Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres (original) (raw)
Abstract
Here we analyze the functional interaction between Ku86 and telomerase at the mammalian telomere by studying mice deficient for both proteins. We show that absence of Ku86 prevents the end-to-end chromosomal fusions that result from critical telomere shortening in telomerase-deficient mice. In addition, Ku86 deficiency rescues the male early germ cell apoptosis triggered by short telomeres in these mice. Together, these findings define a role for Ku86 in mediating chromosomal instability and apoptosis triggered by short telomeres. In addition, we show here that Ku86 deficiency results in telomerase-dependent telomere elongation and in the fusion of random pairs of chromosomes in telomerase-proficient cells, suggesting a model in which Ku86 keeps normal-length telomeres less accessible to telomerase- mediated telomere lengthening and to DNA repair activities.
Keywords: apoptosis/Ku86/telomerase/telomere dysfunction
Introduction
Telomeres are protective structures at the ends of chromosomes composed of repetitive DNA (TTAGGG repeats in vertebrates) and associated proteins (reviewed in Blackburn, 2000, 2001). Additionally, telomeres have a 3′ G-rich single-stranded DNA (ssDNA) overhang (G-strand overhang), which can invade the duplex telomeric repeats, forming telomeric ‘T-loops’ (Griffith et al., 1999). In mammals, TRF1 and TRF2 proteins bind to TTAGGG repeats (Bilaud et al., 1997; van Steensel et al., 1997, 1998; Smogorzewska et al., 2000). TRF1 and TRF2 are also found at the telomeric T-loops, and their effects on telomere length and end protection could be mediated by this property (Griffith et al., 1999). In addition, TRF1 and TRF2 recruit to the telomere various DNA double-strand break (DSB) repair proteins, such as Ku proteins and the Rad50/Mre11/Nbs1 complex (Bianchi and de Lange, 1999; Hsu et al., 1999, 2000; Song et al., 2000; Zhu et al., 2000). A ssDNA-binding protein, named Pot-1, has recently been proposed to bind to the 3′ G-strand overhang (Baumann and Cech, 2001).
Loss of telomere function can result from (i) loss of TTAGGG repeats with age or with increasing cell divisions and/or from (ii) loss or mutation of telomere proteins such as TRF2, Ku86 or DNA-PKcs, in the absence of telomere shortening (van Steensel et al., 1998; Samper et al., 2000; Goytisolo et al., 2001). In turn, loss of telomere function results in end-to-end chromosomal fusions and compromised cell viability. The underlying mechanisms by which TTAGGG repeats and/or telomere-binding proteins maintain the telomeric protective function, however, are largely unknown.
Telomerase synthesizes telomeres de novo, hence preventing telomere shortening in those cells where it is expressed at sufficiently high levels. Telomerase consists of two essential components, a reverse transcriptase known as Tert (telomerase reverse transcriptase) and an RNA molecule or Terc (telomerase RNA component), which contains the template for the synthesis of new telomeric repeats (reviewed in Collins, 2000). Telomerase activity has been proposed to be relevant for both cancer and aging (reviewed in Shay and Bacchetti, 1997). Mice that lack telomerase and have critically short telomeres exhibit severe proliferative defects (Blasco et al., 1997; Lee et al., 1998; Rudolph et al., 1999; Herrera et al., 1999a,b, 2000), and, in the presence of wild-type p53, they are less susceptible to developing tumors (Chin et al., 1999; Greenberg et al., 1999; González-Suárez et al., 2000; Rudolph et al., 2001). In turn, constitutive telomerase expression in adult transgenic tissues makes them more susceptible to tumorigenesis (González-Suárez et al., 2001).
Here, we have impaired both telomerase-mediated telomere maintenance and Ku86 function by generating double knockout Terc–/–/Ku86–/– mice. Our results define a fundamental role for Ku86 in the chromosomal aberrations and apoptosis associated with critical telomere shortening. In particular, we show that Ku86 deficiency prevents the fusion of chromosomes with critically short telomeres, as well as the early germline apoptosis associated with short telomeres in late-generation telomerase-deficient males (Lee et al., 1998; Hemann et al., 2001a). These findings have important implications for understanding the origin of the chromosomal aberrations associated with cancer and aging. Furthermore, we demonstrate a functional interaction between Ku86 and telomerase. In particular, the results described here show that Ku86 deficiency in a Terc+/+ mouse background leads to a reproducible telomere elongation, confirming previous data from our group (Samper et al., 2000). Importantly, we show here that Ku86 deficiency does not result in elongated telomeres when in a telomerase-deficient background, indicating that Ku86 is directly or indirectly impairing telomerase-mediated telomere elongation. Hence, Ku86 is one of the first negative regulators of telomerase-mediated telomere elongation that is demonstrated in the context of a mammalian organism.
Results
Generation of Terc–/–/Ku86–/– mice with progressively shorter telomeres
Double heterozygous Terc+/–/Ku86+/– mice were derived from crossing Terc+/– and Ku86+/– mice in a C57Bl/6 background (Herrera et al., 1999b; Samper et al., 2000). Terc+/–/Ku86+/– intercrosses produced the first generation (G1) of double mutant mice (Figure 1). Successive generations of Terc–/–/Ku86–/– mice, as well as the corresponding Terc–/–/Ku86+/+ littermate controls, were obtained by serial intercrossing of Terc–/–/Ku86+/– mice until the fourth generation (G4). At this point, the G4 Terc–/–/Ku86+/– mice became infertile due to critical telomere shortening and no further generations could be obtained, as described previously for Terc–/– mice in a C57Bl/6 background (Herrera et al., 1999b) (Figure 1; the genotypes used in the current analysis are printed in gray).
Fig. 1. Generation of the different mutant mice. The genotypes printed in gray are those used for the analysis.
