Selective Transcription in Response to an Inflammatory Stimulus (original) (raw)
. Author manuscript; available in PMC: 2011 Mar 19.
Abstract
An inflammatory response is initiated by the temporally controlled activation of genes encoding a broad range of regulatory and effector proteins. A central goal is to devise strategies for the selective modulation of proinflammatory gene transcription, to allow the suppression of genes responsible for inflammation-associated pathologies while maintaining a robust host response to microbial infection. Toward this goal, recent studies have revealed an unexpected level of diversity in the mechanisms by which chromatin structure and individual transcription factors contribute to the selective regulation of inflammatory genes.
Introduction
Inflammation evolved as a rapid and highly beneficial response to microbial infection, tissue injury, and other insults (Nathan 2002; Medzhitov 2008). When host cells capable of innate immune activation, such as tissue macrophages, encounter a microbe or another foreign or host irritant, the inflammatory response initiates within minutes. The host cells first recognize the stimulus through a wide variety of sensing mechanisms, often involving transmembrane receptors. These interactions transmit signals to the nucleus, resulting in the activation of numerous genes via both transcriptional and post-transcriptional mechanisms (Akira et al. 2006; Medzhitov 2007; Ishii et al. 2008; Beutler 2009). The products of these genes carry out diverse physiological functions (Foster and Medzhitov 2009). Some inducible gene products, such as antimicrobial peptides and complement factors, directly target infectious microorganisms. Others, including proinflammatory cytokines and chemokines, activate endothelial cells and recruit cells of both the innate and adaptive immune systems to the site of infection. In addition to their local effects, inducible gene products can act systemically to induce fever, the acute phase response in the liver, and other physiological changes.
Although the beneficial role of the inflammatory response undoubtedly led to its evolution, most researchers study inflammation because of its connection to tissue damage and disease. Acute inflammation can promote tissue repair, but it can also damage host tissues. The detrimental effects are further exacerbated in a chronic inflammatory state, which has been linked to a diverse range of diseases, including inflammatory autoimmune diseases, atherosclerosis, and cancer (Karin et al. 2006; Iscue et al. 2009). Chronic inflammation arises due to the continual presence of a stimulus or to genetic or physiological alterations that disrupt normal feedback mechanisms for attenuating the response.
The critical link between chronic inflammation and disease has led to a search for anti-inflammatory pharmaceuticals that can be tolerated for an extended time period, without significant side effects or the suppression of anti-microbial immunity. One strategy toward this goal is the suppression of individual inflammatory genes or gene products, or specific subsets of genes. Anti-inflammatory drugs have been developed that inhibit the functions of specific proteins, such as the tumor necrosis factor (TNF) receptor and cyclooxygenase-2, but less progress has been made toward the goal of modulating the transcription of specific subsets of proinflammatory genes.
Much has been learned about the recognition of inflammatory stimuli by host receptors, and numerous signal transduction pathways activated by these receptors have been defined and characterized. Signal transduction remains a major focal point for efforts to understand how genes induced by an inflammatory stimulus are differentially regulated. This focus is highly appropriate because differential regulation is dictated largely by differences in the signal transduction pathways required for transcriptional induction. Moreover, many signal transduction pathways rely on enzymes that are attractive therapeutic targets. Nevertheless, molecular events orchestrated in the nucleus by transcription factors and chromatin add additional levels of complexity and may be equally important for an understanding of selectivity.
In this article, I summarize recent progress toward understanding the contributions of transcription factors and chromatin to the selective regulation of inducible proinflammatory genes, with a focus on two major themes. First, although a large number of genes are coordinately induced by a limited set of transcription factors during the primary and secondary responses to a stimulus, detailed studies of specific transcription factors and of the chromatin organization of proinflammatory genes have revealed remarkable diversity in the range of mechanisms employed for transcriptional activation. This mechanistic diversity, which extends well beyond the binding of distinct sets of inducible transcription factors to different promoters and enhancers, probably reflects the need for highly specific regulation of each gene in diverse physiological settings. Second, much of the capacity for selective regulation of genes induced by an inflammatory stimulus appears to be established at the chromatin level during the development of relevant host cells, such as macrophages. Thus, selective regulation is likely to depend just as strongly on the molecular features of proinflammatory loci in cells that have not yet encountered a stimulus as on the properties they acquire upon their induction.
Early Discoveries
Initial efforts to understand the regulation of genes induced by inflammatory stimuli closely followed the cloning of genes encoding key cytokines, including TNF-α, interleukin-1 (IL-1), and interferon-β (IFN-β). Early studies revealed that the genes encoding these cytokines are potently induced at the transcriptional level in macrophages and other cell types, with additional post-transcriptional regulation at the levels of mRNA stability and translation (Zinn et al. 1983; Beutler and Cerami 1986; Caput et al. 1986; Collart et al. 1986; Sariban et al. 1988).
From a transcription perspective, the many genes activated in response to an inflammatory stimulus can be divided at their most fundamental level into two classes: primary response genes are usually activated most rapidly and are formally defined as those genes that can be induced without de novo protein synthesis (Yamamoto and Alberts 1976; Herschman 1991). In other words, the transcription factors required for activation of these genes must be expressed in the unstimulated cell and must be either constitutively active or activated via post-translational mechanisms following cell stimulation. Primary response genes are sometimes referred to as immediate early genes, by analogy to viral genes activated immediately after infection of host cells (Milanesi et al. 1970; Lau and Nathans 1987). Secondary response genes are generally induced more slowly and require new protein synthesis. The transcription of secondary response genes can depend on the de novo synthesis of either transcription factors, signaling molecules needed for the activation of transcription factors, or cytokines that can act in an autocrine fashion to activate additional signaling pathways and transcription factors. Although secondary response genes require newly synthesized proteins, the factors responsible for the activation of primary response genes can also contribute directly to their transcription. Primary and secondary response genes can be distinguished by differences in their sensitivity to inhibitors of protein synthesis, such as cycloheximide (Yamamoto and Alberts 1976).
