Darwinian evolution in the light of genomics (original) (raw)
Abstract
Comparative genomics and systems biology offer unprecedented opportunities for testing central tenets of evolutionary biology formulated by Darwin in the Origin of Species in 1859 and expanded in the Modern Synthesis 100 years later. Evolutionary-genomic studies show that natural selection is only one of the forces that shape genome evolution and is not quantitatively dominant, whereas non-adaptive processes are much more prominent than previously suspected. Major contributions of horizontal gene transfer and diverse selfish genetic elements to genome evolution undermine the Tree of Life concept. An adequate depiction of evolution requires the more complex concept of a network or ‘forest’ of life. There is no consistent tendency of evolution towards increased genomic complexity, and when complexity increases, this appears to be a non-adaptive consequence of evolution under weak purifying selection rather than an adaptation. Several universals of genome evolution were discovered including the invariant distributions of evolutionary rates among orthologous genes from diverse genomes and of paralogous gene family sizes, and the negative correlation between gene expression level and sequence evolution rate. Simple, non-adaptive models of evolution explain some of these universals, suggesting that a new synthesis of evolutionary biology might become feasible in a not so remote future.
INTRODUCTION
Charles Darwin's book On the Origin of Species that appeared in London in 1859 (1) was the first plausible, detailed account of biological evolution ever published, along with the simultaneous and independent brief outlines by Darwin and Alfred Russell Wallace published the previous year (2–3). Of course, Darwin did not discover evolution and did not even offer the first coherent description of evolution—arguably, that honor belongs to Jean-Baptiste Lamarck whose magnum opus Philosophie Zoologique (4) was, uncannily, published in the year of Darwin's birth. However, Lamarck's picture of evolution was based on an innate drive of evolving organisms toward perfection, an idea that cannot be acceptable to a rationalist mind. Besides, Lamarck did not proclaim the universal character of evolution: he postulated multiple acts of creation, apparently, one for each species. Darwin was the first to present a rational, mechanistic, and arguably, magnificent picture of the origin of the entire diversity of life forms ‘from so simple a beginning’, probably, from a single common ancestor (1). Darwin's vision of the evolution of life was sufficiently complete and powerful to win over or, at least, deeply affect the minds of most biologists (and scientists in general, and the educated public at large), so that all research in biology during the last 150 years developed within the framework set by the Origin (even when in opposition to Darwin's ideas).
Darwin's vision lacked the essential foundation in genetics because mechanisms of heredity were unknown in his day (Mendel's work went unnoticed, whereas Darwin's own ideas in this area were less than productive). The genetic basis of evolution was established after the rediscovery of Mendel's laws, with the development of population genetics in the first third of the 20th century, primarily, through the pioneering work of Fisher, Wright and Haldane (5–7). The new, advanced understanding of evolution, informed by theoretical and experimental work in genetics, was consolidated in the Modern Synthesis of evolutionary biology, usually, associated with the names of Dobzhansky, Julius Huxley, Mayr and Simpson (8–11). Apparently, the Modern Synthesis (neo-Darwinism) adopted its mature form during the 1959 centennial celebration for the Origin in Chicago (12–14).
Now, 50 years after the consolidation of the Modern Synthesis, evolutionary biology undoubtedly faces a new major challenge and, at the same time, the prospect of a new conceptual breakthrough (15). If the Modern Synthesis can be succinctly described as Darwinism in the Light of Genetics (often referred to as neo-Darwinism), then, the new stage is Evolutionary Biology in the Light of Genomics. In this article, I attempt to outline the changes to the basic tenets of evolutionary biology brought about by comparative and functional genomics and argue that, in many respects, the genomic stage could be a more radical departure from the neo-Darwinism than the latter was from classic Darwinism. Of course, to do so, it is necessary first to recapitulate the principal concepts of evolution proposed by Darwin and amended by the architects of the Modern Synthesis. In the rest of the article, I return to each of these points.
- Undirected, random variation is the main process that provides the material for evolution. Darwin was the first to allow chance as a major factor into the history of life, and arguably, that was one of his greatest insights.
- Evolution proceeds by fixation of the rare beneficial variations and elimination of deleterious variations: this is the process of natural selection that, along with random variation, is the principal driving force of evolution according to Darwin and the Modern Synthesis. Natural selection which is, obviously, akin to and inspired by the ‘invisible hand’ (of the market) that ruled economy according to Adam Smith, was the first mechanism of evolution ever proposed that was simple, plausible, and did not require any mysterious innate trends. As such, this was Darwin's second key insight. The founders of population genetics, in particular, Sewall Wright, emphasized that chance could play a substantial role in the fixation of changes during evolution not only in their emergence, via the phenomenon of genetic drift that entails random fixation of neutral or even deleterious changes. Population-genetic theory indicates that drift is particularly important in small populations that go through bottlenecks (6,16). However, the Modern Synthesis, in its ‘hardened’ form (13), effectively, rejected drift as an important evolutionary force, and adhered to a purely adaptationist model of evolution (17). This model inevitably leads to the concept of ‘progress’, gradual improvement of ‘organs’ during evolution, an idea that Darwin endorsed as a general trend, despite his clear understanding that organisms are less than perfectly adapted, as strikingly exemplified by rudimentary organs, and despite his abhorrence of any semblance of an innate strive for perfection of the Lamarckian ilk.
- The beneficial changes that are fixed by natural selection are ‘infinitesimally’ small, so that evolution proceeds via the gradual accumulation of these tiny modifications. Darwin insisted on strict gradualism as an essential staple of his theory: ‘Natural selection can act only by the preservation and accumulation of infinitesimally small inherited modifications, each profitable to the preserved being … If it could be demonstrated that any complex organ existed, which could not possibly have been formed by numerous, successive, slight modifications, my theory would absolutely break down.’ [(1), chapter 6]. Even some contemporaries of Darwin believed that was an unnecessary stricture on the theory. In particular, the early objections of Thomas Huxley are well known: even before the publication of the Origin Huxley wrote to Darwin ‘‘You have loaded yourself with an unnecessary difficulty in adopting Natura non facit saltum so unreservedly’ (18).
- An aspect of the classic evolutionary biology that is related but not identical to the principled gradualism is uniformitarianism (absorbed by Darwin from Lyell's geology), that is, the belief that the evolutionary processes remained, essentially, the same throughout the history of life.
- Evolution of life can be presented as a ‘great tree’, as epitomized by the single, famous illustration of the Origin [(1), Chapter 4].
- A corollary of the single tree of life (TOL) concept that, however, deserves the status of a separate principle: all extant diversity of life forms evolved from a single common ancestor [or very few ancestral forms, under Darwin's cautious formula (1), chapter 14] that much later was dubbed the Last Universal Common (Cellular) Ancestor (LUCA) (19).
BETWEEN THE MODERN SYNTHESIS AND EVOLUTIONARY GENOMICS
Obviously, evolutionary biologists did not stay idle during the 40-year span that separated the consolidation of the Modern Synthesis and the coming of age of evolutionary genomics; below I briefly summarize what appear to be the key advances (undoubtedly, this brief account is incomplete and might be considered somewhat subjective).
Molecular evolution and phylogeny
The traditional phylogeny that fleshed out Darwin's concept of the TOL was based on comparisons of diagnostic features of organisms’ morphology, such as, for instance, skeleton structure in animals and flower architecture in plants (20). The idea that the actual molecular substrate of evolution that undergoes the changes acted upon by natural selection (the genes, simply put) could be compared for the purpose of phylogeny reconstructions did not enter the minds of evolutionary biologists for the obvious reason that (next to) nothing was known on the chemical nature of that substrate and the way it encoded the phenotype of an organism. Moreover, the adaptationist paradigm of evolutionary biology seemed to imply that genes, whatever their molecular nature, would not be well conserved between distant organisms, given the major phenotypic differences between them, as emphasized in particular by Mayr, one of the chief architects of the Modern Synthesis (21).
The idea that DNA base sequence could be employed for evolutionary reconstruction seems to have been first expressed in print by Crick, appropriately, in the same seminal article where he formulated the adaptor hypothesis (22). The actual principles and the first implementation of molecular evolutionary analysis were given a few years later by Zuckerkandl and Pauling who directly falsified Mayr's conjecture by showing that the amino-acid sequences of several proteins available at the time, such as cytochrome c and globins, were highly conserved even between distantly related animals (23,24). Zuckerkandl and Pauling also proposed the concept of molecular clock, a relatively constant rate of evolution of the sequence that they predicted to be characteristic of each protein in the absence of functional change. In the next few years, primarily, through the efforts of Dayhoff and coworkers, it has been demonstrated that protein sequence conservation extended to the most diverse life forms, from bacteria to mammals (25–27).
The early phase of molecular evolution research culminated in the work of Woese and coworkers who revealed the conservation of the sequences of certain molecules, above all, ribosomal RNA in all cellular life forms, and their suitability for phylogenetic analysis (28). The crowning achievement in this line of study was the entirely unexpected discovery of the third domain of life—archaea—that includes organisms previously lumped with bacteria but shown to be highly distinct by the phylogenetic analysis of rRNA (29,30). As a result of these studies, a growing tendency developed to equate the phylogenetic tree of rRNA, with its three-domain structure (31), with the ‘TOL’ envisaged by Darwin and first explicated by Haeckel (28,32,33). However, even in the pre-genomic era, it became clear that not all trees of protein-coding genes have the same topology as the rRNA tree; the causes of the discrepancies remained murky but there thought to involve horizontal gene transfer (HGT) (34).
The neutral theory and purifying selection
Arguably, the most important conceptual breakthrough in evolutionary biology after the Modern Synthesis was the neutral theory of molecular evolution that is usually associated with the name of Kimura (35,36) although a similar theory was simultaneously and independently developed by Jukes and King (37). Originally, the neutral theory was derived as a development of Wright's population-genetic ideas on the importance of genetic drift in evolution. According to the neutral theory, a substantial majority of the mutations that are fixed in the course of evolution are selectively neutral so that fixation occurs via random drift. A corollary of this theory is that gene sequences evolve in an approximately clock-like manner (in support of the original molecular clock hypothesis of Zuckerkandl and Pauling) whereas episodic beneficial mutations subject to natural selection are sufficiently rare to be safely disregarded for a quantitative description of the evolutionary process. Of course, the neutral theory should not be taken to mean that selection is unimportant for evolution. What the theory actually maintains is that the dominant mode of selection is not the Darwinian positive selection of adaptive mutations, but stabilizing, or purifying selection that eliminates deleterious mutations while allowing fixation of neutral mutations by drift (17).
Subsequent studies refined the theory and made it more realistic in that, to be fixed, a mutation needs not to be literally neutral but only needs to exert a deleterious effect that is small enough to escape efficient elimination by purifying selection—the modern ‘nearly neutral’ theory (38). Which mutations are ‘seen’ by purifying selection as deleterious critically depends on the effective populations’ size: in small populations, drift can fix even mutations with a significant deleterious effect (16). The main empirical test of the (nearly) neutral theory comes from measurements of the constancy of the evolutionary rates in gene families. Although it was repeatedly observed that molecular clock is significantly over-dispersed (39,40), such tests strongly suggest that the fraction of neutral mutations among the fixed ones is, indeed, substantial (36). The (nearly) neutral theory is a major departure from the Modern Synthesis selectionist paradigm as it explicitly posits that the majority of mutations fixed during evolution are not affected by Darwinian (positive) selection (Darwin seems to have presaged the neutralist paradigm by remarking that selectively neutral characters would serve best for classification purposes (1); however, he did not elaborate on this idea, and it has not become part of the Modern Synthesis).
Importantly, in the later elaborations of the neutral theory, Kimura and others realized that mutations that were (nearly) neutral at the time of fixation were not indifferent to evolution. On the contrary, such mutations comprised the pool of variation that can be tapped into by natural selection under changed conditions, a phenomenon that could be potentially important for macroevolution (17,41).
Selfish genes, junk DNA and mobile elements
Although this was rarely stated explicitly, classic genetics certainly implies that (nearly) all parts of the genome (all nucleotides in more modern, molecular terms) have a specific function. However, this implicit understanding came into doubt in the 1960–70s owing to accumulating data on the lack of a direct correspondence between genomic and phenotypic complexity of organisms. It was shown that organisms of about the same phenotypic complexity often had genomes that differed in size and complexity by orders of magnitude (the so-called _c_-value paradox) (42,43). This paradox was conceptually resolved by two related, fundamental ideas, those of selfish genes and junk DNA. The selfish gene concept was first developed by Dawkins in his eponymous classic book (44). Dawkins realized, in a striking departure from the organism-centric paradigm of the Modern Synthesis, that natural selection could act not only at the level of the organism as a whole but also at the level of an individual gene. Under a somewhat provocative formulation of this view, the genome and the organism are, simply, vehicles for the propagation of genes. This concept was further advanced by Doolittle and Sapienza (45), and by Orgel and Crick (46), who proposed that much if not the most of the genomic DNA (at least, in complex organisms) consisted of various classes of repeats that originate from the replication of selfish elements (ultimate parasites, according to Orgel and Crick). In other words, from the organism's standpoint, much of its genomic DNA should be considered junk. This view of the genome dramatically differs from the picture implied by the selectionist paradigm under which most if not all nucleotides in the genome would be affected by (purifying or positive) selection acting at the level of the organism.
A conceptually related major development was the discovery, first in plants by McClintock in the 1940s (47), and subsequently, in animals (48), of ‘jumping genes’, later known as mobile elements, that is, genetic elements that were prone to frequently changing their position in the genome. The demonstration of the ubiquity of mobile elements suggested the picture of highly dynamic genomes, ever changing genomes even before the advent of modern genomics (49,50).
Evolution by gene and genome duplication
The central tenet of Darwin, the gradualist insistence on infinitesimal changes as the only material of evolution, was challenged by the concept of evolution by gene duplication that was developed by Ohno in his classic 1970 book (51). The idea that duplication of parts of chromosomes might contribute to evolution goes back to some of the founders of modern genetics, in particular, Fisher (52), but Ohno was the first to propose that gene duplication was central to the evolution of genomes and organisms, and to support this proposition by a qualitative theory. Starting from the evidence of a whole-genome duplication early in the evolution of chordates, Ohno hypothesized that gene duplication could be an important, if not the principal, path to the evolution of new biological functions, because after a duplication, one of the gene copies would be free of constraints imposed by purifying selection, and would have the potential to evolve a new function (a phenomenon later named neofunctionalization). Clearly, the emergence of a new gene as a result of a duplication, let alone duplication of a genomic region including multiple genes or whole genome duplication, are far from being ‘infinitesimal’ changes, and if such larger events are indeed important for evolution, the gradualist paradigm comes into jeopardy.
