Smad4 Dependency Defines Two Classes of Transforming Growth Factor β (TGF-β) Target Genes and Distinguishes TGF-β-Induced Epithelial-Mesenchymal Transition from Its Antiproliferative and Migratory Responses (original) (raw)
Abstract
In response to transforming growth factor β (TGF-β), Smad4 forms complexes with activated Smad2 and Smad3, which accumulate in the nucleus, where they both positively and negatively regulate TGF-β target genes. Mutation or deletion of Smad4 is found in about 50% of pancreatic tumors and in about 15% of colorectal tumors. As Smad4 is a central component of the TGF-β/Smad pathway, we have determined whether Smad4 is absolutely required for all TGF-β responses, to evaluate the effect of its loss during human tumor development. We have generated cell lines from the immortalized human keratinocyte cell line HaCaT or the pancreatic tumor cell line Colo-357, which stably express a tetracyline-inducible small interfering RNA targeted against Smad4. In response to tetracycline, Smad4 expression is effectively silenced. Large-scale microarray analysis identifies two populations of TGF-β target genes that are distinguished by their dependency on Smad4. Some genes absolutely require Smad4 for their regulation, while others do not. Functional analysis also indicates a differential Smad4 requirement for TGF-β-induced functions; TGF-β-induced cell cycle arrest and migration, but not epithelial-mesenchymal transition, are abolished after silencing of Smad4. Altogether our results suggest that loss of Smad4 might promote TGF-β-mediated tumorigenesis by abolishing tumor-suppressive functions of TGF-β while maintaining some tumor-promoting TGF-β responses.
Smad4 is a central component of the Smad pathway that transduces signals from transforming growth factor β (TGF-β) superfamily members (55, 59). Upon ligand stimulation, it forms complexes with the receptor-regulated Smads (Smad2 and Smad3 in the case of TGF-β), that are phosphorylated by the activated receptor complex. These Smad complexes accumulate in the nucleus, where they are directly involved in the regulation of transcription of target genes in conjunction with other transcription factors. It is now appreciated that the activated receptor complex can also signal through other pathways, such as those involving extracellular signal-regulated kinase 1/2 (Erk1/2), p38, or Jun N-terminal protein kinase (JNK) (27) or those involving RhoA, phosphatidylinositol 3-kinase (PI 3-kinase), Pak2, or Par6 (4, 16, 44, 65, 69). In many cases, it is not clear whether the activation of these pathways is induced directly by the activated receptors or whether it is indirect and requires the Smad pathway (14). It is also possible that additional Smad-independent pathways exist that are activated directly in response to TGF-β stimulation.
The gene encoding Smad4 was originally identified as a tumor suppressor at 18q21.1 (24). Allelic loss of 18q has been found in about 90% of pancreatic cancers, and indeed Smad4 has now been shown to be mutated or deleted in about 50% pancreatic cancers and 15% of colorectal tumors (1) and recently in 10 out of 41 cervical carcinomas (2). Germ line mutations in Smad4 also occur in a subset of patients with juvenile polyposis (28). Identification of Smad4 as a tumor suppressor supports the evidence that the TGF-β pathway has tumor-suppressive activities, thought to arise primarily from its ability to cause cell cycle arrest and apoptosis of epithelial cells (13). Indeed, somatic inactivating mutations or deletions have also been found in the genes encoding TβRI, TβRII, and Smad2 in various human tumors (1). In mice, the tumor-suppressive function of TGF-β has also been deduced from the observation that tumor incidence is increased in mice with deletion of one allele of the TGF-β1 gene (58) or in mice that express dominant-negative TβRII (5). In addition, TGF-β also has tumor-promoting activities. These can occur through effects of TGF-β on the tumor cells themselves. TGF-β, in conjunction with the Ras/ERK pathway, induces an epithelial-mesechymal transition (EMT) (43), which makes cells more migratory and invasive (60) and might contribute to invasion and metastasis. For example, in transgenic mice overexpressing TGF-β1 either in skin (8) or in liver (18) the malignant conversion rate is greatly increased. Moreover, a high percentage of human tumors overexpress TGF-β1, and this excess production of TGF-β1 appears to be associated with poor prognosis (62, 64). In addition to direct effects on the tumor cells, TGF-β produced by the tumor cells may have effects on the tumor environment. The high level of TGF-β in tumors is associated with enhanced angiogenesis (10) and immune suppression (61).
It is unclear whether the loss of Smad4 in tumor cells only results in the loss of the tumor-suppressive activities of TGF-β or whether it is also directly involved in TGF-β-driven tumor promotion. Since Smad4 appeared to be a central component of the TGF-β pathway, it was initially accepted that loss of Smad4 would completely abolish TGF-β responses (15, 72). However, there is increasing evidence to suggest that Smad4 is in fact dispensable for some TGF-β responses (7, 21, 29, 33, 36). A number of studies have attempted to address the role of Smad4 in TGF-β signaling, either by overexpression of Smad4 in Smad4-null tumor cell lines (9, 15, 48, 51, 52) or by knockdown or conditional knockout of Smad4 (7, 33, 36, 39, 56, 73). However, the importance of Smad4 in TGF-β signaling and its requirement for different TGF-β-induced functions remain unclear, as these diverse studies have yielded contradictory results.
Here we have sought to identify to what extent Smad4 is required for TGF-β to regulate target gene transcription and functional responses. We have used a tetracycline (Tet)-inducible small interfering RNA (siRNA) approach (63) to inhibit Smad4 function in epithelial cells and to investigate the consequence of the loss of Smad4 on TGF-β responses. A major advantage of this system, which distinguishes it from previous knockdown/knockout approaches, is that the cells do not have to grow for long periods without Smad4. This means that they will not have the opportunity to accumulate mutations or adjust expression levels of genes that might compensate for the loss of Smad4. We have generated Smad4-siRNA Tet-inducible clonal cell lines in both human immortalized keratinocytes (HaCaT) and human pancreatic tumor cells (Colo-357), which exhibit a very strong silencing of Smad4 after 48 h of treatment with Tet. Large-scale microarray analysis of TGF-β-induced target genes in the Smad4-siRNA Tet-inducible HaCaT cells has enabled us to identify different categories of TGF-β target genes with respect to their dependence on Smad4. Indeed, many TGF-β target genes are not affected by the knockdown of Smad4, while others display a dramatic Smad4 dependency. Functional analysis also indicates that Smad4 is involved in only a subset of TGF-β responses. We show that Smad4 is necessary for TGF-β-induced cell cycle arrest and migration but is not involved in the TGF-β-induced EMT. We propose that the loss of Smad4 in cancer might contribute to tumorigenesis by modulating the TGF-β response so that tumor-suppressive activities are lost but responses such as EMT that could be involved in tumor promotion are retained.
MATERIALS AND METHODS
Plasmids.
The pTER siRNA Tet-inducible vector was obtained from Hans Clevers, and cloning was performed as described previously (63). Briefly, the Smad4 siRNA Tet-inducible expression vector pTER-S4 was generated by cloning the following annealed oligonucleotides into pTER: 5′GATCCCCGGTGGAGAGAGTGAAACATTTCAAGAGAATGTTTCACTCTCTCCACCTTTTTGGAAA3′ and 5′AGCTTTTCCAAAAAGGTGGAGAGAGTGAAACATTCTCTTGAAATGTTTCACTCTCTCCACCGGG3′. This generates an siRNA directed against the sequence GGTGGAGAGAGTGAAACAT, corresponding to nucleotides 85 to 103 of Smad4 relative to the ATG. The Tet-inducible hemagglutinin (HA)-tagged Smad4 siRNA-resistant expression vector pcDNA5/TO-HA-S4res was generated by cloning an HA-tagged Smad4, in which the GAGT sequence recognized by the siRNA (at positions 93 to 96 relative to the ATG) was changed to ATCC, into pcDNA5/TO (Invitrogen). These changes do not change the amino acid sequence of Smad4. The reporters CAGA12-Luc, ARE-Luc, and DE-Luc; the XFoxH1a and Mixer expression vectors; and the EF-LacZ control plasmid were described previously (12, 46). The c-junSBR6-Luc reporter was constructed by replacing the three activin response elements (AREs) in ARE-Luc with six copies of the c-jun SBR (30).
Cell culture, transfections, luciferase assays, and establishment of Tet-inducible Smad4 siRNA cell lines.
The HaCaT and Colo-357 cell lines were grown in Dulbecco's modified Eagle's medium and RPMI medium, respectively, supplemented with 10% fetal calf serum. The HaCaT-TR or Colo-TR parental cell lines—which are HaCaT and Colo-357 cells, respectively, that stably express the Tet repressor (TR); the HaCaT-TRS4 and Colo-TRS4 cell lines that stably express the Tet-inducible Smad4 siRNA in addition to the Tet repressor; and the HaCaT-TRS4-rescue cell line in which Smad4 expression is rescued by a Tet-inducible HA-Smad4, which is resistant to the siRNA—were established as described elsewhere (see experimental procedures in the supplemental material). For Tet induction, cells were grown for 48 h in medium containing 10% Tet-approved serum (BD Biosciences) in the presence or absence of Tet (2 μg/ml). TGF-β inductions were carried out at indicated times by adding 2 ng/ml of TGF-β (PeproTech). Transfections were performed using the FuGENE6 transfection reagent (Roche Diagnostics) according to the manufacturer's protocol. For luciferase assays, Tet was added during transfection and cells were incubated for 40 h followed by TGF-β induction for 8 h. Cells were then lysed and assayed for luciferase activity with the Promega luciferase assay system and normalized to β-galactosidase activity expressed from a cotransfected LacZ plasmid control.
Microarray procedures and analysis.
Total RNA was extracted with Trizol (Invitrogen) followed by a purification step on an RNeasy mini column (QIAGEN). Biotinylated cRNA probes were generated from 10 μg total RNA according to the standard Affymetrix GeneChip protocol. Total RNA was prepared from 4 × 106 cultured cells that were untreated or treated with Tet and were either not treated or treated with TGF-β for different times. Two independent cell induction experiments were performed to generate independent duplicates.
Microarray analysis was performed on the U133A and U133B GeneChips that allow the analysis of the expression levels of 39,000 transcripts corresponding to more than 33,000 well-characterized human genes. Analysis of each chip and comparative analyses were carried out using GeneSpring software (Silicon Genetics, Redwood City, CA). For each duplicate, HaCaT-TR (+Tet), HaCaT-TRS4 (−Tet), and HaCaT-TRS4 (+Tet) samples treated for 1 h and 6 h with TGF-β were compared with the corresponding untreated cells (0-h TGF-β) in order to generate a list of gene induction/repression ratios at early (1 h) and late (6 h) times of TGF-β stimulation. Genes whose expression level was changed by more than twofold upon TGF-β stimulation with P < 0.001 in both replicates of the control parental HaCaT-TR (+Tet) cells were scored as TGF-β-regulated genes. To determine to what extent the TGF-β regulation depended on Smad4, these ratios for TGF-β-regulated genes in the control parental HaCaT-TR (+Tet) cells were compared to the equivalent ratios generated in the HaCaT-TRS4 (+Tet) cells in which Smad4 is silenced. Genes were considered Smad4 dependent when (i) the induction/repression ratio at either 1 h and/or 6 h of TGF-β stimulation in the HaCaT-TRS4 (+Tet) cells was twofold or more reduced compared with the control parental HaCaT-TR (+Tet) cells in both replicates and/or (ii) the induction/repression ratio at 1 h of TGF-β stimulation and/or 6 h of TGF-β stimulation in the HaCaT-TRS4 (+Tet) cells was less than or equal to 1.5 in both replicates. This last filter was used to determine the Smad4 dependency of the genes whose TGF-β induction/repression ratio in the control HaCaT-TR cells was low (around twofold). Genes for which the induction/repression ratios in the control parental HaCaT-TR (+Tet) cells and in the HaCaT-TRS4 (+Tet) cells differed by less than twofold in both replicates were considered Smad4 independent. Genes that did not behave consistently in the two replicates were eliminated from the analysis.
