Adaptive Evolution and Antiviral Activity of the Conserved Mammalian Cytidine Deaminase APOBEC3H (original) (raw)
Abstract
The APOBEC3 genes encode cytidine deaminases that act as components of an intrinsic immune defense that have potent activity against a variety of retroelements. This family of genes has undergone a rapid expansion from one or two genes in nonprimate mammals to at least seven members in primates. Here we describe the evolution and function of an uncharacterized antiviral effector, APOBEC3H, which represents the most evolutionarily divergent APOBEC3 gene found in primates. We found that APOBEC3H has undergone significant adaptive evolution in primates. Consistent with our previous findings implicating adaptively evolving APOBEC3 genes as antiviral effectors, APOBEC3H from Old World monkeys (OWMs) has efficient antiviral activity against primate lentiviruses, is sensitive to inactivation by the simian immunodeficiency virus Vif protein, and is capable of hypermutating retroviral genomes. In contrast, human APOBEC3H is inherently poorly expressed in primate cells and is ineffective at inhibiting retroviral replication. Both OWM and human APOBEC3H proteins can be expressed in bacteria, where they display significant DNA mutator activity. Thus, humans have retained an APOBEC3H gene that encodes a functional, but poorly expressed, cytidine deaminase with no apparent antiviral activity. The consequences of the lack of antiviral activity of human APOBEC3H are likely to be relevant to the current-day abilities of humans to combat retroviral challenges.
The members of the APOBEC3 family of proteins are effectors of an antiviral defense that may play an important role in protecting primates and other mammals from a variety of genomic invasions (reviewed in reference 1). While only a single APOBEC3 gene has been found in rodents, this gene family has undergone a large expansion and diversification in primates and consists of a cluster of at least seven genes on chromosome 22 in humans (5, 14, 37). APOBEC3G, the first member of the family to be identified because it is the target of the human immunodeficiency virus type 1 (HIV-1) Vif protein (31), has been shown to inhibit the replication of a diverse array of retroviral elements, including lentiviruses (15) (43), gammaretroviruses (7), deltaretroviruses (27), and spumaretroviruses (17, 26), as well as murine endogenous retroviruses (9) and yeast retrotransposons (8, 30). Although APOBEC3G is the most well-characterized member of the family, other primate APOBEC3 genes, including APOBEC3B, APOBEC3C, and APOBEC3F, have also been shown to possess antiviral activities (38, 43). The ability of APOBEC3G and other APOBEC family members to inhibit retroviral replication is at least partially due to their cytidine deaminase activity, causing hypermutation and degradation of invading retroviral genomes. To successfully infect APOBEC-expressing cells, retroviruses must be able to counteract this host barrier. In the case of primate lentiviruses, this is achieved by the Vif protein, which causes the degradation of APOBEC proteins via the proteasome, thereby allowing efficient viral replication (21, 31, 45).
Positive selection is often observed for host proteins that are directly involved in pathogen defense. This type of genetic conflict between host and pathogen drives rapid change in interacting host and pathogen proteins as they attempt to increase or decrease interactions with one another in the battle for dominance. Such conflict has been proposed for the APOBEC3 genes found in primates and can be observed in the large expansion of this gene family in primates and the rapid evolution of several APOBEC3 family members that must keep pace with rapidly evolving pathogens (28).
APOBEC3H, which lies distal to APOBEC3G on chromosome 22, was not identified as a member of the APOBEC3 family until the human genome project was completed (5, 37) and therefore was not examined in our genetic study (28) or in other functional studies (2). The evolutionary history of APOBEC3H differs from that of the other APOBEC3 genes known to exist in primates as it is derived from an ancestral cytidine deaminase domain that is evolutionarily distinct (5). Despite its unique evolutionary history, the presence of a conserved cytidine deaminase domain encoded by APOBEC3H and its close chromosomal proximity to the other known antiviral APOBEC3 family members suggested that APOBEC3H may also play a role in genome defense.
In this study, we found that an _APOBEC3H_-like gene has been conserved throughout mammalian evolution, implying that APOBEC3H is likely to mediate an important function that predates primate evolution. APOBEC3H, like most other primate APOBEC3 genes, has been subject to episodes of positive selection for at least 33 million years of primate evolution. Further, the evolutionary divergence of the APOBEC3H gene from Old World monkeys (OWMs) to humans has profoundly altered the ability of this protein to be expressed and, potentially, the function of the protein in these primates. Thus, while OWM APOBEC3H is a potent antiviral effector that still participates in these primates' arsenal of antiretroviral defense, humans appear to have lost the services of this important retroviral defense gene.
MATERIALS AND METHODS
APOBEC3H sequences.
Genomic DNA was obtained from Coriell (Camden, New Jersey), with the exception of the sooty mangabey sample, which was a kind gift from Cristian Apetrei (Tulane National Primate Research Center). Exons were amplified from genomic DNA with PCR Supermix High Fidelity (Invitrogen), and PCR products were either sequenced directly or cloned into TA vector (Invitrogen), in which case multiple clones were sequenced. The human APOBEC3H sequence was obtained from the Ensembl database (ENSG00000100298) and from cDNA clones. Exon-intron boundaries are conserved across the species that we sampled. Primers were designed by using alignments of human and rhesus monkey (Macaca mulatta) genomic sequences obtained from a BLASTN search of the National Center for Biotechnology Information Trace Archive sequences. Individual exons were amplified from the various primate genomic DNAs with the following primers: exon 2 (first coding exon), forward primer 5′-GAAACACGATGGCTCTGTTAACAG or 5′-TGAGCTGAGATCGGGAGAATGAG and reverse primer 5′-CAAGCACCCGCTTCCTGCC; exons 3 and 4, forward primer 5′-GAAGTGGGTGCTTGCCAGGC and reverse primer 5′-GGGTTGAAAAACTACCTATTGGGTG; exon 5, forward primer 5′-AGCACCCAATAGGTAGTTTTTCAACCC and reverse primer 5′-GGCAACTGACATGCCCCAGGG. For most primate genomes, we only obtained sequences from exons 2 through 4, which cover all except the last two codons (including termination) of the human APOBEC3H gene. Sequences of other APOBEC3 family members were obtained from GenBank and by searching expressed sequence tag and genomic databases with TBLASTN searches.
Alignments and sequence analysis.
APOBEC3 protein sequences were aligned with CLUSTAL_X (34), followed by a manual adjustment of some gaps, and a neighbor-joining phylogeny was reconstructed (with bootstrapping analysis) by ignoring all gapped positions in the PAUP* suite of programs (33). A Bayesian phylogenetic analysis was done with the MrBayes program, version 3.1 (25). Clade credibility was obtained from a consensus of 80,000 trees after a “burn-in” period of 20,000 trees; these values were largely congruent with those obtained by the neighbor-joining bootstrap analysis. Maximum-likelihood analysis was performed with the PAML software package (41). Global dN/dS ratios for the tree were calculated by a free-ratio model, which allows this parameter to vary along different branches. To detect selection, the multiple alignments were fitted to the F61 model of codon frequencies (the F3x4 model gave similar results). We then compared the log-likelihood ratios of the data with different NSites models—model 1 (two states, neutral, dN/dS > 1 disallowed) to model 2 (similar to model 1 but dN/dS > 1 allowed) and model 7 (fit to a beta distribution, dN/dS > 1 disallowed) to model 8 (similar to model 7 but dN/dS > 1 allowed). In both cases, permitting sites to evolve under positive selection gave a much better fit to the data (P < 0.0001) with a significant fraction of the sites (>25%) predicted to evolve at average dN/dS ratios close to 5. A Bayes empirical Bayes analysis also identified certain amino acid residues with high posterior probabilities (>0.95) of having evolved under positive selection.
