Development of a BACarray translational profiling approach for the molecular characterization of CNS cell types (original) (raw)
. Author manuscript; available in PMC: 2009 Jun 16.
Summary
The cellular heterogeneity of the brain confounds efforts to elucidate the biological properties of distinct neuronal populations. We have now developed a new ‘BACarray’ methodology, based on affinity purification of polysomal mRNAs from genetically defined cell populations. The utility of this approach is illustrated by the comparative analysis of four types of neurons, revealing hundreds of genes that distinguish these four cell populations. Even two morphologically indistinguishable subclasses of MSNs display vastly different translational profiles. Striatopallidal neurons are characterized by a strong and cell-specific release of intracellular Ca2+ in response to sphingosine 1-phosphate, consistent with their selective expression of Gpr6. In contrast, striatonigral neurons demonstrate a selective cell-specific increase in GABAA receptor subunits in response to chronic cocaine treatment. BACarray translational profiling is a generalizable method useful for the identification of molecular changes in any genetically defined cell type in response to genetic alterations, disease, or pharmacological perturbations.
Introduction
A century ago, Ramón y Cajal used Golgi staining to show that distinct cells called neurons comprise the mammalian central nervous system (CNS) (Ramón y Cajal et al., 1899). Today, hundreds of neuronal subtypes have been defined, but a molecular description of each subtype has been hampered by the same problems faced by Ramón y Cajal and his contemporaries: neuronal subtypes are highly heterogeneous and intermixed. Numerous attempts to extend microarray analysis of gene expression to defined cell populations in the CNS have relied upon the physical enrichment of target cell populations using laser-capture microdissection (LCM) or Fluorescence Activated Cell Sorting (FACS) of acutely dissociated primary neurons. Unfortunately, these studies have been limited by stresses introduced during cellular isolation procedures, adaptations which occur upon loss of tissue-intrinsic signals, and the technical challenges associated with RNA purification from fixed tissue. To circumvent these problems, we have developed a direct, rapid affinity purification strategy for isolation of polysomal RNA from genetically targeted cell types.
We describe here a new BACarray methodology, which readily and reproducibly identifies translated mRNAs in any cell type of interest. This methodology involves expression of an EGFP-L10a ribosomal transgene, which enables tagging of polysomes for immunoaffinity purification of mRNA, in specific cell populations using Bacterial Artificial Chromosome (BAC) transgenic mice, allowing CNS translational profiling from whole animals. We illustrate the power of this approach in a study of four distinct neuronal populations. These include striatonigral and striatopallidal medium spiny neurons (MSNs), which are intermixed, indistinguishable in somato-dendritic morphology, and of major interest due to their role in the etiology of various neurological and psychiatric diseases, including Parkinson’s disease, schizophrenia, attention deficit hyperactivity disorder, drug addiction, and Huntington’s disease. In the accompanying paper (Doyle, Dougherty, et al., 2008), we describe the generation, characterization, and analysis of multiple additional BACarray lines as a resource for studies on a wide variety of CNS cell types.
Results
The BACarray approach and methodology
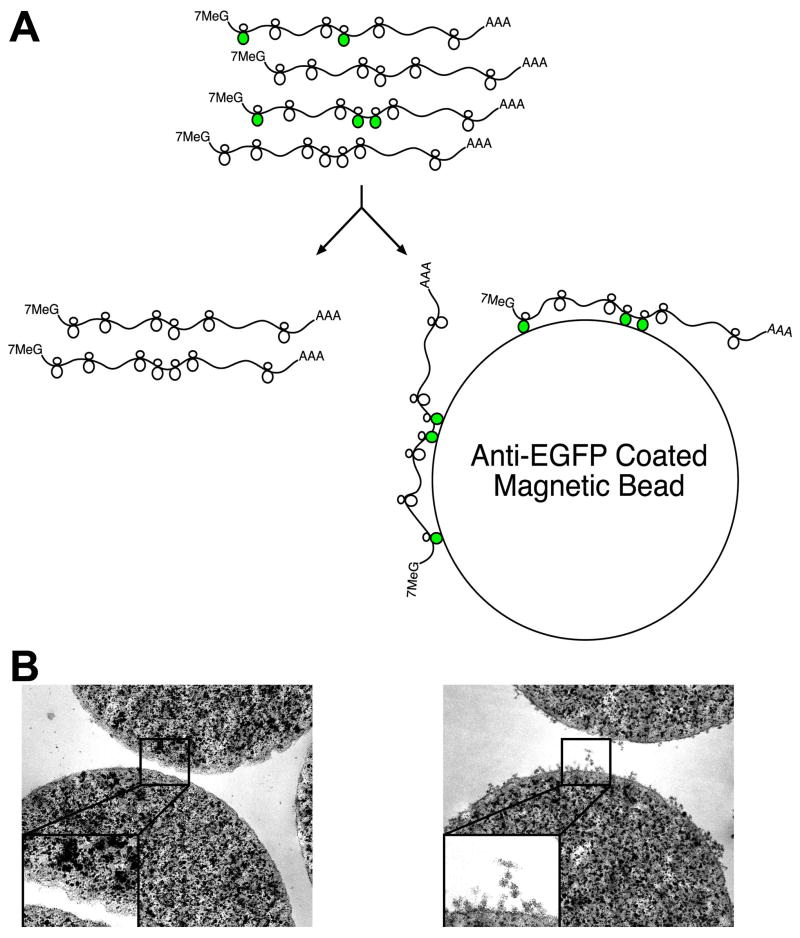
Because all mRNAs translated into protein are at one point attached to a ribosome or polyribosome complex (polysome), we reasoned that an affinity tag fused to a ribosomal protein would allow isolation of bound mRNAs. We therefore screened fusions of ribosomal proteins with Enhanced Green Fluorescent Protein (EGFP) for efficient incorporation into polysomes to provide an immunoaffinity tag for all translated cellular mRNAs (Schematic: Figure 1A). EGFP was chosen because preliminary screens using small epitope tags were unsatisfactory, and because visualization of EGFP fluorescence provides a simple assay for proper expression and localization of the fusion protein. After testing dozens of candidate ribosomal protein fusions, EGFP fused to the N-terminus of the large subunit ribosomal protein L10a (EGFP-L10a) was chosen because its nucleolar and cytoplasmic localization was consistent with incorporation into intact ribosomes, and because immunoelectron microscopy data demonstrated its presence on polysomes (Figure S1 and data not shown). Prior to the production of BACarray transgenic mice, preliminary studies in HEK293T cells transfected with EGFP-L10a achieved rapid and specific immunoaffinity purification of polysomes (Figure 1B), with cultures in which approximately 30% of cells expressed EGFP-L10a yielding an approximate 10% overall co-purification of untagged ribosomal proteins and ribosomal RNA, and the recovery of translated, but not untranslated, mRNAs (Table S1 and Figure S2). As a further benchmark of the technique, measurements of the well-documented shift in translational efficiency of Ferritin mRNA in response to iron treatment were comparable using the BACarray methodology or traditional polysome gradient methods (Figure S2).
Figure 1. The BACarray Methodology.
(A) Schematic of affinity purification of EGFP-tagged polysomes (originating from the target cell population; green polysomes) using anti-GFP antibody-coated beads. (B) Transmission electron micrographs of anti-GFP coated magnetic beads after incubation with extracts taken from HEK293T cells transfected with an empty vector (left panel) or the EGFP-L10a construct (right panel); images acquired at 50,000x magnification, inserts enlarged by a factor of 2.3x.
