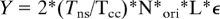Genome-wide studies highlight indirect links between human replication origins and gene regulation (original) (raw)
Abstract
To get insights into the regulation of replication initiation, we systematically mapped replication origins along 1% of the human genome in HeLa cells. We identified 283 origins, 10 times more than previously known. Origin density is strongly correlated with genomic landscapes, with clusters of closely spaced origins in GC-rich regions and no origins in large GC-poor regions. Origin sequences are evolutionarily conserved, and half of them map within or near CpG islands. Most of the origins overlap transcriptional regulatory elements, providing further evidence of a connection with gene regulation. Moreover, we identify c-JUN and c-FOS as important regulators of origin selection. Half of the identified replication initiation sites do not have an open chromatin configuration, showing the absence of a direct link with gene regulation. Replication timing analyses coupled with our origin mapping suggest that a relatively strict origin-timing program regulates the replication of the human genome.
Keywords: chromatin structure, DNA replication origin, ENCODE regions, genome-wide mapping, CpG island
Controlling the number of origins from which replication begins in a given chromosome is necessary to protect it from instability (1, 2). Mapping DNA replication starting points, known as “origins of replication,” would make a large contribution to understanding how genome replication is coordinated. Identification of a large number of replication origins is necessary to decipher the rules of origin specification. However, fewer than 30 origins have been identified in human cells (3), and they were mapped mostly in well-characterized transcribed regions, leaving gene-poor regions unexplored. The Encyclopedia of DNA Elements (ENCODE) project (4), launched to develop high-throughput methods for identifying functional elements, now provides a comprehensive view of gene expression and chromatin structure along 1% of the human genome (30 Mb, 44 regions) (5); it thus provides a powerful model for studying interactions among chromosome organization, gene regulation, and initiation of DNA replication. Origin selection is initiated by the binding of the origin recognition complex (ORC) to origin-proximal DNA sequences (6). In contrast to Saccharomyces cerevisiae, characterized metazoan origins do not conform to a clear consensus sequence, and the ORCs from higher eukaryotes exhibit no sequence specificity in vitro (7). Transcription factors at sites of replication initiation have been shown to stimulate replication in many systems, including viruses, yeast, Drosophila, and Xenopus (8, 9). This stimulation may be a consequence of direct interaction with components of the replication machinery or of facilitating the access of the replication complexes to DNA through recruitment of chromatin remodeling complexes (10–12). Here we present a high-resolution map of replication origins in HeLa cells based on hybridization of short nascent strands (SNS) on DNA microarrays covering ENCODE regions. To construct this map we use one of the most stringent methods for isolating origins of replication based on the resistance of SNS to λ-exonuclease digestion (13). Because only small numbers of short nascent strands can be recovered, they must be amplified before hybridization to the microarray. We chose a method of amplification that gives low biases, the T7-based DNA linear amplification (TLAD). We thereby mapped 282 origins of replication and confirmed 1 already-known origin, increasing by more than 10-fold the number of currently known origins and constituting a statistically relevant dataset for studying general rules for origin selection.
Results
Identification of 283 Origins Along ENCODE Regions.
Because small bubbles of replication are scarce and genome-wide studies based on DNA microarrays require at least 2 μg of material for hybridization, the development of large-scale studies on the positioning of DNA replication origins is a difficult task. We calculated that if 30,000 origins were activated inside a given cell, we should obtain ≈ 10 ng of SNS from 108 exponentially growing cells (see Experimental Procedures). Two stringent methods have been used successfully to purify sequences located at replication initiation sites. One method relies on the trapping of bubble-shaped structures, and the other is based on isolation of transitory RNA-DNA SNS molecules (13, 14). In the latter method, SNS between 1.5 kb and 2 kb can be purified specifically from an asynchronous population of cells because their RNA primers protect them from λ-exonuclease treatment, whereas broken genomic DNA is digested.
We prepared 5 independent samples of SNS from asynchronous HeLa cells. We coupled the stringent preparation of SNS with the TLAD method, a technique of linear amplification that can generate several μg of amplified material from 10–20 ng of input DNA (15). We amplified 2 preparations of SNS by TLAD (experiments A and B) for microarray hybridization and cross-checked the results against those obtained by real-time quantitative PCR (qPCR) using the other 3 samples (experiments C, D, and E). The quality of each SNS preparation was tested systematically by qPCR; this test calculated the relative enrichment of an amplicon located near, but not within, the c-myc origin (considered as background) relative to an amplicon located inside the c-myc origin (defined as 100%). We observed a variation of enrichments that were caused by the accumulation of many critical steps in the SNS purification protocol, namely genomic DNA extraction, polynucleotide kinase treatment, and finally λ-exonuclease digestion. The results were 3%, 15%, 12%, 14%, and 19% for experiments A, B, C, D, and E, respectively. We co-hybridized genomic DNA and SNS amplified by the same technique to reduce possible bias caused by linear amplification. Results obtained on 4 ENCODE regions are shown in supporting information (SI) Figs. S1-S3. To limit the number of false positives, stringent selection criteria were applied for hybridization peak detection (see Experimental Procedures). Typical peaks selected from Fig. S1 are presented in Fig. S2. On the 2 DNA microarrays performed, we identified 277 peaks with SNS preparation A and 228 peaks with SNS preparation B; 187 peaks were common to the 2 preparations, and the other 131 peaks were preparation specific. In total, we detected 318 peaks including 187 overlapping peaks. The difference in the quality of SNS preparations A and B (3% versus 15% of background relative to the c-myc origin) may explain why origin conservation was only 59% when automatic stringent criteria were applied for peak detection. We then checked each of the 131 preparation-specific peaks individually using slightly less stringent criteria: enriched profile with a similar 1.5-kb width containing among 4 enriched probes at least 2 probes with a p-value <10−4. Ninety-six of these 131 peaks were validated manually on the array where peaks were not automatically detected. Thus, we identified a total of 283 clearly visible peaks on the 2 microarrays (Table S1), corresponding to a final reproducibility of ≈ 90%.