RT-PCR.
Total RNA (5 μg) was reverse transcribed using Superscript II RNase H− reverse transcriptase (Invitrogen) with oligo(dT) primers. PCR was carried out in the exponential phase (20 to 35 cycles) to allow a semiquantitative comparison of RNA levels. PCR products were analyzed in 1.5% agarose gels. The housekeeping gene coding for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control. Specific primers were used for analysis of candidate genes, for which the sequences are given elsewhere (see the experimental procedures in the supplemental material).
Western blotting and immunofluorescence.
Whole-cell extracts were prepared using radioimmunoprecipitation assay buffer (50 mM Tris, pH 8, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate). Western blotting was performed using standard procedures. Quantification of Western blots was performed using the Odyssey Infrared Imaging Technology (LI-COR; Biosciences) according to the manufacturer's instructions. The following antibodies were used: Smad4 (B8; Santa Cruz), Smad2/3 (BD Biosciences), phospho-Smad2 (Cell Signaling), phospho-Smad3 (a gift from Ed Leof), p21 (C19; Santa Cruz), p15 (C20; Santa Cruz), Smurf1 (H60; Santa Cruz), PAI-1 (C9; Santa Cruz), Grb2 (BD Biosciences), and HA (monoclonal HA antibody conjugated with peroxidase; Roche). Immunofluorescence was carried out as described previously (41). Antigen detection was performed using anti-E-cadherin (BD Biosciences) or antivimentin (Sigma) antibodies followed by Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin G antibody (Alexa). F-actin was detected with Texas red-phalloidin (Alexa), and cell nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI). Cells were mounted with Mowiol (Calbiochem), and fluorescence was observed with a Zeiss confocal LSM 510 microscope.
Cell cycle analysis.
Cells (5 × 105) were plated in 10-cm dishes, and Tet was added for 48 h. Cells were synchronized in G0/G1 by serum starvation (0.2% serum) for 24 h. The amount of cells in each stage of the cell cycle was analyzed after induction with 10% serum for 22 h in the presence or absence of TGF-β. Cells were fixed in 70% ethanol, treated with RNase (100 μg/ml) for 5 min at room temperature, stained with propidium iodide (50 μg/ml) for 5 min, and analyzed by flow cytometry.
EMT and in vitro scratch assays.
To observe EMT in Colo-357 cells and their derivatives, 5 × 103 cells/well were plated in Lab-Tek chamber glass slides (Nunc) previously coated with (5 to 10 μg/cm2) collagen type I (BD Biosciences). After 48 h of Tet induction, either 4 ng/ml TGF-β or 10 μM SB-431542 was added. Cells were grown for 6 days, and media and supplements were replaced every 48 h. To observe EMT in HaCaT cells and their derivatives, cells were grown to confluence for 48 h in the presence or absence of Tet and then were serum starved (0.2% serum) for 24 h prior to addition of TGF-β for 48 h.
For in vitro scratch assays, confluent cells previously induced or not with Tet were scratched with a 10-μl pipette tip to generate a “wound” of ∼600 μm in width. Images were taken after 48 h, when wound closure due to cell migration was observed in the control cells. In the case of HaCaT cells, cells were serum deprived for 24 h prior to scratching. To quantitate cell migration, cells were monitored by time-lapse video microscopy for 48 h after scratching using a low-light inverted Zeiss microscope. Acquired cell migration movies were then analyzed with the Tracker software by tracking individual random cells localized at the leading edge of the scratch. Mean speeds were then determined and statistical analysis (analysis of variance) under the different conditions was then performed using the “Mathematica” software.
RESULTS
Characterization of a HaCaT stable cell line expressing a Tet-inducible siRNA targeted against Smad4.
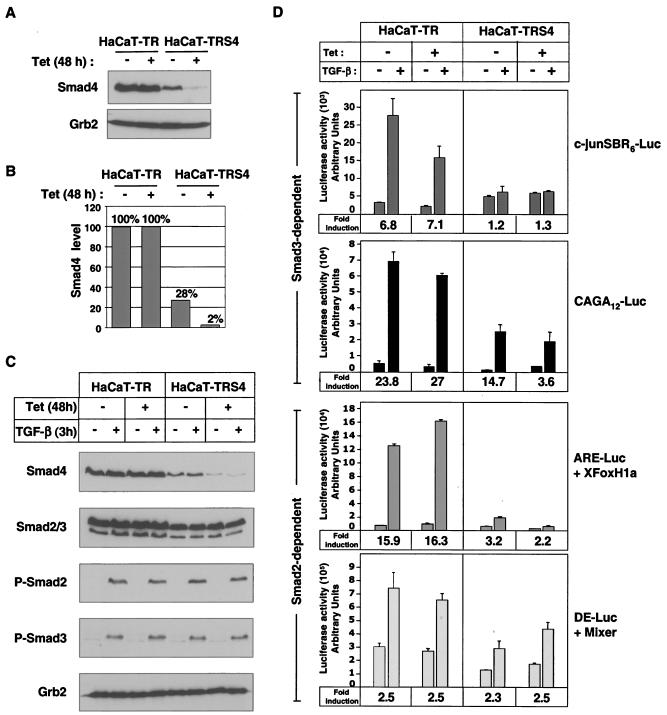
We have generated a clonal HaCaT cell line (HaCaT-TRS4) that expresses a Tet-inducible siRNA against Smad4 and exhibits very strong Smad4 silencing after 48 h of Tet treatment (Fig. 1A). Interestingly, we also detect a small decrease in Smad4 levels (about threefold) in these cells in the absence of Tet, when compared to the parental HaCaT-TR cell line that only expresses the Tet repressor. We took advantage of the “leakiness” of this inducible system to study the requirement for different doses of Smad4 by comparing three different cell lines or conditions: HaCaT-TR parental cells that contain wild-type levels of Smad4 (100%), HaCaT-TRS4 cells in the absence of Tet that contain reduced Smad4 levels (28% of the wild-type Smad4 levels), and HaCaT-TRS4 cells treated with Tet that exhibit a very strong silencing of Smad4 (2% of wild-type Smad4 levels) (Fig. 1B). The siRNA against Smad4 is specific, as it had no effect on the levels of endogenous Smad2 or Smad3, and moreover the knockdown of Smad4 had no effect on Smad2 or Smad3 phosphorylation upon TGF-β stimulation (Fig. 1C). Knockdown of Smad4 also had no effect on TGF-β-induced nuclear localization of Smad2 or Smad3 (data not shown), consistent with previous observations in naturally occurring Smad4-null tumor cell lines (20, 41).
FIG. 1.
Characterization of a stable HaCaT cell line expressing a Tet-inducible siRNA targeted against Smad4. (A and B) Knockdown of Smad4 is strongly induced by Tet in HaCaT-TRS4 cells. HaCaT-TR parental cells and HaCaT-TRS4 cells were treated with Tet for 48 h, and equal amounts of protein were analyzed by Western blotting using antibodies against Smad4 and Grb2 as a loading control (A). Smad4 levels were quantified using the Odyssey detection system (B). (C) Knockdown of Smad4 has no effect on Smad2/3 phosphorylation after TGF-β stimulation. HaCaT-TR parental cells and HaCaT-TRS4 cells were treated with Tet for 48 h and then with TGF-β for 3 h. Equal amounts of protein were analyzed by Western blotting using antibodies against Smad4, Smad2/3, phosphorylated Smad2 (P-Smad2), phosphorylated Smad3 (P-Smad3), and Grb2 as a loading control. (D) Knockdown of Smad4 has differential effects on TGF-β-induced transcription from Smad3-dependent and Smad2-dependent synthetic reporters. Cells were transfected with the Smad3-dependent reporters CAGA12-Luc or c-junSBR6-Luc and the Smad2-dependent reporters ARE-Luc and a plasmid expressing XFoxH1a/XFast-1 or DE-Luc and a plasmid expressing Mixer. Where appropriate, Tet was added at the time of the transfection. Cells were then incubated for 40 h, and TGF-β was added 8 h before assaying luciferase activity. The TGF-β fold inductions are given at the bottom of each graph.
To determine whether the knockdown of Smad4 that we observe in these cell lines was sufficient to abolish known Smad4-dependent transcriptional responses, we investigated the activity of a selection of Smad2/Smad4- and Smad3/Smad4-dependent luciferase reporters in the HaCaT-TR or HaCaT-TRS4 cell lines in the absence and presence of Tet (Fig. 1D). CAGA12-Luc and c-junSBR6-Luc reporters were used to analyze Smad3-dependent transcription. To assay Smad2-dependent transcription, we used the ARE-Luc and DE-Luc reporters, which require FoxH1 and Mixer transcription factors, respectively, to recruit activated Smad complexes to DNA (22). Knockdown of Smad4 resulted in almost complete loss of TGF-β-induced transcription from the c-junSBR6-Luc reporter and dramatically decreased TGF-β induction of the CAGA12-Luc and ARE-Luc reporters. In contrast, the TGF-β inducibility of DE-Luc was unaffected by the level of Smad4 in the cell.
Taken together these results suggest that the silencing of Smad4 upon induction of the siRNA is sufficient to strongly inhibit some Smad-dependent transcriptional responses. It is also clear that these luciferase reporters are differentially affected by the knockdown of Smad4.
Microarray analysis indicates that knockdown of Smad4 only affects a subset of TGF-β gene responses.
We next extended the investigation of Smad4 involvement in TGF-β-regulated transcription by performing a large-scale microarray analysis. cDNA probes were generated from total RNA extracted from HaCaT-TR cells in the presence of Tet and from HaCaT-TRS4 cells in both the absence and presence of Tet after 0 h, 1 h, and 6 h of TGF-β stimulation. The probes were hybridized to Affymetrix U133A and U133B GeneChips. We first generated a list of the TGF-β-regulated genes in the HaCaT-TR control cell line, which expresses wild-type levels of Smad4, by comparing the expression profiles generated after 1 h or 6 h of TGF-β stimulation with that generated in uninduced cells (0 h). Genes whose expression level was changed by more than twofold with P < 0.001 in two independent experiments were scored as TGF-β-regulated genes. This analysis enabled us to identify a panel of 114 early (1 h) and/or late (6 h) TGF-β-inducible target genes in HaCaT cells that were either up- or down-regulated by TGF-β (Table 1). Among these TGF-β-induced genes, we noted about 25 known TGF-β-responsive genes, for example, PAI-1, ITGB6, SMURF1, c-JUN, MADH7 (Smad7), SKIL (Sno), CDKN1A (p21), and CDKN2B (p15), many of which were also identified in another Affymetrix analysis using HaCaT cells (35). We also found 89 potentially novel TGF-β target genes.
TABLE 1.