Spandrels, exaptation, tinkering and the deficiency of the Panglossian paradigm of evolution
A spirited, sweeping critique of the adaptationist program of evolutionary biology was mounted by Gould and Lewontin in the famous ‘Spandrels of San Marco’ paper (53). Gould and Lewontin sarcastically described the adaptationist worldview as the Panglossian paradigm, after the notorious character in Voltaire's Candide who insisted that ‘everything was to the better in this best of all worlds’ (even major disasters). Gould and Lewontin emphasized that, rather than hastily concoct ‘just so stories’ of plausible adaptations, evolutionary biologists should seek explanations of the observed features of biological organization under a pluralist approach that takes into account not only selection but also intrinsic constraints, random drift and other factors. The spandrel metaphor holds that many functionally important elements of biological organization did not evolve as specific devices to perform their current functions but rather are products of non-adaptive architectural constraints—much like spandrels that inevitably appear at arches of cathedrals and other buildings, and can be employed for various functions such as housing key elements of the imagery adorning the cathedral. The process of utilization of spandrels for biological functions was given the special name exaptation and was propounded by Gould as an important route of evolution (54).
In an even earlier, conceptually related development, Jacob promoted the metaphor of evolution as tinkering (55). Jacob's argument, based, primarily, of the results of comparative analysis of developmental mechanisms, that evolution did not act as an engineer or designer but rather as a tinkerer that is heavily dependent on previous contingencies for solving outstanding problems and whose actions, therefore, are unpredictable and unexplainable without detailed knowledge of preceding evolution.
Evolution in the world of microbes and viruses
Perhaps, the development in biology that had the most profound effect on the changes in our understanding of evolution was the extension of evolutionary research into the realm of bacteria (and archaea) and viruses. Darwin's account of evolution and all the developments in evolutionary biology in the subsequent few decades dealt exclusively with animals and plants, with unicellular eukaryotes (Protista) and bacteria (Monera) nominally placed near the root of the TOL by Haeckel and his successors (56). Although by 1950s, genetic analysis of bacteriophages and bacteria was well advanced, making it obvious that these life forms had evolving genomes (57), the Modern Synthesis made no notice of these developments. That bacteria (let alone viruses) would evolve under the same principles and by the same mechanisms as animals and plants, is by no means obvious given all their striking biological differences from multicellular organisms, and specifically, because they lack regular sexual reproduction and reproductive isolation that is crucial for speciation in animals and plants.
Effectively, prokaryotes became ‘visible’ to evolutionary biologists in 1977, with the groundbreaking work of Woese and colleagues on rRNA phylogeny that led to the identification of archaea and major groups of bacteria (28,29,58). Shortly afterward, the field of comparative and evolutionary genomics was born as multiple, complete genome sequences of diverse small viruses became available. Despite the fast sequence evolution that is characteristic of viruses, this early comparative-genomic research was successful in the delineation of sets of genes that are conserved in large groups of viruses (59–62). Moreover, a general principle became apparent: whereas some genes were conserved across an astonishing variety of viruses, genome architectures, virion structures, and biological features of viruses showed much greater plasticity, so that gene exchange, even between highly dissimilar viruses, emerged as a major factor of evolution (62).
Endosymbiosis
The hypothesis that certain organelles of eukaryotic cells, in particular, the plant chloroplasts, evolved from bacteria is not that much younger than the Origin: it was proposed by several researchers in the late 19th century on the basis of microscopic study of plant cells that revealed conspicuous structural similarity between chloroplasts and cyanobacteria (then known as blue-green alga) and was presented in a coherent form by Mereschkowsky in the beginning of the 20th century (63). For the first two-thirds of the 20th century, this hypothesis of endosymbiosis remained a fringe speculation. However, this perception changed shortly after the appearance of the seminal 1967 publication of Sagan (Margulis) who summarized the then available data on the similarity between certain organelles and bacteria, in particular, the striking discovery of organellar genomes, and came to the conclusion that not only chloroplasts but also the mitochondria evolved from endosymbiotic bacteria (64). Subsequent work, in particular, phylogenetic analysis of both genes contained in the mitochondrial genome and genes encoding proteins that function in the mitochondria and apparently were transferred form the mitochondrial to the nuclear genome turned the endosymbiosis hypothesis into a well-established fact (65). Moreover, these phylogenetic studies convincingly demonstrated the origin of mitochondria from a particular group of bacteria, the α-proteobacteria (66,67). The major evolutionary role assigned to effectively unique events like endosymbiosis is, of course, incompatible with both gradualism and uniformitarianism.
EVOLUTIONARY BIOLOGY IN THE AGE OF GENOMICS
The treasure trove of genomic, metagenomic and post-genomic data
The fundamental principles of molecular evolution were established, and many specific observations of major importance and impact on the fundamentals of neo-Darwinism were made in the pre-genomic era, the rRNA-based phylogeny being the premier case in point. However, the advent of full-fledged genome sequencing qualitatively changed the entire enterprise of evolutionary biology. The importance of massive amounts of sequences for comparison is obvious because this material allows researchers to investigate mechanisms and specific events of evolution with the necessary statistical rigor and to reveal even subtle evolutionary trends. In addition, it is worth emphasizing that collections of diverse complete genomes are enormously useful beyond the sheer amount of sequence data. Indeed, only by comparing complete genomes, it is possible to clearly disambiguate orthologous (common descent from a single ancestral gene) and paralogous (gene duplication) relationship between genes; to convincingly demonstrate the absence of a particular gene in a genome, and to pinpoint gene loss events; to perform a complete comparison of genome organizations and reconstruct genome rearrangement events (68–71). Furthermore, for the maximum benefit of evolutionary biology, it is crucial to sample the genome space both deeply (that is, obtain genome sequences of multiple, closely related representatives of the same taxon) and broadly (obtain representative sequences for as many diverse taxa as possible). Genomes separated by different evolutionary distances are most suitable for different tasks, e.g., to reveal the range of the conservation of a particular gene or to attempt reconstruction of major evolutionary events, distantly related genomes have to be compared, whereas for the quantitative characterization of the selection process affecting genomes, sets of closely related genomes are indispensable (72–75). The collection of completely sequenced genomes that is available on Darwin's 200th anniversary consists of thousands of viral genomes, close to 1000 genomes of bacteria and archaea, and close to 100 eukaryotic genomes (76,77). Although, certainly, not all major taxa are adequately represented, this rapidly growing collection increasingly satisfies the demands of both microevolutionary and macroevolutionary research.
Complementary to the advances of traditional genomics is the more recent accumulation of extensive metagenomic data. Although metagenomics typically does not yield complete genomes, it provides invaluable information on the diversity of life in various environments (78,79).
Beyond genomics and metagenomics, one of the hallmarks of the first decade of the new millennium is the progress of research in functional genomics and systems biology. These fields now yield high quality, genome-wide data on gene expression, genetic and protein–protein interactions, protein localization within cells, and more, opening new dimensions of evolutionary analysis, what is sometimes called Evolutionary Systems Biology (80–82). This new field of research has the potential to yield insights into the genome-wide connections between sequence evolution and other variables, such as the rate of expression, and to illuminate the selective and neutral components of the evolution of these aspects of genome functioning.
Below I attempt to briefly synthesize the main insights of evolutionary genomics, with an emphasis on the ways in which these new findings affect the central tenets of evolutionary biology, in particular, with regard to the relative contributions of selective and neutral, random processes.
The evolutionary conservation of gene sequences and structures versus the fluidity of gene composition and genome architecture
A fundamental observation supported by the entire body of evidence amassed by evolutionary genomics is that the sequences and structures of genes encoding proteins and structural RNAs are, generally, highly conserved through vast evolutionary spans. With the present collection of sequenced genomes, orthologs in distant taxa are found for the substantial majority of proteins encoded in each genome (83). For instance, recent genome sequencing of primitive animals, sea anemone and Trichoplax, revealed extensive conservation of the gene repertoire compared to mammals or birds, with the implication that the characteristic life span of an animal gene includes (at least) hundreds millions of years (84–86). The results of extensive comparative analysis of plant, fungal and prokaryotic genomes are fully compatible with this conclusion (87,83). Moreover, deep evolutionary reconstructions suggest that ancestors of hundreds of extant genes were already present in LUCA (88–92). Conservative reconstructions of the gene sets of the common ancestors of the two domains of prokaryotes, bacteria and archaea, seem to indicate that these ancestral forms that, probably, existed over 3 billion years ago, were comparable in genetic complexity, at least, to the simpler of modern free-living prokaryotes (88,93). From an evolutionary biology perspective, it appears that the sequences of many genes encoding core cellular functions, especially, translation, transcription, replication and central metabolic pathways, are subject to strong purifying selection that remained in place for extended time intervals, on many occasions, throughout the ∼3.5 billion year history of cellular life.
Remarkably, it is not only the sequence and structure of the encoded proteins but also features of gene architecture that are not necessarily directly relevant to the gene function that are highly conserved across lengthy periods of life history. In particular, the positions of a large fraction of introns are conserved even between the most distant intron-rich genomes of eukaryotes (25–30% conservation in orthologs from plants and chordates) (94–96), and the great majority of intron positions are shared by mammals and basal animals, such as Trichoplax and the sea anemone (84,86).
The striking conservation of gene sequences and structures contrasts the fluidity of the gene composition of genomes of all forms of life that is revealed by comparative genomics and evolutionary reconstruction. The (nearly) universal genes make up but a tiny fraction of the entire gene universe: altogether, this central core of cellular life consists of, at most, ∼70 genes, that is, no more than 10% of the genes in even the smallest of the genomes of cellular life forms, but typically, closer to 1% of the genes or less (90,97,98). Although in each individual genome, the majority of the genes belong to a moderately conserved genetic ‘shell’ that is shared with distantly related organisms, within the entire gene universe, the core and shell genes (or more precisely, sets of orthologous genes) are a small minority (83). Given this distinctive structure of the gene universe, evolutionary reconstructions inevitably yield a dynamic picture of genome evolution, with numerous genes lost and many others gained via HGT (mostly, in prokaryotes), and gene duplication (see below).
Even to a greater extent than the gene composition of the genomes, the genome architecture, that is, arrangement of genes in a genome shows evolutionary instability compared to gene sequences (99). With the exception of the organization of small groups of functionally linked genes in operons that are, in some cases, shared by distantly related bacteria and archaea, in part, probably, owing to extensive HGT (see below), there is, generally, relatively little conservation of gene order even among closely related organisms (100,101). In particular, in prokaryotes, the long range conservation of gene order completely disappears even in some groups of closely related genomes which retain an almost one-to-one correspondence of orthologous genes and over 99% mean sequence identity between orthologous proteins (75). Thus, in prokaryotes, the organization of genes beyond the level of operons is, mostly, determined by extensive random shuffling, in particular, via inversions centered at the origin of replication (75,102,103). Eukaryotes show a somewhat greater conservation of long range genomic synteny but, even in this case, there are few shared elements of genome architecture between, for instance, different animal phyla, and none at all between different kingdoms (99).
The variability of the genome architectures presents an interesting dilemma to evolutionary biologists: do organisms possess unique genome architectures that are specifically adapted to satisfy unique functional demands of the respective organisms, or is evolution of genome architecture a mostly neutral process? Although local clustering of functionally related genes and other patterns suggestive of functionally relevant gene coexpression were repeatedly observed, these trends are relatively weak and by no means ubiquitous (104,105). Thus, the dominant factor in the evolution of genome architecture appears to be random, non-adaptive rearrangement rather than purifying or positive selection.
Horizontal gene transfer, the network of evolution and the Forest replacing of the TOL
Even long before the genomic era, microbiologists realized that bacteria had the capacity to exchange genetic information via HGT, in some cases, producing outcomes of major importance, such as antibiotic resistance (106). Multiple molecular mechanisms of HGT have been elucidated including plasmid exchange, transduction (HGT mediated by bacteriophages) and transformation (107). These discoveries notwithstanding, HGT was generally viewed as a minor phenomenon that is important only under special circumstances and, in any case, was not considered to jeopardize the concept of the TOL that could be reconstructed by phylogenetic analysis of rRNA and other conserved genes. This fundamental belief was challenged by early results of genome comparisons of bacteria and archaea which indicated that, at least, in some prokaryotic genomes, a major fraction of genes were acquired via demonstrable HGT. The pathogenicity islands and similar simbiosis islands that comprise over 30% of the genome in many pathogenic and symbiotic bacteria are the prime case in point (108–110). Moreover, comparative analysis of the genomes of hyperthermophilic bacteria and archaea suggested that even interdomain HGT can be extensive given shared habitats (111,112).
It can be difficult to demonstrate HGT unambiguously, and in particular, to differentiate from extensive gene loss, so the extent of horizontal genetic mobility between prokaryotes is still debated (113–115). Nevertheless, as the genomic database grows, extensive comparative-genomic and phylogenetic analyses increasingly lead to the conclusion that HGT is virtually ubiquitous in the prokaryotic world in the sense that there are very few if any orthologous gene sets whose history is free of HGT (116,117). The rate of HGT substantially differs for different genes depending on the gene functions, in part, according to the so called complexity hypothesis which posits that barriers might exist for HGT of genes encoding subunits of protein complexes because dosage imbalance and mixing of heterologous subunits resulting from such events could be deleterious (118,119). However, phylogenetic analyses indicate that even such genes, for instance, those for ribosomal proteins and RNA polymerase subunits, are not immune to HGT (120–122).
The high prevalence of HGT in prokaryotes might, in part, explain the persistence of the organization of many operons across broad ranges of organisms, under the selfish operon hypothesis (123,124). Although the operons might be initially selected for the beneficial coexpression and coregulation of functionally linked genes, it is likely that they are maintained and disseminated in the prokaryotic world owing to the increased likelihood of fixation of an operon following HGT, compared, e.g. to a non-operonic pair of genes. This scenario presents a notable case of a combination of selective (coregulation) and neutral (HGT) forces contributing to the evolution of a major aspect of genome organization (76,104).
Eukaryotes are different from prokaryotes with respect to the role played by HGT in genome evolution. In multicellular eukaryotes, where germline cells are distinct from the soma, HGT appears to be rare (125) although not impossible (126). Under certain special circumstances, such as persistence of endosymbiotic bacteria in animals, transfer of large segments of bacterial genomes to the genome of the host are indeed common (127,128). Unicellular eukaryotes do seem to acquire bacterial genes and exchange genes between themselves on relatively frequent occasions (129–131). Far more crucial, however, is the major contribution of the genomes of endosymbionts to the gene complements of all eukaryotes. The discovery of mitochondria-like organelles and genes of apparent mitochondrial origin in all thoroughly characterized unicellular eukaryotes, essentially, ascertain that the last common ancestor of the extant eukaryotes already possessed the mitochondrial endosymbiont (132,133). In terms of their apparent phylogenetic affinities, eukaryotic genes that possess readily identifiable prokaryotic orthologs are sharply split into genes of likely archaeal origin (primarily, but not exclusively, components of information processing systems) and those of likely bacterial origin (mostly, metabolic enzyme and components of various cellular structures) (134,135). It is often assumed on general grounds that the majority of ancestral ‘bacterial’ genes in eukaryotes are of mitochondrial origin but this is hard to demonstrate directly because in phylogenetic analysis, these genes cluster with diverse groups of bacteria (134). These findings are difficult to interpret because the gene composition of the endosymbiont and its host are not known, and conceivably, either or both might have already amassed numerous genes from diverse sources (136). An even bigger point of uncertainty is the actual scenario of the origin of eukaryotes [a detailed discussion of this major subject is outside the scope of this article, see recent reviews and discussions (133,137–140)]. In a nutshell, the competing and hotly debated hypotheses are as follows:
- The symbiogenetic scenario according to which the α-proteobacterial ancestor of mitochondria invaded an archaeal host, and this event triggeredeukaryogenesis including the formation of the signature structural features of the eukaryotic cell such as the endomembrane system, the cytoskeleton and the nucleus (138,141).
