Architecture of the yeast Rrp44–exosome complex suggests routes of RNA recruitment for 3′ end processing (original) (raw)
Abstract
The eukaryotic core exosome (CE) is a conserved nine-subunit protein complex important for 3′ end trimming and degradation of RNA. In yeast, the Rrp44 protein constitutively associates with the CE and provides the sole source of processive 3′-to-5′ exoribonuclease activity. Here we present EM reconstructions of the core and Rrp44-bound exosome complexes. The two-lobed Rrp44 protein binds to the RNase PH domain side of the exosome and buttresses the bottom of the exosome-processing chamber. The Rrp44 C-terminal body part containing an RNase II-type active site is anchored to the exosome through a conserved set of interactions mainly to the Rrp45 and Rrp43 subunit, whereas the Rrp44 N-terminal head part is anchored to the Rrp41 subunit and may function as a roadblock to restrict access of RNA to the active site in the body region. The Rrp44–exosome (RE) architecture suggests an active site sequestration mechanism for strict control of 3′ exoribonuclease activity in the RE complex.
Keywords: electron microscopy, single-particle reconstruction, exoribonuclease
Most eukaryotic RNA transcripts undergo regulated maturation before becoming functionally competent, during which aberrant RNA species are turned over in controlled processes. Besides specific endonucleolytic events (1, 2), two major pathways are responsible for RNA maturation and degradation in eukaryotic cells: the Xrn1 in cytoplasm (and its nuclear homolog Rat1)-mediated 5′ to 3′ decay and exosome-mediated 3′ to 5′ degradation in both the nucleus and cytoplasm (3). Taking the best-studied yeast Saccharomyces cerevisiae system as an example, the nuclear exosome is required for the 3′ trimming of structured RNAs such as rRNA, snRNA, and snoRNA and the 3′ degradation of spliced introns, as well as pre-mRNAs that fail the quality control processes (3). In the cytoplasm, after deadenylation and 5′ decapping, the exosome and Xrn1 mediate the 3′ and 5′ degradation of mRNAs, respectively (4). Furthermore, the exosome has been shown to be involved in many nuclear pre-mRNA surveillance pathways (5–7), as well as cytoplasmic mRNA surveillance pathways, including nonsense-mediated decay, nonstop decay, no-go decay, and ARE-mediated decay (8–12). The exosome also has been shown to degrade the endonucleolytic products of the RNAi pathways (13).
The core eukaryotic exosome is composed of nine subunits. Three of them, Rrp4, Rrp40, and Csl4, are predicted or proven to be RNA-binding proteins. The remaining six subunits are homologous to RNase PH domains and can be further classified into Rrp41-like (Rrp41, Rrp46, and Mtr3) and Rrp42-like (Rrp42, Rrp43, and Rrp45) subunits. The eukaryotic core exosome (CE) (14) shares sequence and architectural similarities with the archaeal exosome (15, 16) and bacterial polynucleotide phosphorylase (17). Their crystal structures revealed a similar donut-shaped architecture composed of two tightly stacked rings, an RNA-binding ring consisting of homo- or heterotrimeric RNA-binding motifs, and a catalytic ring composed of six RNase PH domains forming a trimer of Rrp41/Rrp42-like heterodimers (αβ)3. The presence of a central core in these structures brought up an appealing hypothesis that RNA substrates are recruited by the RNA-binding ring and translocated through a tunnel that only allows the passage of single-stranded RNA to be degraded inside the processing chamber of the catalytic ring (16, 18). Because the recruitment pathway is fairly long, structured RNAs are predicted to stall the exosome and generate products with 7- to 9-nucleotide (nt) single-stranded 3′ overhangs, which is indeed observed for the archaeal exosome (19). The yeast Rrp44-bound exosome, however, is able to degrade RNA to much shorter 3′ overhangs (20), suggesting that there might be alternative recruitment pathways where RNAs can gain access to the active sites more directly.
Exosome functions are regulated by many protein cofactors. To list a few examples from the best-studied yeast S. cerevisiae system, the yeast Rrp44/Dis3 protein, an RNase II-type enzyme, is considered the 10th exosome subunit because of its constitutive association with the CE (21). Rrp6 protein, an RNase D-type enzyme, was shown to associate with the exosome in the nucleus and to contribute to the trimming of the last few nucleotides from the 3′ end of many structured RNA substrates (22, 23). The nuclear TRAMP complex, which contains polyadenylation and putative RNA helicase activities, significantly stimulates the yeast nuclear exosome activity (24). Ski7, a putative GTPase that resembles translation elongation factor EF-1A, was shown to associate with the cytoplasmic exosome and, together with the heterotrimeric Ski2/3/8 complex, to facilitate 3′-to-5′ cytoplasmic mRNA decay (25, 26).
