A possible effector role for the pleckstrin homology (PH) domain of dynamin (original) (raw)
Abstract
The large GTPase dynamin plays a key role in clathrin-mediated endocytosis in animal cells, although its mechanism of action remains unclear. Dynamins 1, 2, and 3 contain a pleckstrin homology (PH) domain that binds phosphoinositides with a very low affinity (KD > 1 mM), and this interaction appears to be crucial for function. These observations prompted the suggestion that an array of PH domains drives multivalent binding of dynamin oligomers to phosphoinositide-containing membranes. Although in vitro experiments reported here are consistent with this hypothesis, we find that PH domain mutations that abolish dynamin function do not alter localization of the protein in transfected cells, indicating that the PH domain does not play a simple targeting role. An alternative possibility is suggested by the geometry of dynamin helices resolved by electron microscopy. Even with one phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)_P_2] molecule bound per PH domain, these dynamin assemblies will elevate the concentration of PtdIns(4,5)_P_2 at coated pit necks, and effectively cluster (or sequester) this phosphoinositide. In vitro fluorescence quenching studies using labeled phosphoinositides are consistent with dynamin-induced PtdIns(4,5)_P_2 clustering. We therefore propose that the ability of dynamin to alter the local distribution of PtdIns(4,5)_P_2 could be crucial for the role of this GTPase in promoting membrane scission during clathrin-mediated endocytosis. PtdIns(4,5)_P_2 clustering could promote vesicle scission through direct effects on membrane properties, or might play a role in dynamin's ability to regulate actin polymerization.
Keywords: endocytosis, phosphoinositide, vesicle, GTPase, actin
Large GTPases in the dynamin family are involved in many cellular trafficking, signaling, and morphological processes (1). The dynamins themselves have been most well-studied in clathrin-mediated endocytosis (1–4), where they play important roles in both coated pit invagination and scission of endocytic vesicles from the plasma membrane. All dynamin family members contain a GTPase domain, a “middle” domain and a GTPase effector (or GED) domain. They self-assemble into ring-like and tubular structures in a manner regulated by GTP binding and hydrolysis. Dynamins 1, 2, and 3 also contain a pleckstrin homology (PH) domain that binds phosphoinositides such as phosphatidylinositol-(4,5)-bisphosphate [PtdIns(4,5)_P_2] with low affinity (5), plus a C-terminal proline/arginine rich domain (PRD) that is important for targeting to coated pits (1–3) (see Fig. S1_A_). Mutations in the dynamin GTPase or PH domains that impair dynamin function cause the overexpressed protein to act as a dominant inhibitor of clathrin-mediated endocytosis (6–10).
Because some PH domains target their host proteins to phosphoinositide-containing membranes (11), we were interested in how (or whether) PH domain/phosphoinositide interactions promote dynamin recruitment to coated pit necks. PtdIns(4,5)_P_2 is important for early and late stages of clathrin-mediated endocytosis (12, 13), and its acute depletion causes rapid dissociation of endocytic proteins such as clathrin and dynamin from the plasma membrane (14). Although the isolated PH domain from dynamin-1 or dynamin-2 binds PtdIns(4,5)_P_2 very weakly, with KD values of 1–4 mM (5, 15), dynamin oligomerization (2) allows high-avidity multivalent PH domain-mediated interaction with membranes (11). This can be recapitulated with engineered PH domain oligomers (15). When a dynamin variant with a defective PH domain is overexpressed in cells, its co-oligomerization with endogenous WT dynamin is expected to impair multivalent targeting to PtdIns(4,5)_P_2-containing membranes—effectively “poisoning” the avidity effect mentioned above. We show here that such dominant inhibition of dynamin binding to PtdIns(4,5)_P_2-containing membranes can be reproduced in vitro. However, we could detect no influence of PH domain mutations on dynamin localization in cellular studies, raising the possibility that the PH domain does not contribute to dynamin targeting in vivo. Structures of dynamin helices thought to resemble those at coated pit necks (16, 17) suggest instead that an array of PH domains might laterally recruit PtdIns(4,5)_P_2 to the site of dynamin assembly at the membrane. Thus, instead of recruiting its host protein to phosphoinositide-containing membranes, the dynamin PH domain could play the converse role: recruiting phosphoinositides to dynamin assemblies. Dynamin could thus direct local elevation of PtdIns(4,5)_P_2 density (or “clustering”), which might be important for membrane scission (18) and/or the crucial (19–21) coordination of clathrin-mediated endocytosis with actin filament nucleation and polymerization (22).
Results and Discussion
PH Domain-Mediated PtdIns(4,5)_P_2 Binding in Vitro.
We first tested the hypothesis that PH* dynamin (Fig. S1_A_), which has mutations in the PH domain β6/β7 loop that impair phosphoinositide binding (15), can dominantly inhibit in vitro binding of WT dynamin to PtdIns(4,5)_P_2-containing membranes. Surface plasmon resonance (SPR) studies show that PH* dynamin binds very weakly to PtdIns(4,5)_P_2 (Fig. 1A), whereas WT dynamin binds membranes containing 10% (mole/mole) PtdIns(4,5)_P_2 with an apparent KD of 205 ± 126 nM (Fig. 1A). For comparison, KD values for PtdIns(4,5)_P_2 binding by PH domain monomers and dimers are in the 1 mM and 10 μM ranges respectively (15). Deleting a putative coiled-coil region (23) in Δcc1 dynamin (which lacks residues 652–681) prevents dynamin oligomerization as assessed in analytical ultracentrifugation studies (Fig. S1_B_) and binds PtdIns(4,5)_P_2 with greatly reduced affinity (Fig. 1A). Thus, both a WT PH domain and oligomerization are required for high-affinity phosphoinositide binding by dynamin.