Cloning of APOBEC constructs.
Human, rhesus macaque, and sooty mangabey homologs of APOBEC3H were cloned by reverse transcription (RT)-PCR from testis tissue, spleen tissue, and unstimulated peripheral blood lymphocytes (PBLs), respectively. Each of the APOBEC3H genes was then transferred into the same mammalian expression vector, which drives gene expression under the control of the cytomegalovirus IE94 promoter. An identical hemagglutinin (HA) epitope tag was added at the N terminus of each protein. The human APOBEC3G construct was made by PCR amplification from CEM15HA (31). All constructs were confirmed by sequencing. The human APOBEC3H clone used in our studies matches the one in the National Center for Biotechnology Information Consensus CDS database for human ARP10 (APOBEC3H), except for a single amino acid change at position 140 from K to E. We changed this amino acid back to the consensus and found no difference in function between the two proteins as determined by Western blot analysis and a single-round infectivity assay (data not shown).
Viral infectivity assays.
Wild-type HIV (HIV-WT) and HIVΔ_vif_ were expressed from the pLai3ΔenvLuc2 (40) and pLai3ΔenvLuc2Δvif (constructed by NdeI-StuI deletion in pLai3ΔenvLuc2) proviruses. These proviruses contain a deletion in the env gene and have the firefly luciferase gene inserted into nef. The luciferase-expressing SIVagmTAN provirus was a gift from Ned Landau (20). The vesicular stomatitis virus glycoprotein (VSV-G) was used for pseudotyping. Viruses were produced through lipid transfection (Fugene; Roche) of 293T cells plated in 1 ml in 12-well plates at 3.5 × 105 cells/ml 1 day prior. Cotransfections consisted of 1 μg of total APOBEC plasmid or an empty expression plasmid, 0.5 μg of proviral plasmid, 0.25 μg of L-VSV-G, and 0.1 μg of HIV-1 Tat in 100 μl of serum-free medium. Viral supernatants were harvested at 48 h and clarified by centrifugation for 5 min at 1,000 × g, and the total amount of virus in supernatants was quantified by p24 gag (HIV-1) or p27 gag (SIVagm) enzyme-linked immunosorbent assay (Coulter). Viral stocks were frozen at −80°C until use. One nanogram of virus in 50 μl was used to infect 293T cells in a 96-well plate that were plated at 1.5 × 105 cells/ml in 150 μl 24 h prior to infection. Polybrene at 5 μg/ml and spinoculation of plates at 1,000 × g for 1 h at 24°C were used to increase levels of infection. After 48 h, cells from triplicate infections were lysed in 80 μl of Cell Culture Lysis Reagent (Promega). Ten microliters of lysates was used for quantitation of luciferase activity with the Luciferase Assay Kit (Promega) and read on a luminometer.
Protein and mRNA expression assays.
For mRNA expression studies, a cDNA panel (Primgen) consisting of 10 ng of first-strand cDNA from various human tissues was screened with primers specific for human APOBEC3H or human APOBEC3G (APOBEC3H, forward primer 5′-ATGGCTCTGTTAACAGCCGAAACATTCCG-3′ and reverse primer 5′-CTCTCAAGCCGTCGCTTTATGGC-3′; APOBEC3G, forward primer 5′-ATGCGCTCCACCTCATAACACAG-3′ and reverse primer 5′-TGGAGCCCCTGCACAAAGTG-3′). For mammalian protein expression analysis, plasmids were transiently transfected into 293T cells by Fugene lipid transfection (Roche) and their expression levels were evaluated by Western blot analysis. Briefly, 3.5 × 105 cells were plated in 1 ml in 12-well plates, transfected the next day with 0.5 μg of APOBEC plasmid, lysed 24 h later in NP-40-doc buffer (20 mM Tris [pH 8.0], 120 mM NaCl, 1 mM EDTA, 1% NP-40, 0.2% Na-deoxycholate, protease inhibitors [either phenylmethylsulfonyl fluoride or Roche Complete Cocktail Tablets]), incubated on ice for 5 min, centrifuged at 12,000 × g for 10 min to clear cell debris, and then frozen at −20°C. Equivalent amounts of total protein, as determined by Bradford assay (Bio-Rad), were loaded onto a sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gel and transferred to Immobilon-P polyvinylidene difluoride membrane (Millipore). Membranes were probed with an HA-specific antibody (HA.11; Babco) at a 1:1,000 dilution, followed by horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Santa Cruz Biotechnology) at 1:10,000 and detection with ECL Plus Reagent (Amersham Biosciences). For semiquantitative RT-PCR analyses, cytoplasmic mRNAs were harvested with an RNeasy Kit (QIAGEN) from cells transiently transfected with equivalent amounts of either human APOBEC3H or macaque APOBEC3H expression plasmid. Cytoplasmic RNA preparations were treated with 2 μl of DNase (Invitrogen) for 15 min to degrade contaminating plasmid DNA, followed by inactivation of DNase by adding EDTA to 2.5 mM and heating to 65°C for 10 min. Cytoplasmic transcripts were than reverse transcribed for 30 min at 50°C (QIAGEN One-Step RT-PCR Kit) with an APOBEC3H-specific primer (HSM179 [5′-GAGAGTTCGGCAGCGAAATACCG-3′]). cDNAs were then serially diluted (1:2), and 5 μl was used for 30 cycles of PCR amplification (94°C for 30 s, 55°C for 30 s, 72°C for 1 min) with a 5′ HA-specific primer (HASense [5′-GGATACCCATACGATGTTCCAG-3′]) and the 3′ APOBEC3H-specific primer (HSM179; see above).
Proteasome inhibitor Western blot analysis.
293T cells were plated in 1 ml in a 12-well plate at 3.5 × 105 cells/ml 1 day prior to transfection. Individual wells were transfected with 0.5 μg of APOBEC plasmid and, 24 h after transfection, the medium was replaced with Dulbecco modified Eagle medium containing either 10.5 μM MG-132 (Calbiochem) or an equivalent volume of dimethyl sulfoxide (DMSO) as a control. Lysates were harvested with NP-40-doc buffer (see above) 2 and 20 h after initiation of treatment. The DMSO-only control was harvested at the 20-h time point. Lysates were normalized for total protein content via a Bradford assay, resuspended in SDS sample buffer, and boiled for 5 min, and 10 μg of total cellular protein was loaded per well onto an SDS-PAGE gel for Western blot analysis as outlined above.
Bacterial mutator assay.