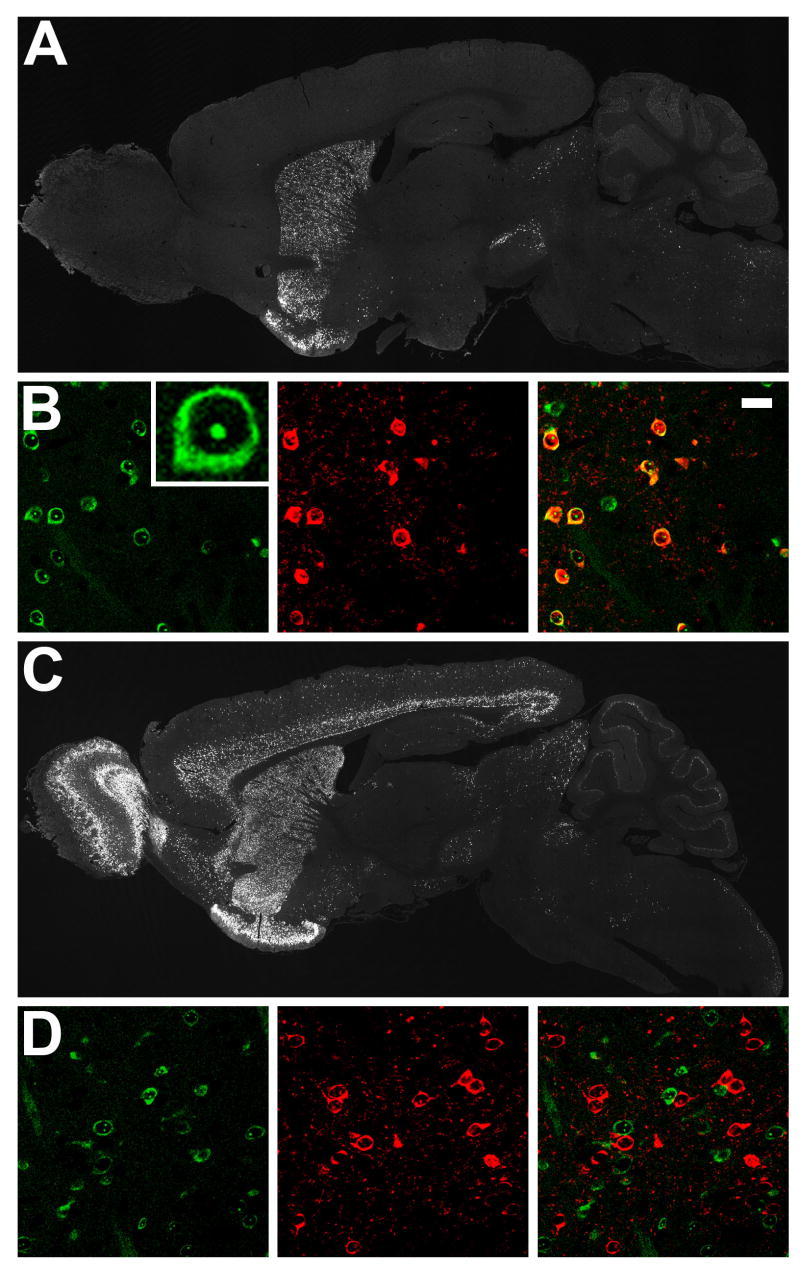
To genetically target expression of EGFP-L10a to defined CNS cell populations, we generated BAC transgenic mice. A large number of BAC vectors driving expression in specific CNS cell types have been characterized as part of the Gene Expression Nervous System Atlas (GENSAT) project (Gong et al., 2003). To tag polysomes in striatonigral and striatopallidal cells of the mouse striatum, homologous recombination in bacteria was used to place EGFP-L10a under the control of either the D2 receptor (striatopallidal) or D1 receptor (striatonigral) loci in the appropriate BACs. Striatonigral MSNs send projection axons directly to the output nuclei of the basal ganglia, i.e. the substantia nigra and the internal segment of the globus pallidus (the entopeduncular nucleus in rodents), while striatopallidal MSNs send projection axons to the external segment of the globus pallidus. Mouse lines bearing the BACarray transgenes were generated and screened by immunohistochemistry for appropriate expression of the transgene, as judged by known D1 and D2 receptor expression patterns. The D2 BACarray line CP101 showed highest transgenic EGFP-L10a expression in the dorsal and ventral striatum, olfactory tubercle, and hippocampus, as well as in the substantia nigra pars compacta and ventral tegmental area, as expected due to D2 autoreceptor expression in dopaminergic cells (Figure 2A). The D1 BACarray line CP73 showed highest transgenic EGFP-L10a expression in the dorsal and ventral striatum, olfactory bulb, olfactory tubercle, and cortical layers 5 and 6 (Figure 2C). As expected for a ribosomal protein fusion, EGFP fluorescence localized to the nucleoli and cytoplasm (Figure 2B). EGFP direct fluorescence coincident with enkephalin immunohistochemical detection (striatopallidal cell marker) was observed in striatal cells from the D2 BACarray line but not the D1 BACarray line (Figure 2B and 2D), verifying correct BAC-mediated cell-type expression. Velocity sedimentation analysis of polysome complexes isolated from striatal extracts of both BACarray lines confirmed incorporation of the EGFP-L10a fusion protein into functional polysomes in vivo (Figure S3 and data not shown).
Figure 2. Expression of EGFP-L10a in D1 and D2 BACarray lines.
(A) Immunohistochemistry to EGFP in adult sagittal sections from the D2 BACarray line CP101. (B) Characterization of D2 BACarray line CP101 striatal MSN cells: direct EGFP fluorescence (left panel with high-magnification image insert); enkephalin immunohistochemical staining (middle panel); merge (right panel, with 20 μm scale bar). (C) Immunohistochemistry to EGFP in adult sagittal sections from the D1 BACarray line CP73. (D) Characterization of D1 BACarray line CP73 striatal MSN cells: direct EGFP fluorescence (left panel); enkephalin immunohistochemical staining (middle panel); merge (right panel).
We next developed procedures for rapid extraction and immunoaffinity purification of the EGFP-tagged polyribosome complexes from intact brain tissue, which proved substantially more challenging than from transfected cells in culture. However, after several optimization steps (see Experimental Procedures), highly purified RNA was consistently obtained from BACarray mouse brain tissue (Figure 3). Key steps of the purification protocol include rapid manual dissection and homogenization of the tissue, inclusion of magnesium and cycloheximide in the lysis buffer to maintain ribosomal subunits on mRNA during purification, inhibition of endogenous RNase activity, solubilization of rough endoplasmic reticulum-bound polysomes under non-denaturing conditions, use of high-affinity anti-EGFP antibodies, and the addition of high-salt washes after immunoaffinity purification to reduce background.
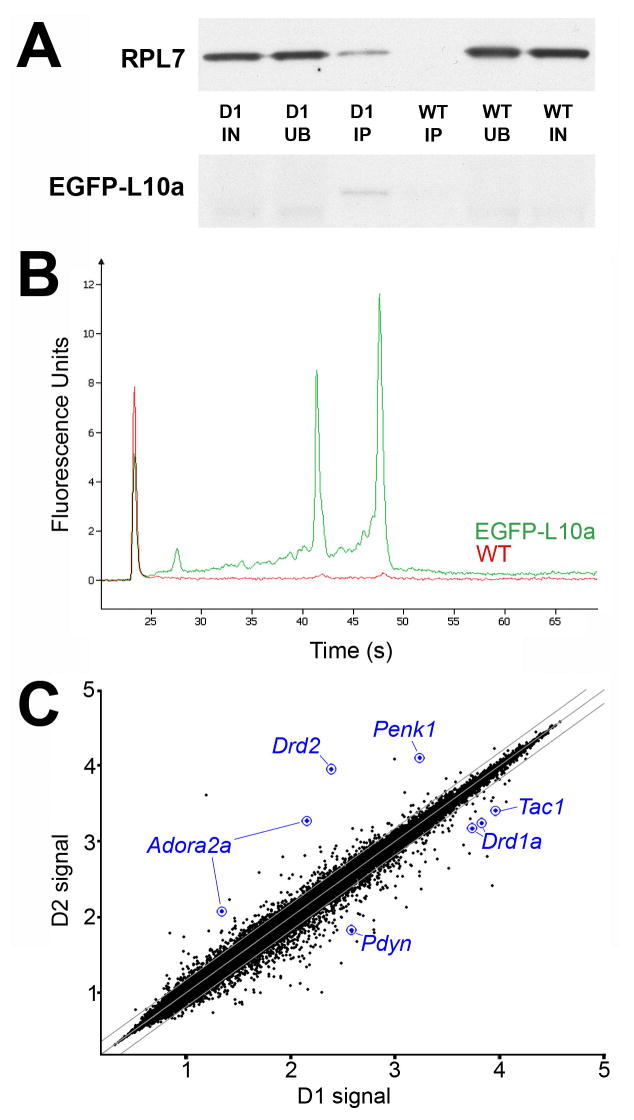
Figure 3. Protein and mRNA purification from BACarray lines.
(A) Representative purification of EGFP-tagged L10a and co-purification of untagged ribosomal protein L7 from D1 BACarray animals but not wild-type littermates (D1, samples from D1 BACarray mice; WT, samples from wild-type littermates; IN, 1% Input; UB, 1% Unbound; IP, 6.5% Immunoaffinity purified sample). EGFP-L10a signal is only present in the D1 IP lane because the IP samples were more concentrated relative to IN and UB. (B) Representative purification of 18S and 28S rRNA from D1 BACarray transgenic animals (green) but not wild type littermates (red) as detected by Bioanalyzer PicoChips (Agilent Technologies). 28S rRNA runs at ~47 sec, 18S rRNA runs at ~43 sec, and the Picochip marker peak runs at ~23 sec. (C) Normalized expression values from Affymetrix Mouse Genome 430 2.0 arrays are plotted for D1 and D2 BACarray samples. Middle diagonal line represents equal expression, and lines to each side represent 1.5-fold enrichment in either cell population. Axes are labelled for expression in powers of 10. The probesets of well-studied genes known to be differentially expressed are represented in blue.