- The archezoan scenario under which the host of the mitochondrial endosymbiont was a primitive eukaryote that already possessed all the principal features of the eukaryotic cell that evolved without any relation to endosymbiosis but facilitated the latter through the phagocytic capability of the protoeukaryote (137,142).
Regardless of the exact role played by endosymbiosis in eukaryogenesis, there is no reasonable doubt that the gene complement of eukaryotes is a chimera comprised of functionally distinct genes of archaeal and bacteria descents (134,143). Moreover, endosymbiosis apparently made substantial contributions to the gene complements of some of the individual major groups of eukaryotes. Thus, strong evidence was presented of massive HGT of thousands of genes from a cyanobacterial endosymbiont (the chloroplast) to the host (plant) genomes (144). Similarly, genes of apparent algal origin were detected in chromalveolates that engulfed a red alga in an act of secondary endosymbiosis (145).
The observations of extensive, ubiquitous and occurring via multiple routes HGT outlined above lead to a fundamental generalization: the genomes of all life forms are collections of genes with diverse evolutionary histories. The corollary of this generalization is that the TOL concept must be substantially revised or abandoned because a single tree topology or even congruent topologies of trees for several highly conserved genes cannot possibly represent the history of all or even the majority of the genes (146–149). Thus, an adequate representation of life's history is a network of genetic exchanges rather than a single tree, and accordingly, the ‘strong’ TOL hypothesis, namely, the existence of a ‘species tree’ for the entire history of cellular life, is falsified by the results of comparative genomics.
Certainly, this conclusion is not to be taken as an indication that the concept of evolutionary tree introduced by Darwin (1) should be abandoned altogether. First, trees have the potential to accurately represent the evolution of individual gene families. Secondly, there exist, beyond doubt, expansive parts of life's history for which congruent trees can be obtained for large sets of orthologous genes, and accordingly, the consensus topology of these trees qualifies as a species tree. Evolution of major groups of eukaryotes, such as animals or plants, is the most obvious case in point but tree-like evolution seems to apply also to many groups of prokaryotes at relatively shallow phylogenetic depths. The question remains open whether evolution of life in its entirety is best depicted as:
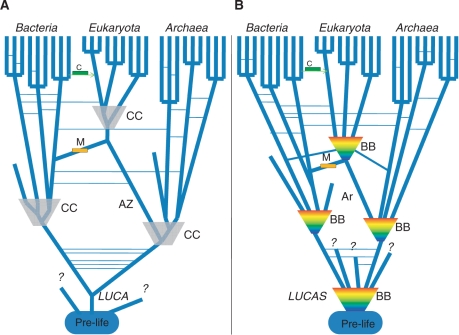
- a consensus tree of highly conserved genes that represents a ‘central trend’ in evolution, with HGT events, including massive ones associated with endosymbiosis, comprising horizontal connections between the tree branches [Figure 1A; (150)], or
- a complex network where phases of tree-like evolution (with horizontal connections) are interspersed with ‘Big Bang’ phases of rampant horizontal exchange of genetic information that cannot be represented as trees in principle [Figure 1B; (151)].
Figure 1.
Two views of life history to replace the Tree of Life. (A) The ‘TOL as a central trend’ model. The history of life is represented as a tree, with connecting lines between branches depicting HGT and shaded trapezoids depicting phases of compressed cladogenesis (276). The origin of eukaryotes is depicted according to the archezoan hypothesis whereby the host of the mitochondrial endosymbiont was a proto-eukaryotes (archezoan). A cellular Last Universal Common Ancestor (LUCA) is envisaged. (B) The ‘Big Bang’ model. The history of life is represented as a succession of tree-like phases accompanied by HGT and non-tree-like, Big Bang phases. Connecting lines between tree branches depict HGT and colored trapezoids depict Big Bang phases (151). The origin of eukaryotes is depicted according to the symbiogenesis model whereby the host of the mitochondrial endosymbiont was an archaeon. A pre-cellular Last Universal Common Ancestral State (LUCAS) is envisaged. Ar, archaeon (host of the mitochondrion in b), AZ, archezoan (host of the mitochondrion in a), BB, Big Bang, C, chloroplast, CC, compressed cladogenesis, M, mitochondrion.
Metagenomics, the expanding world of selfish replicons and replicon fusion
Metagenomics is a major new direction of genomic research that pursues (typically, partial, at this stage) sequencing of the genomes of all life forms that thrive in a certain habitat. Although a young field, metagenomics can already claim major advances in characterizing the bacterial diversity of a variety of habitats, in particular, those in the oceans (152–154). The direction that I would like to emphasize as being of particular conceptual importance for evolutionary biology is metagenomics of viruses (155). The striking conclusion of several viral metagenomic studies is that, at least, in some, particularly, marine habitats, viruses (bacteriophages) are the most abundant biological entities, with the number of viral particles exceeding by an order of magnitude the number of cells (156,157). Although viral genomes are small compared to genomes of cellular life forms, these metagenomic results indicate that viral genomes comprise a major part of the genetic universe that is, at least, comparable in size with the part taken by genomes of cellular organisms. Moreover, given that, in viruses with large genomes, a substantial fraction of genes do not have detectable homologs in current sequence databases (158–160), it seems most likely that viruses encompass most of the genetic diversity on this planet. These findings reverberate with the high prevalence of various classes of mobile elements within the genomes of many cellular organisms. Indeed, in mammalian genomes, sequences derived from mobile elements, primarily, retrotransposons (SINEs and LINEs) appear to constitute, at least, 40% of the genomic DNA (161).
Viruses and various other selfish replicons (defined as genetic elements that do not encode a complete translation system), such as diverse plasmids and transposons, comprise an interconnected genetic pool that is variously known as the mobilome, the virosphere or the virus world (76,162–164). The identity of the virus world is manifested in the existence of a set of ‘hallmark genes’ that encode proteins with key roles in the reproduction of selfish elements (including viral capsid proteins) and are present in extremely diverse elements that propagate in a broad variety of hosts, but not in cellular life forms. The existence of the distinct pool of hallmark genes that includes, among others, RNA-dependent RNA and DNA polymerase, replication enzymes that, probably, antedate large DNA genomes, strongly suggests that the virus world coexists with cellular life forms throughout their history, and possibly, even originates from a primordial, pre-cellular pool of genetic elements (164).
Although distinct, the virus world constantly interacts with the genomic pool of cellular life forms, as illustrated by constant movement of genes between transducing bacteriophages, plasmids and bacterial chromosomes (83), or by the capture of cellular genes (protooncogenes) by animal retroviruses (165). Recent observations of bacteriophage-mediated gene transfer between distantly related bacteria, even without the phage propagation in the recipient organism, suggest that the gene flow mediated by selfish replicons could be more extensive than so far suspected (166). Importantly, parts of mobile elements are frequently recruited (exapted) by host genes as regulatory elements (167,168) and, in some cases, parts of protein-coding sequences (169). Individual cases of exaptation of complete genes from mobile elements are also known as strikingly exemplified by the evolution of the hedgehog gene, a key regulator of animal development, from an intein (170,171).
All prokaryotic genomes, without exception, contain traces of integration of multiple plasmids and phages. Even more revealingly, the archaeal genomes typically carry multiple versions of an operon that encodes key components of the plasmid partitioning machinery, and often possess more than one origin of replication (172). Thus, fusion of distinct replicons appears to routinely occur in prokaryotes, and over the course of evolution, such fusion might have been a major factor in shaping the observed architecture of prokaryotic chromosomes (83,173).
In summary, comparative genomics and metagenomics reveal a vast, dynamic, interconnected world of selfish replicons that interacts with genomes of cellular life forms and, over long spans of evolution, makes major contributions to the composition of chromosomes. In prokaryotes, the interaction between bacterial and archaeal chromosomes and selfish replicons is so intensive, and the distinction between chromosomes and megaplasmids is blurred to such an extent that chromosomes are, probably, best viewed as ‘islands’ of relative stability in the turbulent ‘sea’ of mobile elements (83). In eukaryotes, especially, in multicellular forms that evolved the separation between the germline and soma, the distinction between chromosomes and selfish replicons is sharper. Nevertheless, intragenomic mobility of selfish transposable elements is extensive, and intergenomic mobility, at least, within a species, is actually facilitated by sex, with bursts of transposable element propagation likely marking evolutionary transitions (16). The central role of mobile elements in genome evolution further undermines the TOL concept, although phylogenetic trees of individual hallmark genes can be highly informative for the reconstruction of the evolution of the selfish elements themselves (174,175).
The nature of the Last Universal Common Ancestor and early evolutionary transitions
Comparative genomics vindicates Darwin's conjecture on the origin of all extant life forms from a single common ancestor. Indeed, evolutionary reconstructions suggest that hundreds of conserved genes, most likely, trace back to LUCA (88,89–91). More specifically, these reconstructions indicate that LUCA already possessed a complete system of translation that was not dramatically different from (at least) the simpler versions of the modern translation machinery (that is, consisted of, roughly, 100 RNA and protein molecules) as well as the core transcription system and several central metabolic pathways, such as those for purine and pyrimidine nucleotide biosynthesis (90). However, the sets of genes assigned to LUCA in these reconstructions lack certain essential components of the modern cellular machinery. In particular, the core components of the DNA replication machinery are non-homologous (or, at least, non-orthologous) in bacteria, on the one hand, and archaea and eukaryotes, on the other hand (176). In another sharp divide, the membrane lipids have distinct structures, and the membrane biogenesis enzymes are accordingly non-homologous (non-orthologous) (177).
These major gaps in the reconstructed gene set of LUCA support the idea that different cellular systems ‘crystallized’ asynchronously and are suggestive of ‘phase transitions’ in the early phases of cellular evolution (151,178). One class of hypotheses holds that LUCA was radically different from modern cells, possibly, not a cell at all, but rather a pool of genetic elements that employed diverse replication and expression strategies, and might have populated inorganic compartments like those seen at hydrothermal vents (179,180). Under these scenarios, the modern-type DNA replications systems and membranes evolved at least twice independently in two domains of life (assuming a symbiogenetic origin for eukaryotes). In this case, the very concept of a distinct LUCA becomes ambiguous, and it might be more appropriate to speak of LUCAS, the Last Universal Common Ancestral State (181). The alternative class of scenarios postulate that LUCA was a modern-type cell with either the archaeal or the bacterial varieties of the DNA replication systems and membranes, or even mixed systems (177,182). This class of scenarios implies that there were switches from one type to the other in the evolution of each of these key cellular systems or differential loss of the respective genes.
Regardless of which scenario is preferred, the lack of conservation of central cellular systems among the domains of life indicates that the early stages of cell evolution involved radical changes which are hardly compatible with uniformitarianism.
Genome-wide quantification of selection and junk DNA: distinct evolutionary regimes for different genomes
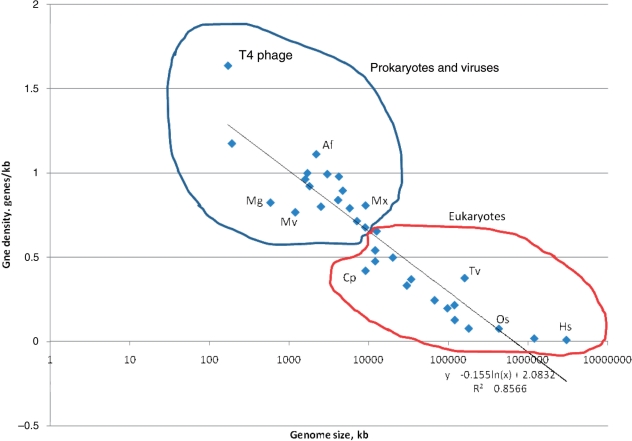
There are major differences in the genome layouts between different lines of life evolution. Prokaryotes and, especially, viruses have ‘wall-to-wall’ genomes that consist, mainly, of genes encoding proteins and structural RNAs, with non-coding regions comprising, with a few exceptions, no more than 10–15% of the genomic DNA. The genomes of unicellular eukaryotes have lower characteristic gene densities but, on the whole, do not depart too far from the prokaryotic principles, with most of the DNA dedicated to protein-coding, despite the distinct, exon–intron gene architecture. The genomes of multicellular eukaryotes are drastically different in that only a minority (a small minority in vertebrates) of the genomic DNA is comprised of sequences encoding proteins or structural RNAs. Generally, across the entire range of life forms, there is a notable negative exponential dependence between the density of protein-coding genes and genome size although significant deviations from this overall dependence are seen as well, particularly, in prokaryotes (Figure 2).
Figure 2.
Dependence between genome size and gene density for large viruses and diverse cellular life forms. The plot is semi-logarithmic. Points corresponding to selected organisms are marked: Af, Archaeoglobus fulgidus (archaeon), Cp, Cryptosporidium parvum (unicellular eukaryote, alveolate), Hs, Homo sapiens, Os, Oryza sativa (rice), Mg, Mycoplasma genitalium (obligate parasitic bacterium), Mv, mimivirus, Tv, Trichomonas vaginalis (unicellular eukaryote, excavate).
This dramatic difference in genome organization between the genomes of (most) unicellular and multicellular organisms demands an explanation, and the simplest, plausible one is given by the population-genetic theory according to which the intensity of purifying selection affecting a population is proportional to the effective population size. Fixation of non-coding sequences, such as introns or mobile elements is, at best, neutral but, more likely, at least, slightly deleterious, even if only because of the extra burden on the replication machinery. Therefore, extensive accumulation of such sequences is possible only in relatively small populations in which the intensity of purifying selection falls below the ‘complexification threshold’. More specifically, theory predicts that all mutations with selection coefficient (s) less than 10–6 would accumulate as neutral in genomes of multicellular eukaryotes, and many cases of insertion of non-coding sequences indeed are associated with such low s values (16,183,184).
Considering the genome-scale study of evolution, the next series of important questions has to do with the distribution of selection coefficients across genomes: how much of the non-coding DNA is actually junk, what is the pressure of purifying selection in different genes, and how common positive (Darwinian) selection actually is? Although measurement of selection for individual genes, let alone individual sites, especially, in non-coding regions is technically challenging (185,186), several genome-wide analyses have been reported. A comprehensive analysis of the human protein set that combined data on pathogenic mutations, non-synonymous SNPs, and divergence in human-chimpanzee orthologs led to the estimate that only ∼12% of the amino-acid residues are associated with s < 10–5, whereas about half of the sites have s values between 10–4 and 10–2 (187). Thus, the majority of the protein sequences seem to be subject to substantial purifying selection. A complementary study on the evolution regimes of multiple groups of closely related bacteria and archaea also revealed typically strong purifying selection, with the genome wide means of the d_N/d_S ratios (the ratio of non-synonymous to synonymous nucleotide substitution rates that is the traditional measure of selection in protein-coding sequences) between 0.02 and 0.2 (d_N/d_S <<1 is the signature of purifying selection) (75).
