A mutation in Irak2c identifies IRAK-2 as a central component of the TLR regulatory network of wild-derived mice (original) (raw)
Abstract
In a phenotypic screen of the wild-derived mouse strain MOLF/Ei, we describe an earlier and more potent toll-like receptor (TLR)–mediated induction of IL-6 transcription compared with the classical inbred strain C57BL/6J. The phenotype correlated with increased activity of the IκB kinase axis as well as p38, but not extracellular signal-regulated kinase or c-Jun N-terminal kinase, mitogen-activated protein kinase (MAPK) phosphorylation. The trait was mapped to the Why1 locus, which contains Irak2, a gene previously implicated as sustaining the late phase of TLR responses. In the MOLF/Ei TLR signaling network, IRAK-2 promotes early nuclear factor κB (NF-κB) activity and is essential for the activation of p38 MAPK. We identify a deletion in the MOLF/Ei promoter of the inhibitory Irak2c gene, leading to an increased ratio of pro- to antiinflammatory IRAK-2 isoforms. These findings demonstrate that IRAK-2 is an essential component of the early TLR response in MOLF/Ei mice and show a distinct pathway of p38 and NF-κB activation in this model organism. In addition, they demonstrate that studies in evolutionarily divergent model organisms are essential to complete dissection of signal transduction pathways.
In vertebrates, Toll-like receptors (TLRs) respond to an array of pathogen-expressed molecules by initiating a series of intracellular signaling events that activate innate immunity and, thus, play a crucial role in the response to infection (Akira et al., 2006; Leulier and Lemaitre, 2008). Although this system is capable of providing a fast and vigorous response, it has to be kept under tight control to avoid hyperresponsiveness that would be detrimental to the host by initiating such pathologies as sepsis, chronic inflammation, and autoimmunity (Nathan, 2002). Key upstream processes in TLR signal transduction include MyD88 and MAL adaptor binding and phosphorylation of the IRAK family kinases. Subsequent ubiquitination of the TRAF6 complex leads to activation of NF-κB and mitogen-activated protein kinase (MAPK) proteins, which ultimately induce the transcription of inflammatory genes (Doyle and O'Neill, 2006; Kawai and Akira, 2006). The remaining innate and adaptive immune responses are highly dependent on the time course and composition of this early inflammatory milieu (Hoebe et al., 2004). Thus, a detailed understanding of the initial events in TLR signaling is critical to predicting the entire nature of a response to pathogen invasion as well as identifying the safest and most effective substrates for targeted antiinflammatory therapies.
IRAK family kinases are centrally positioned in the TLR signaling cascade, and, as such, they have been implicated as key positive and negative inflammatory regulators (Janssens and Beyaert, 2003). Two family members, IRAK-1 and IRAK-4 have been shown to promote inflammation (Cao et al., 1996; Thomas et al., 1999; Swantek et al., 2000; Li et al., 2002; Suzuki et al., 2002), whereas IRAK-M has a demonstrated inhibitory effect (Kobayashi et al., 2002). Recently reported gene targeting of IRAK-2 firmly established its importance in sustaining high levels of inflammation at late time points (Kawagoe et al., 2008; Wan et al., 2009). However, the exact role of IRAK-2 in TLR-mediated signaling remains less clear because other studies imply its involvement in early signaling events (Muzio et al., 1997; Keating et al., 2007). Further elucidation of IRAK-2's role in inflammatory signaling is critical to complete our understanding of the role of all members of the IRAK family. Experimentation is complicated by the fact that there are four isoforms of IRAK-2 in mice, with at least one of them, IRAK-2C, being inhibitory, whereas there is only one human IRAK-2 isoform (Hardy and O'Neill, 2004). Thus, the model system in which the function of IRAK-2 is interrogated may be a critical determinant of the conclusions that are reached.
Classical inbred strains of mice have been instrumental in the genetic analysis of innate immunity. Observations on the molecular nature of the immune response in laboratory mice are highly reproducible because of their genetic homogeneity. Many, though not all, of these features have been shown to recapitulate parallel human processes, an expected outcome given the high level of homology between the genomes (Waterston et al., 2002; Mestas and Hughes, 2004). However, because most of the classical inbred mouse strains descend from a relatively restricted number of founder animals largely within the Mus mus domesticus subspecies (Frazer et al., 2007; Yang et al., 2007), their genetic diversity is limited, thereby restricting their utility for phenotypic screening. It is therefore not surprising that the genetics of innate immunity has been increasingly studied by other means, such as genome-wide ENU mutagenesis, which has resulted in numerous remarkable discoveries of gene functions (Beutler et al., 2006).
Wild-derived mice, which diverged from the common ancestor of classical strains more than one million years ago, provide an additional reservoir of genetic diversity, which, unlike that resulting from laboratory mutagenesis, has arisen in an evolutionary context (Guenet and Bonhomme, 2003). As a result of their early divergence, many of the wild-derived strains have large genomic regions originating from the subspecies M. m. musculus and M. m. castaneus (Frazer et al., 2007; Yang et al., 2007). When compared with commonly used inbred strains, they exhibit polymorphisms every 100–200 bp (Ideraabdullah et al., 2004). This genetic diversity is a rich substrate for experimental interrogation. The likelihood of phenotypic variation in these strains is increased compared with that observed between classical strains. Furthermore, novel phenotypes that are revealed likely have important biological consequences because they have been selected naturally over the course of subspeciation. Wild-derived mice are an emerging, though still largely unexplored, model for the identification and study of novel immunological genes. Their usefulness as genetic models is exemplified by studies exploring response to pathogen infections as well as lethality mediated by TNF (Urosevic et al., 1999; Staelens et al., 2002; Sancho-Shimizu and Malo, 2006).
To identify novel factors that regulate TLR signaling, we initiated a genetic screen in wild-derived and classical inbred mice and identified a novel dominant phenotype of hyperresponse to LPS, peptidoglycan, and lipoteichoic acid (LTA) in several wild-derived strains. We have focused on two loci, termed Why1 and Why2 (wild-derived hyperresponse 1 and 2), with opposite effects on the trait. Our previous study identified a novel antiinflammatory role for IRAK-1BP1 (IRAK-1 binding protein or SIMPL) that is encoded by Why2 (Conner et al., 2008). In this paper, we provide evidence that IRAK-2 is the product of Why1. The effect is caused by a 10-bp deletion in the promoter area of the inhibitory isoform IRAK-2C, thus resulting in relatively higher levels of expression of the proinflammatory isoform of IRAK-2 in MOLF/Ei as compared with C57BL/6. These findings extend our knowledge of the role and importance of IRAK-2 in TLR-mediated signaling. Furthermore, they emphasize the versatile nature of the innate immune system, which is able to undergo substantial changes in the utilization of core signaling factors even within the course of evolution of a single species.
RESULTS
Hyperresponsiveness to TLR/IL-1R activation in MOLF/Ei mice
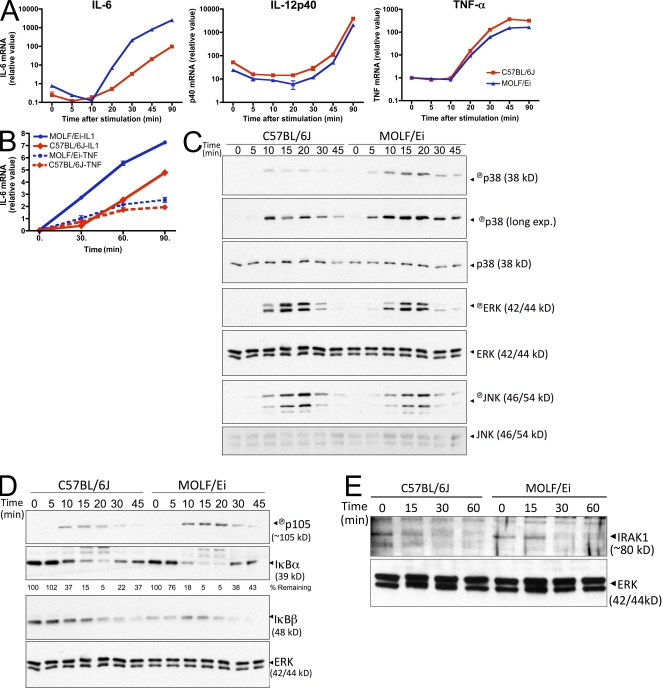
Macrophages from the wild-derived mouse strain MOLF/Ei induce transcription of the proinflammatory cytokine IL-6 faster and in higher quantities upon TLR stimulation when compared with the classical inbred strain C57BL/6J (Conner et al., 2008). In an attempt to gain mechanistic insight into the molecular events underlying this hyperresponsive phenotype, we examined the transcriptional response of additional cytokine genes early after stimulation with the TLR2 agonist LTA. Although MOLF/Ei macrophages displayed an increased amount of IL-6 transcript, two other proinflammatory genes, TNF-α and IL-12p40, were induced to similar levels in MOLF/Ei and C57BL/6J cells (Fig. 1 A).
Figure 1.
Hyperactivation of cytokines and intracellular mediators in TLR2-stimulated MOLF/Ei macrophages. (A) Peritoneal macrophages from C57BL/6J and MOLF/Ei mice were stimulated with 2 µg/ml LTA for the indicated times. Cytokine mRNA was quantified by real-time PCR. Data represent mean ± range from duplicate wells. One of at least three representative experiments is shown. (B) MEFs were prepared from MOLF/Ei and C57BL/6J mice and stimulated with 100 ng/ml human IL-1β or TNF-α for the indicated times. IL-6 mRNA was quantified using real-time PCR. Data are shown as mean ± range from duplicate wells, representative of two independent experiments. (C–E) BMDMs from each strain were stimulated with 2 µg/ml LTA for the indicated times. Cytoplasmic protein lysates were analyzed by Western blot analysis for MAPK (C) and NF-κB (D) pathway activity or IRAK-1 degradation (E). Data are representative of three (C and D) or two (E) independent experiments.