APOBEC cDNAs were cloned into the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible pTrc bacterial expression vector (pTrc-C; Invitrogen) and transformed into Ung−/− Escherichia coli strain BW310 (E. coli Genetic Stock Center, Yale University). Single colonies were used to inoculate independent cultures in Luria broth with 150-μg/ml carbenicillin and 1 mM IPTG and grown overnight at 37°C to saturation. The optical density at 600 nm (OD600) of 1:5 dilutions was measured to assess relative viability, and cultures were then plated onto Luria broth-agar plates containing rifampin (Calbiochem) to select for rifampin resistance. Results were analyzed with GraphPad Prism software. Single rifampin-resistant colonies were picked from plates and used for colony PCR with primers specific to the bacterial RNA polymerase gene rpoB (10). One microliter of each PCR mixture was used to directly sequence products with an internal primer (_rpoB_Seq [5′-ATCTGGATACCCTGATGCCACAG-3′]). For bacterial expression analysis, 3-ml cultures in log phase (as determined by OD600) were induced with 1 mM IPTG, and 100 μl of culture was pelleted after 2.5 h, resuspended in 100 μl of 1× SDS sample buffer (0.06 M Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, 0.0025% bromophenol blue), and boiled for 5 min. Ten microliters of a 1:100 dilution of lysates was loaded onto an SDS-PAGE gel, transferred to Immobilon-P polyvinylidene difluoride membrane, and detected with a c-myc monoclonal antibody (9E10) at a 1:1,000 dilution. Equal loading of total cell lysates was confirmed by Coomassie staining of the polyacrylamide gel after transfer.
Hypermutation assay.
VSV-G-pseudotyped HIVΔ_vif_ was produced in 293T cells in the presence or absence of transiently transfected pCS2HA/APOBEC expression vectors. Ten nanograms of p24 was used to infect 293T cells plated at 8 × 105 cells/ml 1 day prior to infection. Genomic DNA was harvested with the DNeasy Tissue Kit (QIAGEN) 48 h postinfection, followed by treatment with 1 μl of DpnI for 1 h at 37°C to exclude contaminating plasmid DNA from being amplified in subsequent PCR amplifications. A 407-bp region of the HIV-1 pol sequence was amplified with the high-fidelity Pfu DNA polymerase and _pol_-specific primers (pol1721 [5′-GAGCAGACCAGAGCCAAC-3′] and pol2152 [5′-AGTTTCAATAGGACTAATGGG-3′]). The pol fragment was subsequently gel purified (QIAGEN), cloned into the TOPO-TA vector (Invitrogen), and sequenced.
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to GenBank and assigned accession no. DQ408605 to DQ408615.
RESULTS
Evolution of APOBEC3H.
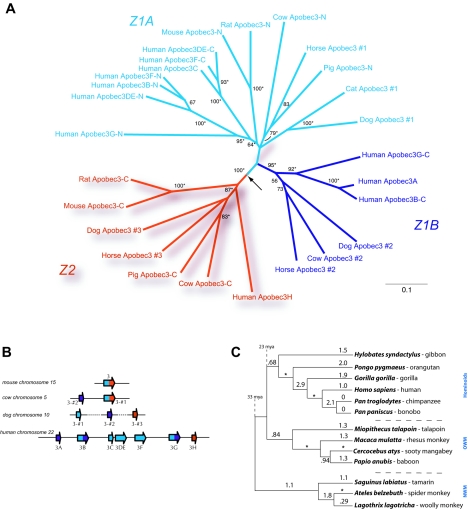
The APOBEC3H gene has an evolutionary history different from that of other primate APOBEC3 genes. To investigate APOBEC3H function in primates, APOBEC3 proteins from a number of species were aligned and a neighbor-joining tree was constructed with CLUSTAL_X (34). Our results are consistent with those recently published by Conticello et al. (5), and we have maintained the same nomenclature for consistency. Based on phylogenetic analyses, mammalian APOBEC3 cytidine deaminase domains can be classified as belonging to one of two types that appear to have been present in the common ancestor of the mammalian lineage (termed Z1A, Z1B [in blue and purple, respectively], and Z2 [in red]; Fig. 1A and B) (5). While the APOBEC3A to -G genes in primates are paralogous genes (related by duplication) of the Z1 type (blue in Fig. 1), the primate APOBEC3H gene is distinct as it is the only primate APOBEC3 of the Z2 type (in red in Fig. 1A and B). Our analysis of available sequences from completed and ongoing genome sequencing projects identified the presence of an _APOBEC3H_-like ortholog throughout the mammalian lineage (Z2-type domains in red in Fig. 1A). While the Z1-like domains have expanded in many mammalian lineages (most dramatically in humans), the _APOBEC3H_-like domain is always present in a single copy, either as part of a fusion gene (rodents, cows, and pigs) or as a stand-alone, single-domain gene (dogs, horses, and humans) (Fig. 1B). While any functional differences that may distinguish Z1- and Z2-type domains have yet to be described, it is clear that the evolutionary constraints acting on the two types of domains are distinct as the Z1 domain has expanded independently in a number of genomes while the Z2 domain has not.
FIG. 1.
APOBEC3H is conserved in mammalian genomes and adaptively evolving in primates. APOBEC3 genes from humans, primates, and other mammals were used for an analysis of the evolutionary history and selective pressures affecting APOBEC3H throughout mammalian evolution. (A) A neighbor-joining tree was constructed in CLUSTAL_X based on a protein alignment of APOBEC sequences from humans and nonprimate eutherian mammals. The N-terminal and C-terminal domains of double-domain APOBEC proteins have been split and are designated -N and -C, respectively. APOBEC3 proteins from mammals in which more than one APOBEC3 was identified are numbered. An APOBEC3H-like domain (red lineages, Z2) has been conserved in a number of mammalian species, including mice, rats, dogs, horses, pigs, cows, and primates. This phylogeny is rooted with the most closely related activation-induced deaminase, cytidine deaminase, as an outgroup lineage; the placement of this root is shown by an arrow. Bootstrap support for the groupings is indicated by numbers next to the relevant branches; those nodes that were supported by a Bayesian maximum-likelihood analysis are indicated with an asterisk. (B) Schematic of APOBEC3 genes identified in representative mammalian genomes, including the mouse, cow, dog, and human genomes. Red denotes Z2 (APOBEC3H like), while blue and purple denote Z1A and Z1B, respectively (the nomenclature system used was suggested in reference 5). While rodents encode only one APOBEC3 gene (a Z1-Z2 fusion), the cow and pig genomes both contain at least two APOBEC3 genes, one whose domain structure mirrors that of the rodent Z1-Z2 fusion gene, as well as an additional Z1-type gene. The horse and dog genomes contain at least three single-domain APOBEC3 genes, two Z1-type genes, and a Z2-type gene; however, the dog genes have not been assigned to a genomic contig yet and hence the chromosomal assignment, and the order of the genes, is still tentative (this uncertainty is represented as a dotted line). (C) APOBEC3H is adaptively evolving in primates. dN/dS ratios were calculated and are indicated on the APOBEC3H phylogeny, which is completely congruent with the accepted primate phylogeny (24). We obtained APOBEC3H sequences from a panel of six hominoids, four OWMs, and three NWMs. dN/dS ratios were calculated along each branch of the phylogeny with the free-ratio model in the PAML package that allows the dN/dS ratio to vary along each branch. A dN/dS ratio of greater than 1 is indicative of positive selection. In some instances, zero synonymous substitutions lead to an apparent dN/dS ratio of infinity (shown with an asterisk). mya, million years ago.