Summary of Smad4 dependency of genes regulated by TGF-β in HaCaT cellsa
| TGF-β target gene | Description | Regulationb |
|---|---|---|
| Early (1 h) targets | ||
| Smad4 dependent | ||
| BMP2K | BMP2-inducible kinase | Up |
| CLU | Clusterin | Up |
| DLX2 | Distal-less homeo box 2 | Up |
| GADD45B | Growth arrest and DNA-damage-inducible, beta | Up |
| PRKD2 | Protein kinase D2 | Up |
| TGM4 | Transglutaminase 4 | Up |
| TIEG | TGF-β-inducible early growth response | Up |
| COPA | Coatomer protein complex, subunit alpha | Down |
| LDLR | Low-density lipoprotein receptor | Down |
| Smad4 independent | ||
| CLC | Cardiotrophin-like cytokine | Up |
| EDN1 | Endothelin-1 | Up |
| HES1 | Hairy and enhancer of split 1. | Up |
| IER3 | Immediate-early response 3 | Up |
| SNAI2 (Slug)c | Snail homolog 2 | Up |
| Molecule possessing ankyrin repeats | Down | |
| TPD52 | Tumor protein D52 | Down |
| Early (1 h) and late (6 h) targets | ||
| Smad4 dependent | ||
| AKT3 | v-akt murine thymoma viral oncogene homolog 3 | Up |
| ATF3 | Activating transcription factor 3 | Up |
| BHLHB2 | Basic helix-loop-helix domain containing, class B2 | Up |
| CXCL11 | Chemokine (C-X-C motif) ligand 11 | Up |
| NEDD9 (Hef1) | Neural precursor developmentally down-regulated 9 | Up |
| PAI1c | Plasminogen activator inhibitor type 1 | Up |
| SERTAD4 | SERTA domain containing 4 | Up |
| TGM1 | Transglutaminase 1 | Up |
| Smad4 independent | ||
| CDKN1A (p21)c | Cyclin-dependent kinase inhibitor 1A | Up |
| c-JUNc | v-jun sarcoma virus 17 oncogene homolog | Up |
| JUNB | junB proto-oncogene | Up |
| MADH7 (Smad7)c | MAD, mothers against decapentaplegic homolog 7 | Up |
| PTHLH | Parathyroid hormone-like hormone | Up |
| RDC1 | G protein-coupled receptor | Up |
| SKIL (sno) | SKI-like | Up |
| TCF8 | Transcription factor 8 | Up |
| TMEPAIc | Transmembrane, prostate androgen-induced RNA | Up |
| VMP1 | Likely ortholog of rat vacuote membrane protein 1 | Up |
| CITED2 | Cbp/p300-interacting transactivator 2 | Down |
| RGS2 | Regulator of G-protein signalling 2, 24 kDa | Down |
| Late (6 h) targets | ||
| Smad4 dependent | ||
| AKAP12c | A kinase (PRKA) anchor protein (gravin) 12 | Up |
| ARNTL | Aryl hydrocarbon receptor nuclear translocator-like | Up |
| _BJ-TSA-_9 | Hypothetical protein MGC14128 | Up |
| CKLFSF3 | Chemokine-like factor super family 3 | Up |
| ETS2 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | Up |
| IRAK2 | Interleukin-1 receptor-associated kinase 2 | Up |
| ITGB6c | Integrin, beta 6 | Up |
| LAMC2 | Laminin, gamma 2 | Up |
| MASP2 | Mannan-binding lectin serine protease 2 | Up |
| PHACTR3 | Phosphatase and actin regulator 3 | Up |
| POLR2J2 | DNA directed RNA polymerase II polypeptide J-related gene | Up |
| SMURF1c | E3 ubiquitin ligase SMURF1 | Up |
| ZNF198 | Zinc finger protein 198 | Up |
| AMSH-LP | Associated molecule with the SH3 domain of STAM-like protein | Down |
| CLDN8 | Claudin 8 | Down |
| FLG | Filaggrin | Down |
| FOXQ1 | Forkhead box Q1 | Down |
| GALNT5 | UDP-_N_-acetyl-α-d-galactosamine | Down |
| GLMN | Glomulin, FKBP-associated protein | Down |
| GPR126 | G protein-coupled receptor 126 | Down |
| IL1RN | Interleukin-1 receptor antagonist | Down |
| PARD6B | Par-6 partitioning defective 6 homolog beta (Caenorhabditis elegans) | Down |
| PBX1 | Pre-B-cell leukemia transcription factor 1 | Down |
| PCDH7 | BH-protocadherin 7 (brain-heart) | Down |
| PLEKHK1 | Pteckstrin homology domain containing, family K member 1 | Down |
| RAB27B | RAB27B, member RAS oncogene family | Down |
| RASSF5 | Similar to protein interacting with GEF | Down |
| RDH10 | Retinol dehydrogenase 10 (all-trans) | Down |
| TBX3 | Transcription factor Tbx3 | Down |
| TIMP3 | Tissue inhibitor of metalloproteinase 3 | Down |
| TNFSF10 | Tumor necrosis factor (ligand) superfamily, member 10 | Down |
| ZNF488 | Zinc finger protein 488 | Down |
| Smad4 independent | ||
| ACTN1 | Actinin, alpha 1 | Up |
| ADAM19 | A disintegrin and metalloproteinase domain 19 | Up |
| AMIGO2 | Amphoterin-induced gene 2 | Up |
| CDKN2B (p15)c | Cyclin-dependent kinase inhibitor 2B | Up |
| CHST11 | Carbohydrate (chondroitin 4) sulfotransferase 11 | Up |
| COL4A1 | Collagen, type IV, alpha 1 | Up |
| CST6 | Cystatin E/M | Up |
| DLC1 | Deleted in liver cancer 1 | Up |
| ELK3 | ELK3, ETS-domain protein (SRF accessory protein 2) | Up |
| FHL2 | Four and a half LIM domains 2 | Up |
| FSTL3 | Follistatin-like 3 | Up |
| HS3ST2 | Heparan sulfate (glucosamine) 3-_O_-sulfotransferase 2 | Up |
| IVL | Involucrin | Up |
| KAL1 | Kallmann syndrome 1 sequence | Up |
| LBH | Likely ortholog of mouse limb-bud and heart gene | Up |
| LIPG | Lipase, endothelial | Up |
| LTBP2 | Latent transforming growth factor beta binding protein 2 | Up |
| MGC29643 | Hypothetical protein MGC29643 | Up |
| NAV2 | Neuron navigator 2 | Up |
| NET1 | Neuroepithelial cell transforming gene 1 | Up |
| P114-RHO-GEF | Rho-specific guanine nucleotide exchange factor p114 | Up |
| PAPPA | Pregnancy-associated plasma protein A, pappalysin 1 | Up |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | Up |
| PTPRB | Protein tyrosine phosphatase, receptor type, B | Up |
| RUNX1 | Runt-related transcription factor 1 | Up |
| SEC14L2 | SEC14-like 2 (Saccharomyces cerevisiae) | Up |
| SEMA3C | Sema domain, immunoglobulin domain (semaphorin) 3C | Up |
| SOX4 | SRY (sex-determining region Y)-box 4 | Up |
| TAGLN | Transgelin | Up |
| TGFB2 | TGF-β2 | Up |
| TP73L | Tumor protein p73-like | Up |
| ANKRD22 | Ankyrin repeat domain 22 | Down |
| BCMP11 | Breast cancer membrane protein 1 | Down |
| CLDN1 | Claudin 1 | Down |
| CLECSF2 | C-type lectin, superfamily member 2 | Down |
| CYP1A1 | Cytochrome P450, family 1A, polypeptide 1 | Down |
| DOCK8 | Dedicator of cytokinesis 8 | Down |
| GATA3 | GATA binding protein 3 | Down |
| GBP2 | Guanylate binding protein 2, interferon-inducible | Down |
| ID2 | Inhibitor of DNA binding 2 | Down |
| ID3 | Inhibitor of DNA binding 3 | Down |
| ID4 | Inhibitor of DNA binding 4 | Down |
| IFIT2 | Interferon-induced protein with tetratricopeptide repeats 2 | Down |
| PPFIBP2 | PTPRF-Interacting protein, binding protein 2 (liprin beta 2) | Down |
| SASH1 | SAM and SH3 domain containing 1 | Down |
| TGFBR3 | TGF-β receptor III | Down |
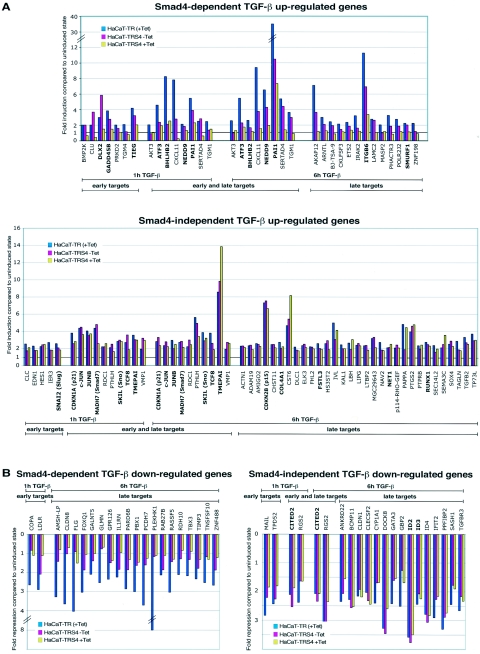
We next sought to determine which TGF-β target genes were dependent on Smad4 for their induction. To that end, we compared their TGF-β inducibility at 1 h and 6 h after TGF-β stimulation in the control cells, HaCaT-TR, to their TGF-β inducibility in the Smad4-silenced cells, HaCaT-TRS4 (+Tet) (see Materials and Methods for details of the criteria used for this analysis). The results clearly indicate that TGF-β target genes can be divided into two subpopulations depending on their requirement for Smad4. The Smad4-dependent and independent genes are listed in Table 1, and their activation/repression in response to TGF-β in the HaCaT-TR cells in the presence of Tet and in the HaCaT-TRS4 cells in the absence and presence of Tet in one of the two replicate experiments are shown in Fig. 2A and B. Genes classified as Smad4-dependent TGF-β target genes (Fig. 2A, upper panel and B, left panel) exhibit a dramatic dependency on the level of Smad4 for their activation/repression upon TGF-β stimulation, whereas target genes classified as Smad4 independent (Fig. 2A, lower panel, and B, right panel) are still fully regulated by TGF-β after silencing of Smad4. Strikingly, many of the known TGF-β target genes that contain Smad4 binding elements in their promoters such as c-JUN, JUNB, CDKN1A (p21), CDKN2B (p15), and SMAD7 (11, 19, 34, 53, 57, 67) appear to be substantially Smad4 independent for their TGF-β induction, while others, such as PAI-1, ATF3, and SMURF1, are strongly dependent on Smad4. Interestingly, genes in the Smad4-dependent category can be further classified based on their ability to be regulated by TGF-β in the cell lines with different Smad4 levels. Considering the up-regulated genes, some genes clearly require high levels of Smad4 for their induction, since their TGF-β inducibility is strongly impaired even in the HaCaT-TRS4 (−Tet) cells that have reduced Smad4 levels. Their induction is not further affected when Smad4 is silenced under the HaCaT-TRS4 (+Tet) condition. Examples of such genes are ATF3 or BHLHB2 at the 1-h time point (Fig. 2A). Other genes, in contrast, show a clear dose-dependent response according to the level of Smad4. Their inducibility is decreased when the Smad4 level is reduced under the HaCaT-TRS4 (−Tet) condition and then further decreased or abolished under the HaCaT-TRS4 (+Tet) condition, when Smad4 is silenced. Examples of such genes are CXCL11 or PAI-1 at both the 1-h and 6-h time points (Fig. 2A). Similar results are seen in the Smad4-dependent down-regulated genes (Fig. 2B).
