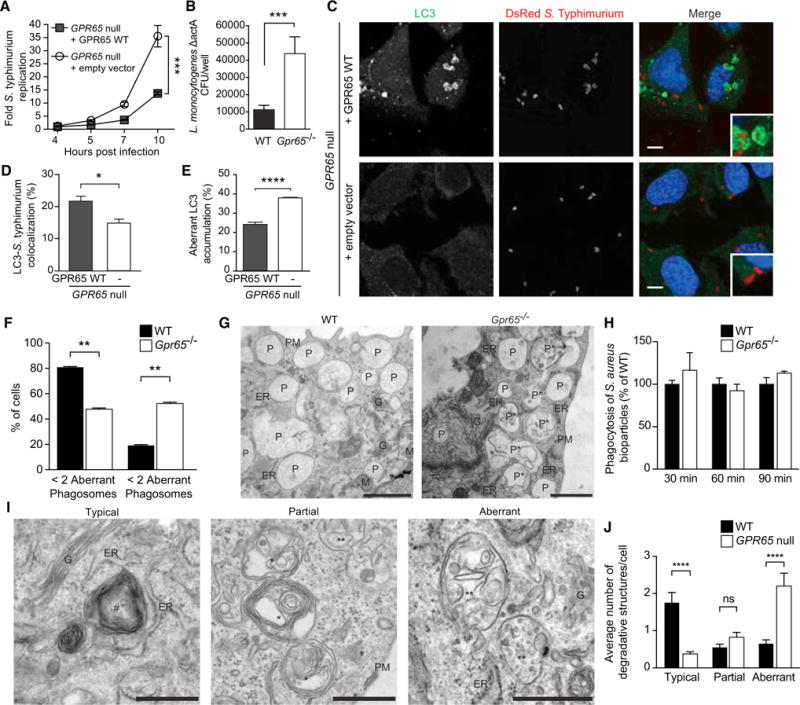
(A) S. Typhimurium intracellular replication in _GPR65_-null HeLa cells stably expressing either an empty vector or GPR65 WT. Bars represent means ± SD from n = 4. ***p < 0.001 (unpaired t test).
(B) CFU of intracellular Listeria ΔactA recovered from WT or _Gpr65_−/− BMDMs at 3 hr after infection. ***p < 0.001 (unpaired t test).
(C) Confocal micrographs of endogenous LC3-S. Typhimurium colocalization in _GPR65_-null HeLa cells stably expressing either an empty vector or GPR65 WT at 1 hr after infection. Scale bars represent 7.5 μm.
(D) Quantification of colocalization of S. Typhimurium and endogenous LC3 in indicated cells 1 hr after infection. Data shown represent means + SEM from n = 3 independent experiments. *p < 0.05 (unpaired t test).
(E) Quantification of aberrant endogenous LC3 accumulation at sites of S. Typhimurium-LC3 colocalization in cells shown in Figure S3D. Data shown represent means + SEM of n = 3 independent experiments. ****p < 0.0001 (unpaired t test).
(F) Quantification of the percentage of BMDMs displaying more than two aberrant phagosomes as shown in (B). Results are expressed as percent of cells with a defined type of phagosome in the cell population + SEM. **p < 0.01 (t test).
(G) Representative EM micrographs of WT and _Gpr65_−/− BMDMs showing phagosomal morphology. Abbreviations are as follows: ER, endoplasmic reticulum; G, Golgi; M, mitochondria; P, phagosomes; P*, aberrant phagosomes; PM, plasma membrane. Scale bars represent 1 μm.
(H) WT and _Gpr65_−/− BMDMs were treated with _S. aureus_-coated bioparticles conjugated to the pH indicator dye for the indicated times. Number of puncta were normalized to WT levels at each time point. n = 3 independent experiments.
(I) Representative EM micrographs of the lysosomal subpopulations observed in WT and _GPR65_-null HeLa cells. Abbreviations and symbols are as follows: #, typical lysosome; *, lysosome with a partial degradation defect; **, aberrant lysosome with a degradative defect; ER, endoplasmic reticulum; G, Golgi; PM, plasma membrane. Scale bars represent 500 nm.
(J) Quantification of each lysosome subpopulation shown in (I). Results are expressed as the number lysosomes per cell + SEM. ****p < 0.0001 (t test).
See also Figure S4.
GPR65 deficiency also resulted in decreased LC3-S. Typhimurium colocalization in HeLa cells (Figures 4C and 4D), and decreased LC3-Listeria colocalization was observed in _Gpr65_−/− BMDMs at 1 hr after infection, suggesting differences in intracellular bacterial targeting by autophagy in these cells (Figures S4A and S4B). Examination of LC3-S. Typhimurium autophagosomal structures by confocal microscopy revealed not only a decrease in LC3-containing membranes around S. Typhimurium, but also aberrant LC3 accumulation near S. Typhimurium within _GPR65_-null cells (arrowheads, Figure S4C). These LC3 accumulations presented as either empty circular structures or globular concentrations of LC3 adjacent to the bacteria without the characteristic engulfment observed during antibacterial autophagy (Figure S4D). This aberrant LC3 accumulation was reduced when GPR65 was re-expressed, showing that this phenotype was specific to GPR65 disruption (Figures 4E and S4D). Aberrant LC3 accumulation was also observed in _Listeria_-infected _Gpr65_−/− BMDMs and in S. Typhimurium-infected HeLa cells treated with GPR65 siRNA, suggesting that GPR65 functions similarly in pathogen targeting in multiple cell types (Figures S4E–S4H). The impaired recruitment of LC3 to bacteria as well as aberrant LC3 accumulations around bacteria indicated a potential block in autophagic turnover in _GPR65_-null cells.