Sensitivity and Specificity of Our Genome-Wide Method.
To validate the specificity and the sensitivity of our mapping study, we randomly chose 29 of the 283 peaks and 8 background regions and quantified their relative enrichment by qPCR with a third and fourth SNS preparation (experiments C and D) (Fig. S3). All but 1 origin (ori7, 26.5%) had a significant enrichment relative to the c-myc background signal (13%; Fig. S3). Amplicons located in the 8 background regions had a very low enrichment, between 2% and 20%. The previously described G6PD origin (16) was found at the same location on our microarrays (Table S1, ori G6PD). For further validation, we analyzed the profile of SNS enrichment of the 5′ part of the HoxA locus using qPCR with 32 primer pairs and on a fifth preparation confirmed 5 enriched peaks detected by our microarray approach (Fig. S3). These validations demonstrate the specificity and sensitivity of our genome-wide mapping of DNA replication origins. Finally, among 1067 replication origins that have been predicted computationally in the human genome (17), 7 are located within ENCODE regions. Two of these computational predictions fall less than 1 kb from our experimentally identified origins. Although the sample of predicted origins is limited, the overlaps with experimental data are highly significant (P = 10−4, Fig. S4).
Primer pairs located in the middle of peaks detected on microarrays were selected for qPCR quantification. The level of enrichment observed by qPCR therefore should indicate origin strength. We observed SNS enrichment ranging from 40% to 330% (Fig. S3), suggesting that our method can detect origins activated in only 12% of cell cycles. This result shows that our microarray approach is sensitive enough to detect weak origins. However, the stringent method applied for peak selection may have missed between 5% and 10% of true positives.
Small ssDNA Are Not Enriched in Short Nascent Strands.
A recent paper described a high-throughput mapping of origins of replication in human cells based on a rapid, non-PCR-based hybridization of short ssDNA extracted from asynchronous human cells (18). In this paper, the authors obtained 6–8 μg of 300- to 1000-bp ssDNA from 108 cells. This amount is between 2 and 3 orders of magnitude higher than the predicted amount of SNS (see Experimental Procedures), suggesting that in this preparation true SNS are swamped by a mass of irrelevant broken genomic DNA. In our study, after λ-exonuclease digestion, we repeatedly obtained ∼ 10 ng of 1.5- to 2-kb SNS molecules from 108 exponentially growing cells. This amount fits well with the theoretical amount of SNS. In our experiments, before λ-exonuclease treatment, we obtained a much higher yield of single-stranded molecules, ≈2 μg. At this step of purification, we could not detect enrichment of c-myc origin by qPCR, showing that, although SNS were in the preparation, the excess of irrelevant broken genomic DNA masks them.
To analyze a preparation of short ssDNA on a larger scale, we co-hybridized on an ENCODE microarray a preparation of 300- to 1000-bp ssDNA with genomic DNA extracted from G1 cells. We applied for peak detection stringent criteria similar to those used for our SNS study (Experimental Procedures). Nine highly significant peaks (Table S2) were identified, and none overlaps with peaks detected with 1.5- to 2-kb SNS. With less stringent criteria (Experimental Procedures), we identified 229 peaks (Table S3), including 6 peaks in common with our 283 origin segments. This co-localization is not significant (P = 0.08). Moreover, the G6PD origin previously identified is not detected on this microarray. In conclusion, our data show that regions of short ssDNA enrichment do not overlap with peaks obtained with a preparation of SNS stringently purified by λ-exonuclease treatment of 1.5 to 2 kb ssDNA. This result provides a clear demonstration that the identification of replication origins from short ssDNA requires λ-exonuclease digestion.
Origin Density Is Correlated with GC Richness, and Only Half of the Origins Co-Localize with CpG Islands.
