Protrudin-mediated ER–endosome contact sites promote MT1-MMP exocytosis and cell invasion (original) (raw)
Cancer cell dissemination is facilitated by small actin-rich plasma membrane protrusions called invadopodia. Pedersen et al. now show that invadopodia maturation and function depend on contact site formation between the endoplasmic reticulum and late endosomes, which promotes translocation of the latter to growing invadopodia.
Abstract
Cancer cells break tissue barriers by use of small actin-rich membrane protrusions called invadopodia. Complete invadopodia maturation depends on protrusion outgrowth and the targeted delivery of the matrix metalloproteinase MT1-MMP via endosomal transport by mechanisms that are not known. Here, we show that the ER protein Protrudin orchestrates invadopodia maturation and function. Protrudin formed contact sites with MT1-MMP–positive endosomes that contained the RAB7-binding Kinesin-1 adaptor FYCO1, and depletion of RAB7, FYCO1, or Protrudin inhibited MT1-MMP–dependent extracellular matrix degradation and cancer cell invasion by preventing anterograde translocation and exocytosis of MT1-MMP. Moreover, when endosome translocation or exocytosis was inhibited by depletion of Protrudin or Synaptotagmin VII, respectively, invadopodia were unable to expand and elongate. Conversely, when Protrudin was overexpressed, noncancerous cells developed prominent invadopodia-like protrusions and showed increased matrix degradation and invasion. Thus, Protrudin-mediated ER–endosome contact sites promote cell invasion by facilitating translocation of MT1-MMP–laden endosomes to the plasma membrane, enabling both invadopodia outgrowth and MT1-MMP exocytosis.
Introduction
Tumor cells can change phenotype over time and activate cellular pathways that make them able to breach basement membranes and migrate into the underlying mesenchymal tissue. This behavior leads to the development of cancer, and the escaping cells can eventually metastasize to distant organs (Chambers et al., 2002; Rowe and Weiss, 2008; Paterson and Courtneidge, 2018). One important characteristic of disseminating cancer cells is that they develop cellular protrusions called invadopodia. Invadopodia are actin-rich plasma membrane protrusions, which secrete matrix metalloproteinases (MMPs) to degrade the ECM. Whereas cancer cells use invadopodia for dissemination, invadopodia-like structures called podosomes are found in a variety of normal cells. Podosomes are used for attachment and invasion in tissue development, and in the immune system. The formation of invadopodia and podosomes largely depends on the same molecular machinery, but podosomes are thought to be more transitory and less protrusive than invadopodia (Eddy et al., 2017; Paterson and Courtneidge, 2018; Murphy and Courtneidge, 2011; Jacob and Prekeris, 2015; Castro-Castro et al., 2016 and references therein).
Growth factor signaling initiates the assembly of invadopodia precursors like actin, cortactin, and the Src substrate and scaffold protein Tyrosine kinase substrate with five SH3 domains (TKS5). This typically happens close to focal adhesion sites, where integrins or other cell-matrix adhesion receptors connect the cell to the ECM. In addition to growth factor signaling, degradation products of the ECM as well as substrate rigidity can stimulate the formation of invadopodia (Di Martino et al., 2016; Beaty and Condeelis, 2014; Parekh and Weaver, 2016; Eddy et al., 2017; Siqueira et al., 2016; Seals et al., 2005). Precursor stabilization allows invadopodia maturation, which occurs through a two-pronged mechanism. On one hand, actin polymerization and cortactin-dependent branching allow the invadopodium to expand and elongate. On the other hand, MMP-containing vesicles fuse with the invadopodial plasma membrane, leading to ECM degradation. Interestingly, both steps of invadopodia maturation depend on membrane plasticity and vesicle transport. Whereas lysosomes have been suggested to contribute membrane for invadopodium expansion (Naegeli et al., 2017), late endosomes and lysosomes (hereafter collectively called LE/Lys) have an established role in the delivery of the transmembrane MMP MT1-MMP (also known as MMP14) to the invadopodial plasma membrane (Castro-Castro et al., 2016).
The local high concentration of MT1-MMP in the invadopodial plasma membrane is thought to be important for its potency in ECM remodeling. The internalization of MT1-MMP into endosomes is a central mechanism in this respect, since recycling from endosomal pools can ensure efficient and targeted delivery of MT1-MMP to invadopodia (Castro-Castro et al., 2016). A high concentration of MT1-MMP in invadopodia can be maintained by the anchoring of MT1-MMP to the actin/cortactin invadopodial core (Yu et al., 2012). Furthermore, dystroglycan and matrix adhesion proteins can form barriers at the base of invasive protrusions, which could inhibit the lateral diffusion of MT1-MMP (Naegeli et al., 2017; Branch et al., 2012). To increase its potency even further, MT1-MMP is released to the ECM via exosomes, which derive from the fusion of late multivesicular endosomes with the plasma membrane (Hoshino et al., 2013). Both early and late endosomes are implicated in the endocytic circuit of MT1-MMP (Frittoli et al., 2014; Sneeggen et al., 2019; Castro-Castro et al., 2016). However, LE/Lys are particularly important for the targeting of MT1-MMP to invadopodia (Chevalier et al., 2016; Hoshino et al., 2013; Macpherson et al., 2014; Monteiro et al., 2013; Rossé et al., 2014; Steffen et al., 2008; Williams and Coppolino, 2011; Yu et al., 2012), and LE/Lys accumulate at the invadopodia base (Monteiro et al., 2013). It is not known how LE/Lys are guided to the forming protrusions, but studies in Caenorhabditis elegans have shown that local signaling through netrin receptors can cause polarization of lysosomes at the site of invasive protrusion formation (Naegeli et al., 2017; Hagedorn et al., 2013).
Despite the growing evidence that endocytic recycling of LE/Lys is important for invadopodia maturation and function, less is known about the cellular pathways that contribute to the delivery of LE/Lys to the base of forming invadopodia. This likely involves microtubule based transport, since the plus-end microtubule motors kinesin-1 and kinesin-2 have been implicated in MT1-MMP translocation and invadopodia function, and drugs affecting microtubule stability inhibit invadopodia elongation (Schoumacher et al., 2010; Kikuchi and Takahashi, 2008; Marchesin et al., 2015). In addition, endosomal WASH- and JIP4-dependent tubulation, as well as endosomal cortactin and coronin, facilitate MT1-MMP transport to invadopodia (Marchesin et al., 2015; Castagnino et al., 2018).
To be able to target invadopodia in cancer, it is of great importance to identify the underlying cellular pathways leading to their formation and function, such as LE/Lys translocation. We have previously identified a molecular pathway for anterograde LE/Lys translocation and neurite outgrowth, which depends on the transmembrane ER protein Protrudin (hereafter called the Protrudin pathway; Raiborg et al., 2015). Protrudin forms ER-LE/Lys contact sites by interacting with phosphatidylinositol 3-phosphate (PtdIns3P) and RAB7 in the endosomal membrane. Docking of the LE/Lys to Protrudin in the ER enables the transfer of kinesin-1 from Protrudin to the RAB7-binding endosomal kinesin-1 adaptor FYCO1. The LE/Lys, loaded with kinesin-1, are then released from the Protrudin-ER docking site, allowing their translocation along microtubules toward the cell periphery. Synaptotagmin VII (SYT7)–dependent fusion of the LE/Lys with the plasma membrane facilitates protrusion formation and neurite outgrowth. Since the proteins involved in the Protrudin pathway show a wide tissue expression, we hypothesized that this pathway might function in different cell types and processes beyond neurite outgrowth.
Given the apparent similarities between invadopodia maturation and neurite outgrowth in the requirement for anterograde LE/Lys translocation and fusion with the plasma membrane, we set out to investigate whether the Protrudin pathway is involved in invadopodia maturation and MT1-MMP trafficking. We show here that the Protrudin pathway facilitates LE/Lys translocation to invadopodia and that this is important for invadopodia growth and MT1-MMP exocytosis, leading to increased ECM degradation and invasive migration in 3D cell culture models.
Results
FYCO1-positive LEs localize in close apposition to invadopodia
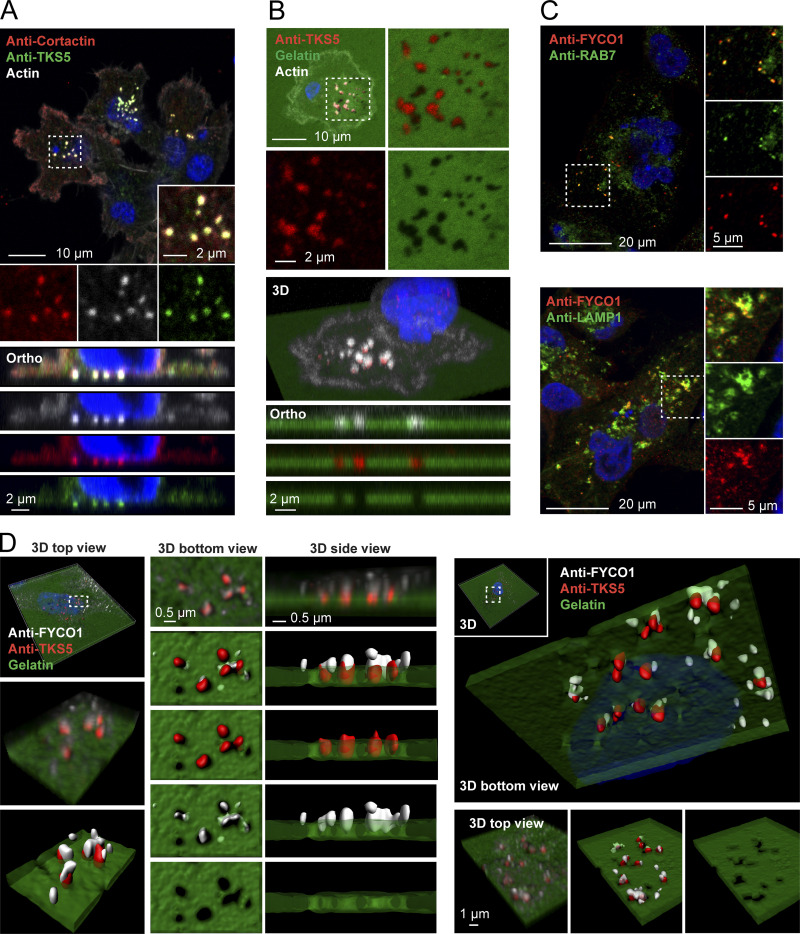
To investigate whether Protrudin-mediated endosome translocation plays a role in invadopodia formation and function, we chose to study the highly invasive breast cancer cell line MDA-MB-231, which is known to form robust invadopodia (Wen-Tien Chen et al., 1994). Indeed, we could detect prominent invadopodia at the ventral side of the cells by examining the endogenous invadopodia markers filamentous actin, cortactin, and TKS5 in MDA-MB-231 cells grown on glass coverslips with or without gelatin coating (Fig. 1, A and B). As it is generally believed that invadopodia form in contact with a matrix substrate, the presence of prominent invadopodia in cells grown on glass was surprising. One explanation for this observation could be that MDA-MB-231 cells likely produce their own ECM.
Figure 1.
FYCO1-positive LE/Lys localize to invadopodia. (A) MDA-MB-231 cells were grown on coverslips, stained with antibodies against TKS5 and cortactin, and analyzed by confocal microscopy. Phalloidin/Alexa Fluor 647 was used to detect F-actin. A section from a confocal z-stack shows invadopodia. Orthographic sections show invadopodia on the ventral side of the cell. (B) MDA-MB-231 cells were grown on coverslips coated with Oregon Green gelatin for 4 h, stained with anti-TKS5 and phalloidin/Alexa Fluor 647 (actin) to visualize invadopodia, and analyzed by confocal microscopy. A section from a confocal z-stack shows invadopodia correlating with degraded gelatin (black areas). 3D view and orthographic sections of the same cell are shown. (C) Colocalization of FYCO1 with LE/Lys markers in MDA-MB-231 cells. Cells were grown on coverslips; stained with antibodies against FYCO1, RAB7, or LAMP1; and analyzed by confocal microscopy. (D) MDA-MB-231 cells were grown on coverslips coated with Oregon Green gelatin for 4 h, stained with antibodies against TKS5 and FYCO1, and analyzed by superresolution microscopy (Airyscan). Z-stacks and Imaris surface 3D renderings from two independent cells show FYCO1-positive LE/Lys in close apposition to TKS5-positive invadopodia correlating with degraded gelatin. Data are representative of at least 16 captures.
FYCO1 localized to RAB7- and LAMP1-positive LE/Lys in MDA-MB-231 cells, as previously found in other cell types (Chen et al., 1994; Raiborg et al., 2015; Fig. 1 C). Intriguingly, we could observe FYCO1-positive endosomes in close apposition to TKS5-positive invadopodia, and some vesicles even localized within the invadopodia (Fig. 1 D). This is consistent with a possible role of Protrudin-mediated LE/Lys translocation to invadopodia.