To investigate the observed phenotypes at the ultrastructural level, we performed electron microscopy (EM) analysis on primary WT and _Gpr65_−/− BMDMs as well as our _GPR65_-null HeLa cells. WT BMDMs exhibited classical morphology with numerous phagosomes distributed throughout the cytoplasm (Figure 4F). A significant proportion of the phagosomes in _Gpr65_−/− BMDMs lost their luminal homogeneity and accumulated membranous structures in their interiors, suggesting a defect in the hydrolytic processing of endocytosed material (Figures 4F and 4G). The observed defect in phagosome biogenesis did not affect phagocytic uptake of _Staphylococcus aureus_-coated bioparticles, indicating that this phenotype was restricted to intracellular pathways (Figure 4H).
Lysosomes from WT and _Gpr65_−/− BMDMs differed in their appearance; however, the number of total lysosomes in BMDMs was inadequate for quantitative analysis of these differences (data not shown). HeLa cells, on the other hand, contained numerous lysosomes, allowing quantitative analysis of the major ultrastructural differences between lysosomes of cells with or without GPR65. In WT HeLa cells, most of the lysosomes had a typical morphology with onion-like membranous conformations in their lumen (Figures 4I and 4J). Two minor subpopulations of these degradative organelles were also observed. The first included lysosomes that were partially filled with unprocessed subvacuolar material as well as onion-like membranes (middle panel, Figure 4I). The second subpopulation included aberrant lysosomes that displayed accumulated undigested material in their lumen (right panel, Figure 4I). In contrast to WT cells, _GPR65_-null cells contained significantly fewer typical lysosomes and an increase in lysosomes with a degradation defect (Figures 4I and 4J). Taken together, the accumulation of unprocessed endolysosomal content in _GPR65_-null cells supports a partial lysosomal degradative defect in these cells.
GPR65 Crohn’s Disease Risk Variant Alters Lysosomal Activity
Consistent with our observation of aberrant lysosome accumulation in _GPR65_-null cells, an unbiased genome-wide analysis of gene expression differences between WT and _Gpr65_−/− BMDMs previously identified differential regulation of genes implicated in lysosomal function (Atp6v1d and Atp6v1e1) and vesicular transport (Snx10), suggesting potential dysregulation of these processes (Figure S5A; Efeyan et al., 2012; Qin et al., 2006). Atp6v1d and Atp6v1e1, two of the genes downregulated in _Gpr65_−/− cells, encode subunits of the H+ transporting vacuolar ATPase (V-ATPase) that functions to acidify lysosomes as well as other intracellular compartments. Additionally, SNX10 is known to directly bind the V-ATPase complex, controlling the subcellular localization and function of the V-ATPase complex (Chen et al., 2012). To determine whether these genes were also downregulated in non-immune cell types, we performed quantitative RT-PCR on _GPR65_-null HeLa cells and cells expressing either GPR65 WT or the IBD-risk variant GPR65 I231L (rs3742704). Reduced mRNA expression of ATP6V1D, ATP6V1E1, and SNX10 was also observed in _GPR65_-null HeLa cells and GPR65 I231L-expressing cells relative to WT cells, suggesting that this is a common feature of cells lacking GPR65 and not limited to specialized cell types (Figure S5B). Together, these data suggest that GPR65 signaling could alter V-ATPase complex trafficking and lysosomal function.
Expression of GPR65 WT and GPR65 I231L was equivalent in the generated cell lines and both proteins showed similar subcellular localization (Figures S6A and S6B). Knockout of GPR65 or expression of GPR65 I231L each resulted in an accumulation of the autophagy protein LC3-I and the autophagy target p62, consistent with a late block in autophagy and impairment of autophagic turnover (Figure S6C). Consistent results were obtained with _Gpr65_−/− BMDMs (Figure S6D). Similar protein levels of LC3-I and p62 were observed in the presence of the autophagy inducer, Torin 1, and the lysosomal protease inhibitors E64d and pepstatin A, and these data are consistent with impaired lysosomal processing in both _GPR65_-null and GPR65 I231L cells (Figures S6C and S6D).
Loss of GPR65 resulted in an increase in lysosomal pH compared to cells expressing GPR65 WT as measured by a decrease in the relative fluorescence intensity of the pH-sensitive LysoTracker Green probe in live cells, which marks acidic organelles (Figure 5A). Expression of GPR65 I231L also resulted in abnormal organelle pH (Figure 5A). Cells with or without GPR65 were similarly sensitive to treatment with the specific V-ATPase inhibitor, bafilomycin A1, which increases lysosomal pH and thus abolishes LysoTracker fluorescence. Imaging of LysoTracker-stained cells also revealed peripheral localization and accumulation of lysosomes in _GPR65_-null cells (Figures 5B and 5C). This aberrant accumulation of lysosomes was also observed to a lesser extent in cells expressing GPR65 I231L compared to GPR65 WT (Figures 5B and 5C). To determine whether this aberrant subcellular distribution of lysosomes was associated with defects in lysosomal function, cells were treated with the protease inhibitors pepstatin A and E64d to impair lysosomal proteolytic activity without perturbing lysosomal pH, thereby allowing tracking of lysosomal localization with pH-sensitive dyes. Treatment of cells with these inhibitors resulted in aberrant lysosome localization and accumulation in GPR65 I231L and GPR65 WT cells, but no change in the distribution of lysosomes in _GPR65_-null cells (Figures 5B and 5C). These findings align with a previous study demonstrating that lysosomal positioning is critical for lysosome function (Korolchuk et al., 2011).
Figure 5. GPR65 I231L-Expressing Cells Exhibit Aberrant Lysosomal Function and Increased Bacterial Replication.