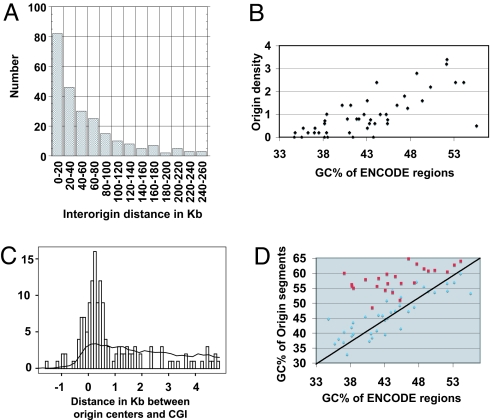
Whether strong sites of replication initiation are distributed regularly along the human genome remains an open question. The interorigin distances in our sample are diverse (Fig. 1A),ranging from ≈1 kb to 500 kb. The mean interorigin distance (∼63 kb) and most interorigin distances (75% < 81 kb) are within the range of the human replicon size (19). Surprisingly, we found 500-kb regions lacking origins. The base composition of mammalian genomes is very heterogeneous, and gene density is much higher in GC-rich than in GC-poor genomic regions (20, 21). Interestingly, we observe the same pattern for replication origins: replication origin density within ENCODE regions is strongly correlated with the GC content (R2 = 52%, P < 10−7; Fig. 1B). Previous studies suggested that many origins in human cells are within CpG islands (22, 23). We compared the distribution of distances between the origin center and the edge of the nearest CpG island in our sample with that in a bootstrap group (i.e., DNA segments of the same size as origins, randomly sampled from ENCODE regions; see Experimental Procedures). Origin centers were more frequently within and close to CpG islands than expected (P < 2.2 × 10−16) (Fig. 1C). More than one-third of origin segments overlap a CpG island (7 times more than bootstrap segments), and half are less than 1 kb away from the edge of a CpG island. In contrast to this subset, origin segments not within CpG islands are neither enriched nor depleted in G+C relative to the G+C richness of the surrounding region (Fig. 1D). Therefore origin specification presumably is under the control of at least 2 distinct types of _cis_-element.
Fig. 1.
Strong origins are not evenly distributed along the genome, and a subset of origins is associated with CpG islands. (A) Histogram of the interorigin distances (median = 36080; n = 245). Four interorigin distances greater than 260 kb (333, 411, 453, and 480 Kb) are not shown on the graph. (B) Origin density within ENCODE regions correlates with GC richness. The density of origins within each ENCODE region (number of origins per 100 kb) is plotted against the GC percent of the whole region. (C) Origins are enriched in and near CpG islands. The histogram shows the distribution of origin centers relative to CpG islands (CGI). Negative values correspond to origin centers located within CpG islands. The line shows the distribution of the bootstrap sample. (D) Segments containing non-CpG island origins are not enriched in GC. For each ENCODE region, the GC percentage of replication origin segments within CpG islands (red) and outside CpG islands (blue) is plotted against the GC percentage of the whole ENCODE region.
Replication Origins Are Evolutionarily Conserved.
To determine whether replication origins contain sequence motifs under selective constraints, we compared the position of origins with the positions of evolutionary conserved regions (CRs) identified in mammalian genomes by the ENCODE project (24) (see SI Text). We observed that 70% of origins overlap with CRs, much more than would be expected by chance (43%, P < 10−6). This overlap is lower than the intersection between protein-coding exons and CRs (86%) but is similar to that obtained with promoter regions (72%). Thus, origins seem to contain a proportion of constrained sequence motifs comparable to promoters. As expected from our previous results, we found that many origins overlap with promoter regions (34%, compared with 15% of bootstrap regions; P < 10−6). A large fraction (79%) of those promoter origins are associated with a CpG island. Interestingly, promoter origins seem to be more constrained than other origins and promoters (85%), indicating that combining the 2 activities (promoter and replication origin) requires additional sequence motifs. The finding that the large majority of origins overlap with CRs suggests that the location of replication origins remained evolutionarily conserved, at least among mammals. Thus, the conservation of predicted replication domains (17) seems to result from the conservation of specific _cis_- elements at replication origins.
Open Chromatin Structure Does Not Regulate Replication Initiation.
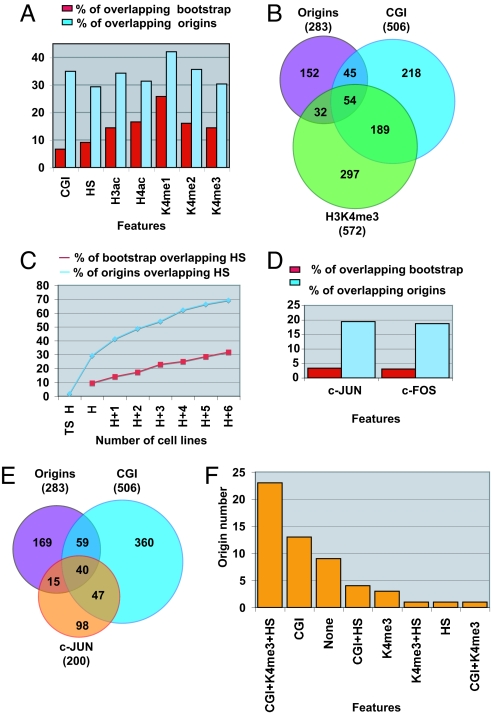
A simple way to explain the lack of an origin consensus sequence in metazoa is that chromatin structure rather than DNA sequence regulates origin selection. We analyzed the distribution of distances between origin centers and previously identified Dnase I hypersensitive sites (HS) and histone H4 acetylation (H4ac), histone H3 acetylation (H3ac), and histone H3 mono-methylation on lysine 4 (K4me1), di-methylation (K4me2), and tri-methylation (K4me3) segments in HeLa cells (25, 26). The positioning of origins correlated strongly with HS, H3ac, H3K4me2, and H3K4me3 sites (P < 10−16) and more weakly with H4ac and H3K4me1 sites (P = 5.35 × 10−11 and 6.46 × 10−13, respectively). These findings are consistent with the replication machinery preferentially recognizing open chromatin structures found near promoters of actively transcribed genes (8). Such an association, however, seems to be favored but not necessary: 29%, 34%, 36%, and 30% of origin segments overlap with HS, H3ac, H3K4me2, and H3K4me3 sites, respectively (Fig. 2A). These modifications typically co-localize and collectively are good indicators of promoters near highly transcribed genes (25). Consequently, 21% of origin segments overlap with sites displaying all 4 of these features. However, 47% of origins have no histone modifications, and 44% have neither histone modifications nor an HS site. Therefore, although an open chromatin structure seems to be a preferential substrate for replication initiation, nearly half of the identified origins do not have these modifications and may be defined and recognized as replication origins by another mechanism.