Invadopodia outgrowth depends on Protrudin and SYT7
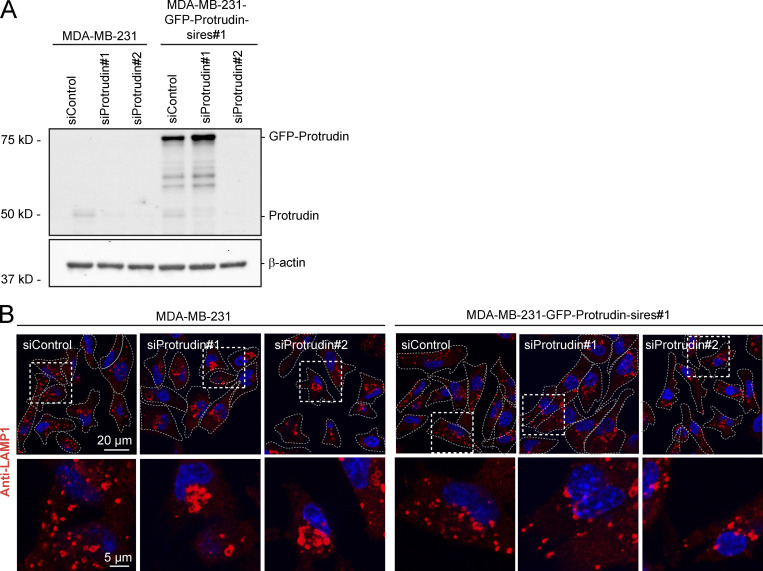
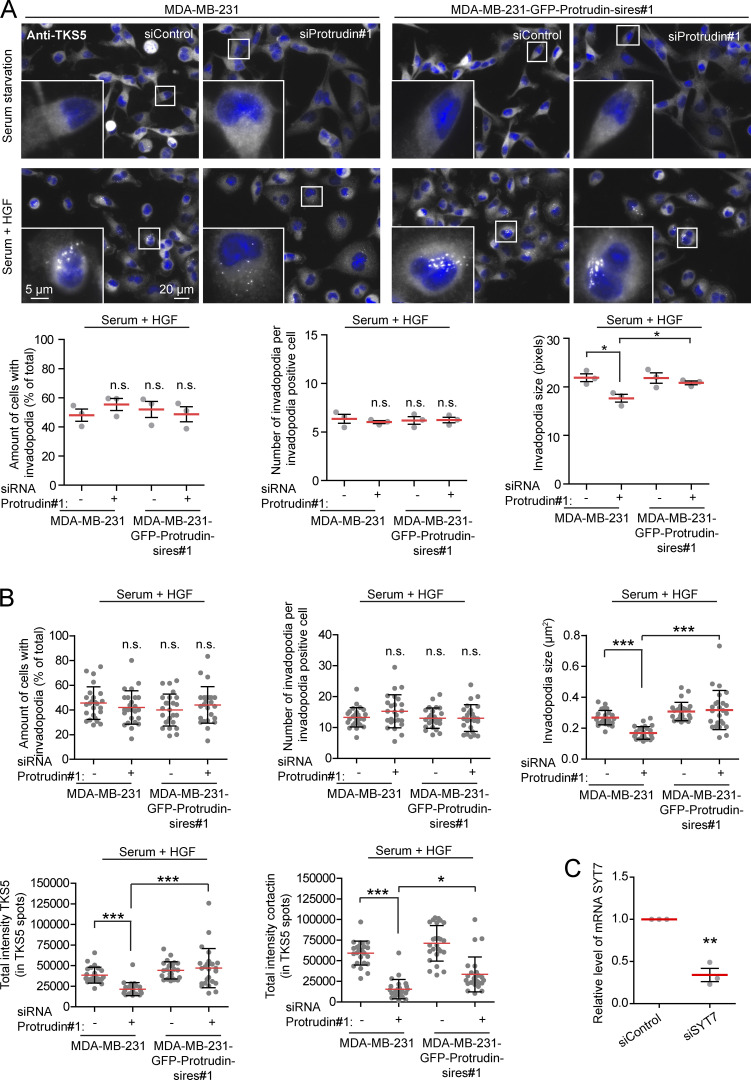
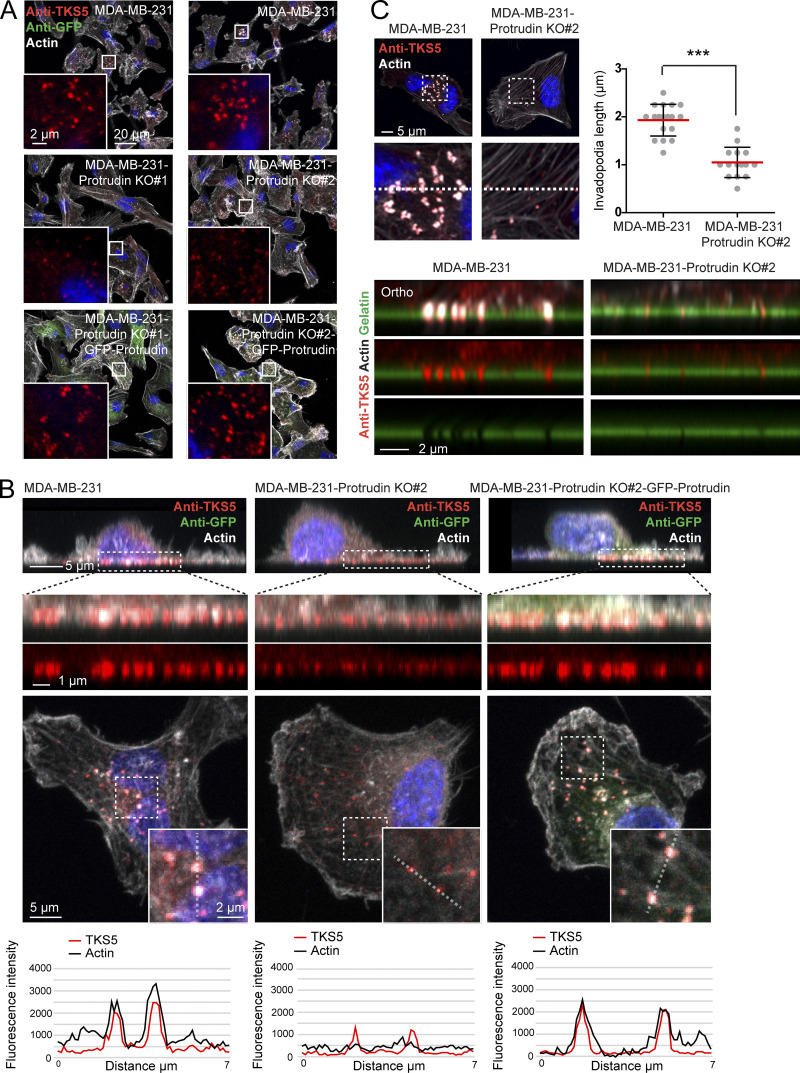
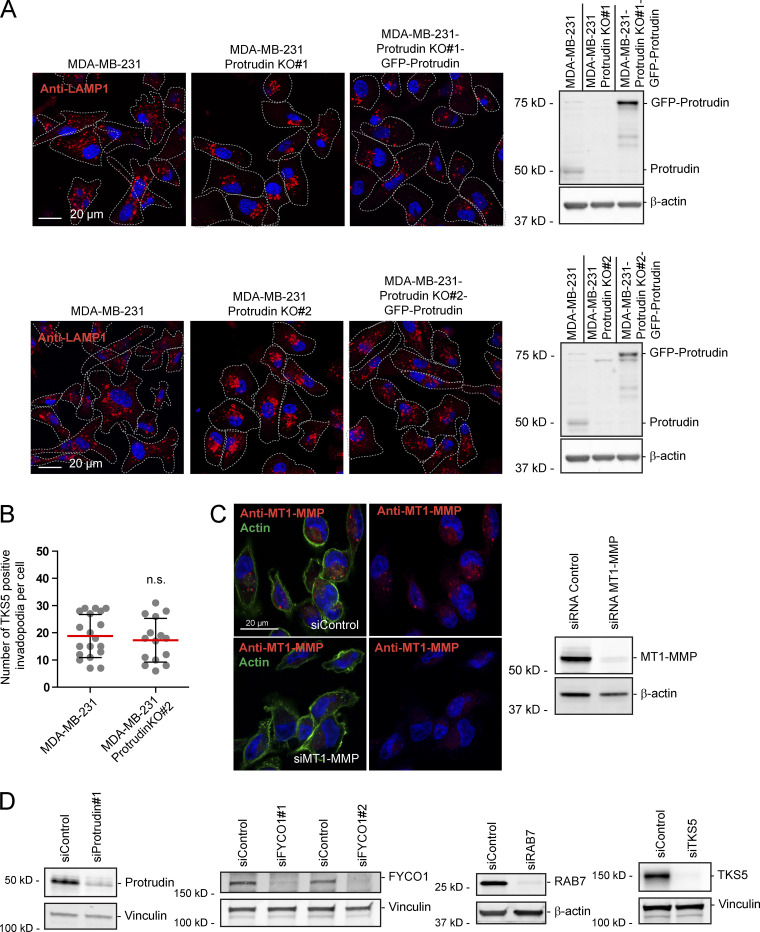
Since neurite outgrowth depends on LE/Lys translocation to the tip of the growth cone (Raiborg et al., 2016), we asked whether invadopodia formation would be affected in cells where LE/Lys translocation to the cell periphery is inhibited. To this end, we depleted MDA-MB-231 cells for Protrudin by siRNA transfection. As expected, in Protrudin-depleted cells, LAMP1-positive LE/Lys clustered perinuclearly, a phenotype that was rescued by stable expression of siRNA-resistant GFP-Protrudin (Fig. S1, A and B; Raiborg et al., 2015). To measure the ability to form invadopodia, the cells were first depleted of invadopodia by serum removal and Src inhibition and then stimulated or not with serum- and hepatocyte growth factor (HGF)-containing medium for 1 h to allow invadopodia reformation. The cells were stained for TKS5 and analyzed by high-content microscopy. Whereas serum starvation prevented invadopodia reformation, TKS5-positive invadopodia could be detected in ∼50% of the serum and HGF-stimulated cells (Fig. S2 A). The number of cells with invadopodia and the number of invadopodia per cell were similar between control-treated and Protrudin-depleted cells and in the GFP-Protrudin–expressing cell line. However, it was evident from the automated measurements that the sizes of the individual TKS5-positive invadopodia were significantly reduced in Protrudin-depleted cells, and the TKS5-positive dots were often more scattered on the ventral cell surface. In siRNA-treated cells stably expressing siRNA-resistant GFP-Protrudin, the invadopodia size was similar to control cells. These results point to a specific role of Protrudin in defining the size of invadopodia as they form (Fig. S2 A).
Figure S1.
Characterization of MDA-MB-231 cells stably expressing GFP-Protrudin. (A) MDA-MB-231 cells stably expressing GFP-Protrudin siRNA resistant to oligo #1 (sires#1) was generated by lentiviral transduction. MDA-MB-231 and MDA-MB-231-GFP-Protrudin sires#1 cells were transfected with two different siRNAs targeting Protrudin (#1 and #2) or control siRNA for 4 d and subjected for Western blotting to detect protein levels of Protrudin, GFP-Protrudin, and actin (loading control) using anti-Protrudin and anti-actin antibodies, respectively. (B) Cells transfected as in A were seeded on coverslips, immunostained for LAMP1, and analyzed by confocal microscopy. Note that LAMP1-positive LE/Lys cluster perinuclearly in Protrudin-knockdown cells as expected, but not in Protrudin/GFP-Protrudin–positive cells, verifying the functionality of GFP-Protrudin.
Figure S2.
Invadopodia reformation is impaired in Protrudin-depleted cells. (A) MDA-MB-231 and MDA-MB-231-GFP-Protrudin-sires#1 cells were transfected with siRNA targeting Protrudin (oligo #1) or control siRNA. 4 d after transfection, cells were serum starved for 4 h and treated with Src inhibitor (10 µM PP2) for the last 30 min to remove invadopodia. Cells were stimulated for 1 h with serum containing medium supplemented with HGF (50 ng/ml) to allow reformation of invadopodia or serum free medium (SFM) as a negative control. Cells were stained with antibodies against TKS5 and analyzed by high-content microscopy. Note that TKS5-positive invadopodia only reform in the serum and HGF-treated cells and that cells expressing siRNA-resistant GFP-Protrudin reform invadopodia comparable to control cells. Graphs represent quantifications of different features of invadopodia reformation. Each plotted point represents the mean value of one experiment. Values represent mean ± SD. *, P < 0.05, one-way ANOVA, Tukey’s post hoc test. _n_ = 3 independent experiments. In total, >2,800 cells were analyzed per condition. (B) Graphs represent quantifications of different features of invadopodia reformation from Fig. 2 B. Each plotted point symbolizes one image representing the average value of typically 10–15 cells. Values represent mean ± SD. *, P < 0.05; ***, P < 0.001, one-way ANOVA, Tukey’s post hoc test. n = 25 images per condition from three independent experiments. (C) Real-time PCR for verification of SYT7 knockdown from three independent experiments in MDA-MB-231 cells. **, P < 0.01, one-sample t test.
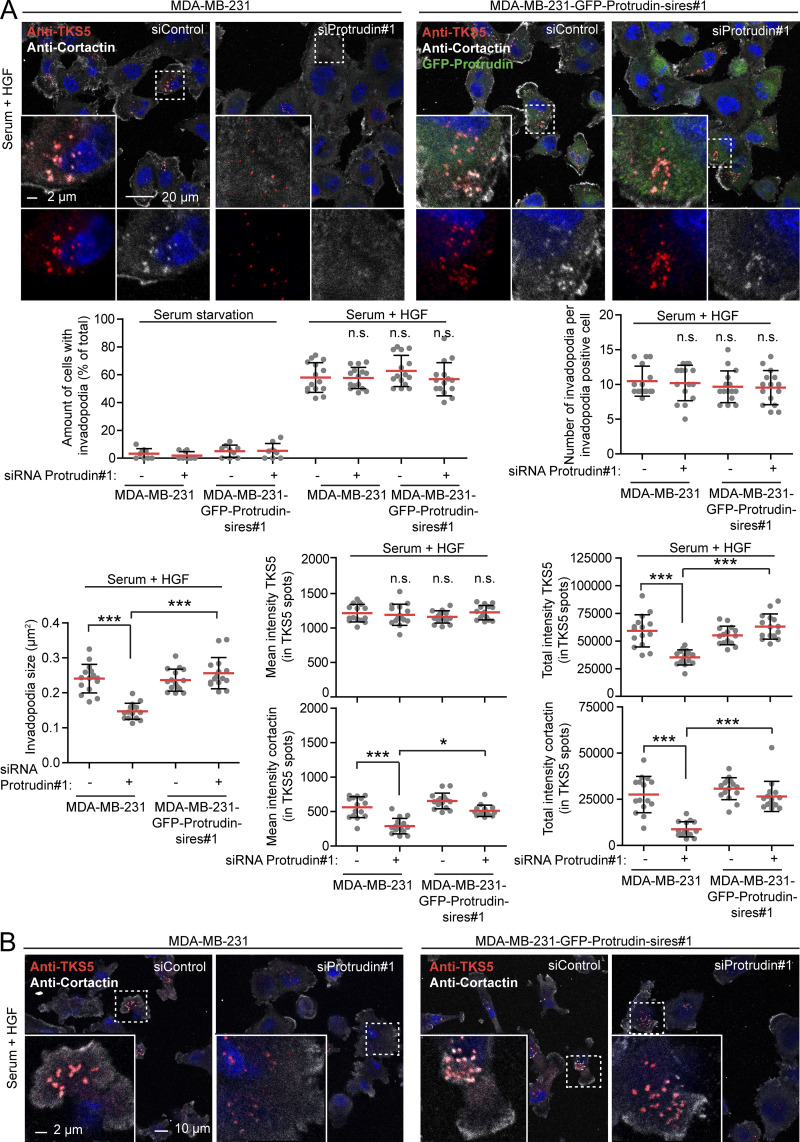
We next analyzed the invadopodia recovery and size in more detail by confocal microscopy of MDA-MB-231 cells stained with antibodies against TKS5 and cortactin. Both TKS5 and cortactin accumulate in invadopodia as they stabilize (Paterson and Courtneidge, 2018; Eddy et al., 2017; Artym et al., 2006), and the amount of TKS5 and cortactin in invadopodia was measured to assess their size and stabilization status. This analysis reproduced the findings from the high-content image analyses regarding numbers and sizes of invadopodia (Fig. 2 A). Whereas the mean fluorescence intensity of TKS5 in the smaller invadopodia found in Protrudin-depleted cells was similar to invadopodia in control cells, the mean intensity of cortactin in invadopodia was reduced in Protrudin-depleted cells. Moreover, the total intensities of TKS5 and cortactin per invadopodium were reduced in Protrudin-depleted cells, consistent with their reduced size. This suggests that invadopodia formation is still initiated in the absence of Protrudin, whereas they fail to mature properly. Similar results were obtained in Protrudin-depleted cells grown on gelatin coated coverslips (Figs. 2 B and S2 B) and in Protrudin knockout (KO) cells generated by CRISPR/Cas9-mediated genome editing, where the amount of TKS5 and actin was clearly reduced in invadopodia (Fig. 3, A and B; and Fig. S3 A). The number of TKS5-positive invadopodia per cell was similar between parental and Protrudin-KO cells (Fig. S3 B), supporting the notion that Protrudin is dispensable for their initiation. Analyses of z-stacks from confocal microscopy revealed that the small invadopodia found in Protrudin-KO cells were significantly shorter than those observed in control cells, with a typical length of 2 µm in parental cells and 1 µm in Protrudin-KO cells (Fig. 3 C). Importantly, the invadopodia outgrowth defects could be rescued in both siRNA-treated cells and KO cells by stable expression of GFP-Protrudin (Fig. 2; Fig. 3, A and B; Fig. S2, A and B).
Figure 2.
Protrudin-depleted cells fail to regrow invadopodia. (A) MDA-MB-231 and MDA-MB-231-GFP-Protrudin-sires#1 cells grown on coverslips were transfected with siRNA targeting Protrudin (oligo #1) or control siRNA. 4 d after transfection, cells were serum starved for 4 h and treated with Src inhibitor (10 µM PP2) for the last 30 min to remove invadopodia. Cells were stimulated for 1 h with serum containing medium supplemented with HGF (50 ng/ml) to allow reformation of invadopodia or SFM as a negative control. Cells were stained with antibodies against TKS5 and cortactin and analyzed by confocal microscopy. Micrographs show reformation of TKS5-positive invadopodia in serum- and HGF-treated cells. Note that cells expressing siRNA-resistant GFP-Protrudin reform invadopodia as in control cells. Graphs represent quantifications of different features of invadopodia reformation. Each plotted point symbolizes one image representing the average value of typically 15–20 cells. Values represent mean ± SD. *, P < 0.05; ***, P < 0.001, one-way ANOVA, Tukey’s post hoc test. n = 9 (serum starvation) or n = 15 images per condition from three independent experiments. (B) MDA-MB-231 and MDA-MB-231-GFP-Protrudin-sires#1 cells were siRNA transfected with siRNA targeting Protrudin (oligo #1) or control siRNA. 4 d after transfection, the cells were seeded on unconjugated-gelatin coated coverslips in SFM for 2 h to allow attachment before treating with Src inhibitor (10 µM PP2) for 30 min. Serum containing medium with HGF (50 ng/ml) was added and cells were allowed to reform invadopodia for 4 h, immunostaining and confocal imaging as in A. Note that cells expressing siRNA-resistant GFP-Protrudin reform invadopodia as in control cells (GFP not shown). For quantifications of micrographs, see Fig. S2 B. n.s, not statistically significant.
Figure 3.
Protrudin-KO cells display small and short invadopodia. (A) Cell lines as indicated in the figure were grown on coverslips; stained with anti-TKS5, anti-GFP, and phalloidin/Alexa Fluor 647 (actin); and analyzed by confocal microscopy. Note the undersized invadopodia in cells lacking Protrudin. (B) Cell lines as indicated in the figure were grown on coverslips, stained with anti-TKS5, anti-GFP and phalloidin/Alexa Fluor 647 (actin) and analyzed by confocal microscopy. Micrographs show orthographic and parallel maximum projections of z-stacks. In the parallel projections, the fluorescent intensity of TKS5 and actin were measured along a dotted line overlaying two invadopodia in each cell line, showing a reduced intensity of bothTKS5 and actin in cells lacking Protrudin as indicated in the intensity plots. (C) MDA-MB-231 and MDA-MB-231-Protrudin KO#2 cells were grown on coverslips coated with Oregon Green gelatin for 4 h, stained with anti-TKS5 and phalloidin/Alexa Fluor 647 (actin), and examined by superresolution microscopy (Airyscan). Z-stacks of individual cells were captured. Micrographs show section #9 from the parallel plane, and orthographic sections of the areas indicated by the dotted line. The graph shows the average length of TKS5-positive invadopodia per cell, quantified from Airyscan confocal z-stacks as described in Materials and methods. Each plotted point represents one cell, mean ± SD; parental, n = 19 cells; Protrudin KO, n = 15 cells from three independent experiments. ***, P < 0.0001, unpaired two-sided t test.
Figure S3.
Characterization of Protrudin-KO cell lines with or without stable expression of GFP-Protrudin, antibody validation, and verification of siRNA-mediated protein depletion. (A) KO of Protrudin in MDA-MB-231 cells was performed by CRISPR/Cas9-mediated genome editing. Stable expression of GFP-Protrudin in two KO clones was generated by lentiviral transduction. The indicated cell lines were either seeded on coverslips for confocal microscopy or lysed and subjected for Western blotting to detect protein expression levels of Protrudin. Cells grown on coverslips were immunostained for LAMP1. For Western blotting, anti-Protrudin and anti-actin (loading control) were used. Note that LAMP1-positive LE/Lys cluster perinuclearly in Protrudin-KO cells as expected, but not in Protrudin/GFP-Protrudin positive cells, verifying the functionality of GFP-Protrudin. (B) MDA-MB-231 and MDA-MB-231-Protrudin-KO#2 cells were grown on coverslips coated with Oregon Green gelatin for 4 h, stained with anti-TKS5 and phalloidin/Alexa Fluor 647 (actin), and examined by confocal microscopy (Airyscan; same dataset as in Fig. 3 C). The number of TKS5-positive invadopodia per cell was quantified. Each plotted point represents one cell, and values represent mean ± SD. Parental, n = 19 cells; Protrudin KO#2, n = 15 cells from three independent experiments, unpaired two-sided t test. (C) Verification of MT1-MMP antibody specificity by confocal microscopy and Western blotting. MDA-MB-231 cells were transfected with siRNA targeting MT1-MMP or control siRNA for 3 d. Cells were either lysed and subjected for Western blotting or seeded on coverslips and stained with anti-MT1-MMP and Alexa Fluor 488/Phalloidin (actin). Note that the anti-MT1-MMP signal is virtually gone in knockdown cells. (D) Western blots verifying knockdown of Protrudin, TKS5, RAB7, or FYCO1 in MDA-MB-231 cells using lysates with the indicated siRNA-transfected cells and the respective siRNA control treatments. Actin or vinculin was used as a loading control.