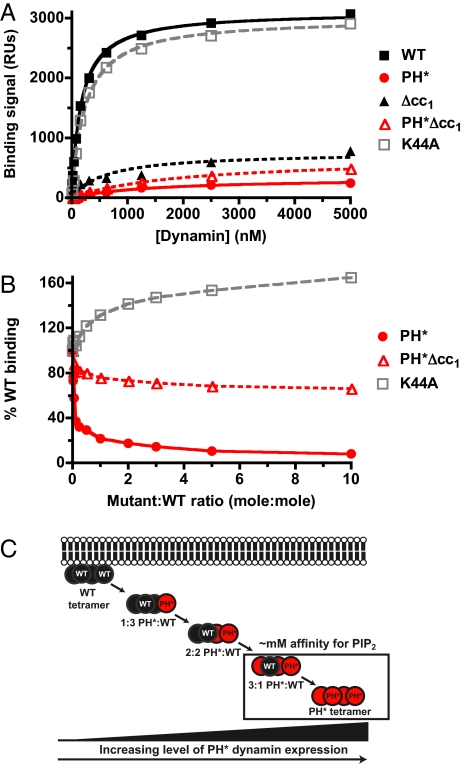
Fig. 1.
PH* dynamin has a dominant-negative effect on binding of WT protein to PtdIns(4,5)_P_2-containing membranes in vitro. (A) SPR binding curves for association of WT and mutated dynamins with membranes containing 10% PtdIns(4,5)_P_2 in a DOPC background. (B) Adding increasing amounts of PH* dynamin to a fixed concentration (400 nM) of WT dynamin dominantly inhibits its binding to a PtdIns(4,5)_P_2-containing membrane. Adding oligomerization-defective PH*Δcc1 dynamin does not. Adding K44A dynamin increases total membrane binding, saturating at ≈160% WT binding levels. A representative experiment from at least three repeats is shown. (C) Hypothetical scheme for the dominant-negative effect of PH* dynamin on PtdIns(4,5)_P_2 binding by the WT protein.
When PH* dynamin was added to a fixed concentration (400 nM) of WT protein, the total SPR binding signal fell dramatically (Fig. 1B). Thus, PH* dynamin exerts a potent dominant-negative effect on in vitro binding of WT dynamin to PtdIns(4,5)_P_2-containing membranes. Adding just one PH* dynamin molecule per WT tetramer (to give a PH*:WT ratio of 0.25) was sufficient to reduce the binding signal 3-fold. By contrast, adding oligomerization-defective PH*Δcc1 had only a small effect (Fig. 1B). These in vitro data argue that the dominant-negative effect of PH* dynamin overexpression in cells (8–10) could reflect impaired membrane-binding of tetramers (or higher oligomers) that incorporate PH* dynamin (Fig. 1C).
Subcellular Localization of Dynamin Is Unaffected by PH Domain Mutation.
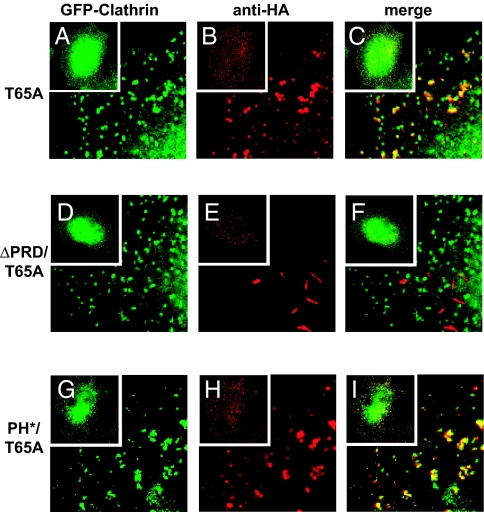
If PH* dynamin exerts its dominant-negative effect in cells by interfering with membrane targeting of endogenous WT protein, this should be reflected in its subcellular localization. We took advantage of a specific GTPase-defective (T65A) dynamin variant to investigate this. Over-expressing dynamin with a T65A (or T65F) mutation leads to the accumulation of coated pits with elongated necks (24, 25). T65A dynamin accumulates in electron-dense tubules at these elongated necks, apparently trapped in a prescission step because it cannot hydrolyze GTP (25). If multivalent PH domain-mediated binding to membrane PtdIns(4,5)_P_2 is required for targeting dynamin to coated pit necks, a T65A variant that also contains the PH* mutation should fail to accumulate at this location. However, Fig. 2 shows that this is not the case. HA-tagged T65A dynamin colocalizes with GFP-clathrin at the periphery of tTA-HeLa cells (Fig. 2 A–C), consistent with previous studies (25). Removing the PRD from T65A dynamin (by deleting residues 751–851) abolishes colocalization with GFP-clathrin (Fig. 2 D–F), and tubular structures form in the cytoplasm (Fig. 2E). This reflects the importance of the PRD for targeting dynamin to endocytic structures (2). By contrast, mutating the PH domain (in PH*/T65A dynamin) has no effect on colocalization of T65A dynamin with clathrin at the cell periphery (Fig. 2 G–I).
Fig. 2.