A genome-wide search for positive selection (measured as the gene-specific d_N/d_S ratio) in protein-coding genes from six mammalian species revealed ∼400 genes (∼2.5%) that seem to have experienced positive selection in at least one branch of the phylogenetic tree of the analyzed species; the values for most of the individual branches were very small (188). These estimates, although conservative, show that, at least, in mammals, positive selection affecting entire gene sequence is quite rare although many genes that are, generally, subject to purifying selection are likely to include positively selected sites. Comprehensive analyses of amino-acid coding sites in 12 Drosophila genomes yielded very different results, suggesting that a substantial fraction and, perhaps, the majority of amino-acid replacements are driven by positive selection although the beneficial effects of most of these replacements seem to be quite small (189,190). Notably, the distribution of positively selected sites is strongly non-random among functional categories of genes, with genes involved in immunity and other defense functions, reproduction, and sensory perception being particularly amenable to positive selection; this distribution seemed to be stable among widely different animals including mammals, flies, and nematodes (188,189,191).
A burning question in genome-wide evolutionary studies, especially, for mammals with their huge genomes, what fraction of the non-coding DNA is ‘real’ junk, and how much is subject to yet unknown functional constraints. The possibility that, despite the lack of detectable evolutionary conservation, a large fraction if not most of the human DNA is, in fact, functionally important and hence maintained by selection is often discussed, especially, in the light of the demonstrations that a very large fraction of the genome is transcribed (192–194). The discovery of the so-called ultraconserved sequences that appear to be subject to an exceptionally strong purifying selection (195,196) is compatible with this idea. Furthermore, a considerable fraction of the ‘junk’ DNA could be involved in functional roles that entail only limited sequence conservation but nevertheless are important, in particular, for chromatin structure maintenances and remodeling such as scaffold/matrix attachment regions (SARs/MARs) (197,198). Nevertheless, a recent genome-wide analysis of the distribution of insertion and deletions (in comparisons of human, mouse and dog genomes) suggests that only ∼3% of the human euchromatin DNA is under selective constraints (199). Given that protein-coding sequences comprise only ∼1.2% of the euchromatin, these results indicate that the majority of functionally important DNA sequences in mammals do not code for proteins, but also vindicate the early conjectures that most of the human genome is non-functional that is, after all, junk (45,46). Of course, it should be kept in mind that any definition of junk is conditional in that yesterday's garbage tomorrow can be recruited for a functional role. In contrast, interspecies comparisons of non-coding genomic regions in Drosophila indicate that the majority (70% or more of the nucleotides) of these sequences evolve under selective constraints, and a significant fraction (up to 20%) seems to be affected by positive selection (200–202). Certainly, these studies are based on different simplifying assumptions (that cannot be here discussed in detail), so the conclusion on major differences in selective regimes between different lineages should be assessed with caution and is subject to further validation. However, the very fact that organisms with comparable sizes of the gene sets and levels of organizational complexity, such as insects, on the one hand, and mammals, on the other hand, differ so dramatically in terms of gene density and the amount of the apparent genomic ‘junk’ (Figure 2) suggests that their genomes evolve under different selective pressures.
The study of the interplay between neutral processes, purifying selection, and positive selection is still in its early stages. The collection of sets of closely related genomes from diverse taxa that is essential for this analysis is currently small, although rapidly growing, and the methods for discriminating different modes of evolution are still under active development. Nevertheless, even the already available results make it abundantly clear that the contributions of each of these factors are highly variable among organisms, depending on the effective population size, the characteristic rates of mutation and recombination, and probably, other factors that are not yet elucidated.
Gene and genome duplication: the principal route of genomic innovation
Analysis of the numerous sequenced genomes vindicated Ohno's vision of gene duplication as a major evolutionary mechanism (51), perhaps, even to a greater extent than the originator of the concept could anticipate. The majority of the genes in most genomes of cellular life forms (except for the smallest genomes of obligate parasites) possess paralogs indicative of duplication at some point during evolution (16,69), and many genes belong to large families of paralogs which form a characteristic power-law distribution of the number of members [(203,204); see discussion below]. With regard to the contribution of duplication to the origin of new genes, it is important to note that there is little compelling evidence of de novo emergence of genes from non-coding sequences; although genes can expand by recruiting small adjacent segments of non-coding sequence [for instance, from an intron (205), birth of a complete novel gene via this route seems to be an exceptional event (206)]. Hence it is tempting to generalize that gene duplication is not just an important but indeed the dominant route that leads to the origin of new genes, with the important addition that duplication is often followed by accelerated sequence evolution as well as rearrangement of a gene, an evolutionary mode that obliterates detectable connections to the original source.
Ohno's idea on the elimination or relaxation of selection following a gene duplication, allowing accelerated evolution that has the potential to produce functional novelty, also was supported by comparative-genomic data, albeit with a significant twist. It was argued theoretically and then demonstrated by empirical measurement of the selection pressure on recently duplicated gene sequences that relaxation of purifying selection was more likely to be symmetrical, to affect both duplicates more or less equally (207,208). Thus, the more common path of evolution of duplicated genes might not be neofunctionalization postulated by Ohno but rather subfunctionalization whereby new paralogs retain distinct subsets of the original functions of the ancestral gene whereas the rest of the functions differentially deteriorate (209,210). More sophisticated analyses seem to suggest that both regimes of evolution could realize at different stages of the history of paralogous genes, with fast subfunctionalization immediately after duplication succeeded by subsequent, slower neofunctionalization (211–213).
Gene duplications occurs throughout the evolution of any lineage but the rate of duplication is not uniform on large evolutionary scales, so that organizational transitions in evolution seem to be accompanied by bursts of gene duplication, conceivably, enabled by weak purifying selection during population bottlenecks (see below). Perhaps, the most illustrative case in point is the emergence of eukaryotes that was accompanied by a wave of massive duplication, yielding the characteristic many-to-one co-orthologous relationship between eukaryotic genes and their prokaryotic ancestors (214). Similarly, differential duplication of Hox gene clusters and other developmental regulators is thought to have played a pivotal role in the differentiation of animal phyla (215,216). Arguably, the most dramatic cases of ‘saltatory’ gene duplication involve whole-genome duplication (WGD) events (217). Following the original hypothesis of Ohno, genome analysis revealed traces of independent WGD events retained in the size distribution of paralogous gene families and/or genomic positions of paralogous regions, despite the extensive loss of genes after WGD, in yeasts (218,219), chordates (220–223) and plants (224,225). Mechanistically, the high prevalence of WGD in eukaryotes might not be particularly surprising because it results from a well known, widespread genetic phenomenon, polyploidization. However, evolutionary consequences of WGD appear to be momentous as these events create the possibility of rapid sub/neofunctionalization simultaneously in the entire gene complement of an organism (226). In particular, WGD is thought to have played a central role in the primary radiation of chordates (220). It is difficult to rule out the possibility that more ancient WGD events are no longer readily detectable owing to numerous gene losses that obscure the WGD signal; in particular, the burst of duplications that followed eukaryogenesis but antedates the last common ancestor of extant eukaryotes might have been brought about by the first WGD in eukaryotic evolution (214).
Considering the wide occurrence of WGD in multiple eukaryotic lineages, it is notable that so far no such events were detected by analysis of the numerous available prokaryotic genomes although transient polyploidy was repeatedly observed (227,228). Conceivably, the absence of detectable WGD in prokaryotes is due to the efficient purifying selection that acts in large prokaryotic population (see below) and leads to rapid elimination of duplicate genes that would obliterate traces of WGD should such an event occur.
At the level of general concepts of evolutionary biology with which I am primarily concerned here, genomic studies on gene duplication lead to, at least, two substantial generalizations. First, the demonstration of the primary evolutionary significance of duplications including duplications of large genome regions and whole genomes is a virtual death knell for Darwinian gradualism: even a single gene duplication hardly qualifies as an infinitesimally small variation whereas WGD qualifies as a bona fide saltatory event. Secondly, the primacy of gene duplication with the subsequent (sometimes, rapid) diversification of the paralogs as the route of novel gene origin reinforces the metaphor of evolution as a tinkerer: evolution clearly tends to generate new functional devices by tinkering with the old ones after making a backup copy rather than create novelty from scratch.
Emergence and evolution of genomic complexity: the non-selective paradigm and the fallacy of evolutionary progress
Undoubtedly, multicellular eukaryotes, such as animals and plants, are characterized by a far greater organizational complexity than unicellular life forms, and in the spirit of the Modern Synthesis, this complexity is generally seen as a result of numerous adaptive changes driven by natural selection, and, being so regarded, can be viewed as a manifestation of ‘progress’ in evolution. The correspondence between the organizational complexity and genomic complexity is an open issue, in part, because genomic complexity is not easy to define. A simple and plausible definition can be the number of nucleotides that carry functionally relevant information, that is, are affected by selection (229,230). Under this definition, genomes of multicellular eukaryotes, of course, are much more complex than genomes of unicellular forms, and this higher genomic complexity translates into functional complexity as well.
A striking case in point is alternative splicing that is a crucial functional device in complex organisms like mammals where it creates several-fold more proteins than there are protein-coding genes (231–233) (thus, the fact that humans have ∼20 000 genes compared to ∼10 000 genes in the bacterium Myxococcus xanthus should not be translated into the claim that ‘the human proteome is twice as complex as that of a bacterium’: the real difference is greater owing to alternative splicing). Alternative splicing is made possible by weak splice signals that are processed or skipped by the spliceosome with comparable frequencies (234). In a sense, functionally important alternative splicing events are encoded in these splice junctures and, to some extent, also in additional intronic sequences. However, did alternative splicing evolve as a functional adaptation? In all likelihood, no. Indeed, it was shown that intron-rich genomes typically possess weak splice signals whereas intron-poor genomes (mostly, those of unicellular eukaryotes) have tight splice junctions, presumably, ensuring high fidelity of splicing (235). Recent detailed studies demonstrated low splicing fidelity in intron-rich organisms, so that numerous misspliced variants are produced and are, mostly, destroyed by the nonsense-mediated decay (NMD) system (236). Evolutionary reconstructions strongly suggest that ancient eukaryotes including the last common ancestor of extant forms possessed high intron densities comparable to those in the most intron-rich modern genomes, such as vertebrates (237–239) and, by inference, had weak splice signals yielding numerous alternative transcripts (235). The conservation of the NMD machinery in all eukaryotes (240) is fully compatible with this hypothesis. Thus, it appears that alternative splicing emerged as a ‘genomic defect’ of which the respective organisms could not get rid, presumably, because of weak purifying selection, and evolved a special mechanism to cope with, namely, NMD. Gradually, they also evolved ways to utilize this spandrel for multiple functions.
The above account of the origin of alternative splicing could epitomize the non-adaptationist population-genetic theory of evolution of genomic complexity that was recently expounded by Lynch (16,183,184). As already alluded to in the preceding section, the central tenet of the theory is that genetic changes leading to an increase of complexity, such as gene duplications or intron insertions are slightly deleterious, and therefore can be fixed at an appreciable rate only when purifying selection in a population is weak. Therefore, given that the strength of purifying selection is proportional to the effective population size, substantial increase in the genomic complexity is possible only during population bottlenecks. Under this concept, genomic complexity is not, originally, adaptive but is brought about by neutral evolutionary processes when purifying selection is ineffective. In other words, complexification begins as a ‘genomic syndrome’ although complex features (spandrels) subsequently are co-opted for various functions and become subject to selection. By contrast, in highly successful, large populations, like those of many prokaryotes, purifying selection is so intense that no increase in genomic complexity is feasible, and indeed, genome contraction is more likely.
Of course, there are exceptions to these principles, such as bacterial genomes with more than 12 000 genes (241), viral genomes with extensive proliferation of duplicated genes (158), and genomes of unicellular eukaryotes [e.g. _Chlamydomonas_ (242) or Trichomonas (243)] that, by most criteria, are as complex as the genomes of multicellular animals or plants. Furthermore, some prokaryotic genomes [e.g. the crenarchaeon _Sulfolobus solfataricus_ (244)] and genomes of unicellular eukaryotes [e.g. _Trichomonas vaginalis_ (243)] are among those with the highest content of transposable elements. Apparently, the outcome of genome evolution depends on the balance between the pressure of purifying selection, itself dependent on the population size and mutation rate, the intensity of recombination processes, the activity of selfish elements, and adaptation to specific habitats (99). An attractive hypothesis is that, at least, in prokaryotes, the upper bound for the number of genes in a genome, a good proxy for genomic complexity, is determined by the ‘regulatory (bureaucratic) overhead’ (83,245,246). The existence of such an overhead is implied by the notable observation that different functional classes of genes scale differently with respect to the total number of genes in a genome, and in particular, regulatory genes (such as transcription repressors and activators) show a (nearly) quadratic scaling (83,245,247,248). Conceivably, at some ratio of the number of regulators to the number of regulated genes, perhaps, close to 1:1, the burden of regulators becomes unsustainable. Thus, evolution of genome complexity, undoubtedly, depends on a complex combination of stochastic (neutral) and adaptive processes. It appears, however, that at present, the most consistent, simple null hypothesis of genomic evolution is that genome expansion, a pre-requisite for complexification, is not a result of adaptation but rather a consequence of weak purifying selection.
The next big question that begs to be asked with regard to complexity, both organizational and genomic, is: was there a consistent trend towards increasing complexity during the ∼3.5 billion years of life evolution on earth? The most likely answer is, no. Even very conservative reconstructions of ancestral genomes of archaea and bacteria indicate that these genomes were comparable in size and complexity to those of relatively simple modern forms (88,89,91,93). Furthermore, reconstructions for some individual groups, and not only parasites, point to gene loss and genome shrinking as the prevailing mode of evolution (249). Considering that numerous prokaryotic groups undoubtedly have gone extinct in the course of life history, there is every reason to believe that, even prior to the radiation of all major lineages known today, the distribution of genome sizes and the mean complexity in prokaryotes was (nearly) the same as it is now. Of course, it is conceivable that the most complex forms known evolved relatively late in evolution but, should that be the case, it could be accounted for by purely stochastic processes, given that life, in the pre-LUCAS stages of its evolution, must have started ‘from so simple a beginning’ (1250).
In the same vein, the discovery of large and complex genomes in stem animals (that is, animals with radial symmetry, such as Cnidaria, that branched off the trunk of metazoan evolution prior to the origin of the Bilateria) (84–86) suggests that there was little if any increase in genomic complexity during the evolution of the metazoa (although organizational complexity did increase); instead, recurrent gene loss in different lineages was the most prevalent evolutionary process.
Certainly, episodes of major increase in complexity are known, such as the origin of eukaryotes, and the origin of multicellular forms, to mention obvious examples. However, these seem not to be parts of a consistent, gradualist trend, but rather singular, more or less catastrophic events triggered by rare, chance occurrences such as the domestication of the endosymbiont in the case of the origin of eukaryotes.
On the whole, the theoretical and empirical studies on the evolution of genomic complexity suggest that there is no trend for complexification in the history of life and that, when complexity does substantially increase, this occurs not as an adaptation but as a consequence of weak purifying selection, in itself, paradoxical as this might sound, a telltale sign of evolutionary failure. It appears that these findings are sufficient to put to rest the notion of evolutionary ‘progress’, a suggestion that was made previously on more general grounds.
