Array-Based Comparative Genomic Hybridization Identifies Localized DNA Amplifications and Homozygous Deletions in Pancreatic Cancer (original) (raw)
Abstract
Pancreatic cancer, the fourth leading cause of cancer death in the United States, is frequently associated with the amplification and deletion of specific oncogenes and tumor-suppressor genes (TSGs), respectively. To identify such novel alterations and to discover the underlying genes, we performed comparative genomic hybridization on a set of 22 human pancreatic cancer cell lines, using cDNA microarrays measuring ∼26,000 human genes (thereby providing an average mapping resolution of <60 kb). To define the subset of amplified and deleted genes with correspondingly altered expression, we also profiled mRNA levels in parallel using the same cDNA microarray platform. In total, we identified 14 high-level amplifications (38–4934 kb in size) and 15 homozygous deletions (46–725 kb). We discovered novel localized amplicons, suggesting previously unrecognized candidate oncogenes at 6p21, 7q21 (SMURF1, TRRAP), 11q22 (BIRC2, BIRC3), 12p12, 14q24 (TGFB3), 17q12, and 19q13. Likewise, we identified novel polymerase chain reaction-validated homozygous deletions indicating new candidate TSGs at 6q25, 8p23, 8p22 (TUSC3), 9q33 (TNC, TNFSF15), 10q22, 10q24 (CHUK), 11p15 (DKK3), 16q23, 18q23, 21q22 (PRDM15, ANKRD3), and Xp11. Our findings suggest candidate genes and pathways, which may contribute to the development or progression of pancreatic cancer.
Keywords: Pancreatic cancer, array CGH, comparative genomic hybridization, expression profiling, DNA amplification
Introduction
Pancreatic cancer is the fourth leading cause of cancer death in the United States. With a 5-year survival rate of less than 5%, mortality rates closely mirror incidence rates, reflecting the ineffectiveness of current treatment regimens [1]. An improved understanding of the molecular pathogenesis of pancreatic cancer is urgently needed to identify new targets and strategies for effective therapy [2].
As with other solid tumor types, the amplification of oncogenes and deletion of tumor-suppressor genes (TSGs) play a critical role in the development and progression of pancreatic cancer. Activating mutations—and less frequently amplification [3,4]—of the KRAS2 oncogene (12p12), for example, have been identified as early events in nearly all pancreatic adenocarcinomas [2]. Likewise, the CDKN2A, TP53, and SMAD4 TSGs are frequently deleted or inactivated by mutation or promoter hypermethylation [2]. Indeed, the discovery of homozygous deletions first led to the identification of CDKN2A and SMAD4 as important TSGs [5,6].
Pancreatic cancers likely harbor additional localized DNA amplifications and deletions that are not apparent by conventional cytogenetic techniques such as comparative genomic hybridization (CGH) [7]. Array-based CGH (aCGH) methods provide an alternative higher-resolution approach for identifying these lesions [8–10]. In the current study, we have performed a cDNA microarray-based CGH analysis to identify localized DNA amplifications and deletions in a set of pancreatic cancer cell lines. In parallel, we have measured mRNA levels using the same microarray platform, thereby defining the subset of amplified/deleted genes displaying correspondingly altered expressions.
Materials and Methods
Pancreatic Cancer Cell Lines
AsPC-1, BxPC-3, Capan-1, Capan-2, CFPAC-1, HPAC, HPAF-II, Hs 766T, MIA PaCa-2, MPanc96, PANC-1, Panc 02.03, Panc 02.13, Panc 03.27, Panc 08.13, Panc 10.05, PL45, SU.86.86, and SW 1990 were obtained from the American Type Culture Collection (ATCC, Manassas, VA). PL5 and PL8 were obtained from Dr. Anirban Maitra (Johns Hopkins University, Baltimore, MD). Colo-357 was a kind gift of Dr. Caroline Hill (Cancer Research UK London Research Institute, London, UK). Cell lines (Table W1) were grown to 80% confluence in RPMI 1640 medium supplemented with 10% fetal bovine serum. Genomic DNA was isolated using the Qiagen (Valencia, CA) Blood and Cell Culture DNA Maxi Kit, and RNA was isolated using the Trizol (Invitrogen, Carlsbad, CA) method.