Individual inflammatory stimuli, such as TNFR, IL-1R, and TLR agonists, use a combination of shared and stimulus-specific downstream mediators to induce cytokine gene transcription (Verstrepen et al., 2008). To gain insight into which signaling components are hyperresponsive in MOLF/Ei mice, we tested mouse embryonic fibroblasts (MEFs) from MOLF/Ei and C57BL/6J strains for their response to IL-1β and TNF-α stimulation. Whereas MOLF/Ei MEFs displayed increased IL-6 production relative to C57BL/6J in response to IL-1β, equivalent levels of IL-6 were produced upon TNF-α stimulation (Fig. 1 B). These findings suggest that the factor promoting increased IL-6 production in MOLF/Ei cells is shared between the MyD88-dependent TLR and IL-1R pathways but is not common to the TNFR pathway.
Complete induction of responsive genes upon TLR stimulation requires a combination of downstream signaling events, including transcriptional activation through the NF-κB family factors, chromatin remodeling, and activity of the MAPK proteins p38, extracellular signal-regulated kinase (ERK), and c-Jun N-terminal kinase (JNK; Kawai and Akira, 2006). Individual inflammatory genes exhibit different requirements for each of these activities, a feature which aids in conferring specificity to immune responses (Saccani et al., 2002; Park et al., 2005; Sanjabi et al., 2005; Foster et al., 2007). To assess the activation status of upstream molecular mediators in MOLF/Ei and C57BL/6J macrophages, Western blot analysis was performed. Examination of phosphorylation status for all three MAPK family members revealed that p38, but not ERK or JNK, was activated faster and at higher levels in MOLF/Ei cells (Fig. 1 C). Furthermore, assessment of the IκB kinase (IKK) axis of TLR signaling was performed by observing the direct phosphorylation of the IκB family member p105 as well as the IKK-dependent process of proteasome-mediated degradation of IκBα and IκBβ. Each of these processes was enhanced in MOLF/Ei macrophages relative to C57BL/6J (Fig. 1 D). Thus, MOLF/Ei mice exhibit a specific enhancement in p38 and IKK activity after TLR2 stimulation.
We next asked if more receptor proximal and regulatory signaling events also displayed increased activity in the MOLF/Ei TLR response. Stimulus-induced degradation of IRAK-1 is a well characterized outcome of IL-1R/TLR signaling (Yamin and Miller, 1997), which is believed to be an important inhibitory event (Siedlar et al., 2004). An examination of IRAK-1 protein concentration after TLR2 stimulation revealed that MOLF/Ei macrophages did not degrade IRAK-1 more quickly than C57BL/6J but, rather, exhibited a slightly more prolonged IRAK-1 half-life (Fig. 1 E). Together, these findings imply that the hyperresponsiveness in MOLF/Ei TLR signaling is biased toward specific proinflammatory outcomes, with regulatory functions such as IRAK-1 protein degradation showing equivalent or decreased activity.
Genetic mapping of high IL-6 production in MOLF/Ei
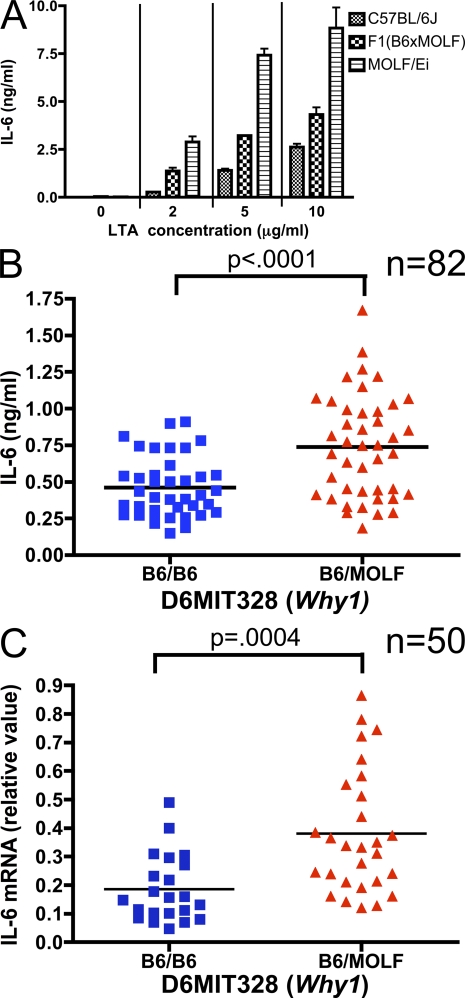
We next sought to elucidate the genetic basis for the difference in TLR-stimulated MOLF/Ei and C57BL/6J macrophages. We previously reported that genome-wide linkage analysis in a panel of N2(C57BL/6Jx(C57BL/6JxMOLF/Ei)) mice identified two loci associated with LTA-stimulated IL-6 secretion (Conner et al., 2008). The MOLF/Ei allele at one of these loci, Why2, was negatively associated with IL-6 production, which was the result of a cis-acting gene regulatory element allowing TLR-inducible expression of the negative regulator IRAK-1BP1. We therefore focused on the Why1 locus, a 21-Mb region on chromosome 6 between markers D6MIT36 and TNFRSF1a. Using secreted IL-6 protein 6 h after activation with LTA, a codominant mode of inheritance was observed as indicated by the intermediate phenotype of F1(B6xMOLF) mice (Fig. 2 A). Evaluation of 82 N2 backcross mice by this assay revealed a significantly increased amount of IL-6 secretion associated with a copy of the MOLF Why1 allele (Fig. 2 B), indicating that a factor promoting TLR-stimulated IL-6 production in MOLF/Ei mice is contained within this genomic interval.
Figure 2.
TLR2-stimulated IL-6 production is associated with the Why1 locus. (A) Codominant inheritance of IL-6 protein secretion. Peritoneal macrophages from parental strains and F1(C57BL/6JxMOLF/Ei) hybrid mice were plated and stimulated for 6 h with the indicated concentrations of LTA. IL-6 secretion was measured by ELISA. Results are ± SEM from triplicate wells, representative of three independent experiments. (B) Peritoneal macrophages from 82 N2 (C57BL/6Jx(C57BL/6JxMOLF/Ei)) mice were plated and stimulated with 2 µg/ml LTA as in A. Mean IL-6 production from triplicate wells is plotted for each mouse, grouped by genotype at D6MIT328, a marker near the peak of the Why1 locus. (C) Peritoneal macrophages from 50 independent N2 backcross mice were stimulated for 1 h with 2 µg/ml LTA. IL-6 mRNA was quantified by real time PCR. Mean mRNA production from two wells is plotted, grouped by genotype at D6MIT328. P-values were calculated in B and C using a two-tailed Student's t test.
We assessed IL-6 protein secretion after only 6 h of LTA stimulation in an effort to maximize the effect of the early signaling differences depicted in Fig. 1, C and D. However, the possibility remained that late-acting effects, such as differences in protein translation, stability, or secretion, were playing a significant role in the phenotypic outcome. We therefore assessed IL-6 transcript 1 h after stimulation to confirm that the underlying polymorphism at Why1 was functioning within the initial pathway leading to TLR-induced gene transcription. Increased IL-6 messenger RNA (mRNA) accumulation 1 h after stimulation was significantly associated with the MOLF Why1 allele (Fig. 2 C). Thus, the Why1 locus interacts with TLR pathways within the first hour after activation.
In both association studies (Fig. 2, B and C), all mice that were homozygous for the C57BL/6J allele of Why1 were low producers of IL-6, indicating that the MOLF/Ei Why1 allele is necessary for N2 mice to generate a heightened response. In contrast, mice with a heterozygous genotype for Why1 exhibited a wide range of secreted IL-6. These data indicate that heterozygosity for Why1 alone is not sufficient to confer high levels of IL-6 but probably acts in combination with additional MOLF/Ei alleles.
To confirm that the Why1 locus is a critical heritable determinant of IL-6 production in TLR-stimulated macrophages, we assessed its inheritance through five generations of mice chosen for serial backcrossing to C57BL/6J on a phenotypic basis. At each generation, peritoneal macrophages were isolated from each mouse using a nonlethal method and high-responding mice were identified for continued backcrossing. Mice producing high levels of IL-6 in each generation were significantly more likely to carry a MOLF allele of the Why1 locus (Table I) as determined by a transmission disequilibrium test (Beebe et al., 1997). Together, these data demonstrate that the Why1 genomic interval is required for the MOLF/Ei TLR hyperresponsive phenotype.
Table I.
Transmission disequilibrium test of the Why1 locus in high IL-6–secreting mice through the N6 generation
| Total mice | High mice used for breeding | χ2 | P-value | |||
|---|---|---|---|---|---|---|
| Why1 genotype | ||||||
| Generation | Hom | Het | Total | |||
| N2 | 41 | 1 | 11 | 12 | ||
| N3 | 67 | 0 | 7 | 7 | ||
| N4 | 28 | 0 | 6 | 6 | ||
| N5 | 32 | 0 | 3 | 3 | ||
| N6 | 32 | 1 | 6 | 7 | ||
| Total | 200 | 2 | 33 | 35 | 12.86 | 0.0003 |
Genetic background complements the effects of Why1
The detected genetic linkage to the Why1 locus suggests that the MOLF/Ei allele of Why1 is uniquely capable of mediating the hyperresponsive phenotype depicted in Fig. 1. However, the extent to which this locus depends upon the activity of other MOLF/Ei genes is not clear. Indeed, haplotype grouping of TLR2-mediated IL-6 production revealed that N2 backcross mice homozygous for the C57BL/6J allele of Why1 were universally low responders, whereas a wide distribution of responses was observed for mice containing a copy of the MOLF/Ei allele (Fig. 2, B and C). These data imply that the MOLF/Ei allele of Why1 is necessary but not sufficient for the earlier and more robust activation of TLR signaling.