Ku86 impairs telomerase-dependent telomere elongation
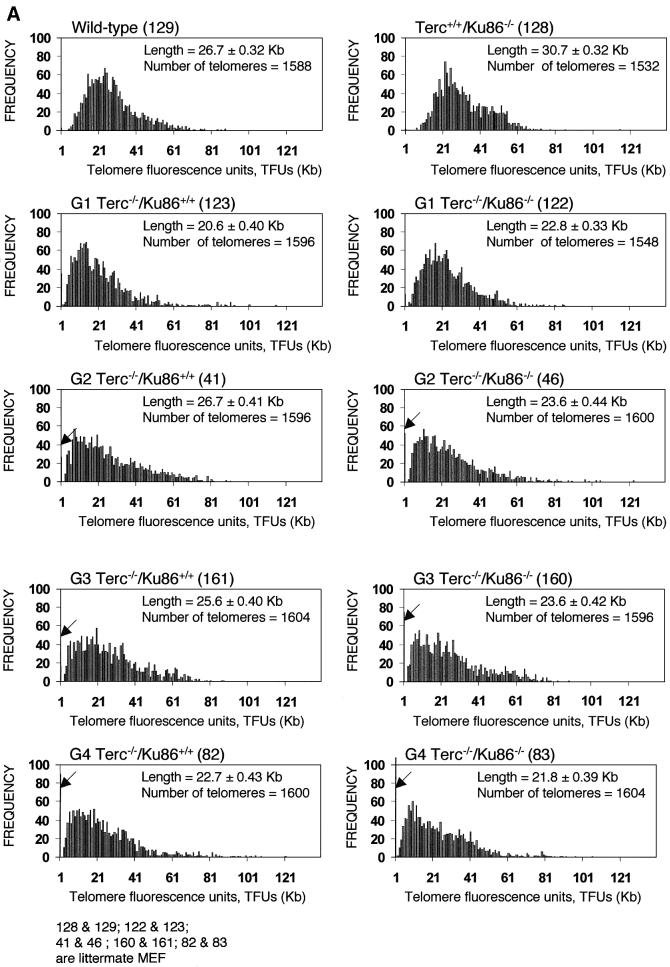
We observed previously that Ku86-deficient cells showed a reproducible telomere elongation compared with wild-type controls despite normal telomerase activity in these cells (Samper et al., 2000). To confirm these preliminary observations and to study the mechanism by which Ku86 deficiency results in telomere elongation, we measured telomere length in mice that lack both Ku86 and telomerase. For this, quantitative fluorescence in situ hybridization (Q-FISH) of telomeres was carried out on metaphases from passage 1 mouse embryonic fibroblasts (MEFs). Average telomere lengths and standard errors were 26.7 ± 0.32 and 30.7 ± 0.32 kb for wild-type and Terc+/+/Ku86–/– MEFs, respectively, indicating that Ku86 deficiency results in a reproducible and significant telomere elongation. This telomere elongation is illustrated by increased frequencies of longer telomeres in Terc+/+/Ku86–/– MEFs compared with the wild-type controls (see histograms in Figure 2A). Student’s _t_-test including >1000 telomere length values from each genotype indicated that the difference was highly significant: P = 0.02 (Figure 2A). As expected, increasing generations of Terc–/–/Ku86+/+ MEFs showed progressive telomere shortening as indicated by both (i) increasingly higher frequencies of short telomeres (see histograms in Figure 2A; arrows indicate telomeres <1 kb) and (ii) a higher percentage of chromosome ends with undetectable TTAGGG signals (Figure 2B). These two parameters are better indicators of critical telomere shortening than the average telomere length values, as illustrated in Figure 2A–C and as also shown previously (Blasco et al., 1997; Hemann et al., 2001b; Samper et al., 2001b). Importantly, Terc–/–/Ku86–/– MEFs showed a similar rate of telomere loss as the corresponding Terc–/–/Ku86+/+ controls (Figure 2A and B), indicating that Ku86 deficiency does not result in elongated telomeres when in a telomerase-deficient background. Hence, Ku86 acts as a negative regulator of telomerase-dependent telomere elongation (see Discussion).
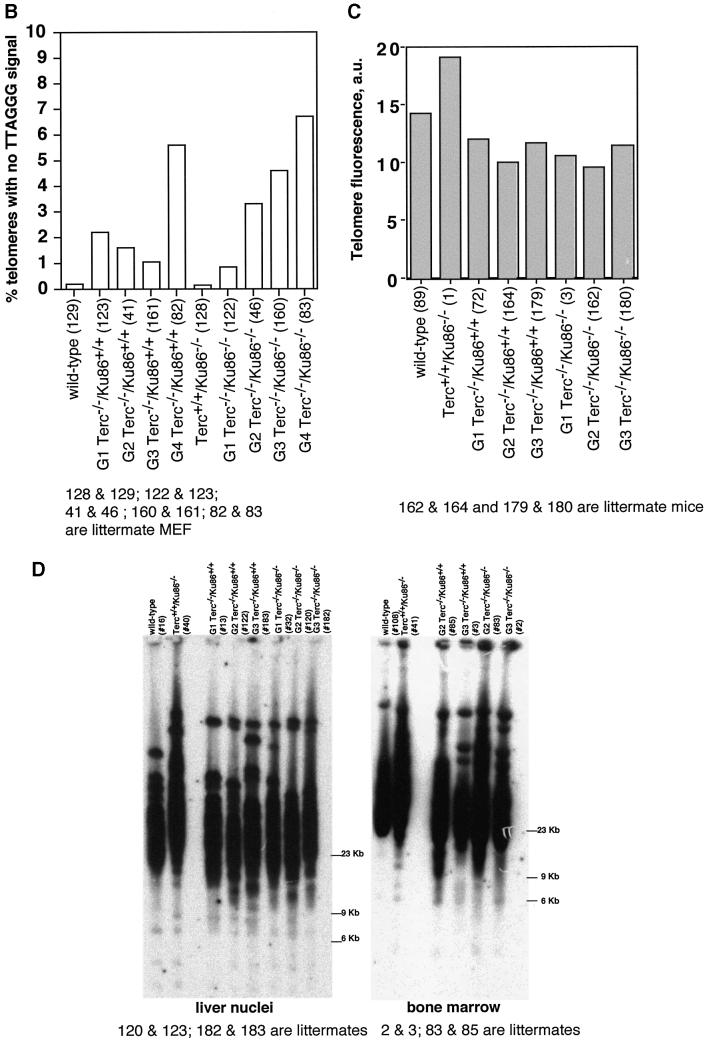
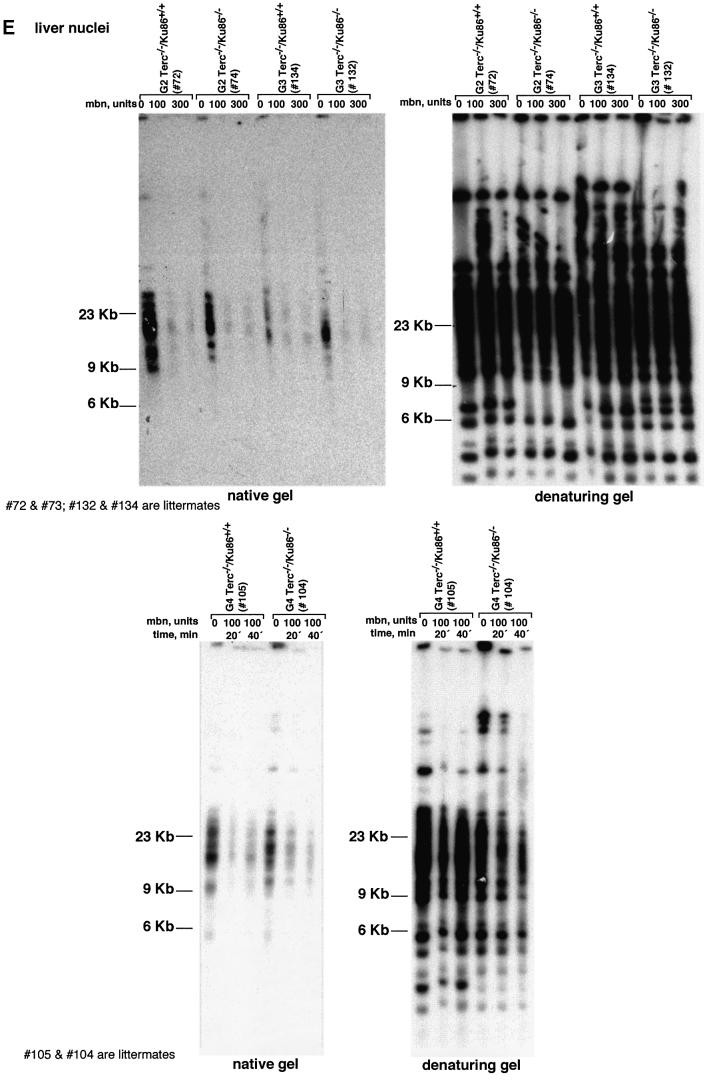
Fig. 2. (A) Telomere length distribution in primary MEFs. One TFU corresponds to 1 kb of TTAGGG repeats (Zijlmans et al., 1997). Arrows indicate telomeres with <1 kb of TTAGGG repeats. Average telomere length and the standard error, as well as the total number of telomeres analyzed, are indicated. Note that standard error and not standard deviation is shown. The distribution of the telomere length frequencies for each MEF is an indication of the standard deviation. Littermate MEFs are shown. (B) Quantification using Q-FISH of the percentage of telomeres with undetectable TTAGGG repeats out of the total number of telomeres shown in (A). Littermate MEFs are indicated. (C) Telomeric fluorescence (in arbitrary units, a.u.) using flow cytometry combined with telomeric Q-FISH (Flow-FISH). Littermate mice are indicated. (D) TRF analysis of primary BM cells or liver nuclei, as indicated, from different genotypes. Notice the upwards shift of TRF fragments in Terc+/+/Ku86–/– compared with the wild type. Littermate mice are indicated. (E) G-strand overhangs using liver nuclei visualized in the native gel after hybridization with a (CCCTAA)4 probe. Notice that upon treatment with mung bean nuclease (mbn), the G-strand-specific signal decreases. As control, the same gel was denatured and reprobed with the (CCCTAA)4 probe to visualize telomeres. Table I shows quantification of the G-strand overhang signals corrected by the denaturing gel hybridization signal for the 0 units of mbn lanes. Littermate mice are indicated.