PH domain mutations do not impair localization of dynamin at endocytic structures. (A–F) GFP-clathrin (A) and HA-tagged T65A dynamin (B) colocalize at the plasma membrane in transfected tTA-HeLa cells. PRD deletion relocalizes ΔPRD/T65A dynamin to perinuclear aggregates that do not colocalize with clathrin (D–F). (G–I) By contrast, impairing PtdIns(4,5)_P_2 binding in PH*/T65A dynamin does not affect clathrin colocalization. Insets show whole cell images. Merges are shown in C, F, and I. Data represent at least three independent experiments in which >50 cells coexpressing GFP-clathrin and the HA-tagged dynamin were analyzed. More than 80% of punctae formed by T65A and PH*/T65A dynamin colocalized with GFP-clathrin, whereas <5% of structures formed by ΔPRD/T65A also containing GFP-clathrin.
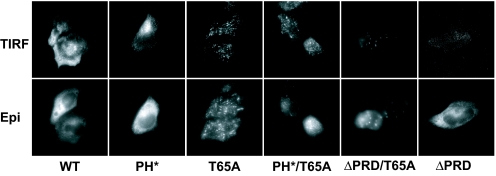
These epifluorescence studies show that PH*/T65A dynamin is targeted to endocytic structures despite its very low affinity for PtdIns(4,5)_P_2, and does not differ from a T65A variant with a WT PH domain. To determine whether PH*/T65A dynamin is located close to the plasma membrane in these structures (as it would be in coated pit necks), we used total internal reflection fluorescence (TIRF) microscopy as described in ref. 26. Fig. 3 compares epifluorescence and TIRF images for each HA-tagged dynamin variant. A significant amount of WT and T65A dynamin is found within the TIRF zone (i.e., within ≈100–150 nm of the cell surface), and mutating the PH domain (in PH* and PH*/T65A) has little effect on the proportion of fluorescent label excited by the evanescent wave. By contrast, PRD deletion significantly reduces the amount of WT or T65A dynamin within the TIRF zone. These data argue that eliminating the ability of the dynamin PH domain to bind phosphoinositides (in PH* dynamin) does not impair its targeting to endocytic sites. Although it is possible that PH*/T65A dynamin fails to form WT-like helices, the TIRF studies indicate that it is likely to be at the coated pit neck.
Fig. 3.
TIRF microscopy indicates that PH domain mutations do not affect dynamin targeting. Localization of WT and mutated dynamin was determined by total internal reflection fluorescence (TIRF) and epifluorescence (Epi) microscopy in HeLa cell transfectants, imaging at the same plane and exposure with both methods.
An Effector Role for the Dynamin PH Domain?
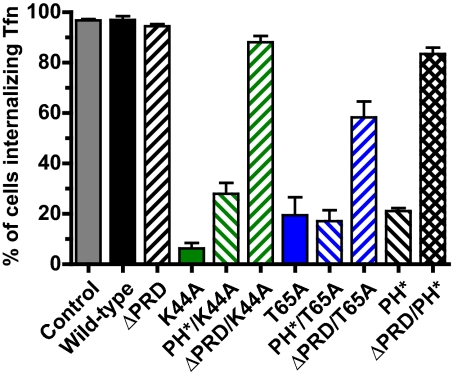
The cellular results above suggest that the dynamin PH domain does not simply target the protein. Because it is clearly essential for function, might the PH domain instead play an effector role? As shown in Fig. 4 and Fig. S2, the dominant-negative effect of PH* dynamin on clathrin-mediated endocytosis in HeLa cells resembles that of the GTPase-defective T65A or K44A mutants (6, 7, 24, 25), whereas ΔPRD dynamin (targeting-defective) has no such influence (7). Moreover, the dominant-negative effect of T65A, K44A, and PH* dynamin variants is lost when the PRD is removed (Fig. 4 and Fig. S2), arguing that normal targeting is required for inhibition. By contrast, introducing the PH* mutation into T65A or K44A dynamin does not block dominant-negative activity, suggesting that the PH domain is not required for their targeting. Thus, in this analysis the PH* mutation has more in common with T65A and K44A (GTPase) mutations than with PRD deletion—consistent with an effector role for the PH domain.
Fig. 4.
Double mutants suggest that the PH* mutation affects action, rather than targeting, of dynamin. HeLa cells transfected with HA-tagged dynamin constructs were tested for their ability to internalize biotinylated transferrin (B-Tfn), and scored as described in Methods. Each experiment involved at least 100 cells (see Fig. S2 for images) and data are quoted as the mean ± SD for at least three independent experiments.
EM studies of negatively stained samples also support this suggestion. PH* dynamin retains its ability to form 50-nm-diameter tubes when incubated with phosphatidylserine (PS)-containing liposomes (Fig. S3). These tubes were indistinguishable (Fig. S3 D–G) from those seen for WT dynamin (27, 28), and underwent a similar constriction (28) upon GTP addition (Fig. S3 H and I). Although PH* dynamin has reduced affinity for PS-containing liposomes (Fig. S3_A_), it is clearly capable of forming the same tubular structures as WT dynamin, consistent with the unchanged subcellular localization of PH*/T65A dynamin in our immunofluorescence studies. It is true that PH domain mutations might impair the kinetics of dynamin assembly and/or constriction at clathrin-coated pits. However, it seems equally likely that that the PH domain plays a direct part in executing dynamin action at the necks of coated pits.
Geometric Arguments Suggest that Dynamin Clusters PtdIns(4,5)_P_2.