BACarray profiling of striatonigral and striatopallidal MSNs
Translational profiling analysis was performed with immunoaffinity-purified mRNA from adult striatonigral or striatopallidal BACarray mice. Following two rounds of in vitro transcription, biotin-labeled antisense RNA (cRNA) was used to interrogate Affymetrix GeneChip Mouse Genome 430 2.0 arrays. Replicate BACarray samples collected from each line gave nearly identical genome-wide translational profiles (average Pearson correlation of 0.982 and 0.985 for striatonigral and striatopallidal samples, respectively) For each cell type, data were collected from three independent biological replicates, each prepared from a cohort of 7 animals. Analysis of immunoaffinity-purified samples revealed no bias for mRNA length or abundance (Figure S4). Comparative analysis of these data (see Experimental Procedures) revealed that all of the well-characterized, differentially expressed MSN markers (Gerfen, 1992) were enriched using the BACarray approach: D2 (Drd2) (36.6x), adenosine 2a receptor (Adora2a) (13.2x), and enkephalin (Penk) (7.5x) were enriched in the striatopallidal BACarray sample, while D1 (Drd1a) (3.9x), substance P (Tac1) (3.6x), and dynorphin (Pdyn) (5.6x) were enriched in the striatonigral BACarray sample (Figure 3C and Table S2). We also confirmed four striatopallidal-enriched mRNAs (Adk, Plxdc1, BC004044, and Hist1h2bc), as well as six striatonigral-enriched mRNAs (Slc35d3, Zfp521, Ebf1, Stmn2, Gnb4, and Nrxn1) reported in a microarray study of FACS isolated MSNs (Lobo et al., 2006) (Table S2). We further identified ~70 additional striatopallidal-enriched transcripts and >150 additional striatonigral-enriched transcripts (Table S2). To initially verify our data, we performed quantitative PCR assays using independent biological BACarray D1 and D2 samples and a different cDNA amplification procedure (see Supplemental Experimental Procedures). Differential translation of Eya1, Isl1, Gng2, and Crym in striatonigral MSNs, and Gpr6, Lhx8, Gpr88, Trpc4, and Tpm2 in striatopallidal MSNs was confirmed (Tables S3 and S4). These genes were selected because they represent both highly and moderately enriched messages.
Given the apparent enhanced sensitivity of the BACarray method, we were next interested in large-scale validation of the data using publicly available gene expression databases. Since in situ hybridization (ISH) alone cannot distinguish MSN subtypes, we pooled data from D1 and D2 BACarray experiments and compared them to data collected from the total RNA of one whole brain (minus striatum) (Table S5). This analysis would be expected to identify most MSN-enriched transcripts, which can be evaluated in the ISH databases, whether or not they are differentially expressed in striatopallidal or striatonigral projection neurons. The analysis resulted in detection of several thousand translated mRNAs enriched in striatum relative to whole brain, including all previously well-known striatal-enriched genes: Ppp1r1b/Darpp-32 (Walaas and Greengard, 1984), Ptpn5/Step (Lombroso et al., 1993), Arpp-19 (Girault et al., 1990), Arpp-21/RCS (Ouimet et al., 1989), Gnal/Golf (Herve et al., 1993), Rhes/Rasd2 (Falk et al., 1999), Rgs9 (Gold et al., 1997), Adcy5 (Glatt and Snyder, 1993), Gng7 (Watson et al., 1994), Rasgrp2 (Kawasaki et al., 1998), Pde1b (Polli and Kincaid, 1992), Pde10a (Fujishige et al., 1999), Gpr88 (Mizushima et al., 2000), Rarb (Krezel et al., 1999), and Strn4 (Castets et al., 1996) as well as the transcription factors Foxp1, Foxp2 (Ferland et al., 2003), Ebf1 (Lobo et al., 2006), and Zfp503/Nolz (Chang et al., 2004) (Table S5). Of the first 100 genes appearing in our MSN-enriched dataset, 26 were present in both of two major gene expression databases (the GENSAT/Brain Gene Expression Map (BGEM) and Allen Brain Atlas (ABA) in situ hybridization (ISH) databases (http://www.ncbi.nlm.nih.gov/projects/gensat/; http://www.stjudebgem.org/) (Gong et al., 2003; Lein et al., 2007; Magdaleno et al., 2006)), and enriched striatal expression is evident for 22 of these genes (Table S5; Figure S5). Thus, BACarray translational profiling can provide a sensitive tool for discovery of large sets of translated messages in defined CNS cell populations.
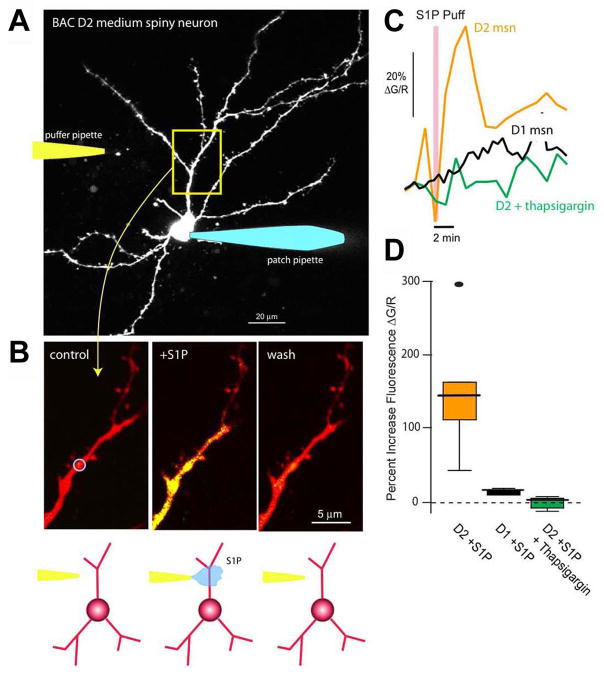
To group striatonigral- and striatopallidal-enriched genes according to biological function, we looked for statistically over-represented associations with Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway terms (Table S6–S11). GO terms delineate the known molecular function, biological process, as well as the cellular localization (component) for a particular gene (Ashburner et al., 2000), while KEGG pathways summarize known molecular interaction and reaction networks (Kanehisa, 1997). Some differentially translated mRNAs immediately predicted physiological differences between striatonigral and striatopallidal cells. For example, among striatopallidal-enriched mRNAs is Gpr6 (Lobo et al., 2007), which encodes a G-protein-coupled receptor for the lysophospholipid sphingosine 1-phosphate (S1P) (Ignatov et al., 2003). In heterologous expression systems, S1P activation of Gpr6 receptors induces intracellular Ca2+ release. As intracellular Ca2+ is a crucial regulator of neuronal physiology, we investigated whether striatopallidal enrichment of Gpr6 reflects a differential response to S1P. To this end, BAC D2 striatopallidal or BAC D1 striatonigral MSNs (expressing soluble EGFP) were identified in brain slices, patch-clamped (Day et al., 2006), loaded with Alexa 594, to visualize dendrites, and with Fluo-4 to monitor intracellular Ca2+ concentration (Figure 4A). A second pipette was then brought into close physical proximity to a dendrite, 60–80 microns from the soma, and used to apply S1P (Figure 4A, B). With the somatic membrane potential clamped at −70 mV, focal application of S1P consistently and reversibly increased dendritic Ca2+ levels in BAC D2 striatopallidal neurons (Kruskal-Wallis ANOVA, p<0.01, n=6) but not in BAC D1 striatonigral neurons (Kruskal-Wallis ANOVA, p>0.01, n=4) (Figure 4C, D). Depletion of intracellular Ca2+ stores with the Ca2+-ATPase inhibitor thapsigargin abolished this response to S1P (Kruskal-Wallis ANOVA, p>0.01, n=4; Figure 4C, D). This striatopallidal-specific S1P response would be predicted to result in a decrease in threonine 34 (T34) phosphorylation of the centrally important regulatory protein DARPP-32, due to activation of the Ca2+ and calmodulin-dependent phosphatase calcineurin (Nishi et al., 1999) and/or inhibition of adenylyl cyclase type 5 (AC5; Adcy5) (Ishikawa et al., 1992; Glatt and Snyder, 1993). Indeed, a decrease in T34 DARPP-32 phosphorylation was seen after 5 minutes of S1P treatment of striatal slices (1.04 ± 0.17 normalized units at 0 minutes; 0.58 normalized units ± 0.24 at 5 minutes after S1P addition, one-tailed Mann-Whitney test, p=0.05, n=12). These data demonstrate the utility of BACarray translational profiling in identifying previously unrecognized functional differences in signalling responses between cell types.
