The impact of transposable elements on mammalian development (original) (raw)
. Author manuscript; available in PMC: 2018 Feb 28.
Published in final edited form as: Development. 2016 Nov 15;143(22):4101–4114. doi: 10.1242/dev.132639
Summary
Despite often being classified as selfish or junk DNA, transposable elements (TEs) are a group of abundant genetic sequences that significantly impact on mammalian development and genome regulation. In recent years, our understanding of how pre-existing TEs affect genome architecture, gene regulatory networks and protein function during mammalian embryogenesis has dramatically expanded. In addition, the mobilization of active TEs in selected cell types has been shown to generate genetic variation during development and in fully differentiated tissues. Importantly, the ongoing domestication and evolution of TEs appears to provide a rich source of regulatory elements, functional modules and genetic variation that fuels the evolution of mammalian developmental processes. Here, we review the functional impact that TEs exert on mammalian developmental processes and how the somatic activity of TEs can influence gene regulatory networks.
Keywords: Retrotransposon, endogenous retrovirus, LINE-1, genome regulation, genetic variation
Introduction
Genomes are the habitat in which genes reside, and their complexity is an indication of the number of biological processes that are required during the life of an organism. Comparative genomic studies have revealed that the proportion of the genome occupied by genes decreases as biological complexity increases (Boeke and Devine, 1998). Intriguingly, the opposite is observed for transposable elements (TEs) (Boeke and Devine, 1998), which are pieces of DNA that can move within genomes. Copies of these elements are typically interspersed throughout the genomes of most organisms examined to date. Up to 70% of the human genome is derived from TEs (de Koning et al., 2011), while genes occupy less than 2% (Lander et al., 2001). These data imply that there has been a significant activity of TEs during evolution but that most of the genetic changes caused by TEs are not detrimental. Rather, the high percentage of TE-derived sequences in mammalian genomes could indicate their inherent potential to create and diversify biological processes, as proposed 60 years ago by McClintock, Britten and Davidson (Britten and Davidson, 1969; McClintock, 1956). The proposal that TEs have a present day function in host genomes to provide _cis_-regulatory elements that co-ordinate the expression of groups of genes (Britten and Davidson, 1969) is starting to be tested on a genome-wide scale with the advent of next generation sequencing. Furthermore, recent findings showing that TEs mobilize much more frequently in development than previously anticipated suggest that these sequences may have additional present-day functions in host genomes (recently reviewed in (Hancks and Kazazian, 2016; Munoz-Lopez et al., 2016; Richardson et al., 2015)).
In this Review, we cover the functional impact that TEs exert on gene regulatory networks operating during mammalian embryogenesis but also in somatic adult tissues. We also review some of the recent evidence outlining the myriad of ways that TEs can further increase functional variability in mammalian genomes, which may shed some light on why these elements have become so abundant in mammalian genomes.
Types of TEs in the mammalian genome
There are several classes of TEs (Fig. 1) that vary with regards to their structure, impact and regulation in mammalian genomes (as reviewed in (Hancks and Kazazian, 2016; Munoz-Lopez et al., 2016; Richardson et al., 2015)). These include DNA transposons as well as retrotransposons, which can be further sub-divided into long terminal repeat (LTR) retrotransposons and non-LTR retrotransposons (Hancks and Kazazian, 2016; Munoz-Lopez et al., 2016; Richardson et al., 2015).
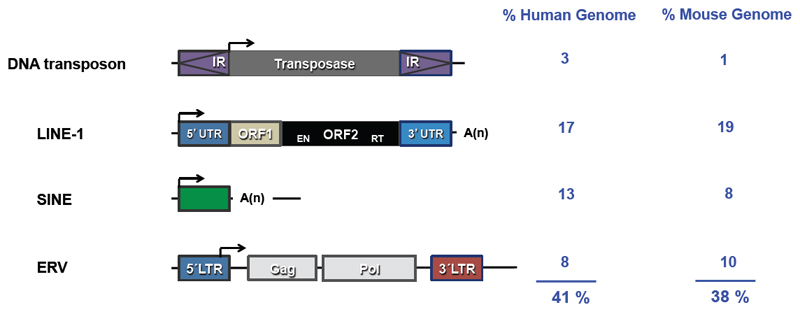
Figure 1. The structures of key classes of TEs found in mammalian genomes.
Different types of TEs (DNA transposons; LINE-1, long interspersed element class 1; SINE, short interspersed element; ERV, endogenous retrovirus) found in mammalian genomes are represented. The percentage of the human and mouse genomes occupied by each TE is indicated in blue. Abbreviations: IR, inverted repeat; UTR, untranslated region; EN, endonuclease; RT, reverse transcriptase; LTR, long terminal repeat; ORF, open reading frame.
Briefly, approximately 3% of a typical mammalian genome is made of DNA transposons (Lander et al., 2001; Waterston et al., 2002). However, with the exception of some bat species, DNA transposons no longer mobilize in mammals (Mitra et al., 2013; Ray et al., 2007). In contrast, LTR retrotransposons and non-LTR retrotransposons comprise more than 40% of a typical mammalian genome and are still active in most mammalian species (Hancks and Kazazian, 2016; Lander et al., 2001; Richardson et al., 2015). LTR retrotransposons are similar to retroviruses in terms of their structure and mechanism of retrotransposition (Fig. 2A) and are hence often called endogenous retroviruses (ERVs; reviewed in (Mager and Stoye, 2015)). Full-length ERVs are flanked by LTRs that promote the transcription and maturation of ERV RNAs, and they also contain functional Gag and Pol genes, which encode structural proteins and enzymes involved in retrotransposition. However, ERVs often lack a functional Env gene, which encodes the envelope protein that retroviruses typically use to exit cells (Lee and Bieniasz, 2007). Furthermore, recombination between LTRs occurs frequently, deleting the intervening internal ERV sequence and generating solo LTRs (Belshaw et al., 2007). The mobilization of active ERVs involves an RNA intermediate and a copy-and-paste mechanism that is similar to the initial steps of retroviral infection (Fig. 2A). ERV mobilization in mice is responsible for nearly 10% of spontaneous mutations in this species; by contrast, ERVs generally no longer mobilise in humans (Mager and Stoye, 2015; Maksakova et al., 2006). However, recent reports have identified polymorphic HERV-K insertions in humans ((Wildschutte et al., 2016), reviewed in (Hohn et al., 2013)) suggesting recent mobilisation activity and the possibility that some HERV-K copies might retain capability to mobilize in present day humans. Hundreds of different types of ERVs are present in a typical mammalian genome, some of which are autonomous and encode the functional retroviral proteins required for their mobilization (Fig. 2A), while others are non-autonomous and rely on retroviral proteins encoded by active ERVs to mobilize (Mager and Stoye, 2015; Maksakova et al., 2006). Notably, two types of ERVs – IAP elements and MusD/ETn - appear to be the most active ERVs in mice and their activity can vary among inbred mouse strains (Maksakova et al., 2006).
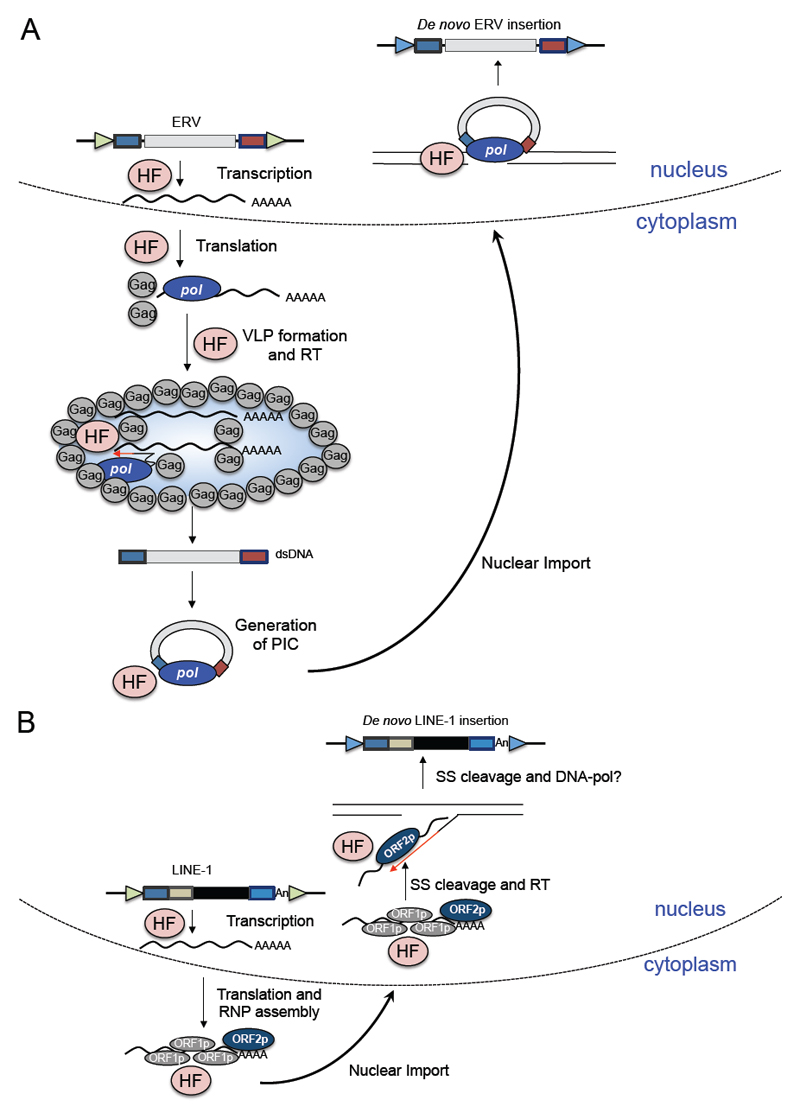
Figure 2. Modes of TE mobilization in mammalian genomes.