FIG. 2.
Quantitative analysis of the microarray results. Comparison of the fold induction (A) or fold repression (B) ratios of the early (1 h) and late (6 h) TGF-β target genes reveals two different categories of TGF-β-regulated genes dependent on or independent of Smad4. Fold inductions or repressions of one of the replicates for HaCaT-TR (+Tet), HaCaT-TRS4 (−Tet), and HaCaT-TRS4 (+Tet) cells are shown. A significant dependency on the level of Smad4 is observed for TGF-β target genes scored as Smad4 dependent (A, upper panel; B, left panel), whereas the TGF-β target genes scored as Smad4 independent (A, lower panel, B, right panel) display no significant differences in activation/repression under the three conditions. Known TGF-β target genes are shown in bold.
Validation of the microarray data.
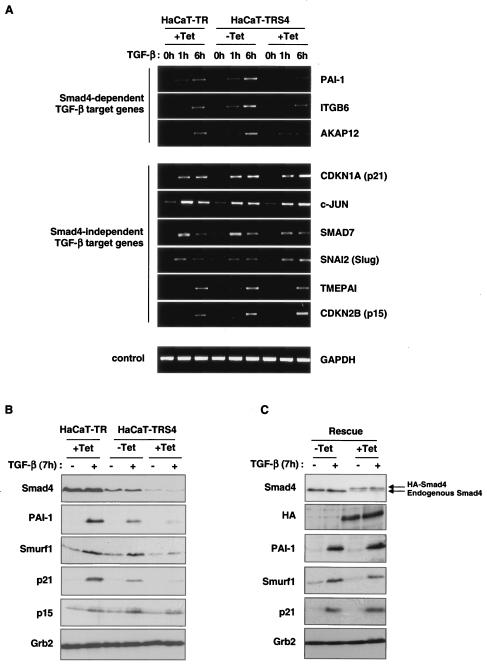
We validated the Affymetrix data by both RT-PCR (Fig. 3A) and RNase protection (see Fig. S1 in the supplemental material), focusing on some representative genes in the Smad4-dependent and -independent categories of TGF-β targets. We have confirmed that the TMEPAI, c-JUN, CDKN1A (p21), CDKN2B (p15), SNAI2 (Slug), and SMAD7 genes are induced by TGF-β independently of Smad4, but that PAI-1, ITGB6 and the newly-discovered TGF-β target gene, AKAP12, require Smad4 for their induction.
FIG. 3.
Validation of the microarray data. (A) Differences in transcriptional activation upon TGF-β stimulation were confirmed by RT-PCR. HaCaT-TR or HaCaT-TRS4 cells were incubated with or without Tet for 48 h and treated with TGF-β for 0 h, 1 h, or 6 h. Representative candidates of the two different categories of TGF-β-induced genes were analyzed. PAI-1, ITGB6, and AKAP12 were confirmed as Smad4-dependent target genes, and CDKN1A (p21), c-JUN, SMAD7, SNAI2 (Slug), TMEPAI, and CDKN2B (p15) were confirmed as Smad4-independent TGF-β target genes. GAPDH was used as a control. (B) Gene inductions assayed at the protein level. HaCaT-TR or HaCaT-TRS4 cells were incubated with or without Tet for 48 h and treated with TGF-β for 7 h. Whole-cell extracts were prepared, and equal amounts of protein were analyzed by Western blotting using antibodies against p21 and p15 as representatives of Smad4-independent TGF-β targets, against PAI-1 and Smurf1 as representative products of Smad4-dependent TGF-β target genes, and against Smad4 to confirm the Smad4 protein levels in the different conditions. Grb2 serves as a loading control. In all cases except p21, the protein expression in response to TGF-β in the different cell lines/conditions mirrored the RNA expression. (C) The loss of induction of the Smad4-dependent genes is rescued when an siRNA-resistant HA-Smad4 is expressed in the HaCaT-TRS4 cells. HaCaT-TRS4-rescue cells were incubated with or without Tet for 48 h and then treated with TGF-β for 7 h. Whole-cell extracts were prepared, and equal amounts of protein were analyzed by Western blotting. Tet induction of the HA-tagged siRNA-resistant Smad4 in the rescue clone is observed in the HA blot and in the Smad4 blot, where it is detected as a band running with slightly lower mobility compared to endogenous Smad4 (see arrows). Induction of PAI-1, Smurf1, and p21 by TGF-β is rescued when the Tet-induced silencing of the endogenous Smad4 is rescued by the induction of the siRNA-resistant form of Smad4.
We next tested whether the differences in gene induction observed were also seen at the protein level. Western blot analysis confirmed that protein induction in response to TGF-β of PAI-1 and Smurf1 was severely impaired in the absence of Smad4, whereas induction of p15 was not affected (Fig. 3B), in agreement with our RNA analysis. Curiously, TGF-β induction of CDKN1A (p21) RNA was Smad4 independent, whereas its induction at the protein level appears strongly dependent on Smad4. This suggests that upon TGF-β signaling, Smad4 might be required for either p21 stability or for the translation of p21 (see Discussion).
We then verified that the effects on gene expression observed upon activation of the Smad4 siRNA were really due to loss of Smad4 and not to off-target effects of the siRNA (6, 32), by reexpressing Smad4 in the HaCaT-TRS4 cells. We generated a HaCaT-TRS4 rescue clone, HaCaT-TRS4-rescue, that expresses an HA-tagged siRNA-resistant form of Smad4 upon stimulation with Tet (Fig. 3C). In this clone, addition of Tet results in decreased expression of endogenous Smad4 due to the siRNA induction and a roughly comparable increase in expression of HA-Smad4. Although expression of the resistant form of Smad4 is relatively weak, it is able to rescue the loss of induction of TGF-β target genes that display a dose dependency for Smad4, such as those coding for PAI-1 and Smurf1, and is even able to rescue p21 protein induction after treatment with Tet. This result confirms that the loss of TGF-β responses that we observe is specifically due to the knockdown of Smad4.
Knockdown of Smad4 abolishes TGF-β-induced cell cycle arrest in G1.
We have shown that the knockdown of Smad4 impairs the expression of only a subset of TGF-β target genes. We therefore extended our analysis to investigate whether TGF-β-mediated biological functions that might be relevant to tumor development and progression, such as cell cycle arrest, EMT, or migration, are differentially affected by the knockdown of Smad4.
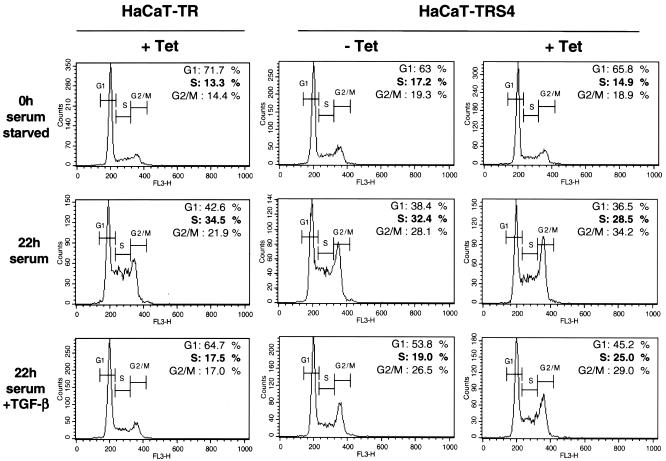
We first examined the consequence of the knockdown of Smad4 on TGF-β-induced cell cycle arrest. HaCaT cells that have been serum starved for 24 h accumulate in the G0/G1 phase of the cell cycle. Addition of serum allows reentry into the cell cycle, which is inhibited by TGF-β. Thus, HaCaT-TR parental cells (left panel) synchronized in G0/G1 (0-h serum starved) accumulate in S phase after 22 h of serum induction but not in the presence of TGF-β (Fig. 4, left panels; 34.5% of cells are in S phase after 22 h of serum induction compared to only 17.5% in the presence of TGF-β). We found that this effect of TGF-β is dependent on Smad4 since TGF-β cannot inhibit the G1-to-S-phase transition when Smad4 is silenced in the presence of Tet in the HaCaT-TRS4 cell line (Fig. 4, right panels, with 28.5% of cells in S phase after 22 h of serum induction compared to 25% in the presence of TGF-β). This result demonstrates that TGF-β-induced cell cycle arrest is dependent on Smad4 and suggests that the loss of Smad4 during tumorigenesis would abolish this tumor suppressor function of TGF-β.
FIG. 4.
Smad4 is necessary for TGF-β-induced cell cycle arrest in HaCaT cells. HaCaT-TR or HaCaT-TRS4 cells were incubated with or without Tet for 48 h as indicated and synchronized by serum starvation (0.2% serum for 24 h). Cells were then collected before (0h serum starved) or after treatment with 10% serum for 22 h in the absence (22h serum) or presence of TGF-β (22h serum + TGF-β). Samples were analyzed by fluorescence-activated cell sorting to determine the number of cells in the G1, S, and G2/M phases. FL3-H on the x axis corresponds to propidium iodide fluorescence.
Knockdown of Smad4 does not affect EMT induced by TGF-β.
In contrast to its tumor suppressor function, which is important to maintain the epithelial identity of cells, TGF-β has also been shown to promote tumor progression at late stages of tumorigenesis. One mechanism whereby TGF-β might act as a tumor promoter is through its ability to induce EMT (13). EMT is the process in which epithelial cells lose their polarity and cell-cell contacts and acquire a fibroblastoid and invasive phenotype.
HaCaT cells have been shown to manifest some of the phenotypic features of EMT, such as loss of cell-cell contacts and actin reorganization, when quiescent serum-deprived cells are treated with TGF-β (70). Indeed, after 48 h of TGF-β treatment, the HaCaT-TR parental cells treated in this manner were less tightly organized and displayed disrupted adherens junctions characterized by loss of actin and E-cadherin colocalization at the membrane (see Fig. S2 in the supplemental material). The same morphological changes were observed in the HaCaT-TRS4 cells in the presence or absence of Tet, indicating that Smad4 is not involved in this process.
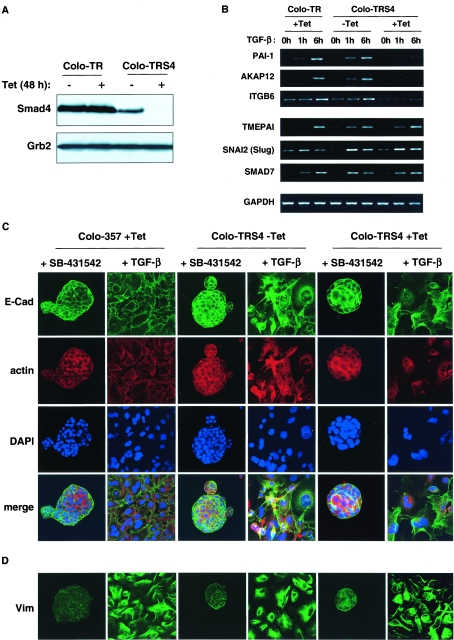
Although HaCaT cells display a decrease in cell-cell interactions and actin reorganization after TGF-β stimulation, they do not acquire a fibroblastoid morphology or express mesenchymal markers. To investigate the role of Smad4 in full EMT, we used another cell line, the pancreatic tumor cell line Colo-357, that expresses normal levels of Smad4 and undergoes a full EMT upon TGF-β stimulation (17). We generated a Colo-357 cell line that expresses the Tet repressor (Colo-TR) and one that expresses the Tet repressor together with the Tet-inducible Smad4-siRNA (Colo-TRS4), which exhibits very strong Smad4 silencing after 48 h of Tet treatment (Fig. 5A). As observed in the HaCaT-TRS4 cell line, upon TGF-β stimulation, the Colo-TRS4 cell lines treated with Tet lose the induction of some representative Smad4-dependent target genes (PAI-1, AKAP12, and ITGB6) when compared to the parental Colo-TR cell line, but are still able to induce some Smad4-independent genes (TMEPAI, SNAI2 [Slug], and SMAD7) (Fig. 5B).
