Neuronal Activity Rapidly Induces Transcription of the CREB-Regulated microRNA-132, in vivo (original) (raw)
. Author manuscript; available in PMC: 2011 Apr 1.
Published in final edited form as: Hippocampus. 2010 Apr;20(4):492–498. doi: 10.1002/hipo.20646
Abstract
Activity-dependent changes in gene-expression are believed to underlie the molecular representation of memory. In this study, we report that in vivo activation of neurons rapidly induces the CREB-regulated microRNA miR-132. To determine if production of miR-132 is regulated by neuronal activity its expression in mouse brain was monitored by quantitative RT-PCR (RT-qPCR). Pilocarpine-induced seizures led to a robust, rapid, and transient increase in the primary transcript of miR-132 (pri-miR-132) followed by a subsequent rise in mature microRNA (miR-132). Activation of neurons in the hippocampus, olfactory bulb, and striatum by contextual fear conditioning, odor-exposure, and cocaine-injection, respectively, also increased pri-miR-132. Induction kinetics of pri-miR-132 were monitored and found to parallel those of immediate early genes, peaking at 45 minutes and returning to basal levels within two hours of stimulation. Expression levels of primary and mature-miR-132 increased significantly between postnatal days 10 and 24. We conclude that miR-132 is an activity-dependent microRNA in vivo, and may contribute to the long-lasting proteomic changes required for experience-dependent neuronal plasticity.
Keywords: MicroRNA, CREB, Plasticity, Experience-dependent, Immediate-early, mir-132
Introduction
MicroRNAs (miRNA) are small (20–25 nt) single-stranded RNA molecules that bind to messenger RNA and block subsequent protein production by either inhibiting translation machinery or inducing message degradation, depending on the degree of complementarity (Ambros et al., 2003). Genes encoding miRNA produce a primary transcript (pri-miRNA), which is cotranscriptionally cleaved by a complex containing the double-stranded RNA-binding protein Pasha and its RNAse counterpart Drosha (Lee et al., 2003; Morlando et al., 2008). The resulting stem-loop pre-miRNA is exported to the cytoplasm by Exportin 5 (Yi et al., 2003), where it is further processed by the endonuclease Dicer into a 20–25 nt double-stranded RNA molecule (Hutvagner et al., 2001). One of these strands, known as the guide strand, is selected by Argonaut and incorporated into a RNA-induced silencing complex (RISC) for base-pairing with target mRNA, while the other strand, dubbed the passenger, is degraded (Gregory et al., 2005; Martinez et al., 2002). Based on sequence homology, each miRNA has the potential to regulate the translation of hundreds of different genes (Lim et al., 2005), and greater than 30% of all mammalian genes may be regulated by miRNAs (Lewis et al., 2003).
Recently, using an unbiased genome-wide screen for CREB-bound transcripts in vitro, Impey et al. identified 16 non-coding miRNA that are induced by CREB-mediated transcription (Impey et al., 2004). Further characterization of one of these, miR-132, has recently revealed that it is induced in culture by neurotrophins and neuronal activity, and it is able to modulate dendritic morphology via suppression of a specific target, p250 GTPase-activating protein (p250GAP) (Vo et al., 2005; Wayman et al., 2008). Thus, miR-132 may play an important role in the development or plasticity of synaptic architecture and networks. MiR-132 expression is circadian-regulated in the superchaismatic nucleus (SCN), where it is important for proper clock-resetting responses to light (Cheng et al., 2007). It is also expressed in the periphery, and LPS stimulation of immune responses in the human acute moncytic leukemia cell line THP-1 stimulates its expression (Taganov et al., 2006).
Since every miRNA can potentially regulate the translation of hundreds of proteins, these molecules may contribute to the vast experience-dependent changes in protein expression believed to be necessary for neuronal plasticity. Here we show, in vivo, that neuronal activation via pilocarpine-induced seizures, contextual fear conditioning, cocaine injection, and exposure to odorants all induce rapid and transient increases in miR-132 expression.