(A) The LTR retrotransposition cycle (adapted from (Mager and Stoye, 2015)). The ERV RNA is transcribed from the 5’ LTR and transported to the cytoplasm, where Gag and Pol are translated and processed into mature proteins including protease, integrase and reverse transcriptase (not shown in the figure but see (Mager and Stoye, 2015) for further details). These ERV proteins and RNAs are then packaged into a virus-like particle (VLP; blue opal surrounded by Gag molecules) and reverse-transcribed (RT) by Pol. The resulting ERV dsDNA is then processed by the integrase activity of Pol to generate a pre-integration complex (PIC), which is imported to the nucleus. Here, the integrase activity of Pol inserts the ERV dsDNA into the genome. New ERV insertions are often flanked by target site duplications (blue or green arrowheads). Host factors (HF; pink circles) also participate in several steps of the retrotransposition process. (B) The non-LTR retrotransposition cycle (adapted from (Macia et al., 2015)). The full-length active LINE-1 RNA is transcribed and transported to the cytoplasm where ORF1p and ORF2p are translated (Alisch et al., 2006; Dmitriev et al., 2007). These proteins preferentially bind to their encoding mRNA, generating a ribonucleoprotein particle (RNP), which is imported into the nucleus. The EN activity of ORF2p generates a single strand (SS) break in genomic DNA that is used by the RT activity of ORF2p to generate the first strand cDNA (red arrow). How second strand synthesis occurs is not well understood. New LINE-1 insertions are often flanked by TSDs (blue or green arrowheads) and are also often 5’-truncated (not shown). As is the case for the LTR retrotransposition cycle, host factors (pink circles) are involved in several steps of the non-LTR retrotransposition process.
Mammalian non-LTR retrotransposons are exemplified by long interspersed element class 1 (LINE-1 or L1) retrotransposons, which are the only active autonomous retrotransposons in the human genome (Hancks and Kazazian, 2016; Munoz-Lopez et al., 2016; Richardson et al., 2015). LINE-1 retrotransposons make up 17% of the human genome and, although most LINE-1s are molecular fossils that have lost their ability to move, 80-100 LINE-1 copies retain retrotransposition potential (Beck et al., 2010; Brouha et al., 2003). In mice, LINE-1 elements comprise a similar fraction of the genome to humans (Waterston et al., 2002) but a few thousand LINE-1 elements may retain the capacity to retrotranspose (reviewed in (Richardson et al., 2015)). Mammalian genomes also contain numerous short interspersed element (SINE) non-LTR retrotransposons, exemplified by Alu and SVA (SINE-VNTR-_Alu_s) in the human genome (Lander et al., 2001). SINEs are non-autonomous retrotransposons that use LINE-1 proteins in trans to mobilise (Dewannieux et al., 2003; Dewannieux and Heidmann, 2005; Hancks et al., 2011; Raiz et al., 2011). Non-LTR retrotransposons also move by a copy-and-paste mechanism (Fig. 2B), but one that is fundamentally different from that used by LTR-retrotransposons (Richardson et al., 2015). Notably, active LINE-1 elements code for two protein products termed Open Reading Frame (ORF) 1 and ORF2 that are strictly required for LINE-1 mobilization (Moran et al., 1996). While ORF1 encodes an RNA binding protein with nucleic acid chaperone activity (Hohjoh and Singer, 1997; Martin and Bushman, 2001), ORF2 codes for a protein with endonuclease and reverse transcriptase activity (Feng et al., 1996; Mathias et al., 1991) (Figs. 1 and 2).
The regulation of TE activity in the mammalian genome
The activity of TEs is tightly regulated in mammals to control the number of insertions accumulated in genomes (Hancks and Kazazian, 2016; Munoz-Lopez et al., 2016; Richardson et al., 2015). Mechanisms that restrict TE expression and mobilization are likely to be particularly important in germ cells, as well as in the pluripotent cells in early embryos that act as germ cell precursors, as new TE insertions in these cells can potentially be transmitted to the next generation and increase TE copy number during evolution (Crichton et al., 2014). However, the differential activity of these restriction mechanisms in different cell types can influence the ability of TEs to impact gene regulatory networks. Given that these topic have been extensively reviewed recently (Goodier, 2016; Hancks and Kazazian, 2016; Heras et al., 2014; Munoz-Lopez et al., 2016; Pizarro and Cristofari, 2016; Richardson et al., 2015), below we provide just an overview of some of the main mechanisms used to control TE activity.
Transcriptional repression of TEs
Transcriptional repression is a major mechanism of defense against retrotransposons. In mice, the transcriptional regulation of LINE-1 and ERVs is dynamic during development, and different mechanisms contribute to the repression of these TEs in different cell types. Histone modifications and DNA methylation, for example, both play important roles, although the relative importance of each of these mechanisms may depend on both the TE and the cell type (Crichton et al., 2014; Gerdes et al., 2016; Rowe and Trono, 2011; Schlesinger and Goff, 2015). DNA methylation plays a role in repressing both mouse and human LINE-1 elements, and some mouse ERVs including IAP elements (Bourc'his and Bestor, 2004; Karimi et al., 2011; Walsh et al., 1998). Multiple histone modifications, including methylation at histones H3K4, H3K9, H2A/H4R3 and H3K27 as well as histone acetylation, have also been implicated in TE transcriptional repression (Brunmeir et al., 2010; Di Giacomo et al., 2014; Karimi et al., 2011; Kim et al., 2014; Leeb et al., 2010; Macfarlan et al., 2011; Matsui et al., 2010; Reichmann et al., 2012). One of the major histone modifications used to repress a large number of TEs in pluripotent mouse embryonic stem cells (ESCs) is H3K9me3 (Karimi et al., 2011; Matsui et al., 2010; Rowe et al., 2010). This modification is largely deposited at TE sequences by the histone methyltransferase Setdb1, which is recruited to TEs by Krüppel-associated box-containing zinc-finger proteins (KRAB-ZFPs) and their associated co-repressor KAP1 (Castro-Diaz et al., 2014; Ecco et al., 2016; Karimi et al., 2011; Matsui et al., 2010; Rowe et al., 2010; Wolf and Goff, 2009; Wolf et al., 2015b). KRAB-ZFPs provide the sequence specificity to target repression to TEs, and some KRAB-ZFPs that target specific types of TE have been identified (Castro-Diaz et al., 2014; Ecco et al., 2016; Wolf and Goff, 2009; Wolf et al., 2015a). However, as a reflection of the parasite/host battleground, young and presumably active TEs escape KAP1-mediated silencing as KRAB-ZFPs have not yet evolved to target these sequences (Castro-Diaz et al., 2014; Jacobs et al., 2014). Thus, KAP1 does not control expression of all mammalian TEs and alternate mechanisms exist to control the expression of young and active TEs (Castro-Diaz et al., 2014; Jacobs et al., 2014). Intriguingly, tissue-specific expression of some KRAB-ZFPs may underlie tissue-specific host gene expression in somatic tissues through their effects on TEs (see below) (Ecco et al., 2016).
Co-transcriptional repression of TE expression
In addition to transcriptional repression, splicing has been shown to regulate LINE-1 mobilization in cultured cell lines and somatic tissues by generating non-functional LINE-1 transcripts (Belancio et al., 2006; Perepelitsa-Belancio and Deininger, 2003). In addition, Microprocessor – a complex that naturally processes structured pre-microRNAs (pre-miRNAs) to generate miRNAs - can also process LINE-1 and SINE RNAs, thereby reducing their retrotransposition activity (Heras et al., 2014; Heras et al., 2013).
Post-transcriptional control of TEs
A number of viral restriction factors have also been shown to act post-transcriptionally to regulate the activity of retrotransposons (reviewed in (Goodier, 2016; Pizarro and Cristofari, 2016)). Some factors such as RNaseL (Zhang et al., 2014), SAMHD1 (Zhao et al., 2013), hnRNPL and nucleolin (Peddigari et al., 2013), MOV10 (Goodier et al., 2012), UPF1 (Taylor et al., 2013), Pin1 (Cook et al., 2015) and ZAP (Goodier et al., 2015) has been implicated in post-transcriptionally regulating TE RNAs or post-translationally modifying TE proteins. Others including SAMHD1 (Zhao et al., 2013) and PCNA (Taylor et al., 2013) have been implicated in modulating reverse transcription. Finally, APOBEC proteins (Richardson et al., 2014b; Schumann, 2007) and PCNA (Taylor et al., 2013) have been shown to interfere with later steps of the retrotransposition cycle. We speculate that the pattern of expression of TE-restriction mechanisms may impact human biology, as this will establish a level of TE activity in different cell types. Furthermore, the use of proteomics has resulted in a list of host factors that interact with LINE-1 and may regulate its retrotransposition (Goodier et al., 2013; Moldovan and Moran, 2015; Taylor et al., 2013). However, the role of most of these identified LINE-1 interactors remains to be determined and future studies will help to understand the dynamic interaction between TEs and cellular host factors.
TEs can impact developmental processes via various mechanisms
When expressed, TEs can affect developmental processes either via their gene products, which can influence the behavior of host cells, or through new insertions that cause genetic changes in the host genome (Fig. 3). Not surprisingly, therefore, the dysregulated expression of TEs has been linked with defects in various developmental processes in mice, including aberrant proliferation of male germ cells (Galli et al., 2005), defects in oogenesis (Malki et al., 2014; Su et al., 2012), disruption of homologous chromosome synapsis during meiosis (reviewed in (Crichton et al., 2014; Ollinger et al., 2010)), activation of the unfolded protein response during differentiation of B lymphocytes (Pasquarella et al., 2016), and inappropriate activation of innate immune responses (Herquel et al., 2013; Stetson et al.,2008). On the other hand, new TE insertions into genes can act as insertional mutagens in mammalian genomes and interfere with gene function (Fig. 3, reviewed in (Hancks and Kazazian, 2016; Heras et al., 2014; Munoz-Lopez et al., 2016; Pizarro and Cristofari, 2016; Richardson et al., 2015)). Such insertions can, for example, introduce actively transcribing promoters into genes and cause transcriptional interference. They can also induce premature termination of transcription via the incorporation of TE-derived polyadenylation sites (Perepelitsa-Belancio and Deininger, 2003). In addition, inefficient transcriptional elongation through the AT-rich LINE-1 sequence can modulate gene expression levels (Han et al., 2004). TE insertions can also introduce TE-derived splice acceptor or donor sites that alter splicing, generating non-functional or nonsense transcripts (Belancio et al., 2006) (Fig. 3A), or can be incorporated into mRNAs and introduce frameshifts or premature termination codons (Fig. 3A). However, it should be noted that many of these mechanisms can also potentially confer new properties and functions to a host gene rather than simply inactivate it (Fig. 3B-D). Below, we examine how the present day functions of TEs can affect developmental genes and processes in mammals.