FIG. 5.
Smad4 is not required for EMT induced by TGF-β. (A) A Colo-357-derived cell line that expresses a Tet-inducible Smad4 siRNA shows a strong silencing of Smad4. Colo-TR parental cells and Colo-TRS4 cells that express the Smad4 siRNA were treated with or without Tet for 48 h, and equal amounts of protein were analyzed by Western blotting using antibodies against Smad4 and Grb2 as a loading control. (B) The Smad4 dependency of some representative Smad4-dependent and independent TGF-β target genes is validated in the Colo-TRS4 cells. Colo-357 or Colo-TRS4 cells were incubated with or without Tet for 48 h and treated with TGF-β for 0 h, 1 h, or 6 h. PAI-1, ITGB6, and AKAP12 were confirmed as Smad4-dependent target genes, and SMAD7, SNAI2 (Slug), and TMEPAI were confirmed as Smad4-independent TGF-β target genes. GAPDH was used as a loading control. (C and D) Smad4 is not involved in TGF-β-induced EMT in Colo-357 cells. Colo-TR parental cells and Colo-TRS4 cells were grown on collagen type I and treated with Tet for 48 h. TGF-β or SB-431542 was then added for 6 days. In the presence of SB-431542, cells display an epithelial-like phenotype, whereas growth in the presence of TGF-β induces a mesenchymal-like phenotype independently of the presence of Smad4. Cells were processed for immunofluorescence using an anti-E-cadherin (E-Cad) antibody, to analyze adherens junctions (B) or an antivimentin (Vim) antibody to analyze mesenchymal marker expression (C). Actin reorganization was visualized with Texas red-conjugated phalloidin, and nuclei were visualized with DAPI.
When grown at very low density on fibronectin (data not shown) or collagen type I (Fig. 5C), we observed that Colo-357 cells display a strongly polarized epithelial morphology and organize themselves in structures resembling pancreatic acini (3) with strong cell-cell interactions characterized by the colocalization of E-cadherin and actin at the plasma membrane. We observe this phenotype in untreated cells (data not shown) and also in the presence of the TGF-β type I receptor inhibitor (SB-431542) (31, 37), which blocks any signaling arising from autocrine production of TGF-β (Fig. 5C). In the presence of TGF-β, the polarized epithelial cell architecture is completely abolished and cells grow individually on the collagen. Adherens junctions are disrupted probably more as a consequence of E-cadherin delocalization rather than E-cadherin down-regulation, as the E-cadherin protein level is not affected by TGF-β treatment (see Fig. S3 in the supplemental material and Fig. 5C). Moreover, expression of the mesenchymal marker vimentin is detected both by Western blotting analysis and in immunofluorescence (see Fig. S3 in the supplemental material and Fig. 5D), suggesting that the cells have undergone a full EMT. We found that knockdown of Smad4 in the Colo-TRS4 (+Tet) cells had no effect on this TGF-β-induced EMT.
Knockdown of Smad4 abolishes TGF-β-induced migration.
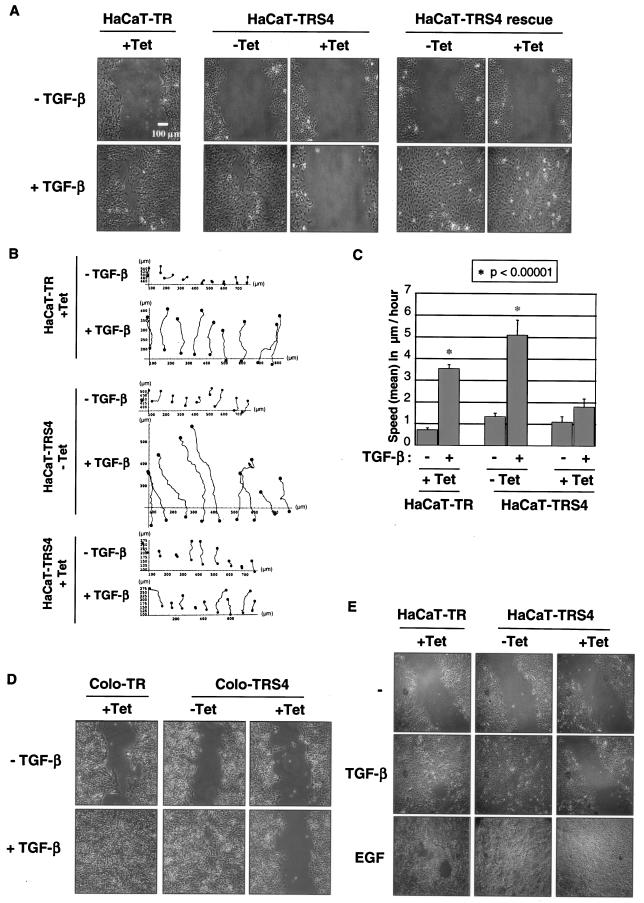
One of the consequences of the transition of epithelial cells to a more mesenchymal phenotype during tumorigenesis is to allow cells to become more prone to migrate and to become more invasive (60). We thus next measured the migratory potential of the cells by performing an in vitro scratch assay in which TGF-β induces cell migration after a “wound” is introduced in a confluent serum-deprived HaCaT cell monolayer. Surprisingly, silencing of Smad4 completely abolished TGF-β-induced migration in HaCaT cells, and this defect in motility is specific to the loss of Smad4, as it is rescued in the HaCaT-TRS4-rescue cell line (Fig. 6A). Time-lapse microscopy followed by cell tracking analysis (Fig. 6B) indicated that in the absence of Smad4, the defect in migration is associated with a significant decrease in cell speed (Fig. 6C) and therefore is not due to a lack of cell orientation during migration. Moreover, we did not observe any differences in Golgi or MTOC polarization toward the migration front in the absence of Smad4 (data not shown). TGF-β-induced migration during in vitro “wound healing” was also abolished when Smad4 was silenced in the pancreatic tumor cell line Colo-357 (Fig. 6D). Altogether, these results indicate that Smad4 is a key effector of TGF-β-induced migration.
FIG. 6.
Smad4 is necessary for TGF-β-dependent migration in a scratch assay. (A, B, and C) The Tet-induced silencing of Smad4 in HaCaT-TRS4 cells abolishes TGF-β-induced migration in a scratch assay. HaCaT-TR, HaCaT-TRS4, or HaCaT-TRS4-rescue cells were grown to confluence with or without Tet for 48 h and were then serum starved for 24 h. A scratch was introduced into the confluent cell monolayer, and TGF-β-induced migration was monitored with time-lapse microscopy for 48 h. A photograph of the cells after 48 h of migration is shown in panel A. Tracking of cells at the leading edge of the scratch was performed on the acquired images using the Tracker software for the HaCaT-TR cells (+Tet) and HaCaT-TRS4 cells (±Tet) (B). Mean speeds were calculated from the cell tracking analysis using the Mathematica software (C). The mean speeds of cells in different condition are represented, and the asterisk indicates conditions that are significantly different from the others with a P value of <0.00001. (D) Smad4 is also necessary for TGF-β-induced migration in pancreatic cells. Colo-TR and Colo-TRS4 cells were grown to confluence with or without Tet for 48 h. A scratch was introduced into the monolayer, and TGF-β-induced “wound” closure was photographed after 48 h of cell migration. (E) EGF-induced migration is still functional in the absence of Smad4. HaCaT-TR or HaCaT-TRS4 cells were grown to confluence with or without Tet for 48 h and then serum starved for 24 h. After scratching, cells were incubated with or without TGF-β or EGF for 48 h, at which time the images were taken.
Finally, to determine whether the knockdown of Smad4 affected all cell migration, we analyzed epidermal growth factor (EGF)-induced migration in a scratch assay in HaCaT-TR and HaCaT-TRS4 cells. We found that knockdown of Smad4 had no effect on EGF-induced migration, indicating that Smad4 is specifically required for TGF-β-induced migration (Fig. 6E). This suggests that Smad4-null tumor cells might still be able to migrate in response to growth factors other than TGF-β.
DISCUSSION
An inducible siRNA approach to investigate the role of Smad4 in TGF-β-induced gene regulation and functional responses.
In this study, we have addressed the role of Smad4 in TGF-β-induced gene regulation using a large-scale microarray approach and have assessed its role in TGF-β-induced functional responses. We have used an inducible system to knock down levels of Smad4 in either the human immortalized keratinocyte cell line HaCaT or a pancreatic tumor cell line, Colo-357. This inducible knockdown system has several advantages over previous approaches used to investigate the role of Smad4. Cells are not grown for extended periods in the absence of Smad4. Thus they will not accumulate mutations or adjust expression levels of other genes that might compensate for the loss of Smad4, which might occur when Smad4-null cells are grown from knockout mice, when Smad4-null tumor cells are cultured in vitro, or when an siRNA is constitutively expressed. Also, for both of our cell lines, by comparing the parental line with the cell line containing the siRNA in the presence or absence of Tet, we had the opportunity to compare the effects of wild-type, intermediate, and very low levels of Smad4. Thus, we have been able to correlate the degree of TGF-β inducibility of a subset of target genes with levels of Smad4, strengthening our interpretation that these genes are dependent on Smad4 for their transcriptional regulation. Finally, using this cell system it has been possible to perform a rescue experiment using a cell line that inducibly expresses an siRNA-resistant Smad4 under Tet control. This has been used to confirm the Smad4 dependency of some of the TGF-β target genes and functional responses.
Identification of Smad4-dependent and -independent TGF-β target genes and functions.
The major finding from our microarray analysis is that there are two main categories of TGF-β-regulated genes with respect to their Smad4 requirement. Some genes are dramatically dependent on Smad4 for their induction or repression by TGF-β, while others are still fully induced or repressed by TGF-β or only weakly affected in their induction or repression after silencing of Smad4. Out of the 114 genes that we scored as TGF-β target genes, 49 were Smad4 dependent and 65 were Smad4 independent. To validate the screen, we performed RT-PCR and RNase protection analysis of representative genes and have confirmed that, at the mRNA level, PAI-1, AKAP12, and ITGB6 are Smad4-dependent TGF-β target genes, while TMEPAI, CDKN1A (p21), CDKN2B (p15), SMAD7, SNAI2 (Slug), and c-JUN are induced by TGF-β independently of Smad4. For simplicity, we have divided the TGF-β-inducible genes into two categories: Smad4 dependent and Smad4 independent. However, it should be emphasized that some Smad4-dependent target genes, such as PAI-1, still display a residual induction by TGF-β after Smad4 silencing, while other genes, such as SMAD7, which we have classified as Smad4 independent, also have a small Smad4-dependent contribution. We conclude therefore that genes such as these are regulated by both Smad4-dependent and Smad4-independent pathways. We have classified these genes according to which pathway is dominant.