Materials and Methods
Subjects
Adult male (8–12 weeks old) C57/BL/6 mice (Taconic Farms, Hudson, NY) were group-housed 5 mice per cage, and food was available ad libitum. Lights in the room were on a 12-hr light-dark schedule. For all biochemical experiments, mice were sacrificed by cervical dislocation then decapitation, and tissue was dissected, stored in cryo tubes, snap frozen in liquid nitrogen, and stored at −80°C until RNA isolation.
Primer Design
We first analyzed two different primer sets designed for the detection of the miR-132/miR-212 primary transcript (pri-mir-132). The first set was designed as published (Vo et al., 2005), and the second we designed using Primer Express (Applied Biosystems, Foster City, CA), with default settings except the following changes: %GC: 40–60; minimum primer Tm: 58–60°C; minimum primer length: 18–30, optimal 20; minimum amplified region Tm: 0–100°C; minimum amplified region length: 90–130bp. Highest scoring sets were analyzed for duplexing in Vector NTI (Invitrogen, Carlsbad, CA) and sets with minimal duplexing were BLAST analyzed. Both primer sets produced cDNA amplicons of the expected size, and melting curves showed no sign of primer-dimers (data not shown). Furthermore, sequence analysis of the cDNAs resulting from a round of RT-qPCR verified amplification of the predicted primary transcript amplicon (data not shown). The standard curve efficiency from our primer set was more reproducible than that of the previously published primers, and thus we chose to use our primers for this study.
Pharmacologic Neuronal Activation
Mice were injected with pilocarpine (300 mg/kg i.p.) and sacrificed 45 minutes or 8 hours later. Control and experimental animals in the 8-hour group also received diazepam (4 mg/kg ip) 60 min after pilocarpine to terminate subsequent seizure activity. For the cocaine experiments, animals were injected intraparitoneal with 20 mg/kg cocaine diluted in sterile water, and animals were sacrificed at appropriate times.
Behavioral Neuronal Activation
For odor-induced activity, animals were group housed. Two cotton swabs were soaked with 200ul of 5 uM citralva, dabbed dry on a paper towel, and placed on opposite sides of a mouse cage, suspended through a wire cage top, such that the odor-soaked tip was approximately 1.5 inches up from the bottom of the cage for 5 minutes. Mice were sacrificed at appropriate times post-exposure. Contextual fear conditioning was performed as previously reported (Eckel-Mahan et al., 2008). Unpaired mice were placed in a context and immediately given a 2 second, 0.7 mA foot-shock, before being returned to their home cage. Unpaired mice from our experiments did not show contextual fear memories when placed back into the training context (data not shown). Paired mice were allowed to acclimate to the context for 2 minutes prior to a 2 second, 0.7mA foot-shock, and then 1 minute following the shock these mice were returned to their home-cage. Context mice were allowed to acclimate to the context for 3 minutes without a foot-shock, and then they were returned to their home-cage. As above, mice were sacrificed at appropriate times and tissue was stored at −80°C for later RNA extraction.
SYBR-Green quantitative real-time PCR (Detection of Primary transcript)
For these experiments, we preferred SYBR-green chemistry as this technique is relatively inexpensive and when coupled with the proper controls, affords a high level of sensitivity and specificity for the detection of RNA species. Aliquots of RNA from all control and experimental samples were pooled and serially diluted for the generation of standard curves. For each unknown sample, 50ng of total DNAse-treated (Invitrogen, Carlsbad, CA) RNA was mixed with master mix containing the following reagents: 10ul sybergreen 2x mastermix (ABI), forward and reverse primers, and StrataScript reverse transcriptase (Stratagene, La Jolla, CA). The RT-PCR protocol was as follows: 1. 48°C-30 min. 2. 95°C-10 min. 3. 95°C-15 sec. 4. 58°C-1 min. (repeat 3 and 4, 40 cycles). 5. 95°C-1 min. 6. 55°C ramp 1 degree/sec for 41 sec. Results were normalized to ARBP as its mRNA expression levels were unchanged by our experimental conditions (Sup Fig 2).
Figure 2.