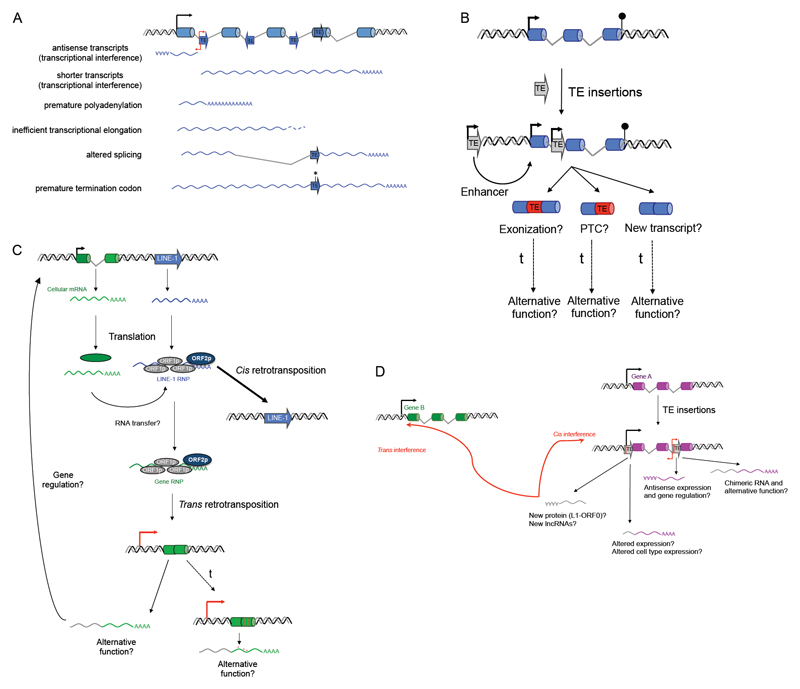
Figure 3. The impact of TEs on mammalian genomes.
(A) Deleterious effects of TE insertions on host gene expression. A cartoon of a host gene containing an upstream promoter (black arrow), exons (blue tubes) and introns (grey lines) is shown. TEs are inserted in sense or antisense orientations (blue arrows). Red arrows on TEs denote promoter sequences. Various RNA transcripts (wavy lines) can be produced, based on the location/orientation of the TE and the promoter used. The left side indicate the type of mechanism that is responsible for the generation of each type of transcript. Asterisk indicates a premature termination codon or frameshift. For full details see main text. (B) De novo TE insertions (grey) can impact genes (blue) in various ways. Shown are examples where a new TE insertion can act as: i) an enhancer; ii) a TE insertion within the gene body can lead to exonization of the TE and this can lead to a new protein product with an alternative function; iii) additionally, exonized TE sequences can induce premature termination of translation (PTC, premature termination codon); and iv) a full-length TE insertion within the gene can generate a shorter gene transcript when the TE promoter is used (new transcript) that could have an alternative function. t, denotes evolutionary time. (C) LINE-1 elements can generate processed pseudogenes. Shown are a cellular gene (green) and an active LINE-1 (blue). Upon transcription (mRNAs, green and blue lines) and translation, the LINE-1 RNP can generate a new insertion by conventional cis retrotransposition. However, the cellular RNA could be occasionally transferred to the LINE-1 RNP, generating a gene RNP that can be inserted into the genome by trans retrotransposition. The potential consequences are indicated below: i) using a nearby promoter (red arrow), a new transcript (grey/green wavy line) can be generated and this can have an alternative function or could also interfere with the regulation of the parental gene; ii) the pseudogene can acquire mutations (red lines and dots in the pseudogene) over evolution and thus acquire a new cellular function. t, denotes evolutionary time. (D) TE insertions can impact gene regulation and chromosome architecture. The cartoon shows the possible outcomes of a gene (A, purple) that accumulated two new TE insertions. The upstream TE, if full-length, could generate antisense transcripts (grey wavy line) that act as long non coding RNAs (New lncRNAs) or that could be translated into a protein product as recently described (New protein (LINE-1 ORF0) (Denli et al., 2015)). Note that these antisense RNAs could also alter chromosome architecture and gene regulation by interfering with: gene A (cis interference) or with a gene located elsewhere in the genome (gene B, green, trans interference). Additionally, the upstream TE can generate new chimeric RNAs (grey/purple wavy line) that can induce deregulation of gene A expression (altered expression) or that could induce expression of gene A in a different cell type (altered cell type expression). Similarly, the TE inserted within the gene could also generate antisense RNAs (purple wavy line) that can interfere with gene A regulation (antisense expression and gene regulation). Finally, the TE inserted within the gene can generate a chimeric RNA using the TE promoter (grey/purple wavy line) that could have an alternative function (chimeric RNA and alternative function).
TEs as promoters that drive the transcription of host genes
TEs contain transcription factor binding sites that promote transcription by RNA polymerase II (in the case of DNA transposons, ERVs, LINE-1s, primate SVAs and even SINEs (Lai et al., 2009)) or RNA polymerase III (in the case of short SINEs such as human Alu, and murine B1 and B2). As such, if a TE integrates into or near host genes, its promoters can drive the expression of novel transcripts that encompass part of the coding region (Fig. 3B-D). The co-option of TE-derived sequences as gene promoters can allow a gene to be expressed in new cell types or contexts (Fig. 3B, D) and can generate truncated or extended protein products, potentially allowing host genes to acquire new functions (Fig. 3B-D). Indeed, LTR sequences frequently act as promoters for host genes (Cohen et al., 2009). One example of a TE-derived promoter generating a cell-type specific isoform of a host gene with novel properties is mouse Dicer1 (Flemr et al., 2013). Dicer1 encodes an RNA endonuclease that generates small regulatory RNAs during oogenesis and in other cell types. However, due to an intragenic insertion of an MT-C ERV retrotransposon, a truncated form of mouse Dicer1 is expressed specifically in oocytes. This truncated ERV-driven form of Dicer1, which is essential for oogenesis, lacks a potentially autoinhibitory helicase domain present in the N-terminal region of the full-length protein and is more enzymatically active than full-length Dicer1 (Flemr et al., 2013). Many genes in mouse oocytes and zygotes are similarly expressed from ERV insertions acting as promoters for nearby genes (Peaston et al., 2004), including Gata4 and Tead4, which encode key transcription factors that drive the specification of primitive endoderm and trophectoderm, respectively (Macfarlan et al., 2012). It will be of interest to investigate if there are additional examples in which co-option of ERV promoters modifies the function of host genes in cells. Interestingly, the expression of selected TE-derived transcripts can be used to track cell types during development and possibly also in adult tissues (see Box 1).
Box 1. Using TEs as markers of cell identity or fate.
The expression of individual TEs can be used to distinguish and/or isolate specific cell types during development. For example, the knowledge that MERVL ERVs are highly expressed in totipotent 2-cell stage mouse zygotes has recently been used to isolate sub-populations of ESCs in culture with totipotent rather than pluripotent developmental potential (Macfarlan et al., 2012). A similar strategy has been used to show that depletion of the chromatin assembly factor CAF1 promotes the generation of 2-cell-like cells from mouse ES cells (Ishiuchi et al., 2015). Similarly, the transcription of select primate-specific ERVs has also been used to isolate populations of naïve-like pluripotent stem cells from human ESCs cultures (Wang et al., 2014). It will be of interest to see whether this approach may facilitate the identification of other transient or low abundance cell populations in developing tissues.
ERVs are also able to drive host gene expression in differentiating somatic tissues. Transcription of Bglap3, which encodes an osteocalcin-related protein, originates from a nearby IAP element in mouse ESCs and in some differentiated somatic tissues (Ecco et al., 2016). The regulation of Bglap3 expression depends on the activity of a KRAB-ZFP that targets KAP1 and H3K9me3 repressive histone modifications to this IAP element, and conditional deletion of either the KRAB-ZFP or KAP1 post-natally in the liver results in transcriptional activation of Bglap3 in a tissue where it is normally repressed (Ecco et al., 2016). In sum, the developmental regulation of KRAB-ZFPs, and potentially other regulators of TE expression, can therefore impact on host gene expression via the regulation of ERVs in embryonic development but also in fully differentiated somatic tissues. Furthermore, some ERVs appear to acquire epigenetic silencing marks early in development that maintain their repression even when KRAB-ZFPs or the KAP1 co-repressor is deleted later in differentiating tissues (Rowe et al., 2013; Rowe et al., 2010; Wolf et al., 2015a).
Variable epigenetic silencing of ERV-derived alternative promoters in somatic tissues can also contribute to the regulation of host genes. This mechanism is exemplified by the mouse Agouti gene. Agouti encodes a signaling molecule that is expressed in hair follicles and inhibits the Mc1r melanocortin receptor in melanocytes (Voisey and van Daal, 2002). During the hair growth cycle, a burst of Agouti expression causes melanocytes to change the type of melanin they produce from black eumelanin to yellow phaeomelanin, resulting in a sub-apical yellow band on an otherwise black hair (Blewitt and Whitelaw, 2013). A naturally occurring mouse mutant exhibiting an insertion of an IAP element immediately 5' and antisense to the first coding exon of Agouti produces mice with yellow coats. A cryptic antisense promoter in this IAP element drives constitutive expression of functional Agouti transcripts in these mutants (Michaud et al., 1994), but variable DNA methylation of this IAP element between individuals means that these genetically identical mice display a continuous spectrum of coat colour phenotypes from yellow to agouti. Offspring arising from yellow mothers, but not yellow fathers, are more likely to have yellow coats suggesting that there can be some trans-generational inheritance of the epigenetic state of this IAP insertion (Morgan et al., 1999). Thus, mechanisms that regulate ERV expression are able to impact on ERV promoter-driven host gene expression and the development of somatic tissues.