Although both the bacterial polynucleotide phosphorylase and the archaeal exosome contain three 3′-to-5′ phosphorolytic exoribonuclease active sites secluded in the processing chamber for controlled RNA degradation (17, 27), the number of active sites in the eukaryotic CE varies from one in human to zero in yeast (14, 20). In yeast, the 10th exosome subunit, Rrp44/Dis3, provides the sole source of processive 3′-to-5′ exoribonuclease activity (14, 20). Although Rrp44 is highly conserved among eukaryotes, it is controversial whether its interaction with the CE is conserved in other eukaryotes (9, 28–31).
In this study, using single-particle EM image analysis of negatively stained samples, we obtained 3D reconstructions of the core and Rrp44-bound yeast exosome at 23- and 19-Å resolution, respectively. This leap is significant compared with previous EM work, which only revealed a rough shape of a mixture of different exosome–cofactor complexes (32). With improved resolution, we were able to locate the two-lobed Rrp44 protein on the exosome by comparison of the two structures. A pseudoatomic Rrp44–exosome (RE) model was generated by automatic docking of the human CE and the Escherichia coli RNase II structures, allowing the unambiguous characterization of the RE interactions. Homology analysis revealed that interactions between exosome subunits and Rrp44 are likely conserved across species. The architecture of the RE complex suggests an active site sequestration mechanism for strict control of 3′ exoribonuclease activity in the RE complex.
Results
Purification and EM Reconstruction of the Yeast Core and Rrp44-Bound Exosome.
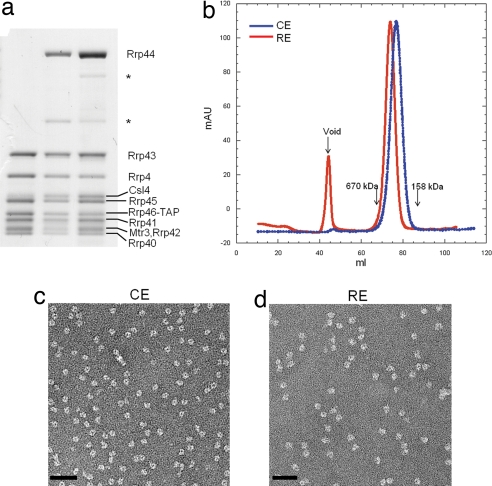
Both the 284-kDa CE and 398-kDa RE complexes were purified from a S. cerevisiae strain lacking RRP6 by using the tandem affinity purification (TAP) scheme, followed by anion-exchange and size-exclusion chromatography to remove endogenous RNA contaminants. All protein components in the purified complexes were confirmed by mass spectrometry. They appear in stoichiometric amounts as judged by SDS/PAGE, with the exception of Csl4, whose association with the exosome is salt-sensitive (data not shown) and which was present substoichiometrically or lost completely during purification (Fig. 1a).
Fig. 1.
Characterization of the yeast CE and RE complexes. (a) SDS/PAGE analysis of the CE (lane 1) and RE (lanes 2 and 3) complexes. The Csl4 protein is near stoichiometric in lane 3. Identity of each exosome subunit, including the two proteolyzed Rrp44 fragments (marked with *) in lanes 2 and 3, was confirmed by using mass spectrometry. (b) Gel-filtration profile showing that RE migrates as a bigger complex than CE. (c and d) Negative-stain EM of CE (c) and RE (d) complexes. (Scale bars: 50 nm.)
The purified complexes appear as well dispersed (Fig. 1b), homogeneous globular particles as observed by negative-stain EM. The particles show an average diameter of ≈150 Å and the characteristic exosome channel [Fig. 1 c and d and supporting information (SI) Fig. 7_a_]. RE particles are, on average, 30% bigger than CEs in dimensions and show an extra density that likely corresponds to the ≈110-kDa Rrp44 protein.
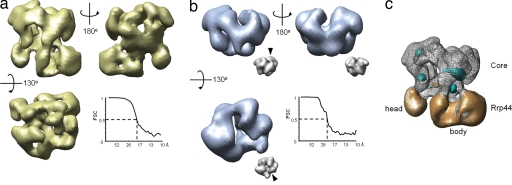
To elucidate the architecture of the RE complex, we carried out single-particle negative-stain EM analysis and 3D reconstructions as a first step toward further studies by cryo-EM. The random conical tilt method was used to generate an initial RE model (33). In all, 3,872 tilt pairs of particles were analyzed, and 50 class volumes were calculated from specimens tilted ≤55°. Two distinct models (with approximately orthogonal missing cones) dominate the class volumes (SI Fig. 7_b_), with both containing a ring with quasi-threefold symmetry and additional asymmetric densities attached to the bottom of the ring. They were used as alternative initial references for projection-matching refinement (34) of a data set of 3,020 untilted particle images. Both gave rise to indistinguishable final structures, with a resolution of 19 Å (Fig. 2a). Although most orientations of the complex are observed in the micrographs (SI Fig. 7_c_), there are preferential orientations (SI Fig. 7_d_) resulting in certain anisotropy of the resolution in the final reconstruction, with 17 Å in the direction of the most populated views and 23 Å in the least observed views (SI Fig. 7_e_).