Figure 4. Functional Gpr6 receptors are found in BAC D2 striatopallidal but not BAC D1 striatonigral medium spiny neurons (MSNs).
(A) Projection of an EGFP-labeled MSN from a BAC D2 mouse. The cell was patched with a pipette containing Alexa 594 (50 μM) for visualization and Fluo-4 (200 μM) for measuring changes in intracellular Ca2+ (right). Cells were voltage clamped at −70 mV. A puffer pipette containing sphingosine 1-phosphate (S1P, 10 μM) was positioned near a dendrite, 60–80 μm from the soma (left/cartoon). (B) High magnification images of a dendritic segment (control, left panel) show an increase in Ca2+ associated with S1P application (S1P puff, center panel) that reversed with washing (wash, right panel). The change in Ca2+ was determined by calculating the percent change in fluorescence of Fluo-4 relative to that of Alexa 594 ( G/R). The blue circle in the first panel indicates the analyzed region of interest (ROI). (C) Time course showing the S1P induced increase in intracellular Ca2+ in the ROI from B (orange trace); similar recordings from BAC D1 MSNs (black trace) or 10 μM thapsigargin loaded BAC D2 MSNs (green trace) did not reveal any significant changes in dendritic Ca2+ levels with S1P application. (D) Box plot summarizing the S1P effects. Percent increase in fluorescence (ΔG/R) in BAC D2 MSNs (median=146%, range 44 to 294%, n=6); BAC D1 MSNs (median=17%, range 13 to 22%, n=4); and thapsigargin-loaded BAC D2 MSNs (median=4%, range –9 to 10%, n=4).
BACarray profiling of cocaine-induced transcriptional changes
Based on the data above, it seemed likely that BACarray profiling could identify molecular responses to genetic, pharmacologic or environmental changes in single cell types. To test this idea, we investigated changes in mRNA translation of MSNs upon pharmacological perturbation of dopaminergic signaling using cocaine, a competitive inhibitor of the dopamine transporter, which acts as a psychostimulant by elevating synaptic dopamine levels (Ritz et al., 1987; Di Chiara and Imperato, 1988). Adult mice were treated acutely or chronically with cocaine or saline and used for BACarray profiling of striatonigral (D1) and striatopallidal (D2) MSNs. From this analysis we identified hundreds of genes whose expression was increased or decreased in each cell type in response to cocaine (Tables S12 and S13). It is often difficult to directly compare gene expression data across drug administration experiments from published studies, due to differences in strain background, dosing regimen, and assay sensitivity or array platform; in spite of this, we were able to identify various genes whose expression has been reported to be affected by cocaine administration, including: Cartpt (Douglass et al., 1995) (up in acute D1; Table S12); Fosb (Hope et al., 1992) (up in acute D1 and D2, and chronic D1; Tables S12 and S13); Homer1 (Brakeman et al., 1997) (up in acute D1 and D2, chronic D1 and D2; Tables S12 and S13); Per2 (Yuferov et al., 2003) (up in acute D2 and chronic D1 and chronic D2; Tables S12 and S13); Vamp2 (McClung and Nestler, 2003) (up in chronic D1; Table S13); Kcnd2 (McClung and Nestler, 2003) (up in chronic D1; Table S13); and Zfp64 (McClung and Nestler, 2003) (up in acute D2, down in chronic D2; Tables S12 and S13).
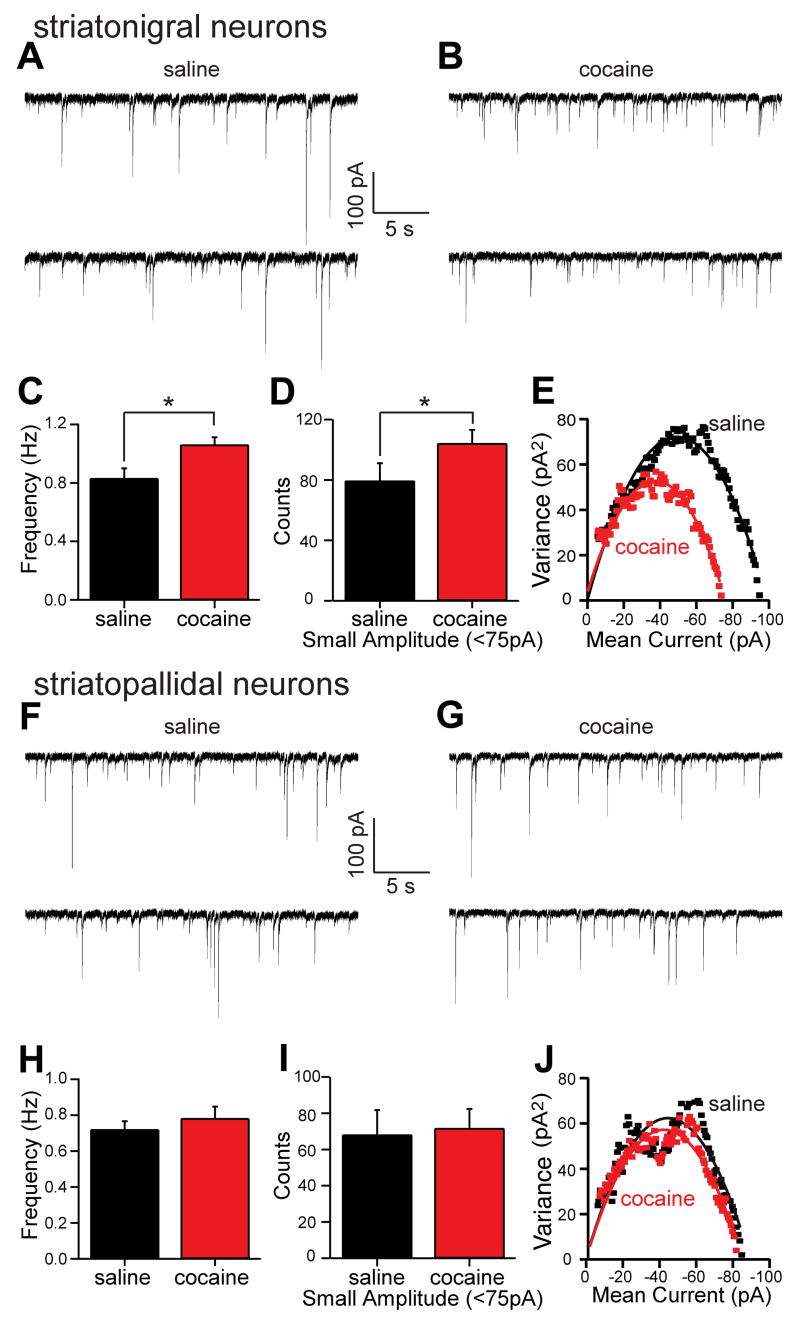
We looked for statistically over-represented GO and KEGG terms (Tables S14–S21), and noted that amongst the most significant GO biological processes altered in D1-expressing striatonigral neurons upon chronic cocaine administration was the gamma-aminobutyric acid (GABA) signaling pathway (Gabrb3, Gabra1, Cacnb4, and Gabra4, Table S20). This finding is particularly interesting in light of positron emission tomography (PET) studies that have documented an enhanced sensitivity to the benzodiazepine lorazepam (binds GABA receptors) among chronic cocaine abusers (Volkow et al., 1998). To evaluate the possible physiological significance of this up-regulation, mice were chronically treated with cocaine or saline, and GABAA receptor function was assayed electrophysiologically. Cocaine treatment significantly increased the frequency of GABAA receptor miniature inhibitory postsynaptic currents (mIPSCs) in striatonigral neurons (Mann-Whitney Rank Sum Test, p<0.05, saline median=0.82 Hz, n=22; cocaine median=1.03 Hz, n=26; Figure 5A–C). Visual inspection of representative records (as shown in Figure 5A, B) suggested that cocaine treatment increased the frequency of small amplitude mIPSCs. To evaluate this possibility, the amplitude distribution of mIPSCs was compiled. The mode of this distribution was 75 pA in control recordings. Cocaine-treatment appeared to shift the mode to smaller amplitudes. To determine if the change in the distribution was significant, equal length records were compared for the number of sub-modal (<75 pA) and supra-modal (≥75 pA) IPSCs. Cocaine treatment significantly increased the number of sub-modal events (saline=79.4±8.2, n=22; cocaine=104.2±6.3, n=26; p<0.05, one-tailed t-test, Figure 5D), but did not change the number of supra-modal mIPSCs (saline=81.9±8.6, n=22; cocaine=80.8±7.0, n=26; p>0.05, one tailed t-test, data not shown). The alterations in frequency and amplitude were not accompanied by a significant change in mIPSC kinetics (Figure S6A–B). Non-stationary noise analysis was also used to estimate single receptor conductance and the number of receptors per synapse (De Koninck and Mody, 1994). In neurons from saline-treated animals, the GABAA single receptor conductance was estimated to be 31 pS (Figure 5E; Figure S6C), a value very near that reported in other cell types (De Koninck and Mody, 1994; Kilman et al., 2002); cocaine treatment did not alter this value (t-test, p>0.05, saline N=31.1±1.6 n=22; cocaine N=30.8±1.2, n=26; Figure 5E; Figure S6C). However, cocaine treatment did significantly reduce the estimated average number of receptors per synapse (t-test, p<0.05, saline N=33.1±1.4, n=22; cocaine N=29.3±1.0, n=26; Figure 5E; Figure S6D). The most parsimonious interpretation of these results is that cocaine treatment leads to 1) the addition of small, dendritic GABAA synapses resulting in an increase in mIPSC frequency and 2) a decrease in the average number of receptors per synapse, due to dendritic synapses having fewer receptors than larger somatic synapses.