SINE and LINE-1 TEs can also act as alternative promoters to drive the expression of host genes (Fig. 3B,D). SINE elements typically contain an internal RNA polymerase III promoter that transcribes these elements, but some also carry an active RNA polymerase II promoter that drives transcription in an anti-sense orientation (Lai et al., 2009). Transcripts originating from the anti-sense RNA polymerase II promoter in these elements can drive the expression of nearby host genes (Ferrigno et al., 2001). Full-length LINE-1s contain conserved sense and an anti-sense promoters (Speek, 2001; Swergold, 1990), and recent studies have shown that the anti-sense promoter in primate LINE-1 drives the expression of a trans-acting polypeptide, ORF0, that can stimulate LINE-1 mobilization in trans (Fig. 3D) (Denli et al., 2015). For some host genes, the majority of their transcripts in induced pluripotent stem cells (iPSCs) originate from a nearby LINE-1 antisense promoter and contain ORF0 peptide sequences that can be spliced on to the host-derived protein (Denli et al., 2015).
The exaptation of TEs as alternative promoters thus appears to be a relatively simple way to alter the pattern or level of expression of host genes during development (Fig. 3). There are also examples of convergent exaptation of TEs as promoters of specific developmental genes. In some mammals, the gene encoding the hormone prolactin is expressed in the pituitary gland but is also expressed from an alternative promoter in the maternal endometrial decidua during pregnancy, regulating immune cells, angiogenesis and invasion of the fetal placenta into maternal tissues (Jabbour and Critchley, 2001). In humans, the transcription of decidual prolactin initiates from an upstream MER39 ERV. In contrast, the transcription of decidual prolactin in mice originates from a MER77 ERV located further upstream, whereas elephants transcribe decidual prolactin from an elephant-specific LINE-1 insertion (Emera et al., 2012; Emera and Wagner, 2012). Thus, the independent evolution of TE-derived prolactin promoters at multiple points in mammalian evolution suggests that TEs provide a rich source of transcription factor binding sites that allows host genes to acquire expression and function in new cell types and tissues (Fig. 3).
TEs as tissue-specific enhancers of host genes
In addition to acting as promoters that drive the expression of alternative isoforms of host genes, transcription factor binding sites within TEs can act as host gene enhancers in specific tissues or developmental contexts (Fig. 3B, D). Indeed, conserved non-exonic TEs in the human genome tend to cluster within 1 Mb of developmental genes and transcriptional regulators, suggesting that this may be a common mechanism for TEs to impact mammalian development (Lowe et al., 2007). Several examples of TEs acting as enhancers have been noted. In trophoblast stem cells, for example, the mouse-specific RLTR13 ERV recruits the trophoblast transcription factors Eomes, Cdx2 and Elf5, and appears to act as a trophoblast enhancer for around 100 host genes (Chuong et al., 2013). ERV-derived enhancers have also been reported in developing primordial germ cells (Liu et al., 2014), and in ESCs (Kunarso et al., 2010). TEs, particularly ERVs, are present in 5-25% of the genomic regions bound by pluripotency-associated transcription factors OCT4 or NANOG in human and mouse ESCs, and in mouse ESCs ERV elements provide binding sites for the pluripotency-associated transcription factor SOX2 (Bourque et al., 2008; Kunarso et al., 2010). Additionally, TEs, especially ERVs, are also able to act as tissue-specific enhancers in the innate immune system (Fig. 3B, D), where they act to mediate part of the interferon response (see Box 2).
Box 2. TEs in the immune system.
TEs can function as tissue-specific enhancers in the innate immune system, acting to mediate the response to interferons, which are pro-inflammatory signalling molecules that are secreted in response to infection, inhibit viral replication in nearby cells and activate immune cells. Specific TEs are enriched close to genes that are activated by interferon (interferon-stimulated genes, ISGs) in human cells (Chuong et al., 2016). One of the most enriched TEs, MER41B ERV, contains interferon-inducible binding sites for the STAT1 transcription factor, which mediates part of the interferon response (Chuong et al., 2016). A MER41B insertion upstream of AIM2, a human ISG, is required for AIM2 expression in response to interferon, and deletion of this element impairs the anti-viral response of human cell lines (Chuong et al., 2016). Interestingly, there is no copy of MER41B upstream of AIM2 in mice, and AIM2 is constitutively expressed rather than interferon-inducible in this species. Notably, RLTR30B ERVs in mice also contain interferon-inducible STAT1 binding sites and are enriched near functionally annotated immunity genes in this species (Chuong et al., 2016). It's possible that the infectious ancestors of these ERVs possessed interferon-inducible STAT1-binding sites in order to exploit the host's innate immune system to promote their own transcription, and that this innovation has now been repeatedly and independently co-opted by mammals in order to drive evolution of the innate immune system (Chuong et al., 2016).
Domesticated TEs also play a role in the generation of antibody repertoires during development of the adaptive immune system (Teng and Schatz, 2015). The breaking and rejoining of DNA molecules that occurs during diversification of antibody genes by V(D)J recombination has some similarities to the cut-and-paste mobilization of DNA transposons (Melek et al., 1998; Teng and Schatz, 2015; van Gent et al., 1996). Indeed, it has been suggested that the RAG1 and RAG2 genes that are required for V(D)J recombination in developing lymphocytes were derived nearly 500 million years ago from a Transib superfamily DNA transposon (Kapitonov and Jurka, 2005). The domestication of TE-derived sequences in this developmental context thus appears to play an important role in increasing genetic diversity in this somatic lineage beyond that encoded by the germline genome.
SINEs are also able to act as enhancers for host genes, particularly during brain development. The gene encoding the LIM homeobox transcription factor ISL1, which is required for motor neuron development, has a nearby conserved exapted LF-SINE TE insertion that can drive gene expression in neural tissues (Bejerano et al., 2006). Similarly, AmnSINE1 TE insertions are associated with genes involved in brain development, such as FGF8, and act as neural-specific enhancers in transgenic mice (Sasaki et al., 2008).
DNA transposons, too, can act as enhancers to influence host gene expression and contribute to gene regulatory networks in development, even though they no longer mobilize in most mammals. The MER130 DNA transposon appears to act as a neocortical enhancer for a number of genes involved in neural development including Robo1 and Id4 (Notwell et al., 2015). Similarly, DNA transposons are strongly represented amongst the large number of TEs that contribute to gene regulatory networks in the mammalian endometrium during pregnancy. The MER121 and MER97C DNA transposons are enriched in regions bound by activated progesterone receptor in human endometrial cells and appear to contribute to progesterone-responses in this tissue (Lynch et al., 2015).
TEs as regulators of chromosome organization
In addition to their more direct roles in regulating host gene expression, TEs can influence the organization of mammalian chromosomes. One of the major regulators of mammalian chromosome organization is CTCF, which can act as an insulator to block the interaction between an enhancer and a promoter, as a barrier to prevent the spreading of chromatin domains, and as an anchor that assembles chromatin into loops or domains within which regulatory elements can interact (reviewed in (Merkenschlager and Nora, 2016)). In combination with cohesin, CTCF plays an important role in allowing developmental enhancers to regulate gene expression, particularly in the context of long-range enhancers (Fig. 3D) (Merkenschlager and Odom, 2013). TEs are strongly enriched within regions of mammalian genomes that bind CTCF, and in mice B2 SINEs are enriched in these regions and carry a CTCF binding motif (Bourque et al., 2008; Schmidt et al., 2012; Sundaram et al., 2014). One example of a context in which a B2 SINE insertion influences developmental gene regulation through effects on chromatin domains occurs during the expression of growth hormone (GH) in the developing pituitary (Lunyak et al., 2007). During the early stages of pituitary development, when GH is not expressed, a repressive chromatin domain extends across the GH locus. However, this domain becomes restricted as pituitary development proceeds. A boundary element located upstream of GH marks the edge of the chromatin domain, and a B2 SINE TE in this region is both necessary and sufficient for the insulating activity that prevents repressive chromatin from extending into the GH domain during late pituitary development (Lunyak et al., 2007). The insulating activity of this SINE TE requires its RNA polymerase II and RNA polymerase III transcripts, suggesting that bidirectional transcription of this element causes a local change in chromatin structure that prevents repressive chromatin from spreading across it (Lunyak et al., 2007; Ponicsan et al., 2010). Notably, additional examples of human and rodent SINEs containing defined transcription factor binding sites have been previously documented (Morales-Hernandez et al., 2016; Roman et al., 2011). Future research will help to define the full-repertoire of effects that TEs can exert on mammalian chromosome organization.
The domestication of TE-derived proteins and processes in development
TEs can also impact on mammalian development through their proteins becoming domesticated, i.e. performing functions for the host organism. The human genome contains around fifty genes that are probably domesticated TEs (Lander et al., 2001). One developing tissue that appears to rely significantly on domesticated TEs is the placenta. Domestication of Gag-Pol regions of _sushi-ishi_-derived ERVs has generated the paternally imprinted Peg10 and Peg11 genes that have essential functions in mouse placenta development. Peg10 plays a role in the development of the trophectoderm-derived spongiotrophoblast and labyrinth layers of the placenta, the latter being the site in which components are exchanged between maternal and fetal circulatory systems (Ono et al., 2006), while Peg11 functions in extraembryonic mesoderm-derived endothelial cells that line the fetal capillaries of the placenta. Domestication of the Env regions of ERVs has also generated genes with essential functions for placenta development. The fetal capillaries in the labyrinth are surrounded by syncytial trophoblast cells that play an important role in allowing the exchange of components of the maternal and foetal circulatory systems. This epithelial layer is formed via inter-cellular fusion between trophoblast cells in a process involving proteins known as syncytins (Dupressoir et al., 2009; Dupressoir et al., 2011). These syncytins are co-opted fusogenic Env proteins derived from ERVs (Blond et al., 2000; Mi et al., 2000). Remarkably, mouse, human and rabbits have all independently co-opted _Env_s from different ERVs to act as syncytins, and independent capture of syncytins has occurred at least six times during mammalian evolution (Dupressoir et al., 2012). Even marsupials, which have a relatively transient placenta that is in contact with the maternal endometrium for a short period of time, may have domesticated a fusogenic placentally-expressed ERV Env protein (Cornelis et al., 2015).
DNA transposons can also undergo domestication, as exemplified by the RAG1 and RAG2 genes that have essential functions during development of the adaptive immune system (see Box 2). Finally, in what may be a form of domestication, recent data suggests that LINE-1 retrotransposons may have a present day function during oogenesis in mice as a quality control mechanism that eliminates defective oocytes, suggesting a function for LINE-1 in regulating the ovarian oocyte pool (Malki et al., 2014). The ovarian oocyte pool gradually declines with age, and the size of the oocyte pool at birth is thought to be a major determinant of reproductive success in older women. It will therefore be of interest to test if LINE-1 plays a similar role in human oogenesis.