The finding that proinflammatory genes are induced at the level of transcription inspired efforts to identify DNA motifs and transcription factors that regulate induction. However, the most important early discoveries emerged from studies of other inducible genes during a prolific period of transcription factor discovery in the mid-1980s, soon after the first eukaryotic sequence-specific DNA-binding proteins were reported (Engelke et al. 1980; Dynan and Tjian 1983). NF-κB, the first transcription factor identified whose sequence-specific DNA-binding activity could be induced by an extracellular stimulus acting via a post-translational mechanism, was discovered as an LPS-induced protein that bound an enhancer for the Ig κ light-chain gene in B-cell extracts (Sen and Baltimore 1986). The DNA-binding activity of NF-κB was rapidly induced by LPS in the absence of new protein synthesis, as demonstrated by robust activation in the presence of cycloheximide.
Soon after the NF-κB discovery, several other transcription factors were reported that can be induced via post-translational mechanisms and are now known to be key regulators of both the primary and secondary responses to inflammatory stimuli. Activator protein-1 (AP-1), a heterodimer of the basic leucine zipper proteins c-Jun and c-Fos, was discovered as a transcription factor that bound sequences in the metallothionine and SV40 promoters (Greenberg and Ziff 1984; Lee et al. 1987; Bohmann et al. 1987; Rauscher et al. 1988). Cyclic-AMP (cAMP) response element binding protein (CREB) was discovered as a cAMP-induced factor that regulates induction of the somatostatin gene in neuroendocrine cells (Montminy and Bilezikjian 1987). E2F was discovered as a DNA-binding protein activated by the adenovirus E1A protein in adenovirus-infected cells (Kovesdi et al. 1986). Serum response factor (SRF) and the associated ternary complex factors (TCFs) were identified through an analysis of the serum induction of Fos transcription (Treisman 1986; Prywes and Roeder 1986; Dalton and Treisman 1992). NFAT was discovered as an inducible DNA-binding activity that binds the Il2 promoter in activated T cells (Shaw et al. 1988). These proteins represent only a subset of the transcription factors that are now known to be induced by inflammatory stimuli via post-translational mechanisms. The post-translational mechanisms often involve direct phosphorylation or dephosphorylation of the factors themselves or of inhibitory proteins. As mentioned above, genes encoding several additional transcription factors and cofactors are induced at the transcriptional level by inflammatory stimuli (Amit et al. 2009 and references therein); these newly synthesized factors can contribute to the secondary response in concert with factors whose activities are induced post-translationally.
The Enhanceosome Model
The selective transcription of each proinflammatory gene is regulated by a promoter that spans the transcription start site and, at most or all genes, by one or more distant enhancers. Each promoter and enhancer contains DNA motifs recognized by a collection of sequence-specific DNA-binding proteins (i.e. transcription factors). Studies of proinflammatory gene transcription have mostly focused on promoters because promoters can easily be identified due to their close proximity to the start site. Moreover, promoters for proinflammatory genes are generally sufficient to support inducible transcription in transfection assays with promoter-reporter plasmids. However, distant enhancers have now been identified for a number of proinflammatory genes and are likely to be essential for proper transcriptional regulation in a native chromatin environment (Carey et al. 2009). In a native environment, transcriptional activation appears to require close physical proximity between the promoter and distant enhancers, with “looping out” of the intervening DNA (Lee et al. 2006; de Laat et al. 2008). Although most studies have focused on long-range interactions between enhancers and promoters, a recent study suggests that inducible factors like NF-κB are delivered to the promoters of target genes by long-range interactions between the promoters and unlinked genomic locations at which the factors accumulate following their initial induction (Apostolou and Thanos 2008).
The finding that numerous transcription factors are induced by an inflammatory stimulus suggests a simple model in which the selective activation of a given gene depends on the induction of a defined set of signal transduction pathways, which activate a defined set of transcription factors capable of binding the DNA motifs present in the promoter and enhancers of that gene. According to this model, the factors will activate transcription synergistically, perhaps by binding cooperatively to the control regions and forming a three dimensional composite surface for the recruitment of the co-activators, chromatin remodeling complexes, and general transcription factors needed for transcription initiation by RNA polymerase II.
Detailed studies of human IFNB activation upon Sendai virus infection have demonstrated the central role of cooperative binding and synergy between multiple sequence-specific DNA-binding proteins in selective transcriptional activation (Thanos and Maniatis 1995; Agalioti et al. 2000). The regulatory region of the IFNB promoter consists of a 55-bp DNA sequence with greater than 90% identity between mouse and human. In extracts from Sendai virus infected cells, a highly stable multiprotein complex, termed an enhanceosome, assembles on this DNA region through the cooperative binding of the inducible transcription factors NF-κB, IRF3/IRF7, and ATF-2/c-Jun (Thanos and Maniatis 1995). Structural studies have revealed that virtually every base-pair within the 55-bp region is directly contacted by at least one of the DNA-binding proteins, explaining the strong sequence conservation through evolution (Panne et al. 2004, 2007). Interestingly, direct protein-protein interactions between the transcription factors play only a minimal role in cooperative binding. Instead, cooperativity is largely due to conformational changes in the DNA upon factor binding that facilitate the binding of other factors to adjacent and overlapping sites (Panne et al. 2004, 2007). Cooperativity may also benefit from the concerted association of multiple enhanceosome factors with common co-activators, such as p300.