One possible complication of our present study is the presence of a very small amount of Smad4 after Tet treatment, which might be sufficient for the regulation of the genes we have scored as Smad4 independent. We think this is very unlikely since we have observed the same Smad4 dependency for representative target genes in both the HaCaT cell line and the Colo-357 cell line. Although residual Smad4 protein was detected after Tet induction in the HaCat-TRS4, we were unable to detect any residual Smad4 in the Colo-TRS4 cell line after Tet induction. Moreover our data are consistent with an analysis of a subset of TGF-β-regulated target genes in the Smad4-null pancreatic cell line, BxPC3, which shows that CDKN1A (p21), SNAI2 (Slug), and TMEPAI, and to a certain extent, JUNB, c-JUN, and SMAD7, are still induced in the absence of Smad4, while PAI-1 is not (41) (see Fig. S4 in the supplemental material). These data strongly support the idea that the genes we have identified here as Smad4 independent can still be regulated by TGF-β, even when Smad4 has been deleted. Notably, in two other Smad4-null tumor cell lines, MDA-MB468 and SW480, we were unable to see inductions of any TGF-β target genes tested, suggesting that these cell lines have other defects in addition to the loss of Smad4 that could affect TGF-β signaling. Indeed, we have previously shown that MDA-MB468 cells have very low levels of Smad3 (41). This result emphasizes the fact that comparison of tumor cells lacking Smad4 with normal epithelial cells with wild-type levels of Smad4 is not a good method by which to determine the relevance of Smad4 in TGF-β signaling.
As a further confirmation of our identification of two distinct categories of TGF-β-regulated target genes defined by their dependency on Smad4, we found that TGF-β-induced functional responses, cell cycle arrest in G1, EMT, and migration were also differentially dependent on Smad4. Our results show that TGF-β-induced cell cycle arrest and cell migration are completely dependent on Smad4, while TGF-β-induced EMT is Smad4 independent. Interestingly, our finding that TGF-β-induced EMT and migration are differentially dependent on Smad4 reveals that these two very closely related processes are regulated by distinct mechanisms.
Two other recent studies have attempted to address the role of Smad4 in TGF-β responses in either transient or constitutive knockdown systems (33, 36). In some respects, our analysis gives similar results to these other studies, but there are also some differences. We have confirmed that TGF-β-induced migration is a Smad4-dependent process (33). However, Jazag and colleagues were not able to detect any effect of Smad4 silencing on the growth arrest induced by TGF-β, while Kretschmer and colleagues detected a partial effect of Smad4 silencing on this process (33, 36). It is possible that the silencing of Smad4 in those studies was not sufficient to fully affect the TGF-β-induced growth arrest. When considering the Smad4 dependence of ratified TGF-β-inducible target genes, we have confirmed that induction of PAI-1 is Smad4 dependent (33), but in our hands, SMAD7 appears to be still induced after Smad4 silencing in both the HaCaT and the Colo-357 cell lines (33). One explanation for this difference may be the different cell lines used. Jazag and colleagues used the pancreatic tumor cell line Panc1, in which many target genes, including SMAD7, are relatively weakly induced by TGF-β compared with HaCaT cells (41). We note that in HaCaTs, even though SMAD7 is still induced after Smad4 silencing, there is a small reduction, indicating that there is a small Smad4-dependent component to the TGF-β induction. It is possible that the weak TGF-β induction of SMAD7 in Panc1 cells is purely the Smad4-dependent component.
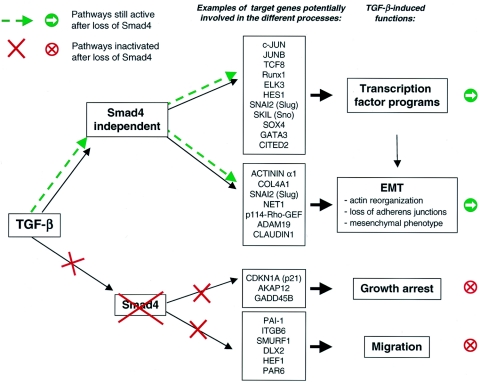
We can use our identification of Smad4-dependent and -independent TGF-β-regulated genes to attempt to understand the Smad4 dependency of these TGF-β-induced functional responses, and in Fig. 7, we indicate some candidate genes that might be expected to play a role in these processes. For example, the loss of the PAI-1 induction could account for the loss of migration, since _PAI-1_−/− keratinocytes have been shown to lose their ability to migrate in an in vitro scratch assay (38, 47). The loss of TGF-β-induced growth arrest may be explained by the loss of p21 protein expression, but also by the loss of AKAP12 or GADD45B induction. Interestingly, CDKN2B (p15), which is still induced after Smad4 silencing, appears insufficient to generate a cell cycle arrest. Finally, it has been shown that TGF-β-induced EMT is mediated through a RhoA-dependent pathway (4) and could therefore be induced by the Rho-specific guanine nucleotide exchange factors p114-RHO-GEF and Net1 (54), which induce RhoA activation and are regulated by TGF-β in a Smad4-independent fashion. Moreover, SNAI2 (Slug) expression, which is maintained after Smad4 silencing, has been shown to induce the first phase of growth factor-induced EMT, characterized by an increase in cell spreading and cell-cell separation (50). Interestingly, the induction of EMT by TGF-β has been proposed to be a Smad3-dependent process, which involves the induction of Slug (49, 71). Since we have found that Slug induction is not dependent on Smad4, this suggests that EMT and Slug induction might be a Smad3-dependent process that does not involve Smad4 (see below). Further work is obviously required to confirm the involvement of these genes in these TGF-β-induced functional responses.
FIG. 7.
Knockdown of Smad4 affects only a subset of TGF-β-regulated target genes and functions. Knockdown of Smad4 in HaCaT cells and in Colo-357 cells reveals that the TGF-β signaling pathway has a Smad4-dependent component and a Smad4-independent component. TGF-β-induced functions that we have shown to be Smad4 dependent or Smad4 independent are shown. Examples of genes that may be involved in these functions, based on the literature, are indicated.
What mechanisms underlie Smad4-independent TGF-β signaling?
One striking result from this study is the identification of a substantial number of Smad4-independent TGF-β target genes. How are these genes regulated in response to TGF-β? One possibility is that at least some Smad4-independent responses could be mediated solely via activated R-Smads, and there is genetic evidence for this. In Drosophila melanogaster, signaling by the TGF-β family member Decapentaplegic (DPP) is completely dependent on the Drosophila R-Smad, MAD, but not all DPP signaling requires the Drosophila Smad4 homolog, MEDEA (66). Similarly during mouse embryogenesis, formation of the node and its derivatives, such as definitive endoderm, is dependent on Smad2 and Smad4, but Nodal-mediated induction of mesoderm derivatives such as somites and lateral plate mesoderm requires Smad2 and Smad3 but is independent of Smad4 (7). “R-Smad only”-mediated transcriptional responses would likely involve recruitment of complexes of activated R-Smads by DNA-binding transcription factors independently of Smad4. Such a mechanism probably accounts for the Smad4-independent transcriptional activation of the DE-Luc reporter, which binds the transcription factor Mixer, which interacts with activated Smad2 (Fig. 1D) (40, 46). Induction of Slug by Smad3, independently of Smad4, may also be an example of this process.
Another possibility is that Smad4-independent transcription might be mediated by R-Smads in conjunction with an alternative Smad4. Although there are no additional Smad family members, as judged by sequence homology, we cannot exclude the possibility of another co-Smad that is unrelated in sequence to Smad4 but has a similar function. Finally, it is also possible that Smad4-independent TGF-β signaling does not require any Smads but instead is mediated by other signaling pathways activated by the TGF-β receptors. Possible candidates would be the pathways involving Erk1/2, p38, or JNK or pathways involving RhoA, PI 3-kinase, Pak2, or PAR6. The activity of all of these signal transducers can be induced by TGF-β in different cell systems (4, 16, 25-27, 44, 65, 69), and many of the pathways involving these signal transducers have been shown to regulate transcription factors in the nucleus. For example in human fibroblasts, fibronectin has been shown to be induced by TGF-β through a Smad4-independent JNK pathway (27).
Most of the Smad4-independent genes that we have identified in this study are novel targets or genes for which the mechanism of TGF-β regulation is unknown. However, some of them, such as SMAD7, CDKN1A (p21), CDKN2B (p15), and c-JUN, have Smad binding elements (SBEs) in their promoters (11, 19, 53, 57, 67). R-Smad/Smad4 complexes have been shown to bind these SBEs by bandshift assays or DNA pulldowns, and in some cases evidence exists for Smad4 being associated with these SBEs in vivo from chromatin immunoprecipitations. It is important to note, however, that evidence of a transcription factor binding to a regulatory element in a gene is not proof that that factor is indispensable for activation. We have shown that a synthetic reporter construct that contains six repeats of the c-Jun SBE (c-junSBR6-Luc) is no longer induced by TGF-β after Smad4 silencing (Fig. 1D), showing that, in isolation, activation of transcription through such an element clearly depends on Smad4. However, in the context of the c-JUN promoter, which contains many other regulatory elements, loss of binding of Smad4 to the one SBE is evidently not sufficient to lose all TGF-β-induced transcription. Indeed, TGF-β induction of the c-JUN promoter is only partially affected when the SBE is mutated, suggesting the existence of other elements targeted by the TGF-β signaling pathway, even in the relatively short promoter fragment tested (67). Thus, for some of its target genes, TGF-β may be able to signal through both Smad4-dependent and -independent pathways targeting the promoter and/or enhancers of the same gene. The balance between these two components would determine the degree of Smad4 dependency of the TGF-β response for these genes.
For the majority of the genes tested, the protein expression matched the mRNA expression in response to TGF-β signaling. One exception to this rule was p21. The Smad dependency status of p21 is still poorly understood and has led to contradictory results in different studies (29, 33, 45, 68). Here, we have shown that CDKN1A (p21) mRNA is induced by TGF-β in a Smad4-independent manner when assayed by microarray, RT-PCR, and RNase protection in accordance with previous studies (29). In contrast we found that p21 protein expression absolutely requires Smad4. Therefore, our results indicate that TGF-β might be able to regulate p21 both at the transcriptional level and at the protein level, by regulating p21 stability or possibly translation. The posttranscriptional regulation is evidently dependent on Smad4. Interestingly, a recent study suggested that TGF-β regulates p21 protein stability during the cell cycle (23). Further analysis is now required to define the role of Smad4 in the regulation of p21 protein stability.
The relevance of Smad4 loss for tumor progression.