Fig. 2.
Origins are strongly associated with regulatory elements but are not favored by open chromatin structure.(A) Chromosomal features corresponding to open chromatin structures are enriched inside origins. This graph shows the percentage of replication origin segments (blue) and bootstrap segments (red) overlapping CpG islands (CGI), HS sites, H3ac, H4ac, H3K4me1 (K4me1), H3K4me2 (K4me2), and H3K4me3 (K4me3) segments. (B) H3K4me3 is not associated preferentially with replication origins. The Venn diagram shows the overlap between replication origin segments, CpG islands (CGI), and H3K4me3 regions. The number of segments found in each group is shown between brackets, and the number of overlapping elements is in intersected regions. (C) Origins of replication are found mostly inside regulatory elements. This graph shows the cumulative percentage of segments overlapping HS sites found in 7 cell lines. The HS sites were added in the following order: HeLa-specific sites (TS H), HeLa (H), GM06990 (H + 1), CD4, HepG2, H9, IMR 90, and K562 cells. The blue and the red lines represent the overlap with origin segments and with 283 random segments, respectively. (D) c-JUN and c-FOS binding sites are enriched inside origins. This graph shows the percentage of replication origin segments (blue) and bootstrap segments (red) overlapping with c-JUN and c-FOS segments. (E) CpG islands (CGI) bound by c-JUN are good substrates for replication initiation. The Venn diagram shows the overlap between replication origin segments, CGI, and c-JUN regions. The number of segments found in each group is shown between brackets, and the number of overlapping elements is given in the intersected regions. (F) c-JUN origins are associated with open and closed chromatin. The 55 c-JUN origins were intersected with CpG islands (CGI), HS sites, and H3K4me3 regions. The histogram shows the number of c-JUN origins in each of the 8 possible classes ranged from the largest to the smallest.
Next, we tested whether the observed correlation between open chromatin structure and origin positioning was a true association or the consequence of the strong association between origins and CpG islands. For this study, we concentrated on H3K4me3, a histone modification characteristic of nucleosomes located near the sites of initiation of many transcribed genes (27). ENCODE regions contain 506 CpG islands, and only 99 of them (19.6%) overlap with replication origins (Fig. 2B). We found that 54% of the CpG islands containing origins also coincided with H3K4me3 sites. Surprisingly, 46% of the CpG islands lacking origins also overlap with H3K4me3 sites. Thus, we did not detect any significant enrichment of H3K4me3 sites in CpG islands linked to replication origins, relative to all CpG islands. This finding suggests that CpG islands associated with highly transcribed genes are not a better substrate for replication initiation than others. We also tested whether origins outside CpG islands are biased toward an association with H3K4me3 sites. Thirty-two of 184 non-CpG island origins coincided with H3K4me3 sites (17%). Again, we did not detect any H3K4me3 enrichment in this group of origins, relative to the bootstrap sample (14%, P = 0.025). From this analysis, we conclude that the link between open chromatin structure and replication initiation sites was only a consequence of the strong association between CpG islands and replication origins. This evidence indicates that although sites of replication initiation are strongly associated with transcription regulatory elements, their use is not favored or impeded by active transcription.
Replication Origins Are Associated Mostly with Regulatory Elements.
HS sites identify regions of open chromatin, which encompass all different types of regulatory elements, including promoters, enhancers, silencers, insulators, and locus control regions. DNase-chip analysis on the same ENCODE regions has been performed using 7 human cell lines including HeLa cells (26). We tested whether most of the origins found in HeLa cells mapped within HS sites identified in at least 1 of the cell lines explored. Twenty percent of origins overlap with HS sites found in both HeLa and at least 1 other cell line, whereas only 2% co-localize with HeLa-specific HS sites (Fig. 2C). As additional cell lines are included, the total percentage of origins covered by HS sites increases gradually, reaching 69% at the seventh cell line. We did not detect any significant leveling off after the addition of the seventh cell type. A similar gradual increase in the total percentage of base pairs of the ENCODE regions covered by HS sites has been described previously (26). These findings suggest that HS sites found in additional cell lines may cover almost all the origins identified in our study. The number of HS sites found in the various cell lines that intersect with bootstrap segments increased much more slowly, indicating the significance of this link (red line in Fig. 2C). This analysis provides further evidence for a connection between transcriptional regulatory elements and replication origins.