Diversification of the mammalian transcriptome by TEs
TEs often insert into introns or untranslated regions of genes, and this can sporadically result in the exonization of TEs (Fig. 3B). Indeed, exonized TEs can expand the mammalian transcriptome and proteome but can also be used to fine tune gene regulation (Piriyapongsa et al., 2007a). The accumulation of TE-derived sequences in cellular mRNAs can lead to their differential regulation due to TE control mechanisms targeting these TE-portions or structures (Heras et al., 2014; Heras et al., 2013). Notably, several classes of TEs have been found inserted within mammalian RNAs (Piriyapongsa et al., 2007a; Piriyapongsa et al., 2007b; Zarnack et al., 2013) and it is likely that their presence may impact gene regulation and function by providing or interfering with regulatory elements in those RNAs (Fig. 3).
Another way that TEs diversify the mammalian transcriptome is by generating long non-coding RNAs (lncRNAs) from their promoters (Fig. 3D). Remarkably, almost 2/3 of all known human lncRNAs contain TE fragments in their sequences (Kapusta et al., 2013; Macia et al., 2011). LncRNAs are more abundant than known genes and are involved in multiple biological processes including gene regulation, maintenance of nuclear architecture and splicing (Macia et al., 2015; Mercer et al., 2009; Moran et al., 2012). The existence of functional lncRNAs was discovered in the early 1990’s, with the characterization of the Xist – a lncRNA involved in X chromosome inactivation ((Brown et al., 1992), reviewed in (Moran et al., 2012)). This X chromosome-encoded lncRNA is thought to interact with LINE-1 elements to bring about X chromosome inactivation in females (Lyon, 2003). LINE-1 elements are over-represented on the X chromosome (Lander et al., 2001), and it has been suggested that evolutionary older silent LINE-1 elements are involved in assembling a Xist-dependent repressive domain on the inactive X chromosome, while evolutionary younger transcribed LINE-1 elements help spread this repressive domain into adjacent chromosomal regions (Chow et al., 2010). The over-representation of LINE-1s on the X chromosome may therefore reflect a present-day function for these sequences in X chromosome inactivation.
Intriguingly, despite a lack of sequence conservation, unexpected roles for multiple human and mouse TE-derived IncRNAs in regulating pluripotency have also recently been described (Fort et al., 2014; Guttman et al., 2011; Lu et al., 2014). Multiple mechanisms may be involved, and some of these TE-derived lncRNAs represent transcripts originating from ERV elements acting as enhancers or promoters in these cells (Fort et al., 2014). However, lncRNAs derived from HERV-H ERVs in human ESCs appear to act in trans via physical association with chromatin modifiers (Guttman et al., 2011; Lu et al., 2014; Wang et al., 2014). Future research will likely reveal how these diverse species-specific TE-derived sequences can regulate conserved developmental processes.
TEs can also influence developmental processes in trans through the LINE-1-dependent generation of processed pseudogenes (Esnault et al., 2000). Although LINE-1 encoded proteins tend to bind to their encoding mRNA in cis (Esnault et al., 2000; Wei et al., 2001) (Fig. 2A), they occasionally bind to cellular host mRNAs in trans and catalyze their insertion into the genome as processed pseudogenes (Fig. 3C). Over evolution, mammalian genomes have accumulated more processed pseudogenes than annotated genes (Zhang et al., 2004). Although most inserted processed pseudogenes lack a functional promoter upon insertion, a promoter can evolve, be captured by a new TE insertion, or be generated by recombination resulting in a new functional gene (Ji et al., 2015) (Fig. 3C). In dogs, the expression of a LINE-1-generated pseudogene derived from the FGF4 gene is strongly associated with a short-legged phenotype selected in dogs (Parker et al., 2009). Thus, the activity of TEs can mediate the duplication and subsequent diversification of key developmental regulators.
Finally, ongoing LINE-1 retrotransposition itself can generate new genes by a mechanism termed exon shuffling (Moran et al., 1999). Exon shuffling occurs when an active LINE-1 within a gene retrotransposes to a new genomic location and delivers nearby coding sequences to the new locus. Indeed, LINE-1-mediated exon shuffling has likely increased the repertoire of the human proteome but, due to frequent 5’ truncation during retrotransposition (Grimaldi et al., 1984; Kazazian et al., 1988), its overall contribution to the human genome remains elusive. Notably, known examples of LINE-1-exon shuffling generated genes have been found in primate species, and include the generation of a new gene product that can restrict HIV infection in new world monkeys (Sayah et al., 2004).
LINE-1 activity in humans
LINE-1 elements, which are the only active autonomous retrotransposons in humans, are thought to mobilize during at least two different developmental contexts: the early embryo and the developing/adult brain (Hancks and Kazazian, 2016; Munoz-Lopez et al., 2016; Richardson et al., 2015). By contrast, the expression and mobilization of TEs in other somatic cells in humans appears to be low.
Retrotransposition in the early embryo
Studies of transgenic mice carrying engineered human LINE-1 retrotransposition reporters suggests that LINE-1 mobilizes in pre-implantation embryos (Kano et al., 2009; Muotri et al., 2005). Some of this LINE-1 mobilization could potentially reflect activity in the trophectoderm, which gives rise to the extra-embryonic component of the placenta (Fig. 4A). Indeed, the placenta has more limited TE restriction mechanisms than other hypomethylated cell types (Reichmann et al., 2013), although LINE-1 mobilization in these cell types has not been directly assessed. However, at least some of the LINE-1 mobilization in pre-implantation embryos occurs in the pluripotent epiblast, which gives rise to all embryonic tissues (Fig. 4A). Notably, such LINE-1 mobilization in pluripotent cells in pre-implantation embryos could result in the new insertion being a mosaic within somatic and germline cells in adults.
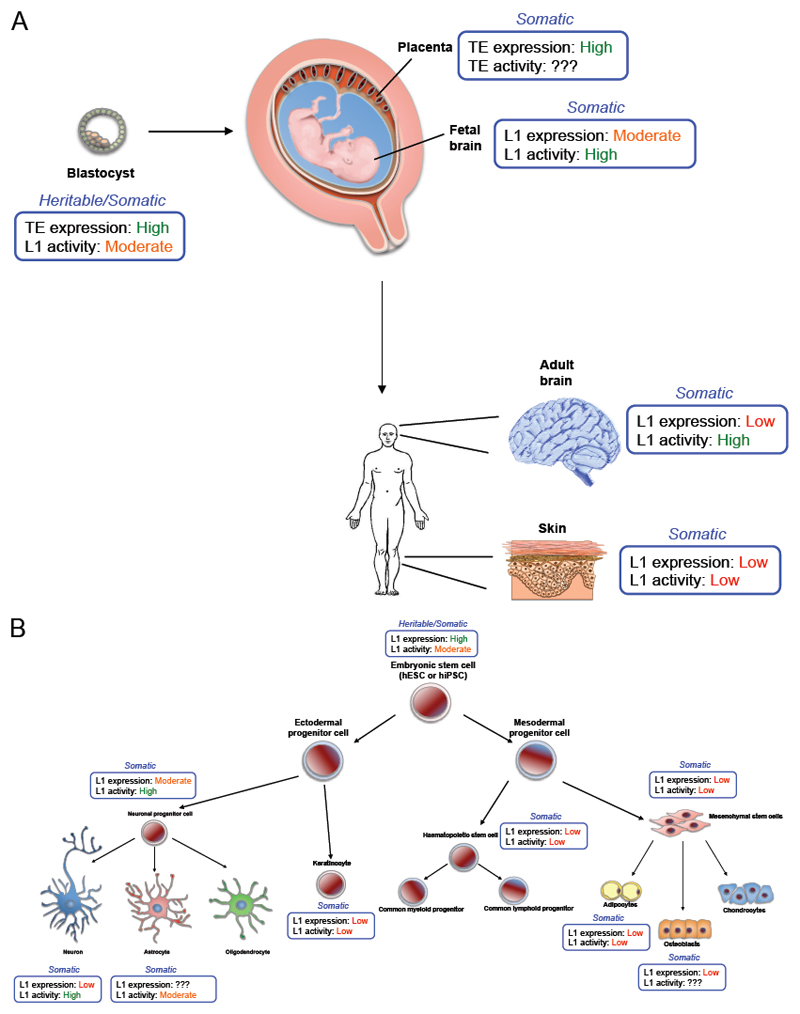
Figure 4. LINE-1 expression and activity in humans: from tissues to cells.
(A) TE and LINE-1 (L1) expression and activity during embryogenesis and in adult humans. Pre-implantation, post-implantation embryos and a scheme of an adult human are shown. (B) LINE-1 (L1) expression and activity in different types of cultured human stem cells are shown. The developmental relationships between these cell types are also shown. In A and B, each blue box indicate the level of TE/L1 expression. On top of each box, it is indicated if new insertions in each tissue/cell type can be transmitted to the next generation (Heritable) or not (Somatic). Data on embryogenesis has been mostly analysed using mouse models, while expression in adult tissues has been analysed using mostly human samples.
LINE-1 is also highly expressed, and new insertions of endogenous LINE-1 and LINE-1 reporters can accumulate, in human pluripotent cell lines that mimic some aspects of embryonic pluripotent cells (Garcia-Perez et al., 2007; Garcia-Perez et al., 2010; Klawitter et al., 2016; Wissing et al., 2011; Wissing et al., 2012). The LINE-1 transcripts expressed in pluripotent cell lines represent a restricted subset of the genomic LINE-1 repertoire, suggesting that the surrounding chromatin environment of a LINE-1 locus might contribute to local activation of these elements in this cell type (Karimi et al., 2011; Macia et al., 2011; Philippe et al., 2016). Notably, the analysis of a new LINE-1 insertion in humans is also consistent with mobilization of this element occurring in pluripotent cells early in embryogenesis (van den Hurk et al., 2007). However, the frequency and specific timing of when retrotransposition takes place in early embryos and during gametogenesis remain to be determined.