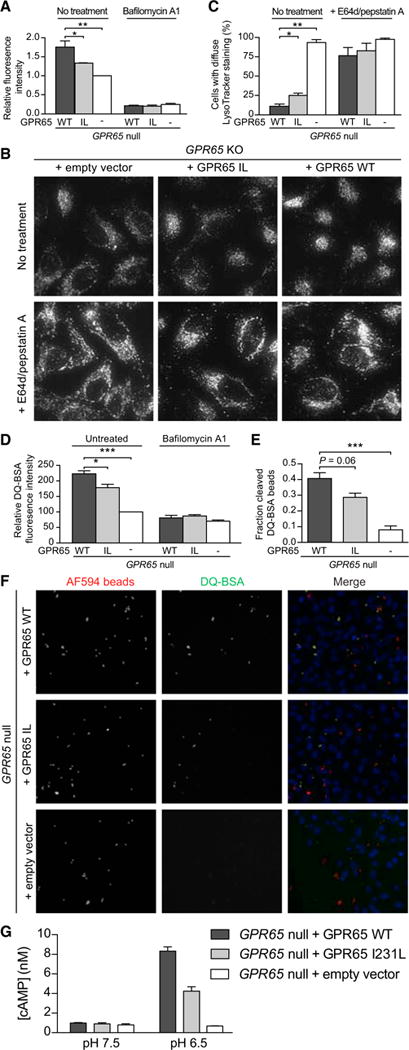
(A) Relative Lysotracker fluorescence in _GPR65_-null HeLa cells stably expressing either an empty vector, GPR65 WT, or GPR65 I231L. Cells were left untreated or treated with 100 nM bafilomycin A1 for 2 hr.
(B) Representative micrographs of _GPR65_-null HeLa cells stably expressing an empty vector, GPR65 WT, or GPR65 I231L stained with Lysotracker. Cells were left untreated or treated with 10 μg/mL E64d and pepstatin A for 3 hr prior to staining.
(C) Quantification of cells imaged in (B) with aberrant lysosomal localization.
(D) Relative DQ-BSA fluorescence in _GPR65_-null, GPR65 WT, or GPR65 I231L HeLa cells. Cells were left untreated or treated with 100 nM bafilomycin A1 for 2 hr.
(E) Fraction of DQ-BSA-positive fluorescent beads compared to total internalized beads in _GPR65_-null, GPR65 WT, or GPR65 I231L HeLa cells.
(F) HeLa cells treated with DQ BSA-conjugated beads. AF594 (red) was used to monitor total uptake of beads into cells. DQ-BSA (green) was used to monitor phagolysosomal activity.
(G) _GPR65_-null HeLa cells stably expressing an empty vector or GPR65-V5 WT were treated with buffers at the indicated pH for 30 min and cAMP levels were measured. Average of n = 3 independent experiments. Error bars represent + SEM.
For all panels, *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA with multiple comparisons. See also Figures S5 and S6.
To specifically test whether endolysosomal activity was disrupted in _GPR65_-null cells, we treated cells with DQ-BSA, a bovine serum albumin derivative that is conjugated to a self-quenched fluorophore that becomes fluorescent upon proteolysis in the lysosome (Reis et al., 1998). DQ-BSA fluorescence was higher in GPR65 WT-expressing cells compared to either _GPR65_-null or GPR65 I231L cells (Figure 5D). To measure phagolysosomal activity, we used 3-μm diameter beads conjugated to DQ-BSA and a pH-insensitive fluorochrome to measure the fraction of internalized beads that underwent lysosomal proteolysis. Consistent with aberrant phagolysosomal function, _GPR65_-null and GPR65 I231L cells contained significantly lower levels of cleaved DQ-BSA beads than did GPR65 WT cells despite similar levels of total internalized beads (Figures 5E, 5F, and S6E). These results suggest that loss of GPR65 or expression of GPR65 I231L increases lysosomal pH and impairs lysosomal function.
Increases in intracellular cAMP (cyclic adenosine monophosphate) regulate V-ATPase trafficking and reduce both phagosomal and lysosomal pH; however, the source of cAMP that regulates these processes is currently undefined (Breton and Brown, 2013; Coffey et al., 2014; Di et al., 2006). We hypothesized that cAMP produced in response to GPR65 activation could stimulate lysosomal acidification. We confirmed that treatment of HeLa cells with forskolin, a cAMP inducer, was sufficient to reduce lysosomal pH (Figure S6F). We next tested whether cAMP production was impaired in _GPR65_-null cells in response to low pH, a known activator of GPR65 (Mogi et al., 2009). _GPR65_-null cells did not produce cAMP in response to low pH, and re-expression of GPR65 rescued this phenotype, consistent with previous reports (Figure 5G). cAMP production was reduced in response to low pH in GPR65 I231L-expressing cells, suggesting that this polymorphism confers reduced activity in response to its protons (Figure 5G). _GPR65_-null HeLa cells were similarly responsive to forskolin independent of GPR65 expression, demonstrating that these cells are capable of producing cAMP and that the defect in cAMP production in response to an acidic buffer is specific to GPR65-dependent signaling (Figure S6G). Given the known role of SNX10 in V-ATPase trafficking, we also tested whether _SNX10_-null cells exhibited increased organelle pH. The pH of organelles in _SNX10_-null cells was higher, suggesting that SNX10 is required to maintain organelle pH (Figure S6H). In sum, these data suggest a model in which impaired GPR65-dependent cAMP production in response to changes in extracellular pH results in alterations to V-ATPase dynamics, potentially through SNX10, and subsequent dysregulation of organelle pH.