Replication Origins Are Enriched in Binding Sites of the Transcription Factors c-JUN and c-FOS.
The binding of several transcription factors was studied along ENCODE regions in HeLa cells by chromatin immunoprecipitation on chip (ChIP-on-chip), allowing us to test for their role in origin selection. We focused our attention on the AP-1 complex, a dimeric transcription factor comprising c-JUN and c-FOS and known to be a strong driver of the cell cycle from G1 to S phase. We observed a highly significant correlation between origins and c-JUN and c-FOS binding sites (P < 2.2 × 10−16) (Fig. 2D). Fifty-five origins are associated with c-JUN, 53 with c-FOS, and 39 with both. Forty c-JUN origins overlap a CpG island (Fig. 2E). We therefore tested whether the observed link between origins and AP-1 binding sites was the result only of the strong association of origins with CpG islands, as previously shown for H3K4me3. ENCODE regions contain 200 c-JUN binding sites, and 55 of them (27.5%) overlap with replication origins (Fig. 2E). We found that 40% of the CpG islands containing origins also coincided with c-JUN binding sites. However, only 13% of the CpG islands lacking origins also overlap with c-JUN binding sites. Thus, in contrast with H3K4me3, we detected a significant enrichment of c-JUN binding sites in CpG islands linked to replication origins, relative to the rest of CpG islands (P = 2.2 × 10−16). This finding suggests that CpG islands associated with the AP-1 complex are a better substrate for replication initiation than others. We also tested whether origins outside CpG islands are biased toward an association with c-JUN binding sites. Fifteen of 184 non-CpG island origins coincided with c-JUN binding sites (8.1%). Again, we detected c-JUN binding site enrichment in this group of origins relative to the bootstrap sample (3.4%, P = 3.5 × 10−4). We analyzed the intersection of the 55 c-JUN origins with HS and H3K4me3 sites. We again found that a large fraction (40%) had neither an HS site nor H3K4me3 modification, as observed previously in the total group of replication origins (Fig. 2F). From this analysis, we conclude that the link between c-JUN and replication initiation sites is highly significant and is independent of the strong association between replication origins and CpG islands. This result indicates that the AP-1 complex might be an important regulator of replication initiation along the human genome.
A Precise Timing Program Regulates the Timing of Origin Firing Independently of Origin Density.
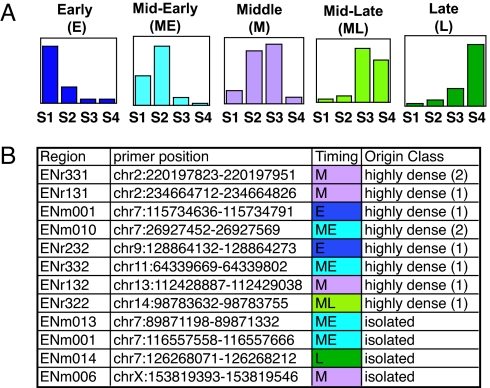
To test whether origin density is correlated with replication timing, we defined 2 classes of origins based on origin density. Isolated origins were defined stringently as those at the center of a 200-kb window, totally included inside ENCODE regions, lacking any other origin (21 origins). Highly dense origins were defined as those having at least 2 origins within 10 kb upstream or downstream from the origin segment's edges (i.e., 3 origins in a 20-kb window). There were 10 such regions. We analyzed replication timing by cell sorting into 4 S phase fractions from early to late (S1-S4) of BrdU pulse-labeled cells. Based on the relative enrichment of nascent DNA in each of the S1-S4 fractions, we defined 5 groups of replication profiles (Fig. 3A).
Fig. 3.
Isolated origins and clusters of closely spaced origins are activated at all stages in S phase. (A) Replication timing profiles obtained after BrdU pulse labeling, cell sorting, and quantitative PCR were classified into 5 groups according to the enrichment found in fractions S1 to S4. Typical early (E), mid-early (ME), middle (M), mid-late (ML), and late (L) profiles are shown. (B) Timing analysis of highly dense and isolated origins. ENCODE regions, primer pair coordinates used for qPCR, replication timing, and origin class are indicated
In this way, we analyzed the timing of the 10 highly dense origins and found 2 early, 3 mid-early, 4 middle, and 1 mid-late replicating clusters (Fig. 3B). Thus, dense clusters of origins do not necessarily fire at the beginning of S phase and can be replicated at any moment during S phase except in late S phase. We then tested replication timing for 4 of the 21 isolated origins: 2 were mid-early, 1 was middle, and 1 was late replicated. Finally, we explored the replication timing of the ENCODE regions lacking origins (six 500-kb regions) to test whether replication forks initiated in surrounding regions passively replicated these regions. If so, we should observe a late, mid-late, or possibly middle replicated pattern in the center of these regions because of the long stretches that must be replicated before these central zones are reached. In agreement with our origin-mapping data, regions ENr113, ENr114, ENr211, and ENm009 showed a late pattern of replication suggesting that they indeed are replicated passively. The centers of ENm003 and ENr122 were replicated in middle S phase. As described for the Ig locus (28), our observations suggest that the human genome contains large chromosomal regions without strong sites of initiation that are passively replicated by forks initiated in flanking regions.