Retrotransposition in the brain
Both endogenous and engineered LINE-1s have also been shown to mobilize in the mammalian brain (Fig. 4A) (Muotri et al., 2005). Surprisingly, LINE-1 mRNAs are expressed in neuronal precursor cells (NPCs) in the mammalian brain and new LINE-1 insertions can accumulate in NPCs, at least in mouse models of human LINE-1 retrotransposition and in cultured human NPCs (Fig. 4A) (Coufal et al., 2009; Muotri et al., 2005). Notably, a study that analyzed LINE-1 expression in fetal NPCs and in other somatic cells (skin) isolated from the same donor demonstrated that a subtle change in LINE-1 promoter DNA methylation levels in brain cells might explain why LINE-1 mRNAs are expressed selectively in NPCs when compared to other tissues such as skin (Fig. 4A) (Coufal et al., 2009). However, the availability of transcription factors can also contribute to this phenomenon (Richardson et al., 2014a; Thomas et al., 2012).
More recently, the development of next generation DNA sequencing and single cell genomics-based studies have allowed researchers to demonstrate that the human brain is in fact made of a mosaic of genomes (Baillie et al., 2011; Erwin et al., 2016; Evrony et al., 2012; Evrony et al., 2015; Upton et al., 2015), although there is an ongoing debate about the frequency of retrotransposition in this tissue (Evrony et al., 2016; Richardson et al., 2014a). Furthermore, while retrotransposition has been proposed to be ubiquitous in the hippocampus of the human brain (Upton et al., 2015), we know little about other brain regions or neuronal cell types that may support elevated levels of retrotransposition. Nonetheless, it is clear that the ongoing activity of LINE-1s in the brain could provide a mechanism to ensure that no two human brains - even those of identical twins - are genetically identical.
The functional impact of LINE-1 retrotransposition in the brain is less clear (Richardson et al., 2014a; Singer et al., 2010). Indeed, it is possible that LINE-1 expression and mobilization in the brain could be a consequence of LINE-1 elements having a present day function as enhancers or promoters for host genes in this tissue. However, it is possible that LINE-1 activity in mammalian brains might be on its way to domestication and that, like the domestication of DNA transposons in the immune system, the ability of TEs to increase genetic diversity beyond that encoded by the germline genome is providing some benefit to the development or function of this highly complex organ. Constructing a map of LINE-1 expression and retrotransposition in the different human brain areas and cellular types will help understand both the magnitude and impact of somatic retrotransposition on brain biology.
LINE-1 retrotransposition in the brain is potentially mutagenic, although de novo somatic LINE-1 insertions disrupting brain development or function in patients remains to be demonstrated. Recent studies suggest that the ongoing activity of LINE-1s in the healthy brain can result in the generation of genomic rearrangements that could delete genomic regions proximal to genes, although any functional significance remains to be determined (Erwin et al., 2016). Similarly, it remains to be elucidated whether dysregulated LINE-1 expression or retrotransposition could contribute to brain disorders, although data acquired in models of Rett syndrome, Ataxia Telangiectasia and Schizophrenia suggest that retrotransposition is indeed associated with these syndrome (Bundo et al., 2014; Coufal et al., 2011; Muotri et al., 2010) (reviewed in (Richardson et al., 2014a)).
The expression and mobilization of TEs in other somatic human cells
The description of LINE-1 activity in the human brain raises another important question: are LINE-1 elements expressed and mobilized in other somatic tissues? Although more research is needed, several studies suggest that the somatic activity of TEs might be restricted to the human brain. Notably, in a recent study it was reported that human tissues including esophagus, prostate, stomach and heart muscle express relatively low levels of LINE-1 mRNAs whereas expression in the adrenal gland, kidney, spleen and cervix was below the detection limit (Belancio et al., 2010). Whether part of the expressed LINE-1 mRNAs corresponds to active retrotransposition-competent LINE-1 mRNAs is unknown. More recently, and exploiting the inherent capability of human ESCs to differentiate into somatic stem cells, the expression level of human LINE-1s as well as putative engineered LINE-1 retrotransposition has been explored in a panel of human somatic stem/progenitor cells, including human NPCs, human mesenchymal stem cells (MSCs), hematopoietic stem cells (HSCs) and progenitor keratinocytes (Macia et al., 2016) (Fig. 4B). These data suggest that NPCs are the only analyzed population of somatic stem/progenitor cells in which LINE-1 expression levels are significant (Macia et al., 2016). In addition, efficient retrotransposition was only detected in NPCs and mature neuronal cells (Macia et al., 2016) (Fig. 4B). These data suggest that retrotransposition in human somatic tissues might be restricted to the brain (Baillie et al., 2011; Erwin et al., 2016; Evrony et al., 2012; Evrony et al., 2015; Macia et al., 2016; Upton et al., 2015). However, additional research is required to define if additional LINE-1-dependent TEs are also mobilising in the human brain, the extent of that mobilization, and to fully understand the phenotypic consequences of TE activity in the human brain.
Conclusions
While our understanding of the precise role of TE expression and mobilization during human development still remains limited, recent studies have provided key insights into the regulation of TEs. Indeed, future studies analysing the impact that the ongoing activity of TEs during embryogenesis and in the adult brain exert, in a healthy or disease genetic background, will shed light into the unknown functions of TEs in normal mammalian development and biology. Recent studies suggest that there are multiple ways in which a TE derived sequence, either at the RNA or DNA level, can affect gene regulatory networks and gene function during mammalian development. Thus, these present day functions of TEs may underlie their abundance in mammalian genomes by providing a convenient mechanism for host genes to evolve new expression patterns and isoforms. In addition, independent convergent exaptation of different TEs as developmentally-regulated gene regulatory sequences has occurred in multiple mammalian species during evolution, and TE domestication has been key in generating a functional placenta. In the future, additional genomics and genome-wide epigenetic maps from multiple cell types across multiple species, in combination with CRISPR/Cas9-driven functional genomics, will more clearly define the role of TEs in mammalian development.
Notably, the existence of active TEs in mammals implies that the mammalian body is a mosaic of gene regulatory networks. On one hand, the mobilization of TEs during early embryogenesis results in the generation of mosaic bodies with respect to their TE content. Whether this mosaicism impacts on human homeostasis or predisposition to disease is currently unknown. Future studies exploiting single-cell genomics and animal models of deregulated retrotransposition will clearly help to define the contribution of active TEs to mammalian biology and disease. On the other hand, in humans LINE-1 mobilization is not equally distributed in the body and seems to be restricted to the brain, suggesting that any functional consequences of LINE-1 mobilization are likely impacting on this tissue. The somatic activity of LINE-1 generates genetic variation in the human brain and it is possible that this activity might be undergoing domestication in mammals. However, many questions remain unanswered: can LINE-1s move equally well in all neuronal and glial cell types present in a vertebrate brain? Do all vertebrate species contain active TEs in their brains? And of course, the most important question in current TE biology: what is the role of the somatic activity of TEs in the vertebrate brain?
Acknowledgements and Funding information
T.J.W. is currently funded by PCIN-2014-115-ERA-NET NEURON II and is a former Marie Curie IEF-fellow (FP7-PEOPLE-2011-IEF-300354). Research in J.L.G.P's. lab is supported by CICE-FEDER-P09-CTS-4980, CICE-FEDER-P12-CTS-2256, Plan Nacional de I+D+I 2008-2011 and 2013-2016 (FIS-FEDER-PI11/01489 and FIS-FEDER-PI14/02152), PCIN-2014-115-ERA-NET NEURON II, the European Research Council (ERC-Consolidator ERC-STG-2012-233764), by an International Early Career Scientist grant from the Howard Hughes Medical Institute (IECS-55007420) and by The Wellcome Trust-University of Edinburgh Institutional Strategic Support Fund (ISFF2). Research in I.R.A.'s lab is supported by an MRC Human Genetics Unit intramural programme grant.
Footnotes
Conflict of Interest
Authors declare that there are no conflicts of interest.