GPR65 I231L Alters Lipid Droplet Levels
Recent reports have suggested an important role for lysosomal degradation and autophagy in formation and turnover of intracellular lipid stores during nutrient deprivation (Rambold et al., 2015; Singh et al., 2009). Aberrant accumulation of lipid can result in cellular toxicity and inflammation, which could have important consequences for intestinal homeostasis. Expression data from _Gpr65_−/− BMDMs revealed differential regulation of genes involved in lipid uptake and/or metabolism (Cd5l, Pla2g7, and Fcrls), suggesting that this pathway might be altered (Figure S5). Cells store neutral lipids in the form of lipid droplets (LDs), which are broken down into free fatty acids during starvation and transported to mitochondria, where they undergo fatty acid oxidation to generate energy for the cell (Hashemi and Goodman, 2015). _GPR65_-null or rescued cells were treated with complete medium or nutrient-depleted medium (HBSS) and stained for LDs using BODIPY, a dye that stains neutral lipids. At steady-state, _GPR65_-null and GPR65 I231L-expressing cells exhibited an accumulation of lipid droplets compared to GPR65 WT cells (Figures 6A and 6B). Under conditions of nutrient depletion, there was a strong increase in the number of LD puncta in GPR65 WT cells. This increase was inhibited by treatment with the lysosomal inhibitor bafilomycin A1, consistent with the reported role for autophagy and lysosomal function in LD growth during starvation (Figure 6C; Rambold et al., 2015). LD growth during starvation was dramatically impaired in _GPR65_-null cells and partially impaired in GPR65 I231L-expressing cells (Figures 6A and 6B). Taken together, these data suggest that GPR65 I231L confers changes to lipid droplet turnover, which could have important consequences for lipid metabolism and cellular energetics.
Figure 6. GPR65 I231L Impairs Lipid Droplet Turnover.
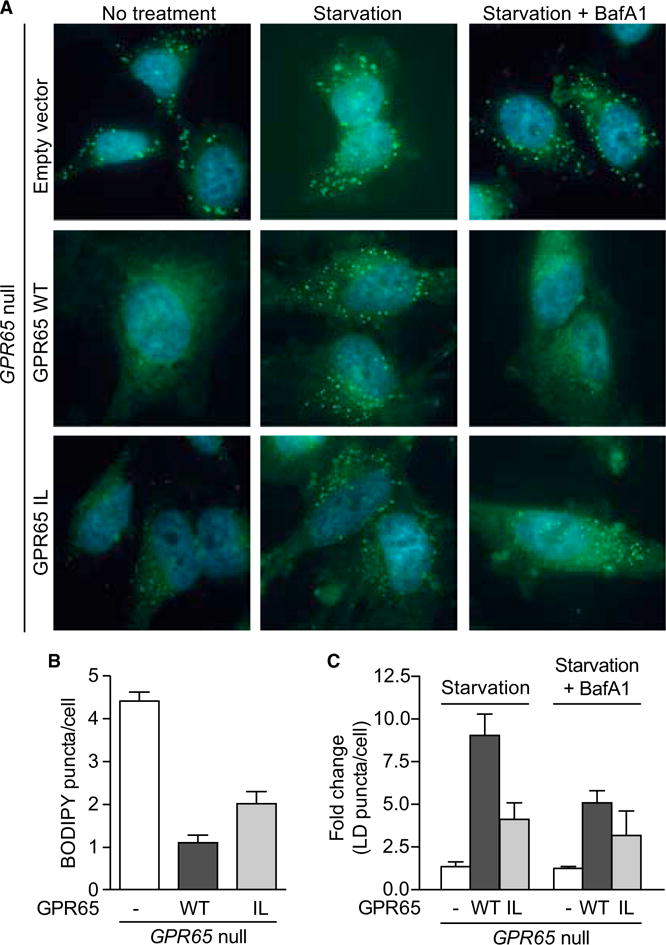
(A) Representative micrographs of cells treated for 4 hr with complete medium, HBSS, or HBSS and 200 nM bafilomycin A1 and stained with BODIPY. n = 4 independent experiments.
(B and C) Quantification of images as shown in (A). Results are representative of n = 4 independent experiments. Bars show mean + SEM.
GPR65 Risk Variant-Associated Lysosomal Dysfunction Disrupts Pathogen Clearance
To determine whether the observed lysosomal impairment in GPR65 I231L-expressing cells results in alterations in pathogen clearance, we performed an intracellular replication assay with S. Typhimurium. Expression of GPR65 I231L in _GPR65_-null cells resulted in an increase in intracellular bacterial replication compared to cells expressing GPR65 WT (Figure 7A). Expression of either of the V-ATPase subunits identified by RNA-seq, ATP6V1D or ATP6V1E1, was sufficient to restore lysosomal function and reduce bacterial replication in GPR65 I231L-expressing cells to WT levels, suggesting that lysosomal dysfunction contributes to the observed deficits in intracellular bacterial defense (Figures 7B and 7C).
Figure 7. Impaired Lysosomal Function in GPR65 I231L-Expressing Cells Results in Increased Bacterial Replication.
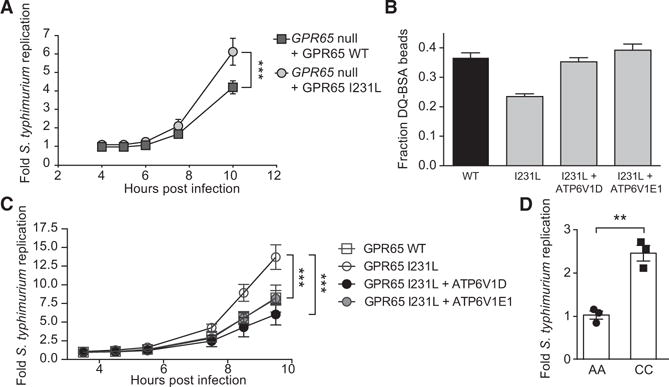
(A) S. Typhimurium intracellular replication in _GPR65_-null HeLa cells stably expressing either GPR65 WT or GPR65 I231L. Bars represent means ± SD from n = 4.