As Smad4 expression is frequently lost or decreased in pancreatic and colon cancer, the functional consequence of the depletion of Smad4 is obviously relevant to tumorigenesis. We were interested in understanding whether Smad4 loss only results in loss of the tumor-suppressive effects of TGF-β or whether it also played a direct role in TGF-β-induced tumor progression. Our results suggest that knockdown of Smad4 results in loss of a tumor-suppressive function of TGF-β, i.e., cell cycle arrest, but has no effect on a tumor-promoting function of TGF-β, EMT, which is required for cells to acquire a fibroblastic phenotype that promotes invasion and metastasis (60). Activation of TGF-β-induced EMT has been shown to also require activation of the Ras/Erk pathway, which synergizes with the TGF-β signaling pathway (42, 43). The loss of Smad4 in human pancreatic tumors occurs after overexpression of EGF ligands and receptors or Ras activation, which all cause activation of the Ras/Erk pathway (3). Thus, it is likely that the Smad4 loss abolishes the antiproliferative effects of TGF-β but does not affect EMT induced by TGF-β in concert with the Ras/Erk pathway. Given that EMT induces a fibroblastoid cell type that is potentially more migratory, we were surprised to find that Smad4 knockdown abolished TGF-β-induced migration. However, EGF-induced migration was unaffected by the loss of Smad4 and given the overexpression of EGF receptors and ligands in Smad4-null pancreatic tumors (3), this is likely to be relevant for tumor progression. Therefore, our results suggest that deletion or degradation of Smad4 in tumors could specifically inhibit the tumor suppressor effect of TGF-β without affecting its tumor promoter activities, hence acting to promote TGF-β-stimulated tumor cells toward a more aggressive phenotype. Future work will focus on testing this idea in vivo.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank Hans Clevers for the pTER siRNA Tet-inducible vector, Ed Leof for the antibody against phosphorylated Smad3, and Mike Howell for the c-junSBR6-Luc construct. We are grateful to the Cancer Research UK Affymetrix Facility at the Paterson Institute, in particular Yvonne Hey, for performing the microarray analysis; to Simon Tomlinson for help in analyzing the microarray data; to the London Research Institute FACS lab for help with cell cycle analysis, in particular Ayad Eddaoudi; and to Debbie Aubyn for help with the video microscopy and the tracking analysis. We thank Julian Downward, Richard Treisman, and members of the Hill lab for useful discussions and comments on the manuscript.
This work was funded by Cancer Research UK.
Footnotes
REFERENCES
- 1.Akhurst, R. J., and R. Derynck. 2001. TGF-β signaling in cancer—a double-edged sword. Trends Cell Biol. 11**:**S44-S51. [DOI] [PubMed] [Google Scholar]
- 2.Baldus, S. E., E. Schwarz, C. Lohrey, M. Zapatka, S. Landsberg, S. A. Hahn, D. Schmidt, H. P. Dienes, W. H. Schmiegel, and I. Schwarte-Waldhoff. 2005. Smad4 deficiency in cervical carcinoma cells. Oncogene 24**:**810-819. [DOI] [PubMed] [Google Scholar]
- 3.Bardeesy, N., and R. A. DePinho. 2002. Pancreatic cancer biology and genetics. Nat. Rev. Cancer 2**:**897-909. [DOI] [PubMed] [Google Scholar]
- 4.Bhowmick, N. A., M. Ghiassi, A. Bakin, M. Aakre, C. A. Lundquist, M. E. Engel, C. L. Arteaga, and H. L. Moses. 2001. Transforming growth factor-β1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol. Biol. Cell 12**:**27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottinger, E. P., J. L. Jakubczak, D. C. Haines, K. Bagnall, and L. M. Wakefield. 1997. Transgenic mice overexpressing a dominant-negative mutant type II transforming growth factor β receptor show enhanced tumorigenesis in the mammary gland and lung in response to the carcinogen 7,12-dimethylbenz-[a]-anthracene. Cancer Res. 57**:**5564-5570. [PubMed] [Google Scholar]
- 6.Bridge, A. J., S. Pebernard, A. Ducraux, A. L. Nicoulaz, and R. Iggo. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34**:**263-264. [DOI] [PubMed] [Google Scholar]
- 7.Chu, G. C., N. R. Dunn, D. C. Anderson, L. Oxburgh, and E. J. Robertson. 2004. Differential requirements for Smad4 in TGFβ-dependent patterning of the early mouse embryo. Development 131**:**3501-3512. [DOI] [PubMed] [Google Scholar]
- 8.Cui, W., D. J. Fowlis, S. Bryson, E. Duffie, H. Ireland, A. Balmain, and R. J. Akhurst. 1996. TGFβ1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell 86**:**531-542. [DOI] [PubMed] [Google Scholar]
- 9.Dai, J. L., M. Schutte, R. K. Bansal, R. E. Wilentz, A. Y. Sugar, and S. E. Kern. 1999. Transforming growth factor-β responsiveness in DPC4/SMAD4-null cancer cells. Mol. Carcinog. 26**:**37-43. [DOI] [PubMed] [Google Scholar]
- 10.de Jong, J. S., P. J. van Diest, P. van der Valk, and J. P. Baak. 1998. Expression of growth factors, growth-inhibiting factors, and their receptors in invasive breast cancer. II. Correlations with proliferation and angiogenesis. J. Pathol. 184**:**53-57. [DOI] [PubMed] [Google Scholar]
- 11.Denissova, N. G., C. Pouponnot, J. Long, D. He, and F. Liu. 2000. Transforming growth factor β-inducible independent binding of SMAD to the Smad7 promoter. Proc. Natl. Acad. Sci. USA 97**:**6397-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennler, S., S. Itoh, D. Vivien, P. ten Dijke, S. Huet, and J. M. Gauthier. 1998. Direct binding of Smad3 and Smad4 to critical TGF β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17**:**3091-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derynck, R., R. J. Ackhurst, and A. Balmain. 2001. TGF-β signaling in tumor suppression and cancer progression. Nat. Genet. 29**:**117-129. [DOI] [PubMed] [Google Scholar]
- 14.Derynck, R., and Y. E. Zhang. 2003. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 425**:**577-584. [DOI] [PubMed] [Google Scholar]
- 15.de Winter, J. P., B. A. Roelen, P. ten Dijke, B. van der Burg, and A. J. van den Eijnden-van Raaij. 1997. DPC4 (SMAD4) mediates transforming growth factor-β1 (TGF-β1) induced growth inhibition and transcriptional response in breast tumour cells. Oncogene 14**:**1891-1899. [DOI] [PubMed] [Google Scholar]
- 16.Edlund, S., M. Landstrom, C. H. Heldin, and P. Aspenstrom. 2002. Transforming growth factor-β-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol. Biol. Cell 13**:**902-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellenrieder, V., S. F. Hendler, W. Boeck, T. Seufferlein, A. Menke, C. Ruhland, G. Adler, and T. M. Gress. 2001. Transforming growth factor β1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 61**:**4222-4228. [PubMed] [Google Scholar]
- 18.Factor, V. M., C. Y. Kao, E. Santoni-Rugiu, J. T. Woitach, M. R. Jensen, and S. S. Thorgeirsson. 1997. Constitutive expression of mature transforming growth factor β1 in the liver accelerates hepatocarcinogenesis in transgenic mice. Cancer Res. 57**:**2089-2095. [PubMed] [Google Scholar]
- 19.Feng, X. H., X. Lin, and R. Derynck. 2000. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-β. EMBO J. 19**:**5178-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fink, S. P., D. Mikkola, J. K. Willson, and S. Markowitz. 2003. TGF-β-induced nuclear localization of Smad2 and Smad3 in Smad4 null cancer cell lines. Oncogene 22**:**1317-1323. [DOI] [PubMed] [Google Scholar]
- 21.Fink, S. P., S. E. Swinler, J. D. Lutterbaugh, J. Massague, S. Thiagalingam, K. W. Kinzler, B. Vogelstein, J. K. Willson, and S. Markowitz. 2001. Transforming growth factor-β-induced growth inhibition in a Smad4 mutant colon adenoma cell line. Cancer Res. 61**:**256-260. [PubMed] [Google Scholar]
- 22.Germain, S., M. Howell, G. M. Esslemont, and C. S. Hill. 2000. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 14**:**435-451. [PMC free article] [PubMed] [Google Scholar]
- 23.Gong, J., S. Ammanamanchi, T. C. Ko, and M. G. Brattain. 2003. Transforming growth factor β1 increases the stability of p21/WAF1/CIP1 protein and inhibits CDK2 kinase activity in human colon carcinoma FET cells. Cancer Res. 63**:**3340-3346. [PubMed] [Google Scholar]
- 24.Hahn, S. A., M. Schutte, A. T. Hoque, C. A. Moskaluk, L. T. da Costa, E. Rozenblum, C. L. Weinstein, A. Fischer, C. J. Yeo, R. H. Hruban, and S. E. Kern. 1996. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 271**:**350-353. [DOI] [PubMed] [Google Scholar]
- 25.Hanafusa, H., J. Ninomiya-Tsuji, N. Masuyama, M. Nishita, J. Fujisawa, H. Shibuya, K. Matsumoto, and E. Nishida. 1999. Involvement of the p38 mitogen-activated protein kinase pathway in transforming growth factor-β-induced gene expression. J. Biol. Chem. 274**:**27161-27167. [DOI] [PubMed] [Google Scholar]
- 26.Hartsough, M. T., and K. M. Mulder. 1995. Transforming growth factor β activation of p44mapk in proliferating cultures of epithelial cells. J. Biol. Chem. 270**:**7117-7124. [DOI] [PubMed] [Google Scholar]
- 27.Hocevar, B. A., T. L. Brown, and P. H. Howe. 1999. TGF-β induces fibronectin synthesis through a c-Jun N-terminal kinase-dependent, Smad4-independent pathway. EMBO J. 18**:**1345-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howe, J. R., S. Roth, J. C. Ringold, R. W. Summers, H. J. Jarvinen, P. Sistonen, I. P. Tomlinson, R. S. Houlston, S. Bevan, F. A. Mitros, E. M. Stone, and L. A. Aaltonen. 1998. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science 280**:**1086-1088. [DOI] [PubMed] [Google Scholar]
- 29.Ijichi, H., M. Otsuka, K. Tateishi, T. Ikenoue, T. Kawakami, F. Kanai, Y. Arakawa, N. Seki, K. Shimizu, K. Miyazono, T. Kawabe, and M. Omata. 2004. Smad4-independent regulation of p21/WAF1 by transforming growth factor-β. Oncogene 23**:**1043-1051. [DOI] [PubMed] [Google Scholar]
- 30.Inman, G. J., and C. S. Hill. 2002. Stoichiometry of active Smad-transcription factor complexes on DNA. J. Biol. Chem. 277**:**51008-51016. [DOI] [PubMed] [Google Scholar]
- 31.Inman, G. J., F. J. Nicolás, J. F. Callahan, J. D. Harling, L. M. Gaster, A. D. Reith, N. J. Laping, and C. S. Hill. 2002. SB-431542 is a potent and specific inhibitor of Transforming Growth Factor-β superfamily type I activin receptor-like kinase receptors, ALK4, ALK5 and ALK7. Mol. Pharmacol. 62**:**65-72. [DOI] [PubMed] [Google Scholar]
- 32.Jackson, A. L., S. R. Bartz, J. Schelter, S. V. Kobayashi, J. Burchard, M. Mao, B. Li, G. Cavet, and P. S. Linsley. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21**:**635-637. [DOI] [PubMed] [Google Scholar]