In vivo and in vitro studies have provided evidence that the conserved IFNB promoter region is nucleosome-free in uninfected cells, but a stable nucleosome encompasses the downstream TATA box (Agalioti et al. 2000; Lomvardas and Thanos 2002). Assembly of the enhancesome upon activation of the relevant transcription factors leads to the sequential recruitment of the p300 and GCN5 histone acetyltransferases and SWI/SNF nucleosome remodeling complex, resulting in sliding of the TATA-associated nucleosome to a downstream location. Nucleosome displacement allows recruitment of the transcription factor IID (TFIID) complex that contains the TATA-binding protein (TBP), along with other general transcription factors and RNA polymerase II.
The IFNB studies exemplify one mechanism by which selective gene activation can be achieved: transcription is activated only by stimuli that activate all transcription factors that associate with the enhanceosome, due to the apparent requirement for highly cooperative binding and synergistic functions. This mechanism helps explain why stimuli acting through Toll-like receptors 3 and 4 (TLR3 and TLR4) lead to IFNB activation, whereas IFNB transcription is not activated by bacterial stimulation of TLR2; TLR2 stimulation by bacteria does not activate IRF3, although it activates NF-κB and ATF-2/c-Jun (Doyle et al. 2002).
The importance of synergy between multiple inducible transcription factors and the signal transduction pathways responsible for their activation cannot be overemphasized, as synergistic activation is likely to play a major role in the selective regulation of many or all genes. Indeed, combinatorial regulation of transcription was postulated more than 40 years ago to be essential for a limited number of transcriptional factors to coordinate the diverse gene expression patterns observed in multicellular organisms (Britten and Davidson 1969; Georgiev 1969; Gierer 1973). However, highly cooperative binding by all inducible transcription factors required for the function of a promoter may be much less common. First, it is rare to find promoters like the IFNB promoter, with greater than 90% sequence conservation between human and mouse through an extended region (Arnosti and Kulkarni 2005). Second, highly stable protein-DNA complexes containing several cooperatively bound proteins have rarely been reported. Third, a growing body of evidence suggests that a subset of key transcription factors, including NF-κB, nuclear hormone receptors, and yeast activators such as Gal4 and Gcn4, associate dynamically or transiently with promoter DNA, with the dynamic association possibly required for transcriptional activation (Lipford et al. 2005; Muratani et al. 2005; Bosisio et al. 2006; Hager et al. 2009). Fourth, even at the IFNB promoter, key transcription factors have been found to associate sequentially rather than simultaneously following Sendai virus infection (Munshi et al. 2001). Thus, an understanding of selective activation cannot be reduced to the goal of isolating stable protein-DNA complexes. As described below, selective activation appears to be achieved through mechanisms that extend well beyond the basic induction of DNA-binding proteins and their differential binding to DNA motifs in promoters and enhancers.
Selective Regulation by NF-κB
Studies of transcriptional regulation by NF-κB have provided considerable insight into the diverse mechanisms that have evolved to regulate distinct sets of inducible genes. The NF-κB family consists of five members: p50, p52, RelA, c-Rel, and RelB (Ghosh et al. 1998). The Rel homology region (RHR) that defines the NF-κB family supports sequence-specific DNA binding and the formation of stable homodimers and heterodimers. RelA, c-Rel, and RelB contain C-terminal activation domains, whereas p50 and p52 lack definable activation domains. Most NF-κB proteins are retained in the cytoplasm of resting cells by ankyrin repeat-containing IκB proteins (Ghosh et al. 1998; Hoffmann et al. 2006; Vallabhapurapu and Karin 2009). Some IκBs are encoded by separate genes. However, p50 and p52 are initially synthesized as the precursor proteins, p105 and p100, respectively, which contain IκB-like ankyrin repeat domains at their C-terminus; these domains are often removed by constitutive proteolytic processing, but they can remain covalently associated with some NF-κB homodimers or heterodimers in the cytoplasm (Vallabhapurapu and Karin 2009). A detailed biochemical analysis recently revealed a surprisingly diverse range of dimeric and multimeric NF-κB/IκB complexes in the cytoplasm of unstimulated cells (Savinova et al. 2009).
NF-κB dimers retained in the cytoplasm can be activated by either of two fundamentally distinct pathways, referred to as the type 1 and type 2 pathways (Vallabhapurapu and Karin 2009). The abundant p50:RelA and p50:c-Rel heterodimers, as well as several other dimeric species, are activated by the classical type 1 pathway, which involves phosphorylation of the associated IκB, leading to its ubiquitylation and proteosome-mediated degradation, thereby releasing the NF-κB dimer to translocate to the nucleus (Ghosh et al. 1998; Hoffmann et al. 2006; Vallabhapurapu and Karin 2009). In contrast, p52:RelB dimers are often activated by the type 2 pathway, which involves inducible proteolytic removal of the ankryin-repeat domain of the p100:RelB heterodimer, releasing p52:RelB to translocate to the nucleus.