(B) Fraction of DQ-BSA-positive fluorescent beads compared to total internalized beads in GPR65 WT cells or GPR65 I231L cells stably expressing either ATP6V1D or ATP6V1E1.
(C) S. Typhimurium intracellular replication in GPR65 WT cells or GPR65 I231L cells stably expressing either ATP6V1D or ATP6V1E1. Bars represent means ± SD from n = 3 independent experiments.
(D) Intracellular S. Typhimurium replication in IBD patient-derived lymphoblasts expressing the ancestral SNP (AA) or the risk allele (CC).
For all panels, **p < 0.01, ***p < 0.001 (unpaired t test in A and D, one-way ANOVA with multiple comparisons in C).
We also performed bacterial replication assays in lymphoblasts derived from IBD patient peripheral blood mononuclear cells homozygous for the tagged GPR65 risk polymorphism and compared these results with age- and disease-matched controls. Enhanced S. Typhimurium replication was observed in lymphoblasts derived from individuals expressing the GPR65 risk polymorphism (Figure 7D). The enhanced intracellular bacterial replication observed with the IBD-associated coding variant in both engineered and patient-derived cells suggests that impaired lysosomal function decreases antimicrobial defense and that this could contribute to disease susceptibility.
DISCUSSION
Functional genetic approaches offer the potential to identify genes of interest and provide key functional clues into pathways involved in pathogenesis. Our findings provide an experimental framework for pinpointing potentially relevant genetic targets from disease-associated risk loci identified through GWASs, an approach that could lead to the identification of relevant therapeutic targets. Using this approach, we identified a number of genes that alter antibacterial autophagy and pathogen defense; however, the majority of these genes are probably not directly altering autophagy, as is the case for ATG16L1. Further assessment of relevant hits will be required to understand the individual contributions of these genes and their associated polymorphisms.
Our data suggest a model in which GPR65 signaling helps maintain appropriate organelle pH and that this signaling is impaired with the IBD-associated I231L polymorphism. This polymorphism in GPR65 is predicted to lie within one of the transmembrane regions of GPR65 (Kyaw et al., 1998). Experimental data suggest that changing an isoleucine to leucine alters the stability of alpha helices in the transmembrane region of proteins (Lyu et al., 1991). Thus, the I231L polymorphism could disrupt the transmembrane helix structure in GPR65, leading to the observed impaired signaling and cAMP induction. To date, the source of cAMP that regulates lysosomal pH is unclear (Rahman et al., 2013). Our data suggest a model in which GPR65 senses changes in pH, inducing cAMP production, which in turn regulates V-ATPase activity, potentially through SNX10. We show that loss of SNX10 is sufficient to alter lysosomal pH. Taken together, this study defines the role of GPR65 signaling in the maintenance of lysosomal pH and function and demonstrates that a single polymorphism could alter this process.
To date, little was known about the role of lysosomal function in IBD susceptibility. Our data suggest that the IBD-associated risk variant GPR65 I231L disrupts lysosomal function and impairs pathogen defense, reinforcing the concept that decreases in bacterial clearance have important consequences for IBD pathogenesis (Lassen et al., 2014; Murthy et al., 2014). However, given the central role of lysosomal function in cellular homeostasis, our finding that GPR65 I231L alters lysosomal function has broad implications for tissue homeostasis beyond autophagy and pathogen clearance. We demonstrate that _Gpr65_−/− mice are highly susceptible to infection with C. rodentium, in contrast to reports using Atg16l1 hypomorphic mice, which have a reduction in autophagy but no effect on lysosome function (Marchiando et al., 2013). Additionally, WT and _Gpr65_−/− mice infected with C. rodentium produced similar levels of IL-1β to WT whereas an impairment of autophagy is known to increase IL-1β production from macrophages (Saitoh et al., 2008). Finally, our data in BM chimeric mice demonstrate an important role for Gpr65 in both the hematopoietic and non-hematopoietic compartments, consistent with the broad role of lysosomes in cellular physiology.
It is likely that GPR65 is playing a critical role in phagocytic cells that require high levels of V-ATPase activity to maintain phagosomal and lysosomal pH, and this activity aids in the direct clearance of enteric pathogens. Recent studies have shed light on the critical role that lysosomes play in the immune response, signaling, tissue repair, and cellular energetics, and it is likely that disruptions in these responses contribute to IBD susceptibility (Ferguson, 2015; Settembre et al., 2013).
Parallels can be drawn between our findings and other diseases known to alter lysosomal function. Hermansky-Pudlak syndrome (HPS) is a rare autosomal-recessive disorder resulting from mutations in genes involved in the formation and trafficking of lysosome-related organelles. Interestingly, a subset of HPS patients develop Crohn’s disease-like colitis, suggesting that genetic alterations to lysosomal function can result in Crohn’s-like pathologies (Hazzan et al., 2006). Although inherited lysosomal disorders are known to cause severe disease, our data suggest that subtle changes to lysosomal function through genetic polymorphisms can alter susceptibility to microbes, particularly in the gut at the host-commensal interface (Ferguson, 2015).
We demonstrated that lysosomal dysfunction in _GPR65_-null cells and GPR65 I231L-expressing cells disrupted lipid droplet formation during acute starvation. Aberrant lipid turnover and metabolism might also directly contribute to IBD pathogenesis. Of note, the fatty acid desaturases FADS1 and FADS2 each scored in both the bacterial replication and LC3-bacteria colocalization assays. FADS1 and FADS2 are rate-limiting enzymes that regulate the unsaturation of ω-3 and ω-6 long-chain polyunsaturated fatty acids. Another gene that scored in our screen, PEX13, underlies the peroxisomal disorder Zellweger syndrome (Shimozawa et al., 1999). Loss of PEX13 in humans or mice is characterized by impaired peroxisomal β-oxidation of very long chain fatty acids and an accumulation of lipid droplets (Maxwell et al., 2003). Recent studies have explored the role of lipid metabolism and lipid signaling in immune activation and pathogen defense (O’Neill and Pearce, 2016; York et al., 2015). For example, fatty acid synthesis and turnover are critical for the Treg/Th17 cell signaling axis as well as M1/M2 macrophage activation (Berod et al., 2014; Huang et al., 2014). Potentially, lipid metabolism in immunity could be a common pathway contributing to IBD pathogenesis.