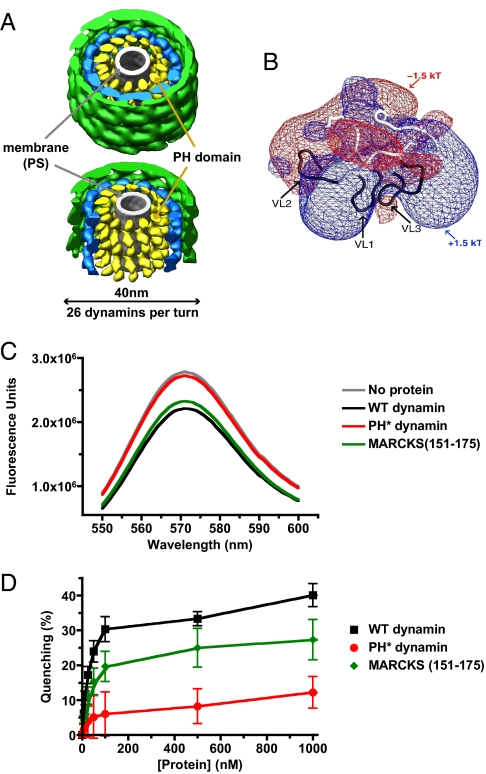
If interactions between the PH domain and PtdIns(4,5)_P_2 do not drive targeting of dynamin to coated pit necks (and its assembly), what might their role be? One possibility is that dynamin modifies phosphoinositide distribution (rather than being recruited by phosphoinositides). For example, recruitment/sequestration of PtdIns(4,5)_P_2 (leading to its clustering) might be important for coordinating dynamin activity with actin polymerization (19, 20). Insight into this possibility comes from cryo-EM-based structural models of dynamin assemblies bound to lipid tubules (16, 17, 29). When treated with the nonhydrolyzable GTP analogue GMP-PCP, ΔPRD dynamin forms well-ordered (constricted) 40-nm-diameter helices (Fig. 5A) with 26 dynamins per turn around a PS tubule (17). Docking structures of dynamin's constituent domains into the EM reconstruction indicated that the PH domain (yellow in Fig. 5A) abuts the (gray) PS tubule (16). The largest tubule that can be enclosed by the dynamin helix has a diameter of ≈130Å. One turn of the helix (26 molecules) extends 94Å along the helical axis, giving a lipid-contact area of 38,390 Å2 per turn, or 1,476 Å2 per dynamin molecule—approximately the surface area of the PH domain lipid-binding face (11). If each PH domain binds just one PtdIns(4,5)_P_2 molecule, a membrane tubule surrounded by a constricted dynamin helix will contain one PtdIns(4,5)_P_2 molecule per 1,476 Å2. This corresponds to a PtdIns(4,5)_P_2 density of 4.8% (mole/mole) – assuming a mean membrane surface area per lipid molecule of ≈70 Å2 [and 90 Å2 for PtdIns(4,5)_P_2]. This significantly exceeds estimates of ≈1% (mole/mole) for plasma membrane PtdIns(4,5)_P_2 density (30). Thus, dynamin (once constricted) can elevate local PtdIns(4,5)_P_2 concentration by up to ≈5-fold even if each PH domain binds just one PtdIns(4,5)_P_2 molecule. Nonconstricted dynamin helices seen without added GTP (17, 29) have a luminal diameter of ≈210 Å, and pitch of 106 Å (with 28 dynamins per helical turn), which would stabilize a local PtdIns(4,5)_P_2 density of ≈2.8% (mole/mole) using the same assumptions. The fact that dynamin's membrane-binding affinity increases in vitro as PtdIns(4,5)_P_2 density is elevated (31) also argues that dynamin assembly on membrane tubules must stabilize local increases in PtdIns(4,5)_P_2 density. Constriction of dynamin helices upon GTP hydrolysis would further increase PtdIns(4,5)_P_2 density, and PtdIns(4,5)_P_2 densities would be significantly higher than these values if each PH domain binds multiple PtdIns(4,5)_P_2 molecules.
Fig. 5.
Dynamin may cluster PtdIns(4,5)_P_2. (A) (Upper) Representation of a constricted dynamin helix surrounding a PS tubule (16). The dynamin helix has a diameter of 40 nm, with 26 dynamin molecules per turn. The region assigned to the PH domain is colored yellow, PS is gray, the GTPase is green, and the middle domain is blue. (Lower) GTPase and middle domains have been removed from the front of the helix to illustrate the PH domain array placed against the membrane surface. (B) Electrostatic potential of the dynamin PH domain, with a positively charged face (blue) that contains the phospholipid binding site made up by loops VL1, VL2 and VL3 (11). (C) Adding WT dynamin or MARCKS (151–175) at 0.5 μM quenches fluorescence of C6-BODIPY-TMR PtdIns(4,5)_P_2 incorporated at 1% (mole/mole) in DOPC vesicles. PH* dynamin has no effect. (D) Concentration dependence of fluorescence quenching (using 0.1 mM total lipid) shown in C.
Like many PH domains (11), the PH domain of dynamin has clear electrostatic sidedness (Fig. 5B), and the PtdIns(4,5)_P_2-binding site lies on the positively charged face (5). If this face lines the inner surface of dynamin helices as suggested (17), dynamin will form a ‘sheath’ of positive charge around the membrane tubule at a coated pit neck. Regions of positive electrostatic potential adjacent to membrane surfaces sequester PtdIns(4,5)_P_2 (32), as has been well-studied for the unstructured basic cluster in MARCKS (myristoylated alanine-rich C kinase substrate), for example (33). Related proteins have been named pipmodulins (34) because they are thought to control levels of free PtdIns(4,5)_P_2 in the plasma membrane through electrostatic sequestration.