Array CGH and Gene Expression Profiling
cDNA microarrays spotted on Corning (Corning, NY) UltraGAPS coated slides were obtained from the Stanford Functional Genomics Facility (Stanford, CA). These arrays contain 39,632 human cDNA, representing approximately 25,856 mapped human genes (22,890 UniGene clusters [11] together with 2966 additional mapped expressed sequence tags not assigned UniGene IDs). aCGH and gene expression profiling were performed essentially as described [12,13]. Briefly, for aCGH, 4 µg of genomic DNA from each cell line was random primer-labeled with Cy5 and cohybridized to the cDNA microarray along with 4 µg of Cy3-labeled sex-matched normal leukocyte reference DNA. For gene expression profiling, 50 µg of total RNA from each cell line and 50 µg of reference RNA (derived from 11 different established human cell lines) were differentially labeled with Cy5 and Cy3, respectively, and cohybridized to cDNA microarrays. Following overnight hybridization and washing, arrays were imaged using a GenePix 4000B scanner (Axon, Union City, CA). Fluorescence ratios were extracted using GenePix Pro 5.0 software, and the data were uploaded into the Stanford Microarray Database [14] for storage, retrieval, and analysis. The complete microarray datasets are available at http://smd.stanford.edu.
Data Analysis
For aCGH, background-subtracted fluorescence ratios were normalized for each array by setting the average fluorescence ratio for all array elements equal to 1. Genes were considered reliably measured if the fluorescence intensity for the Cy3 reference channel was at least 1.4-fold above background. Map positions for arrayed cDNA clones were assigned using the NCBI genome assembly, accessed through the UCSC genome browser (July 2003 freeze). For genes represented by multiple arrayed cDNA, the average fluorescence ratio was used. DNA copy number gains and losses were identified using the CLuster Along Chromosomes (CLAC) method (http://www-stat.stanford.edu/∼wp57/CGH-Miner) [15]. Briefly, the CLAC algorithm builds a hierarchical cluster-style tree along each chromosome, such that neighboring genes with positive and negative ratios are separated into different clusters. Gains and losses are then called significant based on the height and width of clusters, and a false discovery rate is estimated by comparison to normal-normal hybridization data. We defined high-level DNA amplifications and presumptive homozygous deletions as contiguous regions identified by CLAC, with at least 50% of genes displaying fluorescence ratios ≥3 or ≤0.25, respectively. For expression profiling, fluorescence ratios were normalized for each array, and then well-measured genes (fluorescence intensities for the Cy5 or Cy3 channel at least 1.5-fold above background) were subsequently “mean-centered” (i.e., reported for each gene relative to the mean ratio across all samples).
Fluorescence In Situ Hybridization (FISH)
To validate DNA amplification, we performed interphase FISH using a Spectrum Green-labeled bacterial artificial chromosome probe corresponding to the 7q21 locus (RP11-62N3; Children's Hospital Oakland Research Institute, Oakland, CA) and a Spectrum Orange-labeled chromosome 7 centromere probe (CEP 7; Vysis, Downers Grove, IL). FISH was performed exactly according to the Vysis labeling and hybridization protocols, and images were captured using an Olympus (Melville, NY) BX51 microscope and CytoVision 3.0 software (Applied Imaging Corp., San Jose, CA).
Polymerase Chain Reaction (PCR) Validation of Homozygous Deletions
To validate homozygous deletions, we used gene-specific primer pairs to PCR-amplify genomic DNA from cell lines. Primer pairs for genes flanking the regions of homozygous deletion, and designed to have a distinguishable fragment size, were included in the PCR reactions as internal controls. PCR was performed on an Applied Biosystems (Foster City, CA) GeneAmp 9700, using 100 ng of DNA template, 1x PCR buffer (Applied Biosystems), 160 µM dNTPs, 1.5 mM MgCl, 10 pmol of each individual primer (Table W2), and 1 U of Taq DNA polymerase (Applied Biosystems) in a 25-µl reaction for 35 cycles [94°C, 30 seconds; annealing temperature (Table W2), 30 seconds; 72°C, 30 seconds], followed by gel electrophoresis on a 1.8% TAE agarose gel, and visualization using an Alpha Innotech (San Leandro, CA) imaging system.
Results
To identify DNA amplifications and deletions, we performed CGH on a set of 22 pancreatic adenocarcinoma cell lines using cDNA microarrays measuring 25,856 human genes, thereby providing an average mapping resolution of less than 60 kb. We identified numerous chromosomal regions of recurrent gain and loss (Figures W1 and W2), the spectrum of which was consistent with published conventional CGH studies [4,16–20]. Gains were most commonly observed on chromosomes 8q (90%), 11q (75%), 20q (75%), 7q (65%), 3q (60%), 5q (60%), and 7p (60%), whereas losses occurred most often on 18q (95%), 8p (80%), 4q (70%), 6q (65%), 9p (65%), 17p (65%), 3p (60%), 6p (60%), and Xp (60%). Cell lines PL45 and Panc 10.05 displayed highly similar aCGH profiles, consistent with their being established from the same patient. Surprisingly, cell lines AsPC-1 and MPanc96, the latter obtained both from the ATCC repository and from the originator [21], also exhibited nearly identical profiles. Given that AsPC-1 was established 15 years earlier [22], we conclude that both cell lines likely represent AsPC-1.