Discussion
So far fewer than 30 origins have been mapped in human cells. Because only regions containing transcribed genes were analyzed, a genome-wide view of replication initiation was still lacking. The aim of the ENCODE project was to identify every sequence with functional properties in the human genome. For this purpose, regions representing a large range of genomic features characteristic of both gene-dense regions and gene deserts were chosen, providing a comprehensive picture of the whole genome. Using stringent purification of SNS by exonuclease digestion, we have developed a genome-wide mapping of replication origins based on hybridization of SNS to DNA microarrays, covering 30 Mb of the human genome. A previous study MacAlpine, et al. (29) explored replication inside the Drosophila genome on custom-designed microarrays covering a 23-Mb region. Because this mapping was based on the identification of the sequences that replicate at the earliest S phase combined with the mapping of sites of ORC binding by chromatin immunoprecipitation, it did not allow the confident identification of late origins. This study showed that ORC is localized to specific sites that frequently overlap with RNA Pol II-associated sequences, suggesting that origin selection is influenced by transcription. Our results support the notion that there also is a connection between transcription-regulatory elements and replication in human cells. At the same time, we demonstrate that neither CpG island origins nor the other origins are preferentially associated with H3K4me3, a histone modification known to be linked to promoters of expressed genes. This result confirms on a large scale 2 previous studies made at specific loci and shows that there is not a simple direct link between transcription and origin selection (30, 31). Because not all active promoters are efficient sites of replication initiation, active promoters containing an origin must have distinguishing information. The fact that _cis_-elements containing known origins remain strong sites of replication initiation even when inserted ectopically also suggests that specific replication start points are recognized by the replication machinery (32, 33). We speculate that the replication machinery efficiently recognizes specific combinations of transcription factors. Replication origins found in unopened regions also might be bound by transcription factors that do not disturb the chromatin structure. As with transcription, many different combinations of DNA binding factors might regulate replication initiation sites, explaining the lack of consensus sequence for origin selection (12). Our study allowed us to identify c-JUN as a potential regulator of origin selection along the human genome. c-JUN is known to be a positive regulator of cell proliferation, because c-JUN-deficient fibroblasts have a marked proliferation defect in vitro (34). Moreover, c-JUN was shown to activate polyoma virus DNA replication by stimulating the binding of the virus-encoded initiator, large T antigen, to origins through direct protein-protein interaction (35). These data support the hypothesis that c-JUN has a positive role in the regulation of human replication origins. An alternative hypothesis concerning the link between transcriptional regulation and origin positioning is that both molecular mechanisms recognize identical features. DNA topology may be an important signal, because Drosophila ORC displays a high affinity for negatively supercoiled DNA in vitro (36).
The temporal regulation of origin firing is a second way by which chromosome duplication can be regulated precisely. Metazoan genomes replicate with a defined timing program, suggesting that a precise spatiotemporal program controls their duplication. Several genome-wide studies showed that GC-rich regions (on the Mb scale) tend to replicate earlier than GC-poor regions (37). In our study, we found a very strong correlation between regional GC content and the density in origins (Fig. 1B), suggesting that the correlation between isochore organization and replication timing is a consequence of the distribution of origins: regions lacking origins (typically GC-poor regions) are replicated passively, and hence relatively late compared with origin-rich (GC-rich) regions. Note, however, that the density in origin is not sufficient to determine replication timing. Indeed, we found that half of the origin-dense regions are not early replicated (Fig. 3B). This observation is not consistent with the stochastic model of origin firing recently proposed by Rhind (38). This model is based on varying origin efficiency. In this model, individual origins would fire stochastically, but regions that have many efficient origins would almost always replicate early because a random subset of those origins would fire early in each S phase. Our study shows that origin density is not predictive of replication timing. These data are in favor of a model in which strong sites of replication initiation are controlled by a strict origin-timing program rather than by stochastic firing. According to such a model, timing of replication of a chromosomal region would not depend on the density of efficient origins but rather on chromosomal environment.
Methods
Cell Culture.
HeLa S3 cells were cultured with the recommended ATCC complete growth medium.
Theoretical Estimation of Amounts of SNS Extracted from 108 cells.
The estimation of the total SNS between 1.5 and 2 kb in 108 cells was calculated using the following equation:
where Y is the quantity in grams of estimated SNS. Tns is the lifespan of 1.5–2 kb nascent strands (2 minutes). Tcc is the length of the HeLa cell cycle (24 h or 1440 min). N is the number of cells used for SNS extraction (108). Nori is the estimated number of replication origins needed to replicate the whole human genome during S phase (30,000000). L is the average length of isolated SNS (1750 nucleotides). ε is the weight of 1 nucleotide (54.81*10−23g). With this formula, we estimated that around 10 ng of SNS can be isolated from 108 cells.