We have previously shown that genes involved in genome defense, such as APOBEC3G and _TRIM5_α, are frequently subject to positive selection (also called adaptive evolution), indicated by an excess of replacement mutations which alter the encoded amino acid over synonymous mutations in which there is no amino acid change (28). In the case of proposed genome defense genes, the finding of positive selection has proven to be a reliable indicator of antiviral function. Functional studies have shown that of the APOBEC genes tested, the genes that are adaptively evolving in primates (28) are also those that have been shown to possess antiviral activity (APOBEC1, APOBEC3B, APOBEC3C, APOBEC3F, and APOBEC3G) (2, 11, 16, 43). To investigate whether APOBEC3H has also evolved under positive selection (thereby implicating it as an antiviral gene), we sequenced the APOBEC3H coding exons from genomic DNA for a panel of New World monkey (NWM), OWM, and hominoid species. With the PAML (phylogenetic analysis by maximum likelihood) program (41), we calculated the number of observed changes per nonsynonymous site (dN) and the number of observed changes per synonymous site (dS) over the entire length of the APOBEC3H gene and calculated the dN/dS ratio for each branch of the primate phylogeny (Fig. 1C). Despite the fact that whole-gene dN/dS values are often conservative measures of positive selection (due to averaging of values over the entire gene length), many branches show a dN/dS value greater than 1, indicative of positive selection. We found that APOBEC3H has an average dN/dS ratio of 1.39 over primate evolution. Models in which codons are permitted to evolve under positive selection (NsSites models M2 and M8) fit the APOBEC3H data significantly better than those in which positive selection is not permitted (NsSites models M1 and M7, respectively; P < 0.0001). These results are strikingly similar to our finding for APOBEC3G (average primate dN/dS ratio = 1.31) (28), implying that the evolutionary pressures driving the positive selection of APOBEC3H have been just as strong as those affecting APOBEC3G. We found no significant variation in the dN/dS ratio among different branches of the primate phylogeny (data not shown), implying that, like APOBEC3G, a relatively uniform selective constraint has acted on APOBEC3H for the past 33 million years.
Tissue-specific mRNA expression.
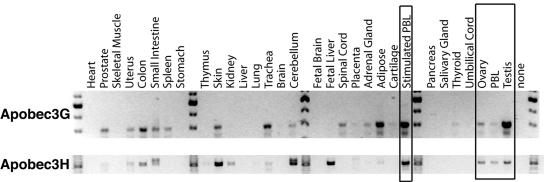
We further asked if APOBEC3H had characteristics of an antiretroviral defense gene through an analysis of the tissue distribution of APOBEC3H mRNA expression. We would expect a gene important for antiretroviral defense to be expressed in tissues relevant to retroviral replication in vivo, similar to what has been found for the other APOBEC3 genes (14, 43). A panel of cDNAs from various human tissues was screened for mRNA expression with primers that are specific for human APOBEC3G and APOBEC3H. Although overlapping in some tissues, the expression patterns of human APOBEC3G and APOBEC3H mRNA transcripts are not identical (Fig. 2). For example, both human APOBEC3H and human APOBEC3G are transcribed in the ovary and testis, potential sites of endogenous retrovirus mobilization, as well as in both stimulated and unstimulated PBLs, sites of lentiviral replication in vivo. In contrast, human APOBEC3G mRNA is expressed at higher levels in the trachea and adipose tissue while human APOBEC3H mRNA is expressed at higher levels in the fetal liver and skin, where _APOBEC3G m_RNA appears to be poorly transcribed. Thus, while the expression pattern of human APOBEC3H in retroviral target tissues supports a role for this gene in antiretroviral defense and is similar to the potent antiretroviral effector human APOBEC3G, there are significant tissue-specific differences, suggesting that these two APOBEC3 genes are not functionally redundant.
FIG. 2.
mRNA expression pattern of APOBEC3H in human tissues. Both APOBEC3G and APOBEC3H were PCR amplified with specific primers from cDNAs made from a panel of different human tissues. Molecular weight markers are shown every ninth lane, and the tissue of origin is marked above each lane. Boxes indicate expression of human APOBEC3G and human APOBEC3H in PBLs and germ line cells (sites relevant to retroviral replication).
APOBEC3H is a retroviral restriction factor.
We next tested the antiretroviral activity of APOBEC3H in single-round viral infectivity assays. First, cDNAs for human (hominoid), macaque (OWM), and sooty mangabey (OWM) APOBEC3H were cloned from testis, spleen, and lymphoid cells, respectively. Plasmids expressing either APOBEC3H (human, macaque, or sooty mangabey) or APOBEC3G (human) were then cotransfected with an HIV-1 (HIV-WT) genome that encodes the luciferase gene along with a VSV-G expression plasmid for pseudotyping of virions. Viral supernatants were collected, normalized for p24 gag, and used to infect 293T cells. As HIV/SIV Vif proteins can counteract the inhibitory effects of other APOBEC3 proteins (20), we also tested APOBEC3H for activity against HIV-1 lacking the vif gene (HIVΔ_vif_).
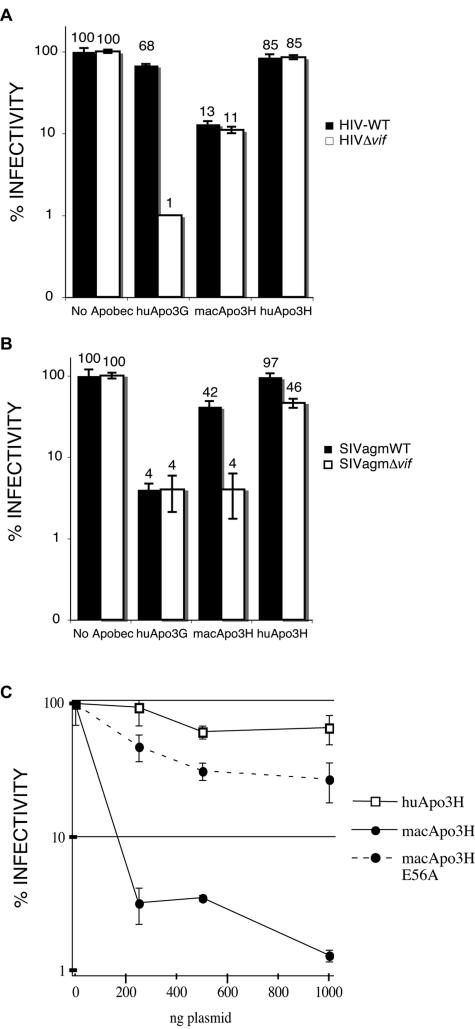
We found macaque APOBEC3H to be a potent inhibitor of both HIV-WT and HIVΔ_vif_ (Fig. 3A). The resistance of macaque APOBEC3H to the effects of HIV-1 Vif is expected due to the known species specificity of Vif-APOBEC interactions (20). For instance, the macaque APOBEC3G protein is also unaffected by HIV-1 Vif but is susceptible to SIV Vif (20). In contrast to this antiretroviral activity of the macaque homolog, we found that human APOBEC3H had almost no effect on HIV replication, even in the absence of the Vif protein (maintenance of 85% infectivity for both HIVΔ_vif_ and HIV-WT) (Fig. 3A). The HA epitope tag at the N terminus was not responsible for the lack of activity of human APOBEC3H, as plasmids expressing human APOBEC3H without the HA tag were also tested and shown to be inactive (data not shown). Human APOBEC3G was used as a control in these experiments, as it has the best-characterized antiviral activity against a broad panel of retroviruses (2, 7-9, 19, 26, 27, 35). As expected, human APOBEC3G was able to efficiently restrict HIVΔ_vif_ but not HIV-WT (human APOBEC3G is known to be potently inhibited by HIV-1 Vif) (31; Fig. 3A). The finding that human APOBEC3H is not able to inhibit HIV-1 despite the potent antiviral activity of the macaque homolog is surprising given that the APOBEC3G homologs from various primate species (humans, chimpanzees, macaques, and African green monkeys) do not demonstrate variable activity against HIV-1 (20).