Figure 5. Cocaine treatment increases the frequency of small amplitude GABAergic mIPSCs in BAC D1 striatonigral medium spiny neurons (MSNs).
(A) Representative spontaneous mIPSCs traces from BAC D1 striatonigral neurons (expressing soluble EGFP under the D1 promoter) taken from mice treated for 15 days with saline or (B) cocaine (20 mg/kg/day). (C) Bar graph summary of mean mIPSC frequency showing a significant increase in BAC D1 striatonigral neuron mIPSCs frequency following cocaine treatment. (D) Bar graph summary showing that the number of small amplitude mIPSCs (<75 pA) in equal length records (7 min) increased in BAC D1 striatonigral neurons following cocaine treatment. (E) Representative variance-mean current plots from saline treated (black symbols) and cocaine treated (red symbols) BAC D1 neurons suggesting that the cocaine-induced small amplitude events arise from synapses that have fewer GABAA receptors (N) per synapse but receptors with an unchanged unitary receptor conductance (N) (saline N=33, N=31pS; cocaine N=29, N=31pS; see Figure S6C -D for means). (F) Representative spontaneous mIPSCs traces from BAC D2 striatopallidal neurons following saline treatment for 15 days and (G) following cocaine treatment for 15 days. (H) Bar graph summary of mean mIPSC frequency in saline and cocaine treated D2 neurons, showing no effect of treatment condition. (I) Bar graph summary showing that the number of small amplitude mIPSCs (<75 pA) in equal length records (7 min) was not altered by treatment condition in BAC D2 neurons. (J) Representative variance-mean current plots showing that cocaine treatment did not change in the number of receptors per synapse or the unitary receptor conductance in BAC D2 neurons (saline N=34, N=31pS; cocaine N=33, N=30pS; see Figure S7C –D for means).
In contrast to D1 striatonigral neurons, D2 striatopallidal neurons displayed no change in the frequency (t-test, p>0.05, saline=0.72±0.05 Hz, n=12; cocaine=0.78±0.07 Hz, n=16; Figure 5H), amplitude (sub-modal mIPSCs, t-test, p>0.05, saline=68.1±9.7, n=12; cocaine=71.7±7.5, n=16; Figure 5I; supra-modal mIPSCs, t-test, p>0.05, saline=59.1±6.5, n=12; cocaine=63.3±4.7, n=16; data not shown) and kinetics (Figure S7A–B) of GABAA mIPSCs following chronic cocaine treatment. Likewise, non-stationary noise analysis suggested that the average receptor conductance and number of receptors per synapse did not change in D2 striatopallidal neurons following cocaine treatment (Figure 5J and Figure S7C–D). Taken together, these results demonstrate the physiological relevance of the selective cocaine-induced up-regulation of mRNAs associated with GABAA receptors specific to D1 striatonigral neurons.
Although alterations in GABA release probability could also cause changes in mIPSC frequency, cocaine treatment altered the amplitude distribution of GABAA mIPSCs selectively in D1 striatonigral neurons, which would not be expected following a blanket increase in GABA release probability. The preferential increase in small amplitude mIPSCs and the suggestion from non-stationary noise analysis that the average number of receptors per synapse fell in striatonigral neurons following cocaine treatment are consistent with the addition of small dendritic synapses, as opposed to larger somatic synapses (Kubota and Kawaguchi, 2000). Dendritic GABAergic synapses arise either from recurrent collaterals of other MSNs or from interneurons (Tepper et al., 2004). Definitively sorting out whether one or both of these classes contribute to the change in synaptic properties will require paired cell recordings and ultrastructural analysis [e.g., (Koos et al., 2004; Kubota and Kawaguchi, 2000)]. Nevertheless, this upregulation may serve as an adaptive response to enhanced excitatory glutamatergic signaling in striatonigral neurons associated with the activation of D1 dopamine receptors by cocaine administration (Flores-Hernandez et al., 2002; Lee et al., 2006; Valjent et al., 2005; Wolf et al., 2004). The increased expression of physiologically active, inhibitory GABAA receptors in striatonigral cells could be a part of the molecular adaptations underlying the well-documented changes in sensitivity to cocaine seen among chronic abusers.
BACarray profiling of cholinergic motor neurons and Purkinje neurons
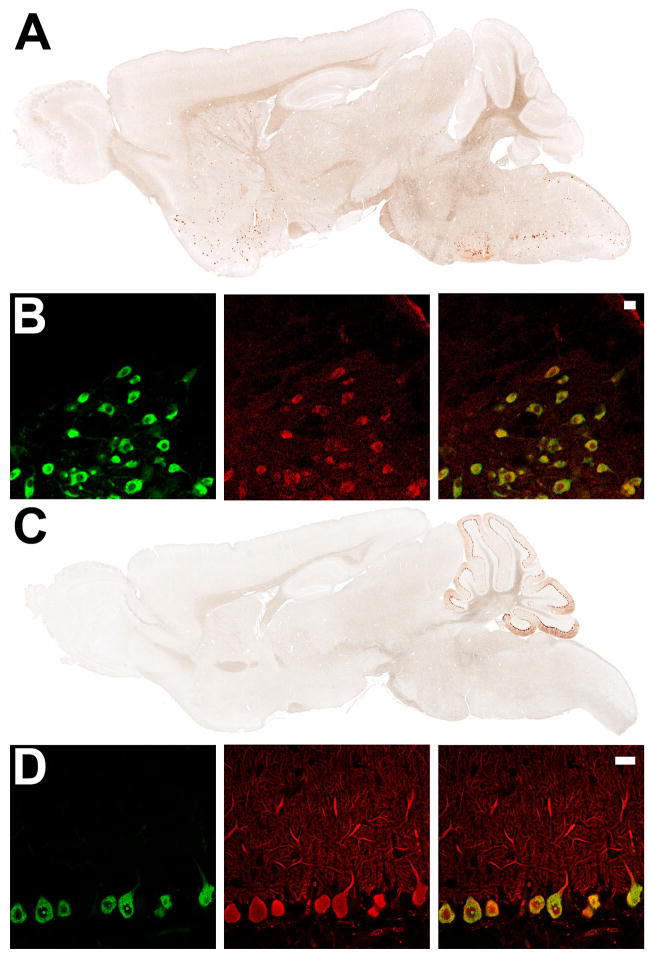
To determine whether the BACarray technique is generalizable, cholinergic and Purkinjie cell-specific BACarray lines were produced. To this end, the EGFP-L10a transgene was placed under the control of the choline acetyltransferase (Chat) locus, specifically expressed in cholinergic cells in the CNS, or the Purkinje cell protein 2 (Pcp2) locus, specifically expressed in cerebellar Purkinje cells of the CNS (Oberdick et al., 1988). Consistent with known expression patterns, (Oh et al., 1992), the Chat BACarray line DW167 showed highest EGFP-L10a expression in cholinergic cells of the dorsal and ventral striatum, basal forebrain, brain stem, spinal cord, and medial habenula (Figure 6A), while the Pcp2 BACarray line DR166 showed EGFP-L10a expression restricted to cells with characteristic Purkinje cell morphology (Figure 6C). Indirect immunofluorescence staining for Chat and Calbindin-D28K confirmed cell-type specific expression of the Chat and Pcp2 lines, respectively (Figure 6B and 6D). One exception was in the pedunculopontine and laterodorsal tegmental nuclei, where only a minority of cholinergic cells were labelled with EGFP (data not shown).
Figure 6. Expression of EGFP-L10a in the Chat and Purkinje cell BACarray lines.
(A) Immunohistochemistry to EGFP in adult sagittal sections from the Chat BACarray line DW167. (B) Indirect immunofluorescent characterization of Chat BACarray line DW167 brain stem facial motor nucelus: EGFP staining (left panel); Chat staining (middle panel) and merge (right panel, with 20 μm scale bar). (C) Immunohistochemistry to EGFP in adult sagittal sections from the Pcp2 BACarray line DR166. (D) Indirect immunofluorescent characterization of Pcp2 BACarray line DR166 Purkinje cell neurons: EGFP staining (left panel); Calbindin-D28K staining (middle panel) and merge (right panel, with 20 μm scale bar).