Fig. 2.
Single-particle reconstruction of the yeast CE and RE complexes. (a) Front, back, and top views of the yeast RE reconstruction. Note that the Fourier shell correlation (FSC) analysis (graph) indicates that the RE reconstruction is at 19-Å resolution by 0.5 criterion. (b) Three views of the CE reconstruction (in the same orientations as the RE complex). The Fourier shell correlation curve (graph) indicates 23-Å resolution for the yeast CE reconstruction. For comparison, the human CE crystal structure was low-pass-filtered to 23-Å resolution and shown in the same views as Insets (scaled at 35% of the yeast CE maps). The arrowhead points to the Csl4 density, which is absent in the yeast CE reconstruction. This result further demonstrates that the projection-matching reconstruction did not introduce model bias. (c) Difference map between the CE and RE (mesh) reconstructions after the alignment of the two. Densities that are present in the RE, but absent in the CE, reconstruction are shown in gold, whereas the densities present in the CE, but not in the RE, reconstruction are shown in cyan. Densities corresponding to the core and Rrp44 protein are indicated, and the head and body region assignment of Rrp44 is marked.
To convincingly locate the Rrp44 density on the CE, we also pursued the 3D reconstruction of the yeast CE by single-particle negative-stain EM analysis. In this case, attempts at random conical tilt reconstruction failed, probably because of the smaller particle size and the somewhat featureless globular particle shape. Taking advantage of the recently available human CE crystal structure, we used a projection-matching reconstruction approach (34) that used the human CE crystal structure low-pass-filtered to 50-Å resolution as the initial reference model (SI Fig. 8_a_). Following this strategy, we were able to obtain a final reconstruction of yeast CE at 23-Å resolution from 2,980 particles (Fig. 2b). As was the case for RE, the CE reconstruction is anisotropic, and the resolution varies from 18 to 25 Å for the orientation of the most and least observed views, respectively (data not shown). In general, our CE and RE reconstructions agree roughly in overall shape with the previously published exosome EM envelope (32), but they revealed more details because of the improved sample-preparation procedure that separates different exosome–cofactor complexes and the reconstruction resolution.
The Two-Lobed Rrp44 Protein Binds to the Bottom of the Exosome PH Ring.
The RNA-binding and PH rings (because the yeast exosome catalytic ring composed of RNase PH homologs is actually not catalytic) of the CE can be easily identified by visual comparison of the CE and RE reconstructions, with the human CE crystal structure filtered to the same resolution as the reconstruction (i.e., 23 Å) (compare Fig. 2 a and b with Fig. 2b Insets). The CE reconstruction can be aligned with the ring structure in the RE complex (Fig. 2c). Difference densities in the CE region are minor (Fig. 2c, cyan and gold), and thus it appears that the yeast CE is structurally robust, and binding of Rrp44 does not result in appreciable conformational changes in the core (at ≈20 Å-resolution).
The Rrp44 density can be unambiguously identified outside the CE region as two large lobes of extra density attached to the bottom of the PH ring in the RE complex (Fig. 2c, gold). The total volume of the additional densities is ≈140,000 Å3, which is in agreement with the presence of a 114-kDa Rrp44 protein (assuming a protein density of 1.34 g/cm3). The larger lobe, which we call the body, accounts for about three-quarters (≈85 kDa) of the total Rrp44 mass and makes multiple contacts with the CE. It contains a narrow channel formed by a clamp-like structure, followed by a globular enlargement containing a cavity. The channel points to the hypothesized RNA-recruiting solvent channel of the CE at a tilted angle (Fig. 3a).
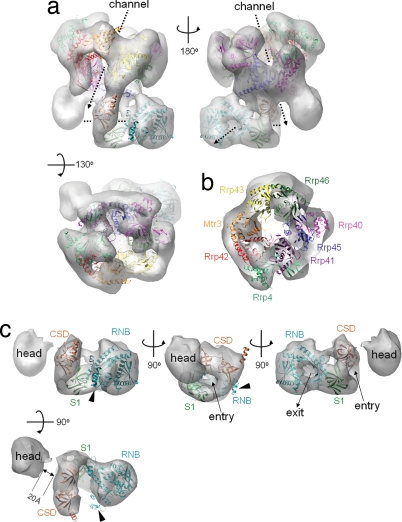
Fig. 3.