Acute cocaine treatment significantly increases striatal primary-miR-132 levels while. Mice were injected with saline or cocaine (20 mg/kg), sacrificed at indicated times, and RNA levels were determined by RT-qPCR. Relative pri-mir-132 normalized to ARBP. Pri-mir-132 significantly increased 45 min post-cocaine injection compared to saline treatment and returned to baseline by 90 min (n=4–5 mice/group, analyzed in triplicate). One-way ANOVA *p < .05 (Tukey’s post-hoc test).
Taqman quantitative real-time PCR (Detection of Mature)
For each unknown sample, 20ng of total DNAse treated RNA was reverse transcribed using the ABI miRNA specific kit with master mix containing the following reagents: dNTPs (100mM), reverse transcriptase (RT) buffer, RNAse inhibitor, and microRNA specific RT primer (ABI), 50 units RT enzyme. The RT protocol was as follows: 1. 16°C-30 minutes 2. 42°C-30 minutes 3. 85°C-5 minutes 4. 4°C-Hold. The PCR protocol was as follows: 1. 95°C-10 minutes 2. 95°C-15 seconds 3. 60°C-1 minute, (repeat 2 and 3, 40 cycles). Results were normalized using the delta-delta ct method, in comparison to sample expression levels of SNO-202, which was found to change very little developmentally or following neuronal activity.
Statistics
Data from experiments in which standard curve efficiency was lower than 80%, or higher than 120%, were not analyzed, and the PCR was repeated. Furthermore, data was analyzed only if the R-squared value was greater than or equal to 0.99. All experimental samples were run in triplicate, and in the rare instance that the standard deviation of these replicates was greater than 0.5 the technical replicate with the greatest variation was removed and the duplicates were analyzed. This threshold was determined prior to all experiments.
Results
In-vivo increases in primary and mature miR-132 following pharmacological activation of neurons
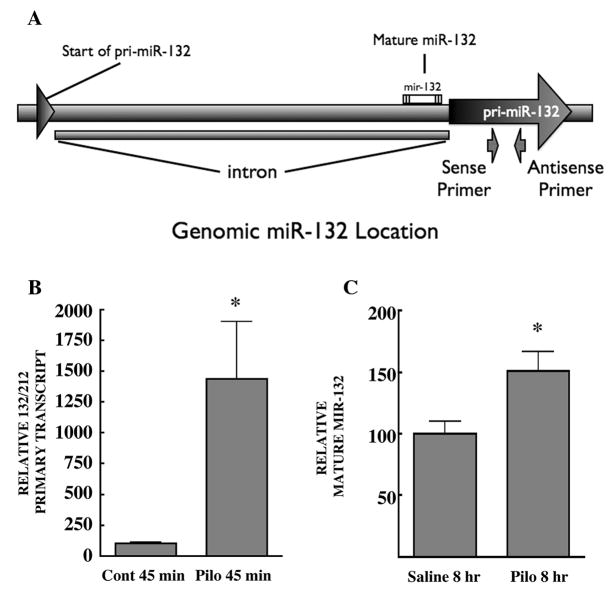
Injection of mice with the muscarinic receptor agonist pilocarpine induces strong generalized seizures due to activation of the M1 muscarinic receptor (Cavalheiro, 1995; Hamilton et al., 1997). Seizure-mediated global activation of neurons in various brain regions, including the hippocampal formation, induces the rapid and transient phosphorylation and activation of both the mitogen-activated protein kinase ERK (Houser et al., 2008) and the transcription factor CREB, ultimately culminating in CRE-mediated transcription (Lee et al., 2007). We tested if pilocarpine-induced activation of neurons might increase production of the primary and mature form miR-132. The primers designed to examine pri-miR-132 produce a 90 bp amplicon and were verified for use in RT-qPCR. Genomic location of miR-132, and primer hybridization sites, are indicated in figure 1a. Mature miR-132 was analyzed as described in the methods section.
Figure 1.