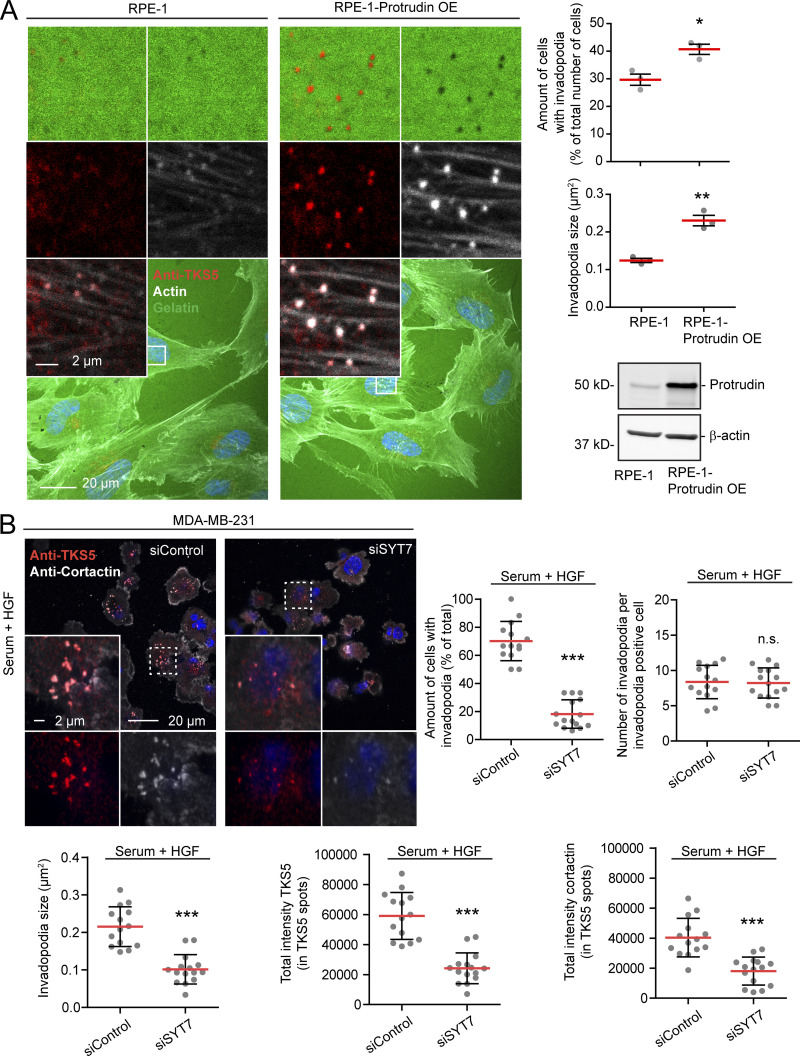
Moreover, stable overexpression of Protrudin in noncancerous RPE-1 cells induced the formation of TKS5- and actin-positive invadopodia-like structures. The amount of cells with detectable TKS5 and actin copositive spots increased from 30% in parental RPE-1 cells to 40% in Protrudin-overexpressing cells, and these cells had more prominent invadopodia, which corresponded to areas of degraded gelatin (Fig. 4 A). Taken together, these results indicate that Protrudin is required for invadopodia growth and elongation but dispensable for the initiation of small TKS5-positive puncta, likely representing invadopodia precursors (Eddy et al., 2017).
Figure 4.
Invadopodia formation is stimulated by Protrudin overexpression and requires SYT7-dependent membrane fusion. (A) RPE-1 cells with or without stable overexpression (OE) of Protrudin were grown on coverslips coated with Oregon Green gelatin for 4 h in serum containing medium and stained with anti-TKS5 and phalloidin/Alexa Fluor 647 (actin). Confocal micrographs show TKS5- and actin-positive invadopodia correlating with degraded gelatin. The graphs show the amount of cells with invadopodia and the size of invadopodia. Each plotted point represents the average of one independent experiment. Shown is mean ± SEM, n = 3. Total number of cells analyzed per condition: percentage of cells with invadopodia, >200 cells; invadopodia size, >45 cells. *, P < 0.05; **, P < 0.01, unpaired two sided t test. The Western blot shows the expression level of Protrudin in the two cell lines. (B) MDA-MB-231 cells grown on coverslips were transfected with siRNA targeting SYT7 or control siRNA. 1 d after transfection, cells were serum starved for 4 h and treated with Src inhibitor (10 µM PP2) for the last 30 min to remove invadopodia. Cells were stimulated for 1 h with serum containing medium supplemented with HGF (50 ng/ml) to allow reformation of invadopodia. Cells were stained with antibodies against TKS5 and cortactin and analyzed by confocal microscopy. Micrographs show reformation of TKS5-positive invadopodia. Graphs represent quantifications of different features of invadopodia reformation. Each plotted point symbolizes one image representing the average value of typically 15 cells. Values represent mean ± SD. ***, P < 0.001, unpaired two-sided t test. n = 15 images per condition from three independent experiments.
Neurite outgrowth is facilitated by SYT7-dependent fusion of LE/Lys with the plasma membrane at the tip of forming protrusions (Arantes and Andrews, 2006; Raiborg et al., 2015). To investigate whether this is also the case for invadopodia outgrowth, we measured invadopodia reformation in cells depleted for SYT7 by siRNA transfection. Indeed, we observed small TKS5-positive invadopodia in SYT7-depleted cells, similar to Protrudin-depleted cells (Fig. 4 B and Fig. S2 C). In addition, only 18% of the SYT7 depleted cells reformed invadopodia after starvation, compared with 70% in control treated cells. In summary, we conclude that invadopodia growth and elongation depend on Protrudin-mediated LE/Lys translocation to the cell periphery and subsequent SYT7-mediated fusion of LE/Lys with the plasma membrane.
Protrudin promotes anterograde translocation of MT1-MMP–containing LEs
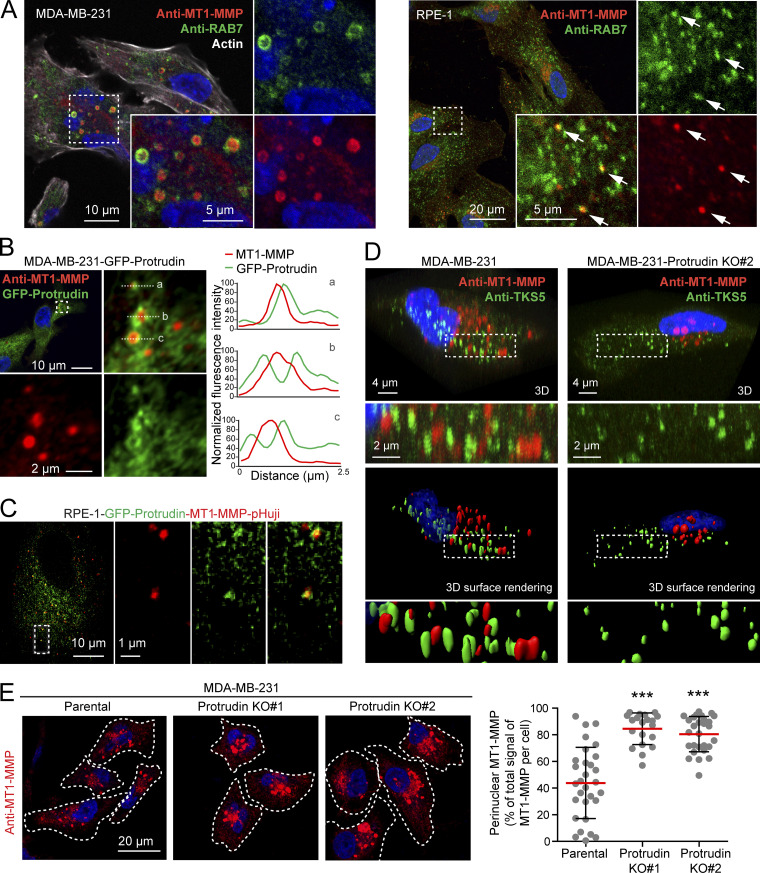
Since LE/Lys have been implicated in the transport of MT1-MMP to invadopodia, which is important for ECM degradation and completion of invadopodia maturation (Castro-Castro et al., 2016), we next asked whether MT1-MMP could be a cargo in Protrudin-associated endosomes. As expected, we could detect a significant portion of endogenous MT1-MMP in RAB7-positive LE/Lys in both MDA-MB-231 cells and RPE-1 cells (Fig. 5 A). The specificity of the MT1-MMP antibody was demonstrated by a loss of staining in cells silenced for this protein (Fig. S3 C). Protrudin is a transmembrane ER protein that localizes in VAMP-Associated Protein A (VAP-A)–positive areas of the ER (Saita et al., 2009; Chang et al., 2013; Raiborg et al., 2015). Protrudin-mediated anterograde LE/Lys translocation initiates in ER–endosome contact sites, where LE/Lys are loaded with kinesin-1 by Protrudin. Importantly, we observed MT1-MMP–containing LE/Lys in contact with GFP-Protrudin–positive areas of the ER in both MDA-MB-231 and RPE-1 cells (Fig. 5, B and C; Video 1). Protrudin depletion prevents contact site formation, resulting in perinuclear clustering of LE/Lys (Fig. S1 and Fig. S3 A; Raiborg et al., 2015). Indeed, in Protrudin-KO cells, MT1-MMP–containing LE/Lys clustered perinuclearly, as opposed to parental cells, where MT1-MMP–positive endosomes spread throughout the cell and were found in close proximity to TKS5-positive invadopodia (Fig. 5, D and E).
Figure 5.
Protrudin forms contact sites with MT1-MMP–positive LE/Lys and regulates their intracellular localization. (A) Confocal micrographs of MDA-MB-231 and RPE-1 cells showing that endogenous MT1-MMP localizes to RAB7-positive endosomes. Arrows show colocalization of MT1-MMP and RAB7 in RPE-1 cells. Manders overlap coefficient shows that 56% (±12 SD) of MT1-MMP positive pixels overlap with Rab7-positive pixels in MDA-MB-321 cells (n = 15 cells), and 33% (±8 SD) in RPE-1 cells (n = 13 cells). (B) Confocal micrographs of MDA-MB-231-GFP-Protrudin cells showing contact sites between GFP-Protrudin and endogenous MT1-MMP. Graphs show three examples of contact sites between GFP-Protrudin in the ER and MT1-MMP–positive endosomes visualized by fluorescence intensity profiles. (C) Still image from Video 1 of RPE-1-GFP-Protrudin-MT1-MMP-pHuji cells showing that GFP-Protrudin forms contact sites with MT1-MMP-pHuji–positive vesicles. (D) MDA-MB-231 and MDA-MB-231-Protrudin KO#2 cells were grown on coverslips, immunostained for MT1-MMP and TKS5, and analyzed by confocal microscopy. Z-stacks and Imaris surface 3D renderings show the subcellular localization of MT1-MMP in relation to TKS5-positive invadopodia. Note the perinuclear clustering of MT1-MMP and the undersized TKS5-positive invadopodia in Protrudin-KO#2 cells. (E) MDA-MB-231 and MDA-MB-231-Protrudin-KO cells were grown on coverslips, immunostained for MT1-MMP, and analyzed by confocal microscopy. Images and graph show the perinuclear clustering of MT1-MMP–positive endosomes in Protrudin-KO cells. Each plotted point represents one cell. Values represent mean ± SD. Number of cells analyzed: parental, n = 30; KO#1, n = 20; KO#2, n = 28 from three independent experiments. ***, P < 0.001, Kruskal-Wallis, Dunn’s post hoc test. Note that the big variation in MT1-MMP localization seen between parental cells is diminished in Protrudin-KO cells where MT1-MMP vesicles mainly cluster perinuclearly.
Video 1.
GFP-Protrudin makes contact with pHuji-MT1-MMP–positive endosomes. RPE-1-GFP-Protrudin sires#1-MT1-MMP-pHuji cells were seeded in MatTek dishes and imaged using a Deltavision OMX V4 microscope with a 60× objective. Simultaneous dual-color live-cell imaging was done at 1 Hz. The video shows one cell, where two examples of MT1-MMP-pHuji–positive LE/Lys making contact with GFP-Protrudin–positive ER are highlighted. The upper MT1-MMP–positive vesicle stays in contact for 12 s (yellow circle) before contact is lost (red circle), leading to its fast anterograde movement. The lower MT1-MMP–positive vesicle stays in contact with Protrudin-positive ER throughout the course of the video (yellow circle).
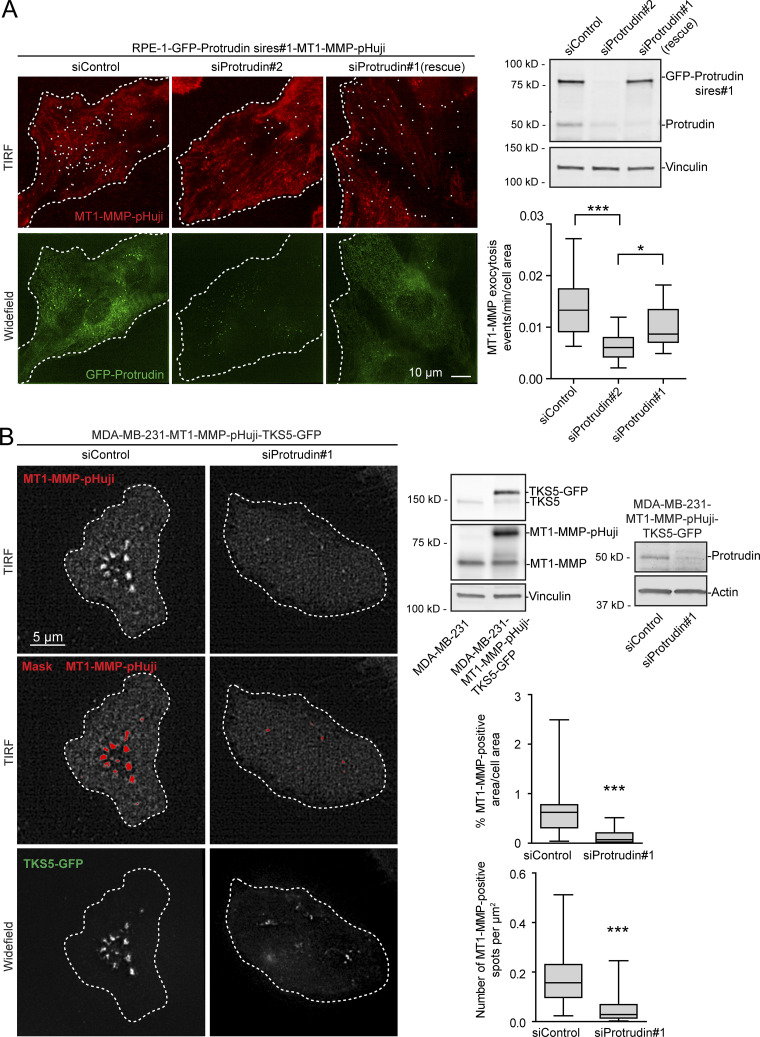
Consistent with this, total internal reflection fluorescence (TIRF) imaging of both MDA-MB-231 and RPE-1 cells revealed that Protrudin depletion reduced the cell surface exposure of MT1-MMP (Fig. 6, A and B). Cells were imaged twice per second for 2 min. In Protrudin-depleted RPE-1 cells, we observed a reduced number of flashes of MT1-MMP-pHuji, indicating fewer fusion events with the plasma membrane. This effect could be partially rescued by stable expression of siRNA-resistant GFP-Protrudin (Fig. 6 A). In control-treated MDA-MB-231 cells, the surface signal of MT1-MMP-pHuji was rather stable in TKS5-GFP positive invadopodia for the whole imaging period, presumably due to stabilized fusion pores, which can occur between late endosomal tubules and the invadopodial plasma membrane, as described previously (Monteiro et al., 2013). We therefore chose to perform an endpoint analysis of the TIRF images, which revealed that siRNA-mediated Protrudin depletion reduced both the total area and the number of MT1-MMP-pHuji puncta per cell surface (Fig. 6 B). Since the number of TKS5-positive invadopodia per cell was not reduced in Protrudin-depleted cells (Fig. 2 and Fig. S2), the reduced number of MT1-MMP-pHuji puncta from the TIRF analysis is consistent with a reduced fusion rate of MT1-MMP–containing endosomes in invadopodia. Taken together, the results indicate that Protrudin promotes anterograde translocation of MT1-MMP–containing LE/Lys to invadopodia for subsequent exocytosis.
Figure 6.