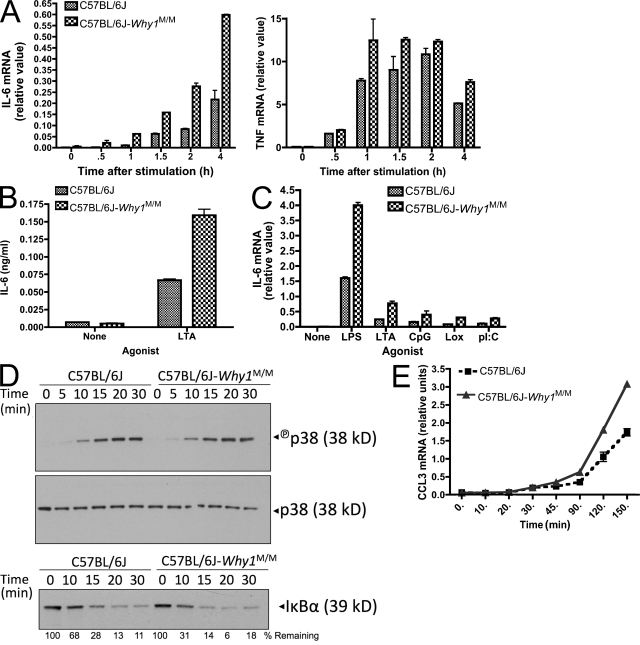
To test the effect of the Why1 locus on TLR signaling in the absence of other MOLF/Ei genes, we generated a congenic strain, which contained a MOLF/Ei haplotype for the entire Why1 interval on a C57BL/6J background. TLR2-stimulated IL-6 transcription and protein secretion were increased in C57BL/6J-_Why1_MOLF/MOLF macrophages, although not to the same extent as in pure MOLF/Ei mice, compared with wild-type C57BL/6J (Fig. 3, A and B). In contrast, a much smaller effect was observed for TNF-α transcription. This effect was not TLR2 specific, as macrophages from congenic mice were hyper-responsive to TLR3, TLR4, TLR9, and TLR7 stimulation as well (Fig. 3 C).
Figure 3.
Increased IL-6 production in C57BL/6J-Why1MOLF/MOLF congenic mice. (A and B) Peritoneal macrophages were isolated from wild-type and Why1 congenic mice and stimulated with 2 µg/ml LTA. IL-6 or TNF mRNA was analyzed by real-time PCR at the indicated times (A) and IL-6 protein secretion was analyzed by ELISA 6 h after stimulation (B). (C) Cells were stimulated for 2 h with 100 ng/ml LPS, 2 µg/ml LTA, 200 nM CpG, 100 µM Loxoribine, or 25 µg/ml poly(I:C), and IL-6 mRNA was analyzed by real-time PCR. Data shown as mean ± range of duplicate wells (A and C) or ± SEM of triplicate wells (B). (D) Cells from wild-type and Why1 congenic mice were stimulated with 2 µg/ml LTA for the indicated times and protein was extracted for Western blot analysis of p38 activation and IκBα degradation. Total p38 served as a loading control. (E) Peritoneal macrophages from the indicated strains were stimulated with 2 µg/ml LTA for the indicated times and CCL3 mRNA was measured by real-time PCR analysis. Error bars indicate SEM. All experiments are representative of two to three independent trials.
We next addressed the upstream signaling effects of the Why1 locus isolated on a C57BL/6J background. In contrast to pure MOLF/Ei cells, Why1 congenic macrophages did not display an increase in p38 activation. A slightly more rapid degradation of IκBα was observed in peritoneal macrophages, which is consistent with increased IKK activity (Fig. 3 D). The observed rate of IκBα degradation by Western blot analysis was only incrementally increased in peritoneal macrophages and was not observed in BM-derived macrophages (BMDMs; Fig. 3 D and not depicted). We reasoned that this may be caused by the low sensitivity of Western blot analysis in detecting slight differences in NF-κB pathway activity. To assess more subtle effects on NF-κB in Why1 congenic macrophages, we examined TLR-induced transcription of CCL3 (MIP-1α). The CCL3 promoter has been shown to be predominantly dependent on NF-κB activity and to contain fewer additional elements than the IL-6 promoter (Grove and Plumb, 1993). We observed increased CCL3 transcription in TLR2-stimulated Why1 congenic macrophages relative to those from wild-type C57BL/6J mice (Fig. 3 E), which is consistent with increased NF-κB activity in these cells. We conclude that MOLF/Ei background alleles, in addition to Why1, are important in conferring the robust levels of phenotypic differences in p38 and NF-κB activity, which is detectable by Western blot analysis in Fig. 1 (C and D). However, the Why1 locus itself is sufficient to partially recapitulate the hyperresponsiveness, as shown by increased cytokine transcription and the small but detectable effects on IκBα degradation.
Essential role of IRAK-2 in the MOLF/Ei early TLR response
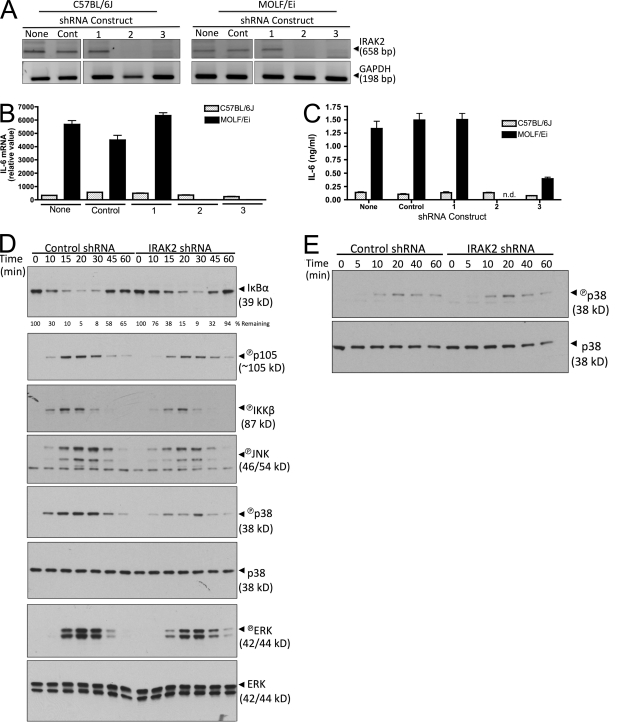
Of the genes located within the Why1 interval (Table S1), Irak2 emerged as a leading candidate to underlie the detected linkage because of the IRAK family's known role in TLR signal transduction. Previous studies have demonstrated that this gene is not required for early activation of NF-κB and early cytokine transcription on a classical inbred background (Kawagoe et al., 2008; Wan et al., 2009). Consistent with these findings, IRAK-2 knockdown in BMDMs using two effective short hairpin RNA (shRNA) targeting constructs had no effect on the relatively small amount of IL-6 produced by C57BL/6J mice. In contrast, knockdown of IRAK-2 in MOLF/Ei BMDMs severely attenuated TLR2-induced IL-6 mRNA and protein accumulation 4 and 6 h after stimulation, respectively (Fig. 4, A–C). Thus, IRAK-2 is required for IL-6 production in MOLF/Ei but not C57BL/6J macrophages early after TLR stimulation.
Figure 4.
IRAK-2 is required for early TLR2 responses in MOLF/Ei but not C57BL/6J macrophages. (A–C) C57BL/6J and MOLF/Ei BMDMs were infected with lentiviral constructs containing either control or one of three IRAK-2–targeting shRNA sequences. (A) To assess knockdown efficiency, mRNA was harvested and cDNA was amplified with IRAK-2 or GAPDH-specific PCR primers. (B and C) Cells were stimulated with 2 µg/ml LTA and IL-6 was quantified at 4 h using RT-PCR, with error bars indicating SEM (B), or at 6 h using ELISA, with error bars indicating SEM (C). n.d., not detected. Data are representative of three independent experiments. (D and E) MOLF/Ei BMDMs were infected with control or IRAK-2–targeting construct number two and stimulated with 2 µg/ml LTA (D) or under hypertonic NaCl conditions (E) for the indicated times. MAPK and NF-κB pathway signaling events were assessed using the indicated Western blot analyses. Total p38 and ERK serve as loading controls. Data are representative of at least two independent experiments.
To assess the mechanism by which IRAK-2 promotes early transcription of IL-6 in MOLF/Ei peritoneal macrophages, we examined the effect of IRAK-2 knockdown on IKK and MAPK activation status by Western blot analysis (Fig. 4 D). IRAK-2 knockdown impaired the IKK-dependent processes of IκBα degradation and p105 phosphorylation. To more directly assay the activation status of IKK, we measured TLR2-induced phosphorylation of the IKK-β subunit, which is the major IKK component targeted by TLR signal transduction (Bonizzi and Karin, 2004). IKK-β phosphorylation was impaired in IRAK-2 knockdown cells (Fig. 4 D), suggesting that in MOLF/Ei mice, IRAK-2 affects the IKK–NF-κB axis above the level of IKK activation. We next examined the phosphorylation status of the MAPK proteins JNK, p38, and ERK. LTA-induced p38 activity was severely impaired by IRAK2 knockdown (Fig. 4 D). This effect was specific for IRAK-dependent pathways, as osmotically stressed macrophages activated p38 to equal levels in control and IRAK-2–targeted knockdown cells (Fig. 4 E). In comparison with p38 status, JNK was more marginally affected and a slightly delayed onset of ERK phosphorylation was observed (Fig. 4 D). Thus, IRAK-2 knockdown in MOLF/Ei macrophages affects IKK activity, as well as all three MAPKs with the largest effect on p38.