Genomic location of miR-132 and Levels of hippocampal primary- and mature-miR-132 following acute pilocarpine treatment. (A), Genomic location of primers used to recognize the primary sequence of miR-132. In mice, miR-132 is located on chromosome 11. The start of pri-miR-132 resides in an exon and ranges from bp 74986063–74986107. This is followed by an intronic sequence, which separates the remaining pri-miR-132 exon ranging from bp 74987260–74987519. The sense primer binds to bp 74987362–74987383 and the antisense primer binds to 74987430–74987451. All experiments in this study use total RNA that is DNase treated to prevent amplification of genomic sequence during RT-qPCR. (B, C), Mice were injected with saline or pilocarpine (300 mg/kg), sacrificed at indicated times, and RNA levels were determined by RT-qPCR. (B), Relative pri-mir-132 normalized to ARBP transcript levels from same samples (saline: n=4 mice, pilocarpine: n=3 mice, analyzed in triplicate). Two tailed t-test p =0.0182. (C), Both groups received diazepam 60 min after the initial injection. Relative mature-mir-132 normalized to snoRNA-202 (saline: n=10 mice, pilocarpine: n=8 mice, analyzed in triplicate). Two tailed t-test p = .0132.
Mice were injected with pilocarpine and sacrificed 45 minutes or 8 hours later, and hippocampi were processed for real-time RT-qPCR. Within 45 minutes pilocarpine had strongly increased the expression of the primary transcript, pri-mir-132 (Fig. 1b), but had not affected the levels of the mature transcript miR-132 (data not shown). However, 8 hours following induction of pilocarpine-mediated seizures we detected a significant increase in mature miR-132 (Fig. 1c).
To verify that pilocarpine induction of miR-132 is a general response to neuronal activation and is not specific to this drug, we sought another pharmacological activator of neurons. Cocaine has been shown to activate neurons in the striatum and facilitates similar signal transduction, including induction of Erk MAPK/CREB-phosphorylation and activation of CRE-mediated transcription. Therefore, mice were injected with cocaine and real-time RT-qPCR was used to interrogate levels of primary and mature miR-132 over a time course. Indeed, 45 minutes following cocaine injection we detected a significant rise in pri-miR-132 (Fig. 2a), however, in this case, no rise in mature miR-132 was detectable (data not shown).
Physiological activation of neurons induces mir-132
Having established that pharmacological activation of neurons can induce expression of the miR-132 primary transcript, in vivo, we sought to determine if more physiologically relevant activation of neurons was capable of miR-132 induction. Odorant exposure has been shown to activate neurons in the olfactory bulb (Lledo et al., 2005; Martin et al., 2007; McGann et al., 2006; Miwa and Storm, 2005; Pautler and Koretsky, 2002), and contextual fear conditioning is known to do so, as well, in the hippocampus (Atkins et al., 1998; Impey et al., 1998; Sindreu et al., 2007). Five-minute exposure to the odorant citralva (5 uM) induced the primary transcript in the olfactory bulb within 15 minutes, with peak induction occurring at approximately 45 minutes (Fig. 3a), however we did not detect changes in the mature miRNA following odorant exposure (data not shown). A similar profile of activation was observed in the hippocampus following fear conditioning (Fig. 3c), again with no detectable changes in the mature miRNA (data not shown). Notably, context-only exposure resulted in significant induction of the primary transcript, while the unpaired stimulus (shock only) had no effect (Fig. 3e). This suggests the interesting possibility that miR-132 may be involved in the formation of a contextual representation. Interestingly, these regulatory properties for the induction of primary miR-132 are reminiscent of the immediate early genes c-fos and arc, as transient rises in their mRNA following contextual fear conditioning occur on similar timescales, and are also induced by context alone (Huff et al., 2006).
Figure 3.