MT1-MMP exocytosis depends on Protrudin. (A) RPE-1-GFP-Protrudin sires#1-MT1-MMP-pHuji cells were seeded in MatTek dishes and transfected with siRNA against Protrudin (oligo #1 or #2) or control siRNA. 2 d after transfection, cells were imaged twice per second for 2 min by TIRF microscopy. Shown are representative TIRF micrographs where individual exocytic events (summed over a 2-min interval) are indicated by a white dot. Widefield micrographs and Western blot analysis show the expression level of GFP-Protrudin and knockdown efficiencies. The graph shows the number of MT1-MMP exocytic events per cell area per minute. For each condition, n = 18 movies were analyzed from three independent experiments. Boxplot whiskers show minimum to maximum. *, P < 0.05; ***, P < 0.001, one-way ANOVA, Tukey’s post hoc test. Note that siProtrudin#2 depletes both endogenous and exogenous Protrudin and reduces the amount of MT1-MMP exocytosis, whereas siProtrudin#1 only depletes the endogenous pool of Protrudin, leaving GFP-Protrudin (sires#1) to maintain MT1-MMP exocytosis. (B) MDA-MB-231-MT1-MMP-pHuji-TKS5-GFP cells were seeded in MatTek dishes and transfected with siRNA against Protrudin (oligo #1) or control siRNA. Western blots show protein expression levels and verify the Protrudin knockdown in the stable cell line. 4 d after transfection, cells were imaged by TIRF microscopy. Shown are representative TIRF micrographs of MT1-MMP-pHuji surface accumulation. Widefield micrographs show TKS5-GFP–positive invadopodia. Plasma membrane exposure of MT1-MMP-pHuji was quantified by segmenting bright pHuji spots from the TIRF images (mask). Graphs show whiskers (minimum to maximum) from n = 30 (siProtrudin) and n = 33 (siControl) TIRF images. ***, P < 0.001, Mann–Whitney test.
Protrudin, FYCO1, and RAB7 mediate degradation of ECM proteins
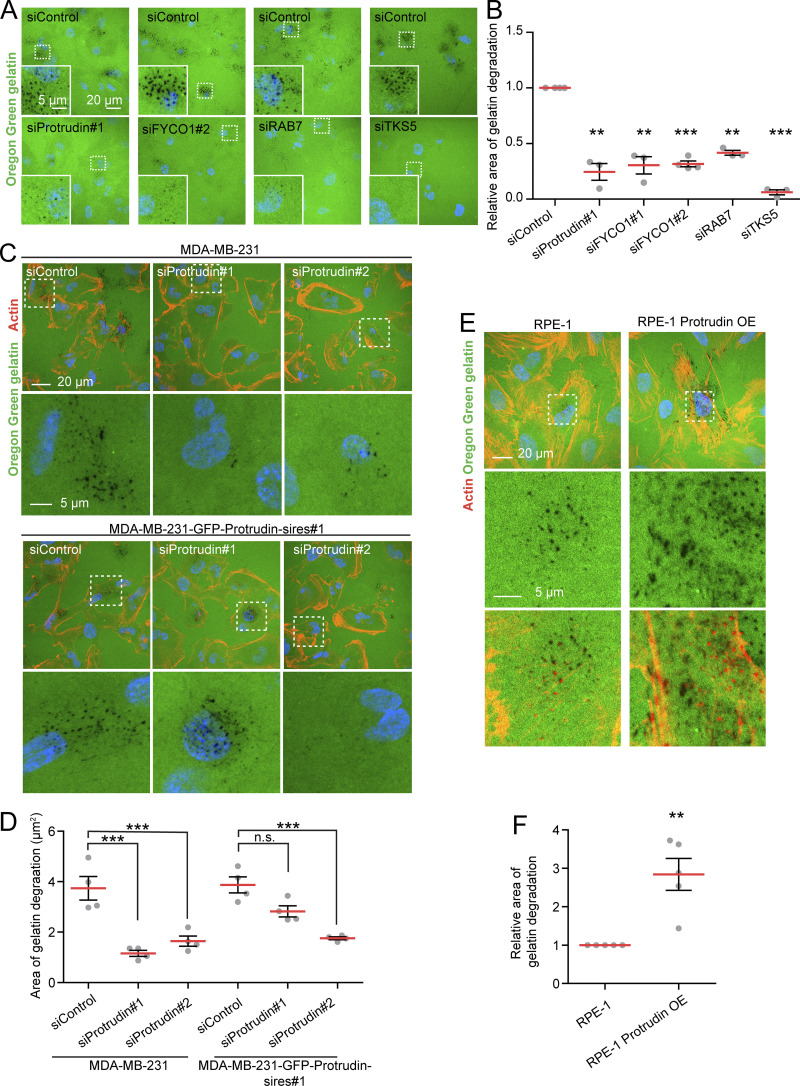
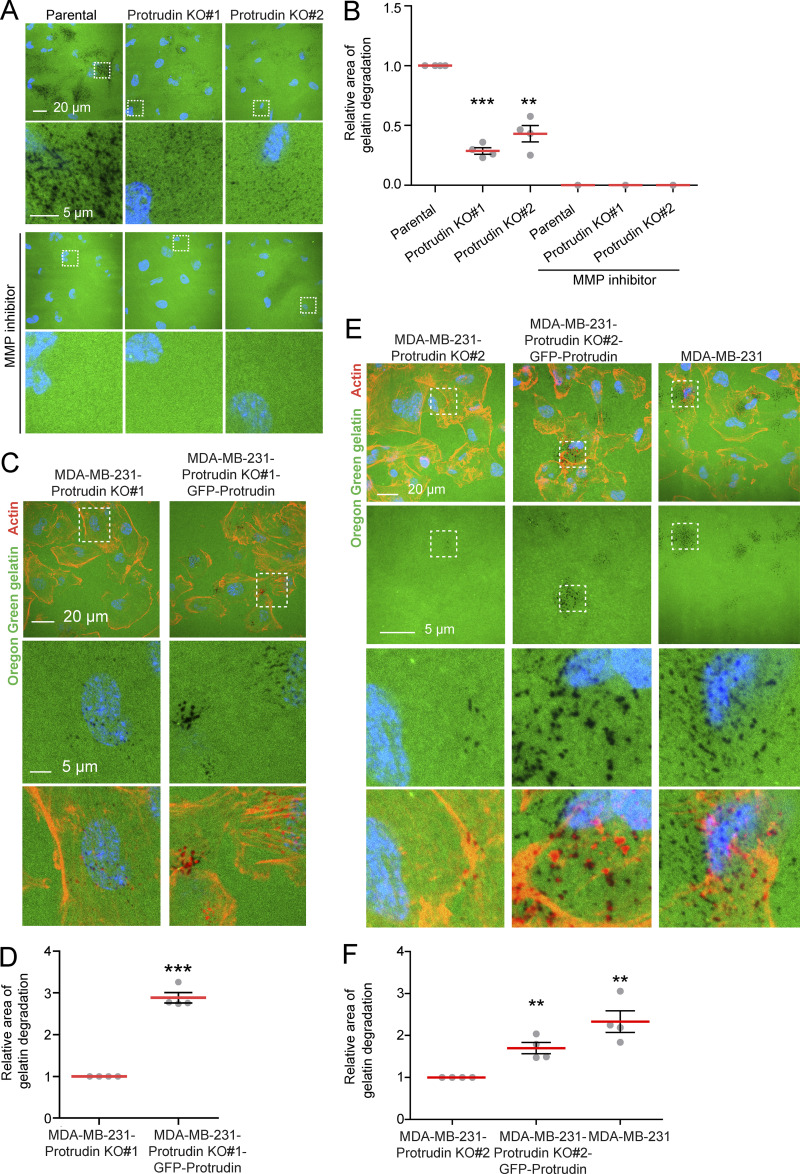
When MT1-MMP is exposed at invadopodia, it promotes degradation of the ECM. Based on the newly identified role of Protrudin in invadopodia outgrowth, MT1-MMP trafficking, and exocytosis, we next assessed whether proteins in the Protrudin pathway were important for the degradation of ECM proteins. To this end, MDA-MB-231 cells, depleted of Protrudin, RAB7, or FYCO1 by siRNA, were plated on coverslips coated with Oregon Green gelatin for 4 h to allow gelatin degradation. In all cases, the area of degraded gelatin per cell was significantly reduced compared with control-treated cells, but not to the same extent as in TKS5-depleted cells, where the degradation was virtually abolished (Fig. 7, A and B; and Fig. S3 D). Moreover, the amount of gelatin degradation was also significantly reduced in Protrudin-KO cells (Fig. S4, A and B). Inhibition of MMP activity by the inhibitor GM6001 completely abolished all gelatin degradation, indicating that the observed degradation was indeed due to MMP activity (Fig. S4, A and B).
Figure 7.
Protrudin promotes degradation of extracellular gelatin in a dose-dependent fashion. (A) MDA-MB-231 cells were transfected with siRNA targeting Protrudin, FYCO1, RAB7, TKS5, or control siRNA. The cells were grown on coverslips coated with Oregon Green gelatin for 4 h and analyzed by confocal microscopy. Representative micrographs show degradation of fluorescent gelatin indicated by black areas. (B) Quantifications of images in A. The graph shows the relative area of gelatin degradation in the different knockdown conditions compared with siControl. Each individual data point represents the average of one independent experiment. Shown is mean ± SEM, n = 3 experiments (Protrudin, FYCO1#1, RAB7, and TKS5), n = 4 experiments FYCO1#2. **, P < 0.01; ***, P < 0.001, one-sample _t_ test. Number of cells in total: siControl(Protrudin/TKS5), 1,145; siProtrudin#1, 800; siTKS5, 633; siControl(FYCO1#1), 1,064; siFYCO1#1, 975; siControl(FYCO1#2), 893; siFYCO1#2, 1,007; siControl(RAB7), 577; siRAB7, 512. **(C)** MDA-MB-231 and MDA-MB-231-GFP-Protrudin-sires#1 cells were transfected with siRNA targeting Protrudin (oligo #1 or #2) or control siRNA. 4 d after transfection, cells were grown on coverslips coated with Oregon Green gelatin for 4 h, stained with Rhodamine-Phalloidin (actin), and analyzed by confocal microscopy (knockdown verification in Fig. S1 A). Representative micrographs show degradation of fluorescent gelatin indicated by black areas. **(D)** Quantifications of images in C. The graph shows area of gelatin degradation in the different knockdown conditions compared with siControl. Note that the reduced gelatin degradation in Protrudin-depleted cells is rescued in the presence of GFP-Protrudin. Each individual data point represents the average of one independent experiment. Values represent mean ±SEM, _n_ = 4. ***, P < 0.001, one-way ANOVA, Tukey’s post hoc test. In total, >700 cells were analyzed per condition. (E) RPE-1 cells or RPE-1-Protrudin-overexpressing cells were grown on coverslips coated with Oregon Green gelatin for 6 h with addition of HGF (50 µm) for the last 2 h, stained with Rhodamine-Phalloidin (actin), and analyzed by confocal microscopy. Representative micrographs show degradation of fluorescent gelatin indicated by black areas. (F) Quantification of images in E. The graph shows relative area of gelatin degradation. Each individual data point represents the average of one independent experiment. Values represent mean ± SEM, n = 5. **, P < 0.01, one sample _t_ test. In total, >1,600 cells were analyzed per condition.
Figure S4.
GFP-Protrudin rescues the loss of gelatin degradation in Protrudin-KO cells. (A) MDA-MB-231 parental or MDA-MB-231-Protrudin-KO (KO#1 and KO#2) cells were grown on coverslips coated with Oregon Green gelatin for 4 h in the absence or presence of the MMP inhibitor GM6001 (20 µM; characterization of cell lines in Fig. S3 A). Confocal micrographs show degradation of the fluorescent gelatin indicated by black areas. (B) Quantifications of experiments displayed in A. The graph shows the relative area of gelatin degradation. Each individual data point represents the average of one independent experiment. Values represent mean ± SD, n = 4 experiments. **, P < 0.01; ***, P < 0.001, one-sample _t_ test. Number of cells in total: parental, 1,156; Protrudin KO#1, 1,146; Protrudin KO#2, 1,174. With MMP inhibitor: _n_ = 1; number of cells: parental, 175; Protrudin KO#1, 163; Protrudin KO#2, 206. **(C)** MDA-MB-231-Protrudin-KO#1 and MDA-MB-231-Protrudin-KO#1-GFP-Protrudin cells were grown on coverslips coated with Oregon Green gelatin for 4 h, stained with Rhodamine-Phalloidin (actin), and analyzed by confocal microscopy (characterization of cell lines in Fig. S3 A). Representative micrographs show degradation of fluorescent gelatin indicated by black areas. **(D)** Quantification of images in C. The graph shows relative area of gelatin degradation. Each individual data point represents the average of one independent experiment. Values represent mean ± SD, _n_ = 4. ***, P < 0.001, one-sample _t_ test. In total, >900 cells were analyzed per condition. (E) The indicated cell lines were grown on coverslips coated with Oregon Green gelatin for 4 h, stained with Rhodamine-Phalloidin (actin), and analyzed by confocal microscopy (characterization of cell lines in Fig. S3 A). Representative micrographs show degradation of fluorescent gelatin indicated by black areas. (F) Quantification of images in E. The graph shows relative area of gelatin degradation. Each individual data point represents the average of one independent experiment. Values represent mean ± SD, n = 4. **, P < 0.01, one-sample _t_ test. In total, >750 cells were analyzed per condition.
Importantly, the reduced gelatin degradation observed upon Protrudin depletion in siRNA cells and Protrudin-KO cells was rescued by stable expression of GFP-Protrudin (Fig. 7, C and D; and Fig. S4, C–F). In addition, in GFP-Protrudin cells, the fully developed actin-positive invadopodia colocalized with areas of degraded gelatin, consistent with a complete invadopodia maturation and function (Fig. S4, C and E). Finally, when Protrudin was overexpressed in RPE-1 cells, a significantly increased amount of gelatin was degraded compared with parental RPE-1 cells, which exhibited very little degradation, and the area of gelatin degradation often colocalized with actin-positive invadopodia indicating that they were fully matured (Fig. 7, E and F). This was consistent with the increased amount of invadopodia observed earlier in Protrudin-overexpressing RPE-1 cells (Fig. 4 A). Whereas these results show that the Protrudin pathway clearly promotes gelatin degradation, TKS5 and MMP activity is indispensable, consistent with the fundamental role of TKS5 in invadopodia formation (Seals et al., 2005).
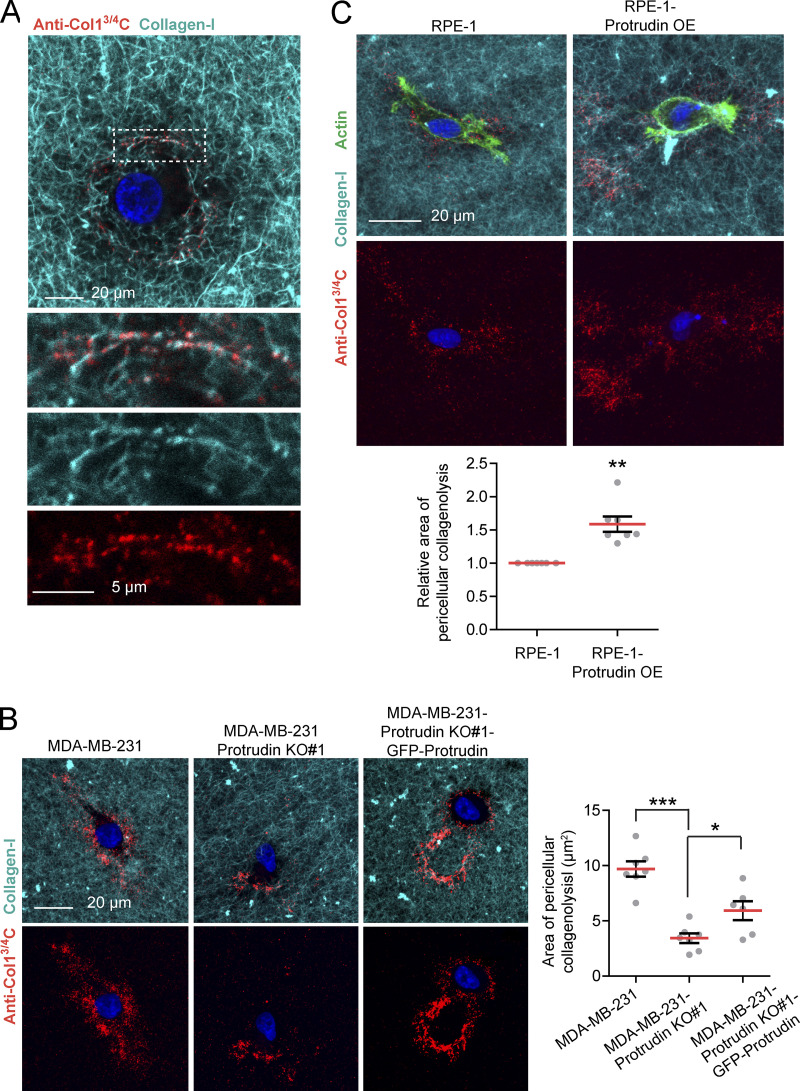
To more specifically test for MT1-MMP activity, we cultured cells in droplets of polymerized collagen-I and assessed pericellular collagenolysis by an antibody that recognizes the MT1-MMP–dependent C-terminal 3/4 cleavage site (Monteiro et al., 2013; Fig. 8 A). Whereas Protrudin KO in MDA-MB-231 cells decreased the amount of collagen degradation in a rescuable manner (Fig. 8 B), Protrudin overexpression in RPE-1 cells increased collagen cleavage compared with parental cells (Fig. 8 C). Taken together, Protrudin and proteins related to the Protrudin pathway facilitate MT1-MMP–dependent degradation of ECM proteins.
Figure 8.
Protrudin mediates cleavage of collagen-I. (A) MDA-MB-231 cells were embedded in fluorescent type I collagen (cyan, 2.0 mg/ml) for 24 h and stained for the cleaved collagen neo epitope using anti-Col-3/4C. The confocal micrograph shows the presence of collagen cleavage sites lining the collagen-I fibrils indicating the specificity of the antibody. (B) Protrudin KO decreases pericellular collagenolysis in MDA-MB-231 cells. MDA-MB-231, MDA-MB-231-Protrudin KO#1, and MDA-MB-231-Protrudin KO#1-GFP-Protrudin cells were embedded in fluorescent type I collagen (cyan, 2.0 mg/ml) for 24 h and stained with anti-Col-3/4C. Representative micrographs show the sum projections of confocal z-stacks from each condition. The graph shows quantification of the area of pericellular collagenolysis (µm2) from sum projections of wide field images. Each plotted point represent the average of one collagen droplet. n = 7 droplets (10 z-stacks per droplet) were analyzed from five different experiments. Values represent mean ± SEM. *, P < 0.05; ***, P < 0.001, one-way ANOVA, Tukey’s post hoc test. In total, >700 cells were analyzed per condition. (C) RPE-1 cells or RPE-1-Protrudin-overexpressing cells were embedded in fluorescent type I collagen (cyan, 2.0 mg/ml) for 24 h and stained with anti-Col-3/4C and Alexa488-Phalloidin (actin). Representative micrographs show the sum projections of confocal z-stacks from each condition. The graph shows quantification of the relative area of pericellular collagenolysis from sum projections of wide-field images. Each plotted point represents the average of one collagen droplet. n = 7 droplets (10 z-stacks per droplet) were analyzed from three different experiments. Values represent mean ± SEM. **, P < 0.01, one-sample _t_ test. In total, >290 cells were analyzed per condition.