In addition to these broad regions of chromosomal gain and loss, we also identified numerous localized high-level DNA amplifications (i.e., fluorescence ratios ≥3, corresponding to at least five-fold amplification [9]; Table 1) and presumptive homozygous deletions (i.e., fluorescence ratios ≤0.25; Table 2). All together, we identified 14 high-level amplifications in eight different cell lines, each spanning 38–4934 kb in size (median 747 kb), and 15 homozygous deletions in 13 cell lines, spanning 46–725 kb (median 183 kb).
Table 1.
High-Level DNA Amplifications Identified by aCGH.
| Cytoband | P Border (nt) | Q Border (nt) | Size (kb) | Cell Line | Remaining Lines with Low-Level Gain | Selected Candidate Oncogenes* |
|---|---|---|---|---|---|---|
| 6p21 | 32,043,837 | 32,410,885 | 367 | SW 1990 | 4/19 | STK19, TNXB, PBX2, NOTCH4 |
| 6p21 | 32,827,514 | 32,865,810 | 38 | SW 1990 | 0/19 | TAP1 |
| 7q21 | 93,671,907 | 98,605,497 | 4934 | AsPC-1 | 9/19 | TRRAP, SMURF1, ARPC1A, ARPC1B |
| 11q13 | 69,241,850 | 70,008,988 | 767 | Colo-357 | 14/19 | CCND1, EMS1 |
| 11q22 | 101,406,590 | 102,133,375 | 727 | Colo-357 | 4/19 | YAP1, BIRC2, BIRC3, MMP7, MMP27 |
| 12p12 | 14,926,093 | 15,186,368 | 260 | Su.86.86 | 4/19 | |
| 12p12 | 25,253,402 | 26,380,345 | 1127 | Su.86.86 | 5/19 | KRAS2 |
| 12p11 | 25,253,402 | 27,366,920 | 2114 | HPAF-II | 5/19 | KRAS2, FGHR1OP2, STK38L |
| 14q24 | 74,414,776 | 74,540,126 | 125 | Panc 08.13 | 6/19 | TGFB3 |
| 17q12 | 36,065,530 | 36,106,569 | 41 | CFPAC-1 | 5/19 | |
| 19q13 | 39,957,195 | 40,808,091 | 851 | Su.86.86 | 8/19 | USF2 |
| 19q13 | 43,616,179 | 45,546,133 | 1930 | PANC-1 | 7/19 | eIF3k, AKT2 |
| 19q13 | 43,997,907 | 46,073,560 | 2076 | Su.86.86 | 7/19 | AKT2 |
| 19q13 | 55,171,535 | 55,539,817 | 368 | Su.86.86 | 6/19 | ZNF473 |
Table 2.
Homozygous Deletions Identified by aCGH.
| Cytoband | P Border | Q Border (nt) | Size (kb) | Cell Lines(s) | Remaining Lines with Single-Copy Loss | Gene Deletions Confirmed by PCR |
|---|---|---|---|---|---|---|
| 6q25 | 157,305,093 | 157,562,885 | 258 | MIA PaCa-2 | 0/19 | ARID1B |
| 8p23 | 1,717,413 | 1,894,213 | 177 | MIA PaCa-2 | 13/19 | CLN8, ARHGEF10 |
| 8p22 | 15,108,999 | 15,639,496 | 530 | MIA PaCa-2 | 14/19 | TUSC3 |
| 9p21*,† | 21,845,793 | 21,984,872 | 139 | BxPC-3, Capan-1, MIA PaCa-2, PANC-1, Panc 02.13, PL5, Su.86.86 | 6/13 | CDKN2A |
| 9q33 (a*) | 112,922,979 | 113,392,575 | 470 | BxPC-3 | 6/19 | TNFSF15‡, TNFSF8, TNC, DEC1‡ |
| 10q22* | 72,655,409 | 72,861,236 | 206 | BxPC-3 | 4/19 | CDH23‡ |
| 10q24 | 101,574,438 | 101,620,286 | 46 | PL8 | 4/19 | CHUK |
| 11p15 | 11,942,981 | 12,118,344 | 175 | BxPC-3 | 4/19 | DKK3 |
| 16q23 | 78,558,946 | 78,688,790 | 130 | HPAF-II | 4/19 | WWOX |
| 18q21† | 46,766,091 | 46,863,399 | 97 | BxPC-3, CFPAC-1, Hs 766T, Panc 03.27, PL8 | 14/15 | SMAD4 |
| 18q21* | 49,277,732 | 50,003,145 | 725 | MIA PaCa-2 | 18/19 | DCC, MBD2 |
| 18q23 | 75,921,728 | 76,104,374 | 183 | Colo-357 | 13/19 | PARD6G |
| 21q22 | 41,433,815 | 41,822,285 | 388 | Panc 02.13 | 6/19 | BACE2, MXI1, ANKRD3, PRDM15 |
| 21q22 | 41,822,119 | 42,524,687 | 703 | BxPC-3 | 6/19 | ANKRD3, PRDM15, ZNF295‡ |
| Xp11 | 42,636,923 | 42,787,663 | 151 | BxPC-3, MIA PaCa-2 | 6/18 | MAOA |
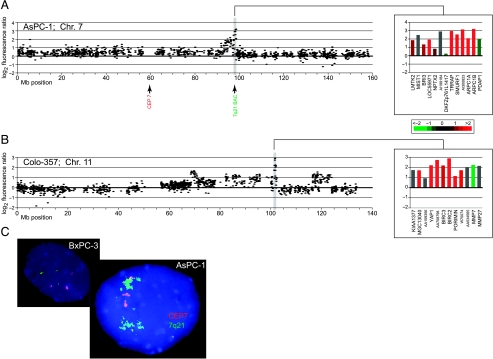