FIG. 3.
Restriction of HIV and SIV by macaque but not human APOBEC3H. Single-round viral infectivity assays were used to assess the relative abilities of APOBEC3 proteins to inhibit retroviral replication, as described in Materials and Methods. (A) Effects of human APOBEC3G (huApo3G), macaque APOBEC3H (macApo3H), and human APOBEC3H (huApo3H) on HIV-1 infectivity. Black boxes are HIV-WT, and white boxes are HIVΔ_vif_. Results are shown as percent infectivity of the control infection in which no APOBEC expression plasmid was included. Results from one representative experiment are shown (error bars reflect variation between triplicate infections within one experiment). (B) The same as panel A, except that APOBEC genes were assayed for activity against SIVagm, a related nonhuman primate lentivirus. Of note, the twofold decrease in SIVagmΔ_vif_ infectivity in the presence of human APOBEC3H was not reproducible in independent experiments. (C) Single-round viral infectivity assay of HIVΔ_vif_ produced in the presence of increasing amounts of APOBEC3H expression plasmids. Human APOBEC3H (open squares) plasmids decreased HIV-1 infectivity less than 2-fold at all of the concentrations of transfected plasmid used, while macaque APOBEC3H (solid circles, solid lines) decreased infectivity almost 50-fold. A catalytic-site mutant form of macaque APOBEC3H (macApo3H E56A; solid circles, dotted lines) was severely impaired in its ability to inhibit HIV-1 replication.
Since the antiviral activity of APOBEC3 family members is sometimes specific to certain viral species (43), the macaque and human APOBEC3H proteins were also tested alongside human APOBEC3G for the ability to inhibit a related nonhuman primate lentivirus, SIVagm. SIVagm endemically infects African green monkeys but also is capable of replicating in the rhesus macaque, suggesting that SIVagm Vif is sufficient to counteract APOBEC3 proteins encountered in this host. We found macaque APOBEC3H to be able to potently restrict SIVagmΔ_vif_ but not SIVagmWT (Fig. 3B). Thus, macaque APOBEC3H is specifically inhibited by SIVagm Vif but not by HIV Vif (compare Fig. 3A with Fig. 3B), similar to what has been shown for macaque APOBEC3G (20). In contrast to macaque APOBEC3H, human APOBEC3H was unable to significantly inhibit either SIVagmWT or SIVagmΔ_vif_, again suggesting that human APOBEC3H is incapable of inhibiting lentiviral replication. Consistent with previous findings, human APOBEC3G inhibited SIVagm and was not neutralized by SIVagm Vif (Fig. 3B). We also cloned the sooty mangabey APOBEC3H cDNA and found that its restriction pattern was essentially identical to that of the macaque homolog (data not shown), as expected given the relatively small evolutionary distance between the two primates.
We also tested the abilities of the macaque and human APOBEC3H proteins to inhibit HIVΔ_vif_ in a dose-response experiment by increasing the amount of APOBEC3H plasmid in the transfection from 250 ng to 1,000 ng. Even at the lowest levels of transfected plasmid, macaque APOBEC3H was able to reduce the infectivity of HIVΔ_vif_ to less than 5% of the control (no APOBEC) infection (Fig. 3C). In contrast, human APOBEC3H had a less-than-twofold effect on HIVΔ_vif_, even at the highest level of transfected plasmid. In addition, we introduced a mutation into the predicted active site of the macaque APOBEC3H cytidine deaminase domain by changing amino acid 56 from glutamate to alanine (macApo3H E56A in Fig. 3C). This mutant was severely impaired in its ability to inhibit HIVΔ_vif_, although even the residual antiviral activity of macApo3H E56A was greater than that of human APOBEC3H (Fig. 3C). We also found that macaque, but not human, APOBEC3H could restrict murine leukemia virus (data not shown).
We conclude, therefore, that APOBEC3H in these OWM species is able to restrict divergent primate lentiviruses with an expected species-specific sensitivity to the lentiviral vif gene, while the human APOBEC3H homolog is incapable of restricting any of the viruses tested here. Our findings suggest an unexpected and fundamental difference in activity between APOBEC3H proteins from OWMs and humans.
APOBEC3H protein expression in primate cells.
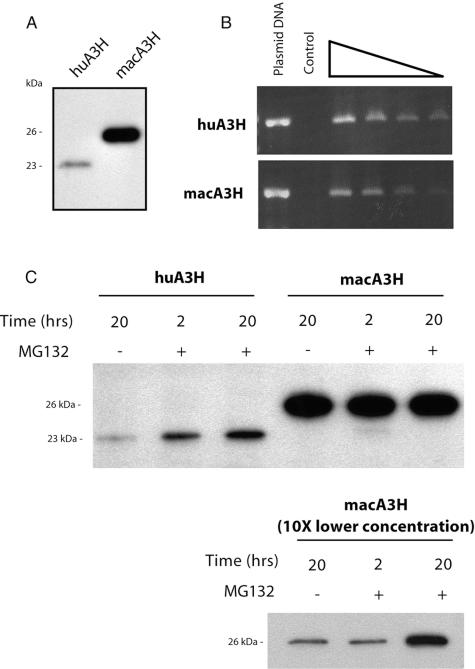
To understand the difference in antiviral activity between the macaque and human APOBEC3H proteins, we examined the steady-state expression levels of the proteins in transiently transfected cells. Western blot analysis of lysates from 293T cells (a human cell line) that were transfected with equivalent amounts of macaque APOBEC3H and human APOBEC3H expression plasmids revealed a large discrepancy between the steady-state levels of the two proteins (Fig. 4A). At levels of transfected plasmid that correspond to efficient inhibition of primate lentiviruses by macaque APOBEC3H (the same levels used for the infectivity assays in Fig. 3A and B), we saw high levels of macaque APOBEC3H protein but were able to detect only low levels of human APOBEC3H protein. Through semiquantitative RT-PCR analysis, we found equivalent amounts of mRNAs to be present in cells transiently transfected with either APOBEC3H homolog (Fig. 4B). Thus, the inability of human APOBEC3H to restrict retroviral replication appears to reflect low protein levels in the virus-producing cell and that this low steady-state level of expression is not due to decreased transcriptional activity or mRNA instability. Further, we looked for expression of stable human APOBEC3H protein in a variety of other human cells (primary human fibroblasts, HeLa cells, and Jurkat T cells), as well as nonhuman mammalian cells (mouse 3T3 and feline CRFK cells) by retroviral transduction and found that there was low to undetectable human APOBEC3H protein expression in all of these cell types (data not shown).
FIG. 4.
Steady-state expression levels of APOBEC3H proteins are drastically different. Lysates of 293T cells transiently transfected with APOBEC expression plasmids were analyzed by Western blotting and semiquantitative RT-PCR to determine relative levels of protein and mRNA, respectively. (A) APOBEC3H protein levels were detected in cell lysates 48 h after transfection. The predicted size of HA-tagged human APOBEC3H is ∼23 kDa, and that of macaque APOBEC3H is ∼26 kDa. Identical results were obtained with an alternate expression vector (data not shown), ruling out an effect specific to the pCS2HA construct used in these experiments. (B) Cytoplasmic mRNAs were reverse transcribed with an _APOBEC3H_-specific primer, and serial dilutions (1:2) were used as a template for PCR amplification. Amplification of APOBEC3H transcripts is equivalent for both the human and macaque APOBEC3H transfections. The control (no reverse transcriptase) was included to rule out the possibility of plasmid DNA contamination. (C) The proteasome inhibitor MG-132 was added to transient transfections of human APOBEC3H and macaque APOBEC3H, and lysates were used for Western blot analysis. Samples harvested at the 2- and 20-h time points revealed approximately fivefold stabilization of the human APOBEC3H protein, even at the earliest time point. In the bottom part, 10-fold less macaque APOBEC3H was transfected.