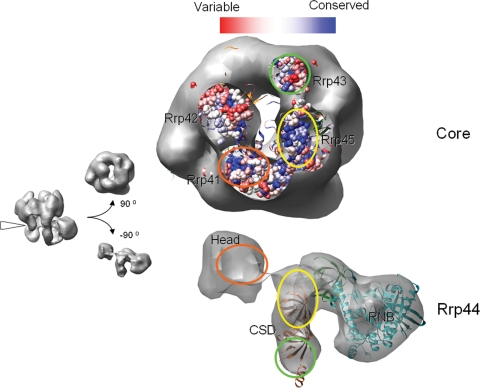
Docking of atomic models into the EM reconstructions. Crystal structures of the human CE and the E. coli RNase II were docked into the yeast RE EM envelope. See text for docking procedure details. (a) Front, back, and top views of the RE EM envelope with the docked CE and RNase II models. The central channels inside the core and Rrp44 protein are marked with dashed arrows. (b) A slice at the CE region perpendicular to the central solvent-accessible channel. The structures of the eight CE subunits are colored the same as their labels. (c) The EM envelope of the Rrp44 protein with the docked E. coli RNase II model at the Rrp44 body region. Landmarks on the RNase II (the CSD, RNB, and S1 domains) are labeled with the same color as their ribbon diagram. The entry and exit of RNA substrate of RNase II are indicated by arrows. Helices 9–11 of RNase II, which protrude partially from the density map, are indicated by arrowheads. Note the 20-Å gap between the head and body, which we believe restricts the access of the RNA substrates.
The smaller Rrp44 lobe, which we call the head, accounts for one-fourth of the mass (≈28 kDa). Its molecular envelope is globular and somewhat featureless. Although it makes significant contacts with the CE, it is barely connected with the body region (visible only at low thresholds, as shown in SI Fig. 9).
The Yeast CE Is Architecturally Similar to the Human Counterpart, and Csl4 Is Dispensable for Structural Integrity.
To deduce the architectural details of the yeast CE complex, we carried out molecular docking of the 3.35-Å resolution nine-subunit human CE crystal structure (PDB ID code 2NN6) into our EM envelope because the human exosome was found to adopt the same subunit arrangement as the yeast exosome (14, 35). To avoid user bias, we relied on an automated molecular-docking procedure by Situs program (36). The best docking solutions for CE and RE complexes, with normalized correlation coefficients of 0.727 and 0.559, respectively, fit well into the 3D density maps and mutually agreed. This procedure allowed the unambiguous identification of EM densities for all exosome subunits (Fig. 3 a and b). Although the CE and RE reconstructions agreed well with the docked models, the density corresponding to the Csl4 was obvious lacking (SI Fig. 10_a_). This result was to be expected, given that Csl4 is either absent or substoichiometric in our samples (Fig. 1a). We tried an alternative purification protocol to obtain RE complexes containing closer to stoichiometric amounts of Csl4 protein (Fig. 1a, lane 3). The EM reconstruction of this sample showed the presence of weak Csl4 density at the predicted position (SI Fig. 10_b_). Thus, it is clear that the Csl4 protein is dispensable for the structural integrity of the CE because its absence causes few changes on other parts of the core structure, and it has a propensity to dissociate from the exosome. However, Csl4 may play an important role in recruiting other exosome cofactors.
The central solvent-accessible channel in the CE and RE complexes appears wider in our EM reconstructions than in the human crystal structure, especially on the Rrp45/Rrp41 heterodimer side (Fig. 3b). This finding could be due to the presence of positive charges inside the channel (particularly abundant on the Rrp45/Rrp41 side) (14), trapping more of the negatively charged stain and, thus, resulting in a reduced apparent local density in the EM reconstructions. Minor docking discrepancies (Fig. 3) may reflect structural differences between the two species. Overall, the docking results strongly support the idea that the yeast exosome adopts the same overall subunit arrangement and similar 3D structure as the human exosome.
The Body Region of Rrp44 Contains the Exoribonuclease Active Site.
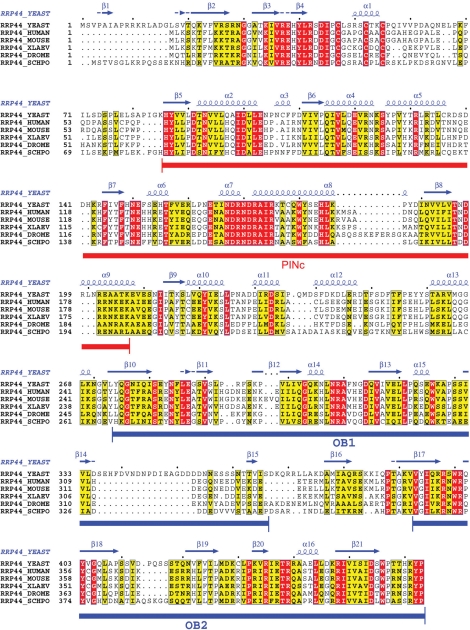
To further analyze our RE structure, we carried out the docking of a 2.7-Å E. coli RNase II structure (37), the closest homolog of Rrp44 with known structure, into the EM 3D map corresponding to the entire Rrp44 protein. In RNase II, the exoribonuclease active site within the RNB catalytic domain is buried in a pocket formed by four conserved elements from the N to the C terminus: two cold-shock domains (CSDs), one RNB domain, and one S1 domain. The CSD and S1 domains, which all belong to the OB (oligonucleotide/oligosaccharide-binding) fold, cuddle to form an RNA recruitment pathway wide enough to admit single-stranded RNA (37). Although the RNB and S1 domains are well conserved between E. coli RNase II and yeast Rrp44 (amino acids 475-1001) (SI Fig. 11) (37), the two CSD-equivalent (OB-fold) domains were detected in Rrp44 (amino acids 274–366 and 393–470) (Fig. 4) only after extensive PSI-BLAST search (OB-fold proteins are frequently found to be conserved three dimensionally, but not at the primary sequence level). The two OB folds in Rrp44 are homologous to many bacterial RNase II/R enzymes (E value of <10−16). Thus, it appears that the E. coli RNase II structure would be a good model for the C-terminal portion of the Rrp44.