Self-Quenching of Fluorescent PtdIns(4,5)_P_2 upon Dynamin Binding.
To test the hypothesis that dynamin clusters PtdIns(4,5)_P_2, we analyzed its ability to promote self-quenching of fluorescently labeled PtdIns(4,5)_P_2 incorporated into vesicles at a low spatial density. This approach was previously used to study PtdIns(4,5)_P_2 sequestration by the MARCKS (151–175) peptide (33), myelin basic protein (35), and NAP-22 (36). We used PtdIns(4,5)_P_2 labeled in the sn-1 position with the 26-carbon BODIPY-TMR label attached via a 6-carbon linker. Either caproic (C6) or palmitic acid (C16) was esterified to the sn-2 position. Although the volume of the acyl-chain region of C6-BODIPY-TMR-PtdIns(4,5)_P_2 most closely resembles that of natural PtdIns(4,5)_P_2, similar results were obtained with both forms.
As shown in Fig. 5C, adding 0.5 μM dynamin to 0.1-μm vesicles containing 1% (mole/mole) C6-BODIPY-TMR-PtdIns(4,5)_P_2 in a DOPC background caused significant fluorescence quenching, to a maximum of 40 ± 3% (Fig. 5D). A similar effect was seen with 0.5 μM MARCKS (151–175) peptide (33), whereas PH* dynamin caused no quenching. At 1% (mole/mole), the mean distance between PtdIns(4,5)_P_2 molecules is ≈100 Å. Because this exceeds _R_o for the BODIPY label (≈50 Å), no self-quenching is expected under these conditions. The mean distance between PtdIns(4,5)_P_2 molecules falls below 50 Å if local density exceeds 2.8% (mole/mole). Thus, the observed dynamin-induced quenching of BODIPY-TMR fluorescence is consistent with the PtdIns(4,5)_P_2 densities likely to be stabilized by dynamin binding/assembly.
Although these data support PtdIns(4,5)_P_2 quenching by dynamin, there are two important caveats. First, the PtdIns(4,5)_P_2-specific PH domain from phospholipase C-δ1 also quenched BODIPY-TMR fluorescence (Fig. S4_A_). This PH domain binds PtdIns(4,5)_P_2 in a well-characterized 1:1 interaction (11), and is thought not to cluster phosphoinositides (33). Quenching in this case could reflect PH domain oligomerization on the membrane surface. Alternatively, this PH domain might bind actually bind multiple PtdIns(4,5)_P_2 molecules. Indeed, binding of multiple PtdIns(4,5)_P_2 molecules to a single PH domain would also explain why a dimeric form of the isolated dynamin PH domain (15) quenches BODIPY-TMR-PtdIns(4,5)_P_2 fluorescence (Fig. S4_B_). However, we cannot currently exclude an alternative explanation: that that the observed fluorescence quenching actually reflects changes in fluor environment arising from binding-induced alterations in membrane structure. One control to test this hypothesis used BODIPY-TMR labeled phosphatidylinositol (PtdIns) at 1% (mole/mole) in DOPC membranes also containing 1% (mole/mole) unlabeled PtdIns(4,5)_P_2. Surprisingly, both WT dynamin and the MARCKS peptide promoted significant (but reduced) quenching of the labeled PtdIns (Fig. S4_C_), which is difficult to ascribe straightforwardly [in the presence of unlabeled PtdIns(4,5)_P_2] to phosphoinositide clustering.
We have worked extensively to study PtdIns(4,5)_P_2 clustering by dynamin and MARCKS (151–175) more directly with 2-color FRET approaches, but without success. However, several approaches, including EPR (37), FCS (38), phospholipase-C accessibility (33), and fluorescence self-quenching studies (33) all argue that the MARCKS (151–175) peptide clusters PtdIns(4,5)_P_2. In our studies using BODIPY-TMR-labeled PtdIns(4,5)_P_2, dynamin promotes a degree of self-quenching that is equal or greater than that seen with the MARCKS peptide. Thus, within the limitations of current technology our data indicate that dynamin may induce PtdIns(4,5)_P_2 clustering.
Possible Implications of PtdIns(4,5)_P_2 Clustering by Dynamin.
If dynamin clusters PtdIns(4,5)_P_2, what might be the functional significance? Elevated PtdIns(4,5)_P_2 levels are well known to activate actin assembly, and could contribute to dynamin's role in regulating actin assembly during clathrin-mediated endocytosis (19, 20, 39).
Local increases in PtdIns(4,5)_P_2 levels can arise when synthesis of this phosphoinositide is increased (and/or degradation is reduced) or from sequestration of PtdIns(4,5)_P_2 by proteins such as MARCKS (32) and other pipmodulins (22, 34). PtdIns(4,5)_P_2 activates WASP family proteins either indirectly (through effects on Cdc42 activation) or directly, and a polybasic PtdIns(4,5)_P_2-binding region in N-WASP has been suggested to function as a ‘sensor’ of local PtdIns(4,5)_P_2 density (40). SNX9 may have similar capability (41). One speculative possibility is that dynamin helices constrict and rapidly disassemble after GTP hydrolysis, and transiently expose a region in the coated pit neck with elevated PtdIns(4,5)_P_2 density. This could promote initiation of actin polymerization by the actin-regulatory complexes known to be preassembled in this region (19) through association with dynamin. Indeed, time-lapse TIRF microscopy of live cells (39) indicates that actin recruitment to coated pit necks follows disassembly of dynamin helices. Actin polymerization may provide force for movement of the coated pit or vesicle into the cytosol (21, 39), with dynamin coordinating this process both through influences on actin regulatory proteins and on local PtdIns(4,5)_P_2 concentration mediated by its PH domain.