This conclusion was supported further when telomere fluorescence was measured by flow cytometry (Flow-FISH) in fresh bone marrow (BM) cells. Again, the average telomere fluorescence values were 135% greater in the Terc+/+/Ku86–/– mice than in control wild-type mice (Figure 2C). This effect was not observed when comparing Terc–/–/Ku86–/– mice with the corresponding Terc–/–/Ku86+/+ controls (Figure 2C), indicating again that Ku86 deficiency does not result in elongated telomeres in telomerase-deficient cells.
Telomere length was also evaluated by terminal restriction fragment (TRF) analysis using both hepatocyte nuclei and BM cells. In the two cell types, TRF analysis showed that Ku86 deficiency results in extended telomeres in a Terc+/+ background, as shown by a clear shift towards larger TRF bands in Terc+/+/Ku86–/– cells compared with the wild-type controls (Figure 2D). This, however, was not observed when comparing different generation Terc–/–/Ku86–/– and Terc–/–/Ku86+/+ mice (Figure 2D), again showing that Ku86 deficiency does not result in elongated telomeres in cells that lack telomerase activity. All together, these results indicate that Ku86 negatively regulates telomerase-dependent telomere elongation.
TRF analysis using native gel conditions (Materials and methods) was used to examine the integrity of the telomeric G-strand overhang as a possible mechanism by which simultaneous Ku86 and Terc deficiencies may affect telomere length and function. However, all different generations of the double mutant Terc–/–/Ku86–/– mice showed G-strand-specific signals that were not significantly different from those of the corresponding Terc–/–/Ku86+/+ controls (Figure 2E), as also demonstrated previously for the single Terc–/– or Ku86–/– mutant mice (Hemann and Greider, 1999; Samper et al., 2000). Table I shows quantification of G-strand signals, which were similar in intensity in the different generation Terc–/–/Ku86–/– mice compared with the corresponding Terc–/–/Ku86+/+ controls; G-strand intensity values in Terc–/–/ Ku86+/+ controls were normalized to 100% (Materials and methods). These observations suggest that the length of the G-strand overhang does not depend on telomerase and Ku86 activities. This is in contrast to what has been reported for Ku-deficient Saccharomyces cerevisiae mutants, which show significantly elongated G-strand overhangs (Gravel et al., 1998).
Table I. G-strand signal quantification.
| Genotype | G-strand (%) |
|---|---|
| G1 Terc–/–/Ku86+/+ (187)a | 100d |
| G2 Terc–/–/Ku86+/+ (72)b | 100d |
| G3 Terc–/–/Ku86+/+ (134)b | 100d |
| G4 Terc–/–/Ku86+/+ (105)c | 100d |
| G1 Terc–/–/Ku86–/– (151)a | 105 |
| G2 Terc–/–/Ku86–/– (74)b | 77 |
| G3 Terc–/–/Ku86–/– (132)b | 100 |
| G4 Terc–/–/Ku86–/– (104)c | 121 |
Ku86 mediates the fusion of chromosomes with critically short telomeres
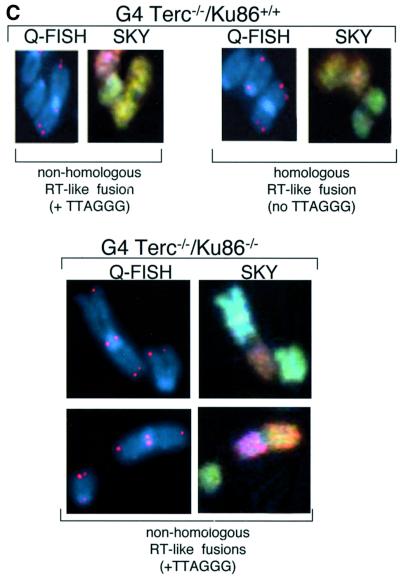
Telomerase deficiency results in loss of telomeric repeats at chromosome ends and in end-to-end fusions (Blasco et al., 1997). These end-to-end fusions have been speculated to result from telomere-exhausted chromosomes being recognized as damaged DNA and ‘fused’ by the DNA repair machinery (Hemann et al., 2001b). This is supported by the fact that late-generation telomerase-deficient mice have been shown to have normal non-homologous end joining (NHEJ) and homologous recombination (HR) activities (Goytisolo et al., 2000). However, the precise mechanism that originates the end-to-end fusions in late-generation telomerase-deficient mice has not been demonstrated to date. In turn, Ku86 deficiency also results in telomere end-to-end fusions in the absence of telomere shortening (Hsu et al., 2000; Samper et al., 2000). To study the mechanisms underlying the end-to-end fusions associated with either Terc–/– or Ku86–/– deficiencies, we performed combined Q-FISH and spectral karyotyping (SKY) on chromosomal aberrations spontaneously arising in passage 1 MEFs from all the different genotypes (Table II; Figure 3). End-to-end fusions through the p-arms (Robertsonian-like, RT-like) were divided into two groups: (i) those with TTAGGG signals at the fusion point (TTAGGG in Table II and white bars in Figure 3A; see Figure 3C for examples); and (ii) those that completely lacked TTAGGG signals at the fusion point (no TTAGGG in Table II and black bars in Figure 3A; see Figure 3C for examples).
Table II. Combined Q-FISH and spectral cytogenetic analysis (SKY) of primary MEFs from all different genotypes.