To test the possibility that human APOBEC3H protein is actively degraded, we used a well-described proteasome inhibitor, MG-132, to determine if blocking proteasome-mediated protein degradation would lead to higher steady-state levels of human APOBEC3H in transiently transfected 293T cells. Indeed, we found that proteasome inhibitor treatment partially stabilized intracellular levels of human APOBEC3H at both the 2- and 20-h time points (a fivefold increase over the DMSO-only control) (Fig. 4C), suggesting that human APOBEC3H is actively degraded by the proteasome. No increase was seen for macaque APOBEC3H when it was transfected at plasmid levels similar that of the human APOBEC3H plasmid (Fig. 4C, middle), but we did see an increase in macaque APOBEC3H in response to MG-132 when 10-fold lower amounts of plasmid were transfected (Fig. 4C, bottom). However, the response of human APOBEC3H differed from that of macaque APOBEC3H since human APOBEC3H could be induced by a 2-h incubation with proteasome inhibitor while induction of macaque APOBEC3H required a 20-h incubation (Fig. 4C, compare middle and bottom parts). Thus, both proteins are likely subject to proteasome-mediated degradation but the human protein may be more sensitive to this degradation. Similar results were obtained with Cos7 cells (data not shown), an African green monkey (OWM) cell line, ruling out the possibility that this degradation is specific to expression of the proteins in a human cell line. Despite the fact that cells treated with a proteasome inhibitor contain increased levels of human APOBEC3H, the levels are still lower than that of macaque APOBEC3H (Fig. 4C), and we found that human APOBEC3H produced at these levels had no antiviral effect on HIVΔ_vif_ (data not shown). These results suggest that the significantly different expression levels of the APOBEC3H proteins may explain the observed disparity in their antiviral activities.
APOBEC3H encodes a conserved cytidine deaminase activity.
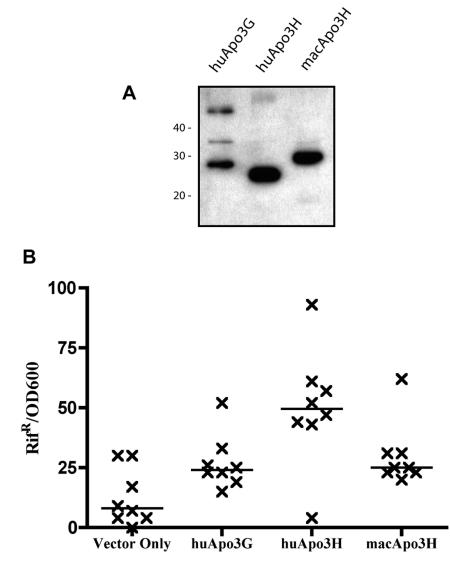
The residues necessary for cytidine deamination, such as the consensus His-X-Glu-X23-28-Pro-Cys-X2-4-Cys cytidine deaminase motif (12), are conserved in both the human and macaque APOBEC3H homologs. To determine whether either of the APOBEC3H homologs has functional enzymatic activity, we took advantage of a well-described bacterial mutator assay (13, 22). In this assay, induced APOBEC proteins catalyze cytidine deamination of the bacterial genome. Mutations that occur at well-characterized positions of the RNA polymerase gene rpoB confer resistance to the antibiotic rifampin, allowing us to screen for DNA mutator activity by assaying for an increased frequency of rifampin resistance. In contrast to our results obtained with primate cells, human APOBEC3H can be stably expressed in the bacterial system (Fig. 5A). In this mutator assay, expression of either APOBEC3H homolog resulted in an increase in the median Rifr frequency (human APOBEC3H, ∼6.1-fold over the background; macaque APOBEC3H, ∼3.1-fold over the background in this experiment; P < 0.05), suggesting that both APOBEC3H homologs have conserved DNA mutator activity. Similar to what other groups have shown (10), we indeed saw an elevated median mutation frequency (about threefold) when comparing human APOBEC3G to the vector-only control (Fig. 5B). To further this analysis, we sequenced a small region of the E. coli RNA polymerase gene isolated from Rifr colonies. If cytidine deamination is the major mechanism through which mutations are conferring a resistance phenotype, sequencing should reveal more C-to-T and G-to-A transition mutations in the APOBEC-expressing samples compared to the vector-only control. As expected, we found that the mutations in the rpoB gene from human APOBEC3H, macaque APOBEC3H, and human APOBEC3G were transition mutations 93%, 95%, and 95% of the time, respectively, whereas transitions occurred only 84% of the time for the vector-only control. We conclude, therefore, that both APOBEC3H homologs have conserved cytidine deaminase activity that is capable of, although perhaps not limited to, DNA mutation.
FIG. 5.
Cytidine deaminase activity is conserved in APOBEC3H homologs. (A) Macaque and human APOBEC3H homologs were expressed in a bacterial system. Whole-cell lysates were used for Western blot analysis to evaluate expression levels of APOBEC proteins in bacteria. Sizes of myc-tagged APOBEC proteins are ∼48 kDa for human APOBEC3G, ∼23 kDa for human APOBEC3H, and ∼26 kDa for macaque APOBEC3H. All of the APOBEC proteins were stably expressed in this system. (B) APOBEC proteins were evaluated for the ability to affect rates of acquired resistance to rifampin in a bacterial cytidine deamination assay. Results are shown as the number of rifampin-resistant (Rifr) colonies normalized for cell number (OD600). One representative experiment is shown in which eight independent cultures were evaluated. Of note, both the human APOBEC3H and macaque APOBEC3H mutation frequencies were higher than the control (Vector Only) in independent experiments (P < 0.05). However, the potency of the macaque and human APOBEC3H proteins relative to one another varied in independent experiments and should not be compared.
G-to-A hypermutation of retroviral genomes.