Fig. 4.
Sequence alignment for the Rrp44 N-terminal region. Rrp44 sequences from budding yeast, fission yeast (Schizosaccharomyces pombe, SCHPO), human, mouse, Xenopus laevis, and Drosophila melanogaster were aligned by using ClustalW (red for identical residues, yellow for similar ones). The secondary structure predictions also are shown. Regions corresponding to the PINc domain (amino acids 86–207) and two OB folds (amino acids 277–366 and 393–464, equivalent to the two CSDs in E. coli RNase II) are highlighted in red and blue closed bars, respectively.
Indeed, the atomic models of the E. coli RNase II can be easily docked into the body region of the Rrp44 density map with a normalized correlation coefficient of 0.771 after local adjustment of individual domains (Fig. 3c). The RNB domain fits into the globular enlargement at the distal end of the body. The S1 domain occupies the lower half of the clamp structure, whereas the two CSDs occupy the upper clamp structure, mediating contacts with the exosome. Interestingly, the channel from the CSD-S1 clamp to the RNB active site in the RNase II structure agrees perfectly with the channel inside the Rrp44 body part, suggesting that the RNA substrate recruitment mechanism is likely conserved. There is no clashing of the docked structures, and little unfilled space is left in the density. The only noticeable discrepancy is the protrusion of the region in E. coli RNase II comprising helices 9–11 from the EM envelop, which happens to be the least conserved region between Rrp44 and RNase II in the RNB domain (Fig. 3c and SI Fig. 11).
The RNA recruitment channel of Rrp44 opens toward the bottom of the exosome solvent-accessible channel at an angle of ≈90°. The arrangement is suggestive of a possible hand-over mechanism between the exosome and Rrp44, by which the RNA substrate is recruited by the exosome through its solvent-accessible channel and then passed to Rrp44.
The Head of Rrp44 Restricts Substrate Entry into the Body.
With all densities for the body region assigned, it follows that the head region must correspond to the N-terminal portion of the Rrp44 sequence (amino acids 1–270). The molecular mass occupied by this region is ≈30 kDa, close to the 28 kDa calculated from the 3D volume of the head region. The region of amino acids 86–203 in the Rrp44 N-terminal portion belongs to the 15-kDa PINc domain, which may contribute to RNA binding (Fig. 4) (38). However, we were unable to dock it into the density unambiguously because of the fairly featureless globular shape of the head part at the present resolution.
The head region is anchored to the exosome through extensive contacts with the Rrp41 subunit, burying a surface area of 1,000 Å2. It connects to the CSD domains through a linker (visible in the EM map at lower contour levels), which is consistent with Rrp44's topological arrangement. Interestingly, the head is positioned ≈20 Å away from the RNA recruitment channel in the body (Fig. 3c), thus blocking straightforward RNA access to the body. The cleft between the head and body accommodates a single-stranded RNA, barely fits a double-stranded RNA substrate, but would be too small for tertiary RNA structures. Such RNAs (i.e., the rRNA precursor) will likely stall the RE complex, thus avoiding overtrimming.
The Rrp44 Body Interacts with the CE Through a Conserved Interface.
The body of Rrp44 is anchored to the CE mainly through interactions between its CSDs and the Rrp45 subunit, burying a continuous surface area of ≈1,800 Å2 (Fig. 5). Many surface loops on Rrp45 are involved in the interactions with Rrp44 CSDs, according to the docked human CE model. Several minor contacts also were observed between the Rrp44 RNB domain and Rrp45 in the RE reconstruction, but they are probably too weak to contribute much to the total binding energy. These findings are consistent with previous reports (35) that Rrp45 is the major point of contact between Rrp44 and the CE. The body of Rrp44 also makes contact with Rrp43 to a lesser extent, burying a surface area of ≈400 Å2.
Fig. 5.
Conservation analysis of the RE interacting interface. The RE reconstruction is sliced at the interface, and the Rrp44 and the CE were rotated 90° to reveal the interaction surface. Based on the multiple sequence alignment of the exosome subunits by using ClustalW, a conservation map for the bottom of the CE was rendered by using the Multialign viewer function in Chimera with the conservative histogram calculated by using program AL2CO (42). The correlated interacting surfaces on the core and Rrp44 are marked with circles of the same color. The conservation map of the Rrp42 surface, which does not contact Rrp44, is shown for comparison.