Functional genomics, systems biology and the determinants of gene evolution rate
Just like the final decade of the 20th century was the age of genomics when the quantity of genome sequences was transformed into a new quality, allowing novel generalization, such as the ‘uprooting’ the TOL, the first decade of the new century became the age of functional genomics and systems biology. These disciplines yielded increasingly reliable data of a new kind that start to fill the previously glaring gap between the genome and the phenotype of an organism (hereinafter denoted phenomic variables). The phenomic variables include genome-wide profiles of gene expression levels, comprehensive maps of protein–protein and genetic interactions, information on the effects of gene knockout (gene dispensability, typically, defined as essentiality of a gene for growth on rich media), and more (81,82). The first comparative analyses that became possible when sufficient information on gene expression became available for multiple organisms revealed an interplay between neutral and selective processes. Although the levels of expression between orthologous genes in human and mouse show significant conservation (compared to random gene pairs), the divergence in expression is more pronounced than that between protein sequences of the orthologs (251,252). Thus, although, in general terms, evolution of gene expression is similar to sequence evolution in that purifying selection is the principal constraining force (253), the genuinely neutral, unconstrained component is likely to contribute more to the evolution of expression.
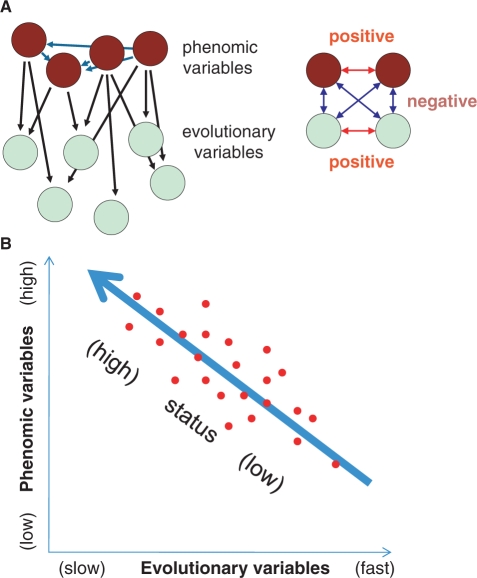
Joint analysis of the novel class of phenomic variables characterized by systems biology and the measures of gene evolution such as sequence evolution rate and propensity for gene loss revealed a rather unexpected structure of correlations [(81,254,255); Figure 3A]. Despite the intuitive link between the rate of evolution and gene dispensability [‘important’ genes would be expected to evolve slower than less important ones (256)], only a weak link (at best) between these characteristics was detected (257–259). The link between evolution rate and functional importance of a gene deserves further investigation because comprehensive analysis reveals a measurable phenotypic effect of knockout of virtually each yeast gene under some conditions (260). However, regardless of the outcome of such studies, clearly, this link is subtle, even if it turns out to be robust. In contrast, the strongest correlation in all comparisons between evolutionary and phenomic variables was seen between gene expression level and sequence evolution rate or propensity for gene loss: highly expressed genes, indeed, tend to evolve substantially slower than lowly expressed genes (254,261). This finding is buttressed by the observations of a positive correlation between sequence divergence and the divergence of expression profiles among human and mouse orthologous genes (252) and the comparatively low rates of expression profile divergence in highly expressed genes (262).
Figure 3.
Evolutionary genomics and systems biology. (A) Evolutionary and phenomic variables. The phenomic variables are viewed as mutually dependent and affecting evolutionary variables (left). Positive correlations are shown by red arrows and negative correlations are shown by blue arrows. (B) The concept of gene status. The red points schematically denote data scatter.
The overall structure of the correlations between evolutionary and phenomic variables is succinctly captured in the concept of a gene's ‘status’ in a genome (255). High-status genes evolve slow, are rarely lost during evolution and are, typically, highly expressed, with numerous protein–protein and genetic interactions, and many paralogs (Figure 3B). It should be noted, however, that despite this appearance of order in the correlation structure, all correlations are relatively weak, and do not seem to significantly increase with the improvement of the data quality (254,255). These observations point to the multiplicity of the determinants of the course of a gene's evolution and suggest that truly random, stochastic noise could be an important factor.
The emergence of the link between sequence evolution rate as the most prominent connection between evolutionary and phenomic variable led to a new concept of the principal determinants of protein evolution. In the pre-genomic era, it was generally assumed that the sequence evolution rate should be a function of, firstly, the intrinsic structural-functional constraints that affect the given protein and, secondly, the importance of the biological role of the protein in the organism (256). With the advent of the systems biology data, it was realized that phenomic variables, in particular, gene expression could be equally or even more important than the traditionally considered factors (263,264). This realization led to the Mistranslation-Induced Misfolding (MIM) hypothesis according to which expression level or, more precisely, the rate of translational events is indeed the dominant determinant of the sequence evolution rate. The cause of the covariation between the sequence evolution rate and expression level is thought to be selection for robustness to protein misfolding that is increasingly important for highly expressed proteins owing to the toxic effects of misfolded proteins (265,266). The MIM hypothesis could additionally explain the rather puzzling but consistent and strong positive correlation between the rates of evolution in synonymous and non-synonymous positions (d_N_ and d_S_, respectively) of protein-coding sequences (267). Indeed, this correlation is likely to be a consequence of the slow evolution in both classes of sites in highly expressed genes which, in the case of synonymous sites, is likely to be caused by selection for codons that minimize mistranslation (268,269). Detailed computer simulations of protein evolution suggest that the toxic effect of protein misfolding, indeed, could suffice to explain the observed covariation of expression level and sequence evolution rate (269). An analysis of the evolution of multidomain proteins revealed substantial homogenization of the domain-specific evolutionary rates compared to the same pair of domains in separate proteins, conceivably, attributable to the equalized translation rates, but significant differences between domain-specific evolution rates persisted even in multidomain proteins (270). Hence the generalized MIM hypothesis according to which the rate of protein evolution, primarily, depends on two factors:
- Intrinsic misfolding robustness that depends on the characteristic stability and designability of the given protein (domain).
- Translation rate that can be viewed as an amplifier of the fitness cost of misfolding and, accordingly, of the selection for the robustness to amino-acid misincorporation.
Evolutionary systems biology revealed a new layer of connections between the evolution and functioning of the genome. It is becoming clear that processes that link the genome and the phenotype of an organism, in particular, gene expression exert a substantial feedback on gene evolution. The rate of evolution of protein-coding genes might depend more on constraints related to the prevention of deleterious effects of misfolding than on constraints associated with the specific protein function.
Universals of genome evolution
Comparative genomics and systems biology yield enormous amounts of data, and this wealth of information begs for a search for patterns and regularities. Indeed, several such regularities that are widespread and could even be universal for the entire course of life evolution were discovered. In the preceding section, I discussed one of such apparent universals, the negative correlation between gene sequence evolution rate and expression level that seems to hold in all organisms for which the data are available and leads to a reappraisal of the factors that affect gene evolution (269).
Other potentially important regularities come in the form of conserved distributions of evolutionary and functional variables. Strikingly, the distributions of the sequence evolution rates of orthologous genes between closely related genomes were found to be highly similar in distant taxa (271); when standardized, these distributions are virtually indistinguishable in bacteria, archaea and eukaryotes and are best approximated by a log-normal distribution (Figure 4A). Considering the dramatic differences in the genomic complexity and architecture (see above) as well as the biology of these organisms, the near identity of the rate distributions is surprising and demands an explanation in terms of universal factors that affect genome evolution. Robustness to protein misfolding discussed above seems to be a good candidate for such a universal factor although quantitative models explaining the rate distribution remain to be developed.
Figure 4.
Universals of evolution. (A) Distributions of evolutionary rates between orthologs in pairs of closely related genomes of bacteria, archaea and eukaryotes. The evolutionary distances between aligned nucleotide sequences of orthologous genes were calculated using the Jukes–Cantor correction and standardized so that the mean of each distribution equaled to 0, and the standard deviation equaled to 1. The plot is semi-logarithmic. Metma—Methanococcus maripaludis C5 versus M. maripaludis C7 (Euryarchaeota); Bursp—Burkholderia cenocepacia MC0-3 versus B. vietnamiensis G4 (Proteobacteria); Salsp—Salinispora arenicola CNS-205 versus S. tropica CNB-440 (Actinobacteria). All sequences were from the NCBI RefSeq database. The probability density curves were obtained by Gaussian-kernel smoothing of the individual data points. (B) Fit of empirical paralogous gene family size distributions to the balanced birth-and-death model. The results are shown for yeast Saccharomyces cerevisiae (Sc, left) and humans (Hs, right). Upper panels, binned distributions of paralogous family sizes; middle panels, paralogous family size distributions in double logarithmic coordinates; bottom panels, cumulative distribution function of paralogous family sizes. The lines show the predictions the balanced birth-and-death model. The figure is from (204).
As discussed above, gene duplication shapes all genomes, and the distribution of family size in all sequenced genomes follows a power-law-like distribution, with the only appreciable difference being the exponent (203,204), so this distribution comes across as a universal of genomic evolution. This distribution is closely fit by a simple birth-and-death model of gene evolution with balanced birth and death rates and without direct involvement of any form of selection [(204,272); Figure 4B].
The differential scaling of functional classes of genes with genome size that is mentioned above suggests the existence of an entire set of fundamental constants of evolution. The ratios of the duplication rates to gene elimination rates that determine the exponents of the power laws for each class of genes appear to be the same for all tested lineages of prokaryotes and invariant with respect to time, so the functional classes of genes appear to possess universal ‘evolutionary potentials’ (245,273).
The apparent universality of these and other central characteristics of genome evolution suggests that relatively simple, non-selective models might be sufficient to form the framework of a general evolutionary theory with respect to which purifying selection would provide boundary conditions (constraints) whereas positive, Darwinian selection (adaptation) would manifest itself as a quantitatively modest, even if functionally crucial modulator of the evolutionary process.
CONCLUSIONS
Two centuries after Darwin's birth, 150 years after the publication of his ‘Origin of Species’, and 50 years after the consolidation of the Modern Synthesis, comparative analysis of hundreds of genomes from many diverse taxa offers unprecedented opportunities for testing the conjectures of (neo)Darwinism and deciphering the mechanisms of evolution. Comparative genomics revealed a striking diversity of evolutionary processes that was unimaginable in the pre-genomic era. In addition to point mutations that can be equated with Darwin's ‘infinitesimal changes’, genome evolution involves major contributions from gene and whole genome duplications, large deletions including loss of genes or groups of genes, horizontal transfer of genes and entire genomic regions, various types of genome rearrangements, and interaction between genomes of cellular life forms and diverse selfish genetic elements. The emerging landscape of genome evolution includes the classic, Darwinian natural selection as an important component but is by far more pluralistic and complex than entailed by Darwin's straightforward vision that was solidified in the Modern Synthesis (16,184). The majority of the sequences in all genomes evolve under the pressure of purifying selection or, in organisms with the largest genomes, neutrally, with only a small fraction of mutations actually being beneficial and fixed by natural selection as envisioned by Darwin. Furthermore, the relative contributions of different evolutionarily forces greatly vary between organismal lineages, primarily, owing to differences in population structure.
Evolutionary genomics effectively demolished the straightforward concept of the TOL by revealing the dynamic, reticulated character of evolution where HGT, genome fusion, and interaction between genomes of cellular life forms and diverse selfish genetic elements take the central stage. In this dynamic worldview, each genome is a palimpsest, a diverse collection of genes with different evolutionary fates and widely varying likelihoods of being lost, transferred, or duplicated. So the TOL becomes a network, or perhaps, most appropriately, the Forest of Life that consists of trees, bushes, thickets of lianas, and of course, numerous dead trunks and branches. Whether the TOL can be salvaged as central trend in the evolution of multiple conserved genes or this concept should be squarely abandoned for the Forest of Life image remains an open question (274).
Table 1 outlines the status of the central tenets of classical evolutionary biology in the age of evolutionary genomics and systems biology. All the classical concepts have undergone transformation, turning into much more complex, pluralistic characterizations of the evolutionary process (15). Depicting the change in the widest strokes possible, Darwin's paramount insight on the interplay between chance and order (introduced by natural selection) survived, even if in a new, much more complex and nuanced form, with specific contributions of different types of random processes and distinct types of selection revealed. By contrast, the insistence on adaptation being the primary mode of evolution that is apparent in the Origin, but especially in the Modern Synthesis, became deeply suspicious if not outright obsolete, making room for a new worldview that gives much more prominence to non-adaptive processes (184).
Table 1.
The status of the central propositions of Darwinism-Modern Synthesis in the light of evolutionary genomicsa
| Proposition | Current status |
|---|---|
| The material for evolution is provided, primarily, by random, heritable variation | True. The repertoire of relevant random changes greatly expanded to include duplication of genes, genome regions, and entire genomes; loss of genes and, generally, genetic material; HGT including massive gene flux in cases of endosymbiosis; invasion of mobile selfish elements and recruitment of sequences from them; and more |
| Fixation of (rare) beneficial changes by natural selection is the main driving force of evolution that, generally, produces increasingly complex adaptive features of organisms; hence progress as a general trend in evolution | False. Natural (positive) selection is an important factor of evolution but is only one of several fundamental forces and is not quantitatively dominant; neutral processes combined with purifying selection dominate evolution. Genomic complexity, probably evolved as a ‘genomic syndrome’ cause by weak purifying selection in small population and not as an adaptation. There is no consistent trend towards increasing complexity in evolution, and the notion of evolutionary progress is unwarranted |
| The variations fixed by natural selection are ‘infinitesimally small’. Evolution adheres to gradualism | False. Even single gene duplications and HGT of single genes are by no means ‘infinitesimally small’ let alone deletion or acquisition of larger regions, genome rearrangements, whole-genome duplication, and most dramatically, endosymbiosis. Gradualism is not the principal regime of evolution |
| Uniformitarianism: evolutionary processes remained, largely, the same throughout the evolution of life | Largely, true. However, the earliest stages of evolution (pre-LUCA), probably, involved distinct processes not involved in subsequent, ‘normal’ evolution. Major transition in evolution like the origin of eukaryotes could be brought about by (effectively) unique events such as endosymbiosis |
| The entire evolution of life can be depicted as a single ‘big tree’ | False. The discovery of the fundamental contributions of HGT and mobile genetic elements to genome evolution invalidate the TOL concept in its original sense. However, trees remain essential templates to represent evolution of individual genes and many phases of evolution in groups of relatively close organisms. The possibility of salvaging the TOL as a central trend of evolution remains |
| All extant cellular life forms descend from very few, and probably, one ancestral form (LUCA) | True. Comparative genomics leaves no doubt of the common ancestry of cellular life. However, it also yields indications that LUCA(S) might have been very different from modern cells |
Beyond the astonishing, unexpected diversity of genome organization and modes of evolution revealed by comparative genomics, is there any chance to discover underlying general principles? Or, is the only such principle the central role of chance and contingency in evolution, elegantly captured by Jacob (55) in his ‘evolution as tinkering’ formula? In a somewhat tongue-in-cheek manner, one is inclined to ask: is a Postmodern Synthesis conceivable and, perhaps, even in sight?