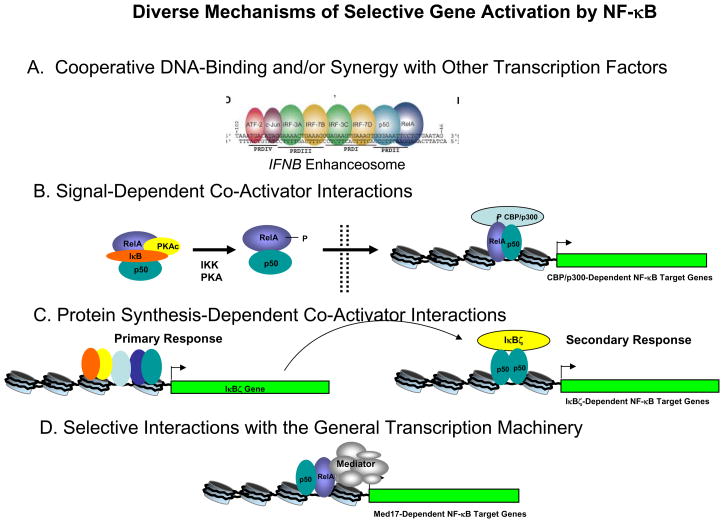
Although inducible nuclear translocation is critical for NF-κB activation, NF-κB’s capacity to activate transcription depends on additional layers of regulation, with different regulatory layers at different NF-κB target genes (Fig. 1). As one example, activation of some target genes depends on the inducible phosphorylation of RelA (Fig. 1B). Mouse RelA is phosphorylated by cyclic AMP-dependent protein kinase (PKAc) on serine 276 (S276), which results in a conformational change that exposes an interaction surface for the transcriptional co-activators p300 and CBP (Zhong et al. 1998, 2002). Importantly, targeted mutation of RelA S276 led to defective activation of some but not all NF-κB target genes in TNFα-stimulated fibroblasts; Cxcl2 and Tnf transcription was reduced, but transcription of Il6, Ptgs2, and Nfkbia was unaffected (Dong et al. 2008). Thus, NF-κB-mediated recruitment of the p300/CBP co-activators, which contain an acetyltransferase domain capable of acetylating core histones, is differentially required at NF-κB target genes. This differential requirement may allow some genes to be activated by any stimulus that promotes IκB phosphorylation and degradation, whereas other genes will be activated only when IκB phosphorylation is accompanied by the activation of PKAc or other kinases that can phosphorylate RelA S276. Interestingly, transcriptional activation by c-Rel, whose RHR is highly homologous to that of RelA, does not appear to be influenced by p300/CBP (Wang et al. 2007). This difference may restrict the activation of p300/CBP-dependent NF-κB target genes to RelA homodimers or heterodimers.
Figure 1. Diverse Mechanisms of Selective Gene Activation by NF-κB.
(A) The highly cooperative binding and synergistic function of multiple transcription factors has been found to play a major role in the selective activation of the human IFNB gene (Thanos and Maniatis 1995; Agalioti et al. 2000). Synergy between multiple transcription factors activated by diverse signal transduction pathways is likely to be critical for the selective activation of all genes induced by inflammatory stimuli.
(B) Analysis of mice containing a mutation in the RelA S276 phosphoacceptor site have revealed that a select subset of NF-κB target genes depend on S276 phosphorylation (Dong et al. 2008), which promotes an interaction between NF-κB and the p300/CBP co-activators (Zhong et al. 1998, 2002).
(C) The Nfkibz gene, which encodes the nuclear IκB protein, IκBζ, is activated at the transcriptional level during the primary response to LPS and other inflammatory stimuli (Yamamoto et al. 2006; Motoyama et al. 2005). IκBζ is subsequently required for the activation of a select subset of secondary response genes. Activation of these genes therefore depends on all signal transduction pathways needed for Nfkibz transcriptional activation.
(D) An interaction between RelA and the Med17 subunit of the Mediator complex is needed for activation of a select subset of NF-κB target genes (van Essen et al. 2009). Other inducible transcription factors may be responsible for recruitment of the Mediator complex to other NF-κB target genes, thereby conferring a requirement for these factors for transcriptional activation.
Recently, methylation of RelA lysine 37 (K37) by the Set9 methyltransferase was identified as another post-translational modification that can contribute to the selective regulation of NF-κB target genes (Ea and Baltimore 2009). Both Set9 and RelA K37 were found to be important for activation of the Tnf and Cxcl10 genes, but not the Nfkbia gene, in TNFα-treated cells. Microarray experiments performed in an independent study suggested that Set9 is important for the activation of approximately 25% of NF-κB target genes in TNFα-treated monocytes (Li et al. 2008). K37 methylation appears to stabilize NF-κB binding to some recognition motifs and may therefore promote the transcription of a select subset of target genes that benefit from this enhanced stability (Ea and Baltimore 2009). Notably, all NF-κB family members appear to extensively modified at a post-translational level (Perkins 2006); the modified residues will need to be disrupted by targeted mutagenesis in mice (similar to the S276 mutagenesis experiments) to evaluate the importance of each modification for NF-κB function.
NF-κB target genes also exhibit differential dependence on nuclear IκB proteins, which function as transcriptional co-activators by interacting with NF-κB dimers associated with target genes (Yamamoto and Takeda 2008). One example is IκBζ, whose gene, Nfkbiz, is activated at the transcriptional level in macrophages during the primary response to an inflammatory stimulus (Yamamoto et al. 2004; Motoyama et al. 2005). Because IκBζ expression is induced during the primary response, all NF-κB target genes that rely on this factor for their activation will be secondary response genes (Fig. 1C). Indeed, an analysis of _Nfkbiz_−/− mice revealed that primary response genes, such as Cxcl1, Cxcl2, and Il23a, were induced in an _Nfkbiz_-independent manner, whereas a subset of secondary response genes, including Il12b, Il6, and Lcn2, exhibited Nfkbiz dependence (Yamamoto et al. 2004). Following its inducible synthesis, IκBζ appears to associate with NF-κB p50 homodimers associated with inactive genes, thereby facilitating their transcriptional activation (Yamamoto et al. 2004). The molecular mechanism by which IκBζ promotes the activation of NF-κB target genes remains unknown, although it appears to act prior to preinitiation complex assembly and histone H3K4 trimethylation at target gene promoters (Kayama et al. 2008). Most importantly, IκBζ-dependent NF-κB target genes are likely to be activated in a highly selective manner, as activation of these genes will require all signal transduction pathways needed for transcriptional induction of the Nfkbiz gene, in addition to signaling pathways required for nuclear translocation of NF-κB. The two other nuclear IκB proteins, Bcl3 and IκBNS, appear to make additional contributions to the selective regulation of inducible NF-κB target genes (Leung et al. 2004; Yamamoto and Takeda 2008).