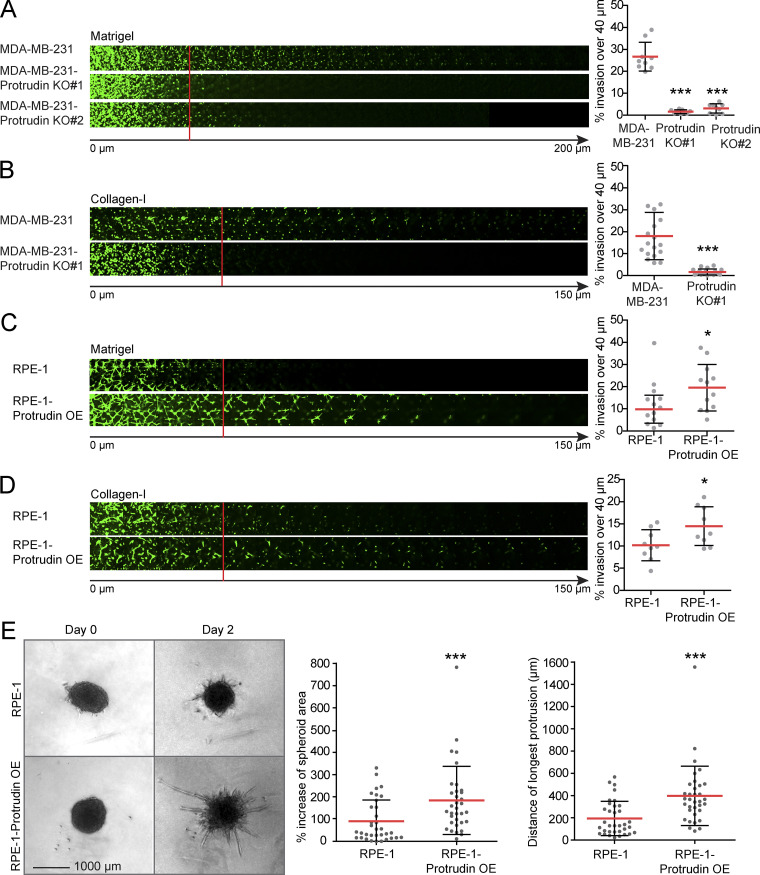
Protrudin is required for cell invasion into Matrigel and collagen-I
Cancer cells use invadopodia to breach basement membranes and invade mesenchymal tissues. To investigate whether Protrudin plays a role in cell invasion, we performed inverted invasion assays, where cells were allowed to invade into a plug composed of either Matrigel or collagen-I. Whereas Matrigel is a solubilized basement membrane preparation containing laminin and collagen-IV, fibrillar collagen-I is more representative of mesenchymal tissues (Kalluri, 2003; Mouw et al., 2014). Importantly, Protrudin-KO cells were unable to invade efficiently into Matrigel or collagen-I (Fig. 9, A and B), and stable expression of GFP-Protrudin in the KO cells reversed this effect (Fig. S5 A). Conversely, when Protrudin was overexpressed in noninvasive RPE-1 cells, the cells became more invasive (Fig. 9, C and D). This was also the case for Protrudin-overexpressing RPE-1 cells that were grown as spheroids in Matrigel (Fig. 9 E). In conclusion, our data suggest that the Protrudin pathway promotes cell invasion by translocating MT1-MMP–laden LE/Lys to the plasma membrane, thereby enabling both invadopodia outgrowth and MT1-MMP exocytosis.
Figure 9.
Protrudin promotes cell invasion. The cell lines indicated were allowed to invade into plugs of fibronectin-supplemented Matrigel or collagen I for 4 d, stained with Calcein, and imaged by confocal microscopy. Optical sections (Δ_z_ = 10 µm) are shown for each condition. Graphs show the percentage of cell invasion over 40 µm. Each plotted point represents the average invasion of one plug. (A) In total n = 9 plugs (five z-stacks per plug) were analyzed from three independent experiments. Values represent mean ± SD. ***, P < 0.001, one-way ANOVA, Dunnett’s post hoc test. (B) In total, n = 18 plugs (five z-stacks per plug) were analyzed from six independent experiments. Values represent mean ± SD. ***, P < 0.001, one-sample t test. (C) In total, n = 12 plugs (five z-stacks per plug) were analyzed from four independent experiments. Values represent mean ± SD. *, P < 0.05, one-sample t test. (D) In total n = 9 plugs (5 z-stacks per plug) were analyzed from 3 independent experiments. Values represent mean ± SD. *, P < 0.05, one-sample t test. (E) Single tumor spheroids from RPE-1 and RPE-1-Protrudin–overexpressing cells were embedded in Matrigel and allowed to invade. Phase contrast micrographs show spheroids from days 0 and 2. Graphs represent percent increase of spheroid area and distance of the longest protrusion from the outer rim of the spheroid dense core. Values represent mean ± SD. Each plotted point represents one spheroid. In total, n = 34 spheroids from six different experiments. ***, P < 0.001, Mann–Whitney. OE, overexpression.
Figure S5.
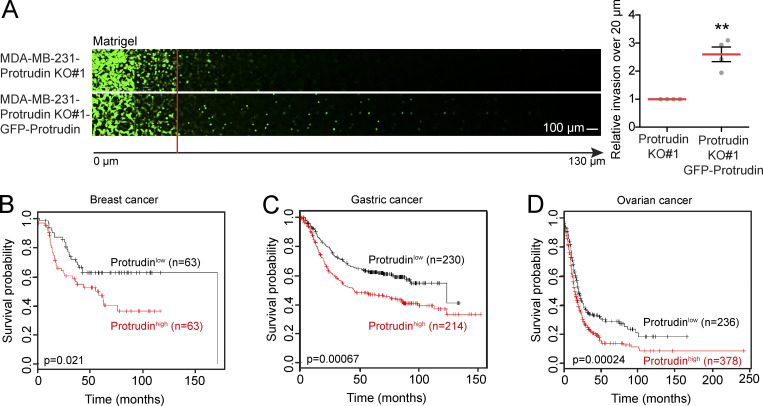
GFP-Protrudin expression rescues cell invasion of Protrudin-KO cells, and the prognostic value of Protrudin in different cancer types. (A) The indicated cell lines were allowed to invade into plugs of fibronectin-supplemented Matrigel for 4 d, stained with Calcein, and imaged by confocal microscopy. Optical sections (Δ_z_ = 10 µm) are shown for each condition. Graphs show the relative cell invasion over 20 µm. Each plotted point represents the average invasion of one independent experiment, n = 4. In total, 12 plugs (five z-stacks per plug) were analyzed. Values represent mean ± SD. **, P < 0.01, one-sample t test. (B–D) Survival curves are plotted for breast, gastric, and ovarian cancer using the publicly available database Kaplan-Meier Plotter for high and low expression of Protrudin (ZFYVE27 Affymetrix ID 225281_at). (B) Analysis of lymph node–positive, estrogen receptor–negative, progesterone receptor–negative breast cancer patient cohorts (hazard ratio [HR], 1.88; 95% confidence interval [CI], 1.09–3.24. (C) Analysis of metastasis-free gastric cancer patient cohorts (HR, 1.62; 95% CI, 1.22–2.14). (D) Analysis of ovarian cancer patient cohorts (HR, 1.44; 95% CI, 1.19–1.76).
To investigate whether the Protrudin pathway is important in human cancers, we compared the survival probabilities for cancer patients with high or low expression of Protrudin. Cohorts from ovarian, gastric, and breast cancer patients were analyzed using the publicly available database Kaplan-Meier Plotter (Györffy et al., 2010, 2012; Szász et al., 2016). The results showed that high expression of Protrudin is associated with reduced survival (Fig. S5, B–D), supporting our experimental findings and underscoring the significance of the Protrudin pathway in invadopodia formation and function.
Discussion
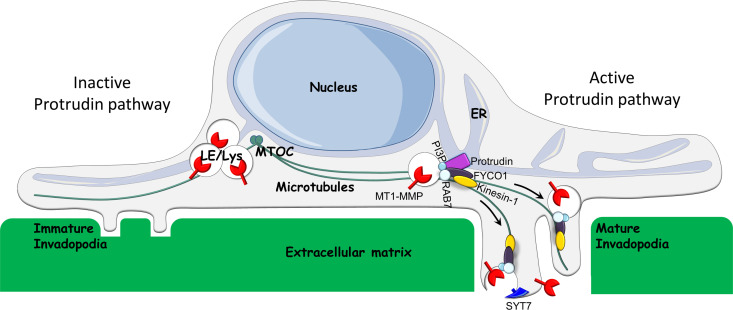
Invadopodia maturation and function depend on endosomal trafficking (Castro-Castro et al., 2016). We show here that Protrudin–mediated ER–endosome contact sites, previously implicated in anterograde endosome translocation and neurite outgrowth, play an important role in cancer cell invasion in vitro by controlling the transport of MT1-MMP–laden endosomes to invadopodia. This facilitates both invadopodia maturation and exocytosis of MT1-MMP (Fig. 10).
Figure 10.
Model for the function of the Protrudin pathway in invadopodia formation and exocytosis of MT1-MMP. When the Protrudin pathway is active (right), the ER protein Protrudin makes contact sites with RAB7 and phosphatidylinositol 3-phosphate(PI3P)–positive LE/Lys, which contain MT1-MMP. In the ER–LE/Lys contact sites, the microtubule motor kinesin-1 is handed over from Protrudin to the RAB7-binding kinesin-1 adaptor protein FYCO1. This enables the translocation of the LE/Lys along microtubules toward immature invadopodia at the plasma membrane. SYT7-dependent fusion of LE/Lys with the invadopodial plasma membrane provides membrane for the maturing invadopodia and ensures that MT1-MMP is exposed at the cell surface. This enables invadopodia growth and degradation of the ECM and facilitates cell invasion. Upon depletion of any of the components of the Protrudin pathway (left), MT1-MMP–containing LE/Lys cluster perinuclearly. This prevents MT1-MMP exocytosis, invadopodia maturation, and cell invasion. MTOC, microtubule organizing center.
The main protein players of the Protrudin pathway are the ER protein Protrudin, endosomal FYCO1 and RAB7, and the plus-end microtubule motor kinesin-1 (Raiborg et al., 2015). We show here that FYCO1, RAB7, or Protrudin depletion inhibits gelatin degradation, and we conclude that the Protrudin pathway is required for invadopodia elongation and ECM degradation but dispensable for invadopodia precursor formation. Overexpression of kinesin-1 has been shown to dramatically increase gelatin degradation, and drugs affecting microtubule dynamics inhibit the elongation of invadopodia, but not invadopodia precursor initiation, in line with our results targeting the Protrudin pathway (Marchesin et al., 2015; Kikuchi and Takahashi, 2008). Whereas we show that FYCO1 and Protrudin are novel mediators of invadopodia formation and function, RAB7 has been implicated before. MT1-MMP colocalizes with RAB7-positive LE/Lys (Steffen et al., 2008), and overexpression of a dominant negative mutant of RAB7 inhibits invadopodia function (Williams and Coppolino, 2011). However, to the best of our knowledge, our work is the first to show that siRNA mediated depletion of RAB7 indeed inhibits ECM degradation, and its connection to Protrudin provides a mechanistic explanation for the involvement of RAB7 in invadopodia.
When invadopodia elongate, the plasma membrane needs to expand as the actin cytoskeleton polymerizes. The source of this extended membrane could simply be the folding of the plasma membrane. Alternatively, vesicles from intracellular membrane sources could fuse with the invadopodial plasma membrane to provide new membrane locally. Different types of internal membranes have been implicated in various plasma membrane extensions (Lecuit and Wieschaus, 2000; Dyer et al., 2007; Reddy et al., 2001). We have shown previously that fusion of LE/Lys with the plasma membrane facilitates neurite outgrowth (Raiborg et al., 2015). Here, we find that invadopodia outgrowth depends on Protrudin-mediated LE/Lys translocation and SYT7-dependent fusion, consistent with LE/Lys being a membrane source for invadopodia elongation. This is in line with experiments with anchor cells in C. elegans, which ruled out plasma membrane folding as a membrane source and showed that exocytosis of lysosomes is important for invadopodia elongation (Naegeli et al., 2017). In their study, the authors observed a reduced invadopodia growth rate and length in cells where they specifically depleted the pool of LAMP1-positive lysosomes by RNAi knockdown of ppk-3 (PIKfyve in mammals). RAB7-positive late endosomes were, however, still present in these cells. The Protrudin pathway translocates LE/Lys containing PtdIns3P, RAB7, and LAMP1, suggesting that different subpopulations of LE/Lys contribute membrane to forming invadopodia in different cell types or organisms.
The exposure of MT1-MMP at the cell surface and the consequent ECM degradation can further facilitate invadopodia maturation (Kumar et al., 2018; Eddy et al., 2017; Castro-Castro et al., 2016). The Protrudin pathway mediates LE/Lys translocation to invadopodia and subsequent fusion and exposure of MT1-MMP, providing both membrane and ECM degradation. It can be difficult to distinguish the contribution of these mechanisms for invadopodia outgrowth. Since invadopodia were shorter in Protrudin-depleted cells grown on uncoated coverslips as well as gelatin, we conclude that fusion of LE/Lys can contribute to invadopodia elongation in the absence of ECM degradation. This is in line with studies in C. elegans, where invasive protrusions could form in the absence of MMPs, although less efficiently than in the presence of MMPs (Kelley et al., 2019). The Protrudin pathway is likely to accelerate invadopodia outgrowth by a combination of membrane growth and MT1-MMP–dependent digestion of the ECM.
Following exocyst-mediated tethering (Monteiro et al., 2013), the fusion of LE/Lys with the invadopodial plasma membrane is mediated by the late-endosomal and RAB7-dependent v-SNARE VAMP7 and its cognate SNAREs Syntaxin4 and SNAP23 (Williams and Coppolino, 2011; Steffen et al., 2008). VAMP7-dependent fusion depends on the Ca2+ adaptor protein SYT7 (Rao et al., 2004), and SYT7 has been implicated in exosome secretion and invadopodia activity in squamous cell carcinoma cells (Hoshino et al., 2013). We show here that SYT7 is important for invadopodia formation and growth, adding SYT7 as an important component of the machinery that mediates LE/Lys fusion with the invadopodial plasma membrane in MDA-MB-231 cells. Interestingly, we observed that SYT7 depletion had a stronger effect on invadopodia reformation than Protrudin depletion in MDA-MB-231 cells. These results indicate that SYT7 regulates the fusion of different populations of LE/Lys and multivesicular endosomes and suggest that several possibly independent pathways for anterograde LE/Lys translocation are being used for invadopodia formation and MT1-MMP exocytosis.
Many cellular functions depend on the localization of LE/Lys within the cell (Ballabio and Bonifacino, 2020; Pu et al., 2016; Raiborg, 2018). Whereas lysosomes close to the plasma membrane can be involved in plasma membrane repair and mTORC1 signaling, perinuclear LE/Lys perform degradation of endocytic or autophagosomal content. Since the targeted delivery of MT1-MMP to invadopodia depends on endosomal recycling routes, mechanisms that control LE/Lys positioning are expected to be involved. For anterograde LE/Lys translocation, two seemingly parallel pathways have been discovered, the BORC/Arl8/SKIP pathway (Pu et al., 2015) and the Protrudin pathway (Raiborg et al., 2015). There are no existing studies on the possible role of the BORC complex in invadopodia formation and MT1-MMP exocytosis, but Arl8 has been implicated in invasive growth of prostate cancer cells (Dykes et al., 2016), suggesting that also the BORC/Arl8/SKIP pathway might be involved. We show here that the Protrudin pathway indeed plays a role in the targeted delivery of MT1-MMP–positive LE/Lys to invadopodia and that this is important for invadopodia maturation and function. The BORC/Arl8/SKIP pathway is triggered by growth factor signaling and amino acids (Filipek et al., 2017, Pu et al., 2017), whereas the Protrudin pathway needs supply of amino acids or glucose (Hong et al., 2017; Palomo-Guerrero et al., 2019). The extracellular environment and nutrient availability are therefore likely to influence the level of MT1-MMP exposure at the invadopodial plasma membrane via different LE/Lys translocation mechanisms.