Several recent developments in evolutionary genomics can be candidates for the roles of high-level generalizations underlying the diversity of evolutionary processes. Perhaps, the most far-reaching of these is the population-genetic concept of genome evolution developed by Lynch (16). According to this concept, the principal features of genomes are shaped not by adaptation but by stochastic evolutionary processes that critically depend on the intensity of purifying selection in the which, in turn, is determined by the effective population size and mutation rate of the respective organisms. In particular, the complexity of the genomes of multicellular eukaryotes is interpreted as evolving, primarily, not as an adaptation ensuring organizational and functional complexity but as a ‘genomic syndrome’ caused by inefficient purifying selection in small populations. Some of the sequence elements accumulated via neutral processes are then recruited for biological functions that collectively, indeed, provide for the evolution of structurally and functionally complex organisms. Conversely, the compact genomes of prokaryotes and some unicellular eukaryotes might not be shaped by selection for ‘genome streamlining’ but rather by effective amelioration of even slightly deleterious sequences in large populations (83). The non-adaptive view of the evolution of genomic complexity by no means implies that no complex features ever evolve as direct adaptations or that genome streamlining can never be a major driving force of genome evolution. However, I believe that the evidence amassed by evolutionary genomics is sufficient to necessitate the change of the central null hypothesis of genome evolution from adaptationist to neutral, with the burden of proof shifted to the adepts of pervasive adaptation (230).
The concept of the substantially non-adaptive character of genome evolution indeed seems to affect our basic understanding of the meaning of conservation of genomic features. As a case in point, the rather enigmatic conservation of the positions of a large fraction of intron positions throughout the evolution of eukaryotes might not be a consequence of strong purifying selection that would cause elimination of variants in which the respective introns were lost (the default interpretation implied by the very notion of purifying selection and fully compatible with the neutral theory). On the contrary, the conservation of introns and other genomic features without obvious functions could be the consequence of weak purifying selection in small populations of complex organisms that is insufficient to efficiently remove these elements. This is not meant to claim that many genomic characters (such as individual genes, amino-acid residues or nucleotides) are not conserved during evolution owing to their functional importance but to suggest that even this ‘sacred’, central tenet of evolutionary biology—‘what is conserved is functionally relevant’—is not an absolute, and the non-adaptive alternative is to be taken seriously. Together with the realization that genome contraction is at least as common in evolution as genome expansion, and the increase of genomic complexity is not a central evolutionary trend, the concept of non-adaptive genome evolution implies that the idea of evolutionary progress can be safely put to rest.
It is sometimes argued that recent developments in genomics and systems biology produce a maze of connections between different type of data that is intractable in any explicit form, thus eliminating any hope for the discovery of simple, ‘law-like’ regularities and reducing much of the research in these areas to the development of predictive algorithms (275). However, it is exactly this type of simple and apparently universal regularities that emerge from the joint analysis of comparative-genomic and systems biology data. The distribution of evolutionary rates across sets of orthologous genes, the distribution of the sizes of paralogous gene families, the negative correlation between the expression level and the sequence evolution rate of a gene, and other relationships between key evolutionary and phenomic variables seem to be genuine universals of evolution. The simplicity of these universal regularities suggests that they are shaped by equally simple, fundamental evolutionary processes, rather than by selection for specific functions. In some cases, explicit models of such processes have already been developed and shown to fit the data. These models either do not include selection at all or give selection a new interpretation. A good case in point is the generalized mistranslation-induced misfolding hypothesis that explains the covariation of gene expression and sequence evolution rate by selection for robustness to misfolding that comes across as a major determinant of protein evolution. The unexpected corollary of this model is that the primary driving force of purifying selection might not be the maintenance of a biological function but rather prevention of non-specific deleterious effects of a misfolded protein.
Collectively, the developments in evolutionary genomics and systems biology outlined here seem to suggest that, although at present only isolated elements of a new, ‘postmodern’ synthesis of evolutionary biology are starting to be formulated, such a synthesis is indeed feasible. Moreover, it is likely to assume definitive shape long before Darwin's 250th anniversary.
FUNDING
DHHS (NIH, National Library of Medicine) intramural funds. Funding for open access charge: DHHS (NIH, National Library of Medicine) intramural funds.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
I thank Valerian Dolja, Allan Drummond, David Lipman, Michael Lynch, Tania Senkevich, Claus Wilke and Yuri Wolf for many helpful discussions, Tania Senkevich for critical reading of the manuscript, and Yuri Wolf for help with the preparation of the figures.
REFERENCES
- 1.Darwin C. On the Origin of Species. London: Murray; 1859. [Google Scholar]
- 2.Darwin C. On the tendency of species to form varieties; and on the perpetuation of vareities and species by natural means of selection. I. Extract from an unpublished work on species, II. Abstract of a letter from C. Darwin, esq., to Prof. Asa Gray. J. Proc. Linn. Soc. London. 1858;3:45–53. [Google Scholar]
- 3.Wallace AR. On the tendency of species to form varieties; an don the perpetuation of varieties and species by natural mean of selection. III. On the tendency of varieties to depart indefinitely from the original type. J. Proc. Linn. Soc. London. 1858;3:53–62. [Google Scholar]
- 4.Lamarck J-B. Philosophie zoologique, ou exposition des considérations relatives à l'histoire naturelle des animaux. Dentu, Paris: 1809. [Google Scholar]
- 5.Fisher RA. The Genetical Theory of Natural Selection. Oxford: Clarendon Press; 1930. [Google Scholar]
- 6.Wright S. Evolution: Selected papers. Chicago: University of Chicago Press; 1986. [Google Scholar]
- 7.Haldane JBS. The Causes of Evolution. Green & Co, London: Longmans; 1932. [Google Scholar]
- 8.Dobzhansky T. Genetics and the Origin of Species. New York: Columbia University Press; 1937. [Google Scholar]
- 9.Huxley JS. Evolution: The Modern Synthesis. London: Allen and Unwin; 1942. [Google Scholar]
- 10.Mayr E. Systematics and the Origin of Species. New York: Columbia University Press; 1944. [Google Scholar]
- 11.Simpson GG. Tempo and Mode in Evolution. New York: Columbia University Press; 1944. [Google Scholar]
- 12.Tax S, Callender C, editors. Evolution after Darwin; the University of Chicago Centennial. Chicago: University of Chicago Press; 1960. [Google Scholar]
- 13.Gould SJ. The Structure of Evolutionary Theory. Cambrdige, MA: Harvard University Press; 2002. [Google Scholar]
- 14.Browne J. Birthdays to remember. Nature. 2008;456:324–325. doi: 10.1038/456324a. [DOI] [PubMed] [Google Scholar]
- 15.Rose MR, Oakley TH. The new biology: beyond the Modern Synthesis. Biol. Direct. 2007;2:30. doi: 10.1186/1745-6150-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch M. The Origins of Genome Architecture. Sunderland, MA: Sinauer Associates; 2007. [Google Scholar]
- 17.Kimura M. Recent development of the neutral theory viewed from the Wrightian tradition of theoretical population genetics. Proc. Natl Acad. Sci. USA. 1991;88:5969–5973. doi: 10.1073/pnas.88.14.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyons S. Thomas Henry Huxley: The Evolution of A Scientist. Maherst-New York: Prometheus; 2000. [Google Scholar]
- 19.Lazcano A, Forterre P. The molecular search for the last common ancestor. J. Mol. Evol. 1999;49:411–412. [PubMed] [Google Scholar]
- 20.Futuyma D. Evolution. Sunderland, MA: Sinauer Associates; 2005. [Google Scholar]
- 21.Mayr E. In: The Evolution of Life: Evolution after Darwin. Tax S, editor. Vol. 1. Chicago: University Chicago Press; 1959. pp. 349–380. [Google Scholar]
- 22.Crick FH. On protein synthesis. Symp. Soc. Exp. Biol. 1958;12:138–163. [PubMed] [Google Scholar]
- 23.Zuckerkandl E, Pauling L. In: Horizons in Biochemistry. Kasha M, Pullman B, editors. New York: Academic Press; 1962. pp. 189–225. [Google Scholar]
- 24.Zuckerkandl E, Pauling L. In: Evolving Gene and Proteins. Bryson V, Vogel HJ, editors. New York: Academic Press; 1965. pp. 97–166. [Google Scholar]
- 25.Dayhoff MO, Barker WC, McLaughlin PJ. Inferences from protein and nucleic acid sequences: early molecular evolution, divergence of kingdoms and rates of change. Orig. Life. 1974;5:311–330. [PubMed] [Google Scholar]
- 26.Eck RV, Dayhoff MO. Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science. 1966;152:363–366. doi: 10.1126/science.152.3720.363. [DOI] [PubMed] [Google Scholar]
- 27.Dayhoff MO, Barker WC, Hunt LT. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- 28.Woese CR. Bacterial evolution. Microbiol. Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl Acad. Sci. USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woese CR, Magrum LJ, Fox GE. Archaebacteria. J. Mol. Evol. 1978;11:245–251. doi: 10.1007/BF01734485. [DOI] [PubMed] [Google Scholar]
- 31.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 33.Pace NR. Time for a change. Nature. 2006;441:289. doi: 10.1038/441289a. [DOI] [PubMed] [Google Scholar]
- 34.Syvanen M. Molecular clocks and evolutionary relationships: possible distortions due to horizontal gene flow. J. Mol. Evol. 1987;26:16–23. doi: 10.1007/BF02111278. [DOI] [PubMed] [Google Scholar]
- 35.Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- 36.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- 37.King JL, Jukes TH. Non-Darwinian evolution. Science. 1969;164:788–798. doi: 10.1126/science.164.3881.788. [DOI] [PubMed] [Google Scholar]
- 38.Ohta T, Gillespie JH. Development of neutral and nearly neutral theories. Theor. Popul. Biol. 1996;49:128–142. doi: 10.1006/tpbi.1996.0007. [DOI] [PubMed] [Google Scholar]
- 39.Takahata N. On the overdispersed molecular clock. Genetics. 1987;116:169–179. doi: 10.1093/genetics/116.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cutler DJ. Understanding the overdispersed molecular clock. Genetics. 2000;154:1403–1417. doi: 10.1093/genetics/154.3.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner A. Robustness, evolvability, and neutrality. FEBS Lett. 2005;579:1772–1778. doi: 10.1016/j.febslet.2005.01.063. [DOI] [PubMed] [Google Scholar]
- 42.Thomas CA., Jr. The genetic organization of chromosomes. Annu. Rev. Genet. 1971;5:237–256. doi: 10.1146/annurev.ge.05.120171.001321. [DOI] [PubMed] [Google Scholar]
- 43.Hartl DL. Molecular melodies in high and low C. Nat. Rev. Genet. 2000;1:145–149. doi: 10.1038/35038580. [DOI] [PubMed] [Google Scholar]
- 44.Dawkins R. The Selfish Gene. Oxford: Oxford University Press; 1976. [Google Scholar]
- 45.Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 46.Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 47.McClintock B. The origin and behavior of mutable loci in maize. Proc. Natl Acad. Sci. USA. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Georgiev GP, Ilyin YV, Ryskov AP, Tchurikov NA, Yenikolopov GN, Gvozdev VA, Ananiev EV. Isolation of eukaryotic DNA fragments containing structural genes and the adjacent sequences. Science. 1977;195:394–397. doi: 10.1126/science.401545. [DOI] [PubMed] [Google Scholar]
- 49.Georgiev GP. Mobile genetic elements in animal cells and their biological significance. Eur. J. Biochem. 1984;145:203–220. doi: 10.1111/j.1432-1033.1984.tb08541.x. [DOI] [PubMed] [Google Scholar]
- 50.Finnegan DJ. Transposable elements in eukaryotes. Int. Rev. Cytol. 1985;93:281–326. doi: 10.1016/s0074-7696(08)61376-5. [DOI] [PubMed] [Google Scholar]
- 51.Ohno S. Evolution by Gene Duplication. Berlin-Heidelberg-New York: Springer-Verlag; 1970. [Google Scholar]
- 52.Fisher RA. The possible modification of the response of the wild type to recurrent mutations. Am. Nat. 1928;62:115–126. [Google Scholar]
- 53.Gould SJ, Lewontin RC. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proc. R. Soc. Lond. B Biol. Sci. 1979;205:581–598. doi: 10.1098/rspb.1979.0086. [DOI] [PubMed] [Google Scholar]
- 54.Gould SJ. The exaptive excellence of spandrels as a term and prototype. Proc. Natl Acad. Sci. USA. 1997;94:10750–10755. doi: 10.1073/pnas.94.20.10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 56.Haeckel E. The Wonders of Life: A Popular Study of Biological Philosophy. London: Watts & Co; 1904. [Google Scholar]
- 57.Cairns J, Stent GS, Watson JD, editors. Phage and the Origins of Molecular Biology. Cold Spring Harbor, NY: CSHL Press; 1966. [Google Scholar]
- 58.Woese CR. There must be a prokaryote somewhere: microbiology's search for itself. Microbiol. Rev. 1994;58:1–9. doi: 10.1128/mr.58.1.1-9.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Argos P, Kamer G, Nicklin MJ, Wimmer E. Similarity in gene organization and homology between proteins of animal picornaviruses and a plant comovirus suggest common ancestry of these virus families. Nucleic Acids Res. 1984;12:7251–7267. doi: 10.1093/nar/12.18.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kamer G, Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984;12:7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goldbach R. Genome similarities between plant and animal RNA viruses. Microbiol. Sci. 1987;4:197–202. [PubMed] [Google Scholar]
- 62.Koonin EV, Dolja VV. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 63.Mereschkowsky C. Uber Natur und Ursprung der Chromatophoren im Pflanzenreiche. Biol. Centralbl. 1905;25:593–604. [Google Scholar]
- 64.Sagan L. On the origin of mitosing cells. J. Theor. Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 65.Martin W, Hoffmeister M, Rotte C, Henze K. An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol. Chem. 2001;382:1521–1539. doi: 10.1515/BC.2001.187. [DOI] [PubMed] [Google Scholar]
- 66.Gray MW. The endosymbiont hypothesis revisited. Int. Rev. Cytol. 1992;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- 67.Gray MW, Burger G, Lang BF. The origin and early evolution of mitochondria. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-reviews1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koonin EV, Mushegian AR. Complete genome sequences of cellular life forms: glimpses of theoretical evolutionary genomics. Curr. Opin. Genet. Dev. 1996;6:757–762. doi: 10.1016/s0959-437x(96)80032-3. [DOI] [PubMed] [Google Scholar]
- 69.Koonin EV, Mushegian AR, Rudd KE. Sequencing and analysis of bacterial genomes. Curr. Biol. 1996;6:404–416. doi: 10.1016/s0960-9822(02)00508-0. [DOI] [PubMed] [Google Scholar]
- 70.Fraser CM, Eisen JA, Salzberg SL. Microbial genome sequencing. Nature. 2000;406:799–803. doi: 10.1038/35021244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eisen JA, Fraser CM. Phylogenomics: intersection of evolution and genomics. Science. 2003;300:1706–1707. doi: 10.1126/science.1086292. [DOI] [PubMed] [Google Scholar]
- 72.Nierman WC, Eisen JA, Fleischmann RD, Fraser CM. Genome data: what do we learn? Curr. Opin. Struct. Biol. 2000;10:343–348. doi: 10.1016/s0959-440x(00)00094-4. [DOI] [PubMed] [Google Scholar]