In conclusion, our investigation used an unbiased screening approach to identify GPR65 as a previously uncharacterized key gene controlling susceptibility to colitis. Defects in lysosomal activity associated with GPR65 I231L will negatively impact autophagy, a known IBD-associated pathway; however, therapeutic approaches aimed to boost overall levels of autophagy in these cells could exacerbate disease in these individuals. By increasing the flux of materials to the lysosome, autophagy modulators could stress lysosomes that are functioning at capacity and further disrupt cellular homeostasis. Potentially, a GPR65 agonist could help restore lysosomal function in individuals who harbor GPR65 risk polymorphisms. For this reason, development of personalized therapeutic strategies will require an understanding of not only the pathogenic contributions of disease-associated genes, but also the function of specific polymorphisms.
EXPERIMENTAL PROCEDURES
siRNA Screens
HeLa cells (parental strain or stably transduced with LC3-GFP) were reverse transfected with 5 pmol pooled siRNA (three siRNAs targeting one specific human gene per well) against selected genes within IBD susceptibility loci from the Silencer Select Human Druggable Genome siRNA Library (Ambion) available in 96-well glass-bottomed plates at a density of 6 × 103 cells per well for imaging analysis. S. Typhimurium overnight cultures were subcultured 1:33 for 3 hr and then infected at MOI 100:1 for DsRed S. Typhimurium and bioluminescent S. Typhimurium (Perkin Elmer). See the Supplemental Experimental Procedures for further details.
Antibacterial Autophagy Assays
S. Typhimurium infection of HeLa cells and LC3 quantification were performed as previously described (Lassen et al., 2014). In brief, 1 × 105 HeLa cells were plated on glass coverslips in 12-well plates. Cells were infected with SL1344 DsRed-Salmonella enterica serovar Typhimurium for 1 hr, fixed in ice-cold methanol, and stained with rabbit anti-LC3 (Sigma), goat anti-CSA (S. Typhimurium), and Hoechst. See the Supplemental Experimental Procedures for further details.
Lysosomal Function Assays
To measure the pH of acidic intracellular compartments, cells were loaded with 75 nM Lysotracker Green DND-26 (Invitrogen) for 5 min. Cells were analyzed on a BD FACSVerse instrument and mean fluorescence intensity (MFI) was calculated via FlowJo software. MFI was normalized to _GPR65_-null cells. For DQ-BSA assays, 10,000 cells/well were plated in 96-well plates. The next day, cells were starved for 1 hr with HBSS and then treated with 10 μg/mL DQ-BSA Green dye (Life Technologies) in complete media to increase endocytosis. Cells were incubated for 1 hr and then washed. Cells were stained with Hoechst 33342 (Molecular Probes) for 5 min to visualize nuclei and imaged with the ImageXpress Micro Widefield High-Content Screening System with a 20× lens. Fluorescence intensity of internalized DQ-BSA was quantified with MetaExpress software. See the Supplemental Experimental Procedures for further details.
Lipid Droplet Analysis
Cells were plated in 96-well plates as described above. For starvation experiments, cells were starved for 3 hr in HBSS. Cells were washed in PBS and fixed in 4% PFA with Hoechst for 15 min. Cells were washed and stained for 15 min with BODIPY 493/503 in PBS at a final concentration of 1 μg/mL. For each well, 6–9 fields were imaged and analyzed to obtain an average number of LDs/cell for each well. For each experiment, 3–6 replicate wells were analyzed. See the Supplemental Experimental Procedures for further details.
Mouse Experiments
All experiments involving mice were carried out according to protocols approved by the relevant animal ethics committees. _Gpr65_−/− mice, in which exon 2 coding sequences are replaced by promoterless IRES-EGFP sequences, were obtained from Jax laboratories and backcrossed on a C57BL/6 background to 13 generations. All experiments were performed on age- and gender-matched animals. C. rodentium infections were performed as described previously. C. rodentium were grown overnight at 37°C in 10 mL LB with 100 μg/mL nalidixic acid or in LB with ampicillin for bioluminescent C. rodentium. Overnight C. rodentium cultures were harvested by centrifugation and resuspended in PBS. Mice were orally gavaged with 1–4 × 109 colony-forming units (CFUs) of the bacterial suspension. CFUs from the indicated organs were determined by plating on LB agar containing nalidixic acid. Histological slides of colon were scored blinded for C. rodentium infection, with a maximum possible score of 10. See the Supplemental Experimental Procedures for further details.
Supplementary Material
1
2
3
Highlights.
- Genomic screen identifies role for nine genes in IBD susceptibility loci in autophagy
- _Gpr65_-deficient mice are more susceptible to bacteria-induced colitis
- Loss of GPR65 results in accumulation of aberrant phagosomes and lysosomes
- GPR65 IBD risk variant confers defects in lysosomal function and pathogen defense
Acknowledgments
We thank Natalia Nedelsky for editorial assistance, Jenny Tam for help with the generation of DQ-BSA-conjugated beads, and Rushika C. Wirasinha for support with _Gpr65_−/− mice. We thank Skip Virgin and Beth Levine for comments. This work was supported by funding from The Leona M. and Harry B. Helmsley Charitable Trust, Crohn’s and Colitis Foundation of America, and NIH grants DK097485 and AI109725 to R.J.X. F.R. is supported by ALW Open Program (822.02.014), DFG-NWO cooperation (DN82-303), SNF (CRSII3_154421), and ZonMW VICI (016.130.606) grants.