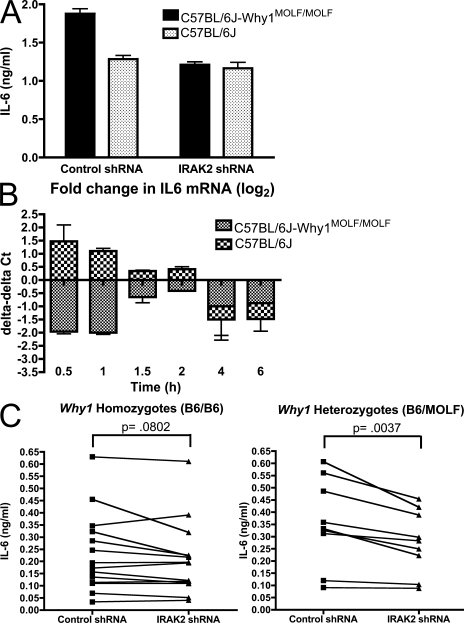
The data depicted in Fig. 4 imply that IRAK-2 occupies a critical position in MOLF/Ei TLR signal transduction. However, they do not address the question of whether allelic variation in IRAK-2 itself is responsible for the increased dependence on this gene or, alternatively, whether other MOLF/Ei gene products function to promote hyperresponsiveness through a requisite interaction with IRAK-2. To address this question, we assayed the effect of IRAK-2 shRNA knockdown in C57BL/6J-Why1 congenic macrophages, which contain the MOLF/Ei allele of Irak2 on a >98% C57BL/6J background. Why1 congenic BMDMs transduced with a control shRNA hairpin construct secreted an increased amount of IL-6 protein relative to C57BL/6J BMDMs 6 h after LTA stimulation. In contrast, IL-6 accumulation was similar between cells from both strains that were transduced with an IRAK-2–targeting hairpin (Fig. 5 A). We next examined the effect of IRAK2 knockdown on IL-6 transcription at various time points after TLR2 activation. The largest difference between strains in the response to IRAK-2 knockdown was at the earliest time points tested, whereas by later times both strains showed a similar decrease in IL-6 mRNA levels. Surprisingly, IRAK-2 knockdown was associated with increased IL-6 accumulation in C57BL/6J cells at 30 and 60 min after LTA stimulation, whereas a notable decrease was occurring in Why1 congenic macrophages at this time (Fig. 5 B). These data demonstrate that the MOLF/Ei allele of IRAK-2 is particularly important for early TLR activity and suggest that the C57BL/6J allele may have a net inhibitory effect on IL-6 production at these time points.
Figure 5.
The effect of IRAK-2 knockdown in macrophages is allele specific. (A and B) BMDMs from C57BL/6J and C57BL/6J-_Why1_MOLF/MOLF mice were transduced with control or an IRAK-2–targeting shRNA construct. Cells were stimulated with 2 µg/ml LTA for 6 h followed by IL-6 ELISA analysis (A) or for the indicated times followed by IL-6 mRNA quantification by real-time PCR (B). Real-time PCR data are shown as the fold change (in Ct values) observed in IRAK-2 shRNA treated relative to control-treated cells. Data shown as mean ± SEM from triplicate wells (A) or ± range from duplicate wells (B), representative of two independent experiments. (C) Peritoneal macrophages from 14 Why1 homozygous or 9 heterozygous N2 backcross mice were plated and infected with control or IRAK-2–targeting shRNA constructs. 3 d after infection, cells were stimulated with 2 µg/ml LTA and IL-6 protein secretion was measured by ELISA. Mean values of three wells for each mouse are plotted. P-values were calculated using a paired Student's t test analysis.
Because we originally identified the Why1 locus through meiotic recombination events in N2 (C57BL/6Jx[C57BL/6JxMOLF/Ei]) mice (Fig. 2, B and C), we hypothesized that genetically assorted progeny inheriting the MOLF/Ei Irak2 allele would demonstrate increased sensitivity to knockdown compared with mice homozygous for the C57BL/6J allele. To test this hypothesis, we lentivirally infected peritoneal macrophages from a small panel of N2 backcross mice with IRAK-2–targeting shRNA construct 2, which showed the greatest efficiency of knockdown in BMDMs (Fig. 4 A). Although peritoneal macrophages are terminally differentiated and nondividing, we have observed up to 30% infection efficiency with lentiviral constructs in these cells (Fig. S1). Consistent with an increased dependence for IRAK-2 in mice inheriting a MOLF/Ei Why1 allele, Why1 heterozygotes, but not C57BL/6J homozygotes, produced significantly less IL-6 when expressing an IRAK-2 targeting shRNA compared with a nonhomologous control construct (Fig. 5 C). Thus, Irak2 is directly involved in the linkage of the trait to Why1.
Differential expression of IRAK-2 isoforms in MOLF/Ei and C57BL/6J mice
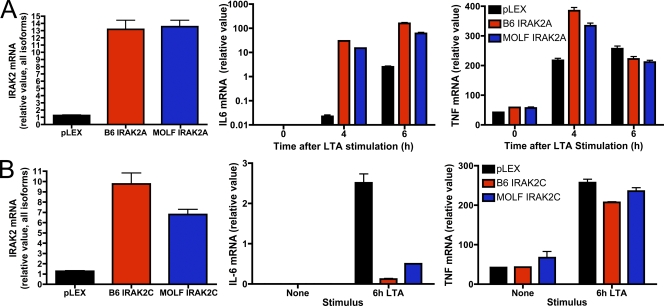
Sequence analysis of the IRAK-2 open reading frame revealed seven nonsynonymous mutations between MOLF/Ei and C57BL/6J. Four of these mutations were also present in two other wild-derived strains with similar TLR-responsive phenotypes (Fig. S2). The Irak2 gene consists of four isoforms, of which two, IRAK-2C and IRAK-2D, are believed to negatively regulate inflammatory signaling (Hardy and O'Neill, 2004). We examined the potential contribution of the amino acid differences between strains by expressing IRAK-2A or IRAK-2C in the macrophage cell line RAW264.7. Consistent with previous findings (Hardy and O'Neill, 2004), IRAK-2A from both strains potently increased TLR-mediated transcription of IL-6, whereas IRAK-2C had an inhibitory effect. However, no functional differences were detected between the alleles of either isoform (Fig. 6, A and B). Therefore, we conclude that polymorphisms in the amino acid sequence of MOLF/Ei and C57BL/6J IRAK-2 do not underlie the observed phenotype.
Figure 6.
Ectopic expression of MOLF/Ei or C57BL/6J IRAK-2 isoforms results in similar effects on TLR2 activity. RAW264.7 macrophages were transduced with a lentiviral construct encoding the MOLF/Ei or C57BL/6J alleles of IRAK-2A (A) or IRAK-2C (B). Cells were stimulated for the indicated times with 2 µg/ml LTA. Total levels of IRAK-2 in unstimulated cells as well as IL-6 and TNF mRNA accumulation were assessed by real-time PCR. Data are depicted as mean ± range of duplicate wells, representative of three independent experiments.
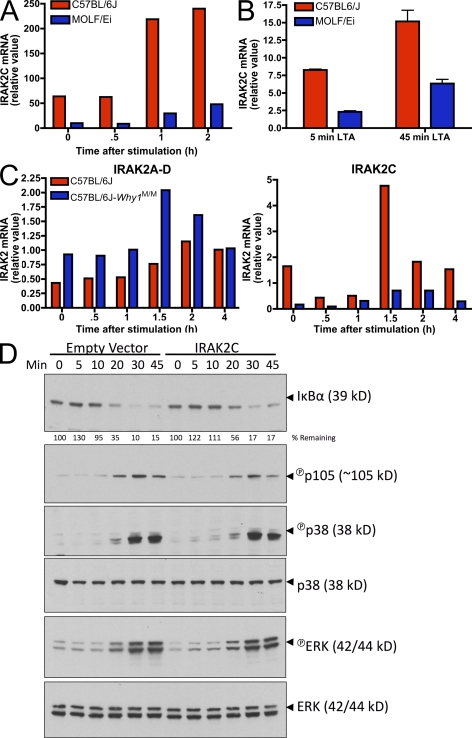
Gene expression polymorphisms are a common determinant of quantitative traits (Schadt et al., 2005). We therefore performed real-time PCR analysis to examine the mRNA levels of IRAK-2 isoforms in MOLF/Ei and C57BL/6J macrophages. We focused specifically on IRAK-2A as well as IRAK-2C, because the former is believed to be the major gene product of Irak2 and the latter is the only isoform that originates from a unique promoter within the third intron (Hardy and O'Neill, 2004). IRAK-2C expression demonstrated the largest difference between strains, with C57BL/6J expressing 5–10-fold higher levels of this antiinflammatory mediator in both BMDMs and peritoneal macrophages (Fig. 7, A and B). To assess IRAK-2 transcriptional regulation by cis-acting elements in the absence of trans-acting differences, we compared C57BL/6J and C57BL/6J-Why1MOLF/MOLF peritoneal macrophages. Why1 congenic mice expressed higher levels of total IRAK-2 mRNA but lower amounts of IRAK-2C, both basally and upon TLR stimulation (Fig. 7 C).
Figure 7.
IRAK-2C is differentially expressed between MOLF/Ei and C57BL/6J macrophages and inhibits NF-κB activity. (A) BMDMs from MOLF/Ei and C57BL/6J mice were stimulated for the indicated times with 2 µg/ml LTA and IRAK-2C mRNA was assessed using real-time PCR primers with a forward primer located within the unique 5′ untranslated region of IRAK-2C and a reverse primer located within IRAK-2 exon 6. Primer specificity was confirmed by agarose gel electrophoresis and sequencing (not depicted). (B and C) Peritoneal macrophages from the indicated strains were stimulated for the times shown with 2 µg/ml LTA. IRAK-2C mRNA was quantified as in A and total IRAK-2 mRNA was quantified using primers common to all isoforms (C). Error bars indicate SEM. (D) RAW264.7 cells were transduced with a lentiviral construct coding for the MOLF/Ei allele of IRAK-2C. Cells were stimulated for the indicated times with 2 µg/ml LTA, and NF-κB and MAPK pathways were assessed by Western blot analysis. Total p38 and ERK served as loading controls. All data are representative of two to four independent experiments.