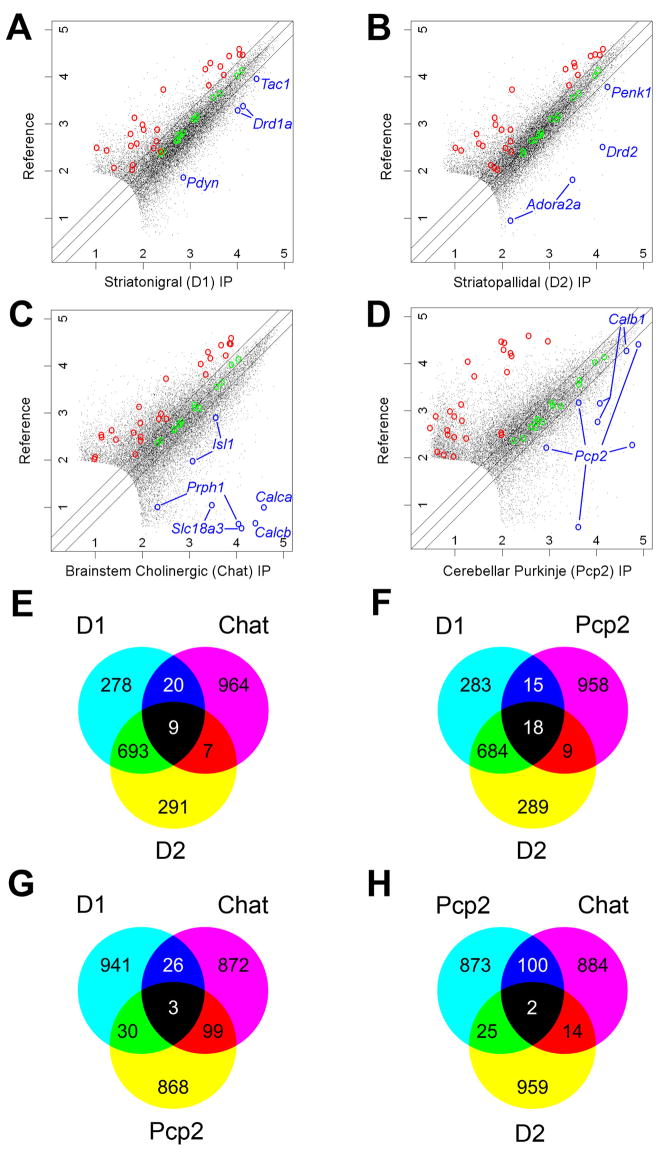
Array data were collected from brain stem cholinergic motor neurons and from Purkinje cells using the Chat and Pcp2 BACarray lines, respectively. Replicate BACarray samples gave nearly identical genome-wide translational profiles (average Pearson correlation of 0.982 and 0.997, respectively). We identified mRNAs enriched in each IP sample compared to an arbitrary reference sample (unbound fractions of the IPs) (Figures 7A–D). Enrichment of cell-specific positive-control genes and exclusion of known negative-control genes (glial genes), were evident for each comparison (Figure 7A–D). Venn diagrams constructed from the top 1,000 enriched probesets from this analysis (Tables S22–25) showed that the translational profiles of striatonigral and striatopallidal cells are more similar to each other than to translational profiles of either brain stem cholinergic cells or Purkinje cells (Figure 7E–H).
Figure 7. BACarray profiles recapitulate known cell-specific markers and reveal new ones for four distinct cell types.
Scatterplots of D1, D2, Chat, and Pcp2 BACarray data compared to a reference mRNA sample reveal hundreds of genes enriched in each cell type (A–D). Green circles indicate Affymetrix biotinylated spike-in controls; blue circles indicate known cell-specific markers; and red circles indicate probesets for known glial genes (negative controls; Table S26). Lines on either side of the diagonal mark 2-fold enrichment. Axes are labelled for expression in powers of 10. Venn diagrams of the top 1,000 enriched probesets (with expression value cut-off >100) for each cell type reveal that each cell type has a unique pattern of enriched genes (E–H).
Discussion
BACarray translational profiling combines genetic targeting of an EGFP-L10a ribosomal fusion protein to specific cell populations; simple, rapid, affinity purification of mRNA; and microarray technology to identify translated mRNAs in situ. Unlike traditional approaches, it combines coincident detection of all translated mRNAs with cell-type specificity; thus, one quantitative BACarray experiment can replace thousands of high-throughput qualitative in situ hybridization runs and, further, can be repeated easily for each experimental condition. Second, the information obtained is cell-type specific while abrogating the physiologic adaptations and mRNA degradation which can occur during the lengthy cell separation procedures used in other approaches that sever neuronal axons and dendrites and disrupt tissue-intrinsic signalling. Third, because polysomes are stabilized with cycloheximide in the extraction buffers and during the affinity purification steps, redistribution of the affinity tag within the mRNA pool during polysome isolation is prevented, obviating the problems inherent in using less stable mRNA binding proteins. Fourth, the use of established BACarray lines ensures that mRNA translation profiles can be reproducibly obtained and directly compared from the same cell population across experimental conditions. Crossing of these and other established BACarray lines to mouse models should overcome one of the major obstacles to the study of many neurodegenerative diseases, such as Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease, which display both vulnerable and non-vulnerable cell populations. Fifth, the use of EGFP as the affinity tag greatly facilitates anatomic studies of candidate BACarray mouse lines, as well as electrophysiological studies of each cell type. Finally, the profiling of only translated mRNA pools will more accurately reflect actual protein levels in a cell than does conventional gene expression profiling, which relies on total mRNA pools.
The BACarray approach thus overcomes limitations of previous mRNA-tagging methods which have targeted other RNA-binding proteins, of which only Poly A–Binding Protein (PABP) has been used in vivo for tissue-specific studies (Roy et al., 2002; Kunitomo et al., 2005), and which requires crosslinking of the tagged PABP to mRNA (and its inherent artifacts) due to its loose association with mRNA. The use of tagged ribosomal proteins to purify translating polysomes from yeast as well as plant cells has recently been reported (Inada et al., 2002; Zanetti et al., 2005), although in these cases cell-specific targeting to obtain cell-specific translational profiles was not attempted. From our own experience, we know that adapting this methodology to cell-specific targeting is not trivial, particularly for applications in the mammalian CNS, with its far more difficult conditions of low expression, limited material, and contamination by blood, myelin, and other biological factors. As a result of the optimization reported here, the BACarray methodology is now extremely robust to these conditions and broadly applicable (Doyle, Dougherty, et al., 2008; accompanying paper).
Furthermore, the BACarray approach was able to identify all previously known, well-studied striatonigral- and striatopallidal-enriched genes [with one exception: the striatonigral-enriched muscarinic receptor M4 (_Chrm4_) (Ince et al., 1997), for which probesets on the Affymetrix Mouse 430 2.0 GeneChips gave very little signal; however, real-time PCR analysis of Chrm4 expression demonstrated clear enrichment of Chrm4 mRNA in our striatonigral BACarray cell sample (Table S3)]. This stands in distinction to a recent microarray study of FACS-sorted MSNs (Lobo et al., 2006), in which well-known positive-control genes (e.g. Chrm4, Pdyn, Drd1a, and Drd2) were not identified as differentially expressed on microarrays and 16 of the 21 mRNAs reported in that study as striatopallidal-enriched in adult MSNs could not be confirmed in our analysis. Furthermore, we report here hundreds of distinguishing transcripts not identified in that study.
More importantly, our BACarray analysis of striatal MSN subtypes has identified novel physiological differences between striatonigral and striatopallidal cells, which may provide new therapeutic targets for various neurological diseases associated with pathophysiology in the striatum. As one example, we demonstrate that striatopallidal cells selectively express Gpr6, and that they correspondingly display a cell-type specific release of intracellular Ca2+ in response to sphingosine 1-phosphate. A second example of the distinct properties of these medically important cell types is provided by our studies of psychostimulant drug action, which give molecular and physiological evidence for upregulation of GABAA receptor subunits in striatonigral neurons following chronic cocaine administration.
The results presented here demonstrate that the BACarray methodology provides an enabling technology for studies of the biology of specific cell types in even the most heterogeneous cell populations, such as those that occur in the CNS. We have identified numerous distinguishing molecular characteristics among four distinct neuronal populations, including two closely related, critical cell types and have demonstrated that the BACarray methodology can be employed to analyze physiological adaptations of specific cell types in vivo. An accompanying study (Doyle, Dougherty, et al., 2008) has demonstrated the general applicability of the BACarray methodology by characterizing additional CNS cell types, each of which exhibits an enormously complex and cell-type specific molecular phenotype.