Several localized high-level amplifications corresponded to known oncogenes, including KRAS2 (12p12) [3,23] and AKT2 (19q13) [24]; each of these genes was amplified in two pancreatic cancer cell lines. In addition, we identified several novel high-level amplifications, suggesting the location of as yet uncharacterized oncogenes. Nonrecurrent amplified loci included 6p21, 7q21, 11q22, 12p12 (proximal to KRAS2), 14q24, 17q12 (proximal to ERBB2), and 19q13 (proximal and distal to AKT2) (Table 1). Of the genes residing within these amplicons, several displayed correspondingly elevated expression and have plausible roles in tumorigenesis. Among these, the 7q21 amplicon “peak” [25] in AsPC-1 (Figure 1_A_) harbors SMURF1, an E3 ubiquitin ligase and negative regulator of TGFβ signaling [26]. Also residing in this amplicon are ARPC1A and ARPC1B, subunits of the ARP2/3 complex that controls actin polymerization and cell motility [27], and TRRAP, an essential cofactor for the transcriptional oncoproteins Myc and E2F [28]. We independently validated the 7q21 amplicon by interphase FISH (Figure 1_C_). Within the 11q22 amplicon in Colo-357 (Figure 1_B_), the apoptotic inhibitors BIRC2 and BIRC3 [29] were highly expressed. TGFB3, a ligand for TGFβ signaling [30], was found amplified at 14q24 in Panc 08.13.
Figure 1.
Array CGH identifies localized DNA amplifications in pancreatic cancer. (A and B) Graphic displays of DNA copy number alteration for selected localized amplifications identified in pancreatic cancer cell lines. Test/reference fluorescence ratios are plotted on a log2 scale according to chromosome nucleotide (Megabase) position. Shaded regions highlight localized high-level amplifications. Insets display genes within highlighted amplicons, ordered by map position and color-coded according to mean-centered expression levels (log2 ratio scale indicated). (A) 7q21 amplicon in AsPC-1. (B) 11q22 amplicon in Colo-357. Complete genomewide profiles of DNA copy number alteration for the 22 pancreatic cancer cell lines are viewable in Figure W2. (C) FISH validation of 7q21 amplification in AsPC-1. Spectrum Orange chromosome 7 centromere probe detects three signals, whereas Spectrum Green 7q21 locus probe identifies multiple signal clusters indicative of DNA amplification. Nonamplified cell line BxPC-3 (triploid for chromosome 7) is shown for comparison.
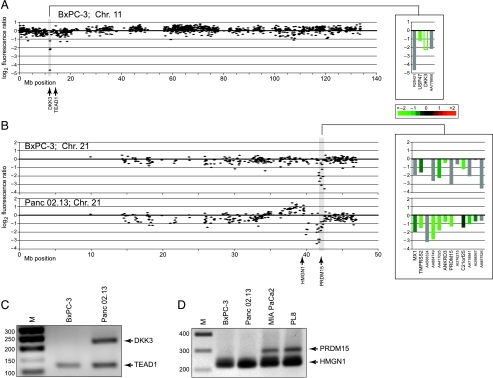
We also identified numerous localized, presumptive homozygous deletions. Several such alterations corresponded to known TSGs, including CDKN2A (9p21), homozygously deleted in seven cell lines, and SMAD4 (18q21), homozygously deleted in five cell lines (Table 2). Interestingly, the MIA PaCa-2 cell line harbors a homozygous deletion in the DCC gene, adjacent to but not affecting SMAD4, supporting a potential tumor-suppressor function for this gene [31]. In addition, we discovered several novel PCR-validated, localized homozygous deletions, suggesting new candidate TSGs at 6q25, 8p23, 8p22, 9q33, 10q22, 10q24, 11p15, 16q23, 18q23, 21q22, and Xp11 (Table 2). Among these, the 8p22 deletion in MIA PaCa-2 harbors TUSC3, a poorly characterized gene also homozygously deleted in a metastatic prostate cancer [32]. TNC, which modulates cell adhesion [33], and TNFSF15, a vascular endothelial inhibitor [34], reside within the 9q33 deletion in BxPC-3. The 10q23 deletion in PL8 harbors CHUK (also called IKKA), an activator of NF-κB signaling [35]. DKK3 [36], an inhibitor of Wnt signaling [37], resides within the 11p15 deletion in BxPC-3 (Figure 2, A and C); an additional four cell lines displayed single-copy loss at this site. The 21q22 locus (Figure 2, B and D), homozygously deleted in BxPC-3 and Panc 2.13 (with an additional six lines displaying single-copy loss), includes PRDM15, a putative histone methyltransferase, a class of enzymes frequently deregulated in human cancer [38], and ANKRD3 (also called RIP4), another activator of NF-κB signaling [39].