Analysis of primary-miR-132 in the olfactory bulb following odorant-exposure, and in the hippocampus following contextual fear conditioning (CFC). (A), Mice were exposed to a cotton swab soaked in water or citralva (5 μM), sacrificed at indicated times, and RNA levels were determined by RT-qPCR. Odor exposure significantly increases pri-mir-132 (n=4–5 mice/group, analyzed in triplicate). One-way ANOVA *p < .01 (Tukey’s post-hoc test). (B–D), Relative pri-miRNA levels in hippocampus following CFC, compared to naive mice. (B), Significant increase in primary-miR-132 following CFC (All groups n=5–6; except naive, n=12; paired-30, n=11, analyzed in triplicate). One-way ANOVA *p < .05, **p < .001 (Tukey’s post-hoc test). (C), Expression levels were analyzed under naive, unpaired, context alone, or paired (p30) conditions. Mice were sacrificed 30 minutes after exposure to each condition and reported as relative levels. (naive, n=12; unpaired, n=6; context, n=5; p30, n=11) One-way ANOVA, **p < .001 (Tukey’s post-hoc test). (D) Relative levels of primary-miR-132 expressed in hippocampus following CFC across a full time course (All groups n=5–6; except naive, n=12; paired-30, n=11, analyzed in triplicate). One-way ANOVA *p < .05, **p < .001 (Tukey’s post-hoc test).
Induction Kinetics
Since we could accurately record the stimulus timing in contextual fear conditioning, we used this paradigm to determine the kinetics for primary miRNA induction and the peak response. Mice were trained for contextual fear conditioning and sacrificed immediately following the conclusion of training (p=0), as well as 15, 30, 45, 90, 240, and 360 minutes later. Real-time RT-qPCR was used to compare levels of the primary miRNA to those of the naive controls. Primary miRNA transcript increased rapidly, reaching significance within 15 minutes, and peaking around 45 minutes before rapidly declining to basal levels (Fig. 3f). This rapid decrease likely represents processing to the pre-miRNA, as this step appears to occur co-transcriptionally (Morlando et al., 2008).
Developmental regulation of miR-132 expression
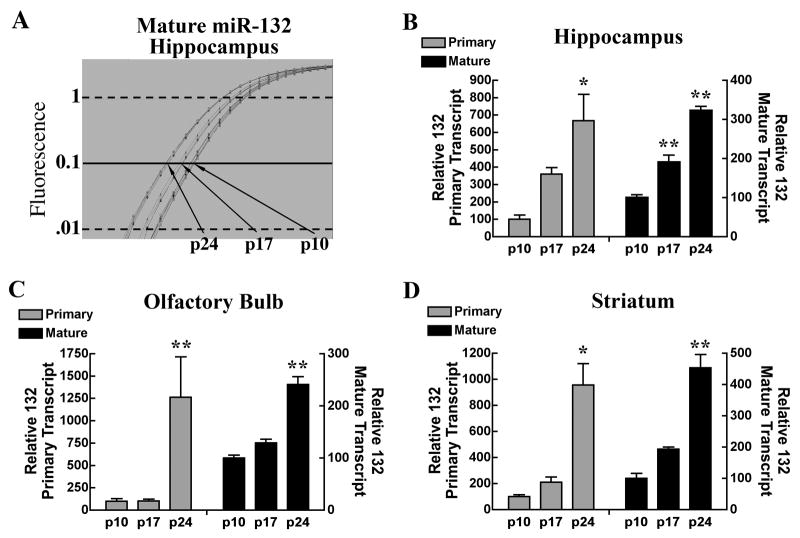
Given that expression of miR-132 is activity dependent, and experience-dependent synaptic refinement occurs in early post-natal development, we hypothesized that its expression may be developmentally regulated. Mice were sacrificed 10, 17, and 24 days after birth, and levels of primary- and mature miR-132 were determined by real-time RT-qPCR. Expression of both of these miR-132 transcripts in the hippocampus (Fig. 4a and 4b), olfactory bulb (Fig. 4c), and striatum (Fig. 4d) increased significantly with age.
Figure 4.
Transcripts of primary and mature-miR-132 progressively increase during post-natal development in various brain regions. (A) Typical qPCR amplification plot of hippocampal mature-miR-132 levels expressed during post-natal days 10, 17 and 24. (B–D) Relative levels of primary (left y axis) and mature mir-132 (right y axis) expression levels in the hippocampus, olfactory bulb, and striatum are depicted for the indicated developmental time points. n=5–6 mice/group, analyzed in triplicate. One-way ANOVA **p < .001 (Tukey’s post-hoc test).