| Genotype | No. of meta | RT-like fusionsa | DCa | RT + DCa | Breaks at Chrb | Gaps at Chrb | No. of fragments | Breaks + gaps + | |
|---|---|---|---|---|---|---|---|---|---|
| phases | TTAGGG | No TTAGGG | fragmentsa | ||||||
| Terc+/+/Ku86+/+ (129) | 54 | 0.018a (8–10)c | 0.0 | 0.0 | 0.018 | 0.0 | |||
| G1 Terc–/–/Ku86+/+ (123) | 40 | 0.0 | 0.0 | 0.0 | 0.0 | 6 | 4 | 0.125 | |
| G2 Terc–/–/Ku86+/+ (41) | 26 | 0.0 | 0.04 (16–16) | 0.0 | 0.04 | 7 | 1 | 0.077 | |
| G3 Terc–/–/Ku86+/+ (161) | 51 | 0.0 | 0.02 (?–? H) | 0.0 | 0.02 | 0.0 | |||
| G4 Terc–/–/Ku86+/+ (82) | 44 | 0.045 (9–15; 19–15) | 0.068 (5–5; 17–17; 14–14) | 0.0 | 0.113 | 1 | 0.023 | ||
| Terc+/+/Ku86–/– (126) | 61 | 0.066 (3–5; 9–12; 10–13; 17–19) | 0.0 | 0.016 (10–10) | 0.082 | 17; 13; 16 | 0.049 | ||
| G1 Terc–/–/Ku86–/– (122) | 28 | 0.18 (1–6; 7–13; 7–13; 7–13; 12–15) | 0.0 | 0.036 (4–11) | 0.216 | 12; 16; ? | 7 | 0.36 | |
| G2 Terc–/–/Ku86–/– (43) | 69 | 0.029 (11–16; ?–? NH) | 0.015 (15–15) | 0.0 | 0.054 | 10; ? | 8 | 0.145 | |
| G2 Terc–/–/Ku86–/– (46) | 50 | 0.06 (17–16; 6–17; 7–16) | 0.0 | 0.0 | 0.06 | 3 | 0.06 | ||
| G3 Terc–/–/Ku86–/– (160) | 35 | 0.057 (7–11; 9–13) | 0.0 | 0.0 | 0.057 | 4 | 0.114 | ||
| G4 Terc–/–/Ku86–/– (50) | 61 | 0.066 (9–7; 1–9; 5–12; 10–14) | 0.0 | 0.016 (7–7) | 0.082 | 2; 4; 7; 8; 3; 19 | 4; 7; 9; 10; 16 | 9 | 0.328 |
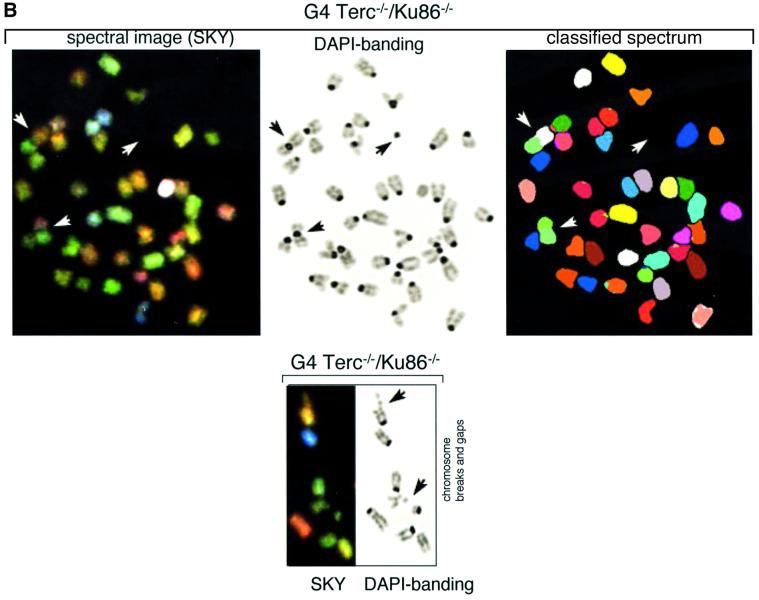
Fig. 3. (A) Frequencies of RT-like fusions per metaphase in primary MEFs from different genotypes. White bars, fusions showing TTAGGG signals at the fusion point and involving non-homologous pairs of chromosomes (NH). Black bars, fusions with undetectable TTAGGG signals at the fusion point and involving homologous pairs of chromosomes (H); these fusions are largely rescued in the absence of Ku86 in late-generation G3 and G4 telomerase-deficient mice. Littermate MEFs are indicated. (B) Top: complete SKY of a metaphase from a G4 Terc–/–/Ku86–/– primary MEF. The corresponding spectral image, the DAPI banding and the classified spectrum are shown for illustrative purposes. Arrowheads point to two RT-like fusions with involvement of non-homologous chromosomes and to one acentric fragment. Bottom: detail from a G4 Terc–/–/Ku86–/– metaphase showing chromosome breaks and gaps from a G4 Terc–/–/Ku86–/– primary MEF. (C) Representative examples of chromosomal aberrations detected by combined Q-FISH and SKY analyses on primary MEFs from the indicated genotypes. Notice that the fusions associated with Ku86 deficiency always show TTAGGG signals at the fusion point and involve random pairs of chromosomes, whereas a fraction of the fusions associated with critical telomere shortening are characterized by lacking TTAGGG signals at the fusion point and involving homologous pairs of chromosomes.
Increasing generations of Terc–/–/Ku86+/+ mice showed an increased frequency of end-to-end fusions compared with the wild-type controls (RT + DC in Table II), a total of 0.113 and 0.018 fusions per metaphase for G4 Terc–/–/Ku86+/+ and wild-type MEFs, respectively. Combined Q-FISH and SKY analyses indicated that a significant fraction (>50%) of the RT-like fusions detected in G4 Terc–/–/Ku86+/+ MEFs were characterized by lacking detectable TTAGGG repeats at the fusion point and by involving homologous pairs of chromosomes (no TTAGGG in Table II and black bars in Figure 3A; see Figure 3C for an example). These fusions are likely to derive from the fusion of chromosomes with critically short telomeres, since they lack detectable TTAGGG signals at the fusion point. The rest of the RT-like fusions detected in G4 Terc–/–/Ku86+/+ MEFs showed at least one detectable TTAGGG signal at the fusion point and involved random pairs of chromosomes (TTAGGG in Table II and white bars in Figure 3A; see Figure 3C for an example). Primary MEFs from Terc+/+/Ku86–/– showed a similarly elevated frequency of end-to-end fusions, 0.082 fusions per metaphase (RT + DC in Table II). However, in the case of the Terc+/+/Ku86–/– MEFs, all the RT-like fusions detected showed very strong TTAGGG signals at the fusion point and involved random pairs of chromosomes (TTAGGG in Table II and white bars in Figure 3A; see Figure 3C for examples). This confirms that the fusions resulting from Ku86 deficiency are not mediated by telomere shortening, as proposed previously by us (Samper et al., 2000). In addition, these chromosomal end-to-end fusions involve random pairs of telomeres. Importantly, combined Q-FISH and SKY analyses of the double Terc–/–/Ku86–/– mice revealed that Ku86 deficiency suppressed the occurrence of RT-like fusions with no TTAGGG at the fusion point in G3 and G4 Terc–/–/Ku86–/– MEFs (Table II; Figure 3A, black bars), despite the fact that these cells show similarly shortened telomeres as the corresponding Terc–/–/Ku86+/+ controls (see above). This finding was confirmed by scanning of hundreds of metaphases using Giemsa staining, which indicated a significant rescue of the RT-like fusion phenotype in the G4 Terc–/–/Ku86–/– MEFs compared with the sum of fusions in the single mutant G4 Terc–/–/Ku86+/+ and Terc+/+/Ku86–/– MEFs (Table III). These observations demonstrate for the first time that Ku86 activity mediates the fusion of chromosome ends with undetectable TTAGGG repeats, hence indicating that these fusions are the result of NHEJ events.