Although our findings (Fig. 6 B), as well as a previous study (Hardy and O'Neill, 2004), demonstrate an inhibitory role for IRAK-2C in the TLR response, the position at which it interacts with the signal transduction pathway is not known. To address this, we examined MAPK and NF-κB status in RAW264.7 cells ectopically overexpressing IRAK-2C. IRAK-2C overexpression attenuated IκBα degradation and p105 phosphorylation but exerted only a minimal effect on p38 MAPK activation and no effect on ERK (Fig. 7 D). These findings suggest that differences in IRAK-2C expression alone are sufficient to affect NF-κB activation in a macrophage cell line. However, the incremental effect on IκBα activation compared with the larger decrease in IL-6 production observed in Fig. 6 B suggests that, in addition to the pathways investigated here, other IRAK-2C–dependent mechanisms of TLR signaling inhibition may also exist.
Identification of IRAK2C promoter mutations
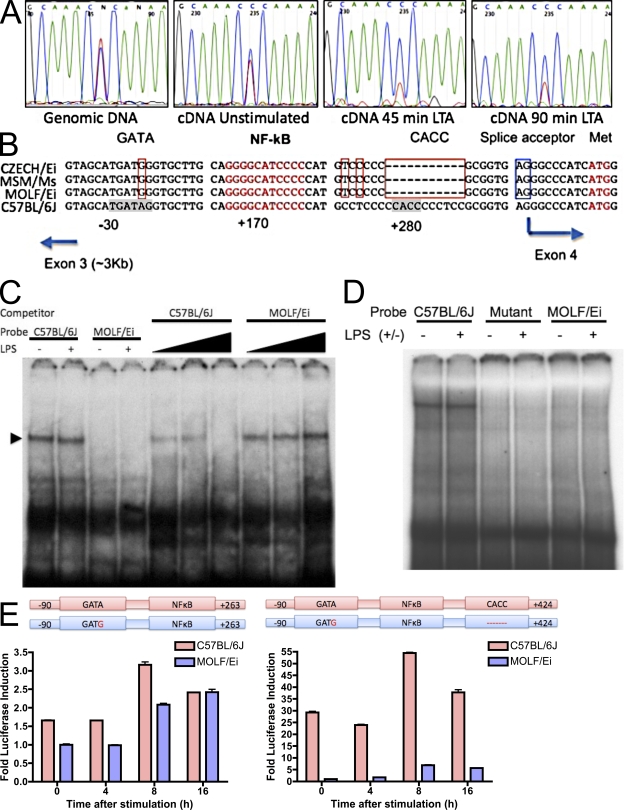
We next asked if differences in IRAK-2C expression mediated by the Why1 locus link the mechanistic observations to the mapping association studies. To confirm that cis-acting regulatory elements within the Why1 interval mediate the IRAK-2C expression differences between strains, we examined the allelic bias of mRNA transcripts produced in F1(C57BL/6JxMOLF/Ei) macrophages. Allelic gene expression analysis based on a C→T polymorphism revealed a larger proportion of C57BL/6J allelic transcript in F1 cells (Fig. 8 A). This difference was amplified upon TLR stimulation, which is consistent with previous studies demonstrating that IRAK-2C is under the transcriptional control of inducible regulatory elements (Hardy and O'Neill, 2004).
Figure 8.
Identification of cis-acting regulatory elements conferring differential IRAK-2C expression. (A) Genomic DNA or cDNA from F1(C57BL/6JxMOLF/Ei) macrophages was PCR amplified and analyzed by sequence analysis to determine allelic bias in IRAK-2C transcript. (B) Sequence alignment of the genomic region of IRAK-2C from three wild-derived strains (top three lanes) and C657BL/6J (bottom lane). Positions of the elements are given in base pairs with respect to the transcription initiation site (at 0 bp). Red boxes, mutations common for all wild-derived strains; Blue box, splice acceptor site for IRAK-2A isoforms; red text, NF-κB consensus and translation start of the IRAK-2C protein. Consensus sites were identified using MatInspector software (Genomatix Software GmbH). (C) EMSA was performed using nuclear extracts of LPS-activated RAW cells and a double-stranded 32P-labeled oligonucleotide corresponding to the sequence containing the GATA-binding site. Cold competition assay was performed with 5×-, 25×-, or 100×-fold excess of either B6 or MOLF unlabeled oligonucleotide. (D) Nuclear extracts from LPS-activated RAW cells were incubated with radiolabeled probe derived from C57BL/6J sequence or a sequence with an (A/T) substitution in the CACC-binding site (mutant). Alternatively, a MOLF-derived probe from the region corresponding to the 10-bp deletion was used. (E) RAW264.7 cells were stably transfected with constructs containing the indicated IRAK-2C promoter regions from MOLF/Ei or C57BL/6J fused to luciferase. Equivalent levels of transfection and integration were confirmed using real-time PCR analysis of genomic DNA (not depicted). Cells were stimulated with 100 ng/ml LPS for the indicated times and luciferase was quantified as a measure of promoter activity. Values are expressed relative to unstimulated MOLF/Ei promoter constructs and are shown as mean ± SEM of three wells. Data are representative of three independent experiments.
We then proceeded to sequence the 4-kb intronic region, which contains the IRAK-2C promoter. Sequence comparison revealed two prime candidate mutations: an A→G point mutation in a GATA transcription factor consensus binding site and an 11-bp deletion in a region including a CACC SP1/KLF binding site (Philipsen and Suske, 1999). These mutations were also shared by two other wild-derived strains, Czech/Ei and MSM/Ms (Fig. 8 B), which are also hyperresponsive to TLR activation (Conner et al., 2008). To assess the effect of each mutant sequence on transcription factor binding, we conducted electromobility shift analysis (EMSA). The C57BL/6J, but not the MOLF/Ei, GATA element was bound by protein from a nuclear lysate prepared from the RAW264.7 macrophage cell line (Fig. 8 C). The CACC element was also capable of binding factors in macrophage nuclei, as the C57BL/6J oligonucleotide, but not the contiguous MOLF/Ei or a C57BL/6J version with mutations in place of the deleted region, was shifted by incubation with RAW264.7 nuclear extract (Fig. 8 D).
Finally, we examined the functional consequence of these mutations on gene expression in a macrophage cell line. Fusion of the luciferase gene to the promoter region containing the GATA site, as well as an NF-κB element present in both strains, revealed a difference of approximately one to twofold between the MOLF/Ei and C57BL/6J sequences. Introduction of the CACC element–containing sequence, however, resulted in a difference of 10–30-fold between strains (Fig. 8 E). Thus, although the GATA binding site alone may be responsible for a small fraction of the expression difference, the CACC site is required for the major increase in C57BL/6J IRAK-2C transcription relative to MOLF/Ei.
DISCUSSION
This study demonstrates the ability of an unbiased phenotypic screen in evolutionarily diverse mouse subspecies to refine our model of immune signaling pathways. Although the genetic diversity of these mouse strains has been appreciated previously, it has also limited their use in forward genetic analyses because of the often polygenic nature underlying observed traits. In addition, high levels of polymorphism in these mice, as compared with classical inbred strains, have made it difficult to establish a causative link between a candidate-gene and a phenotype. In our mapping of Why1, we were able to overcome these problems by performing knockdown of the candidate gene directly on a panel of backcross mice and showing an allele-specific effect of silencing (Fig. 5 C). This method, which was previously applied in describing the product of the Why2 locus, IRAK-1BP1 (Conner et al., 2008), greatly facilitated our ability to assess gene candidates in the context of genetic assortment.
IRAK-2 emerged as the likely candidate in our mapping studies because of the known role of IRAK family kinases in TLR-mediated signaling pathways. Its candidacy was further supported by the strain-specific effect of IRAK-2 silencing, which preferentially inhibited the signaling pathways that were hyperresponsive in MOLF/Ei and C57BL/6J-Why1 congenic mice. The mutation identified in the promoter of the IRAK-2C isoform is a 10-bp deletion covering a CACC consensus element that lies in the 5′ untranslated region. This element has a predicted ability to bind Kruppel-like and Sp1 transcription factors (Philipsen and Suske, 1999). Together with a GATA-specific site that is present in C57BL/6J but not MOLF/Ei mice and located 5′ to the deletion, the CACC site is able to provide constitutively high as well as stimulus-dependent expression of IRAK-2C in C57BL/6J but not MOLF/Ei mice. The presence of both sites within the C57BL/6J promoter is especially pertinent, given previous studies on synergistic gene transactivation by elements binding each of these sites independently (Merika and Orkin, 1995; Gregory et al., 1996).
The outcome of this differential expression of IRAK-2 isoforms is a higher ratio of proinflammatory IRAK-2A relative to the inhibitory IRAK-2C in MOLF/Ei macrophages, which, in turn, allows IRAK-2A to assume a more central role in TLR activation early after stimulation (Fig. 9). Our model considers a critical function for IRAK-2A in MOLF/Ei mice, which provides additional stimulation for NF-κB and p38-mediated signaling. In contrast, the highly expressed IRAK-2C isoform in classical inbred mice potentially offsets the proinflammatory effect of IRAK-2A during the early phase of activation. Our data show that IRAK-2A is required but not sufficient for signaling in MOLF/Ei, suggesting that other gene products are almost certainly involved. In addition to the data presented here, our initial mapping revealed strong epistatic interactions between Why1 and other loci in the genome (Conner et al., 2008). Furthermore, there may be additional recessively inherited loci that promote inflammation but that would only be revealed using a reciprocal (MOLF/Eix[MOLFEixC57BL/6J]) backcross.