Footnotes
ACCESSION NUMBERS
The RNA-seq data reported in this paper are archived at GEO under accession number GSE69445.
AUTHOR CONTRIBUTIONS
K.G.L., J.B., M.M., C.I.M., T.M., L.A.B., E.J.V., and S.-Y.K. performed experiments. K.G.L., J.B., M.M., C.I.M., T.M., L.A.B., E.J.V., G.G., S.-Y.K., H.H., L.M., and A.K.B. analyzed data. M.B. provided reagents. K.G.L., J.B., C.I.M., L.A.B., G.G., H.H., M.J.D., F.R., C.R.M., and R.J.X. designed research. K.G.L., J.B., E.J.V., L.M., A.K.B., M.B., M.J.D., F.R., C.R.M., and R.J.X. provided intellectual contributions throughout the project. K.G.L. and R.J.X. wrote the paper.
References
- Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Böck J, Martinez-Naves E, Glickman JN, Tschurtschenthaler M, Hartwig J, Hosomi S, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berod L, Friedrich C, Nandan A, Freitag J, Hagemann S, Harmrolfs K, Sandouk A, Hesse C, Castro CN, Bähre H, et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat Med. 2014;20:1327–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- Breton S, Brown D. Regulation of luminal acidification by the V-ATPase. Physiology (Bethesda) 2013;28:318–329. doi: 10.1152/physiol.00007.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wu B, Xu L, Li H, Xia J, Yin W, Li Z, Shi D, Li S, Lin S, et al. A SNX10/V-ATPase pathway regulates ciliogenesis in vitro and in vivo. Cell Res. 2012;22:333–345. doi: 10.1038/cr.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey EE, Beckel JM, Laties AM, Mitchell CH. Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer’s disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience. 2014;263:111–124. doi: 10.1016/j.neuroscience.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Kimura T, Timmins G, Moseley P, Chauhan S, Mandell M. Immunologic manifestations of autophagy. J Clin Invest. 2015;125:75–84. doi: 10.1172/JCI73945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di A, Brown ME, Deriy LV, Li C, Szeto FL, Chen Y, Huang P, Tong J, Naren AP, Bindokas V, et al. CFTR regulates phagosome acidification in macrophages and alters bactericidal activity. Nat Cell Biol. 2006;8:933–944. doi: 10.1038/ncb1456. [DOI] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends Mol Med. 2012;18:524–533. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM. Beyond indigestion: emerging roles for lysosome-based signaling in human disease. Curr Opin Cell Biol. 2015;35:59–68. doi: 10.1016/j.ceb.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geremia A, Arancibia-Cárcamo CV, Fleming MP, Rust N, Singh B, Mortensen NJ, Travis SP, Powrie F. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med. 2011;208:1127–1133. doi: 10.1084/jem.20101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LC, Dikic I. Autophagy in antimicrobial immunity. Mol Cell. 2014;54:224–233. doi: 10.1016/j.molcel.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Hashemi HF, Goodman JM. The life cycle of lipid droplets. Curr Opin Cell Biol. 2015;33:119–124. doi: 10.1016/j.ceb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzan D, Seward S, Stock H, Zisman S, Gabriel K, Harpaz N, Bauer JJ. Crohn’s-like colitis, enterocolitis and perianal disease in Hermansky-Pudlak syndrome. Colorectal Dis. 2006;8:539–543. doi: 10.1111/j.1463-1318.2006.01046.x. [DOI] [PubMed] [Google Scholar]
- Huang SC, Everts B, Ivanova Y, O’Sullivan D, Nascimento M, Smith AM, Beatty W, Love-Gregory L, Lam WY, O’Neill CM, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol. 2014;15:846–855. doi: 10.1038/ni.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Niederreiter L, Blumberg RS. Genetically determined epithelial dysfunction and its consequences for microflora-host interactions. Cell Mol Life Sci. 2011;68:3643–3649. doi: 10.1007/s00018-011-0827-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino S, Takeuchi M, Park SB, Hirata A, Kitamura N, Kunisawa J, Kiyono H, Iwamoto R, Isobe Y, Arita M, et al. Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc Natl Acad Sci USA. 2013;110:17808–17813. doi: 10.1073/pnas.1312937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut. 2013;62:1505–1510. doi: 10.1136/gutjnl-2012-303954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13:453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyaw H, Zeng Z, Su K, Fan P, Shell BK, Carter KC, Li Y. Cloning, characterization, and mapping of human homolog of mouse T-cell death-associated gene. DNA Cell Biol. 1998;17:493–500. doi: 10.1089/dna.1998.17.493. [DOI] [PubMed] [Google Scholar]
- Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM, Villablanca EJ, Norman JM, Liu TC, Heath RJ, et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci USA. 2014;111:7741–7746. doi: 10.1073/pnas.1407001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman RS, Diehl GE, Victorio DA, Huh JR, Galan C, Miraldi ER, Swaminath A, Bonneau R, Scherl EJ, Littman DR. CX3CR1+ mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J Exp Med. 2014;211:1571–1583. doi: 10.1084/jem.20140678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu PC, Sherman JC, Chen A, Kallenbach NR. Alpha-helix stabilization by natural and unnatural amino acids with alkyl side chains. Proc Natl Acad Sci USA. 1991;88:5317–5320. doi: 10.1073/pnas.88.12.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, Ian McKenzie C, Hijikata A, Wong C, et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun. 2015;6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- Marchiando AM, Ramanan D, Ding Y, Gomez LE, Hubbard-Lucey VM, Maurer K, Wang C, Ziel JW, van Rooijen N, Nuñez G, et al. A deficiency in the autophagy gene Atg16L1 enhances resistance to enteric bacterial infection. Cell Host Microbe. 