Table III. Ku86 deficiency reduces RT-like fusions in increasing generations of Terc–/–/Ku86–/– MEFs as determined by conventional Giemsa staining.
| Genotype | RT-like fusionsa | Summed RT-like fusionsb | No. of cells scored |
|---|---|---|---|
| Wild type | 3.5 | – | 200 |
| Terc+/+/Ku86–/– | 15 | – | 100 |
| G1 Terc–/–/Ku86+/+ | 1 | – | 100 |
| G2 Terc–/–/Ku86+/+ | 4.5 | – | 200 |
| G4 Terc–/–/Ku86+/+ | 33 | – | 100 |
| G1 Terc–/–/Ku86–/– | 28 (+12)c | 1 + 15 = 16 | 100 |
| G2 Terc–/–/Ku86–/– | 10 (–9.5)c | 4.5 + 15 = 19.5 | 100 |
| G4 Terc–/–/Ku86–/– | 21 (–27)c | 33 + 15 = 48 | 100 |
Increased frequencies of chromosome breaks, gaps and fragments were detected in all MEFs lacking Ku86 compared with wild-type or increasing generations of Terc–/–/Ku86+/+ MEFs, in agreement with the role of Ku86 in NHEJ (see Table II and Figure 3B for examples).
Rescue of early germ cell apoptosis in telomerase-deficient mice lacking Ku86
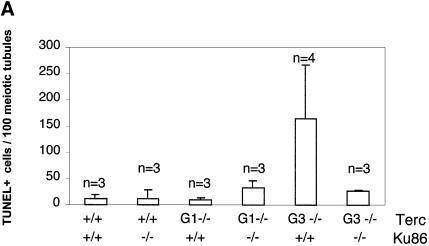
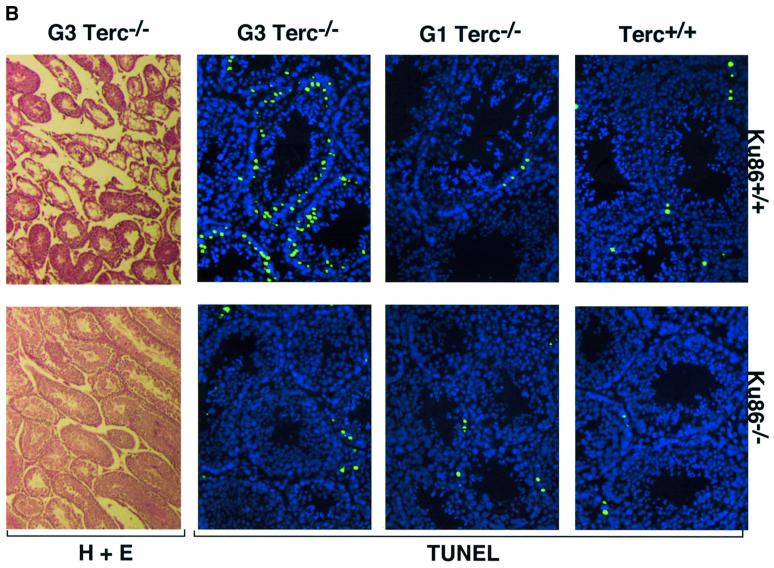
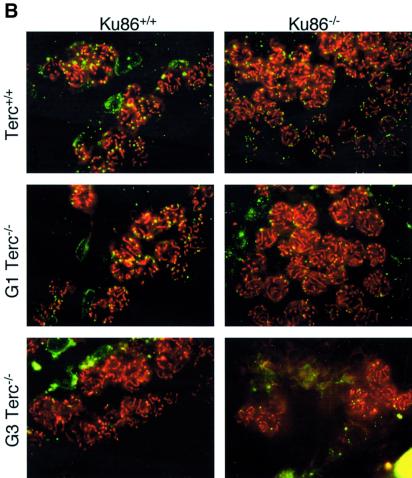
Germ cells from late-generation Terc–/– mice undergo developmentally regulated apoptosis at the early onset of meiosis triggered by critical telomere shortening (Hemann et al., 2001a). To investigate whether Ku86 deficiency also rescues the effects of critical telomere shortening in the context of a mouse, we evaluated the apoptotic index of Ku86+/+ and Ku86–/– germ cells from all Terc–/– generations. Terc+/+/Ku86–/– germ cells showed no increased apoptosis compared with wild-type cells (11.4 ± 7.6 and 12.3 ± 16.8 TUNEL-positive cells/100 tubules for three Terc+/+/Ku86+/+ mice and three Terc+/+/Ku86–/– mice, respectively) (see Figure 4A for quantification and Figure 4B for examples). This indicates that Ku86 deficiency does not result in increased apoptosis of male early germ cells. Lack of telomerase activity in the absence of significant telomere shortening in the G1 Terc–/–/Ku86+/+ mice did not result in increased apoptosis either (10.0 ± 4.2 and 33.6 ± 18.6 TUNEL-positive cells/100 tubules for three G1 Terc–/–/Ku86+/+ mice and three G1 Terc–/–/Ku86–/– mice, respectively) (Figure 4). However, G3 Terc–/–/Ku86+/+ testes with short telomeres displayed a marked apoptosis (167 ± 126.3 TUNEL-positive cells/100 tubules, n = 3) and a variable reduction of tubule cellularity, in agreement with prior reports (Lee et al., 1998; Hemann et al., 2001a) (Figure 4). Sertoli cell-only tubules were also present in some G3 Terc–/–/Ku86+/+ testes (Figure 4B) (Herrera et al., 1999b). Strikingly, apoptosis was rescued in three independent G3 Terc–/–/Ku86–/– testes, with an average apoptotic index similar to that of wild-type mice (26.0 ± 2.0 TUNEL-positive cells/100 tubules) (Figure 4). In agreement with this, germ cell maturation was also preserved in the G3 Terc–/–/Ku86–/– testis (Figure 4B). Nevertheless, these mice were still infertile due to Ku86 deficiency (Vogel et al., 1999; Samper et al., 2000). The rescue of apoptosis in G3 Terc–/–/Ku86–/– testes is not due to rescue of short telomeres in these mice, as shown above. Therefore, these results indicate that Ku86 function directly or indirectly mediates the early apoptosis that occurs in male germ cells as a consequence of critically short telomeres, and that lack of Ku86 rescues this apoptotic phenotype.
Fig. 4. (A) Quantification of TUNEL+ cells per 100 tubules of a total of 3–4 different mice of each of the indicated genotypes. (B) Apoptosis in DAPI-stained (blue) Ku86-deficient and Ku86-proficient Terc–/– testis cryosections with progressive telomere shortening. Fluorescein isothiocyanate- labeled TUNEL+ cells (in green) are detected at low frequency in testes from wild-type or G1 Terc–/– mice, irrespective of their Ku86 status. Increased TUNEL+ cells are seen in G3 Terc–/– testes with critically short telomeres. In contrast, the frequency of TUNEL+ cells in G3 Terc–/–/Ku86–/– testis is similar to that of wild-type testis. Magnification, ×20. Hematoxylin–eosin staining of G3 Terc–/– seminiferous tubules is also shown. Notice the paucity of germ cells and the presence of Sertoli-only tubules in a G3 Terc–/–/Ku86+/+ testis, while germ cell maturation is preserved in a G3 Terc–/–/Ku86–/– counterpart. Magnification, ×10.