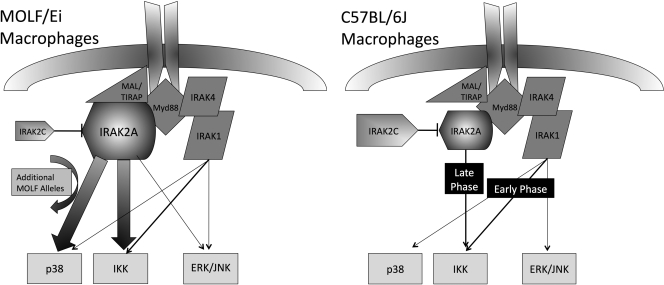
Figure 9.
Model of IRAK-2 usage in MOLF/Ei compared with C57BL/6J TLR signal transduction. In MOLF/Ei macrophages, knockdown experiments reveal that IRAK-2 plays a central role in activation of p38 and IKK, two pathways which show increased activity relative to C57BL/6J mice. Neither of these phenotypes can be completely recapitulated with the MOLF/Ei allele on a classical inbred background, suggesting that additional MOLF/Ei alleles are important for the complete levels of hyperresponsiveness observed in comparing parental strains. The MOLF/Ei allele of Irak2 expresses less of the IRAK-2C isoform, which may underlie the phenotype by decreasing interference with IRAK-2A activity. In C57BL/6J macrophages, which express an increased amount of IRAK-2C, previous studies have demonstrated that early TLR signaling events are not controlled by IRAK-2, although it is important for late activation of the NF-κB axis.
Previous analysis of mice with a targeted null mutation in IRAK-2 revealed its role in sustained levels of NF-κB–mediated gene activation (Kawagoe et al., 2008; Wan et al., 2009). Our analysis extends these findings by showing that in a wild-derived mouse model, IRAK-2 is indispensable for the early response. Silencing of IRAK-2 dramatically down-regulates IL-6 production in these mice, bringing it to levels even lower than those in C57BL/6J macrophages. In addition, the mechanistic basis for the phenotype provides a potential explanation for why targeting IRAK-2 in classical inbred strains does not provide an immediate effect on signaling: the balance between pro- and antiinflammatory isoforms of IRAK-2 in these strains is such that IRAK-2C may be capable of effectively inhibiting IRAK-2A. As the peak of IRAK-2C mRNA accumulation occurs at 1.5 h after stimulation, the proinflammatory nature of IRAK-2A may only become apparent at later time points. For the same reason, we observed that silencing of IRAK-2 in C57BL/6J macrophages does not alter signaling in the first hours after activation because the proinflammatory isoform is checked by high levels of IRAK-2C at these times. This model is further supported by our findings that at very early time points (30 and 60 min) after TLR stimulation, IRAK-2 knockdown is associated with a slight increase in IL-6 production in C57BL/6J mice (Fig. 5 B).
Several negative regulators of TLR-mediated signaling have been described, which exert their inhibitory effect at different positions within the pathway. Some of these inhibitors are products of alternative splicing, such as MyD88s (Janssens et al., 2002; Burns et al., 2003), soluble TLR2 (LeBouder et al., 2003), and soluble TLR4 (Iwami et al., 2000); others, like IRAK-M, are sole gene products (Kobayashi et al., 2002). In this paper, we propose that the previously described inhibitory molecule IRAK-2C, which originates from a unique promoter within the Irak2 gene, plays a role in the regulation of the TLR response of classical inbred mouse strains. The mechanism by which IRAK-2C inhibits TLR signaling remains to be elucidated. However, the domain structure of IRAK-2 offers some important clues. The death domain, positioned at the N terminus of full-length IRAK-2 is required for its interaction with MyD88 (Muzio et al., 1997) and likely with the MyD88 like adaptor MAL (Fitzgerald et al., 2001). The complete absence of the death domain in IRAK-2C suggests that it would not interact with these signaling components at the membrane. In contrast, IRAK-2 is proposed to interact with the key cytoplasmic adaptor molecule TRAF6 through its C terminus, leading to ubiquitination and activation of TRAF6 (Keating et al., 2007). Because the C terminus of IRAK-2C is identical to full-length IRAK-2, it may still interact with TRAF6 and potentially sequester this molecule away from IRAK-2A and other critical membrane signaling components. Our results demonstrate that knockdown of all IRAK-2 isoforms in the low IRAK-2C–expressing MOLF/Ei strain has an overall inhibitory effect, whereas no major effect is seen in the high IRAK-2C–expressing C57BL/6J strain. These findings suggest that direct inhibition of IRAK-2A will be an essential aspect IRAK-2C's mechanism.
One of the most surprising findings that emerged from our studies of the TLR response in MOLF/Ei mice was the observed specificity in affected early upstream signaling mediators. In addition to activation of the NF-κB pathway, MOLF/Ei mice displayed specific hyperactivity in p38, but not ERK or JNK, MAPK phosphorylation. These data are consistent with our observation that not all cytokines are increased in MOLF/Ei TLR responses relative to classical inbred mouse strains. The largest effect was observed for IL-6 mRNA produced at early time points after stimulation. C57BL/6J and MOLF/Ei macrophages displayed similar levels of TNF-α and IL-12p40 transcription, which, like IL-6 are known to be NF-κB–dependent genes (Shakhov et al., 1990; Sanjabi et al., 2005). The underlying mechanism for this specificity remains incompletely characterized. p38 has been shown to play a critical role in IL-6 transcription through histone H3 phosphorylation (Saccani et al., 2002), as well as through activation of the transcription factor CREB, whereas it has a smaller effect on TNF-α and IL-12p40 transcription (Park et al., 2005). Other signaling mediators may also be hyperresponsive in MOLF/Ei macrophages, which could synergize with p38-dependent pathways to specifically enhance the transcription of IL-6.
Remarkably, IRAK-2 knockdown on a MOLF/Ei background had its largest effects on the same signaling molecules that were hyperresponsive relative to C57BL/6J. Furthermore, IRAK-2A and IRAK-2C overexpression in a macrophage cell line vastly affected IL-6 mRNA accumulation, whereas effects on TNF-α were less substantial. These results imply that IRAK-2 not only amplifies the signal from TIR domain–containing molecules but also provides specificity for such amplification. Such discrimination between different targeted MAPKs has not yet been reported for any of the upstream components of the TLR-mediated pathway. This effect of IRAK-2 will be important to investigate further.
Although ubiquitously expressed pathogen-associated molecules activate a relatively small number of TLRs, the effects of TLR-responsive genes are quite specific. Their individual actions can heavily bias the remaining immune response by, for example, promoting the development of T effector subsets toward a Th1, Th2, Th17, or T reg phenotype (Iwasaki and Medzhitov, 2004). In considering the effect of the early innate immune system on subsequent processes, the evolutionary context under which wild-derived mice have recently existed is critical. It is remarkable that incorporation of such TLR-proximal factors as IRAK-2 has arisen to shape differential expression of specific cytokine genes.
Whereas the MOLF/Ei allele of Why2 was associated with an inhibitory effect on TLR-induced IL-6 production (Conner et al., 2008), the findings reported in this paper show that MOLF/Ei Why1 promotes a faster and more robust IL-6 response. The coevolution of these two opposing loci may have been critical, allowing ancestral wild-derived species to develop uniquely potent yet, at the same time, highly regulated innate immune functions. Previous studies have described both increased susceptibility and resistance of wild-derived strains in various infectious models (Urosevic et al., 1999; Sebastiani et al., 2002). Although neither of these phenotypes has been genetically mapped to the Why1 or Why2 loci from our studies, the unique cytokine response profile that we observe in vitro will likely have corresponding outcomes in the context of a live infection. Under the circumstances of persistent pathogen replication and the formation of an adaptive response, the Why1 and Why2 loci may help to identify important effects of the time course and specificity of cytokines produced by the innate immune system on the development of infectious or host mediated pathology.
The scientific approach of studying individual genes in isolation is increasingly giving way to the appreciation of networked genes working together to determine complex phenotypes (Oda and Kitano, 2006). Wild-derived mice are ideal models for comparison of different gene networks. Our data so far support the hypothesis that innate immune networks in classical and wild-derived inbred mice are organized differently and will be rich in unique gene–gene interactions. In addition to identifying single components of these interactions (which could themselves be novel), the biggest potential of this work is the ability to establish new signaling pathways, which would otherwise be difficult to predict. Our data demonstrate that IRAK-2A is positioned in the center of the TLR scaffold in MOLF/Ei and that it is likely much less affected by the inhibitory isoform IRAK-2C. Interestingly, there is only one proinflammatory human homologue of IRAK-2, which suggests a greater similarity between organization of TLR signaling elements in humans and wild-derived mice compared with classically used strains. This possibility strongly supports further characterization of immunological responses in wild-derived mice for their potential to model critical elements of human biology.
MATERIALS AND METHODS
Mice and isolation of MEFs.
C57BL/6J and MOLF/Ei parental strains were obtained from The Jackson Laboratory. All mice were housed in a pathogen-free facility at the Tufts University School of Medicine. MEFs were isolated from 13.5-d mouse embryos by trypsination after removal of the head and liver. All mouse procedures were performed under a protocol approved by the Tufts University/Tufts Medical Center Institutional Animal Care and Use Committee.
Cells.
Primary cells were harvested for phenotypic analysis from all mice at 6–10 wk of age. Peritoneal macrophages were obtained by injection with 1 ml of 3% thioglycollate. 3 d after priming, the peritoneal cavity was washed with cold PBS and cells were suspended in DMEM, 10% FBS (Atlanta Biologicals), and 1% Pen-Strep (Invitrogen). Cells were plated and incubated overnight at 37°C in 5% CO2 before phenotypic analysis. BMDMs were matured and maintained as previously described (Conner et al., 2008). The RAW264.7 mouse macrophage cell line (American Type Culture Collection) was grown in DMEM, 10% FBS, and 1% Pen-Step.