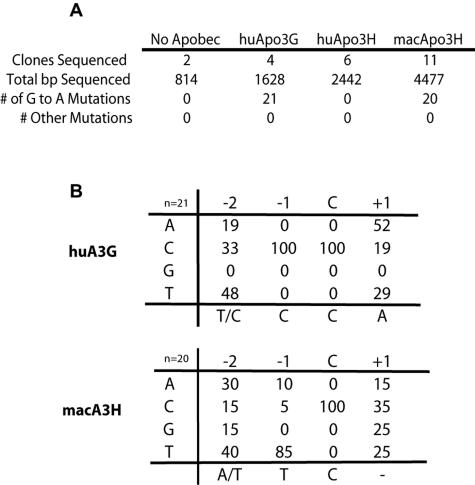
APOBEC_3-mediated antiviral activity has been shown to be due, at least in part, to cytidine deamination of nascently transcribed retroviral cDNAs following infection of target cells. Such cytidine-to-uracil deamination in minus-strand DNA is detected as replacement of guanine with adenine in integrated proviral genomes. We found that a mutation in the putative active site of macaque APOBEC3H disrupted most (but not all) of its antiviral activity (Fig. 3C). To further define the role of deamination in APOBEC3H-mediated antiviral activity, we also looked for direct evidence of APOBEC3H-induced hypermutation of viral genomes. After infection of cells with HIVΔ_vif produced in the presence or absence of APOBEC expression plasmids, a region of the viral genome was amplified, sequenced, and analyzed for evidence of hypermutation. Significant G-to-A hypermutation was observed for both human APOBEC3G and macaque APOBEC3H (Fig. 6A), implying that both enzymes actively deaminate cytidines in minus-strand viral DNA. As might be expected from our viral infectivity and protein expression data, human APOBEC3H infections showed no evidence of hypermutation. Further analysis of the sequences allowed us to delineate the target site preference of macaque APOBEC3H. Macaque APOBEC3H demonstrated a clear target site preference for thymine at the −1 position relative to the deaminated cytidine, an 85% TC dinucleotide preference (Fig. 6B). This dinucleotide preference is similar to what has been reported for human APOBEC3B (6) and human APOBEC3F (2, 16) but is in contrast to the CC dinucleotide motif preferred by human APOBEC3G (Fig. 6B) (44). Our results suggest hypermutation of retroviral genomes as a likely mechanism of _APOBEC3H_-mediated antiviral activity.
FIG. 6.
APOBEC3H induces G-to-A hypermutation of retroviral genomes. Retroviral hypermutation induced by APOBEC proteins was evaluated by sequencing HIVΔ_vif_ proviral genomes following infection of a target cell line, as described in Materials and Methods. (A) Sequences were obtained from infections with viral supernatants from transfections containing human APOBEC3G, macaque APOBEC3H, human APOBEC3H, or no APOBEC expression plasmid. G-to-A changes were the only mutations observed in ∼9,000 bp of the sequenced genomes, ruling out the likelihood of generalized reverse transcriptase errors or experimentally introduced mutations. (B) Hypermutated viral sequences were analyzed to determine the sequence context of the deaminated cytidine. The number of deamination target sites evaluated for each APOBEC is noted (huA3G = 21, macA3H = 20). Shown are percentages of each nucleotide found at the −2, −1, and +1 positions relative to the deaminated cytidine (C). Macaque APOBEC3H demonstrates a clear TC dinucleotide preference.
DISCUSSION
In this study, we have characterized a unique component of the primate APOBEC3 family of antiretroviral effectors, APOBEC3H. We found APOBEC3H orthologs to be conserved in mammalian genomes and undergoing sustained adaptive evolution in primates. As expected given the signal of positive selection acting on APOBEC3H in primates, we found OWM APOBEC3H to be a potent inhibitor of lentiviral infectivity through a mechanism that includes, at least in part, deamination of nascent viral minus-strand DNA.
The fact that SIV Vif allows escape from OWM APOBEC3H-mediated restriction suggests that APOBEC3H plays a significant role in restriction of retroviral infection in these primates since Vif proteins must have evolved to recognize APOBEC3H in OWMs. Moreover, our analysis of positive selection indicates that APOBEC3H may be as much an active participant in host defense as the prototypic antiviral gene APOBEC3G. Thus, in order to productively infect OWMs like rhesus macaques, SIV Vif must be able to inactivate a panel of APOBEC3 proteins, including APOBEC3F, APOBEC3G, and APOBEC3H. Thus, the ability of lentiviral Vif proteins to neutralize APOBEC3 proteins encountered in the host must involve complex interactions that allow Vif proteins to recognize diverse substrates (32), including divergent APOBEC3 proteins.
While orthologous APOBEC3 genes from different primates may encode similar antiviral functions (APOBEC3G, for example), it is clear from our study that other members of the APOBEC3 family, including APOBEC3H, may encode proteins that are not necessarily functionally redundant in different primates. Our study therefore emphasizes the importance of testing orthologs from various primate species. While multiple human APOBEC3 genes have been tested for antiviral function, only APOBEC3G orthologs from other primates have been tested (20). Our APOBEC3H study suggests that a survey limited to human genes may lead to an incomplete picture of antiviral potency, or lack thereof, encoded by the APOBEC3 cluster in primates.
APOBEC3H is a potent restriction factor likely to be important in preventing lentiviral infection in vivo in certain primate species, including the rhesus macaque and sooty mangabey but likely not in humans. Despite its DNA mutator activity and in contrast to the antiretroviral activity of the macaque APOBEC3H homolog, we found that human APOBEC3H is poorly expressed at the protein level and has no detectable antiretroviral activity. The differential stability and subsequent change in antiviral activity of APOBEC3H homologs suggest that the ∼35 million years of evolution that separates OWMs and hominoids has resulted in significant functional divergence of this conserved antiviral cytidine deaminase. Although we found the low expression of human APOBEC3H in a number of cell types, we still cannot rule out the possibility that the protein is differentially regulated in different cells (such as stabilization of the human homolog in an undetermined cell type or cellular environment). The mechanism by which APOBEC proteins are degraded by lentiviral Vif proteins (45) could be a descendant of such a regulatory control system. This model supposes, however, that regulated degradation, a possible mechanism of protein regulation (36), serves as a major regulator of APOBEC3H stability.
Alternatively, our results suggest that an antiviral effector that is active in some primates has become inactive in humans. While this may seem incongruous with the finding of positive selection of APOBEC3H in primates (Fig. 1D), we cannot determine if positive selection has occurred specifically along the evolutionary branch leading to humans. Why an unstable antiviral APOBEC3 gene would evolve specifically in humans, but not in OWMs, could be due to several reasons, but we discuss two strong possibilities. The first possibility is that the loss of function of human APOBEC3H is the result of relaxed selective pressures occurring specifically along the human lineage. Although it is hard to reconcile this possibility with the finding of robust antiviral activities of other human APOBEC3 genes, it is plausible that APOBEC3H became specialized to inhibit a particular retroviral invader and extinction of this retrovirus specifically in humans resulted in decreased selective pressure to maintain activity and allowed for the subsequent loss of APOBEC3H activity. Evidence for differential pressure from retroelements among primates is accumulating, particularly in the case of endogenous retroviruses (18, 42). Given relaxed pressure to maintain antiviral activity, it may even have been advantageous for humans to evolve an unstable APOBEC effector, as this would alleviate the cost associated with the maintenance of a full repertoire of potentially hypermutagenic cytidine deaminases in the cell (4, 23, 39). Similar relaxation of constraints may have led to the high-frequency persistence of an impaired _TRIM5_α allele in the human population (29).
A second possibility is that the original function of APOBEC3H has been assumed by another APOBEC3 family member in humans. The rapid expansion of APOBEC3 genes in primate genomes supports this possibility. APOBEC3A, for example, appears to be a recent arrival in hominoids, revealed in the short evolutionary distance separating it from the C-terminal domain of APOBEC3B (Fig. 1A). This evolutionary distance is less than that between the human and macaque APOBEC3H genes, implying its recent evolution. Our genomic surveys suggest that there is no APOBEC3A ortholog in the parts of the rhesus macaque genome sequenced thus far (H. S. Malik, unpublished data). Since both APOBEC3A and APOBEC3H encode single-domain cytidine deaminases, it is possible that APOBEC3A now performs a function in humans (3) that is usually undertaken by APOBEC3H in other primates.