Isolation of SNS and Short ssDNA.
Nascent strands were prepared as described previously (31). The quality of each preparation was tested by real-time qPCR with primer pairs located in and around the c-myc origin (32) shown in Table S4. For short ssDNA, fragments of 300-1000 bp of single-stranded DNA were selected after sucrose gradient separation.
Amplification of SNS.
The approach used for amplification of SNS is an adaptation of the TLAD amplification described by Liu, et al. (15). Details are described in Supporting Information.
cRNA Labeling and Fragmentation.
The aminoallyl-modified UTPs incorporated into the RNA were coupled with mono-reactive Cy3 or Cy5 dyes (Amersham Biosciences) as follows. One vial of mono-reactive dye (40,000 pmol) was dissolved in 12 μl of DMSO (Sigma) and divided into 4 μl aliquots. Purified RNAs were concentrated into a 5-μl volume, and 11 μl of 0.1 M sodium bicarbonate pH = 8.7 were added. Then, an aliquot of mono-reactive dye was added to the sample, and the coupling reaction allowed to proceed for 90 min at room temperature in the dark. The coupled reaction mix was purified with 5 Microcon YM30 columns washing steps (Milipore). Then, labeled RNA was concentrated to 9 μl and fragmented according to the manufacturer's instructions (Ambion Fragmentation Kit #8740).
Hybridization with DNA Microarrays.
Hybridization of the Human ENCODE ChIP-on-chip microarrays (build hg17) (Agilent Technologies) was performed according to the manufacturer's instructions.
Scanning, Feature Extraction, and Analysis.
Microarrays were scanned with a GenePix 4000B scanner (Axon Instruments) under the control of GenePix Pro 4.1 software (Axon Instruments). Feature Extraction 9.1 software (Agilent Technologies) was used for feature extraction. Analysis was performed with Agilent Chip Analytics 1.3 software without normalization, and peaks were detected by application of the following criteria: (i) maximum distance (in bp) for 2 probes to be considered as neighbors = 500; 2) a probe is considered bound if P(X bar) <10−5 and either central probe of the peak has P(X) < 10−5 and at last 1 neighboring probe has P(X) <10−4 or at least 2 of the neighbors have P(X) < 10−4. For short ssDNA microarray, the less stringent criteria were (i) maximum distance (in bp) for 2 probes to be considered as neighbors = 500; (ii) a probe is considered bound if P(X bar) <10−3 and either central probe of the peak has P(X) < 10−3 and at least 1 neighboring probe has P(X) <10−1 or at least 1 of the neighbors have P(X) < 5.10−3.
Bioinformatics Analysis.
Origin location data were loaded into a MySQL database with the data found and downloaded from the University of California at Santa Cruz ENCODE Website (http://genome.ucsc.edu/ENCODE/encode.hg17.html). We generated a bootstrap sample to calculate statistical significance for contingencies results between origin positions and ENCODE features. For each of the 283 origin segments, we randomly drew a segment of identical size from the same ENCODE region. We repeated this procedure 100 times to produce a set of 28,300 random segments. Then we performed a goodness of fit χ2 test with theoretical expectation under a null hypothesis (i.e., the random position of origin) calculated from the bootstrap sample. All these χ2 tests have 1 degree of freedom. The Galaxy2ENCODE program (http://main.g2.bx.psu.edu/) was used for other simple manipulations such as intersection, subtraction, concatenation, and merging (39).
Replication Timing Analysis.
Timing analyses were made as previously described (31) excepted that S phase was divided into 4 fractions ranging from early to late S phase and designated S1 to S4.
Supplementary Material
Supporting Information
Acknowledgments.
We thank T. Strick and A. West for critical reading of the manuscript and David Gilbert for helpful discussions. We thank D. Goidin from Agilent Technologies and M-C. Gendron from the Jacques-Monod Institute flow-cytometry platform for technical help. We thank A. Necsulea for help in studying correlations between computationally predicted origins and our origin segments. We thank M. Gerstein for allowing us to use ENCODE data on c-JUN and c-FOS binding sites (http://tiling.gersteinlab.org/home/datasets.html) and (http://genome.ucsc.edu/ENCODE/encode.hg17.html). Research in the laboratory of M-N.P. is supported by the Agence Nationale pour le Recherche Grant ANR 05-JCJC-0110, the Association pour la Recherche sur le Cancer, and the Ligue contre le Cancer.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data Deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO), www.ncbi.nlm.nih.gov/geo (accession no. GSE10217).