Reagents.
CpG, loxoribine, and purified LTA from Staphylococcus aureus (tlrl-pslta) were obtained from InvivoGen. Salmonella Minnesota Re595 LPS was obtained from Enzo Biochem., Inc. Poly I:C was obtained from GE Healthcare. Rabbit polyclonal antibodies to phosphorylated and total p38, ERK, and JNK, as well as phosphorylated IKK-α/β and p105, were obtained from Cell Signaling Technology. Polyclonal antibodies to IRAK-1, IκBα, and IκBβ were obtained from Santa Cruz Biotechnology, Inc.
TLR stimulation and phenotypic assays.
All cells were plated and incubated overnight before stimulation. For ELISA analysis, 200,000 cells were plated per well in a 48-well tissue culture-treated dish. Cells were stimulated with prewarmed media containing the TLR agonist for 6 h, after which supernatants were harvested. IL-6 was measured using anti–mouse Duo-Set IL-6 ELISA (DY-406; R&D Systems). For mRNA analysis, cells were plated at a density of 700,000 per well in a 12-well dish. After stimulation, cells were lysed using TRIZOL (Invitrogen). Complementary DNA (cDNA) was synthesized using random primers and Mu-MLV reverse transcription (New England Biolabs, Inc.), followed by real-time PCR analysis using mouse IL-6 or TNF-α along with GAPDH Taqman probes (Applied Biosystems).
Western blot analysis.
After TLR stimulation, cells were lysed on ice with a cytoplasmic lysis buffer (50 mM Tris, pH 8, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 1 mM NaVanadate, and 10 mM NaF) supplemented with Halt protease inhibitor cocktail (Thermo Fisher Scientific) for 10 min. Lysates were then centrifuged at 13,000 rpm at 4°C for 10 min. Cleared lysates were resolved on a 4–12% gradient Bis-Tris SDS gel (NuPAGE; Invitrogen) and transferred to a nitrocellulose membrane. After incubation with specific antibodies, chemiluminescence was detected using ECL substrate (Thermo Fisher Scientific).
Lentiviral transduction.
IRAK-2 targeting shRNA constructs in the pLKO.1 vector were obtained from Open Biosystems (Constructs 1, 2, and 3 represent clone IDs TRCN000022499, TRCN000022502, and TRCN000022503, respectively). IRAK-2A and IRAK-2C cDNAs were cloned using PCR from C57BL/6J or MOLF/Ei cDNA into the pLEX (Open Biosystems) lentiviral expression vector for ectopic expression experiments. Lentiviral particles were generated by transfection (FuGENE) of the expression construct into 293-T cells along with the packaging constructs pSPAX2 and pMD2.G (Addgene). Supernatant from 293-T cells was harvested on days 2 and 3 after transfection and was 0.45-µm filtered. All cells were infected by incubation with a 1:1 ratio of viral supernatant for 24 h. BMDMs were infected on day 4 of maturation and selected with 3 µg/ml puromycin beginning on day 6. Peritoneal macrophages were infected immediately upon plating for 24 h and recovered for 48 h with no selection before assays were performed. RAW264.7 macrophages were infected for 24 h, recovered for 24 h, and subsequently selected in 3 µg/ml puromycin.
IRAK-2 mRNA quantification.
For quantification of IRAK-2 isoforms, real-time PCR primers were designed to amplify all isoforms or IRAK-2C specifically by using a forward primer in the 4′ exon, which has been previously described as unique to IRAK-2C (Hardy and O’Neill, 2004). Specificity of primers was assessed using agarose gel electrophoresis and sequencing of the products. Cells were stimulated and cDNA was prepared as described in TLR stimulation and phenotypic assays, followed by amplification with SYBR Green (Applied Biosystems). All cells were normalized with a GAPDH-specific primer set.
DNA sequencing.
After PCR amplification from genomic DNA or cDNA, products were isolated on an agarose gel and extracted (QIAGEN). DNA was sequenced at the Tufts University Core Facility using a 3130XL sequencer (Applied Biosystems).
EMSA.
RAW264.7 cells were left untreated or stimulated with 100 ng/ml of LPS in DMEM for 1 h, after which nuclear proteins were extracted using NucBuster (EMD) and incubated with Klenow-labeled double-stranded oligonucleotide containing the indicated binding sites from the IRAK-2C promoter. The sequences used to assess the GATA site were 5′-TGTCTGCAAAGTAGCATGATAGGTGCTTGATTCAG-3′ (wild type) and 5′-TGTCTGCAAAGTAGCATGATGGGTGCTTGATTCAG-3′ (mutant). The CACC binding site was assessed using the following C57BL/6J sequence or sequence with an (A/T) mutation: 5′-AGCTGGCCTCCCCCACCCCCTCCGCGGTGTCAGGAGGTG-3′ (wild type) and 5′-AGCTGGCCTCCCCCTCCCCCTCCGCGGTGTCAGGAGGTG-3′ (mutant). Alternatively, the following MOLF/Ei-derived probe from the region corresponding to the 10-bp deletion was used: 5′-AGCTGGCCTCCCCCTCCCCCTCCGCGGTGTCAGGAGGTG-3′. Samples were resolved on a nondenaturing 6% polyacrylamide gel.
Luciferase promoter assays.
The indicated regions of the IRAK-2C promoter were amplified by PCR from C57BL/6J or MOLF/Ei genomic DNA and cloned into the pGL4.20 (Promega) firefly luciferase vector. Vector was linearized using BamHI and transfected into RAW264.7 cells (FuGENE). Cells were selected in 3 µg/ml puromycin for 3 wk to generate stably expressing lines. To confirm equivalent levels of transfection, genomic DNA was isolated from each cell line and amplified with vector specific primers using SYBR green (Applied Biosystems) and normalized to genomic (intronic) GAPDH sequences. Cells were stimulated and lysed using Passive Lysis buffer and luciferase was read using Dual Luciferase reagent (Promega) in a single tube luminometer.
Online supplemental material.
Fig. S1 shows efficiency of lentiviral infection in mouse peritoneal macrophages. Fig. S2 shows amino acid polymorphisms in Irak2 in several wild-derived mouse strains and the C57BL/6J strain. Table S1 shows a list of genes in the Why1 genomic area. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20090490/DC1.
Acknowledgments
We thank Dr. Henry Wortis for critical review of the manuscript and ongoing support.
This study was supported by grants R01AI056234 and R56 AI056234-07 from the National Institutes of Health (to A. Poltorak). J. Conner is the recipient of a Dean’s Fellowship award from the Sackler School of Graduate Biomedical Sciences.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used: BMDM, BM-derived macrophage; cDNA, complementary DNA; EMSA, electromobility shift analysis; ERK, extracellular signal-regulated kinase; IKK, IκB kinase; JNK, c-Jun N-terminal kinase; LTA, lipoteichoic acid; MAPK, mitogen-activated protein kinase; MEF, mouse embryonic fibroblast; mRNA, messenger RNA; shRNA, short hairpin RNA; TLR, Toll-like receptor.