Alternatively, without disassembly, the ability of constricted dynamin helices to concentrate PtdIns(4,5)_P_2 could contribute to an effective phase separation of lipids within the coated pit neck – together with BAR domain-containing proteins such as amphiphysins and endophilins (42). It was proposed that the line tension at the interface between such a lipid phase within the coated pit neck and the adjacent bulk lipid facilitates membrane fusion (and thus fission of the membrane tubule), so that the coated vesicle is “pinched off” (18). Indeed, Roux et al. (43) proposed that proteins involved in endocytosis—including dynamin—might function to change the local lipid composition, triggering phase separation and fission of membrane tubules. In vitro studies of dynamin-coated PtdIns(4,5)_P_2-containing lipid tubules have shown that constriction after GTP hydrolysis is not sufficient for their fission unless longitudinal tension is applied (44). In cells, this tension might be provided by actin polymerization, possibly promoting fission of the coated pit neck at a lipid phase boundary induced by dynamin-mediated PtdIns(4,5)_P_2 clustering.
Conclusions
The data presented here suggest that the PH domain of dynamin plays a functional role that is distinct from targeting the protein to phosphoinositide-containing membranes. Although a PH domain mutation (PH*) impairs PtdIns(4,5)_P_2 binding by dynamin in vitro, it does not alter subcellular localization of dynamin expressed in HeLa cells—despite abolishing function. If dynamin is targeted to the necks of coated pits through other interactions (such as those mediated by its PRD), it has the capacity to cluster PtdIns(4,5)_P_2 at this location. Significant PtdIns(4,5)_P_2 clustering is predicted from simple considerations of EM-based structures of dynamin assemblies, and we present fluorescence quenching data that are consistent with this possibility. We therefore hypothesize that PH domain of dynamin plays a direct part in executing dynamin action on the membrane at the coated pit neck. Clustering of PtdIns(4,5)_P_2 by dynamin could contribute directly to a lipid phase separation proposed to facilitate membrane scission (18). Alternatively (or in addition), dynamin may regulate actin polymerization through alterations in local PtdIns(4,5)_P_2 density. Many PH domains with low affinities for phosphoinositides are found in proteins capable of self-assembly, and it is possible that many of these will function as pipmodulins as suggested here for dynamin.
Materials and Methods
Constructs and Cell Culture.
Mammalian expression constructs for HA-tagged human WT or K44A dynamin-1 in the tet-regulatable vector pUHD10.3 were a gift from Dr. Sandra Schmid. Mutagenesis and cell manipulations were as described in ref. 9. For Sf9 cell expression, dynamin-1 cDNA was subcloned—with an N-terminal hexahistidine tag—into pFastBac-1 (Invitrogen), using _Eco_RI/_Hin_dIII sites.
Protein Production and Purification.
Histidine-tagged dynamin was expressed in baculovirus-infected Sf9 cells. Washed cells were lysed by sonication in 15 mM Tris, pH 8.5, and 225 mM NaCl, supplemented with 2 mM β-mercaptoethanol, 1 mM PMSF, and an EDTA-free protease inhibitor mixture (Roche). Dynamin was purified by metal-affinity chromatography, followed by anion-exchange and gel-filtration in 20 mM Hepes, pH 7.2, 250 mM NaCl, and 2 mM DTT. PH domains were purified as described in ref. 15.
Antibodies.
HA-tagged dynamin was detected with mouse monoclonal anti-HA (Covance), used at 1/1,000 dilution. Secondary antibodies were FITC- or Texas Red-conjugated IgG1 (Santa Cruz Biotechnology) at a dilution of 1/40, or (for TIRF experiments) Alexa Fluor 488 conjugated IgG1 (Molecular Probes/Invitrogen) at a 1/40 dilution.
Immunofluorescence Microscopy.
HeLa (or tTA-HeLa) cells were seeded onto glass coverslips (Fisher) 24 h before transfection. For transferrin uptake and TIRF experiments, cells were cotransfected (using Lipofectamine 2000) with the appropriate dynamin constructs (in pUHD10.3) and pUHD15–1 (which directs tTA expression). For clathrin colocalization experiments, tTA-HeLa (Tet-Off) cells were cotransfected with GFP-clathrin (45) and the noted dynamin construct (in pUHD10.3). Dynamin expression was limited by including 100 ng/mL tetracycline in the medium. After 36 h, cells were washed with PBS and formaldehyde-fixed/permeabilized for immunofluorescence using standard procedures. Coverslips were mounted onto slides with Fluoromount G (Electron Microscopy Sciences). For epifluorescence studies, digital images were acquired using a Hamamatsu Orca CCD camera on a Leica DM IRBE microscope. Serial Z sections were imaged and deconvoluted using OpenLab (Improvision) software. For TIRF microscopy, coverslips were mounted in PBS containing Mg2+ and Ca2+ and sealed with clear nail polish. Cells were then viewed using a Leica Microscope and single wavelength laser apparatus. Epifluorescence and TIRF for each sample were viewed in the same focal plane with the same exposure, as described in ref. 26.
Transferrin Uptake Assays.