Discussion
It is well established that experience-dependent increases and decreases in protein expression are necessary for long-term neuronal plasticity and development. The transcription factor CREB is one of several molecules that have been demonstrated as key regulators of this response, and many genes rapidly induced following neuronal activation are known to be under CREB control. The recent discovery of CREB regulated-miRNA suggests the intriguing possibility that some of the activity-dependent changes in protein expression may be mediated by miRNA control. That miR-132 is responsive to activity in vitro (Vo et al., 2005; Wayman et al., 2008), and is stimulated by light in the SCN (Cheng et al., 2007) supports this theory. To better understand the physiological relevance of miR-132, we asked if general neuronal activation induced its expression, in vivo.
Seizures initiated by pilocarpine strongly activate neurons throughout the brain (Qian et al., 1993), and lead to induction of CRE-mediated transcription in the hippocampus (Lee et al., 2007). 45 minutes after pilocarpine injection, a 14-fold induction of primary miR-132 was observed in the hippocampus. This led to a 40 percent increase in mature-miR-132 eight hours later. More targeted pharmacological activation of striatal neurons with cocaine also increased pri-miR-132 but to a lesser degree. Physiologic activation of neurons in the olfactory bulb and hippocampus via odor exposure and contextual fear conditioning, respectively, also significantly induced pri-miR-132, confirming the behavioral relevance of these activity-dependent miR-132 increases. Production of the primary miRNA was found to be rapid and transient, with levels peaking 45 minutes following neuronal activation and returning to basal within two hours. Finally, analysis of both primary and mature-miR-132 expression at postnatal days 10, 17, and 24 revealed significant developmental regulation in the olfactory bulb, striatum, and hippocampus.
At first glance, it may seem strange that we have only observed increases in mature miR-132 developmentally and following pilocarpine treatment. We interpret this as a limit in our detection sensitivity. Previous work investigating miRNA processing has shown that primary transcripts are expressed at very low levels (Lee et al., 2002), whereas the mature transcripts are far more stable and therefore accumulate at much higher levels. A small increase in the number of primary transcripts is therefore predicted to be easily detectable in whole cell homogenates since the background expression is low. High basal expression of the mature transcript, however, assures that only very large increases in copy number will result in measurable changes. Therefore, although induction of the primary transcript by cocaine, odorant exposure, or contextual fear conditioning did not correspond with measurable increases in mature miRNA, it is likely that it was induced, but to a degree which is below our detection level. Nevertheless, the fact that expression levels of mature miR-132 increased following seizure, unequivocally demonstrates that neuronal activation can control levels of miR-132, in vivo.
To our knowledge, this is the first report demonstrating production of a miRNA following general pharmacological and physiological neuronal activation in vivo. Similar to the global regulatory effect of immediate-early transcription factors, activity-dependent translational regulation via miRNA could represent vital, previously unrecognized players in the control of protein expression. For example, it is possible that activity-dependent production of miRNA could serve as a proteome switch, capable of broad modulation of specific protein expression necessary to establish or maintain long-lasting long-term potentiation (L-LTP) and long-term memory. Indeed, over-expression of miR-124 or miR-1 in HeLa cells inhibits the expression of hundreds of different genes, and promotes distinctive expression patterns with neuron or muscle cell signatures, respectively (Conaco et al., 2006; Lim et al., 2005). Additionally, forced expression of miR-203 drives cells from a proliferative stem-like state into terminal differentiation by inducing exit from the cell cycle (Yi et al., 2008). Perhaps neuronal stimuli induce miR-132, which in turn directs the proteome towards mnemonic changes in dendritic morphology or synaptic tone. Previous work looking at miR-134 has demonstrated that this miRNA is localized in the dendrites of synapsing hippocampal neurons, where it regulates the size of spines (Schratt and Greenberg, 2006). Localized miR-132 induction specifically at active synapses may promote long-term synaptic activity and contribute to memory traces. Indeed, work in cultured cortical neurons has demonstrated that overexpression of miR-132 leads to a significant potentiation of cellular excitability (Cheng et al., 2007), perhaps via down-regulation of potassium channels it is predicted to regulate.