- 73.Brown JR. Genomic and phylogenetic perspectives on the evolution of prokaryotes. Syst. Biol. 2001;50:497–512. doi: 10.1080/10635150117729. [DOI] [PubMed] [Google Scholar]
- 74.Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Microevolutionary genomics of bacteria. Theor. Popul. Biol. 2002;61:435–447. doi: 10.1006/tpbi.2002.1588. [DOI] [PubMed] [Google Scholar]
- 75.Novichkov PS, Wolf YI, Dubchak I, Koonin EV. Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. J. Bacteriol. 2009;191:65–73. doi: 10.1128/JB.01237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2008;36:D475–479. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DeLong EF, Karl DM. Genomic perspectives in microbial oceanography. Nature. 2005;437:336–342. doi: 10.1038/nature04157. [DOI] [PubMed] [Google Scholar]
- 79.Karl DM. Microbial oceanography: paradigms, processes and promise. Nat. Rev. Microbiol. 2007;5:759–769. doi: 10.1038/nrmicro1749. [DOI] [PubMed] [Google Scholar]
- 80.Medina M. Genomes, phylogeny, and evolutionary systems biology. Proc. Natl Acad. Sci. USA. 2005;102(Suppl. 1):6630–6635. doi: 10.1073/pnas.0501984102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koonin EV, Wolf YI. Evolutionary systems biology: links between gene evolution and function. Curr. Opin. Biotechnol. 2006;17:481–487. doi: 10.1016/j.copbio.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 82.Koonin EV, Wolf YI. In: Evolutionary Genomics and Proteomics. Pagel M, Pomiankowski A, editors. Inc., Sunderland, MA: Sinauer Associates; 2008. pp. 11–25. [Google Scholar]
- 83.Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging generalizations after 13 years. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 85.Miller DJ, Ball EE. Cryptic complexity captured: the Nematostella genome reveals its secrets. Trends Genet. 2008;24:1–4. doi: 10.1016/j.tig.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 86.Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 87.Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Snel B, Bork P, Huynen MA. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 2002;12:17–25. doi: 10.1101/gr.176501. [DOI] [PubMed] [Google Scholar]
- 89.Mirkin BG, Fenner TI, Galperin MY, Koonin EV. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol. Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koonin EV. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nature Rev. Microbiol. 2003;1:127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- 91.Kunin V, Ouzounis CA. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 2003;13:1589–1594. doi: 10.1101/gr.1092603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mushegian A. Gene content of LUCA, the last universal common ancestor. Front. Biosci. 2008;13:4657–4666. doi: 10.2741/3031. [DOI] [PubMed] [Google Scholar]
- 93.Makarova KS, Sorokin AV, Novichkov PS, Wolf YI, Koonin EV. Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea. Biol. Direct. 2007;2:33. doi: 10.1186/1745-6150-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fedorov A, Merican AF, Gilbert W. Large-scale comparison of intron positions among animal, plant, and fungal genes. Proc. Natl Acad. Sci. USA. 2002;99:16128–16133. doi: 10.1073/pnas.242624899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 2003;13:1512–1517. doi: 10.1016/s0960-9822(03)00558-x. [DOI] [PubMed] [Google Scholar]
- 96.Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 97.Harris JK, Kelley ST, Spiegelman GB, Pace NR. The genetic core of the universal ancestor. Genome Res. 2003;13:407–412. doi: 10.1101/gr.652803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Charlebois RL, Doolittle WF. Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res. 2004;14:2469–2477. doi: 10.1101/gr.3024704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Koonin EV. Evolution of genome architecture. Int. J. Biochem. Cell Biol. 2009;41:298–306. doi: 10.1016/j.biocel.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mushegian AR, Koonin EV. Gene order is not conserved in bacterial evolution. Trends Genet. 1996;12:289–290. doi: 10.1016/0168-9525(96)20006-x. [DOI] [PubMed] [Google Scholar]
- 101.Itoh T, Takemoto K, Mori H, Gojobori T. Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol. 1999;16:332–346. doi: 10.1093/oxfordjournals.molbev.a026114. [DOI] [PubMed] [Google Scholar]
- 102.Eisen JA, Heidelberg JF, White O, Salzberg SL. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-6-research0011. RESEARCH0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tillier ER, Collins RA. Genome rearrangement by replication-directed translocation. Nat. Genet. 2000;26:195–197. doi: 10.1038/79918. [DOI] [PubMed] [Google Scholar]
- 104.Lawrence JG. Gene organization: selection, selfishness, and serendipity. Annu. Rev. Microbiol. 2003;57:419–440. doi: 10.1146/annurev.micro.57.030502.090816. [DOI] [PubMed] [Google Scholar]
- 105.Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- 106.Syvanen M, Kado CI, editors. Horizontal Gene Transfer. San Diego: Academic Press; 2002. [Google Scholar]
- 107.Bushman F. Lateral DNA Transfer: Mechanisms and Consequences. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 108.Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 109.Ochman H, Moran NA. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science. 2001;292:1096–1099. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- 110.Perna NT, Plunkett G., 3rd, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 111.Aravind L, Tatusov RL, Wolf YI, Walker DR, Koonin EV. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 1998;14:442–444. doi: 10.1016/s0168-9525(98)01553-4. [DOI] [PubMed] [Google Scholar]
- 112.Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 113.Lawrence JG, Hendrickson H. Lateral gene transfer: when will adolescence end? Mol. Microbiol. 2003;50:739–749. doi: 10.1046/j.1365-2958.2003.03778.x. [DOI] [PubMed] [Google Scholar]
- 114.Koonin EV. Horizontal gene transfer: the path to maturity. Mol. Microbiol. 2003;50:725–727. doi: 10.1046/j.1365-2958.2003.03808.x. [DOI] [PubMed] [Google Scholar]
- 115.Kurland CG, Canback B, Berg OG. Horizontal gene transfer: A critical view. Proc. Natl Acad. Sci. USA. 2003;100:9658–9662. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 117.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 118.Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl Acad. Sci. USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wellner A, Lurie MN, Gophna U. Complexity, connectivity, and duplicability as barriers to lateral gene transfer. Genome Biol. 2007;8:R156. doi: 10.1186/gb-2007-8-8-r156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brochier C, Philippe H, Moreira D. The evolutionary history of ribosomal protein RpS14: horizontal gene transfer at the heart of the ribosome. Trends Genet. 2000;16:529–533. doi: 10.1016/s0168-9525(00)02142-9. [DOI] [PubMed] [Google Scholar]
- 121.Makarova KS, Ponomarev VA, Koonin EV. Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-9-research0033. RESEARCH 0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Iyer LM, Koonin EV, Aravind L. Evolution of bacterial RNA polymerase: implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer. Gene. 2004;335:73–88. doi: 10.1016/j.gene.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 123.Lawrence J. Selfish operons: the evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr. Opin. Genet. Dev. 1999;9:642–648. doi: 10.1016/s0959-437x(99)00025-8. [DOI] [PubMed] [Google Scholar]
- 124.Lawrence JG. Selfish operons and speciation by gene transfer. Trends Microbiol. 1997;5:355–359. doi: 10.1016/S0966-842X(97)01110-4. [DOI] [PubMed] [Google Scholar]
- 125.Andersson JO. Lateral gene transfer in eukaryotes. Cell Mol. Life Sci. 2005;62:1182–1197. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kondrashov FA, Koonin EV, Morgunov IG, Finogenova TV, Kondrashova MN. Evolution of glyoxylate cycle enzymes in Metazoa: evidence of multiple horizontal transfer events and pseudogene formation. Biol. Direct. 2006;1:31. doi: 10.1186/1745-6150-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hotopp JC, Clark ME, Oliveira DC, Foster JM, Fischer P, Torres MC, Giebel JD, Kumar N, Ishmael N, Wang S, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- 128.Nikoh N, Tanaka K, Shibata F, Kondo N, Hizume M, Shimada M, Fukatsu T. Wolbachia genome integrated in an insect chromosome: evolution and fate of laterally transferred endosymbiont genes. Genome Res. 2008;18:272–280. doi: 10.1101/gr.7144908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.de Koning AP, Brinkman FS, Jones SJ, Keeling PJ. Lateral gene transfer and metabolic adaptation in the human parasite Trichomonas vaginalis. Mol. Biol. Evol. 2000;17:1769–1773. doi: 10.1093/oxfordjournals.molbev.a026275. [DOI] [PubMed] [Google Scholar]
- 130.Rogers MB, Watkins RF, Harper JT, Durnford DG, Gray MW, Keeling PJ. A complex and punctate distribution of three eukaryotic genes derived by lateral gene transfer. BMC Evol. Biol. 2007;7:89. doi: 10.1186/1471-2148-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Andersson JO, Sjogren AM, Horner DS, Murphy CA, Dyal PL, Svard SG, Logsdon J.M., Jr., Ragan MA, Hirt RP, Roger AJ. A genomic survey of the fish parasite Spironucleus salmonicida indicates genomic plasticity among diplomonads and significant lateral gene transfer in eukaryote genome evolution. BMC Genomics. 2007;8:51. doi: 10.1186/1471-2164-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Embley TM. Multiple secondary origins of the anaerobic lifestyle in eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006;361:1055–1067. doi: 10.1098/rstb.2006.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 134.Esser C, Ahmadinejad N, Wiegand C, Rotte C, Sebastiani F, Gelius-Dietrich G, Henze K, Kretschmann E, Richly E, Leister D, et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 2004;21:1643–1660. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- 135.Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV. The deep archaeal roots of eukaryotes. Mol. Biol. Evol. 2008;25:1619–1630. doi: 10.1093/molbev/msn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Esser C, Martin W, Dagan T. The origin of mitochondria in light of a fluid prokaryotic chromosome model. Biol. Lett. 2007;3:180–184. doi: 10.1098/rsbl.2006.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kurland CG, Collins LJ, Penny D. Genomics and the irreducible nature of eukaryote cells. Science. 2006;312:1011–1014. doi: 10.1126/science.1121674. [DOI] [PubMed] [Google Scholar]
- 138.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentation. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 139.Poole AM, Penny D. Evaluating hypotheses for the origin of eukaryotes. Bioessays. 2007;29:74–84. doi: 10.1002/bies.20516. [DOI] [PubMed] [Google Scholar]
- 140.Dagan T, Martin W. Testing hypotheses without considering predictions. Bioessays. 2007;29:500–503. doi: 10.1002/bies.20566. [DOI] [PubMed] [Google Scholar]
- 141.Martin W, Muller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 142.Poole A, Penny D. Eukaryote evolution: engulfed by speculation. Nature. 2007;447:913. doi: 10.1038/447913a. [DOI] [PubMed] [Google Scholar]
- 143.Rivera MC, Lake JA. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature. 2004;431:152–155. doi: 10.1038/nature02848. [DOI] [PubMed] [Google Scholar]
- 144.Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl Acad. Sci. USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nosenko T, Bhattacharya D. Horizontal gene transfer in chromalveolates. BMC Evol. Biol. 2007;7:173. doi: 10.1186/1471-2148-7-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 147.Bapteste E, Susko E, Leigh J, MacLeod D, Charlebois RL, Doolittle WF. Do orthologous gene phylogenies really support tree-thinking? BMC Evol. Biol. 2005;5:33. doi: 10.1186/1471-2148-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dagan T, Martin W. The tree of one percent. Genome Biol. 2006;7:118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Doolittle WF, Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc. Natl Acad. Sci. USA. 2007;104:2043–2049. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wolf YI, Rogozin IB, Grishin NV, Koonin EV. Genome trees and the tree of life. Trends Genet. 2002;18:472–479. doi: 10.1016/s0168-9525(02)02744-0. [DOI] [PubMed] [Google Scholar]
- 151.Koonin EV. The biological Big Bang model for the major transitions in evolution. Biol. Direct. 2007;2:21. doi: 10.1186/1745-6150-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Langer M, Gabor EM, Liebeton K, Meurer G, Niehaus F, Schulze R, Eck J, Lorenz P. Metagenomics: an inexhaustible access to nature's diversity. Biotechnol. J. 2006;1:815–821. doi: 10.1002/biot.200600111. [DOI] [PubMed] [Google Scholar]
- 153.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 154.Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Delwart EL. Viral metagenomics. Rev. Med. Virol. 2007;17:115–131. doi: 10.1002/rmv.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Edwards RA, Rohwer F. Viral metagenomics. Nat. Rev. Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 158.Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 159.Prangishvili D, Garrett RA, Koonin EV. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 2006;117:52–67. doi: 10.1016/j.virusres.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 160.Glazko G, Makarenkov V, Liu J, Mushegian A. Evolutionary history of bacteriophages with double-stranded DNA genomes. Biol. Direct. 2007;2:36. doi: 10.1186/1745-6150-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Goodier JL, Kazazian H.H., Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 162.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 163.Forterre P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 2006;117:5–16. doi: 10.1016/j.virusres.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 164.Koonin EV, Senkevich TG, Dolja VV. The ancient Virus World and evolution of cells. Biol. Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Swain A, Coffin JM. Mechanism of transduction by retroviruses. Science. 1992;255:841–845. doi: 10.1126/science.1371365. [DOI] [PubMed] [Google Scholar]
- 166.Chen J, Novick RP. Phage-mediated intergeneric transfer of toxin genes. Science. 2009;323:139–141. doi: 10.1126/science.1164783. [DOI] [PubMed] [Google Scholar]
- 167.Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 168.Polavarapu N, Marino-Ramirez L, Landsman D, McDonald JF, Jordan IK. Evolutionary rates and patterns for human transcription factor binding sites derived from repetitive DNA. BMC Genomics. 2008;9:226. doi: 10.1186/1471-2164-9-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Piriyapongsa J, Rutledge MT, Patel S, Borodovsky M, Jordan IK. Evaluating the protein coding potential of exonized transposable element sequences. Biol. Direct. 2007;2:31. doi: 10.1186/1745-6150-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hall TM, Porter JA, Young KE, Koonin EV, Beachy PA, Leahy DJ. Crystal structure of a Hedgehog autoprocessing domain: homology between Hedgehog and self-splicing proteins. Cell. 1997;91:85–97. doi: 10.1016/s0092-8674(01)80011-8. [DOI] [PubMed] [Google Scholar]
- 171.Burglin TR. Evolution of hedgehog and hedgehog-related genes, their origin from Hog proteins in ancestral eukaryotes and discovery of a novel Hint motif. BMC Genomics. 2008;9:127. doi: 10.1186/1471-2164-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.McGeoch AT, Bell SD. Extra-chromosomal elements and the evolution of cellular DNA replication machineries. Nat. Rev. Mol. Cell. Biol. 2008;9:569–574. doi: 10.1038/nrm2426. [DOI] [PubMed] [Google Scholar]
- 174.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Koonin EV, Wolf YI, Nagasaki K, Dolja VV. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 2008;6:925–939. doi: 10.1038/nrmicro2030. [DOI] [PubMed] [Google Scholar]