- 33.Jazag, A., H. Ijichi, F. Kanai, T. Imamura, B. Guleng, M. Ohta, J. Imamura, Y. Tanaka, K. Tateishi, T. Ikenoue, T. Kawakami, Y. Arakawa, M. Miyagishi, K. Taira, T. Kawabe, and M. Omata. 2005. Smad4 silencing in pancreatic cancer cell lines using stable RNA interference and gene expression profiles induced by transforming growth factor-β. Oncogene 24**:**662-671. [DOI] [PubMed] [Google Scholar]
- 34.Jonk, L. J., S. Itoh, C. H. Heldin, P. ten Dijke, and W. Kruijer. 1998. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J. Biol. Chem. 273**:**21145-21152. [DOI] [PubMed] [Google Scholar]
- 35.Kang, Y., C. R. Chen, and J. Massagué. 2003. A self-enabling TGFβ response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell 11**:**915-926. [DOI] [PubMed] [Google Scholar]
- 36.Kretschmer, A., K. Moepert, S. Dames, M. Sternberger, J. Kaufmann, and A. Klippel. 2003. Differential regulation of TGF-β signaling through Smad2, Smad3 and Smad4. Oncogene 22**:**6748-6763. [DOI] [PubMed] [Google Scholar]
- 37.Laping, N. J., E. Grygielko, A. Mathur, S. Butter, J. Bomberger, C. Tweed, J. Fornwald, R. Lehr, J. D. Harling, L. M. Gaster, J. F. Callahan, and B. A. Olson. 2002. Inhibition of transforming growth factor (TGF)-β1-induced extracellular matrix with a novel inhibitor of the TGF-β type I receptor kinase activity. Mol. Pharmacol. 62**:**58-64. [DOI] [PubMed] [Google Scholar]
- 38.Li, F., J. Goncalves, K. Faughnan, M. G. Steiner, I. Pagan-Charry, D. Esposito, B. Chin, K. M. Providence, P. J. Higgins, and L. Staiano-Coico. 2000. Targeted inhibition of wound-induced PAI-1 expression alters migration and differentiation in human epidermal keratinocytes. Exp. Cell Res. 258**:**245-253. [DOI] [PubMed] [Google Scholar]
- 39.Li, W., W. Qiao, L. Chen, X. Xu, X. Yang, D. Li, C. Li, S. G. Brodie, M. M. Meguid, L. Hennighausen, and C. X. Deng. 2003. Squamous cell carcinoma and mammary abscess formation through squamous metaplasia in Smad4/Dpc4 conditional knockout mice. Development 130**:**6143-6153. [DOI] [PubMed] [Google Scholar]
- 40.Maurice, D., C. E. Pierreux, M. Howell, R. E. Wilentz, M. J. Owen, and C. S. Hill. 2001. Loss of Smad4 function in pancreatic tumors: C-terminal truncation leads to decreased stability. J. Biol. Chem. 276**:**43175-43181. [DOI] [PubMed] [Google Scholar]
- 41.Nicolás, F. J., and C. S. Hill. 2003. Attenuation of the TGFβ-Smad signaling pathway in pancreatic tumor cells confers resistance to TGFβ-induced growth arrest. Oncogene 22**:**3698-3711. [DOI] [PubMed] [Google Scholar]
- 42.Oft, M., K. H. Heider, and H. Beug. 1998. TGFβ signaling is necessary for carcinoma cell invasiveness and metastasis. Curr. Biol. 8**:**1243-1252. [DOI] [PubMed] [Google Scholar]
- 43.Oft, M., J. Peli, C. Rudaz, H. Schwarz, H. Beug, and E. Reichmann. 1996. TGF-β1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 10**:**2462-2477. [DOI] [PubMed] [Google Scholar]
- 44.Ozdamar, B., R. Bose, M. Barrios-Rodiles, H. R. Wang, Y. Zhang, and J. L. Wrana. 2005. Regulation of the polarity protein Par6 by TGFβ receptors controls epithelial cell plasticity. Science 307**:**1603-1609. [DOI] [PubMed] [Google Scholar]
- 45.Pardali, K., A. Kurisaki, A. Moren, P. ten Dijke, D. Kardassis, and A. Moustakas. 2000. Role of Smad proteins and transcription factor Sp1 in p21(Waf1/Cip1) regulation by transforming growth factor-β. J. Biol. Chem. 275**:**29244-29256. [DOI] [PubMed] [Google Scholar]
- 46.Pierreux, C. E., F. J. Nicolás, and C. S. Hill. 2000. Transforming growth factor β-independent shuttling of Smad4 between the cytoplasm and nucleus. Mol. Cell. Biol. 20**:**9041-9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Providence, K. M., and P. J. Higgins. 2004. PAI-1 expression is required for epithelial cell migration in two distinct phases of in vitro wound repair. J. Cell Physiol. 200**:**297-308. [DOI] [PubMed] [Google Scholar]
- 48.Ramachandra, M., I. Atencio, A. Rahman, M. Vaillancourt, A. Zou, J. Avanzini, K. Wills, R. Bookstein, and P. Shabram. 2002. Restoration of transforming growth factor β signaling by functional expression of Smad4 induces anoikis. Cancer Res. 62**:**6045-6051. [PubMed] [Google Scholar]
- 49.Saika, S., S. Kono-Saika, Y. Ohnishi, M. Sato, Y. Muragaki, A. Ooshima, K. C. Flanders, J. Yoo, M. Anzano, C. Y. Liu, W. W. Kao, and A. B. Roberts. 2004. Smad3 signaling is required for epithelial-mesenchymal transition of lens epithelium after injury. Am. J. Pathol. 164**:**651-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Savagner, P., K. M. Yamada, and J. P. Thiery. 1997. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J. Cell Biol. 137**:**1403-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwarte-Waldhoff, I., S. Klein, S. Blass-Kampmann, A. Hintelmann, C. Eilert, S. Dreschers, H. Kalthoff, S. A. Hahn, and W. Schmiegel. 1999. DPC4/SMAD4 mediated tumor suppression of colon carcinoma cells is associated with reduced urokinase expression. Oncogene 18**:**3152-3158. [DOI] [PubMed] [Google Scholar]
- 52.Schwarte-Waldhoff, I., O. V. Volpert, N. P. Bouck, B. Sipos, S. A. Hahn, S. Klein-Scory, J. Luttges, G. Kloppel, U. Graeven, C. Eilert-Micus, A. Hintelmann, and W. Schmiegel. 2000. Smad4/DPC4-mediated tumor suppression through suppression of angiogenesis. Proc. Natl. Acad. Sci. USA 97**:**9624-9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seoane, J., H. V. Le, L. Shen, S. A. Anderson, and J. Massagué. 2004. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117**:**211-223. [DOI] [PubMed] [Google Scholar]
- 54.Shen, X., J. Li, P. P. Hu, D. Waddell, J. Zhang, and X. F. Wang. 2001. The activity of guanine exchange factor NET1 is essential for transforming growth factor-β-mediated stress fiber formation. J. Biol. Chem. 276**:**15362-15368. [DOI] [PubMed] [Google Scholar]
- 55.Shi, Y., and J. Massagué. 2003. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113**:**685-700. [DOI] [PubMed] [Google Scholar]
- 56.Sirard, C., S. Kim, C. Mirtsos, P. Tadich, P. A. Hoodless, A. Itie, R. Maxson, J. L. Wrana, and T. W. Mak. 2000. Targeted disruption in murine cells reveals variable requirement for Smad4 in transforming growth factor β-related signaling. J. Biol. Chem. 275**:**2063-2070. [DOI] [PubMed] [Google Scholar]
- 57.Stopa, M., D. Anhuf, L. Terstegen, P. Gatsios, A. M. Gressner, and S. Dooley. 2000. Participation of Smad2, Smad3, and Smad4 in transforming growth factor β (TGF-β)-induced activation of Smad7. The TGF-β response element of the promoter requires functional Smad binding element and E-box sequences for transcriptional regulation. J. Biol. Chem. 275**:**29308-29317. [DOI] [PubMed] [Google Scholar]
- 58.Tang, B., E. P. Bottinger, S. B. Jakowlew, K. M. Bagnall, J. Mariano, M. R. Anver, J. J. Letterio, and L. M. Wakefield. 1998. Transforming growth factor-β1 is a new form of tumor suppressor with true haploid insufficiency. Nat. Med. 4**:**802-807. [DOI] [PubMed] [Google Scholar]
- 59.ten Dijke, P., and C. S. Hill. 2004. New insights into TGF-β-Smad signalling. Trends Biochem. Sci. 29**:**265-273. [DOI] [PubMed] [Google Scholar]
- 60.Thiery, J. P. 2002. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2**:**442-454. [DOI] [PubMed] [Google Scholar]
- 61.Torre-Amione, G., R. D. Beauchamp, H. Koeppen, B. H. Park, H. Schreiber, H. L. Moses, and D. A. Rowley. 1990. A highly immunogenic tumor transfected with a murine transforming growth factor type β1 cDNA escapes immune surveillance. Proc. Natl. Acad. Sci. USA 87**:**1486-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsushima, H., S. Kawata, S. Tamura, N. Ito, Y. Shirai, S. Kiso, Y. Imai, H. Shimomukai, Y. Nomura, Y. Matsuda, and Y. Matsuzawa. 1996. High levels of transforming growth factor β1 in patients with colorectal cancer: association with disease progression. Gastroenterology 110**:**375-382. [DOI] [PubMed] [Google Scholar]
- 63.van de Wetering, M., I. Oving, V. Muncan, M. T. Pon Fong, H. Brantjes, D. van Leenen, F. C. Holstege, T. R. Brummelkamp, R. Agami, and H. Clevers. 2003. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 4**:**609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wikstrom, P., P. Stattin, I. Franck-Lissbrant, J. E. Damber, and A. Bergh. 1998. Transforming growth factor β1 is associated with angiogenesis, metastasis, and poor clinical outcome in prostate cancer. Prostate 37**:**19-29. [DOI] [PubMed] [Google Scholar]
- 65.Wilkes, M. C., S. J. Murphy, N. Garamszegi, and E. B. Leof. 2003. Cell-type-specific activation of PAK2 by transforming growth factor β independent of Smad2 and Smad3. Mol. Cell. Biol. 23**:**8878-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wisotzkey, R. G., A. Mehra, D. J. Sutherland, L. L. Dobens, X. Liu, C. Dohrmann, L. Attisano, and L. A. Raftery. 1998. Medea is a Drosophila Smad4 homolog that is differentially required to potentiate DPP responses. Development 125**:**1433-1445. [DOI] [PubMed] [Google Scholar]
- 67.Wong, C., E. M. Rougier-Chapman, J. P. Frederick, M. B. Datto, N. T. Liberati, J.-M. Li, and X.-F. Wang. 1999. Smad3-Smad4 and AP-1 complexes synergize in transcriptional activation of the c-Jun promoter by transforming growth factor β. Mol. Cell. Biol. 19**:**1821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan, Z., G. Y. Kim, X. Deng, and E. Friedman. 2002. Transforming growth factor β1 induces proliferation in colon carcinoma cells by Ras-dependent, Smad-independent down-regulation of p21cip1. J. Biol. Chem. 277**:**9870-9879. [DOI] [PubMed] [Google Scholar]
- 69.Yi, J. Y., I. Shin, and C. L. Arteaga. 2005. Type I transforming growth factor β receptor binds to and activates phosphatidylinositol 3-kinase. J. Biol. Chem. 280**:**10870-10876. [DOI] [PubMed] [Google Scholar]
- 70.Zavadil, J., M. Bitzer, D. Liang, Y. C. Yang, A. Massimi, S. Kneitz, E. Piek, and E. P. Bottinger. 2001. Genetic programs of epithelial cell plasticity directed by transforming growth factor-β. Proc. Natl. Acad. Sci. USA 98**:**6686-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zavadil, J., L. Cermak, N. Soto-Nieves, and E. P. Bottinger. 2004. Integration of TGF-β/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 23**:**1155-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, Y., X. Feng, R. We, and R. Derynck. 1996. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature 383**:**168-172. [DOI] [PubMed] [Google Scholar]
- 73.Zhou, S., P. Buckhaults, L. Zawel, F. Bunz, G. Riggins, J. L. Dai, S. E. Kern, K. W. Kinzler, and B. Vogelstein. 1998. Targeted deletion of Smad4 shows it is required for transforming growth factor β and activin signaling in colorectal cancer cells. Proc. Natl. Acad. Sci. USA 95**:**2412-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]