References
- Akira S., Uematsu S., Takeuchi O. 2006. Pathogen recognition and innate immunity.Cell. 124:783–801 [DOI] [PubMed] [Google Scholar]
- Beebe A.M., Mauze S., Schork N.J., Coffman R.L. 1997. Serial backcross mapping of multiple loci associated with resistance to Leishmania major in mice.Immunity. 6:551–557 [DOI] [PubMed] [Google Scholar]
- Beutler B., Jiang Z., Georgel P., Crozat K., Croker B., Rutschmann S., Du X., Hoebe K. 2006. Genetic analysis of host resistance: toll-like receptor signaling and immunity at large.Annu. Rev. Immunol. 24:353–389 [DOI] [PubMed] [Google Scholar]
- Bonizzi G., Karin M. 2004. The two NF-kappaB activation pathways and their role in innate and adaptive immunity.Trends Immunol. 25:280–288 [DOI] [PubMed] [Google Scholar]
- Burns K., Janssens S., Brissoni B., Olivos N., Beyaert R., Tschopp J. 2003. Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4.J. Exp. Med. 197:263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Henzel W.J., Gao X. 1996. IRAK: a kinase associated with the interleukin-1 receptor.Science. 271:1128–1131 [DOI] [PubMed] [Google Scholar]
- Conner J.R., Smirnova I.I., Poltorak A. 2008. Forward genetic analysis of Toll-like receptor responses in wild-derived mice reveals a novel antiinflammatory role for IRAK1BP1.J. Exp. Med. 205:305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S.L., O'Neill L.A. 2006. Toll-like receptors: from the discovery of NFkappaB to new insights into transcriptional regulations in innate immunity.Biochem. Pharmacol. 72:1102–1113 [DOI] [PubMed] [Google Scholar]
- Fitzgerald K.A., Palsson-McDermott E.M., Bowie A.G., Jefferies C.A., Mansell A.S., Brady G., Brint E., Dunne A., Gray P., Harte M.T., et al. 2001. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction.Nature. 413:78–83 [DOI] [PubMed] [Google Scholar]
- Foster S.L., Hargreaves D.C., Medzhitov R. 2007. Gene-specific control of inflammation by TLR-induced chromatin modifications.Nature. 447:972–978 [DOI] [PubMed] [Google Scholar]
- Frazer K.A., Eskin E., Kang H.M., Bogue M.A., Hinds D.A., Beilharz E.J., Gupta R.V., Montgomery J., Morenzoni M.M., Nilsen G.B., et al. 2007. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains.Nature. 448:1050–1053 [DOI] [PubMed] [Google Scholar]
- Gregory R.C., Taxman D.J., Seshasayee D., Kensinger M.H., Bieker J.J., Wojchowski D.M. 1996. Functional interaction of GATA1 with erythroid Kruppel-like factor and Sp1 at defined erythroid promoters.Blood. 87:1793–1801 [PubMed] [Google Scholar]
- Grove M., Plumb M. 1993. C/EBP, NF-kappa B, and c-Ets family members and transcriptional regulation of the cell-specific and inducible macrophage inflammatory protein 1 alpha immediate-early gene.Mol. Cell. Biol. 13:5276–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenet J.L., Bonhomme F. 2003. Wild mice: an ever-increasing contribution to a popular mammalian model.Trends Genet. 19:24–31 [DOI] [PubMed] [Google Scholar]
- Hardy M.P., O'Neill L.A. 2004. The murine IRAK2 gene encodes four alternatively spliced isoforms, two of which are inhibitory.J. Biol. Chem. 279:27699–27708 [DOI] [PubMed] [Google Scholar]
- Hoebe K., Janssen E., Beutler B. 2004. The interface between innate and adaptive immunity.Nat. Immunol. 5:971–974 [DOI] [PubMed] [Google Scholar]
- Ideraabdullah F.Y., de la Casa-Esperon E., Bell T.A., Detwiler D.A., Magnuson T., Sapienza C., de Villena F.P. 2004. Genetic and haplotype diversity among wild-derived mouse inbred strains.Genome Res. 14:1880–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwami K.I., Matsuguchi T., Masuda A., Kikuchi T., Musikacharoen T., Yoshikai Y. 2000. Cutting edge: naturally occurring soluble form of mouse Toll-like receptor 4 inhibits lipopolysaccharide signaling.J. Immunol. 165:6682–6686 [DOI] [PubMed] [Google Scholar]
- Iwasaki A., Medzhitov R. 2004. Toll-like receptor control of the adaptive immune responses.Nat. Immunol. 5:987–995 [DOI] [PubMed] [Google Scholar]
- Janssens S., Beyaert R. 2003. Functional diversity and regulation of different interleukin-1 receptor-associated kinase (IRAK) family members.Mol. Cell. 11:293–302 [DOI] [PubMed] [Google Scholar]
- Janssens S., Burns K., Tschopp J., Beyaert R. 2002. Regulation of interleukin-1- and lipopolysaccharide-induced NF-kappaB activation by alternative splicing of MyD88.Curr. Biol. 12:467–471 [DOI] [PubMed] [Google Scholar]
- Kawagoe T., Sato S., Matsushita K., Kato H., Matsui K., Kumagai Y., Saitoh T., Kawai T., Takeuchi O., Akira S. 2008. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2.Nat. Immunol. 9:684–691 [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. 2006. TLR signaling.Cell Death Differ. 13:816–825 [DOI] [PubMed] [Google Scholar]
- Keating S.E., Maloney G.M., Moran E.M., Bowie A.G. 2007. IRAK-2 participates in multiple toll-like receptor signaling pathways to NFkappaB via activation of TRAF6 ubiquitination.J. Biol. Chem. 282:33435–33443 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Hernandez L.D., Galan J.E., Janeway C.A., Jr., Medzhitov R., Flavell R.A. 2002. IRAK-M is a negative regulator of Toll-like receptor signaling.Cell. 110:191–202 [DOI] [PubMed] [Google Scholar]
- LeBouder E., Rey-Nores J.E., Rushmere N.K., Grigorov M., Lawn S.D., Affolter M., Griffin G.E., Ferrara P., Schiffrin E.J., Morgan B.P., Labeta M.O. 2003. Soluble forms of Toll-like receptor (TLR)2 capable of modulating TLR2 signaling are present in human plasma and breast milk.J. Immunol. 171:6680–6689 [DOI] [PubMed] [Google Scholar]
- Leulier F., Lemaitre B. 2008. Toll-like receptors–taking an evolutionary approach.Nat. Rev. Genet. 9:165–178 [DOI] [PubMed] [Google Scholar]
- Li S., Strelow A., Fontana E.J., Wesche H. 2002. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase.Proc. Natl. Acad. Sci. USA. 99:5567–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika M., Orkin S.H. 1995. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF.Mol. Cell. Biol. 15:2437–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestas J., Hughes C.C. 2004. Of mice and not men: differences between mouse and human immunology.J. Immunol. 172:2731–2738 [DOI] [PubMed] [Google Scholar]
- Muzio M., Ni J., Feng P., Dixit V.M. 1997. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling.Science. 278:1612–1615 [DOI] [PubMed] [Google Scholar]
- Nathan C. 2002. Points of control in inflammation.Nature. 420:846–852 [DOI] [PubMed] [Google Scholar]
- Oda K., Kitano H. 2006. A comprehensive map of the toll-like receptor signaling network.Mol. Syst. Biol. 2:2006–0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.M., Greten F.R., Wong A., Westrick R.J., Arthur J.S., Otsu K., Hoffmann A., Montminy M., Karin M. 2005. Signaling pathways and genes that inhibit pathogen-induced macrophage apoptosis–CREB and NF-kappaB as key regulators.Immunity. 23:319–329 [DOI] [PubMed] [Google Scholar]
- Philipsen S., Suske G. 1999. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors.Nucleic Acids Res. 27:2991–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S., Pantano S., Natoli G. 2002. p38-Dependent marking of inflammatory genes for increased NF-kappa B recruitment.Nat. Immunol. 3:69–75 [DOI] [PubMed] [Google Scholar]
- Sancho-Shimizu V., Malo D. 2006. Sequencing, expression, and functional analyses support the candidacy of Ncf2 in susceptibility to Salmonella typhimurium infection in wild-derived mice.J. Immunol. 176:6954–6961 [DOI] [PubMed] [Google Scholar]
- Sanjabi S., Williams K.J., Saccani S., Zhou L., Hoffmann A., Ghosh G., Gerondakis S., Natoli G., Smale S.T. 2005. A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation.Genes Dev. 19:2138–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E.E., Lamb J., Yang X., Zhu J., Edwards S., Guhathakurta D., Sieberts S.K., Monks S., Reitman M., Zhang C., et al. 2005. An integrative genomics approach to infer causal associations between gene expression and disease.Nat. Genet. 37:710–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani G., Blais V., Sancho V., Vogel S.N., Stevenson M.M., Gros P., Lapointe J.M., Rivest S., Malo D. 2002. Host immune response to Salmonella enterica serovar Typhimurium infection in mice derived from wild strains.Infect. Immun. 70:1997–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhov A.N., Collart M.A., Vassalli P., Nedospasov S.A., Jongeneel C.V. 1990. κ B-type enhancers are involved in lipopolysaccharide-mediated transcriptional activation of the tumor necrosis factor α gene in primary macrophages.J. Exp. Med. 171:35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedlar M., Frankenberger M., Benkhart E., Espevik T., Quirling M., Brand K., Zembala M., Ziegler-Heitbrock L. 2004. Tolerance induced by the lipopeptide Pam3Cys is due to ablation of IL-1R-associated kinase-1.J. Immunol. 173:2736–2745 [DOI] [PubMed] [Google Scholar]
- Staelens J., Wielockx B., Puimege L., Van Roy F., Guenet J.L., Libert C. 2002. Hyporesponsiveness of SPRET/Ei mice to lethal shock induced by tumor necrosis factor and implications for a TNF-based antitumor therapy.Proc. Natl. Acad. Sci. USA. 99:9340–9345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Suzuki S., Duncan G.S., Millar D.G., Wada T., Mirtsos C., Takada H., Wakeham A., Itie A., Li S., et al. 2002. Severe impairment of interleukin-1 and Toll-like receptor signalling in mice lacking IRAK-4.Nature. 416:750–756 [DOI] [PubMed] [Google Scholar]
- Swantek J.L., Tsen M.F., Cobb M.H., Thomas J.A. 2000. IL-1 receptor-associated kinase modulates host responsiveness to endotoxin.J. Immunol. 164:4301–4306 [DOI] [PubMed] [Google Scholar]
- Thomas J.A., Allen J.L., Tsen M., Dubnicoff T., Danao J., Liao X.C., Cao Z., Wasserman S.A. 1999. Impaired cytokine signaling in mice lacking the IL-1 receptor-associated kinase.J. Immunol. 163:978–984 [PubMed] [Google Scholar]
- Urosevic N., Silvia O.J., Sangster M.Y., Mansfield J.P., Hodgetts S.I., Shellam G.R. 1999. Development and characterization of new flavivirus-resistant mouse strains bearing Flv(r)-like and Flv(mr) alleles from wild or wild-derived mice.J. Gen. Virol. 80:897–906 [DOI] [PubMed] [Google Scholar]
- Verstrepen L., Bekaert T., Chau T.L., Tavernier J., Chariot A., Beyaert R. 2008. TLR-4, IL-1R and TNF-R signaling to NF-kappaB: variations on a common theme.Cell. Mol. Life Sci. 65:2964–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Xiao H., Affolter J., Kim T.W., Bulek K., Chaudhuri S., Carlson D., Hamilton T., Mazumder B., Stark G.R., et al. 2009. Interleukin-1 receptor-associated kinase 2 is critical for lipopolysaccharide-mediated post-transcriptional control.J. Biol. Chem. 284:10367–10375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome.Nature. 420:520–562 [DOI] [PubMed] [Google Scholar]
- Yamin T.T., Miller D.K. 1997. The interleukin-1 receptor-associated kinase is degraded by proteasomes following its phosphorylation.J. Biol. Chem. 272:21540–21547 [DOI] [PubMed] [Google Scholar]
- Yang H., Bell T.A., Churchill G.A., Pardo-Manuel de Villena F. 2007. On the subspecific origin of the laboratory mouse.Nat. Genet. 39:1100–1107 [DOI] [PubMed] [Google Scholar]