- 176.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pereto J, Lopez-Garcia P, Moreira D. Ancestral lipid biosynthesis and early membrane evolution. Trends Biochem. Sci. 2004;29:469–477. doi: 10.1016/j.tibs.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 178.Woese C. The universal ancestor. Proc. Natl Acad. Sci. USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Martin W, Russell MJ. On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003;358:59–83. doi: 10.1098/rstb.2002.1183. discussion 83–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Koonin EV, Martin W. On the origin of genomes and cells within inorganic compartments. Trends Genet. 2005;21:647–654. doi: 10.1016/j.tig.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Koonin EV. On the origin of cells and viruses: primordial virus world scenario. Ann. NY Acad. Sci. 2009 doi: 10.1111/j.1749-6632.2009.04992.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Glansdorff N, Xu Y, Labedan B. The last universal common ancestor: emergence, constitution and genetic legacy of an elusive forerunner. Biol. Direct. 2008;3:29. doi: 10.1186/1745-6150-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 184.Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc. Natl Acad. Sci. USA. 2007;104(Suppl. 1):8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kreitman M. Methods to detect selection in populations with applications to the human. Annu. Rev. Genomics Hum. Genet. 2000;1:539–559. doi: 10.1146/annurev.genom.1.1.539. [DOI] [PubMed] [Google Scholar]
- 186.Zhang J. Frequent false detection of positive selection by the likelihood method with branch-site models. Mol. Biol. Evol. 2004;21:1332–1339. doi: 10.1093/molbev/msh117. [DOI] [PubMed] [Google Scholar]
- 187.Yampolsky LY, Kondrashov FA, Kondrashov AS. Distribution of the strength of selection against amino acid replacements in human proteins. Hum. Mol. Genet. 2005;14:3191–3201. doi: 10.1093/hmg/ddi350. [DOI] [PubMed] [Google Scholar]
- 188.Kosiol C, Vinar T, da Fonseca RR, Hubisz MJ, Bustamante CD, Nielsen R, Siepel A. Patterns of positive selection in six Mammalian genomes. PLoS Genet. 2008;4:e1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 190.Sawyer SA, Parsch J, Zhang Z, Hartl DL. Prevalence of positive selection among nearly neutral amino acid replacements in Drosophila. Proc. Natl Acad. Sci. USA. 2007;104:6504–6510. doi: 10.1073/pnas.0701572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Castillo-Davis CI, Kondrashov FA, Hartl DL, Kulathinal RJ. The functional genomic distribution of protein divergence in two animal phyla: coevolution, genomic conflict, and constraint. Genome Res. 2004;14:802–811. doi: 10.1101/gr.2195604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zuckerkandl E. Why so many noncoding nucleotides? The eukaryote genome as an epigenetic machine. Genetica. 2002;115:105–129. doi: 10.1023/a:1016080316076. [DOI] [PubMed] [Google Scholar]
- 193.Pheasant M, Mattick JS. Raising the estimate of functional human sequences. Genome Res. 2007;17:1245–1253. doi: 10.1101/gr.6406307. [DOI] [PubMed] [Google Scholar]
- 194.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 195.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 196.Katzman S, Kern AD, Bejerano G, Fewell G, Fulton L, Wilson RK, Salama SR, Haussler D. Human genome ultraconserved elements are ultraselected. Science. 2007;317:915. doi: 10.1126/science.1142430. [DOI] [PubMed] [Google Scholar]
- 197.Glazko GV, Koonin EV, Rogozin IB, Shabalina SA. A significant fraction of conserved noncoding DNA in human and mouse consists of predicted matrix attachment regions. Trends Genet. 2003;19:119–124. doi: 10.1016/S0168-9525(03)00016-7. [DOI] [PubMed] [Google Scholar]
- 198.Linnemann AK, Platts AE, Krawetz SA. Differential nuclear scaffold/matrix attachment marks expressed genes. Hum. Mol. Genet. 2009;18:645–654. doi: 10.1093/hmg/ddn394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lunter G, Ponting CP, Hein J. Genome-wide identification of human functional DNA using a neutral indel model. PLoS Comput. Biol. 2006;2:e5. doi: 10.1371/journal.pcbi.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- 201.Halligan DL, Keightley PD. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res. 2006;16:875–884. doi: 10.1101/gr.5022906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Haddrill PR, Bachtrog D, Andolfatto P. Positive and negative selection on noncoding DNA in Drosophila simulans. Mol. Biol. Evol. 2008;25:1825–1834. doi: 10.1093/molbev/msn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Huynen MA, van Nimwegen E. The frequency distribution of gene family sizes in complete genomes. Mol. Biol. Evol. 1998;15:583–589. doi: 10.1093/oxfordjournals.molbev.a025959. [DOI] [PubMed] [Google Scholar]
- 204.Karev GP, Wolf YI, Rzhetsky AY, Berezovskaya FS, Koonin EV. Birth and death of protein domains: a simple model of evolution explains power law behavior. BMC Evol. Biol. 2002;2:18. doi: 10.1186/1471-2148-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kondrashov FA, Koonin EV. Evolution of alternative splicing: deletions, insertions and origin of functional parts of proteins from intron sequences. Trends Genet. 2003;19:115–119. doi: 10.1016/S0168-9525(02)00029-X. [DOI] [PubMed] [Google Scholar]
- 206.Long M, Betran E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 207.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 208.Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-research0008. RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lynch M, Katju V. The altered evolutionary trajectories of gene duplicates. Trends Genet. 2004;20:544–549. doi: 10.1016/j.tig.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 211.He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Scannell DR, Wolfe KH. A burst of protein sequence evolution and a prolonged period of asymmetric evolution follow gene duplication in yeast. Genome Res. 2008;18:137–147. doi: 10.1101/gr.6341207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 214.Makarova KS, Wolf YI, Mekhedov SL, Mirkin BG, Koonin EV. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res. 2005;33:4626–4638. doi: 10.1093/nar/gki775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hoegg S, Meyer A. Hox clusters as models for vertebrate genome evolution. Trends Genet. 2005;21:421–424. doi: 10.1016/j.tig.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 216.Wagner GP, Amemiya C, Ruddle F. Hox cluster duplications and the opportunity for evolutionary novelties. Proc. Natl Acad. Sci. USA. 2003;100:14603–14606. doi: 10.1073/pnas.2536656100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Freeling M. The evolutionary position of subfunctionalization, downgraded. Genome Dyn. 2008;4:25–40. doi: 10.1159/000126004. [DOI] [PubMed] [Google Scholar]
- 218.Scannell DR, Butler G, Wolfe KH. Yeast genome evolution–the origin of the species. Yeast. 2007;24:929–942. doi: 10.1002/yea.1515. [DOI] [PubMed] [Google Scholar]
- 219.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 220.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Durand D. Vertebrate evolution: doubling and shuffling with a full deck. Trends Genet. 2003;19:2–5. doi: 10.1016/s0168-9525(02)00008-2. [DOI] [PubMed] [Google Scholar]
- 222.McLysaght A, Hokamp K, Wolfe KH. Extensive genomic duplication during early chordate evolution. Nat Genet. 2002;31:200–204. doi: 10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- 223.Panopoulou G, Hennig S, Groth D, Krause A, Poustka AJ, Herwig R, Vingron M, Lehrach H. New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes. Genome Res. 2003;13:1056–1066. doi: 10.1101/gr.874803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Soltis DE, Bell CD, Kim S, Soltis PS. Origin and early evolution of angiosperms. Ann. N Y Acad. Sci. 2008;1133:3–25. doi: 10.1196/annals.1438.005. [DOI] [PubMed] [Google Scholar]
- 225.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 226.Semon M, Wolfe KH. Consequences of genome duplication. Curr. Opin. Genet. Dev. 2007;17:505–512. doi: 10.1016/j.gde.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 227.Mendell JE, Clements KD, Choat JH, Angert ER. Extreme polyploidy in a large bacterium. Proc. Natl Acad. Sci. USA. 2008;105:6730–6734. doi: 10.1073/pnas.0707522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Tobiason DM, Seifert HS. The obligate human pathogen, Neisseria gonorrhoeae, is polyploid. PLoS Biol. 2006;4:e185. doi: 10.1371/journal.pbio.0040185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Adami C. What is complexity? Bioessays. 2002;24:1085–1094. doi: 10.1002/bies.10192. [DOI] [PubMed] [Google Scholar]
- 230.Koonin EV. A non-adaptationist perspective on evolution of genomic complexity or the continued dethroning of man. Cell Cycle. 2004;3:280–285. [PubMed] [Google Scholar]
- 231.Sorek R, Shamir R, Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 232.Artamonova II, Gelfand MS. Comparative genomics and evolution of alternative splicing: the pessimists' science. Chem. Rev. 2007;107:3407–3430. doi: 10.1021/cr068304c. [DOI] [PubMed] [Google Scholar]
- 233.Park JW, Graveley BR. Complex alternative splicing. Adv. Exp. Med. Biol. 2007;623:50–63. doi: 10.1007/978-0-387-77374-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 235.Irimia M, Penny D, Roy SW. Coevolution of genomic intron number and splice sites. Trends Genet. 2007;23:321–325. doi: 10.1016/j.tig.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 236.Jaillon O, Bouhouche K, Gout JF, Aury JM, Noel B, Saudemont B, Nowacki M, Serrano V, Porcel BM, Segurens B, et al. Translational control of intron splicing in eukaryotes. Nature. 2008;451:359–362. doi: 10.1038/nature06495. [DOI] [PubMed] [Google Scholar]
- 237.Roy SW. Intron-rich ancestors. Trends Genet. 2006;22:468–471. doi: 10.1016/j.tig.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 238.Carmel L, Wolf YI, Rogozin IB, Koonin EV. Three distinct modes of intron dynamics in the evolution of eukaryotes. Genome Res. 2007;17:1034–1044. doi: 10.1101/gr.6438607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Csuros M, Rogozin IB, Koonin EV. Extremely intron-rich genes in the alveolate ancestors inferred with a flexible maximum-likelihood approach. Mol. Biol. Evol. 2008;25:903–911. doi: 10.1093/molbev/msn039. [DOI] [PubMed] [Google Scholar]
- 240.Lynch M, Kewalramani A. Messenger RNA surveillance and the evolutionary proliferation of introns. Mol. Biol. Evol. 2003;20:563–571. doi: 10.1093/molbev/msg068. [DOI] [PubMed] [Google Scholar]
- 241.Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, Bartels D, Bekel T, Beyer S, Bode E, et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat. Biotechnol. 2007;25:1281–1289. doi: 10.1038/nbt1354. [DOI] [PubMed] [Google Scholar]
- 242.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl Acad. Sci. USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Van Nimwegen E. In: Power Laws, Sacle-Free Networks and Genome Biology. Koonin EV, Wolf YI, Karev GP, editors. Georgetown, TX: Landes Bioscience; 2006. pp. 236–253. [Google Scholar]
- 246.Ranea JA, Grant A, Thornton JM, Orengo CA. Microeconomic principles explain an optimal genome size in bacteria. Trends Genet. 2005;21:21–25. doi: 10.1016/j.tig.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 247.van Nimwegen E. Scaling laws in the functional content of genomes. Trends Genet. 2003;19:479–484. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- 248.Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 2005;13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, et al. Comparative genomics of the lactic acid bacteria. Proc. Natl Acad. Sci. USA. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Gould SJ. Full House: The Spread of Excellence from Plato to Darwin. New York: Three Rivers Press; 1997. [Google Scholar]
- 251.Jordan IK, Marino-Ramirez L, Koonin EV. Evolutionary significance of gene expression divergence. Gene. 2005;345:119–126. doi: 10.1016/j.gene.2004.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Liao BY, Zhang J. Evolutionary conservation of expression profiles between human and mouse orthologous genes. Mol. Biol. Evol. 2006;23:530–540. doi: 10.1093/molbev/msj054. [DOI] [PubMed] [Google Scholar]
- 253.Khaitovich P, Enard W, Lachmann M, Paabo S. Evolution of primate gene expression. Nat. Rev. Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- 254.Krylov DM, Wolf YI, Rogozin IB, Koonin EV. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–2235. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Wolf YI, Carmel L, Koonin EV. Unifying measures of gene function and evolution. Proc. Biol. Sci. 2006;273:1507–1515. doi: 10.1098/rspb.2006.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wilson AC, Carlson SS, White TJ. Biochemical evolution. Annu. Rev. Biochem. 1977;46:573–639. doi: 10.1146/annurev.bi.46.070177.003041. [DOI] [PubMed] [Google Scholar]
- 257.Hurst LD, Smith NG. Do essential genes evolve slowly? Curr. Biol. 1999;9:747–750. doi: 10.1016/s0960-9822(99)80334-0. [DOI] [PubMed] [Google Scholar]
- 258.Hirsh AE, Fraser HB. Protein dispensability and rate of evolution. Nature. 2001;411:1046–1049. doi: 10.1038/35082561. [DOI] [PubMed] [Google Scholar]
- 259.Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002;12:962–968. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Pal C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Liao BY, Zhang J. Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during mammalian evolution. Mol. Biol. Evol. 2006;23:1119–1128. doi: 10.1093/molbev/msj119. [DOI] [PubMed] [Google Scholar]
- 263.Pal C, Papp B, Lercher MJ. An integrated view of protein evolution. Nat. Rev. Genet. 2006;7:337–348. doi: 10.1038/nrg1838. [DOI] [PubMed] [Google Scholar]
- 264.McInerney JO. The causes of protein evolutionary rate variation. Trends Ecol. Evol. 2006;21:230–232. doi: 10.1016/j.tree.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 265.Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc. Natl Acad. Sci. USA. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Drummond DA, Raval A, Wilke CO. A single determinant dominates the rate of yeast protein evolution. Mol. Biol. Evol. 2006;23:327–337. doi: 10.1093/molbev/msj038. [DOI] [PubMed] [Google Scholar]
- 267.Makalowski W, Boguski MS. Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences. Proc. Natl Acad. Sci. USA. 1998;95:9407–9412. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Jordan IK, Marino-Ramirez L, Wolf YI, Koonin EV. Conservation and coevolution in the scale-free human gene coexpression network. Mol. Biol. Evol. 2004;21:2058–2070. doi: 10.1093/molbev/msh222. [DOI] [PubMed] [Google Scholar]
- 269.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Wolf MY, Wolf YI, Koonin EV. Comparable contributions of structural-functional constraints and expression level to the rate of protein sequence evolution. Biol. Direct. 2008;3:40. doi: 10.1186/1745-6150-3-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Grishin NV, Wolf YI, Koonin EV. From complete genomes to measures of substitution rate variability within and between proteins. Genome Res. 2000;10:991–1000. doi: 10.1101/gr.10.7.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- 273.Molina N, van Nimwegen E. The evolution of domain-content in bacterial genomes. Biol. Direct. 2008;3:51. doi: 10.1186/1745-6150-3-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.O'Malley MA, Boucher Y. Paradigm change in evolutionary microbiology. Stud. Hist. Philos. Biol. Biomed. Sci. 2005;36:183–208. doi: 10.1016/j.shpsc.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 275.Kelley L, Scott M. The evolution of biology. A shift towards the engineering of prediction-generating tools and away from traditional research practice. EMBO Rep. 2008;9:1163–1167. doi: 10.1038/embor.2008.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Rokas A, Carroll SB. Bushes in the tree of life. PLoS Biol. 2006;4:e352. doi: 10.1371/journal.pbio.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]