Figure 2.
Array CGH identifies localized homozygous DNA deletions in pancreatic cancer. (A and B) Test/reference fluorescence ratios are plotted on a log2 scale according to chromosome nucleotide (Megabase) position. Shaded regions highlight localized homozygous deletions. Insets display genes within highlighted deletions, ordered by map position and color-coded according to mean-centered expression levels (log2 ratio scale provided; unfilled green bar indicates measured intensity less than background). (A) 11p15 deletion in BxPC-3. (B) 21q22 deletion in BxPC-3 and Panc 02.13. (C) PCR validation of homozygous deletion at 11p15. DKK3, located within the homozygous deletion, is PCR-amplified in control cell line Panc 02.13 but not in BxPC-3. TEAD1, a control gene flanking the deletion, is PCR-amplified in both cell lines. (D) PCR validation of homozygous deletion at 21q22. PRDM15, located within the homozygous deletion, is PCR-amplified in control cell lines (MiaPaCa2 and PL8) but not in BxPC-3 and Panc 02.13. HMGN1, a control gene flanking the deletion, is PCR-amplified in all four cell lines.
Discussion
The main objective of our study was to use aCGH to identify localized DNA amplifications and deletions in pancreatic cancer, thereby defining the location of previously unrecognized candidate oncogenes and TSGs. In our aCGH analysis of 22 pancreatic adenocarcinoma cell lines, we identified 14 localized high-level amplifications and 15 localized homozygous deletions. Few of these localized aberrations had been identified earlier by conventional chromosome-based CGH [17,18]. Our findings therefore highlight the usefulness of high-resolution aCGH in discovering previously unrecognized DNA copy number aberrations. Notably, compared to broad chromosomal gains and losses, such highly localized aberrations also provide better opportunities to pinpoint and discover the underlying cancer genes.
As noted, the spectrum of gains and losses we observed was consistent with previous conventional CGH studies on primary pancreatic tumors. Furthermore, the observed subset of localized aberrations harboring known cancer genes (i.e., KRAS2, AKT2, CDKN2A, and SMAD4) has been well described in primary pancreatic tumors, suggesting that most novel aberrations are likely also to be found in primary tumor specimens. Nevertheless, as cells may acquire genetic changes during establishment and passage of cultures, the prevalence of these novel aberrations in primary pancreatic tumors will need to be determined.
Although our findings are, in general, consistent with previous chromosome-based CGH studies of the same pancreatic cell lines, we did not identify the frequent gain of 1q reported by Tirado et al. [40], which may represent an artifact of the chromosome-based CGH method [41]. Three aCGH studies of pancreatic cell lines were also recently published. Holzmann et al. [42] surveyed 13 cell lines (five in common with our study) using BAC arrays with low-resolution coverage (498 clones), providing limited data for comparison. Aguirre et al. [43] profiled 24 cell lines (18 in common) using cDNA arrays (14,160 clones); however, their focus on broad low-level gains/losses, along with their not reporting which alterations were identified in which cell lines, makes meaningful comparisons difficult. Heidenblad et al. [44] studied 31 cell lines (nine in common) using BAC (3565 clones) and cDNA (25,468 clones) arrays, and their data were reported in sufficient detail to permit comparisons. Although our findings are in general agreement for the common set of cell lines investigated, we have identified several additional aberrations and candidate genes (Table W3), including the PCR-verified homozygous deletions at 11q and 21q (Figure 2). Discrepancies likely reflect the higher mapping resolution of our arrays and/or the different thresholds for calling gains and losses.