NF-κB activation mechanisms are further diversified through differential interactions with components of the general transcription machinery. This potential contribution to selective gene activation emerged from an analysis of NF-κB’s interaction with the Mediator complex, which is essential for the activation of most eukaryotic genes by facilitating the recruitment of the general transcription machinery and RNA polymerase II by transcriptional activators (Malik and Roeder 2005). RelA was found to associate with the Med17 (Trap80) subunit of the Mediator complex (van Essen et al. 2009), leading to the initial hypothesis that the RelA-Med17 interaction may be critical for the activation of all NF-κB target genes. Surprisingly, however, Med17 knockdown by RNA interference (RNAi) revealed that this Mediator subunit is needed for activation of only a subset of target genes, including Ptgs2, Il6, and Cxcl10, while the expression of Cxcl2, Nfkbia, and many other inducible genes was unaffected (Fig. 1D). In contrast to the p300/CBP and IκBζ selectivity mechanisms described above, it is not yet known whether the selective requirement for the NF-κB-Med17 interaction leads to a requirement for different signal transduction pathways at Med17-dependent and –independent genes. One possibility is that Med17-independent NF-κB target genes require the induction of another transcription factor that can recruit the Mediator through a direct interaction with a different Mediator subunit.
The above findings reveal a clear theme in which at least some post-translational modifications and co-factors involved in NF-κB activation contribute to the activation of only a small subset of NF-κB target genes. At a mechanistic level, much remains to be learned about these differential contributions. We can speculate that this mechanistic diversity plays an important role in facilitating the differential regulation of NF-κB target genes in different physiological settings. It is tempting to speculate, for example, that the subset of NF-κB target genes that require an inducible interaction with p300/CBP are coordinately and selectivity activated in certain physiological circumstances, while target genes requiring IκBζ will remain inactive under those same conditions due to the absence of Nfkbiz transcriptional induction. Hopefully, these connections between activation mechanisms and physiological responses will become apparent as more detailed knowledge is gained about the specific subsets of genes that rely on each activation mechanism, as well as the specific subsets of genes that are induced in different physiological settings. Finally, although I have focused on NF-κB, other inducible transcription factors are likely to exhibit similar levels of diversity in the mechanisms by which they activate specific sets of target genes.
Role of Chromatin Structure and Development in Selective Regulation
Early advances
Although most studies of gene induction by inflammatory stimuli have focused, quite appropriately, on transcription factors that recognize specific DNA sequences, transcriptional activation of eukaryotic genes is also influenced by chromatin structure. The fundamental repeating unit of chromatin is the nucleosome, which consists of a histone octamer containing two molecules each of the core histones H2A, H2B, H3, and H4 (Luger et al. 1997). Linker histones, histone variants, and many non-histone proteins make additional contributions to the structure of chromatin found in eukaryotic cells. Molecular evidence that chromatin may be more accessible to transcription factors and RNA polymerase at active genes than at inactive genes was obtained in the 1970s through the analysis of locus-specific differences in sensitivity and hypersensitivity to cleavage by nucleases added to cell nuclei or permeabilized cells (Weintraub and Groudine 1976; Wu et al. 1979; Carey et al. 2009). The Il2 cytokine gene was among the initial group of genes found to exhibit changes in nuclease hypersensitivity upon transcriptional induction (Siebenlist et al. 1986). In the 1980s and early 1990s, evidence began to emerge that the N-terminal tails of core histones and the post-translational modification of these tails may contribute to transcriptional regulation (Grunstein 1990). However, conclusive evidence that chromatin plays a direct role in transcriptional regulation was not obtained until the mid-1990s. At that time, Gcn5, a transcriptional co-activator in S. cerevisiae, was found to catalyze the acetylation of histone H3, providing definitive evidence that the covalent modification of histones can contribute to transcriptional control (Brownell et al. 1996). In addition, biochemical experiments revealed that the SWI/SNF complex, originally identified by classical genetics as an important regulator of specific subset of yeast genes (Neigeborn and Carlson 1984; Stern et al. 1984), uses the energy of ATP hydrolysis to catalyze changes in nucleosome conformation (Cote et al. 1994; Imbalzano et al. 1994; Kwon et al. 1994). These conformational changes, referred to as nucleosome remodeling, make genomic DNA more accessible to transcription factor binding, either through the translocation or eviction of nucleosomes or a change in their conformation (Clapier and Cairns 2009).
One of the first clear connections between nucleosome remodeling and the regulation of inducible transcription in cells of the immune system was the observation that mammalian SWI/SNF complexes are recruited rapidly and efficiently to chromatin following the activation of resting T cells (Zhao et al. 1998). SWI/SNF association coincided with global decondensation of the chromatin, consistent with the view that decondensation is needed for T cell activation. However, SWI/SNF binding was not observed until the first wave of inducible transcription had begun (Zhao et al. 1998), providing initial evidence that nucleosome remodeling by SWI/SNF complexes may not be required for the activation of many primary response genes (see below).