In addition to LE/Lys translocation to the cell periphery, tubules emanating from LE/Lys are important for the delivery of MT1-MMP to the tip of invadopodia. MT1-MMP localizes to RAB7- and LAMP1-positive LE/Lys, which tubulate in a JIP3/4- and WASH-dependent manner (Marchesin et al., 2015). In addition, endosomal coronin and the retromer complex, which are important for endosomal tubule formation, facilitate MT1-MMP exocytosis (Castagnino et al., 2018; Sharma et al., 2020). Such endosomal tubules are thought to be anchored to the invadopodial plasma membrane through interaction with the plasma membrane–localized small GTPase ARF6, and the tubular part of the LE/Lys can fuse with the plasma membrane to expose MT1-MMP for ECM remodeling. This has been suggested to form a relatively stable fusion pore, delivering MT1-MMP to the invadopodial plasma membrane of MDA-MB-231 cells (Marchesin et al., 2015; Monteiro et al., 2013). These findings are in agreement with our TIRF data, which show a stable signal of MT1-MMP-pHuji invadopodia over a 2-min period. Perturbation of LE/Lys tubulation inhibits MT1-MMP exocytosis and results in a perinuclear accumulation of LE/Lys, presumably due to a disturbed LE/Lys homeostasis (Castagnino et al., 2018). Alternatively, loss of LE/Lys tubules could possibly activate the JIP4-dependent retrograde LE/Lys translocation pathway (Willett et al., 2017; Castagnino et al., 2018). Clearly, pathways that regulate LE/Lys positioning and tubulation are crucial regulators of MT1-MMP exocytosis. The identification of the Protrudin pathway as an important regulator of MT1-MMP trafficking is a significant contribution to our understanding of this process. Taken together, a picture emerges that the Protrudin pathway can translocate MT1-MMP–positive LE/Lys to the base of invadopodia, whereas the endosomal tubulation machinery is required for the entry of MT1-MMP into the narrow invadopodial core. Interestingly, both Arl8/SKIP and FYCO1 have been implicated in LE/Lys tubulation, which increases LE/Lys motility (Mrakovic et al., 2012), suggesting a possible cross talk between LE/Lys translocation and tubulation that might facilitate invadopodia function.
Although extensively studied in cultured cells, we now know that invadopodia do form in vivo and facilitate metastases (Paterson and Courtneidge, 2018; Lohmer et al., 2014; Ngan et al., 2017; Sutoh Yoneyama et al., 2014; Gligorijevic et al., 2012). There are to date no available treatments for targeting invadopodia in cancer, but there is an increasing interest and effort to investigate this possibility. Ongoing research comprises drug repurposing, as well as inhibition of proteins important for invadopodia such as signaling components, ion channels, or MMPs (Leong et al., 2014; Meirson and Gil-Henn, 2018; Cathcart et al., 2015; Stoletov and Lewis, 2015; Paterson and Courtneidge, 2018). Our identification of the Protrudin pathway in invadopodia formation and function gives us new insight into the mechanism of invadopodia maturation and opens further possibilities for cancer treatment. We find that the overexpression of Protrudin in noninvasive RPE-1 cells induces invadopodia formation and invasion. Conversely, in highly invasive MDA-MB-231 cells, inhibition of the Protrudin pathway reduces their invasive potential. In line with this, survival analysis shows a higher risk for cancer patients with high Protrudin expression. Thus, the expression level of Protrudin and its related protein partners might serve as prognostic markers for the invasive potential of some cancer types.
To target the Protrudin pathway in future cancer treatment, we need to learn more about how this pathway is regulated. The pathway is dependent on endosomal PtdIns3P, which is produced by the PtdI ns3-kinase class III VPS34, in the presence of amino acids. This can in turn facilitate mTORC1 signaling by positioning of mTORC1-positive LE/Lys close to signaling hubs at the plasma membrane (Hong et al., 2017). In addition, the malonyl-coenzyme A sensor CPT1C stimulates the Protrudin pathway in nutrient-rich conditions (Palomo-Guerrero et al., 2019). Importantly, VPS34, CPT1C, and mTORC1 are all implicated in cancer development and progression (Melone et al., 2018; Saxton and Sabatini, 2017; Hirsch et al., 2010). It is reasonable to think that these regulators can influence the activity of the Protrudin pathway in invadopodia formation and function, depending on the extracellular environment of the tumor.
Materials and methods
Antibodies and reagents
The antibodies were obtained from the following resources. Mouse anti-LAMP1 (H4A3, immunofluorescence [IF] 1:400) was from the Developmental Studies Hybridoma Bank. Rabbit anti-Protrudin (12680–1-AP, Western blotting [WB] 1:7,500) was from PTG. Mouse anti-FYCO1 (H00079443-A01, IF 1:300, WB 1:1,000) was from Abnova. Rabbit anti-RAB7 (D95F2) XP (#9367, IF 1:50) was from Cell Signaling Technology. Rabbit anti-RAB7 (sc-10767, WB 1:1,000) was from Santa Cruz Biotechnology. Rabbit anti-LAMP1 (L1418, IF 1:400), mouse anti-β-actin (A5316, WB 1:5,000), mouse anti-vinculin (V9131, WB 1:3,000), rabbit anti-TKS5 (HPA037923, IF 1:100, 1:1,000), mouse anti-cortactin (05–180, IF 1:200), mouse anti-GFP (11814 460001, IF 1:500, WB 1:1,000), and mouse anti-MT1-MMP (MAB3328, IF 1:200, WB 1:500) were from Merck Life Science. Rabbit anti-collagen type I, cleavage site (Col1-3/4C; 0217–050, IF 1:500) was from immunoGlobe. Alexa Fluor 647–NHS-Ester (A20006), Rhodamine-phalloidin (R415), Alexa Fluor 647–phalloidin (A22287), and Alexa Fluor 488–phalloidin (A12379) were from Molecular Probes. Oregon Green 488 conjugate (G13186), Hoechst 33342 (H3570), and Calcein AM cell-permeant dye (C3100MP) were from Thermo Fisher Scientific/Life Technologies. Secondary antibodies were from Jackson ImmunoResearch, Molecular Probes, and LI-COR. HGF (H5791) was obtained from Sigma-Aldrich. The MMP inhibitor GM6001 (J65687.MX; Alfa Aesar, VWR International) and Src Tyr kinase inhibitor (In solution PP2- 529576; Calbiochem) were obtained from Merck Life Science.
Plasmids
pcDNA-pHluorin-MT1-MMP was a gift from Philippe Chavrier, Institut Curie, Paris, France (Lizárraga et al., 2009). The internal pHluorin was exchanged into pHuji by generating PCR fragments consisting of MT1-MMP-N-terminus, MT1-MMP-C-terminus, and pHuji, which were then inserted into a pENTR Gateway vector using Gibson assembly. pEGFP-N1-TKS5 was a gift from Philippe Chavrier. TKS5-GFP was inserted into a pENTR Gateway vector by restriction enzyme–based cloning. pEGFP-C2-Protrudin was based on the human cDNA clone BC030621 from ImaGene (described in Raiborg et al., 2015). GFP-Protrudin was made siRNA resistant by site-directed mutagenesis and then inserted into a pENTR Gateway vector by restriction enzyme–based cloning.
Cell culture
Cell lines were grown according to American Type Culture Collection (ATCC). MDA-MB-231 cells were maintained in DMEM (RPMI R2405; Merck Life Science) supplemented with 10% FCS (F7524; Merck Life Science), 2 mM glutamine (25030–024; Thermo Fisher Scientific), 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C with 5% CO2. hTERT-RPE-1 cells were maintained in DMEM/F12 (31331–028; Gibco-BRL), 10% FCS, 2 mM glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C with 5% CO2 in the presence of 0.01 mg/ml hygromycin B. Cell lines are authenticated by genotyping and regularly tested for mycoplasma contamination.
Generation of stable cell lines
hTERT-RPE1 and MDA-MB-231 cells stably expressing Protrudin, GFP-Protrudin, TKS5-GFP, or MT1-MMP-pHuji were generated using lentiviral transduction as described previously (Campeau et al., 2009). For all cell lines, a PGK promoter was used, which ensures moderate levels of expression. Third-generation lentiviral particles were generated as follows: GFP- and pHuji- fusion proteins were cloned into Gateway pENTR plasmids by conventional restriction enzyme–based cloning or Gibson assembly. From these vectors, lentiviral transfer vectors were generated by recombination into lentiviral Destination vectors derived from pLenti-CMV-Blast-DEST (plasmid number 17451; Addgene) or pCDH-PGK-MCS-IRES-PURO (SystemBioSciences) using Gateway LR reactions (Invitrogen). VSV-G pseudotyped lentiviral particles were packaged using a third-generation packaging system (plasmid numbers 12251, 12253, and 12259; Addgene; Dull et al., 1998). Cells were then transduced with virus particles, and stable expressing populations were generated by antibiotic selection. Some of the stable cell lines were sorted by flow cytometry to obtain pools of cells with suitable levels of expression. Detailed cloning procedures can be requested from the authors. We used the following stable cell lines: MDA-MB-231-GFP-Protrudin-sires#1, MDA-MB-231-Protrudin-KO#1-GFP-Protrudin, MDA-MB-231-Protrudin-KO#2-GFP-Protrudin, MDA-MB-231-MT1-MMP-pHuji-TKS5-GFP, RPE-1-Protrudin, and RPE-1-GFP-Protrudin-sires#1-MT1-MMP-pHuji.
CRISPR/Cas9-mediated deletion of ZFYVE27
gRNAs were designed using Bechling software (www.benchling.com). For deletion of ZFYVE27, a gRNA binding from TG of the start codon ATG in exon2 was chosen (gRNA2A: 5′-TGCAGACATCAGAACGTGAG-3′). The gRNA was cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 plasmid (Addgene). The derived PX459 plasmid was transfected into MDA-MB-231 cells using Lipofectamine LTX reagent (Invitrogen). 48 h after transfection, 1 µg/ml puromycin (Merck Life Science) was added to the cells. 7 h after treatment, cells were seeded in several dilutions to obtain single colonies, which were later picked and characterized. Clones lacking ZFYVE27 were identified by Western blot analysis. Two clones were chosen for further experiments, MDA-MB-231-ProtrudinKO#1 and MDA-MB-231-ProtrudinKO#2. The introduced mutations were characterized by genomic DNA extraction using GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific) followed by PCR, cloning, and sequencing. ZFYVE27 is found on chromosome 10 location 10q24.2. MDA-MB-231 cells have three copies of chromosome 10 (https://cansarblack.icr.ac.uk/cell-line/MDA-MB-231/copy-number). The sequencing results showed that both Protrudin KO clones were affected in all three copies. Two chromosomes had an insertion of the nucleotide T at position 16 after the ATG. This initiates a premature stop codon potentially generating a 6-amino-acid peptide. One chromosome had a deletion of the T in position 15 after the ATG. This initiates a premature stop codon potentially generating a 12-amino-acid peptide. The preference for changes in the nucleotide T and nonrandom outcomes at Cas9-mediated breaks has been described earlier (Taheri-Ghahfarokhi et al., 2018; van Overbeek et al., 2016).
siRNA transfections
RPE-1 cells were seeded and transfected with siRNA duplexes using Lipofectamine RNAiMax transfection reagent (13778; Thermo Fisher Scientific) following the manufacturer’s instructions. For MDA-MB-231 cells, reverse transfection was performed using RNAiMax. For both cell lines, the final concentration of siRNA duplexes was 10–20 nM. Cells were analyzed 24–96 h after transfection as indicated in the figure legends. The following human siRNA targeting sequences were used: Protrudin oligo #1, 5′-AGAAUGAGGUGCUGCGCAG-3′ (J-016349-12); Protrudin oligo #2, custom-made 5′-AACGGGTTCCTGAGCAAGAAT-3′ (Horizon/Dharmacon); FYCO1 oligo #1, Silencer custom-made 5′-CCAGUGACUGGUACUAUGC-3′ (Thermo Fisher Scientific); FYCO1 oligo #2, GCCCTAAGAATCAGAAGTA-3′ (J-014350-10; Horizon/Dharmacon); TKS5, 5′-CGACGGAACUCCUCCUUUA-3′ (J-006657-08; Horizon/Dharmacon); RAB7, 5′-CACGTAGGCCTTCAACACAAT-3′ (S102662240; Qiagen); SYT7, Silencer custom-made 5′-CCCTGAATGTCGAGGATAGTA-3′; and MT1-MMP/MMP14, 5′-GCAACAUAAUGAAAUCACU-3′ (s8879, Silencer select siRNA; Ambion/Thermo Fisher Scientific). Nontargeting control siRNA (for Protrudin and FYCO1#2; D-001810-01) was purchased from Horizon/Dharmacon, Silencer Negative Control (for FYCO1#1 and SYT7) was custom-made 5′-ACUUCGAGCGUGCAUGGCU-3′ (Thermo Fisher Scientific), Silencer Select Negative Control (for MT1-MMP; 4390844) was from Ambion, and AllStars Negative control (for RAB7; 1027281) was from Qiagen.
Quantitative RT-PCR
Total RNA was extracted using an RNeasy Plus mini kit (Qiagen). cDNA was synthesized using an iScript cDNA Synthesis Kit (Bio-Rad). Quantitative PCR was performed using the cDNA, SYBR Green I Master Mix (Roche), LightCycler 480 (Roche), and QuantiTect Primer Assays (QT00195601 for SYT7 and QT00000721 for TATA-binding protein [TBP]; Qiagen). Cycling conditions were 5 min at 95°C followed by 45 cycles for 10 s at 94°C, 20 s at 58°C, and 10 s at 72°C. A standard curve made from serial dilutions of cDNA was used to calculate the relative amount of the different cDNAs in each sample. SYT7 expression was normalized to the expression of the internal standard TBP.
Immunoblotting
Cells were washed with ice-cold PBS and lysed in 2× sample buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 200 mM DTT, and 0.004% bromophenol blue). Cell lysates were then subjected to SDS-PAGE on 10% (567–1034; Bio-Rad) or 4–20% (567–1094; Bio-Rad) gradient gels and blotted onto Immobilon-P membranes (IPVH00010; Merck Millipore). Membranes incubated with fluorescently labeled secondary antibodies (IRDye680 and IRDye800; LI-COR) were developed by Odyssey infrared scanner (LI-COR). Membranes detected with HRP-labeled secondary antibodies were developed using Clarity Western ECL substrate solutions (Bio-Rad) with a ChemiDoc XRS+ imaging system (Bio-Rad).
Immunostaining
Cells were seeded on glass coverslips, fixed with 3% formaldehyde (FA; 18814; Polysciences) for 15 min on ice (room temperature for the detection of TKS5), and permeabilized with 0.05% saponin (S7900; Sigma-Aldrich) in PBS. For cells grown on unlabeled or Oregon Green–labeled gelatin, 0.1% Triton X-100 (Sigma-Aldrich) was used instead of saponin. Fixed cells were then stained with primary antibodies at room temperature for 1 h, washed in PBS/saponin, stained with fluorescently labeled secondary antibody for 1 h, washed in PBS, and mounted with Mowiol containing 2 µg/ml Hoechst 33342 (H3570; Thermo Fisher Scientific). For the detection of endogenous FYCO1 and RAB7, cells were permeabilized for 5 min on ice with 0.05% saponin in PEM buffer (0.1 M Pipes, pH 6.95, P7643; Sigma-Aldrich), 2 mM EGTA, and 1 mM MgSO4 [105886; Merck Millipore]) before fixation in order to decrease the fluorescent signal from the cytosolic pool of the proteins (Simonsen et al., 1998). For Airyscan microscopy, nuclei were stained in PBS/Hoechst 33342 (2 µg/ml) for 10 min and mounted in ProLong Diamond (P36961; Thermo Fisher Scientific).
Confocal fluorescence microscopy, Airyscan microscopy, and image analyses
Confocal micrographs were obtained using an LSM710 or LSM780 confocal microscope (Carl Zeiss) equipped with an Ar-laser multiline (458/488/514 nm), a DPSS-561 10 (561 nm), a continuous-wave laser diode 405–30 CW (405 nm), and an HeNe laser (633 nm). The objective used was a Plan-Apochromat 63×/1.40 oil differential interference contrast (DIC) III (Carl Zeiss). Images were analyzed and adjusted (brightness/contrast) in ImageJ/Fiji (Schindelin et al., 2012) or Zen Blue. For superresolution microscopy, a Zeiss LSM 880 Airyscan (Carl Zeiss) was used with a Zeiss plan-apochromat 63× NA/1.40 oil DIC II objective (Carl Zeiss). The Airyscan detector was either in confocal or super-resolution mode, giving images with voxel size 0.0426 × 0.0426 × 0.1850 µm. Airyscan raw images were processed using Zen Blue and aligned in Zen Black. Images were further processed in ImageJ/Fiji (brightness/contrast; Schindelin et al., 2012) or 3D rendered using Imaris 7.7.2 (Bitplane). All images within one dataset were taken at fixed intensities below saturation, and identical settings were applied for all treatments within one experiment. In general, at least five (but often more) images were taken randomly throughout the coverslips. Manders colocalization coefficient was determined with the ImageJ plugin “JACoP” (Bolte and Cordelières, 2006). Specific analyses are described below.
Analysis of invadopodia reformation
Invadopodia reformation on glass coverslips
Cells were siRNA transfected and seeded onto coverslips. The cells were starved in serum-free medium (SFM) for 4 h. Src inhibitor (10 µm PP2) was added to the SFM the last 30 min to remove all invadopodia. Cells were washed twice in SFM before complete medium containing HGF (50 ng/ml) or SFM (negative control) was added to allow reformation of invadopodia. Upon 1 h of incubation, cells were fixed in room-temperature in 3% FA in PBS before staining with anti-TKS5 and anti-cortactin. Coverslips were either examined by high content microscopy using the Olympus ScanR illumination system with an UPLSAPO 40× objective or confocal microscopy (LSM710/780). Quantification of invadopodia number and size was done using ImageJ/Fiji (Schindelin et al., 2012) or the Olympus ScanR analyses software. Identical imaging and analysis settings were applied for all treatments within one experiment. In brief, ScanR analyses software was used for background correction (“rolling ball”) and automated image analyses. Fluorescent TKS5 dots were segmented by the ScanR software, and the number and area (pixels) of the dots were measured in each cell. The total number of cells was quantified by detection of Hoechst nuclear stain by the software. Due to the relatively low resolution obtained with the ScanR microscope, the number of TKS5-positive invadopodia per cell appears lower than in the analysis from the confocal micrographs. ImageJ/Fiji was used to measure the TKS5 and cortactin-positive invadopodia from confocal micrographs. Images were background subtracted using a rolling ball algorithm and a fixed threshold was used to segment TKS5-positive dots from all images. The number, area, and fluorescent intensities of the dots were measured by the software. The number of cells was quantified manually from the images using Hoechst nuclear stain.