Normal TRF1 binding to meiotic chromosomes in Ku86-deficient mice
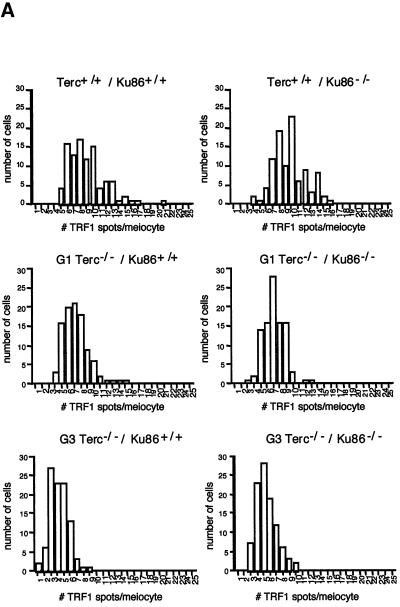
Ku86 has been shown to interact with TRF1 (Hsu et al., 2000). Hence, the study of TRF1 function in Ku86-deficient cells is relevant to understanding the role of Ku86 at the telomere. For this, we have studied binding of TRF1 to meiotic chromosomes using immunofluorescence with an anti-mouse TRF1 antibody (Materials and methods). The average number of telomeric TRF1 spots per meiotic nucleus was 9.6 ± 2.5 and 9.0 ± 3.0 for Terc+/+/Ku86–/– and Terc+/+/Ku86+/+ controls, respectively (Figure 5A shows histograms of TRF1 spots/meiocyte). Importantly, Ku86 deficiency does not alter TRF1 localization and binding to the telomere (Figure 5B). Successive generations of Terc–/–/Ku86–/– and Terc–/–/Ku86+/+ mice showed a similar decrease in the average number of TRF1 spots per meiotic nucleus, 7.3 ± 2.0 and 4.3 ± 1.6 for G1 and G3 Terc–/–/Ku86+/+ compared with 7.2 ± 2.0 and 5.0 ± 1.6 for G1 and G3 Terc–/–/Ku86–/– mice (see Figure 5A for histograms), in agreement with a similar rate of telomere shortening with increasing Terc–/– generations as shown above by direct telomere length analysis.
Fig. 5. (A) The histograms show the frequency distribution of the number of TRF1 spots per meiotic cell (n = 100 cells). (B) TRF1 distribution in pachytene meiocytes. The TRF1 signal (green) localizes at the end of paired meiotic chromosomes at the pachytene stage (red).
Discussion
The existence of a protective structure at the extremities of chromosomes was first deduced in the 1930s by Müller, who showed that X-irradiation of Drosophila rarely resulted in terminal deletions or inversions of chromosomes (Müller, 1938). Strikingly, broken chromosomes and telomeres are not so different in terms of the proteins that they bind, such as the various NHEJ and HR components. These proteins repair DSBs at internal sites of damage; however, when at telomeres, they seem to prevent chromosome end joining (Bailey et al., 1999, 2001; Hsu et al., 2000; Samper et al., 2000; Goytisolo et al., 2001).
Ku86 is an essential component of the NHEJ machinery, and it also binds to telomeres and protects them from fusing (Bailey et al., 1999; Hsu et al., 2000; Samper et al., 2000). Indeed, we show here that Ku86-deficient cells show a degree of telomeric dysfunction (end-to-end fusions) comparable to that of late-generation telomerase-deficient mice. We find, however, different origins for the end-to-end fusions associated with either telomere shortening or Ku86 deficiency. While late-generation telomerase-deficient cells show a significant fraction of end-to-end fusions (∼50%) that lack TTAGGG repeats at the fusion point, these fusions are never found in Ku86-deficient cells. Indeed, Ku86-deficient cells show end-to-end fusions with long tracts of TTAGGG repeats at the fusion point and which involve random pairs of chromosomes. Hence, Ku86 protects normal-length random telomeres from fusing. What happens, however, when telomeres are critically short? Strikingly, the study of late-generation Terc–/–/Ku86–/– mice shows that those end-to-end fusions associated with critical telomere shortening (they lack detectable TTAGGG signals at the fusion point) are prevented in the absence of Ku86. Hence, Ku86 activity mediates the fusion of chromosomes with critically short telomeres, indicating that these fusions are the result of NHEJ events. In addition, Ku86 deficiency abolishes the severe early germline apoptosis associated with critical telomere shortening in late-generation Terc–/– males, suggesting that Ku86 is a direct or indirect mediator of the apoptosis associated with telomere shortening.
Finally, the study of double Terc–/–/Ku86–/– mice demonstrates a functional interaction between Ku86 and telomerase at the telomere. In particular, our data indi cate that Ku86 is interfering directly or indirectly with telomerase-mediated telomere lengthening. In this regard, Ku86 could be directly impairing the access of telomerase to the telomere, or it could be contributing to the formation of a higher order structure at the telomere (i.e. T-loops). Alternatively, Ku86 could also interfere with G-strand overhang processing or with second strand synthesis machinery, recruited directly or indirectly by telomerase. Ku86 is thus the first regulator of telomerase-mediated telomere elongation to have been demonstrated in mammals. Other telomere-binding proteins, such as TRF1, TRF2 and TIN2, have been proposed to regulate the action of telomerase at the mammalian telomere, although this has been not formally demonstrated by using telomerase-deficient cells (Kim et al., 1999; Smogorzewska et al., 2000).
Taken together, the results presented here support a model in which Ku86 contributes to making the normal-length telomere less accessible to telomerase-mediated telomere elongation or to other DNA repair activities, hence preventing further telomere elongation or fusions, respectively. In normal-length telomeres, Ku86 could be contributing to the maintenance of the telomere in a ‘closed’ conformation (Blackburn, 2001). This could be mediated by Ku86’s ability to interact with TRF1 and TRF2, and contribute to the formation of T-loops. However, binding of TRF1 to the telomere is normal in Ku86-deficient meiotic chromosomes. In addition, Ku86 deficiency does not seem to disrupt the G-strand overhang as shown here by looking at the bulk of the telomeres; however, we cannot rule out the possibility that Ku86 deficiency could cause a small percentage of telomeres to have an altered G-strand overhang, which in turn could be responsible for the fused telomeres in these cells. Ku86 could also have a role in the post-replicative processing of the telomeres, as recently suggested for its partner DNA-PKcs (Bailey et al., 2001). Importantly, we show here that Ku86 mediates NHEJ of TTAGGG-exhausted telomeres in somatic cells, as well as the massive germ cell apoptosis associated with critically short telomeres in late-generation telomerase-deficient males. These essential roles of Ku86 at the telomere have important implications in understanding the origin of chromosomal aberrations that occur during cancer and aging.
Materials and methods
Mice and cells
To obtain successive generations of the double Terc–/–/Ku86–/– mutant mice and MEFs, as well as the corresponding Terc–/–/Ku86+/+ controls, Terc–/–/Ku86+/– mice from each generation were intercrossed as shown in Figure 1. For all studies, primary MEFs (passage 1) were used. MEFs were prepared from day 13.5 embryos as described (Blasco et al., 1997). First passage MEFs corresponded to approximately two population doublings (PDL 2). Mice used for all studies were 8–12 weeks old.
Mouse handling
All mice were housed at our barrier area in Madrid, where pathogen-free procedures are employed in all mouse rooms. Quarterly health monitoring reports have been negative for all pathogens in accordance with FELASA (Federation of European Laboratory Animal Science Associations) recommendations.
Scoring of chromosomal abnormalities
Q-FISH. The indicated numbers of metaphases from each MEF culture were scored for chromosomal aberrations by superimposing the telomere image on the 4′,6-diamidino-2-phenylindole (DAPI) chromosome image in the TFL-Telo software (gift from Dr Peter Lansdorp, Vancouver, Canada). End-to-end fusions can be two chromosomes fused by their p-arms (RT-like fusions) or two chromosomes fused by their q-arms (dicentrics).
SKY. Painting probes for each chromosome were generated from flow-sorted mouse chromosomes using sequence-independent DNA amplification. Labeling was performed by incorporating four different dyes in a combination sequence that allows unique and differential identification of each chromosome. Slides were prepared from fixative-stored material and hybridized and washed using the SKY method according to the manufacturer’s protocol (Applied Spectral Imaging, Migdal Ha-Emck, Israel). Chromosomes were counterstained with DAPI. Images were captured and processed as described (Samper et al., 2001). The indicated number of metaphases of each culture were captured and analyzed by SKY, and chromosomal abnormalities were scored as above.