Variable requirements for nucleosome remodeling at inducible genes
A second major conceptual advance was provided by Saccani et al. (2001), who found that NF-κB associates with its target genes in LPS-stimulated macrophages with variable kinetics. In chromatin immunoprecipitation (ChIP) experiments, NF-κB bound the Cxcl2 and Nfkbia promoters as soon as it entered the nucleus, whereas binding to the promoters for Ccl5, Il6, and other genes was substantially delayed. An attractive explanation for this difference was that a chromatin barrier needs to be overcome for NF-κB binding to some but not all target genes. Saccani et al. hypothesized that the nucleosome barrier provides a potential mechanism for selectively regulating NF-κB target genes, as additional transcription factors could control NF-κB access by regulating the recruitment of remodeling factors.
Subsequent studies revealed variable requirements for SWI/SNF nucleosome remodeling complexes at genes induced by LPS in mouse macrophages (Ramirez-Carrozzi et al. 2006). This variability is likely to explain, at least in part, the different kinetics of NF-κB binding. Analysis of macrophages in which the core Brg1 and Brm ATPase subunits of the mammalian SWI/SNF complexes had been depleted by retroviral delivery of short hairpin RNAs (shRNAs) revealed that most primary response genes are induced by LPS in a SWI/SNF-independent manner, with almost all secondary response genes exhibiting SWI/SNF dependence. SWI/SNF dependence was also observed at a subset of primary response genes that were generally induced with delayed kinetics. The promoters of representative SWI/SNF-independent genes were found to be accessible to nuclease cleavage in nuclei from both unstimulated and stimulated cells; in contrast, the promoters of SWI/SNF-dependent genes exhibited low nuclease accessibility prior to cell stimulation, with increased accessibility following stimulation, suggestive of inducible nucleosome remodeling (Ramirez-Carrozzi et al. 2006). SWI/SNF-independent promoters also exhibited constitutively high histone acetylation and histone H3K4 trimethylation, providing further support for the view that these promoters are assembled, prior to stimulation, into a chromatin structure similar to that found at active genes (Ramirez-Carrozzi et al. 2006, 2009).
Regulation of SWI/SNF-independent genes
Importantly, SWI/SNF-independent primary response genes usually contain CpG island promoters, whereas SWI/SNF-dependent primary and secondary response genes almost always contain promoters with a low CpG content (Ramirez-Carrozzi et al. 2009). In vitro nucleosome assembly experiments revealed that nucleosomes assembled on CpG-island promoters are less stable than those assembled on low CpG promoters. This finding is consistent with a large body of evidence that properly spaced AA/TT dinucleotides, which are deficient at most CpG-island promoters, are required for stable nucleosome assembly (Segal et al. 2006, and references therein). This intrinsic instability may facilitate rapid transcriptional activation of primary response genes containing CpG islands in the absence of a nucleosome remodeling requirement. Remodeling-independent activation may also benefit from the high prevalence of binding sites for constitutive transcription factors, such as Sp1, in CpG island promoters; binding of Sp1 and other constitutively expressed factors may contribute to the SWI/SNF-independence of these promoters and may be responsible for their constitutive histone acetylation and H3K4 trimethylation, as well as for the low level of constitutive transcription frequently observed at these promoters (Ramirez-Carrozzi et al. 2009).
Recent studies have elucidated the molecular mechanism by which primary response genes can be efficiently induced in a stimulus-dependent manner, despite their constitutive assembly into a chromatin structure resembling that found at active genes. Although low levels of precursor transcripts are constitutively produced at these genes, NF-κB and possibly other inducible factors are needed to enhance the efficiency of transcription elongation and pre-mRNA processing, in addition to enhancing the frequency of transcription initiation (Amir-Zilberstein et al. 2007; Hargreaves and Medzhitov 2009). These inducible factors promote acetylation of histone H4K5, K8, and K12, with the acetyl lysines then recognized by the bromodomain-containing adaptor protein Brd4. Brd4 recruits P-TEFb, which promotes elongation and pre-mRNA processing through its ability to phosphorylate the C-terminal domain of RNA polymerase II (Hargreaves and Medzhitov 2009, and references therein).
Selective regulation conferred by a nucleosome barrier
As initially proposed by Saccani et al. (2001), the nucleosome barrier found at most secondary response genes and some primary response genes provides an attractive strategy for differentially regulating the induction of specific subsets of genes by inflammatory stimuli. Stimulus-specific transcription factors may facilitate the recruitment of SWI/SNF complexes to distinct sets of remodeling-dependent genes, allowing their selective activation. Consistent with this hypothesis, a substantial subset of SWI/SNF-dependent LPS-induced primary response genes was found to require IRF3 for their activation and the promoters for these genes usually contain consensus IRF3 binding sites (Ramirez-Carrozzi et al. 2009). Inducible nucleosome remodeling at these promoters, analyzed by restriction enzyme accessibility, was absent in LPS-stimulated macrophages from IRF3−/− mice, demonstrating that IRF3 promotes nucleosome remodeling, either directly or indirectly, at this select subset of LPS-induced primary response genes.