Our findings suggest several new candidate genes and pathways in pancreatic cancer. For example, SMURF1 (7q21), amplified and overexpressed in AsPC-1, encodes an E3 ubiquitin ligase, which targets the degradation of TβRI receptor complex through its association with Smad7, thus suppressing the growth-inhibitory effects of TGFβ [26] (more frequently accomplished through SMAD4 disruption). In contrast, our discovery of TGFB3 (14q24) amplification in Panc 08.13 supports a possible tumorigenic role of Smad4-independent TGFβ signaling in pancreatic cancer [45]. Other alterations highlight the importance of inhibiting apoptosis, through amplification and overexpression of the antiapoptotic genes BIRC2 and BIRC3 (11q22), or possible modulation of NF-κB signaling [46] by deletion of CHUK (10q24) or ANKRD3 (21q22). Interestingly, we identified DKK3 (11p15), an inhibitor of Wnt signaling, within a highly localized homozygous deletion in the BxPC-3 cell line. Other cell lines exhibited single-copy loss and/or decreased expression of DKK3, suggesting that aberrant Wnt signaling may be common in pancreatic cancer. Although altered Wnt signaling contributes to the development of human cancers, most notably colorectal cancer [47], few reports to date have implicated Wnt signaling in the pathogenesis of pancreatic adenocarcinoma. However, consonant with our findings, Caca et al. [48] identified constitutive Tcf (the downstream effector of canonical Wnt signaling) transcriptional activity in two pancreatic cell lines.
In conclusion, our high-resolution genomewide aCGH study has led to the identification of numerous previously unrecognized localized DNA amplifications and deletions in pancreatic cancer. The expression levels, along with the known or inferred functions of the genes residing within these aberrations, suggest several new candidate oncogenes and TSGs. Our findings provide insight into the pathogenesis of pancreatic cancer, and may suggest new targets for improved therapies. Additional studies are required to characterize the functional role of identified genes in the development or progression of pancreatic cancer.
Supplementary Material
Supplementary Figures and Tables
Acknowledgements
We thank Mike Fero and the staff of the Stanford Functional Genomics Facility for providing high-quality cDNA microarrays, and Gavin Sherlock and Catherine Ball of the Stanford Microarray Database group for providing outstanding database support.
Abbreviations
aCGH
array-based comparative genomic hybridization
FISH
fluorescence in situ hybridization
TSG
tumor-suppressor gene
Footnotes
1
This work was supported, in part, by a grant from the Lustgarten Foundation. M.D.B. was also supported by a Biotechnology Overseas Associateship from the Department of Biotechnology, Ministry of Science and Technology, Government of India.
*
This article refers to supplementary material which is designated by “W” (ie, Table W1, Figure W1) and is available online at www.bcdecker.com
References
- 1.Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, Ward E, Feuer EJ, Thun MJ. Cancer statistics, 2004. CA Cancer J Clin. 2004;54:8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 2.Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 3.Yamada H, Sakamoto H, Taira M, Nishimura S, Shimosato Y, Terada M, Sugimura T. Amplifications of both c-Ki-ras with a point mutation and c-myc in a primary pancreatic cancer and its metastatic tumors in lymph nodes. Jpn J Cancer Res. 1986;77:370–375. [PubMed] [Google Scholar]
- 4.Solinas-Toldo S, Wallrapp C, Muller-Pillasch F, Bentz M, Gress T, Lichter P. Mapping of chromosomal imbalances in pancreatic carcinoma by comparative genomic hybridization. Cancer Res. 1996;56:3803–3807. [PubMed] [Google Scholar]
- 5.Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 6.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 7.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 8.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 9.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 10.Lucito R, West J, Reiner A, Alexander J, Esposito D, Mishra B, Powers S, Norton L, Wigler M. Detecting gene copy number fluctuations in tumor cells by microarray analysis of genomic representations. Genome Res. 2000;10:1726–1736. doi: 10.1101/gr.138300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuler GD. Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J Mol Med. 1997;75:694–698. doi: 10.1007/s001090050155. [DOI] [PubMed] [Google Scholar]