Transferrin uptake assays were performed exactly as described in ref. 9, monitoring uptake of biotinylated transferrin by staining fixed, permeabilized cells with streptavidin-Texas Red (Molecular Probes). Transfected cells were scored by microscopy for whether they internalized transferrin (indicated by the presence of B-Tfn in intracellular punctae) or displayed an endocytosis defect (with B-Tfn staining diffusely at the cell surface only). Each dataset comprised of a minimum of 100 cells, was repeated at least three times. Data are quoted in Fig. 4 as means (with SD).
PtdIns(4,5)_P_2 Binding.
Analysis of dynamin binding to vesicles composed of dioleoylphosphatidylcholine (DOPC, Sigma) with or without 10% (mole/mole) PtdIns(4,5)_P_2 (Cell Signals) was performed using surface plasmon resonance (SPR), exactly as described in ref. 46. Experiments were performed at 25 °C in 10 mM Hepes, pH 7.2, containing 225 mM NaCl. Dynamin mixtures were incubated for at least 10 min before injection.
Fluorescence Self-Quenching.
Unilamellar vesicles containing DOPC and C6 (or C16) BODIPY-TMR PtdIns(4,5)_P_2 (Echelon) in a 99:1 (mole:mole) ratio were prepared by extrusion although a 0.1-μm polycarbonate membrane. Experiments used a total lipid concentration of 0.1 mM, in 20 mM Hepes, pH 7.2, with 150 mM NaCl. Proteins were added in increasing concentrations up to 1 μM. Fluorescence was measured using a Photon Technology International fluorimeter, with excitation and emission slits at 4 nm. The BODIPY-TMR label was excited at 543 nm and emission was measured at 572 nm.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Stuart McLaughlin (Stony Brook University, Stony Brook, NY) for kindly providing the MARCKS (151–175) peptide, Jim Keen (Thomas Jefferson University, Philadelphia) for the GFP-clathrin construct, and Sandra Schmid (Scripps Research Institute, La Jolla, CA) for dynamin-1 constructs. We also thank Kate Ferguson and members of the Lemmon laboratory for valuable discussions and Jason Mears for help with Fig. 5A. This work was supported in part by National Institutes of Health (NIH) Grants R01-GM57247 (to E.M.O.) and R01-GM078345 (to M.A.L.), Predoctoral Fellowship DAMD17–02-1–0546 from the U.S. Army Breast Cancer Research Program (to M.C.K.), a Predoctoral Fellowship from the Great Rivers Affiliate of the American Heart Association (to K.A.B.), and the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases (J.E.H).
Footnotes
The authors declare no conflict of interest.
References
- 1.Praefcke GJ, McMahon HT. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nature Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 2.Hinshaw JE. Dynamin and its role in membrane fission. Annu Rev Cell Dev Biol. 2000;16:483–519. doi: 10.1146/annurev.cellbio.16.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sever S, Damke H, Schmid SL. Garrotes, springs, ratchets, and whips: Putting dynamin models to the test. Traffic. 2000;1:385–392. doi: 10.1034/j.1600-0854.2000.010503.x. [DOI] [PubMed] [Google Scholar]
- 4.Macia E, et al. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Zheng J, et al. Identification of the binding site for acidic phospholipids on the PH domain of dynamin: Implications for stimulation of GTPase activity. J Mol Biol. 1996;255:14–21. doi: 10.1006/jmbi.1996.0002. [DOI] [PubMed] [Google Scholar]
- 6.van der Bliek AM, et al. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herskovits JS, Burgess CC, Obar RA, Vallee RB. Effects of mutant rat dynamin on endocytosis. J Cell Biol. 1993;122:565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Achiriloaie M, Barylko B, Albanesi JP. Essential role of the dynamin pleckstrin homology domain in receptor-mediated endocytosis. Mol Cell Biol. 1999;19:1410–1415. doi: 10.1128/mcb.19.2.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee A, Frank DW, Marks MS, Lemmon MA. Dominant-negative inhibition of receptor-mediated endocytosis by a dynamin-1 mutant with a defective pleckstrin homology domain. Curr Biol. 1999;9:261–264. doi: 10.1016/s0960-9822(99)80115-8. [DOI] [PubMed] [Google Scholar]
- 10.Vallis Y, Wigge P, Marks B, Evans PR, McMahon HT. Importance of the pleckstrin homology domain of dynamin in clathrin-mediated endocytosis. Curr Biol. 1999;9:257–260. doi: 10.1016/s0960-9822(99)80114-6. [DOI] [PubMed] [Google Scholar]
- 11.Lemmon MA, Ferguson KM. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem J 350 Pt. 2000;1:1–18. [PMC free article] [PubMed] [Google Scholar]
- 12.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 13.Jost M, Simpson F, Kavran JM, Lemmon MA, Schmid SL. Phosphatidylinositol-4,5-bisphosphate is required for endocytic coated vesicle formation. Curr Biol. 1998;8:1399–1402. doi: 10.1016/s0960-9822(98)00022-0. [DOI] [PubMed] [Google Scholar]
- 14.Zoncu R, et al. Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proc Natl Acad Sci USA. 2007;104:3793–3798. doi: 10.1073/pnas.0611733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein DE, Lee A, Frank DW, Marks MS, Lemmon MA. The pleckstrin homology domains of dynamin isoforms require oligomerization for high affinity phosphoinositide binding. J Biol Chem. 1998;273:27725–27733. doi: 10.1074/jbc.273.42.27725. [DOI] [PubMed] [Google Scholar]
- 16.Zhang P, Hinshaw JE. Three-dimensional reconstruction of dynamin in the constricted state. Nat Cell Biol. 2001;3:922–926. doi: 10.1038/ncb1001-922. [DOI] [PubMed] [Google Scholar]