Invadopodia reformation on unconjugated gelatin–coated coverslips
Cells treated with or without siRNA were trypsinized and seeded on coverslips coated with unconjugated gelatin in SFM. 2 h after seeding, the cells were treated with Src inhibitor (PP2, 10 µm) in SFM for 30 min to remove all invadopodia. Cells were washed twice with SFM before complete medium containing HGF (50 ng/ml) was added for 4 h to allow invadopodia reformation. The cells were fixed in 3% FA in PBS and immunostained with anti-TKS5 and anti-cortactin to detect reformed invadopodia. Samples were examined using confocal microscopy, and quantification was done as described for reformation on glass coverslips.
Measurements of invadopodia length
Cells were grown on Oregon Green–coated coverslips for 4 h, fixed in room-temperature in 3% FA for 15 min and stained with anti-TKS5 and phalloidin/Alexa Fluor 647 as described above. Nuclei were stained in PBS/Hoechst 33342 (2 µg/ml) for 10 min, and the cells were mounted in ProLong Diamond. To measure the length of invadopodia, z-stacks of individual cells were acquired using a Zeiss LSM 880 Airyscan microscope in confocal mode, with a step size of 0.24 µm. The average invadopodia lengths from individual cells were calculated by defining the first and the last section from each z-stack that clearly showed a typical invadopodia staining of both TKS5 and actin and multiplying the number of sections with the step size.
Intracellular positioning of MT1-MMP–positive endosomes
Confocal images of MDA-MB-231 parental or Protrudin-KO cells, immunostained with anti-MT1-MMP, were analyzed using ImageJ/Fiji as follows. MT1-MMP–positive vesicles were segmented by applying a manual threshold for each individual cell. The fluorescence intensity of perinuclear MT1-MMP was measured within a circle in the center of the cell, with a fixed size of 120 µm2, and presented as the percentage of the total fluorescent intensity of the segmented MT1-MMP in the whole cell.
MT1-MMP exocytosis
RPE-1 or MDA-MB-231 cells stably expressing GFP-Protrudin and MT1-MMP-pHuji or GFP-TKS5 and MT1-MMP-pHuji, respectively, were transfected with siRNA against Protrudin or control siRNA and seeded in MatTek dishes (Inter Instruments). Imaging was performed on a Deltavision OMX V4 microscope (GE Healthcare) using a 60× TIRF objective. Images were taken at 2 Hz for 2 min in the red channel. To document the expression of GFP- and pHuji-tagged proteins, the whole-cell volume was imaged by epifluorescence before TIRF microscopy. Images were deconvolved using softWoRx software (Applied Precision, GE Healthcare). Images were analyzed using ImageJ/Fiji (Schindelin et al., 2012).
For RPE-1-GFP-Protrudin sires#1-MT1-MMP-pHuji cells, exocytosis events were quantified by scoring bright dots that appeared and disappeared in the TIRF field within a few frames.
MT1-MMP surface accumulation in MDA-MB-231 cells was quantified by segmenting pHuji spots from TIRF images, which ensures visualization of the basal plasma membrane of the cells and exclusion of intracellular MT1-MMP signal. Due to variations in expression levels, the threshold was determined manually for each cell to identify bright MT1-MMP-pHuji–positive punctae. The MT1-MMP–positive area and the number of MT1-MMP–positive spots were quantified relative to cell area.
Coating of coverslips with gelatin
Oregon Green–conjugated gelatin-coated (Life Technologies) coverslips were prepared as previously described (Martin et al., 2012). In short, coverslips (12 mm diameter, No. 1 thickness; VWR International) were precleaned in 4% nitric acid for 30 min. After washing, the coverslips were coated with 50 µg/ml poly-L-lysine (Sigma-Aldrich) for 30 min, washed in PBS, and fixed with cold 0.5% glutaraldehyde (Sigma-Aldrich) in PBS for 15 min on ice. Subsequently, the coverslips were washed in PBS and coated for 20 min with preheated (44°C) 10 mg/ml unlabeled or Oregon Green–conjugated gelatin/2% sucrose in PBS. After coating, the coverslips were washed with PBS and incubated in 5 mg/ml sodium borohydride (Sigma-Aldrich) for 15 min. The coverslips were then washed with PBS, sterilized with 70% ethanol, and equilibrated in serum-containing medium for 1 h at 37°C before addition of cells.
Measurement of gelatin degradation
Cells were suspended in 1 ml culture medium and added to wells containing equilibrated gelatin-coated coverslips followed by incubation at 37°C for 4–6 h (time points indicated in the figure legends). Upon incubation, cells were fixed in 3% FA in PBS for 15 min, permeabilized with 0.1% Triton X-100 in PBS, incubated with Rhodamine-phalloidin and Hoechst 33342 for 15 min, and mounted for examination by confocal microscopy. Cells incubated with MMP inhibitor (GM6001) were seeded in complete growth medium containing the MMP inhibitor. For experiments using siRNA-mediated depletion of Protrudin, FYCO1, RAB7, or TKS5, measurements of gelatin degradation were performed 24–96 h after siRNA transfection, and the knockdown efficiencies were checked by Western blotting. Samples were analyzed using a LSM710 confocal microscope (Carl Zeiss); a 63× objective and zoom 1.0. Cells/field of imaging were chosen on basis of the nuclear staining and gelatin quality, and at least 20 images were randomly taken throughout the coverslips in each experiment, with typically 10–15 cells per image. All images within one experiment were taken with constant gain and pinhole parameters. Images were quantified using ImageJ/Fiji (Schindelin et al., 2012) by measuring the average area of gelatin degradation per cell as judged by black areas in the Oregon Green fluorescent gelatin. Due to variations in gelatin quality, the threshold was determined manually. The number of cells was counted manually based on Hoechst nuclear stain.
Quantification of pericellular collagenolysis
Cells were trypsinized and resuspended (2.5 × 105 cells/ml) in 0.2 ml pH-adjusted Alexa Fluor 647–labeled collagen-I solution (2.2 mg/ml, 734–1085; Corning) before 40 µl of the cell-collagen suspension was loaded on 18-mm glass coverslips (631–1567; VWR International) to make a 3D droplet as previously described (Rey et al., 2011; Monteiro et al., 2013). At least four droplets were made for each condition. Polymerization was induced for 30 min at 37°C before complete medium was carefully added, and collagen-embedded cells were incubated for 24 h at 37°C in 1% CO2. Droplets were fixed in 3% FA in PBS for 30 min at 37°C before samples were incubated with rabbit anti–Col1-3/4C antibody for 2 h at 4°C to detect pericellular cleaved collagen. Droplets were carefully washed with PBS and counterstained with secondary antibody, Alexa Fluor 488–phalloidin to stain actin and Hoechst 33342 to detect the nuclei. Image acquisition was performed using a Delta Vision Deconvolution microscope (Applied Precision, GE Healthcare) equipped with Elite TruLight Illumination System, a CoolSNAP HQ2camera and a 40× plan-apochromat lens. Images (30–50 z-sections 0.5 µm apart) were acquired and deconvolved using the softWoRx software (Applied Precision). For each droplet, ≥10 z-stacks were captured with 5–15 cells per image stack. All images within one experiment were taken with fixed imaging conditions. Maximum projections of z-stacks were quantified using ImageJ/Fiji (Schindelin et al., 2012). Images were background subtracted with a rolling ball algorithm, and a manual threshold was used to segment areas of pericellular collagenolysis based on areas positive for Col1-3/4C. The number of cells was counted manually based on actin and Hoechst nuclear stain and used to calculate the average area of pericellular collagenolysis per cell.
Inverted invasion assay
Matrigel
Inverted invasion assays were performed as described previously (Hennigan et al., 1994). In short, Matrigel (Corning) supplemented with 25 µg/ml fibronectin (Sigma-Aldrich) was added to Transwell filter inserts (8-µm pores; Sigma-Aldrich) and allowed to polymerize for 45 min at 37°C. The inserts were then inverted, and 4 × 104 cells were seeded on top of the filter on the opposite side of the Matrigel. Cells were allowed to adhere for 4 h before the inserts were turned and placed in SFM. The upper chamber was filled with serum-containing medium and HGF (100 ng/ml). 48–72 h after seeding (depending on the cell line, indicated in figure legends), cells were stained with Calcein AM (4 µM; Thermo Fisher Scientific) for 1 h before invading cells were visualized by confocal microscopy (LSM 710, ×20 objective; Carl Zeiss). Cells that did not pass the filter were removed with a tissue paper. Sections of 10-µm intervals were captured at five random locations for each insert, and three inserts were made for each condition. Images were analyzed with ImageJ/Fiji (Schindelin et al., 2012). Invasion is presented as the sum of pixels of all slides from >20 or 40 µm and beyond, divided by the sum of all pixels of all slides.
Collagen
Inverted invasion assays in collagen were performed and analyzed as described for the inverted invasion assay in Matrigel. The collagen matrix was prepared as follows: high-concentrated rat tail collagen I (734–1085; Corning) was mixed with 10× DMEM (Sigma-Aldrich), NaHCO3 (Merck Millipore), and dH2O and then transferred to a tube containing fibronectin (all on ice). Before the collagen matrix was applied to Transwell filter insets, the pH was controlled between 7 and 8, and the matrix was allowed to polymerize for 30–45 min at 37°C before cells were added.
Spheroid assay
RPE-1 and RPE-1-Protrudin–overexpressing cells were seeded (3 × 104 cells per well) in 96-well ultralow-attachment plates from Merck Life Science (CLS7007; Sigma-Aldrich) and allowed to make spheroids for 24 h. Then, Matrigel (Corning) supplemented with fibronectin was added and allowed to polymerize for 1 h before the spheroids were imaged by an OlympusIX81 microscope, using a positive-low (PL) 4× phase contrast objective (day 0). After imaging, complete medium containing HGF (100 ng/ml) was added, and the plates were placed in an incubator at 37°C. Spheroids were inspected and imaged every 24 h. Quantifications were done on images from day 2. Quantification of individual spheroids was done using ImageJ/Fiji (Schindelin et al., 2012). For the calculation of percent increase of spheroid area, the area (µm2) of the whole spheroid (as defined by the growth of the longest protrusions; area 1) and the area (µm2) of the dense spheroid core (area 2) were measured and presented as ((area 1/area 2) − 1) × 100. For the calculation of the distance of the longest protrusion, the length (µm) of the longest protrusion from the outer rim of the dense spheroid core was measured.
Calculation of the prognostic value of the Protrudin/ZFYVE27 gene
The prognostic impact of the Protrudin/ZFYVE27 gene in different cancer types was analyzed using the Kaplan-Meier Plotter (http://kmplot.com/analysis/), an online database containing gene expression profiles and survival data for cancer patients (Nagy et al., 2018). The patients were divided into a high-expression group and a low-expression group according to an auto selected cutoff performed by the Kaplan-Meier Plotter.
Statistical analysis and considerations
The number of individual experiments and the number of cells or images analyzed are indicated in the figure legends. The number of experiments was adapted to the expected effect size and the anticipated consistency between experiments. We tested our datasets for normal distribution by Kolmogorov-Smirnov, D’Agostino and Pearson, and Shapiro–Wilk normality tests, using GraphPad Prism Version 5.01. For parametric data, an unpaired two-sided t test was used to test two samples with equal variance, and a one-sample t test was used in the cases where the value of the control sample was set to 1. For more than two samples, we used one-way ANOVA with a suitable post hoc test. For nonparametric samples, Mann–Whitney test was used to test two samples and Kruskal–Wallis with Dunn’s post hoc test for more than two samples. All error bars denote mean values ± SD or SEM, as indicated in every figure legend (*, P < 0.05; **, P< 0.01; ***, P< 0.001). No samples were excluded from the analysis.
Online supplemental material
Fig. S1 shows a characterization of MDA-MB-231 cells stably expressing GFP-Protrudin. Fig. S2 shows that invadopodia reformation is impaired in Protrudin-depleted cells. Fig. S3 shows a characterization of Protrudin-KO cell lines with or without stable expression of GFP-Protrudin, antibody validation, and verification of siRNA-mediated protein depletion. Fig. S4 shows that GFP-Protrudin rescues the loss of gelatin degradation in Protrudin-KO cells. Fig. S5 shows that GFP-Protrudin expression rescues cell invasion of protrudin-KO cells and the prognostic value of protrudin in different cancer types. Video 1 shows how GFP-Protrudin makes contact with pHuji-MT1-MMP–positive endosomes.
Supplementary Material
Review History
Acknowledgments
We thank Viola Nähse for methodological teaching and supervision regarding the generation of CRISPR/Cas9-mediated deletion of ZFYVE27, Kay O. Schink for help measuring gelatin degradation using ImageJ/Fiji, and Anne Engen for expert help with cell cultures. The Core Facility for Advanced Light Microscopy at Oslo University Hospital is acknowledged for providing access to and training on relevant microscopes.
N.M. Pedersen is a postdoctoral fellow, and L. Wang holds a technical position funded by Kreftforeningen (project number 198140). E.M. Wenzel is a research fellow of InvaCell. This work was partly supported by a donation from Trond Paulsen (InvaCell), by Kreftforeningen (project number 198140), and by Norges Forskningsråd through its Centers of Excellence funding scheme (project number 262652).
The authors declare no competing financial interests.
Author contributions: N.M. Pedersen and E.M. Wenzel contributed to conceptualization, formal analysis, investigation, methodology, validation, visualization, and review and editing. L. Wang contributed to investigation, methodology, validation, visualization, and review and editing. S. Antoine contributed to methodology. P. Chavrier contributed to methodology, supervision, and review and editing. H. Stenmark contributed to conceptualization, funding acquisition, resources, supervision, and review and editing. C. Raiborg conceived the study and contributed to conceptualization, formal analysis, investigation, methodology, validation, visualization, the original draft, review and editing, funding acquisition, supervision, and project administration.