The observation that SWI/SNF complexes are consistently needed for the activation of IRF3-dependent genes suggests that the nucleosome barrier at the promoters for these genes is critical for restricting their activation to stimuli that efficiently induce IRF3. Striking evidence in support of this hypothesis was provided by Lomvardas and Thanos (2002) through their analysis of the IRF3-dependent IFNB promoter, discussed above as a paradigm for the enhanceosome model. Despite strong evidence that IRF3, NF-κB, and ATF-2/c-Jun form a stable enhanceosome at the IFNB promoter, displacement of the TATA box-encompassing nucleosome through the use of an artificial nucleosome positioning sequence resulted in efficient IRF3-independent promoter activity. Thus, NF-κB and ATF-2/c-Jun appear to be quite effective in stimulating IFNB promoter activity in the absence of IRF3 when the nucleosome barrier is eliminated. This suggests that the nucleosome barrier at IRF3-dependent genes is more important than cooperative binding of the full complement of transcription factors for selective regulation. That is, the nucleosome barrier appears to be the main reason IFNB transcription is induced by TLR3 and TLR4, which activate IRF3, but not by TLR2 or TNFα, which do not activate this factor (Doyle et al. 2002).
Interestingly, at representative SWI/SNF-dependent secondary response promoters, including Il12b, Il6, and Nos2, nucleosome remodeling as monitored by restriction enzyme accessibility and the recruitment of SWI/SNF complexes is strongly dependent on new protein synthesis (Weinmann et al. 1999; Ramirez-Carrozzi et al. 2006). Therefore, a major mechanism for restricting the activation of many secondary response genes is through their requirement for specific primary response gene products to promote the recruitment of SWI/SNF complexes. Unfortunately, the identities of the primary response proteins that drive remodeling at specific subsets of secondary response genes have not yet been determined. Notably, nucleosome remodeling by SWI/SNF complexes may be subject to additional regulatory layers that can further enhance the extent to which a remodeling requirement can contribute to selective activation. In particular, nucleosome remodeling at the promoters of LPS-induced genes by SWI/SNF complexes was found to require a calcium signaling pathway, which promotes the activation of SWI/SNF complexes after their recruitment to target genes (Lai et al. 2009).
Taken together, the above results reveal that an understanding of selectivity requires, not only the identification of transcription factors induced by various stimuli, but also an understanding of the chromatin state of inducible loci in differentiated cells that have not yet encountered a stimulus. For example, the studies described above suggest that IRF3 dependence in macrophages is achieved by the assembly of IRF3-dependent promoters into stable nucleosomes during macrophage development, whereas the promoters of most IRF3-independent primary response genes are assembled into constitutively accessible chromatin due to the presence of CpG-island promoters.
Extensive chromatin diversity at the promoters of inducible genes prior to their activation
Several additional studies of chromatin structure at promoters induced by inflammatory stimuli have revealed a surprising degree of variability in mature unstimulated cells (Figure 2). One early example was the finding that histone H3K9 methylation, which generally correlates with transcriptional repression, is readily apparent at the promoters of some but not all inducible genes in unstimulated human dendritic cells (Saccani and Natoli 2002). H3K9 methylation was observed at the IL12B, CCL19, and CCL22 promoters in unstimulated cells and was rapidly lost upon LPS stimulation. However, this repressive mark was not observed at the CCL3, IL8, and NFKBIA promoters. This finding suggests a model in which transcriptional activation of a select subset of LPS-induced genes requires the recruitment and function of an H3K9 demethylase. Although the identity of the H3K9 demethylase remains to be established, the regulation of this demethylase, either through the regulation of its expression or the direct regulation of its catalytic activity, could play a major role in the differential regulation of genes that contain or lack H3K9 methylation in unstimulated cells.
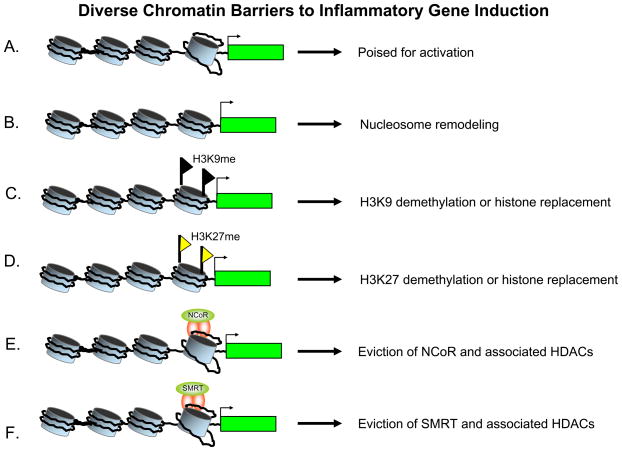
Figure 2. Diverse Chromatin Barriers to Inflammatory Gene Induction.
(A) Most genes activated during the primary response to an inflammatory stimulus are poised for activation by their constitutive assembly into a chromatin structure resembling that found at constitutively active genes (Ramirez-Carrozzi et al. 2009; Hargreaves and Medzhitov 2009). At some of these genes, inducible transcription factors may not need to remove chromatin barriers, but these factors must instead enhance transcription initiation, elongation and pre-mRNA processing. Genes within this class are generally induced promiscuously by a wide range of generic signaling pathways.
(B) The assembly of promoters and other control regions into stable nucleosomes provides a substantial barrier to transcriptional activation (Ramirez-Carrozzi et al. 2009). SWI/SNF complexes are often required for the remodeling of these nucleosomes. Inducible remodeling, as monitored by restriction enzyme accessibility, requires specialized factors that are either directly activated by a stimulus, such as IRF3, or that are encoded by genes expressed during the primary response to the stimulus. The nucleosome remodeling requirement contributes to the tight regulation of SWI/SNF-dependent genes.