Akin to the neuronal plasticity involved in experience encoding, activity-dependent pruning and remodeling of synaptic circuits in the early postnatal brain of mammals is vital for its functional maturation. Our finding that miR-132 expression increases during early post-natal development, coupled with its established ability to induce dendritic outgrowth and branching in vitro, suggests the interesting possibility that in vivo levels of miR-132 might be involved in these critical synaptic refinements. Interestingly, expression of methyl CpG-binding protein 2 (MeCP2), a bonafide miR-132 target (Klein et al., 2007), is also developmentally regulated, with brain expression levels increasing from birth until postnatal day 7 (Shahbazian et al., 2002). Maintaining MeCP2 expression at precisely the right level is critical for proper brain development and function. Gene locus duplication as well as frame-shift, missense, and nonsense mutations are all associated with the developmental disorder Rett syndrome. Characterized by a postnatal regression in brain maturation as well as deficits in synaptogenesis, this disorder can result in a variety of debilitating emotive and cognitive deficiencies as well as seizures and heart irregularities (Moretti and Zoghbi, 2006). It has been proposed that miR-132 acts as a homeostatic regulator of MeCP2, helping to maintain expression in mature neurons at proper levels (Klein et al., 2007), and our finding that miR-132 increases on a similar developmental timescale supports this model.
In summary, we have shown that general in vivo activation of neurons pharmacologically, behaviorally, and during development induce the CREB-regulated miRNA, miR-132. We propose that regulation of protein expression in this way may contribute to the vast changes in protein expression known to support cellular representations of experience.
Supplementary Material
Supp Fig. Supporting Figure 1.
ARBP levels are not altered in experimental conditions that stimulate miR-132. A) Amplification plot of pri-miR-132 transcript in individual samples that have been treated with either pilocarpine or saline. B) Amplification plot of ARBP transcript of the same individual samples from A. (C–D) Values of ARBP transcript levels are obtained from a standard curve run in the same experiment as samples. Pilocarpine treatment, odorant exposure, and cocaine injection do not alter levels of ARBP transcript compared to control groups.
Acknowledgments
This work was supported by the following grants: NIH grant #ES015594, and NIH grant # MH 073601. D.P.D. is supported the Ruth L. Kirschstein NRSA pre-doctoral fellowship # F31NS061429.
The authors would like to thank Brian Van Yserloo and Elizabeth A Rutledge for their help with the RT-qPCR experiments, David T Petrillo for his technical assistance with Primer Express, and Soren Impey for his ongoing assistance with this project.
References
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. Rna. 2003;9(3):277–9. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins CM, Selcher JC, Petraitis JJ, Trzaskos JM, Sweatt JD. The MAPK cascade is required for mammalian associative learning. Nat Neurosci. 1998;1(7):602–9. doi: 10.1038/2836. [DOI] [PubMed] [Google Scholar]
- Cavalheiro EA. The pilocarpine model of epilepsy. Ital J Neurol Sci. 1995;16(1–2):33–7. doi: 10.1007/BF02229072. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54(5):813–29. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103(7):2422–7. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Phan T, Han S, Wang H, Chan GC, Scheiner ZS, Storm DR. Circadian oscillation of hippocampal MAPK activity and cAMP: implications for memory persistence. Nat Neurosci. 2008 doi: 10.1038/nn.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Hamilton SE, Loose MD, Qi M, Levey AI, Hille B, McKnight GS, Idzerda RL, Nathanson NM. Disruption of the m1 receptor gene ablates muscarinic receptor-dependent M current regulation and seizure activity in mice. Proc Natl Acad Sci U S A. 1997;94(24):13311–6. doi: 10.1073/pnas.94.24.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser CR, Huang CS, Peng Z. Dynamic seizure-related changes in extracellular signal-regulated kinase activation in a mouse model of temporal lobe epilepsy. Neuroscience. 