References
- Arantes R.M.E., and Andrews N.W.. 2006. A role for synaptotagmin VII-regulated exocytosis of lysosomes in neurite outgrowth from primary sympathetic neurons. J. Neurosci. 26:4630–4637. 10.1523/JNEUROSCI.0009-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artym V.V., Zhang Y., Seillier-Moiseiwitsch F., Yamada K.M., and Mueller S.C.. 2006. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 66:3034–3043. 10.1158/0008-5472.CAN-05-2177 [DOI] [PubMed] [Google Scholar]
- Ballabio A., and Bonifacino J.S.. 2020. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 21:101–118. 10.1038/s41580-019-0185-4 [DOI] [PubMed] [Google Scholar]
- Beaty B.T., and Condeelis J.. 2014. Digging a little deeper: the stages of invadopodium formation and maturation. Eur. J. Cell Biol. 93:438–444. 10.1016/j.ejcb.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., and Cordelières F.P.. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 224(Pt 3):213–232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Branch K.M., Hoshino D., and Weaver A.M.. 2012. Adhesion rings surround invadopodia and promote maturation. Biol. Open. 1:711–722. 10.1242/bio.20121867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeau E., Ruhl V.E., Rodier F., Smith C.L., Rahmberg B.L., Fuss J.O., Campisi J., Yaswen P., Cooper P.K., and Kaufman P.D.. 2009. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 4 e6529 10.1371/journal.pone.0006529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagnino A., Castro-Castro A., Irondelle M., Guichard A., Lodillinsky C., Fuhrmann L., Vacher S., Agüera-González S., Zagryazhskaya-Masson A., Romao M., et al. . 2018. Coronin 1C promotes triple-negative breast cancer invasiveness through regulation of MT1-MMP traffic and invadopodia function. Oncogene. 37:6425–6441. 10.1038/s41388-018-0422-x [DOI] [PubMed] [Google Scholar]
- Castro-Castro A., Marchesin V., Monteiro P., Lodillinsky C., Rossé C., and Chavrier P.. 2016. Cellular and Molecular Mechanisms of MT1-MMP-Dependent Cancer Cell Invasion. Annu. Rev. Cell Dev. Biol. 32:555–576. 10.1146/annurev-cellbio-111315-125227 [DOI] [PubMed] [Google Scholar]
- Cathcart J., Pulkoski-Gross A., and Cao J.. 2015. Targeting matrix metalloproteinases in cancer: Bringing new life to old ideas. Genes Dis. 2:26–34. 10.1016/j.gendis.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers A.F., Groom A.C., and MacDonald I.C.. 2002. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer. 2:563–572. 10.1038/nrc865 [DOI] [PubMed] [Google Scholar]
- Chang J., Lee S., and Blackstone C.. 2013. Protrudin binds atlastins and endoplasmic reticulum-shaping proteins and regulates network formation. Proc. Natl. Acad. Sci. USA. 110:14954–14959. 10.1073/pnas.1307391110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C., Collin G., Descamps S., Touaitahuata H., Simon V., Reymond N., Fernandez L., Milhiet P.-E., Georget V., Urbach S., et al. . 2016. TOM1L1 drives membrane delivery of MT1-MMP to promote ERBB2-induced breast cancer cell invasion. Nat. Commun. 7:10765 10.1038/ncomms10765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino J., Henriet E., Ezzoukhry Z., Goetz J.G., Moreau V., and Saltel F.. 2016. The microenvironment controls invadosome plasticity. J. Cell Sci. 129:1759–1768. 10.1242/jcs.182329 [DOI] [PubMed] [Google Scholar]
- Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D., and Naldini L.. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463–8471. 10.1128/JVI.72.11.8463-8471.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer N., Rebollo E., Domínguez P., Elkhatib N., Chavrier P., Daviet L., González C., and González-Gaitán M.. 2007. Spermatocyte cytokinesis requires rapid membrane addition mediated by ARF6 on central spindle recycling endosomes. Development. 134:4437–4447. 10.1242/dev.010983 [DOI] [PubMed] [Google Scholar]
- Dykes S.S., Gray A.L., Coleman D.T., Saxena M., Stephens C.A., Carroll J.L., Pruitt K., and Cardelli J.A.. 2016. The Arf-like GTPase Arl8b is essential for three-dimensional invasive growth of prostate cancer in vitro and xenograft formation and growth in vivo. Oncotarget. 7:31037–31052. 10.18632/oncotarget.8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy R.J., Weidmann M.D., Sharma V.P., and Condeelis J.S.. 2017. Tumor Cell Invadopodia: Invasive Protrusions that Orchestrate Metastasis. Trends Cell Biol. 27:595–607. 10.1016/j.tcb.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipek P.A., De Araujo M.E.G., Vogel G.F., De Smet C.H., Eberharter D., Rebsamen M., Rudashevskaya E.L., Kremser L., Yordanov T., Tschaikner P., et al. . 2017. LAMTOR/Ragulator is a negative regulator of Arl8b- and BORC-dependent late endosomal positioning. J. Cell Biol. 216:4199–4215. 10.1083/jcb.201703061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frittoli E., Palamidessi A., Marighetti P., Confalonieri S., Bianchi F., Malinverno C., Mazzarol G., Viale G., Martin-Padura I., Garré M., et al. . 2014. A RAB5/RAB4 recycling circuitry induces a proteolytic invasive program and promotes tumor dissemination. J. Cell Biol. 206:307–328. 10.1083/jcb.201403127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gligorijevic B., Wyckoff J., Yamaguchi H., Wang Y., Roussos E.T., and Condeelis J.. 2012. N-WASP-mediated invadopodium formation is involved in intravasation and lung metastasis of mammary tumors. J. Cell Sci. 125:724–734. 10.1242/jcs.092726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., and Szallasi Z.. 2010. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123:725–731. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- Györffy B., Lánczky A., and Szállási Z.. 2012. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer. 19:197–208. 10.1530/ERC-11-0329 [DOI] [PubMed] [Google Scholar]
- Hagedorn E.J., Ziel J.W., Morrissey M.A., Linden L.M., Wang Z., Chi Q., Johnson S.A., and Sherwood D.R.. 2013. The netrin receptor DCC focuses invadopodia-driven basement membrane transmigration in vivo. J. Cell Biol. 201:903–913. 10.1083/jcb.201301091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennigan R.F., Hawker K.L., and Ozanne B.W.. 1994. Fos-transformation activates genes associated with invasion. Oncogene. 9:3591–3600. [PubMed] [Google Scholar]
- Hirsch D.S., Shen Y., Dokmanovic M., and Wu W.J.. 2010. pp60c-Src phosphorylates and activates vacuolar protein sorting 34 to mediate cellular transformation. Cancer Res. 70:5974–5983. 10.1158/0008-5472.CAN-09-2682 [DOI] [PubMed] [Google Scholar]
- Hong Z., Pedersen N.M., Wang L., Torgersen M.L., Stenmark H., and Raiborg C.. 2017. PtdIns3P controls mTORC1 signaling through lysosomal positioning. J. Cell Biol. 216:4217–4233. 10.1083/jcb.201611073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino D., Kirkbride K.C., Costello K., Clark E.S., Sinha S., Grega-Larson N., Tyska M.J., and Weaver A.M.. 2013. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 5:1159–1168. 10.1016/j.celrep.2013.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A., and Prekeris R.. 2015. The regulation of MMP targeting to invadopodia during cancer metastasis. Front. Cell Dev. Biol. 3:4–4. 10.3389/fcell.2015.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R. 2003. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat. Rev. Cancer. 3:422–433. 10.1038/nrc1094 [DOI] [PubMed] [Google Scholar]
- Kelley L.C., Chi Q., Cáceres R., Hastie E., Schindler A.J., Jiang Y., Matus D.Q., Plastino J., and Sherwood D.R.. 2019. Adaptive F-Actin Polymerization and Localized ATP Production Drive Basement Membrane Invasion in the Absence of MMPs. Dev. Cell. 48:313–328.e8. 10.1016/j.devcel.2018.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., and Takahashi K.. 2008. WAVE2- and microtubule-dependent formation of long protrusions and invasion of cancer cells cultured on three-dimensional extracellular matrices. Cancer Sci. 99:2252–2259. 10.1111/j.1349-7006.2008.00927.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Das A., Barai A., and Sen S.. 2018. MMP Secretion Rate and Inter-invadopodia Spacing Collectively Govern Cancer Invasiveness. Biophys. J. 114:650–662. 10.1016/j.bpj.2017.11.3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T., and Wieschaus E.. 2000. Polarized insertion of new membrane from a cytoplasmic reservoir during cleavage of the Drosophila embryo. J. Cell Biol. 150:849–860. 10.1083/jcb.150.4.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong H.S., Robertson A.E., Stoletov K., Leith S.J., Chin C.A., Chien A.E., Hague M.N., Ablack A.L., Simmen K.C., Mcpherson V.A., et al. . 2014. 578 Invadopodia are required for cancer cell extravasation and are a therapeutic target for metastasis. Eur. J. Cancer. 50:187 10.1016/S0959-8049(14)70704-X [DOI] [PubMed] [Google Scholar]
- Lizárraga F., Poincloux R., Romao M., Montagnac G., Le Dez G., Bonne I., Rigaill G., Raposo G., and Chavrier P.. 2009. Diaphanous-related formins are required for invadopodia formation and invasion of breast tumor cells. Cancer Res. 69:2792–2800. 10.1158/0008-5472.CAN-08-3709 [DOI] [PubMed] [Google Scholar]
- Lohmer L.L., Kelley L.C., Hagedorn E.J., and Sherwood D.R.. 2014. Invadopodia and basement membrane invasion in vivo. Cell Adhes. Migr. 8:246–255. 10.4161/cam.28406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson I.R., Rainero E., Mitchell L.E., van den Berghe P.V.E., Speirs C., Dozynkiewicz M.A., Chaudhary S., Kalna G., Edwards J., Timpson P., et al. . 2014. CLIC3 controls recycling of late endosomal MT1-MMP and dictates invasion and metastasis in breast cancer. J. Cell Sci. 127:3893–3901. 10.1242/jcs.135947 [DOI] [PubMed] [Google Scholar]
- Martin K.H., Hayes K.E., Walk E.L., Ammer A.G., Markwell S.M., and Weed S.A.. 2012. Quantitative Measurement of Invadopodia-mediated Extracellular Matrix Proteolysis in Single and Multicellular Contexts. J. Vis. Exp. 66 e4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesin V., Castro-Castro A., Lodillinsky C., Castagnino A., Cyrta J., Bonsang-Kitzis H., Fuhrmann L., Irondelle M., Infante E., Montagnac G., et al. . 2015. ARF6-JIP3/4 regulate endosomal tubules for MT1-MMP exocytosis in cancer invasion. J. Cell Biol. 211:339–358. 10.1083/jcb.201506002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirson T., and Gil-Henn H.. 2018. Targeting invadopodia for blocking breast cancer metastasis. Drug Resist. Updat. 39:1–17. 10.1016/j.drup.2018.05.002 [DOI] [PubMed] [Google Scholar]
- Melone M.A.B., Valentino A., Margarucci S., Galderisi U., Giordano A., and Peluso G.. 2018. The carnitine system and cancer metabolic plasticity. Cell Death Dis. 9:228–228. 10.1038/s41419-018-0313-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro P., Rossé C., Castro-Castro A., Irondelle M., Lagoutte E., Paul-Gilloteaux P., Desnos C., Formstecher E., Darchen F., Perrais D., et al. . 2013. Endosomal WASH and exocyst complexes control exocytosis of MT1-MMP at invadopodia. J. Cell Biol. 203:1063–1079. 10.1083/jcb.201306162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouw J.K., Ou G., and Weaver V.M.. 2014. Extracellular matrix assembly: a multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 15:771–785. 10.1038/nrm3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrakovic A., Kay J.G., Furuya W., Brumell J.H., and Botelho R.J.. 2012. Rab7 and Arl8 GTPases are necessary for lysosome tubulation in macrophages. Traffic. 13:1667–1679. 10.1111/tra.12003 [DOI] [PubMed] [Google Scholar]
- Murphy D.A., and Courtneidge S.A.. 2011. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12:413–426. 10.1038/nrm3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegeli K.M., Hastie E., Garde A., Wang Z., Keeley D.P., Gordon K.L., Pani A.M., Kelley L.C., Morrissey M.A., Chi Q., et al. . 2017. Cell Invasion In Vivo via Rapid Exocytosis of a Transient Lysosome-Derived Membrane Domain. Dev. Cell. 43:403–417.e10. 10.1016/j.devcel.2017.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Á., Lánczky A., Menyhárt O., and Győrffy B.. 2018. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 8:9227 10.1038/s41598-018-27521-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngan E., Stoletov K., Smith H.W., Common J., Muller W.J., Lewis J.D., and Siegel P.M.. 2017. LPP is a Src substrate required for invadopodia formation and efficient breast cancer lung metastasis. Nat. Commun. 8:15059 10.1038/ncomms15059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomo-Guerrero M., Fadó R., Casas M., Pérez-Montero M., Baena M., Helmer P.O., Domínguez J.L., Roig A., Serra D., Hayen H., et al. . 2019. Sensing of nutrients by CPT1C regulates late endosome/lysosome anterograde transport and axon growth. eLife. 8 e51063 10.7554/eLife.51063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh A., and Weaver A.M.. 2016. Regulation of invadopodia by mechanical signaling. Exp. Cell Res. 343:89–95. 10.1016/j.yexcr.2015.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson E.K., and Courtneidge S.A.. 2018. Invadosomes are coming: new insights into function and disease relevance. FEBS J. 285:8–27. 10.1111/febs.14123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Schindler C., Jia R., Jarnik M., Backlund P., and Bonifacino J.S.. 2015. BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell. 33:176–188. 10.1016/j.devcel.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Guardia C.M., Keren-Kaplan T., and Bonifacino J.S.. 2016. Mechanisms and functions of lysosome positioning. J. Cell Sci. 129:4329–4339. 10.1242/jcs.196287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Keren-Kaplan T., and Bonifacino J.S.. 2017. A Ragulator-BORC interaction controls lysosome positioning in response to amino acid availability. J. Cell Biol. 216:4183–4197. 10.1083/jcb.201703094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C. 2018. How Nutrients Orchestrate Lysosome Positioning. Contact (Thousand Oaks). 1 2515256418756111. [Google Scholar]
- Raiborg C., Wenzel E.M., Pedersen N.M., Olsvik H., Schink K.O., Schultz S.W., Vietri M., Nisi V., Bucci C., Brech A., et al. . 2015. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 520:234–238. 10.1038/nature14359 [DOI] [PubMed] [Google Scholar]
- Raiborg C., Wenzel E.M., Pedersen N.M., and Stenmark H.. 2016. ER-endosome contact sites in endosome positioning and protrusion outgrowth. Biochem. Soc. Trans. 44:441–446. 10.1042/BST20150246 [DOI] [PubMed] [Google Scholar]
- Rao S.K., Huynh C., Proux-Gillardeaux V., Galli T., and Andrews N.W.. 2004. Identification of SNAREs involved in synaptotagmin VII-regulated lysosomal exocytosis. J. Biol. Chem. 279:20471–20479. 10.1074/jbc.M400798200 [DOI] [PubMed] [Google Scholar]
- Reddy A., Caler E.V., and Andrews N.W.. 2001. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 106:157–169. 10.1016/S0092-8674(01)00421-4 [DOI] [PubMed] [Google Scholar]
- Rey M., Irondelle M., Waharte F., Lizarraga F., and Chavrier P.. 2011. HDAC6 is required for invadopodia activity and invasion by breast tumor cells. Eur. J. Cell Biol. 90:128–135. 10.1016/j.ejcb.2010.09.004 [DOI] [PubMed] [Google Scholar]
- Rossé C., Lodillinsky C., Fuhrmann L., Nourieh M., Monteiro P., Irondelle M., Lagoutte E., Vacher S., Waharte F., Paul-Gilloteaux P., et al. . 2014. Control of MT1-MMP transport by atypical PKC during breast-cancer progression. Proc. Natl. Acad. Sci. USA. 111:E1872–E1879. 10.1073/pnas.1400749111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe R.G., and Weiss S.J.. 2008. Breaching the basement membrane: who, when and how? Trends Cell Biol. 18:560–574. 10.1016/j.tcb.2008.08.007 [DOI] [PubMed] [Google Scholar]
- Saita S., Shirane M., Natume T., Iemura S., and Nakayama K.I.. 2009. Promotion of neurite extension by protrudin requires its interaction with vesicle-associated membrane protein-associated protein. J. Biol. Chem. 284:13766–13777. 10.1074/jbc.M807938200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R.A., and Sabatini D.M.. 2017. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 168:960–976. 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher M., Goldman R.D., Louvard D., and Vignjevic D.M.. 2010. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 189:541–556. 10.1083/jcb.200909113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals D.F., Azucena E.F. Jr., Pass I., Tesfay L., Gordon R., Woodrow M., Resau J.H., and Courtneidge S.A.. 2005. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell. 7:155–165. 10.1016/j.ccr.2005.01.006 [DOI] [PubMed] [Google Scholar]
- Sharma P., Parveen S., Shah L.V., Mukherjee M., Kalaidzidis Y., Kozielski A.J., Rosato R., Chang J.C., and Datta S.. 2020. SNX27-retromer assembly recycles MT1-MMP to invadopodia and promotes breast cancer metastasis. J. Cell Biol. 219:219 10.1083/jcb.201812098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Bremnes B., Rønning E., Aasland R., and Stenmark H.. 1998. Syntaxin-16, a putative Golgi t-SNARE. Eur. J. Cell Biol. 75:223–231. 10.1016/S0171-9335(98)80116-7 [DOI] [PubMed] [Google Scholar]
- Siqueira A.S., Pinto M.P., Cruz M.C., Smuczek B., Cruz K.S.P., Barbuto J.A.M., Hoshino D., Weaver A.M., Freitas V.M., and Jaeger R.G.. 2016. Laminin-111 peptide C16 regulates invadopodia activity of malignant cells through β1 integrin, Src and ERK 1/2. Oncotarget. 7:47904–47917. 10.18632/oncotarget.10062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneeggen M., Pedersen N.M., Campsteijn C., Haugsten E.M., Stenmark H., and Schink K.O.. 2019. WDFY2 restrains matrix metalloproteinase secretion and cell invasion by controlling VAMP3-dependent recycling. Nat. Commun. 10:2850 10.1038/s41467-019-10794-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A., Le Dez G., Poincloux R., Recchi C., Nassoy P., Rottner K., Galli T., and Chavrier P.. 2008. MT1-MMP-dependent invasion is regulated by TI-VAMP/VAMP7. Curr. Biol. 18:926–931. 10.1016/j.cub.2008.05.044 [DOI] [PubMed] [Google Scholar]
- Stoletov K., and Lewis J.D.. 2015. Invadopodia: a new therapeutic target to block cancer metastasis. Expert Rev. Anticancer Ther. 15:733–735. 10.1586/14737140.2015.1058711 [DOI] [PubMed] [Google Scholar]
- Sutoh Yoneyama M., Hatakeyama S., Habuchi T., Inoue T., Nakamura T., Funyu T., Wiche G., Ohyama C., and Tsuboi S.. 2014. Vimentin intermediate filament and plectin provide a scaffold for invadopodia, facilitating cancer cell invasion and extravasation for metastasis. Eur. J. Cell Biol. 93:157–169. 10.1016/j.ejcb.2014.03.002 [DOI] [PubMed] [Google Scholar]
- Szász A.M., Lánczky A., Nagy Á., Förster S., Hark K., Green J.E., Boussioutas A., Busuttil R., Szabó A., and Győrffy B.. 2016. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 7:49322–49333. 10.18632/oncotarget.10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri-Ghahfarokhi A., Taylor B.J.M., Nitsch R., Lundin A., Cavallo A.L., Madeyski-Bengtson K., Karlsson F., Clausen M., Hicks R., Mayr L.M., et al. . 2018. Decoding non-random mutational signatures at Cas9 targeted sites. Nucleic Acids Res. 46:8417–8434. 10.1093/nar/gky653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek M., Capurso D., Carter M.M., Thompson M.S., Frias E., Russ C., Reece-Hoyes J.S., Nye C., Gradia S., Vidal B., et al. . 2016. DNA Repair Profiling Reveals Nonrandom Outcomes at Cas9-Mediated Breaks. Mol. Cell. 63:633–646. 10.1016/j.molcel.2016.06.037 [DOI] [PubMed] [Google Scholar]
- Chen W.-T., Lee C.-C., Goldstein L., Bernier S., Liu C.H.L., Lin C.-Y., Yeh Y., Monsky W.L., Kelly T., Dai M., et al. . 1994. Membrane proteases as potential diagnostic and therapeutic targets for breast malignancy. Breast Cancer Res. Treat. 31:217–226. 10.1007/BF00666155 [DOI] [PubMed] [Google Scholar]
- Willett R., Martina J.A., Zewe J.P., Wills R., Hammond G.R.V., and Puertollano R.. 2017. TFEB regulates lysosomal positioning by modulating TMEM55B expression and JIP4 recruitment to lysosomes. Nat. Commun. 8:1580 10.1038/s41467-017-01871-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K.C., and Coppolino M.G.. 2011. Phosphorylation of membrane type 1-matrix metalloproteinase (MT1-MMP) and its vesicle-associated membrane protein 7 (VAMP7)-dependent trafficking facilitate cell invasion and migration. J. Biol. Chem. 286:43405–43416. 10.1074/jbc.M111.297069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Zech T., McDonald L., Gonzalez E.G., Li A., Macpherson I., Schwarz J.P., Spence H., Futó K., Timpson P., et al. . 2012. N-WASP coordinates the delivery and F-actin-mediated capture of MT1-MMP at invasive pseudopods. J. Cell Biol. 199:527–544. 10.1083/jcb.201203025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Review History