The finding that humans now encode a poorly expressed APOBEC3H protein is significant because orthologous genes are conserved in most mammals, implying an important although unknown function for this gene in mammalian cells. If the human homolog of APOBEC3H has indeed lost the ability to serve as an effective lentiviral restriction factor, our current ability to combat retroviral infections must be significantly different from that of other primates, a finding that is both evolutionarily and medically important.
Acknowledgments
We thank Michael Metzger for cloning sooty mangabey APOBEC3H; Shannon Murray and Maxine Linial for rhesus mRNA; Ned Landau for SIVagm-luc; Michael Malim for CEM15HA; and Shari Kaiser, Sara Sawyer, Yegor Veronon, and Masahiro Yamashita for comments on the manuscript.
This work was supported by NIH grant R37 AI30937 (M.E.) and a Searle Scholar Award and Sloan fellowship (H.S.M.). J.A.K. was supported by a Viral Oncology Training Grant (T32-CA09229), and M.O. was supported by a Hearst Foundation Interdisciplinary Fellowship and a National Science Foundation Graduate Research Fellowship.
REFERENCES
- 1.Bieniasz, P. D. 2003. Restriction factors: a defense against retroviral infection. Trends Microbiol. 11**:**286-291. [DOI] [PubMed] [Google Scholar]
- 2.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14**:**1392-1396. [DOI] [PubMed] [Google Scholar]
- 3.Bogerd, H. P., H. L. Wiegand, B. P. Doehle, K. K. Lueders, and B. R. Cullen. 2006. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 34**:**89-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cascalho, M. 2004. Advantages and disadvantages of cytidine deamination. J. Immunol. 172**:**6513-6518. [DOI] [PubMed] [Google Scholar]
- 5.Conticello, S. G., C. J. Thomas, S. K. Petersen-Mahrt, and M. S. Neuberger. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22**:**367-377. [DOI] [PubMed] [Google Scholar]
- 6.Doehle, B. P., A. Schafer, and B. R. Cullen. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339**:**281-288. [DOI] [PubMed] [Google Scholar]
- 7.Doehle, B. P., A. Schafer, H. L. Wiegand, H. P. Bogerd, and B. R. Cullen. 2005. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 79**:**8201-8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutko, J. A., A. Schafer, A. E. Kenny, B. R. Cullen, and M. J. Curcio. 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 15**:**661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannieux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433**:**430-433. [DOI] [PubMed] [Google Scholar]
- 10.Hache, G., M. T. Liddament, and R. S. Harris. 2005. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 280**:**10920-10924. [DOI] [PubMed] [Google Scholar]
- 11.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113**:**803-809. [DOI] [PubMed] [Google Scholar]
- 12.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4**:**868-877. [DOI] [PubMed] [Google Scholar]
- 13.Harris, R. S., S. K. Petersen-Mahrt, and M. S. Neuberger. 2002. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10**:**1247-1253. [DOI] [PubMed] [Google Scholar]
- 14.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79**:**285-296. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi, M., A. Takaori-Kondo, K. Shindo, A. Abudu, K. Fukunaga, and T. Uchiyama. 2004. APOBEC3G targets specific virus species. J. Virol. 78**:**8238-8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14**:**1385-1391. [DOI] [PubMed] [Google Scholar]
- 17.Lochelt, M., F. Romen, P. Bastone, H. Muckenfuss, N. Kirchner, Y. B. Kim, U. Truyen, U. Rosler, M. Battenberg, A. Saib, E. Flory, K. Cichutek, and C. Munk. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. USA 102**:**7982-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Sanchez, P., J. C. Costas, and H. F. Naveira. 2005. Paleogenomic record of the extinction of human endogenous retrovirus ERV9. J. Virol. 79**:**6997-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424**:**99-103. [DOI] [PubMed] [Google Scholar]
- 20.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114**:**21-31. [DOI] [PubMed] [Google Scholar]
- 21.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9**:**1398-1403. [DOI] [PubMed] [Google Scholar]
- 22.Petersen-Mahrt, S. K., R. S. Harris, and M. S. Neuberger. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418**:**99-103. [DOI] [PubMed] [Google Scholar]
- 23.Pham, P., R. Bransteitter, and M. F. Goodman. 2005. Reward versus risk: DNA cytidine deaminases triggering immunity and disease. Biochemistry 44**:**2703-2715. [DOI] [PubMed] [Google Scholar]
- 24.Purvis, A. 1995. A composite estimate of primate phylogeny. Philos. Trans. R. Soc. Lond. B Biol. Sci. 348**:**405-421. [DOI] [PubMed] [Google Scholar]
- 25.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19**:**1572-1574. [DOI] [PubMed] [Google Scholar]
- 26.Russell, R. A., H. L. Wiegand, M. D. Moore, A. Schafer, M. O. McClure, and B. R. Cullen. 2005. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 79**:**8724-8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasada, A., A. Takaori-Kondo, K. Shirakawa, M. Kobayashi, A. Abudu, M. Hishizawa, K. Imada, Y. Tanaka, and T. Uchiyama. 2005. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology 2**:**32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawyer, S. L., M. Emerman, and H. S. Malik. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2**:**E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawyer, S. L., L. I. Wu, J. M. Akey, M. Emerman, and H. S. Malik. 2006. High-frequency persistence of an impaired allele of the retroviral defense gene TRIM5α in humans. Curr. Biol. 16**:**95-100. [DOI] [PubMed] [Google Scholar]
- 30.Schumacher, A. J., D. V. Nissley, and R. S. Harris. 2005. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl. Acad. Sci. USA 102**:**9854-9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418**:**646-650. [DOI] [PubMed] [Google Scholar]
- 32.Simon, J. H., T. E. Southerling, J. C. Peterson, B. E. Meyer, and M. H. Malim. 1995. Complementation of _vif_-defective human immunodeficiency virus type 1 by primate, but not nonprimate, lentivirus vif genes. J. Virol. 69**:**4166-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (* and other methods). Ver. 4.0b10. Sinauer Associates, Sunderland, Mass.
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25**:**4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303**:**1829. [DOI] [PubMed] [Google Scholar]
- 36.Varshavsky, A. 2005. Regulated protein degradation. Trends Biochem. Sci. 30**:**283-286. [DOI] [PubMed] [Google Scholar]
- 37.Wedekind, J. E., G. S. Dance, M. P. Sowden, and H. C. Smith. 2003. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 19**:**207-216. [DOI] [PubMed] [Google Scholar]
- 38.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23**:**2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, X., P. Geraldes, J. L. Platt, and M. Cascalho. 2005. The double-edged sword of activation-induced cytidine deaminase. J. Immunol. 174**:**934-941. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita, M., and M. Emerman. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 78**:**5670-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13**:**555-556. [DOI] [PubMed] [Google Scholar]
- 42.Yohn, C. T., Z. Jiang, S. D. McGrath, K. E. Hayden, P. Khaitovich, M. E. Johnson, M. Y. Eichler, J. D. McPherson, S. Zhao, S. Paabo, and E. E. Eichler. 2005. Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol. 3**:**e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, Q., D. Chen, R. Konig, R. Mariani, D. Unutmaz, and N. R. Landau. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279**:**53379-53386. [DOI] [PubMed] [Google Scholar]
- 44.Yu, Q., R. Konig, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11**:**435-442. [DOI] [PubMed] [Google Scholar]
- 45.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302**:**1056-1060. [DOI] [PubMed] [Google Scholar]