Conventional cytogenetics (Giemsa). Metaphase preparations were made using 0.075 M KCl hypotonic treatment and methanol/acetic acid fixation from MEFs following 2 h treatment with 0.01 µg/ml colcemid. Slides were stained with 5% Giemsa in pH 6.8 buffer for 10 min. A total of 100–200 metaphases were scored.
Statistical analysis
Statistical calculation was performed using Microsoft Excel. For statistical significance, Student’s _t_-test values were calculated.
Telomere length analysis
Q-FISH. First passage MEFs were prepared for Q-FISH and hybridized as described (Samper et al., 2000). To correct for lamp intensity and alignment, images from fluorescent beads (Molecular Probes, USA) were analyzed using the TFL-Telo program. Telomere fluorescence values were extrapolated from the telomere fluorescence of LY-R (R cells) and LY-S (S cells) lymphoma cell lines of known lengths of 80 and 10 kb (McIlrath et al., 2001). There was a linear correlation (_r_2 = 0.999) between the fluorescence intensity of the R and S telomeres with a slope of 38.6. The calibration-corrected telomere fluorescence intensity (ccTFI) was calculated as described (Herrera et al., 1999a).
Images were captured using Leica Q-FISH software at 400 ms integration time in a linear acquisition mode to prevent oversaturation of fluorescence intensity, and recorded using a COHU CCD camera on a Leica Leitz DMRB fluorescence microscope.
TFL-Telo software (gift from Dr P.Lansdorp, Vancouver) was used to quantify the fluorescence intensity of telomeres from at least 10 metaphases of each data point. The images of metaphases from different MEF cultures were captured on the same day, in parallel, and scored blind.
Flow-FISH. Fresh BM cells from the different mice analyzed were prepared as described (Herrera et al., 1999b). Flow-FISH hybridization was performed as described (Rufer et al., 1998). To normalize Flow-FISH data, two mouse leukemia cell lines (LY-R and LY-S, described above) were used as internal controls in each experiment. Telomere fluorescence of at least 2000 cells gated at the G1–G0 cell cycle stage was measured using a Coulter flow EPICS XL cytometer with the SYSTEM 2 software.
TRF analysis. Fresh BM cells and hepatocyte nuclei (Kipling and Cooke, 1990) from the different mice analyzed were isolated as described above, and TRF analysis was performed as described in Blasco et al. (1997).
G-strand overhang assay
Hepatocyte nuclei prepared as described (Kipling and Cooke, 1990) were included in agarose plugs following instructions provided by the manufacturer (Bio-Rad). After overnight digestion in LDS buffer (1% LDS, 100 mM EDTA pH 8.0 and 10 mM Tris pH 8.0), the plugs were digested with 0, 100 or 300 U of mung bean nuclease for 15 min, or with 100 U for 20 or 40 min, as indicated. Then the plugs were digested with _Mbo_I overnight and run on pulse field electrophoresis gels as described (Blasco et al., 1997). The sequential in-gel hybridizations in native and denaturing conditions were carried out as described before (Samper et al., 2000). Quantification of the G-strand overhang radioactive signals was carried out using a STORM 860 PhosphoImager (Molecular Dynamics), using the software provided by the manufacturer. These values were corrected by the TRF signal in denaturing gel conditions. G-strand radioactive signals in the Terc–/–/Ku86+/– mice were normalized to 100%.
Histological analysis
Testes from age-matched mice were either fixed in 10% buffered formalin phosphate, dehydrated and paraffin embedded, or snap frozen in Tissue Freezing Medium (Leica Instruments, Nussloch, Germany). For hematoxylin–eosin staining, 4 µm paraffin-embedded sections were used.
Apoptosis by the TUNEL assay
Single-cell DNA fragmentation was determined in 6 µm frozen sections of testis using the MEBSTAIN Apoptosis Kit II (Immunotech, Marseille, France). Sections were mounted in Vectashield Mounting Medium with DAPI (Vector Laboratories, Burlingame, CA). DAPI-stained TUNEL-stained nuclei were counted in at least 100 tubules and results expressed as TUNEL+ cells per 100 seminiferous tubules.
TRF1 detection
For identification of meiotic cells, the synaptonemal complex (SC) was stained using anti-SCP3 antibodies (gift from Dr R.Benavente, University of Würzburg, Germany). Testes were snap-frozen, cut in 6 µm sections, fixed in cold acetone for 2 min, permeabilized in 0.5% NP-40, blocked in 10% bovine serum albumin (BSA) for 1 h at 37°C and incubated with a pure guinea pig anti-SCP3 antibody for 1 h at room temperature. Then slides were incubated with a secondary Cy3-conjugated anti-guinea pig antiserum for 30 min at room temperature and mounted in Vectashield with DAPI (Vector Laboratories). For simultaneous staining of the SCs and telomere-bound protein TRF1, 6 µm testis cryosections were incubated with both guinea pig anti-SCP3 and rabbit anti-mouse TRF1 antibodies (644) (gift from Titia de Lange, Rockefeller University, New York). Secondary antibodies were Cy-3-conjugated goat anti- guinea pig IgG and Alexa 488-conjugated goat anti-rabbit IgG (Jackson Immunoresearch, West Grove, PA). For quantification of TRF1 spots, 100 single meiotic SCP3-, TRF1-stained cells at the zygotene stage were identified under a fluorescence microscope and the number of TRF1 spots per meiotic cell was counted at the focal plane with the highest observed frequency of TRF1 spots. Images were obtained with a Leica Leitz DMRB microscope equipped with a COHU CCD camera.
Acknowledgments
Acknowledgements
We are indebted to Titia de Lange (Rockefeller University, New York) and Ricardo Benavente (University of Würzburg, Germany) for the TRF1 and SPC3 antibodies, respectively. We thank R.Serrano, E.Santos and E.Tarrent for technical assistance, and M.Serrano and F.Goytisolo for helpful comments. S.E. and S.F. are supported by the European Union (EU). M.A.B.’s laboratory is funded by SWISS BRIDGE AWARD 2000, by the Ministry of Science and Technology (PM97-0133), Spain, by the EU (EURATOM/991/0201, FIGH-CT-1999-00002, FIS5-1999-00055) and by the Department of Immunology and Oncology (DIO). The DIO is funded by the Spanish Council for Scientific Research and by Pharmacia Corporation. S.R.-P. and J.C.C. are supported by the National Cancer Center and the Ministry of Health, Spain. S.D.B.’s laboratory is supported by EU contract FIGH-CT-1999-00009.