We then carried out a sequence conservation analysis to address whether the RE interactions might be conserved among other eukaryotes. At the bottom of the exosome's PH ring, conserved patches of amino acid residues were found on the Rrp45 and Rrp41 surfaces (Fig. 5 Upper) (14). Interestingly, these patches coincide with the major RE interface (Fig. 5 Lower). Because these residues are unlikely conserved for structural reasons (they reside mainly in surface loops according to the human exosome crystal structure), we suspect that they are conserved for protein interaction purposes, in particular for the recruitment of Rrp44. The Rrp44 protein is highly conserved from yeast to human throughout the ORF. This finding includes the two E. coli CSD-equivalent OB folds, which mediate extensive interactions with Rrp45 of the exosome and are >34% identical (55% similar) in primary sequence between yeast and human (Fig. 4).
Discussion
The eukaryotic exosome, the archaeal exosome, and bacterial polynucleotide phosphorylase constitute a class of conserved multisubunit macromolecules essential for 3′ end RNA processing and degradation. Unlike the archaeal and bacterial counterparts, which contain three phosphorolytic ribonuclease-active sites, the eukaryotic CEs studied so far contain either one (in human) or no (in yeast) phosphorolytic active sites at all. This finding suggests that, during evolution, the eukaryotic exosome may have been transformed from an enzymatic machinery to a regulator that achieves its functionality through selective binding of a set of cofactors, including exoribonucleases, polyadenylase, and RNA helicases. The focus of this study, the yeast Rrp44 protein, is considered the 10th yeast exosome subunit because it constitutively associates with the CE and provides the sole source of 3′-to-5′ exoribonuclease activity. Most significantly, its RNase activity is tightly regulated and greatly reduced upon association with the CE. How does binding to the CE regulate Rrp44's RNase activity? Is this regulation mechanism conserved in other eukaryotes? We addressed these questions through the structural inspection of the core and Rrp44-bound yeast exosomes.
At ≈20-Å resolution, the single-particle negative-stain EM reconstructions of the core and Rrp44-bound exosome complexes allowed us to locate the two-lobed Rrp44 protein at the bottom of the yeast CE. The active site of Rrp44 is occluded inside the Rrp44 body region by a single-stranded RNA recruitment channel. Strikingly, the entry of the recruitment channel is not directly accessible from the surrounding solvent. Instead, the channel tilts toward the central solvent channel of the CE at an ≈90° angle. From the dorsal side of Rrp44, only those RNAs emerging from the processing chamber can be further recruited by Rrp44. From the ventral side, accessing Rrp44-active sites is sterically restricted as well. Immediately in front of the substrate channel in the body region lies the ≈30-kDa head of Rrp44. This domain prevents direct RNA access to the body's substrate channel. To enter this channel, single-stranded RNA from the ventral side has to approach at restricted angles and be bent ≈90°. Double-stranded RNAs can barely fit into the gap between the head and body regions of Rrp44, but their 3′ overhang has to be continuously peeled off and then bent ≈90° to enter the Rrp44-active site, an action that may require the help of additional RNA helicases. Any RNA with a strong tertiary structure would not fit into the gap, thus leaving behind products with a longer 3′ overhang. Overall, our RE EM reconstruction suggests a steric hindrance mechanism that down-regulates the Rrp44 exoribonuclease activity.
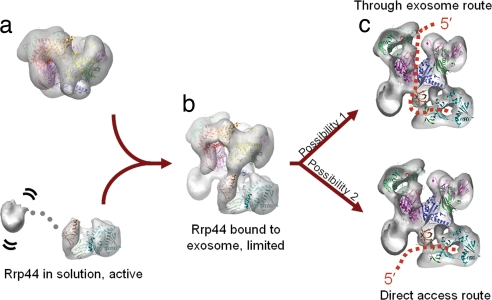
The steric constraints that down-regulate the Rrp44 activity are likely to take place only after Rrp44 is bound by the core exosome because the head and body regions of Rrp44 show little or no contact with each other, as suggested by our EM reconstruction, but are held in place through extensive interactions with the CE subunits. It is conceivable that, in the absence of the CEs, the head of Rrp44 would tumble freely relative to the rest of the protein, no longer interfering with the substrate recruitment by the body (Fig. 6a). Indeed, the purified yeast Rrp44 protein, either washed off from the endogenous exosome preparations or overexpressed in E. coli, is rapidly proteolyzed into two fragments of ≈30 and ≈80 kDa. The proteolyzed fragments do not copurify in subsequent chromatography steps (data not shown). The observation suggests that free Rrp44 is likely a dumbbell-shaped molecule with its head and body regions connected by a flexible linker. In summary, we propose that the yeast CE down-regulates the RNase activity of Rrp44 by sterically restricting the RNA's access to the nucleolytic active site (Fig. 6b).
Fig. 6.