- 17.Mears JA, Ray P, Hinshaw JE. A corkscrew model for dynamin constriction. Structure. 2007;15:1190–1202. doi: 10.1016/j.str.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Kaksonen M, Drubin DG, Oster G. Endocytic vesicle scission by lipid phase boundary forces. Proc Natl Acad Sci USA. 2006;103:10277–10282. doi: 10.1073/pnas.0601045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schafer DA. Regulating actin dynamics at membranes: A focus on dynamin. Traffic. 2004;5:463–469. doi: 10.1111/j.1600-0854.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 20.Yarar D, Waterman-Storer CM, Schmid SL. A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis. Mol Biol Cell. 2005;16:964–975. doi: 10.1091/mbc.E04-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 22.Caroni P. Actin cytoskeleton regulation through modulation of PI(4,5)P(2) rafts. EMBO J. 2001;20:4332–4336. doi: 10.1093/emboj/20.16.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okamoto PM, Tripet B, Litowski J, Hodges RS, Vallee RB. Multiple distinct coiled-coils are involved in dynamin self-assembly. J Biol Chem. 1999;274:10277–10286. doi: 10.1074/jbc.274.15.10277. [DOI] [PubMed] [Google Scholar]
- 24.Damke H, Binns DD, Ueda H, Schmid SL, Baba T. Dynamin GTPase domain mutants block endocytic vesicle formation at morphologically distinct stages. Mol Biol Cell. 2001;12:2578–2589. doi: 10.1091/mbc.12.9.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marks B, et al. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature. 2001;410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- 26.Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Mol Biol Cell. 2006;17:4856–4865. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carr JF, Hinshaw JE. Dynamin assembles into spirals under physiological salt conditions upon the addition of GDP and gamma-phosphate analogues. J Biol Chem. 1997;272:28030–28035. doi: 10.1074/jbc.272.44.28030. [DOI] [PubMed] [Google Scholar]
- 28.Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 29.Chen YJ, Zhang P, Egelman EH, Hinshaw JE. The stalk region of dynamin drives the constriction of dynamin tubes. Nat Struct Mol Biol. 2004;11:574–575. doi: 10.1038/nsmb762. [DOI] [PubMed] [Google Scholar]
- 30.McLaughlin S, Wang J, Gambhir A, Murray D. PIP(2) and proteins: Interactions, organization, and information flow. Annu Rev Biophys Biomol Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 31.Ramachandran R, Schmid SL. Real-time detection reveals that effectors couple dynamin's GTP-dependent conformational changes to the membrane. EMBO J. 2008;27:27–37. doi: 10.1038/sj.emboj.7601961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 33.Gambhir A, et al. Electrostatic sequestration of PIP2 on phospholipid membranes by basic/aromatic regions of proteins. Biophys J. 2004;86:2188–2207. doi: 10.1016/S0006-3495(04)74278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laux T, et al. GAP43, MARCKS, and CAP23 modulate PI(4,5)P(2) at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J Cell Biol. 2000;149:1455–1472. doi: 10.1083/jcb.149.7.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musse AA, Gao W, Homchaudhuri L, Boggs JM, Harauz G. Myelin basic protein as a “PI(4,5)P2-modulin”: A new biological function for a major central nervous system protein. Biochemistry. 2008;47:10372–10382. doi: 10.1021/bi801302b. [DOI] [PubMed] [Google Scholar]
- 36.Epand RM, Vuong P, Yip CM, Maekawa S, Epand RF. Cholesterol-dependent partitioning of PtdIns(4,5)P2 into membrane domains by the N-terminal fragment of NAP-22 (neuronal axonal myristoylated membrane protein of 22 kDa) Biochem J. 2004;379:527–532. doi: 10.1042/BJ20040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rauch ME, Ferguson CG, Prestwich GD, Cafiso DS. Myristoylated alanine-rich C kinase substrate (MARCKS) sequesters spin-labeled phosphatidylinositol 4,5-bisphosphate in lipid bilayers. J Biol Chem. 2002;277:14068–14076. doi: 10.1074/jbc.M109572200. [DOI] [PubMed] [Google Scholar]
- 38.Golebiewska U, Nyako M, Woturski W, Zaitseva I, McLaughlin S. Diffusion coefficient of fluorescent phosphatidylinositol 4,5-bisphosphate in the plasma membrane of cells. Mol Biol Cell. 2008;19:1663–1669. doi: 10.1091/mbc.E07-12-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 40.Papayannopoulos V, et al. A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol Cell. 2005;17:181–191. doi: 10.1016/j.molcel.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 41.Yarar D, Surka MC, Leonard MC, Schmid SL. SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic. 2008;9:133–146. doi: 10.1111/j.1600-0854.2007.00675.x. [DOI] [PubMed] [Google Scholar]
- 42.Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta. 2006;1761:897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 43.Roux A, et al. Role of curvature and phase transition in lipid sorting and fission of membrane tubules. EMBO J. 2005;24:1537–1545. doi: 10.1038/sj.emboj.7600631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roux A, Uyhazi K, Frost A, De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- 45.Gaidarov I, Santini F, Warren RA, Keen JH. Spatial control of coated-pit dynamics in living cells. Nat Cell Biol. 1999;1:1–7. doi: 10.1038/8971. [DOI] [PubMed] [Google Scholar]
- 46.Yu JW, Lemmon MA. All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J Biol Chem. 2001;276:44179–44184. doi: 10.1074/jbc.M108811200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information