References
- Bailey S.M., Meyne,J., Chen,D.J., Kurimasa,A., Li,G.C., Lehnert,B.E. and Goodwin,E.H. (1999) DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl Acad. Sci. USA, 96, 14899–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S.M., Conforth,M.N., Kurimasa,A., Chen,D.J. and Goodwin,E.H. (2001) Strand-specific postreplicative processing of mammalian telomeres. Science, 293, 2462–2465. [DOI] [PubMed] [Google Scholar]
- Baumann P. and Cech,T.R. (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science, 292, 1171–1175. [DOI] [PubMed] [Google Scholar]
- Bianchi A. and de Lange,T. (1999) Ku binds telomeric DNA in vitro. J. Biol. Chem., 274, 21223–21227. [DOI] [PubMed] [Google Scholar]
- Bilaud T., Brun,C., Ancelin,K., Koering,C.E., Laroche,T. and Gilson,E. (1997) Telomeric localization of TRF2, a novel human telobox protein. Nature Genet., 17, 236–239. [DOI] [PubMed] [Google Scholar]
- Blackburn E.H. (2000) Telomere states and cell fates. Nature, 408, 53–56. [DOI] [PubMed] [Google Scholar]
- Blackburn E.H. (2001) Switching and signaling at the telomere. Cell, 106, 661–673. [DOI] [PubMed] [Google Scholar]
- Blasco M.A., Lee,H.-W., Hande,P. Samper,E., Lansdorp,P., DePinho,R. and Greider,C.W. (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell, 91, 25–34. [DOI] [PubMed] [Google Scholar]
- Chin L., Artandi,S.E., Shen,Q., Tam,A., Lee,S.L., Gottlieb,G.J., Greider,C.W. and DePinho,R.A. (1999) p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell, 97, 527–538. [DOI] [PubMed] [Google Scholar]
- Collins K. (2000) Mammalian telomeres and telomerase. Curr. Opin. Cell Biol., 12, 378–383. [DOI] [PubMed] [Google Scholar]
- González-Suárez E., Samper,E., Flores,J.M. and Blasco,M.A. (2000) Telomerase-deficient mice with short telomeres are resistant to skin tumorigenesis. Nature Genet., 26, 114–117. [DOI] [PubMed] [Google Scholar]
- González-Suárez E., Samper,E., Ramírez,A., Flores,J.M., Martín-Caballero,J., Jorcano,J.L. and Blasco,M.A. (2001) Increased epi dermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J., 20, 2619–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goytisolo F., Samper,E., Martín-Caballero,J., Finnon,P., Herrera,E., Flores,J.M., Bouffler,S.D. and Blasco,M.A. (2000) Short telomeres result in organismal hypersensitivity to ionizing radiation in mammals. J. Exp. Med., 192, 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goytisolo F., Samper,E., Edmonson,S., Taccioli,G.E. and Blasco,M.A. (2001) Absence of DNA-PKcs in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol. Cell. Biol., 21, 3642–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Larrivee,M., Labrecque,P. and Wellinger,R.J. (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science, 280, 741–744. [DOI] [PubMed] [Google Scholar]
- Greenberg R.A., Allsopp,R.C., Chin,L., Morin,G. and DePinho,R. (1999) Short dysfunctional telomeres impair tumorigenesis in the INK4aD2/3 cancer-prone mouse. Cell, 97, 515–525. [DOI] [PubMed] [Google Scholar]
- Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A. Moss,H. and de Lange,T. (1999) Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- Hemann M.T. and Greider,C.W. (1999) G-strand overhangs on telomeres in telomerase deficient mouse cells. Nucleic Acids Res., 27, 3964–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann M.T., Rudolph,K.L., Strong,M.A., DePinho,R.A., Chin,L. and Greider,C.W. (2001a) Telomere dysfunction triggers developmentally regulated germ cell apoptosis. Mol. Biol. Cell, 12, 2023–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann M.T., Strong,M.A., Hao,L.Y. and Greider,C.W. (2001b) The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell, 107, 67–77. [DOI] [PubMed] [Google Scholar]
- Herrera E., Samper,E. and Blasco,M.A. (1999a) Telomere shortening in mTR–/– embryos is associated with failure to close the neural tube. EMBO J., 18, 1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera E., Samper,E., Martín-Caballero,J., Flores,J.M., Lee,H.-W. and Blasco,M.A. (1999b) Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J., 18, 2950–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera E., Martinez,A.C. and Blasco,M.A. (2000) Impaired germinal center reaction in mice with short telomeres. EMBO J., 19, 472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.-L., Gilley,D., Backburn,E. and Chen,D.J. (1999) Ku is associated with the telomere in mammals. Proc. Natl Acad. Sci. USA, 96, 12454–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.-L. et al. (2000) Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev., 14, 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Kaminker,P. and Campisi,J. (1999) TIN2, a new regulator of telomere length in human cells. Nature Genet., 234, 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling D. and Cooke,H.J. (1990) Hypervariable ultra-long telomeres in mice. Nature, 347, 400–402. [DOI] [PubMed] [Google Scholar]
- Lee H.-W., Blasco,M.A., Gottlieb,G.J., Horner,J.W., Greider,C.W. and DePinho,R.A. (1998) Essential role of mouse telomerase in highly proliferative organs. Nature, 392, 569–574. [DOI] [PubMed] [Google Scholar]
- McIlrath J. et al. (2001) Telomere length abnormalities in mammalian radiosensitive cells. Cancer Res., 61, 912–915. [PubMed] [Google Scholar]
- Müller H.J. (1938) The remaking of chromosomes. Collect. Net Woods Hole, 13, 181–198. [Google Scholar]
- Rudolph K.L., Chang,S., Lee,H.W., Blasco,M., Gottlieb,G.J., Greider,C. and DePinho,R.A. (1999) Longevity, stress response and cancer in aging telomerase-deficient mice. Cell, 96, 701–712. [DOI] [PubMed] [Google Scholar]
- Rudolph K.L., Millard,M., Bosenberg,M.W. and DePinho,R.A. (2001) Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nature Genet., 28, 155–159. [DOI] [PubMed] [Google Scholar]
- Rufer N., Dragowska,W. Thornbury,G., Roosnek,E. and Lansdorp,P.M. (1998) Telomere length dynamics in human lymphocyte sub populations measured by flow cytometry. Nature Biotechnol., 16, 743–747. [DOI] [PubMed] [Google Scholar]
- Samper E., Goytisolo,F., Slijepcevic,P., van Buul,P. and Blasco,M.A. (2000) Mammalian Ku86 prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO rep., 1, 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper E., Goytisolo,F., Ménissier-de Murcia,J., González-Suárez,E., Cigudosa,J.C., de Murcia,G. and Blasco,M.A. (2001a) Normal telo mere length and chromosomal end-capping in poly(ADP-ribose) polymerase deficient mice and primary cells despite increased chromosomal instability. J. Cell Biol., 154, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper E., Flores,J.M. and Blasco,M.A. (2001b) Restoration of telomerase activity rescues chromosomal instability and premature aging in Terc–/– mice with short telomeres. EMBO rep., 2, 800–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J.W. and Bacchetti,S. (1997) A survey of telomerase activity in human cancer. Eur. J. Cancer, 33, 787–791. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A., van Steensel,B., Bianchi,A., Oelmann,S., Schaefer,M.R., Schnapp,G. and de Lange,T. (2000) Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol., 20, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Jung,D., Jung,Y., Lee,S.G. and Lee,I. (2000) Interaction of human Ku70 with TRF2. FEBS Lett., 481, 81–85. [DOI] [PubMed] [Google Scholar]
- van Steensel B. and de Lange,T. (1997) Control of telomere length by human telomeric protein TRF1. Nature, 385, 740–743. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska,A. and de Lange,T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell, 92, 401–413. [DOI] [PubMed] [Google Scholar]
- Vogel H., Lim,D.-S., Karsenty,G., Finegold,M. and Hasty,P. (1999) Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl Acad. Sci. USA, 96, 10770–10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.D., Kuster,B., Mann,M., Petrini,J.H. and de Lange,T. (2000) Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nature Genet., 25, 347–352. [DOI] [PubMed] [Google Scholar]
- Zijlmans J.M., Martens,U.M., Poon,S., Raap,A.K., Tanke,H.J., Ward,R.K. and Lansdorp,P.M. (1997) Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl Acad. Sci. USA, 94, 7423–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]