Possible RNA processing mechanism by the yeast RE complex. (a) In the absence of the CE, the head of Rrp44 can tumble freely relative to the body and does not interfere with the enzymatic activity in the body region. The CE adopts a rigid conformation regardless of Rrp44's presence. (b) Binding of Rrp44 to the CE positions its head 20 Å away from its body, resulting in down-regulation of Rrp44's activity due to a steric hindrance effect in substrate recruitment. (c) RNA substrates can access the Rrp44 body domain active site either through the central channel of the CE (through exosome route) or directly from the ventral side of Rrp44 (direct access route). The two routes are not mutually exclusive.
Does RNA enter Rrp44 from the exosome or ventral side of Rrp44? Our structural model is compatible with both routes (Fig. 6c). Results from the Conti laboratory suggest that the archaeal exosome likely recruits RNA through the central solvent channel (15). Because the yeast CE is catalytically inactive, the RNA can potentially emerge from the bottom of the processing chamber just to be captured and degraded by the body region of Rrp44 (through exosome route) (Fig. 6c Upper). This route predicts that a highly structured RNA cannot be processed efficiently if its single-stranded 3′ overhang is not long enough to span the distance from the central channel to the Rrp44-active site (at least 15–20 nt), which is roughly consistent with the observations from Liu et al. (14). However, Dziembowski et al. (20) showed that pre-tRNAs can be processed to much shorter 3′ overhangs by the yeast RE complex. This observation is more consistent with a model where RNA enters Rrp44 from the ventral side (direct access route) (Fig. 6c Lower). From this route, RNAs with strong secondary structures can still be recruited at a tilted angle and be processed to an ≈4- to 5-nt 3′ overhang by Rrp44 (37, 39). Because these two routes do not preclude each other, it is possible that both may be used inside cells for different RNA substrates. Structures of the RNA-bound RE complex will help to distinguish the two possibilities.
Although Rrp44 is considered an indispensable component of the yeast exosome, its role in other eukaryotes has not been fully established. Experimental evidence supporting RE interactions includes direct demonstration of yeast Rrp44–Arabidopsis thaliana Rrp41 interaction by using a GST pull-down assay (28), yeast two-hybrid assay showing interactions between the human Rrp44 and Rrp43 proteins (31), and rescue experiments showing that human Rrp44 can complement the yeast Rrp44 mutant phenotype (29). However, others concluded that there are no tight RE interactions in human and Trypanosoma brucei (9, 30). The apparent discrepancy may be due to the high ionic strength used in the biochemical assays or to difficulties in expressing foreign Rrp44s in yeast for functional assays.
Our EM reconstruction supports the notion that the RE interactions are conserved in eukaryotes. It shows that the Rrp44 body region interacts mainly with Rrp45, Rrp41, and, to a lesser extent, Rrp43. Moreover, the interaction surfaces on the Rrp45/Rrp41 heterodimer appear to be the most conserved area at the bottom of the PH ring (Fig. 5) (14). These conserved residues are unlikely conserved for structural purposes because they mainly reside in loop regions. In addition, Rrp44 is highly conserved from yeast to human. Mutagenesis experiments and higher-resolution structural information will further test our hypothesis. Furthermore, functional RE complexes may be generated from heteroassembled yeast CE and human Rrp44, for example, to provide additional support for the conservation hypothesis. If the RE interactions are conserved among other eukaryotes, it is highly likely that the RE architecture and its RNA degradation mechanism, as proposed in this study, also will be conserved.
Materials and Methods
Protein Purification and Characterization.
The yeast core and Rrp44-bound exosome (CE and RE) samples were purified from an Rrp46-TAP-tagged yeast strain with the _RRP6_-knockout by following the standard tandem affinity purification procedure with minor modifications. The CE and RE samples were further purified on a mono-Q column to remove the tightly bound endogenous RNA contaminations. See SI Materials and Methods for details.
Preparation of Negative-Stained Samples.
CE and RE samples were thawed on ice and diluted to ≈0.1 μM in a buffer containing 25 mM Tris·HCl (pH 7.5), 50 mM NaCl, 2 mM DTT, and 10 μM ZnCl2. Four microliters of the protein solution was negatively stained with 2% uranyl formate solution between two thin layers of carbon on a copper grid by using the sandwich method (40).
EM and Image Processing.
Tilted pairs of micrographs of the specimen were taken on a Tecnai-12 EM at a magnification of ×49,000. The single-particle reconstruction of RE was performed by using random conical tilt (33), followed by projection-matching refinement (34), whereas that of CE was performed by using projection-matching refinement with low-pass-filtered human CE crystal structure as the initial model. See SI Materials and Methods for more details.
Docking of the Atomic Model, EM Map Alignment, and Sequence Alignment.
The modified human CE atomic model (PDB ID code 2NN6) and E. coli RNase II atomic model (PDB ID code 2IX0) were docked into the 3D reconstruction models by using Situs 2.0 (36, 41). The long C-terminal tail of Rrp45 (from amino acids 292–449) was deleted from the human CE model because it is not present in the yeast homolog. The detailed procedure for sequence and EM map alignment can be found in SI Materials and Methods.