References
- Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV. Unconventional translation of mammalian LINE-1 retrotransposons. Genes & development. 2006;20:210–224. doi: 10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:1512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Research. 2010;38:3909–3922. doi: 10.1093/nar/gkq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R, Watson J, Katzourakis A, Howe A, Woolven-Allen J, Burt A, Tristem M. Rate of recombinational deletion among human endogenous retroviruses. Journal of virology. 2007;81:9437–9442. doi: 10.1128/JVI.02216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt M, Whitelaw E. The use of mouse models to study epigenetics. Cold Spring Harbor perspectives in biology. 2013;5:a017939. doi: 10.1101/cshperspect.a017939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. Journal of virology. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Devine SE. Yeast retrotransposons: finding a nice quiet neighborhood. Cell. 1998;93:1087–1089. doi: 10.1016/s0092-8674(00)81450-6. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunmeir R, Lagger S, Simboeck E, Sawicka A, Egger G, Hagelkruys A, Zhang Y, Matthias P, Miller WJ, Seiser C. Epigenetic regulation of a murine retrotransposon by a dual histone modification mark. PLoS genetics. 2010;6:e1000927. doi: 10.1371/journal.pgen.1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundo M, Toyoshima M, Okada Y, Akamatsu W, Ueda J, Nemoto-Miyauchi T, Sunaga F, Toritsuka M, Ikawa D, Kakita A, et al. Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron. 2014;81:306–313. doi: 10.1016/j.neuron.2013.10.053. [DOI] [PubMed] [Google Scholar]
- Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes & development. 2014;28:1397–1409. doi: 10.1101/gad.241661.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Rumi MA, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet. 2013;45:325–329. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Cook PR, Jones CE, Furano AV. Phosphorylation of ORF1p is required for L1 retrotransposition. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:4298–4303. doi: 10.1073/pnas.1416869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G, Vernochet C, Carradec Q, Souquere S, Mulot B, Catzeflis F, Nilsson MA, Menzies BR, Renfree MB, Pierron G, et al. Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E487–496. doi: 10.1073/pnas.1417000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Marchetto MC, Muotri AR, Mu Y, Carson CT, Macia A, Moran JV, Gage FH. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crichton JH, Dunican DS, Maclennan M, Meehan RR, Adams IR. Defending the genome from the enemy within: mechanisms of retrotransposon suppression in the mouse germline. Cellular and molecular life sciences : CMLS. 2014;71:1581–1605. doi: 10.1007/s00018-013-1468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS genetics. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Narvaiza I, Kerman BE, Pena M, Benner C, Marchetto MC, Diedrich JK, Aslanian A, Ma J, Moresco JJ, et al. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell. 2015;163:583–593. doi: 10.1016/j.cell.2015.09.025. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Heidmann T. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J Mol Biol. 2005;349:241–247. doi: 10.1016/j.jmb.2005.03.068. [DOI] [PubMed] [Google Scholar]
- Di Giacomo M, Comazzetto S, Sampath SC, Sampath SC, O'Carroll D. G9a co-suppresses LINE1 elements in spermatogonia. Epigenetics & chromatin. 2014;7:24. doi: 10.1186/1756-8935-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev SE, Andreev DE, Terenin IM, Olovnikov IA, Prassolov VS, Merrick WC, Shatsky IN. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5' untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated. Molecular and cellular biology. 2007;27:4685–4697. doi: 10.1128/MCB.02138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Lavialle C, Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta. 2012;33:663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, Heidmann T. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Vernochet C, Harper F, Guegan J, Dessen P, Pierron G, Heidmann T. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E1164–1173. doi: 10.1073/pnas.1112304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, Imbeault M, Rowe HM, Turelli P, Trono D. Transposable Elements and Their KRAB-ZFP Controllers Regulate Gene Expression in Adult Tissues. Developmental cell. 2016;36:611–623. doi: 10.1016/j.devcel.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emera D, Casola C, Lynch VJ, Wildman DE, Agnew D, Wagner GP. Convergent evolution of endometrial prolactin expression in primates, mice, and elephants through the independent recruitment of transposable elements. Molecular biology and evolution. 2012;29:239–247. doi: 10.1093/molbev/msr189. [DOI] [PubMed] [Google Scholar]
- Emera D, Wagner GP. Transformation of a transposon into a derived prolactin promoter with function during human pregnancy. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11246–11251. doi: 10.1073/pnas.1118566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin JA, Paquola AC, Singer T, Gallina I, Novotny M, Quayle C, Bedrosian TA, Alves FI, Butcher CR, Herdy JR, et al. L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nature neuroscience. 2016 doi: 10.1038/nn.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony GD, Lee E, Mehta BK, Benjamini Y, Johnson RM, Cai X, Yang L, Haseley P, Lehmann HS, Park PJ, et al. Cell lineage analysis in human brain using endogenous retroelements. Neuron. 2015;85:49–59. doi: 10.1016/j.neuron.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony GD, Lee E, Park PJ, Walsh CA. Resolving rates of mutation in the brain using single-neuron genomics. Elife. 2016;5 doi: 10.7554/eLife.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Ferrigno O, Virolle T, Djabari Z, Ortonne JP, White RJ, Aberdam D. Transposable B2 SINE elements can provide mobile RNA polymerase II promoters. Nat Genet. 2001;28:77–81. doi: 10.1038/ng0501-77. [DOI] [PubMed] [Google Scholar]
- Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, Svoboda P. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell. 2013;155:807–816. doi: 10.1016/j.cell.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Fort A, Hashimoto K, Yamada D, Salimullah M, Keya CA, Saxena A, Bonetti A, Voineagu I, Bertin N, Kratz A, et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet. 2014;46:558–566. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- Galli UM, Sauter M, Lecher B, Maurer S, Herbst H, Roemer K, Mueller-Lantzsch N. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene. 2005;24:3223–3228. doi: 10.1038/sj.onc.1208543. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, O'Shea KS, Moran JV. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez JL, Morell M, Scheys JO, Kulpa DA, Morell S, Carter CC, Hammer GD, Collins KL, O'Shea KS, Menendez P, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466:769–773. doi: 10.1038/nature09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes P, Richardson SR, Mager DL, Faulkner GJ. Transposable elements in the mammalian embryo: pioneers surviving through stealth and service. Genome biology. 2016;17:100. doi: 10.1186/s13059-016-0965-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL. Restricting retrotransposons: a review. Mobile DNA. 2016;7:16. doi: 10.1186/s13100-016-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Cheung LE, Kazazian HH., Jr MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS genetics. 2012;8:e1002941. doi: 10.1371/journal.pgen.1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Cheung LE, Kazazian HH., Jr Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res. 2013;41:7401–7419. doi: 10.1093/nar/gkt512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Pereira GC, Cheung LE, Rose RJ, Kazazian HH., Jr The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition. PLoS genetics. 2015;11:e1005252. doi: 10.1371/journal.pgen.1005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi G, Skowronski J, Singer MF. Defining the beginning and end of KpnI family segments. The EMBO journal. 1984;3:1753–1759. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum Mol Genet. 2011;20:3386–3400. doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH., Jr Roles for retrotransposon insertions in human disease. Mobile DNA. 2016;7:9. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras SR, Macias S, Caceres JF, Garcia-Perez JL. Control of mammalian retrotransposons by cellular RNA processing activities. Mobile genetic elements. 2014;4:e28439. doi: 10.4161/mge.28439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras SR, Macias S, Plass M, Fernandez N, Cano D, Eyras E, Garcia-Perez JL, Caceres JF. The Microprocessor controls the activity of mammalian retrotransposons. Nature structural & molecular biology. 2013;20:1173–1181. doi: 10.1038/nsmb.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herquel B, Ouararhni K, Martianov I, Le Gras S, Ye T, Keime C, Lerouge T, Jost B, Cammas F, Losson R, et al. Trim24-repressed VL30 retrotransposons regulate gene expression by producing noncoding RNA. Nature structural & molecular biology. 2013;20:339–346. doi: 10.1038/nsmb.2496. [DOI] [PubMed] [Google Scholar]
- Hohjoh H, Singer MF. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. The EMBO journal. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn O, Hanke K, Bannert N. HERV-K(HML-2), the Best Preserved Family of HERVs: Endogenization, Expression, and Implications in Health and Disease. Frontiers in oncology. 2013;3:246. doi: 10.3389/fonc.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T, Enriquez-Gasca R, Mizutani E, Boskovic A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla ME. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nature structural & molecular biology. 2015;22:662–671. doi: 10.1038/nsmb.3066. [DOI] [PubMed] [Google Scholar]
- Jabbour HN, Critchley HO. Potential roles of decidual prolactin in early pregnancy. Reproduction. 2001;121:197–205. doi: 10.1530/rep.0.1210197. [DOI] [PubMed] [Google Scholar]
- Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516:242–245. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, Kazazian HH. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes & development. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS biology. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS genetics. 2013;9:e1003470. doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell stem cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH, Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- Kim S, Gunesdogan U, Zylicz JJ, Hackett JA, Cougot D, Bao S, Lee C, Dietmann S, Allen GE, Sengupta R, et al. PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Molecular cell. 2014;56:564–579. doi: 10.1016/j.molcel.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klawitter S, Fuchs NV, Upton KR, Munoz-Lopez M, Shukla R, Wang J, Garcia-Canadas M, Lopez-Ruiz C, Gerhardt DJ, Sebe A, et al. Reprogramming triggers endogenous L1 and Alu retrotransposition in human induced pluripotent stem cells. Nature communications. 2016;7:10286. doi: 10.1038/ncomms10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Lai CB, Zhang Y, Rogers SL, Mager DL. Creation of the two isoforms of rodent NKG2D was driven by a B1 retrotransposon insertion. Nucleic Acids Res. 2009;37:3032–3043. doi: 10.1093/nar/gkp174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS pathogens. 2007;3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes & development. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Brind'Amour J, Karimi MM, Shirane K, Bogutz A, Lefebvre L, Sasaki H, Shinkai Y, Lorincz MC. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes & development. 2014;28:2041–2055. doi: 10.1101/gad.244848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8005–8010. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Sachs F, Ramsay L, Jacques PE, Goke J, Bourque G, Ng HH. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nature structural & molecular biology. 2014;21:423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, Hutt K, Roy R, Garcia-Diaz A, Zhu X, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Nnamani MC, Kapusta A, Brayer K, Plaza SL, Mazur EC, Emera D, Sheikh SZ, Grutzner F, Bauersachs S, et al. Ancient transposable elements transformed the uterine regulatory landscape and transcriptome during the evolution of mammalian pregnancy. Cell reports. 2015;10:551–561. doi: 10.1016/j.celrep.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. The Lyon and the LINE hypothesis. Seminars in cell & developmental biology. 2003;14:313–318. doi: 10.1016/j.semcdb.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes & development. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia A, Blanco-Jimenez E, Garcia-Perez JL. Retrotransposons in pluripotent cells: Impact and new roles in cellular plasticity. Biochimica et biophysica acta. 2015;1849:417–426. doi: 10.1016/j.bbagrm.2014.07.007. [DOI] [PubMed] [Google Scholar]