2008 doi: 10.1016/j.neuroscience.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff NC, Frank M, Wright-Hardesty K, Sprunger D, Matus-Amat P, Higgins E, Rudy JW. Amygdala regulation of immediate-early gene expression in the hippocampus induced by contextual fear conditioning. J Neurosci. 2006;26(5):1616–23. doi: 10.1523/JNEUROSCI.4964-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119(7):1041–54. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Impey S, Smith DM, Obrietan K, Donahue R, Wade C, Storm DR. Stimulation of cAMP response element (CRE)-mediated transcription during contextual learning. Nat Neurosci. 1998;1(7):595–601. doi: 10.1038/2830. [DOI] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10(12):1513–4. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- Lee B, Dziema H, Lee KH, Choi YS, Obrietan K. CRE-mediated transcription and COX-2 expression in the pilocarpine model of status epilepticus. Neurobiol Dis. 2007;25(1):80–91. doi: 10.1016/j.nbd.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo J. 2002;21(17):4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lledo PM, Gheusi G, Vincent JD. Information processing in the mammalian olfactory system. Physiol Rev. 2005;85(1):281–317. doi: 10.1152/physrev.00008.2004. [DOI] [PubMed] [Google Scholar]
- Martin C, Grenier D, Thevenet M, Vigouroux M, Bertrand B, Janier M, Ravel N, Litaudon P. fMRI visualization of transient activations in the rat olfactory bulb using short odor stimulations. Neuroimage. 2007;36(4):1288–93. doi: 10.1016/j.neuroimage.2007.04.029. [DOI] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110(5):563–74. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- McGann JP, Pirez N, Wachowiak M. Imaging odor coding and synaptic plasticity in the mammalian brain with a genetically-encoded probe. Conf Proc IEEE Eng Med Biol Soc. 2006;1:664–7. doi: 10.1109/IEMBS.2006.259465. [DOI] [PubMed] [Google Scholar]
- Miwa N, Storm DR. Odorant-induced activation of extracellular signal-regulated kinase/mitogen-activated protein kinase in the olfactory bulb promotes survival of newly formed granule cells. J Neurosci. 2005;25(22):5404–12. doi: 10.1523/JNEUROSCI.1039-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Zoghbi HY. MeCP2 dysfunction in Rett syndrome and related disorders. Curr Opin Genet Dev. 2006;16(3):276–81. doi: 10.1016/j.gde.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008 doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautler RG, Koretsky AP. Tracing odor-induced activation in the olfactory bulbs of mice using manganese-enhanced magnetic resonance imaging. Neuroimage. 2002;16(2):441–8. doi: 10.1006/nimg.2002.1075. [DOI] [PubMed] [Google Scholar]
- Qian Z, Gilbert ME, Colicos MA, Kandel ER, Kuhl D. Tissue-plasminogen activator is induced as an immediate-early gene during seizure, kindling and long-term potentiation. Nature. 1993;361(6411):453–7. doi: 10.1038/361453a0. [DOI] [PubMed] [Google Scholar]
- Schratt G, Greenberg ME. A brain-specific microRNA regulates dendritc spine development. Nature. 2006 doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum Mol Genet. 2002;11(2):115–24. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- Sindreu CB, Scheiner ZS, Storm DR. Ca2+-stimulated adenylyl cyclases regulate ERK-dependent activation of MSK1 during fear conditioning. Neuron. 2007;53(1):79–89. doi: 10.1016/j.neuron.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103(33):12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102(45):16426–31. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105(26):9093–8. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452(7184):225–9. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp Fig. Supporting Figure 1.
ARBP levels are not altered in experimental conditions that stimulate miR-132. A) Amplification plot of pri-miR-132 transcript in individual samples that have been treated with either pilocarpine or saline. B) Amplification plot of ARBP transcript of the same individual samples from A. (C–D) Values of ARBP transcript levels are obtained from a standard curve run in the same experiment as samples. Pilocarpine treatment, odorant exposure, and cocaine injection do not alter levels of ARBP transcript compared to control groups.