Supplementary Material
Supporting Information
Acknowledgments
We thank David King, Arnie Falick, and Sharleen Zhou for generous help in the mass spectrometry analysis of the exosome samples; Jianhua Fu and Mincheng Zhang for suggestions in protein purification from yeast; Richard Gillian for small-angle x-ray scattering analysis of the sample homogeneity; Andres Leschziner, Bunpote Siridechadilok, and Vince Ramey for sharing scripts and helpful discussions; and Eric Alani and Linda Nicholson for helpful discussions and comments on the manuscript. This work was supported by National Institutes of Health Public Health Service Grant GM-59898 (to. K.P.C. and J.S.B.). E.N. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
CE
core exosome
RE
Rrp44–exosome.
Note Added in Proof.
While this paper was under review, we learned that, similar to the yeast core exosome, the human core exosome is not enzymatically active (43). This observation seems to futher strengthen our prediction that the RE architecture and its RNA degradation mechanism are conserved across eukaryotes. A relevant study on the substrate preference of the Rrp44 was published (44) while our paper was under review.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Density maps for reconstructions have been deposited in the EM Data Bank at the European Bioinformatics Institute, www.ebi.ac.uk/msd-srv/emsearch/index.html [accession codes 1438 (RE) and 1439 (CE)].
References
- 1.Venema J, Tollervey D. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 2.Gatfield D, Izaurralde E. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 3.Houseley J, LaCava J, Tollervey D. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 4.Parker R, Song H. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 5.Vasiljeva L, Buratowski S. Mol Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 6.Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- 7.Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- 8.Hilleren PJ, Parker R. Mol Cell. 2003;12:1453–1465. doi: 10.1016/s1097-2765(03)00488-x. [DOI] [PubMed] [Google Scholar]
- 9.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 10.Doma MK, Parker R. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell P, Tollervey D. Mol Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 13.Orban TI, Izaurralde E. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q, Greimann JC, Lima CD. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 15.Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E. EMBO Rep. 2007;8:470–476. doi: 10.1038/sj.embor.7400945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buttner K, Wenig K, Hopfner KP. Mol Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Symmons MF, Jones GH, Luisi BF. Structure (London) 2000;8:1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- 18.Lorentzen E, Conti E. Cell. 2006;125:651–654. doi: 10.1016/j.cell.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Lorentzen E, Conti E. Mol Cell. 2005;20:473–481. doi: 10.1016/j.molcel.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 20.Dziembowski A, Lorentzen E, Conti E, Seraphin B. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 22.Briggs MW, Burkard KT, Butler JS. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 23.Phillips S, Butler JS. RNA. 2003;9:1098–1107. doi: 10.1261/rna.5560903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 25.van Hoof A, Staples RR, Baker RE, Parker R. Mol Cell Biol. 2000;20:8230–8243. doi: 10.1128/mcb.20.21.8230-8243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T. EMBO J. 2001;20:4684–4693. doi: 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. Nat Struct Mol Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- 28.Chekanova JA, Dutko JA, Mian IS, Belostotsky DA. Nucleic Acids Res. 2002;30:695–700. doi: 10.1093/nar/30.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiomi T, Fukushima K, Suzuki N, Nakashima N, Noguchi E, Nishimoto T. J Biochem (Tokyo) 1998;123:883–890. doi: 10.1093/oxfordjournals.jbchem.a022020. [DOI] [PubMed] [Google Scholar]
- 30.Estevez AM, Kempf T, Clayton C. EMBO J. 2001;20:3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehner B, Sanderson CM. Genome Res. 2004;14:1315–1323. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aloy P, Ciccarelli FD, Leutwein C, Gavin AC, Superti-Furga G, Bork P, Bottcher B, Russell RB. EMBO Rep. 2002;3:628–635. doi: 10.1093/embo-reports/kvf135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radermacher M, Wagenknecht T, Verschoor A, Frank J. J Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 34.Penczek PA, Grassucci RA, Frank J. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 35.Hernandez H, Dziembowski A, Taverner T, Seraphin B, Robinson CV. EMBO Rep. 2006;7:605–610. doi: 10.1038/sj.embor.7400702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wriggers W, Milligan RA, McCammon JA. J Struct Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 37.Frazao C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Nature. 2006;443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 38.Jeyakanthan J, Inagaki E, Kuroishi C, Tahirov TH. Acta Crystallogr F. 2005;61:463–468. doi: 10.1107/S1744309105012406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuo Y, Vincent HA, Zhang J, Wang Y, Deutscher MP, Malhotra A. Mol Cell. 2006;24:149–156. doi: 10.1016/j.molcel.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 40.Ohi M, Li Y, Cheng Y, Walz T. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chacon P, Wriggers W. J Mol Biol. 2002;317:375–384. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- 42.Pei J, Grishin NV. Bioinformatics. 2001;17:700–712. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- 43.Liu Q, Greimann JC, Lima CD. Cell. 2007;131:188–189. [Google Scholar]
- 44.Schneider C, Anderson JT, Tollervey D. Mol Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information