- Macia A, Munoz-Lopez M, Cortes JL, Hastings RK, Morell S, Lucena-Aguilar G, Marchal JA, Badge RM, Garcia-Perez JL. Epigenetic control of retrotransposon expression in human embryonic stem cells. Molecular and cellular biology. 2011;31:300–316. doi: 10.1128/MCB.00561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia A, Widmann TW, Heras SR, Ayllon V, Sanchez L, Benkaddour-Boumzaouad M, Munoz-Lopez M, Amador-Cubero S, Blanco-Jimenez E, Garcia-Castro J, et al. LINE-1 retrotransposition is lineage-dependent in human stem cells. Genome Res. 2016 doi: 10.1101/gr.206805.116. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager DL, Stoye JP. Mammalian Endogenous Retroviruses. Microbiology spectrum. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0009-2014. MDNA3-0009-2014. [DOI] [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS genetics. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malki S, van der Heijden GW, O'Donnell KA, Martin SL, Bortvin A. A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Developmental cell. 2014;29:521–533. doi: 10.1016/j.devcel.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Molecular and cellular biology. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- McClintock B. Controlling elements and the gene. Cold Spring Harbor Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- Melek M, Gellert M, van Gent DC. Rejoining of DNA by the RAG1 and RAG2 proteins. Science. 1998;280:301–303. doi: 10.1126/science.280.5361.301. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature reviews Genetics. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Merkenschlager M, Nora EP. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annual review of genomics and human genetics. 2016 doi: 10.1146/annurev-genom-083115-022339. [DOI] [PubMed] [Google Scholar]
- Merkenschlager M, Odom DT. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- Michaud EJ, van Vugt MJ, Bultman SJ, Sweet HO, Davisson MT, Woychik RP. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes & development. 1994;8:1463–1472. doi: 10.1101/gad.8.12.1463. [DOI] [PubMed] [Google Scholar]
- Mitra R, Li X, Kapusta A, Mayhew D, Mitra RD, Feschotte C, Craig NL. Functional characterization of piggyBat from the bat Myotis lucifugus unveils an active mammalian DNA transposon. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:234–239. doi: 10.1073/pnas.1217548110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan JB, Moran JV. The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition. PLoS genetics. 2015;11:e1005121. doi: 10.1371/journal.pgen.1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Hernandez A, Gonzalez-Rico FJ, Roman AC, Rico-Leo E, Alvarez-Barrientos A, Sanchez L, Macia A, Heras SR, Garcia-Perez JL, Merino JM, et al. Alu retrotransposons promote differentiation of human carcinoma cells through the aryl hydrocarbon receptor. Nucleic Acids Res. 2016;44:4665–4683. doi: 10.1093/nar/gkw095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Munoz-Lopez M, Vilar-Astasio R, Tristan-Ramos P, Lopez-Ruiz C, Garcia-Perez JL. Study of Transposable Elements and Their Genomic Impact. Methods in molecular biology. 2016;1400:1–19. doi: 10.1007/978-1-4939-3372-3_1. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notwell JH, Chung T, Heavner W, Bejerano G. A family of transposable elements co-opted into developmental enhancers in the mouse neocortex. Nature communications. 2015;6:6644. doi: 10.1038/ncomms7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollinger R, Reichmann J, Adams IR. Meiosis and retrotransposon silencing during germ cell development in mice. Differentiation; research in biological diversity. 2010;79:147–158. doi: 10.1016/j.diff.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, Hino T, Suzuki-Migishima R, Ogonuki N, Miki H, et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet. 2006;38:101–106. doi: 10.1038/ng1699. [DOI] [PubMed] [Google Scholar]
- Parker HG, VonHoldt BM, Quignon P, Margulies EH, Shao S, Mosher DS, Spady TC, Elkahloun A, Cargill M, Jones PG, et al. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;325:995–998. doi: 10.1126/science.1173275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquarella A, Ebert A, Pereira de Almeida G, Hinterberger M, Kazerani M, Nuber A, Ellwart J, Klein L, Busslinger M, Schotta G. Retrotransposon derepression leads to activation of the unfolded protein response and apoptosis in pro-B cells. Development. 2016;143:1788–1799. doi: 10.1242/dev.130203. [DOI] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Developmental cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Peddigari S, Li PW, Rabe JL, Martin SL. hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition. Nucleic Acids Res. 2013;41:575–585. doi: 10.1093/nar/gks1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- Philippe C, Vargas-Landin DB, Doucet AJ, van Essen D, Vera-Otarola J, Kuciak M, Corbin A, Nigumann P, Cristofari G. Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci. Elife. 2016;5 doi: 10.7554/eLife.13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J, Polavarapu N, Borodovsky M, McDonald J. Exonization of the LTR transposable elements in human genome. BMC genomics. 2007a;8:291. doi: 10.1186/1471-2164-8-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J, Rutledge MT, Patel S, Borodovsky M, Jordan IK. Evaluating the protein coding potential of exonized transposable element sequences. Biology direct. 2007b;2:31. doi: 10.1186/1745-6150-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro JG, Cristofari G. Post-Transcriptional Control of LINE-1 Retrotransposition by Cellular Host Factors in Somatic Cells. Frontiers in cell and developmental biology. 2016;4:14. doi: 10.3389/fcell.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponicsan SL, Kugel JF, Goodrich JA. Genomic gems: SINE RNAs regulate mRNA production. Current opinion in genetics & development. 2010;20:149–155. doi: 10.1016/j.gde.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiz J, Damert A, Chira S, Held U, Klawitter S, Hamdorf M, Lower J, Stratling WH, Lower R, Schumann GG. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr863. in pres. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, Pagan HJ, Thompson ML, Stevens RD. Bats with hATs: evidence for recent DNA transposon activity in genus Myotis. Molecular biology and evolution. 2007;24:632–639. doi: 10.1093/molbev/msl192. [DOI] [PubMed] [Google Scholar]
- Reichmann J, Crichton JH, Madej MJ, Taggart M, Gautier P, Garcia-Perez JL, Meehan RR, Adams IR. Microarray analysis of LTR retrotransposon silencing identifies Hdac1 as a regulator of retrotransposon expression in mouse embryonic stem cells. PLoS computational biology. 2012;8:e1002486. doi: 10.1371/journal.pcbi.1002486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann J, Reddington JP, Best D, Read D, Ollinger R, Meehan RR, Adams IR. The genome-defence gene Tex19.1 suppresses LINE-1 retrotransposons in the placenta and prevents intra-uterine growth retardation in mice. Hum Mol Genet. 2013;22:1791–1806. doi: 10.1093/hmg/ddt029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SR, Doucet AJ, Kopera HC, Moldovan JB, Garcia-Perez JL, Moran JV. The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes. Microbiology spectrum. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0061-2014. MDNA3-0061-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SR, Morell S, Faulkner GJ. L1 retrotransposons and somatic mosaicism in the brain. Annual review of genetics. 2014a;48:1–27. doi: 10.1146/annurev-genet-120213-092412. [DOI] [PubMed] [Google Scholar]
- Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. eLife. 2014b:3. doi: 10.7554/eLife.02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman AC, Gonzalez-Rico FJ, Fernandez-Salguero PM. B1-SINE retrotransposons: Establishing genomic insulatory networks. Mobile genetic elements. 2011;1:66–70. doi: 10.4161/mge.1.1.15455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development. 2013;140:519–529. doi: 10.1242/dev.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Trono D. Dynamic control of endogenous retroviruses during development. Virology. 2011;411:273–287. doi: 10.1016/j.virol.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Nishihara H, Hirakawa M, Fujimura K, Tanaka M, Kokubo N, Kimura-Yoshida C, Matsuo I, Sumiyama K, Saitou N, et al. Possible involvement of SINEs in mammalian-specific brain formation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4220–4225. doi: 10.1073/pnas.0709398105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- Schlesinger S, Goff SP. Retroviral transcriptional regulation and embryonic stem cells: war and peace. Molecular and cellular biology. 2015;35:770–777. doi: 10.1128/MCB.01293-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Goncalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148:335–348. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann GG. APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem Soc Trans. 2007;35:637–642. doi: 10.1042/BST0350637. [DOI] [PubMed] [Google Scholar]
- Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Molecular and cellular biology. 2001;21:1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Sun F, Pendola JK, Cox GA, Handel MA, Schimenti JC, Eppig JJ. MARF1 regulates essential oogenic processes in mice. Science. 2012;335:1496–1499. doi: 10.1126/science.1214680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014;24:1963–1976. doi: 10.1101/gr.168872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Molecular and cellular biology. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MS, Lacava J, Mita P, Molloy KR, Huang CR, Li D, Adney EM, Jiang H, Burns KH, Chait BT, et al. Affinity Proteomics Reveals Human Host Factors Implicated in Discrete Stages of LINE-1 Retrotransposition. Cell. 2013;155:1034–1048. doi: 10.1016/j.cell.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng G, Schatz DG. Regulation and Evolution of the RAG Recombinase. Advances in immunology. 2015;128:1–39. doi: 10.1016/bs.ai.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Thomas CA, Paquola AC, Muotri AR. LINE-1 retrotransposition in the nervous system. Annual review of cell and developmental biology. 2012;28:555–573. doi: 10.1146/annurev-cellbio-101011-155822. [DOI] [PubMed] [Google Scholar]
- Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sanchez-Luque FJ, Bodea GO, Ewing AD, Salvador-Palomeque C, van der Knaap MS, Brennan PM, et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell. 2015;161:228–239. doi: 10.1016/j.cell.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hurk JA, Meij IC, Seleme MC, Kano H, Nikopoulos K, Hoefsloot LH, Sistermans EA, de Wijs IJ, Mukhopadhyay A, Plomp AS, et al. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16:1587–1592. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- van Gent DC, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- Voisey J, van Daal A. Agouti: from mouse to man, from skin to fat. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2002;15:10–18. doi: 10.1034/j.1600-0749.2002.00039.x. [DOI] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Wang J, Xie G, Singh M, Ghanbarian AT, Rasko T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516:405–409. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Molecular and cellular biology. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildschutte JH, Williams ZH, Montesion M, Subramanian RP, Kidd JM, Coffin JM. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E2326–2334. doi: 10.1073/pnas.1602336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. The Journal of biological chemistry. 2011;286:36427–36437. doi: 10.1074/jbc.M111.251058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S, Munoz-Lopez M, Macia A, Yang Z, Montano M, Collins W, Garcia-Perez JL, Moran JV, Greene WC. Reprogramming Somatic Cells into iPS Cells Activates LINE-1 Retroelement Mobility. Hum Mol Genet. 2012;21:208–218. doi: 10.1093/hmg/ddr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Greenberg D, Macfarlan TS. Spotting the enemy within: Targeted silencing of foreign DNA in mammalian genomes by the Kruppel-associated box zinc finger protein family. Mobile DNA. 2015a;6:17. doi: 10.1186/s13100-015-0050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Yang P, Fuchtbauer AC, Fuchtbauer EM, Silva AM, Park C, Wu W, Nielsen AL, Pedersen FS, Macfarlan TS. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes & development. 2015b;29:538–554. doi: 10.1101/gad.252767.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnack K, Konig J, Tajnik M, Martincorena I, Eustermann S, Stevant I, Reyes A, Anders S, Luscombe NM, Ule J. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Dong B, Doucet AJ, Moldovan JB, Moran JV, Silverman RH. RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic Acids Res. 2014;42:3803–3820. doi: 10.1093/nar/gkt1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Carriero N, Gerstein M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet. 2004;20:62–67. doi: 10.1016/j.tig.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Zhao K, Du J, Han X, Goodier JL, Li P, Zhou X, Wei W, Evans SL, Li L, Zhang W, et al. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell reports. 2013;4:1108–1115. doi: 10.1016/j.celrep.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]