- 12.Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gollub J, Ball CA, Binkley G, Demeter J, Finkelstein DB, Hebert JM, Hernandez-Boussard T, Jin H, Kaloper M, Matese JC, et al. The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res. 2003;31:94–96. doi: 10.1093/nar/gkg078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang P, Kim Y, Pollack J, Narasimhan B, Tibshirani R. A method for calling gains and losses in array CGH data. Biostatistics. 2005;6:45–58. doi: 10.1093/biostatistics/kxh017. [DOI] [PubMed] [Google Scholar]
- 16.Mahlamaki EH, Hoglund M, Gorunova L, Karhu R, Dawiskiba S, Andren-Sandberg A, Kallioniemi OP, Johansson B. Comparative genomic hybridization reveals frequent gains of 20q, 8q, 11q, 12p, and 17q, and losses of 18q, 9p, and 15q in pancreatic cancer. Genes Chromosomes Cancer. 1997;20:383–391. doi: 10.1002/(sici)1098-2264(199712)20:4<383::aid-gcc10>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 17.Curtis LJ, Li Y, Gerbault-Seureau M, Kuick R, Dutrillaux AM, Goubin G, Fawcett J, Cram S, Dutrillaux B, Hanash S, et al. Amplification of DNA sequences from chromosome 19q13.1 in human pancreatic cell lines. Genomics. 1998;53:42–55. doi: 10.1006/geno.1998.5405. [DOI] [PubMed] [Google Scholar]
- 18.Ghadimi BM, Schrock E, Walker RL, Wangsa D, Jauho A, Meltzer PS, Ried T. Specific chromosomal aberrations and amplification of the AIB1 nuclear receptor coactivator gene in pancreatic carcinomas. Am J Pathol. 1999;154:525–536. doi: 10.1016/S0002-9440(10)65298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schleger C, Arens N, Zentgraf H, Bleyl U, Verbeke C. Identification of frequent chromosomal aberrations in ductal adenocarcinoma of the pancreas by comparative genomic hybridization (CGH) J Pathol. 2000;191:27–32. doi: 10.1002/(SICI)1096-9896(200005)191:1<27::AID-PATH582>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 20.Harada T, Okita K, Shiraishi K, Kusano N, Furuya T, Oga A, Kawauchi S, Kondoh S, Sasaki K. Detection of genetic alterations in pancreatic cancers by comparative genomic hybridization coupled with tissue microdissection and degenerate oligonucleotide primed polymerase chain reaction. Oncology. 2002;62:251–258. doi: 10.1159/000059573. [DOI] [PubMed] [Google Scholar]
- 21.Peiper M, Nagoshi M, Patel D, Fletcher JA, Goedegebuure PS, Eberlein TJ. Human pancreatic cancer cells (MPanc-96) recognized by autologous tumor-infiltrating lymphocytes after in vitro as well as in vivo tumor expansion. Int J Cancer. 1997;71:993–999. doi: 10.1002/(sici)1097-0215(19970611)71:6<993::aid-ijc15>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 22.Chen WH, Horoszewicz JS, Leong SS, Shimano T, Penetrante R, Sanders WH, Berjian R, Douglass HO, Martin EW, Chu TM. Human pancreatic adenocarcinoma: in vitro and in vivo morphology of a new tumor line established from ascites. In Vitro. 1982;18:24–34. doi: 10.1007/BF02796382. [DOI] [PubMed] [Google Scholar]
- 23.Heidenblad M, Jonson T, Mahlamaki EH, Gorunova L, Karhu R, Johansson B, Hoglund M. Detailed genomic mapping and expression analyses of 12p amplifications in pancreatic carcinomas reveal a 3.5-Mb 3.-Mb target region for amplification. Genes Chromosomes Cancer. 2002;34:211–223. doi: 10.1002/gcc.10063. [DOI] [PubMed] [Google Scholar]
- 24.Cheng JQ, Ruggeri B, Klein WM, Sonoda G, Altomare DA, Watson DK, Testa JR. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc Natl Acad Sci USA. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albertson DG, Ylstra B, Segraves R, Collins C, Dairkee SH, Kowbel D, Kuo WL, Gray JW, Pinkel D. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genet. 2000;25:144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- 26.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 27.Welch MD. The world according to Arp: regulation of actin nucleation by the Arp2/3 complex. Trends Cell Biol. 1999;9:423–427. doi: 10.1016/s0962-8924(99)01651-7. [DOI] [PubMed] [Google Scholar]
- 28.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 29.Liston P, Fong WG, Korneluk RG. The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene. 2003;22:8568–8580. doi: 10.1038/sj.onc.1207101. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 31.Hilgers W, Song JJ, Haye M, Hruban RR, Kern SE, Fearon ER. Homozygous deletions inactivate DCC, but not MADH4/DPC4/SMAD4, in a subset of pancreatic and biliary cancers. Genes Chromosomes Cancer. 2000;27:353–357. [PubMed] [Google Scholar]
- 32.MacGrogan D, Levy A, Bova GS, Isaacs WB, Bookstein R. Structure and methylation-associated silencing of a gene within a homozygously deleted region of human chromosome band 8p22. Genomics. 1996;35:55–65. doi: 10.1006/geno.1996.0322. [DOI] [PubMed] [Google Scholar]