2013;14:216–224. doi: 10.1016/j.chom.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell M, Bjorkman J, Nguyen T, Sharp P, Finnie J, Paterson C, Tonks I, Paton BC, Kay GF, Crane DI. Pex13 inactivation in the mouse disrupts peroxisome biogenesis and leads to a Zellweger syndrome phenotype. Mol Cell Biol. 2003;23:5947–5957. doi: 10.1128/MCB.23.16.5947-5957.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi C, Tobo M, Tomura H, Murata N, He XD, Sato K, Kimura T, Ishizuka T, Sasaki T, Sato T, et al. Involvement of proton-sensing TDAG8 in extracellular acidification-induced inhibition of proinflammatory cytokine production in peritoneal macrophages. J Immunol. 2009;182:3243–3251. doi: 10.4049/jimmunol.0803466. [DOI] [PubMed] [Google Scholar]
- Mota LJ, Ramsden AE, Liu M, Castle JD, Holden DW. SCAMP3 is a component of the Salmonella-induced tubular network and reveals an interaction between bacterial effectors and post-Golgi trafficking. Cell Microbiol. 2009;11:1236–1253. doi: 10.1111/j.1462-5822.2009.01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, Roose-Girma M, DeVoss J, Diehl L, Graham RR, van Lookeren Campagne M. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014;506:456–462. doi: 10.1038/nature13044. [DOI] [PubMed] [Google Scholar]
- Nell S, Suerbaum S, Josenhans C. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat Rev Microbiol. 2010;8:564–577. doi: 10.1038/nrmicro2403. [DOI] [PubMed] [Google Scholar]
- Nugent SG, Kumar D, Rampton DS, Evans DF. Intestinal luminal pH in inflammatory bowel disease: possible determinants and implications for therapy with aminosalicylates and other drugs. Gut. 2001;48:571–577. doi: 10.1136/gut.48.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Sumpter R, Jr, Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin B, He M, Chen X, Pei D. Sorting nexin 10 induces giant vacuoles in mammalian cells. J Biol Chem. 2006;281:36891–36896. doi: 10.1074/jbc.M608884200. [DOI] [PubMed] [Google Scholar]
- Rahman N, Buck J, Levin LR. pH sensing via bicarbonate-regulated “soluble” adenylyl cyclase (sAC) Front Physiol. 2013;4:343. doi: 10.3389/fphys.2013.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell. 2015;32:678–692. doi: 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis RC, Sorgine MH, Coelho-Sampaio T. A novel methodology for the investigation of intracellular proteolytic processing in intact cells. Eur J Cell Biol. 1998;75:192–197. doi: 10.1016/S0171-9335(98)80061-7. [DOI] [PubMed] [Google Scholar]
- Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell. 2015;58:586–597. doi: 10.1016/j.molcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich KA, Burkett C, Webster P. Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol. 2003;5:455–468. doi: 10.1046/j.1462-5822.2003.00292.x. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, Garrett N, Alpern JA, Akyildiz A, Theiss AL, Nusrat A, Klapproth JM. Invasive Escherichia coli are a feature of Crohn’s disease. Lab Invest. 2007;87:1042–1054. doi: 10.1038/labinvest.3700661. [DOI] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SY, Tran K, Castoreno AB, Peloquin JM, Lassen KG, Khor B, Aldrich LN, Tan PH, Graham DB, Kuballa P, et al. Selective modulation of autophagy, innate immunity, and adaptive immunity by small molecules. ACS Chem Biol. 2013;8:2724–2733. doi: 10.1021/cb400352d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimozawa N, Suzuki Y, Zhang Z, Imamura A, Toyama R, Mukai S, Fujiki Y, Tsukamoto T, Osumi T, Orii T, et al. Nonsense and temperature-sensitive mutations in PEX13 are the cause of complementation group H of peroxisome biogenesis disorders. Hum Mol Genet. 1999;8:1077–1083. doi: 10.1093/hmg/8.6.1077. [DOI] [PubMed] [Google Scholar]
- Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song-Zhao GX, Srinivasan N, Pott J, Baban D, Frankel G, Maloy KJ. Nlrp3 activation in the intestinal epithelium protects against a mucosal pathogen. Mucosal Immunol. 2014;7:763–774. doi: 10.1038/mi.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terawaki S, Camosseto V, Prete F, Wenger T, Papadopoulos A, Rondeau C, Combes A, Rodriguez Rodrigues C, Vu Manh TP, Fallet M, et al. RUN and FYVE domain-containing protein 4 enhances autophagy and lysosome tethering in response to Interleukin-4. J Cell Biol. 2015;210:1133–1152. doi: 10.1083/jcb.201501059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallance BA, Deng W, Knodler LA, Finlay BB. Mice lacking T and B lymphocytes develop transient colitis and crypt hyperplasia yet suffer impaired bacterial clearance during Citrobacter rodentium infection. Infect Immun. 2002;70:2070–2081. doi: 10.1128/IAI.70.4.2070-2081.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, Maher L, Redinbo MR, Phillips RS, et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JQ, Kon J, Mogi C, Tobo M, Damirin A, Sato K, Komachi M, Malchinkhuu E, Murata N, Kimura T, et al. TDAG8 is a proton-sensing and psychosine-sensitive G-protein-coupled receptor. J Biol Chem. 2004;279:45626–45633. doi: 10.1074/jbc.M406966200. [DOI] [PubMed] [Google Scholar]
- York AG, Williams KJ, Argus JP, Zhou QD, Brar G, Vergnes L, Gray EE, Zhen A, Wu NC, Yamada DH, et al. Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell. 2015;163:1716–1729. doi: 10.1016/j.cell.2015.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3