References
- 1.Dominguez-Sola D, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448(7152):445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 2.Lengronne A, Schwob E. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1) Mol Cell. 2002;9(5):1067–1078. doi: 10.1016/s1097-2765(02)00513-0. [DOI] [PubMed] [Google Scholar]
- 3.Aladjem MI. Replication in context: Dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet. 2007;8(8):588–600. doi: 10.1038/nrg2143. [DOI] [PubMed] [Google Scholar]
- 4.ENCODE pc. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306(5696):636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 5.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357(6374):128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 7.Vashee S, et al. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17(15):1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danis E, et al. Specification of a DNA replication origin by a transcription complex. Nat Cell Biol. 2004;6(8):721–730. doi: 10.1038/ncb1149. [DOI] [PubMed] [Google Scholar]
- 9.Kohzaki H, Murakami Y. Transcription factors and DNA replication origin selection. Bioessays. 2005;27(11):1107–1116. doi: 10.1002/bies.20316. [DOI] [PubMed] [Google Scholar]
- 10.Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430(6997):372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- 11.Calvi BR, Byrnes BA, Kolpakas AJ. Conservation of epigenetic regulation, ORC binding and developmental timing of DNA replication origins in the genus Drosophila. Genetics. 2007;177(3):1291–1301. doi: 10.1534/genetics.107.070862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert DM. In search of the holy replicator. Nat Rev Mol Cell Biol. 2004;5(10):848–855. doi: 10.1038/nrm1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerbi SA, Bielinsky AK. Replication initiation point mapping. Methods. 1997;13(3):271–280. doi: 10.1006/meth.1997.0526. [DOI] [PubMed] [Google Scholar]
- 14.Mesner LD, Crawford EL, Hamlin JL. Isolating apparently pure libraries of replication origins from complex genomes. Mol Cell. 2006;21(5):719–726. doi: 10.1016/j.molcel.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Liu CL, Schreiber SL, Bernstein BE. Development and validation of a T7 based linear amplification for genomic DNA. BMC Genomics. 2003;4(1):19–30. doi: 10.1186/1471-2164-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen SM, Brylawski BP, Cordeiro-Stone M, Kaufman DG. Same origins of DNA replication function on the active and inactive human X chromosomes. J Cell Biochem. 2003;88(5):923–931. doi: 10.1002/jcb.10429. [DOI] [PubMed] [Google Scholar]
- 17.Touchon M, et al. Replication-associated strand asymmetries in mammalian genomes: Toward detection of replication origins. Proc Natl Acad Sci USA. 2005;102(28):9836–9841. doi: 10.1073/pnas.0500577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucas I, et al. High-throughput mapping of origins of replication in human cells. EMBO Rep. 2007;8(8):770–777. doi: 10.1038/sj.embor.7401026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hand R. Eucaryotic DNA: Organization of the genome for replication. Cell. 1978;15(2):317–325. doi: 10.1016/0092-8674(78)90001-6. [DOI] [PubMed] [Google Scholar]
- 20.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 21.Mouchiroud D, et al. The distribution of genes in the human genome. Gene. 1991;100:181–187. doi: 10.1016/0378-1119(91)90364-h. [DOI] [PubMed] [Google Scholar]
- 22.Delgado S, Gomez M, Bird A, Antequera F. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 1998;17(8):2426–2435. doi: 10.1093/emboj/17.8.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ladenburger EM, Keller C, Knippers R. Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol Cell Biol. 2002;22(4):1036–1048. doi: 10.1128/MCB.22.4.1036-1048.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margulies EH, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17(6):760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koch CM, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17(6):691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xi H, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3(8):1377–1388. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Norio P, et al. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell. 2005;20(4):575–587. doi: 10.1016/j.molcel.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 29.MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18(24):3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez M, Brockdorff N. Heterochromatin on the inactive X chromosome delays replication timing without affecting origin usage. Proc Natl Acad Sci USA. 2004;101(18):6923–6928. doi: 10.1073/pnas.0401854101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prioleau MN, Gendron MC, Hyrien O. Replication of the chicken beta-globin locus: Early-firing origins at the 5′ HS4 insulator and the rho- and betaA-globin genes show opposite epigenetic modifications. Mol Cell Biol. 2003;23(10):3536–3549. doi: 10.1128/MCB.23.10.3536-3549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malott M, Leffak M. Activity of the c-myc replicator at an ectopic chromosomal location. Mol Cell Biol. 1999;19(8):5685–5695. doi: 10.1128/mcb.19.8.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paixao S, et al. Modular structure of the human lamin B2 replicator. Mol Cell Biol. 2004;24(7):2958–2967. doi: 10.1128/MCB.24.7.2958-2967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schreiber M, et al. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13(5):607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito K, et al. c-Jun stimulates origin-dependent DNA unwinding by polyomavirus large T antigen. EMBO J. 1996;15(20):5636–5646. [PMC free article] [PubMed] [Google Scholar]
- 36.Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 2004;23(4):897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe Y, et al. Chromosome-wide assessment of replication timing for human chromosomes 11q and 21q: Disease-related genes in timing-switch regions. Hum Mol Genet. 2002;11(1):13–21. doi: 10.1093/hmg/11.1.13. [DOI] [PubMed] [Google Scholar]
- 38.Rhind N. DNA replication timing: Random thoughts about origin firing. Nat Cell Biol. 2006;8(12):1313–1316. doi: 10.1038/ncb1206-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blankenberg D, et al. A framework for collaborative analysis of ENCODE data: Making large-scale analyses biologist-friendly. Genome Res. 2007;17(6):960–964. doi: 10.1101/gr.5578007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information