- 33.Chiquet-Ehrismann R, Chiquet M. Tenascins: regulation and putative functions during pathological stress. J Pathol. 2003;200:488–499. doi: 10.1002/path.1415. [DOI] [PubMed] [Google Scholar]
- 34.Zhai Y, Ni J, Jiang GW, Lu J, Xing L, Lincoln C, Carter KC, Janat F, Kozak D, Xu S, et al. VEGI, a novel cytokine of the tumor necrosis factor family, is an angiogenesis inhibitor that suppresses the growth of colon carcinomas in vivo. FASEB J. 1999;13:181–189. doi: 10.1096/fasebj.13.1.181. [DOI] [PubMed] [Google Scholar]
- 35.Regnier CH, Song HY, Gao X, Goeddel DV, Cao Z, Rothe M. Identification and characterization of an IkappaB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 36.Krupnik VE, Sharp JD, Jiang C, Robison K, Chickering TW, Amaravadi L, Brown DE, Guyot D, Mays G, Leiby K, et al. Functional and structural diversity of the human Dickkopf gene family. Gene. 1999;238:301–313. doi: 10.1016/s0378-1119(99)00365-0. [DOI] [PubMed] [Google Scholar]
- 37.Hoang BH, Kubo T, Healey JH, Yang R, Nathan SS, Kolb EA, Mazza B, Meyers PA, Gorlick R. Dickkopf 3 inhibits invasion and motility of Saos-2 osteosarcoma cells by modulating the Wnt-beta-catenin pathway. Cancer Res. 2004;64:2734–2739. doi: 10.1158/0008-5472.can-03-1952. [DOI] [PubMed] [Google Scholar]
- 38.Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem Sci. 2002;27:396–402. doi: 10.1016/s0968-0004(02)02141-2. [DOI] [PubMed] [Google Scholar]
- 39.Meylan E, Martinon F, Thome M, Gschwendt M, Tschopp J. RIP4 (DIK/PKK), a novel member of the RIP kinase family, activates NF-kappa B and is processed during apoptosis. EMBO Rep. 2002;3:1201–1208. doi: 10.1093/embo-reports/kvf236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tirado CA, Sandberg AA, Stone JF. Identification of a novel amplicon at 1q31 in pancreatic cancer cell lines. Cancer Genet Cytogenet. 1999;113:110–114. doi: 10.1016/s0165-4608(99)00012-6. [DOI] [PubMed] [Google Scholar]
- 41.Kallioniemi OP, Kallioniemi A, Piper J, Isola J, Waldman FM, Gray JW, Pinkel D. Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes Cancer. 1994;10:231–243. doi: 10.1002/gcc.2870100403. [DOI] [PubMed] [Google Scholar]
- 42.Holzmann K, Kohlhammer H, Schwaenen C, Wessendorf S, Kestler HA, Schwoerer A, Rau B, Radlwimmer B, Dohner H, Lichter P, et al. Genomic DNA-chip hybridization reveals a higher incidence of genomic amplifications in pancreatic cancer than conventional comparative genomic hybridization and leads to the identification of novel candidate genes. Cancer Res. 2004;64:4428–4433. doi: 10.1158/0008-5472.CAN-04-0431. [DOI] [PubMed] [Google Scholar]
- 43.Aguirre AJ, Brennan C, Bailey G, Sinha R, Feng B, Leo C, Zhang Y, Zhang J, Gans JD, Bardeesy N, et al. High-resolution characterization of the pancreatic adenocarcinoma genome. Proc Natl Acad Sci USA. 2004;101:9067–9072. doi: 10.1073/pnas.0402932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heidenblad M, Schoenmakers EF, Jonson T, Gorunova L, Veltman JA, van Kessel AG, Hoglund M. Genome-wide array-based comparative genomic hybridization reveals multiple amplification targets and novel homozygous deletions in pancreatic carcinoma cell lines. Cancer Res. 2004;64:3052–3059. doi: 10.1158/0008-5472.can-03-3159. [DOI] [PubMed] [Google Scholar]
- 45.Subramanian G, Schwarz RE, Higgins L, McEnroe G, Chakravarty S, Dugar S, Reiss M. Targeting endogenous transforming growth factor beta receptor signaling in SMAD4-deficient human pancreatic carcinoma cells inhibits their invasive phenotype 1. Cancer Res. 2004;64:5200–5211. doi: 10.1158/0008-5472.CAN-04-0018. [DOI] [PubMed] [Google Scholar]
- 46.Kucharczak J, Simmons MJ, Fan Y, Gelinas C. To be, or not to be: NF-kappaB is the answer—role of Rel/NF-kappaB in the regulation of apoptosis. Oncogene. 2003;22:8961–8982. doi: 10.1038/sj.onc.1207230. [DOI] [PubMed] [Google Scholar]
- 47.Karim R, Tse G, Putti T, Scolyer R, Lee S. The significance of the Wnt pathway in the pathology of human cancers. Pathology. 2004;36:120–128. doi: 10.1080/00313020410001671957. [DOI] [PubMed] [Google Scholar]
- 48.Caca K, Kolligs FT, Ji X, Hayes M, Qian J, Yahanda A, Rimm DL, Costa J, Fearon ER. Beta- and gamma-catenin mutations, but not E-cadherin inactivation, underlie T-cell factor/lymphoid enhancer factor transcriptional deregulation in gastric and pancreatic cancer. Cell Growth Differ. 1999;10:369–376. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Tables