Experimental Procedures
BACarray transgenesis
BAC transgenic mice were produced according to published protocols (Gong et al., 2003), with the exception that the EGFP-L10a transgene was used in place of EGFP.
Purification of mRNA from BACarray mice
For striatonigral or striatopallidal translated mRNA purification, mice were decapitated and the striata of seven BACarray transgenic mice were quickly manually dissected. Pooled striatal tissue was immediately homogenized in ice-cold polysome extraction buffer (10 mM HEPES [pH 7.4], 150 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 100 μg/ml cycloheximide, protease inhibitors, and recombinant RNase inhibitors) using a motor-driven Teflon-glass homogenizer. Homogenates were centrifuged for 10 minutes at 2,000 × g, 4 °C, to pellet large cell debris, and NP-40 (EMD Biosciences, San Diego, CA) and DHPC (Avanti Polar Lipids, Alabaster, AL) were added to the supernatant at a final concentration of 1% and 30 mM, respectively. After incubation on ice for 5 minutes, the clarified lysate was centrifuged for 10 minutes at 13,000 × g to pellet unsolubilized material. Goat anti-GFP (custom made) coated protein G Dynal magnetic beads (Invitrogen Corporation, Carlsbad, CA) were added to the supernatant and the mixture was incubated at 4°C with end-over-end rotation for 30 minutes. Beads were subsequently collected on a magnetic rack, washed three times with high-salt polysome wash buffer (10 mM HEPES [pH 7.4], 350 mM KCl, 5 mM MgCl2, 1% NP-40, 0.5mM dithiothreitol, 100 μg/ml cycloheximide) and immediately placed in TriZol-LS reagent (Invitrogen Corporation, Carlsbad CA) and chloroform to extract the bound rRNA and mRNA from polysomes. After extraction, RNA was precipitated with sodium acetate and Glycoblue (Ambion, Austin, TX) in isopropanol overnight at −80°C, washed twice with 70% ethanol, resuspended in water, and further purified using an Rneasy Micro Kit (Qiagen, Valencia, CA) with in-column DNase digestion. For the purification of translated mRNAs from brain stem cholinergic motor neurons (from the Chat BACarray line) or cerebellar Purkinje neurons (from the Pcp2 BACarray line), nearly identical purifications as outlined above were performed, with a few minor modifications (see Supplemental Experimental Procedures).
Microarray data normalization and analysis
Three biological replicates were performed for each experiment. Quantitative PCR reactions were performed to validate array results using an independent biological source and amplification methodology (see Supplemental Experimental Procedures). Striatonigral and striatopallidal GeneChip CEL files were imported into Genespring GX 7.3.1 (Agilent Technologies, Santa Clara, CA), processed with the GC-RMA algorithm, and expression values on each chip were normalized to that chip’s 50th percentile. Data was filtered to eliminate genes with intensities in the lower range. A moderated two-tailed paired t-test was performed using the Limma package from the Bioconductor project (http://www.bioconductor.org). The p-value of the moderated t-test was adjusted for multiple hypothesis testing, controlling the false discovery rate (FDR) using the Benjamini-Hochberg procedure. We then selected all genes that had an FDR less than 0.1 (10%) and fold change larger than 1.5. Other comparisons were performed as described in detail in Supplemental Experimental Procedures.
Supplementary Material
01
02
03
04
05
06
07
08
09
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
Acknowledgments
We thank Helen Shio for performing electron microscopy, Angus Nairn and Cordelia Stearns for striatal slice experimental data, Joseph Doyle and Joseph Dougherty for BACarray data and analysis, as well as members of the P. Greengard, N. Heintz, GENSAT, G. Blobel, and R. Darnell laboratories and Maxwell Heiman for comments and assistance. This work was supported by NIDA fellowship 5F32DA021487 to M.H.; German Research Foundation fellowship SCHA 1482/1-1 to A.S., NINDS grant NS34696 to D. J. S.; grants from The F. M. Kirby Foundation, The Picower Foundation, The Jerry and Emily Spiegel Family Foundation, The Simons Foundation, The Peter Jay Sharp Foundation, The Michael Stern Foundation, NIMH Conte Center grant MH074866, and NIDA grant DA10044 to P.G.; The Simons Foundation, The Howard Hughes Medical Institute, the Adelson Medical Research Foundation, NIA grant AG09464, and NIMH Conte Center grant MH074866 to N.H. None of the authors of this work have a financial interest related to this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The microarray data can be found in the Gene Expression Omnibus (GEO) of NCBI.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakeman PR, Lanahan AA, O’Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- Castets F, Bartoli M, Barnier JV, Baillat G, Salin P, Moqrich A, Bourgeois JP, Denizot F, Rougon G, Calothy G, Monneron A. A novel calmodulin-binding protein, belonging to the WD-repeat family, is localized in dendrites of a subset of CNS neurons. J Cell Biol. 1996;134:1051–1062. doi: 10.1083/jcb.134.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW, Tsai CW, Wang HF, Tsai HC, Chen HY, Tsai TF, Takahashi H, Li HY, Fann MJ, Yang CW, et al. Identification of a developmentally regulated striatum-enriched zinc-finger gene, Nolz-1, in the mammalian brain. Proc Natl Acad Sci U S A. 2004;101:2613–2618. doi: 10.1073/pnas.0308645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day M, Wang Z, Ding J, An X, Ingham CA, Shering AF, Wokosin D, Ilijic E, Sun Z, Sampson AR, et al. Selective elimination of glutamatergic synapses on striatopallidal neurons in Parkinson disease models. Nat Neurosci. 2006;9:251–259. doi: 10.1038/nn1632. [DOI] [PubMed] [Google Scholar]
- De Koninck Y, Mody I. Noise analysis of miniature IPSCs in adult rat brain slices: properties and modulation of synaptic GABAA receptor channels. J Neurophysiol. 1994;71:1318–1335. doi: 10.1152/jn.1994.71.4.1318. [DOI] [PubMed] [Google Scholar]
- Di Chiara G, Imperato A. Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proc Natl Acad Sci U S A. 1988;85:5274–5278. doi: 10.1073/pnas.85.14.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass J, McKinzie AA, Couceyro P. PCR differential display identifies a rat brain mRNA that is transcriptionally regulated by cocaine and amphetamine. J Neurosci. 1995;15:2471–2481. doi: 10.1523/JNEUROSCI.15-03-02471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk JD, Vargiu P, Foye PE, Usui H, Perez J, Danielson PE, Lerner DL, Bernal J, Sutcliffe JG. Rhes: A striatal-specific Ras homolog related to Dexras1. J Neurosci Res. 1999;57:782–788. [PubMed] [Google Scholar]
- Ferland RJ, Cherry TJ, Preware PO, Morrisey EE, Walsh CA. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J Comp Neurol. 2003;460:266–279. doi: 10.1002/cne.10654. [DOI] [PubMed] [Google Scholar]
- Flores-Hernandez J, Cepeda C, Hernandez-Echeagaray E, Calvert CR, Jokel ES, Fienberg AA, Greengard P, Levine MS. Dopamine enhancement of NMDA currents in dissociated medium-sized striatal neurons: role of D1 receptors and DARPP-32. J Neurophysiol. 2002;88:3010–3020. doi: 10.1152/jn.00361.2002. [DOI] [PubMed] [Google Scholar]
- Fujishige K, Kotera J, Omori K. Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A. Eur J Biochem. 1999;266:1118–1127. doi: 10.1046/j.1432-1327.1999.00963.x. [DOI] [PubMed] [Google Scholar]
- Gerfen CR. The neostriatal mosaic: multiple levels of compartmental organization in the basal ganglia. Annu Rev Neurosci. 1992;15:285–320. doi: 10.1146/annurev.ne.15.030192.001441. [DOI] [PubMed] [Google Scholar]
- Girault JA, Horiuchi A, Gustafson EL, Rosen NL, Greengard P. Differential expression of ARPP-16 and ARPP-19, two highly related cAMP-regulated phosphoproteins, one of which is specifically associated with dopamine-innervated brain regions. J Neurosci. 1990;10:1124–1133. doi: 10.1523/JNEUROSCI.10-04-01124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt CE, Snyder SH. Cloning and expression of an adenylyl cyclase localized to the corpus striatum. Nature. 1993;361:536–538. doi: 10.1038/361536a0. [DOI] [PubMed] [Google Scholar]
- Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Herve D, Levi-Strauss M, Marey-Semper I, Verney C, Tassin JP, Glowinski J, Girault JA. G(olf) and Gs in rat basal ganglia: possible involvement of G(olf) in the coupling of dopamine D1 receptor with adenylyl cyclase. J Neurosci. 1993;13:2237–2248. doi: 10.1523/JNEUROSCI.13-05-02237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope B, Kosofsky B, Hyman SE, Nestler EJ. Regulation of immediate early gene expression and AP-1 binding in the rat nucleus accumbens by chronic cocaine. Proc Natl Acad Sci U S A. 1992;89:5764–5768. doi: 10.1073/pnas.89.13.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatov A, Lintzel J, Kreienkamp HJ, Schaller HC. Sphingosine-1-phosphate is a high-affinity ligand for the G protein-coupled receptor GPR6 from mouse and induces intracellular Ca2+ release by activating the sphingosine-kinase pathway. Biochem Biophys Res Commun. 2003;311:329–336. doi: 10.1016/j.bbrc.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Inada T, Winstall E, Tarun SZ, Jr, Yates JR, 3rd, Schieltz D, Sachs AB. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA. 2002;8:948–958. doi: 10.1017/s1355838202026018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince E, Ciliax BJ, Levey AI. Differential expression of D1 and D2 dopamine and m4 muscarinic acetylcholine receptor proteins in identified striatonigral neurons. Synapse. 1997;27:357–366. doi: 10.1002/(SICI)1098-2396(199712)27:4<357::AID-SYN9>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Ishikawa Y, Katsushika S, Chen L, Halnon NJ, Kawabe J, Homcy CJ. Isolation and characterization of a novel cardiac adenylylcyclase cDNA. J Biol Chem. 1992;267:13553–13557. [PubMed] [Google Scholar]
- Kanehisa M. A database for post-genome analysis. Trends Genet. 1997;13:375–376. doi: 10.1016/s0168-9525(97)01223-7. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Springett GM, Toki S, Canales JJ, Harlan P, Blumenstiel JP, Chen EJ, Bany IA, Mochizuki N, Ashbacher A, et al. A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc Natl Acad Sci U S A. 1998;95:13278–13283. doi: 10.1073/pnas.95.22.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilman V, van Rossum MC, Turrigiano GG. Activity deprivation reduces miniature IPSC amplitude by decreasing the number of postsynaptic GABA(A) receptors clustered at neocortical synapses. J Neurosci. 2002;22:1328–1337. doi: 10.1523/JNEUROSCI.22-04-01328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koos T, Tepper JM, Wilson CJ. Comparison of IPSCs evoked by spiny and fast-spiking neurons in the neostriatum. J Neurosci. 2004;24:7916–7922. doi: 10.1523/JNEUROSCI.2163-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krezel W, Kastner P, Chambon P. Differential expression of retinoid receptors in the adult mouse central nervous system. Neuroscience. 1999;89:1291–1300. doi: 10.1016/s0306-4522(98)00342-x. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Kawaguchi Y. Dependence of GABAergic synaptic areas on the interneuron type and target size. J Neurosci. 2000;20:375–386. doi: 10.1523/JNEUROSCI.20-01-00375.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitomo H, Uesugi H, Kohara Y, Iino Y. Identification of ciliated sensory neuron-expressed genes in Caenorhabditis elegans using targeted pull-down of poly(A) tails. Genome Biol. 2005;6:R17. doi: 10.1186/gb-2005-6-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Kim Y, Kim AM, Helmin K, Nairn AC, Greengard P. Cocaine-induced dendritic spine formation in D1 and D2 dopamine receptor-containing medium spiny neurons in nucleus accumbens. Proc Natl Acad Sci U S A. 2006;103:3399–3404. doi: 10.1073/pnas.0511244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Cui Y, Ostlund SB, Balleine BW, William Yang X. Genetic control of instrumental conditioning by striatopallidal neuron-specific S1P receptor Gpr6. Nat Neurosci. 2007;10:1395–1397. doi: 10.1038/nn1987. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Lombroso PJ, Naegele JR, Sharma E, Lerner M. A protein tyrosine phosphatase expressed within dopaminoceptive neurons of the basal ganglia and related structures. J Neurosci. 1993;13:3064–3074. doi: 10.1523/JNEUROSCI.13-07-03064.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdaleno S, Jensen P, Brumwell CL, Seal A, Lehman K, Asbury A, Cheung T, Cornelius T, Batten DM, Eden C, et al. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat Neurosci. 2003;6:1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- Mizushima K, Miyamoto Y, Tsukahara F, Hirai M, Sakaki Y, Ito T. A novel G-protein-coupled receptor gene expressed in striatum. Genomics. 2000;69:314–321. doi: 10.1006/geno.2000.6340. [DOI] [PubMed] [Google Scholar]
- Nishi A, Snyder GL, Nairn AC, Greengard P. Role of calcineurin and protein phosphatase-2A in the regulation of DARPP-32 dephosphorylation in neostriatal neurons. J Neurochem. 1999;72:2015–2021. doi: 10.1046/j.1471-4159.1999.0722015.x. [DOI] [PubMed] [Google Scholar]
- Oberdick J, Levinthal F, Levinthal C. A Purkinje cell differentiation marker shows a partial DNA sequence homology to the cellular sis/PDGF2 gene. Neuron. 1988;1:367–376. doi: 10.1016/0896-6273(88)90186-9. [DOI] [PubMed] [Google Scholar]
- Oh JD, Woolf NJ, Roghani A, Edwards RH, Butcher LL. Cholinergic neurons in the rat central nervous system demonstrated by in situ hybridization of choline acetyltransferase mRNA. Neuroscience. 1992;47:807–822. doi: 10.1016/0306-4522(92)90031-v. [DOI] [PubMed] [Google Scholar]
- Ouimet CC, Hemmings HC, Jr, Greengard P. ARPP-21, a cyclic AMP-regulated phosphoprotein enriched in dopamine-innervated brain regions. II. Immunocytochemical localization in rat brain. J Neurosci. 1989;9:865–875. doi: 10.1523/JNEUROSCI.09-03-00865.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polli JW, Kincaid RL. Molecular cloning of DNA encoding a calmodulin-dependent phosphodiesterase enriched in striatum. Proc Natl Acad Sci U S A. 1992;89:11079–11083. doi: 10.1073/pnas.89.22.11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón y Cajal S, Pasik P, Pasik T. Texture of the nervous system of man and the vertebrates. Wien ; New York: Springer; 1899. [Google Scholar]
- Ritz MC, Lamb RJ, Goldberg SR, Kuhar MJ. Cocaine receptors on dopamine transporters are related to self-administration of cocaine. Science. 1987;237:1219–1223. doi: 10.1126/science.2820058. [DOI] [PubMed] [Google Scholar]
- Roy PJ, Stuart JM, Lund J, Kim SK. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- Tepper JM, Koos T, Wilson CJ. GABAergic microcircuits in the neostriatum. Trends Neurosci. 2004;27:662–669. doi: 10.1016/j.tins.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Valjent E, Pascoli V, Svenningsson P, Paul S, Enslen H, Corvol JC, Stipanovich A, Caboche J, Lombroso PJ, Nairn AC, et al. Regulation of a protein phosphatase cascade allows convergent dopamine and glutamate signals to activate ERK in the striatum. Proc Natl Acad Sci U S A. 2005;102:491–496. doi: 10.1073/pnas.0408305102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Fowler JS, Hitzemann R, Gatley SJ, Dewey SS, Pappas N. Enhanced sensitivity to benzodiazepines in active cocaine-abusing subjects: a PET study. Am J Psychiatry. 1998;155:200–206. doi: 10.1176/ajp.155.2.200. [DOI] [PubMed] [Google Scholar]
- Walaas SI, Greengard P. DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. I. Regional and cellular distribution in the rat brain. J Neurosci. 1984;4:84–98. doi: 10.1523/JNEUROSCI.04-01-00084.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JB, Coulter PM, 2nd, Margulies JE, de Lecea L, Danielson PE, Erlander MG, Sutcliffe JG. G-protein gamma 7 subunit is selectively expressed in medium-sized neurons and dendrites of the rat neostriatum. J Neurosci Res. 1994;39:108–116. doi: 10.1002/jnr.490390113. [DOI] [PubMed] [Google Scholar]
- Wolf ME, Sun X, Mangiavacchi S, Chao SZ. Psychomotor stimulants and neuronal plasticity. Neuropharmacology. 2004;47(Suppl 1):61–79. doi: 10.1016/j.neuropharm.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Yuferov V, Kroslak T, Laforge KS, Zhou Y, Ho A, Kreek MJ. Differential gene expression in the rat caudate putamen after “binge” cocaine administration: advantage of triplicate microarray analysis. Synapse. 2003;48:157–169. doi: 10.1002/syn.10198. [DOI] [PubMed] [Google Scholar]
- Zanetti ME, Chang IF, Gong F, Galbraith DW, Bailey-Serres J. Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 2005;138:624–635. doi: 10.1104/pp.105.059477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05
06
07
08
09
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24