(C) and (D) ChIP analyses of large panels of promoters and genome-wide ChIP-Seq experiments have revealed that histone H3K9 and histone H3K27 are heavily methylated at small subsets of inducible promoters in mature unstimulated cells (Saccani and Natoli 2002; De Santa 2007, 2009). Transcriptional activation of these genes generally coincides with the loss of histone H3K9 or H3K27 methylation, suggesting the demethylation or histone replacement may be required for activation.
(E) and (F) The NCoR and SMRT co-repressor complexes appear to be associated with distinct subsets of inducible genes in mature, unstimulated cells (Ghisletti et al. 2009; Hargreaves and Medzhitov 2009). Transcriptional activation requires the removal of these co-repressor complexes, which contain histone deaceylases (HDACs) and other subunits that may help maintain a repressive chromatin structure.
H3K27 methylation, another repressive histone modification, which acts by recruiting polycomb complexes, has also been identified at a select subset of inducible genes prior to their activation (De Santa et al. 2007, 2009). Furthermore, a histone demethylase, Jmjd3, was found in genome-wide ChIP experiments to be associated in unstimulated macrophages with a small subset of inducible genes (De Santa et al. 2007, 2009). Interestingly, although Jmjd3 can demethylate methylated H3K27, the subset of genes associated with Jmjd3 differed from the subset marked by methyl H3K27.
Further variability in the chromatin features of inducible genes in resting macrophages was uncovered through analyses of the co-repressors NCoR (nuclear receptor co-repressor) and SMRT (silencing mediator of retinoic acid and thyroid hormone recptors) (Jepsen and Rosenfeld 2002). These co-repressors, which function in part through the recruitment of histone deacetylases, appear to be associated with overlapping subsets of inducible genes in resting cells through their interaction with different DNA-binding proteins (Ghisletti et al. 2009). For example, NCoR is specifically associated with the Mmp13 promoter, SMRT is specifically associated with the Il12b promoter, and both NCoR and SMRT are associated with the Ccl2 promoter in unstimulated macrophages. The Ets-domain protein TEL appears to be responsible for NCoR association, with c-Jun homodimers and NF-κB p50 homodimers contributing to SMRT association (Ghisletti et al. 2009). The binding of these co-repressors to distinct subsets of target genes in resting cells has the potential to contribute to their selective activation.
Role of developmental events in chromatin diversity
The above examples underscore the hypothesis that differences in chromatin structure at the promoters of inducible genes in resting cells are likely to play a critical role in their selective activation. Most likely, these chromatin differences are established during the development of macrophages and other responsive cell types. It is well established that transcription factor interactions with the control regions of inducible and tissue-specific genes, and changes in chromatin structure at these genes, can begin early in development and long before the genes are expressed, sometimes as early as the embryonic stem cell stage (Lefevre et al. 2008; Zaret et al. 2008; Xu et al. 2009; and references therein). However, the developmental events that lead, for example, to H3K9 or H3K27 methylation at a select subset of inducible genes in mature resting cells remain undefined. These chromatin differences must be dictated by specific DNA motifs in the promoters and distal control regions of the inducible genes. However, in most instances, the transcription factors involved, and the developmental stage at which the chromatin state is established, have not been determined.
Consistent with the proposal that developmental events play a major role in selective regulation, developmental differences can lead to variable activation requirements at specific genes in different cell types. As one example, the Il6 promoter is assembled into inaccessible nucleosomes in mouse macrophages and its activation by LPS requires nucleosome remodeling by SWI/SNF complexes, as well as new protein synthesis for the induction of nucleosome remodeling (Ramirez-Carrozzi et al. 2009). In contrast, the same promoter is assembled into open chromatin in fibroblasts, allowing Il6 activation by LPS in a SWI/SNF-independent and protein synthesis-independent manner. These chromatin differences in two different mature cell types, which can have a profound influence on the signal transduction and transcription factor requirements for Il6 activation, must be established during macrophage and fibroblast development.
Physiological Relevance of the Mechanistic Variability
The many examples discussed above of mechanistic variability in the transcriptional response to an inflammatory stimulus provide considerable potential for the selective regulation of inducible genes. However, the precise relationship between this mechanistic diversity and physiological responses remains poorly understood. As mentioned above, physiological responses have not been identified that are characterized by activation of the select subset of genes that require RelA phosphorylation-dependent recruitment of p300/CBP. Similarly, physiological settings have not been identified that are characterized by activation of the select subset of genes possessing methyl H3K9 or methyl H3K27 marks.
Clear connections between mechanistic diversity and selective regulation have been equally difficult to uncover when gene regulation in specific physiological settings is first considered. For example, tolerance to repetitive stimulation of macrophages by LPS is a well-characterized process that is thought to have evolved to protect a host from tissue damage during prolonged exposure to a bacterial pathogen. Strong suppression of transcription occurs primarily at proinflammatory genes, whose products can lead to tissue damage, while robust activation of anti-microbial genes is maintained (Foster et al. 2007). The selective suppression of some proinflammatory genes involves changes in chromatin structure and histone modifications (Foster et al. 2007). However, susceptibility or resistance to LPS tolerance cannot be connected to a specific NF-κB activation mechanism or to genes containing a specific chromatin signature in unstimulated cells. As another example, the key anti-inflammatory cytokine IL-10 inhibits a select subset of LPS-induced genes, but without a clear connection to any of the selective regulatory mechanisms discussed above (Lang et al. 2002). Most likely, LPS tolerance and the anti-inflammatory effects of IL-10 involve additional layers of regulation that are not dependent on a specific NF-κB activation mechanism or chromatin signature.
Although much remains to be learned about the connections between mechanistic diversity and physiological regulation, a few glimpses of the underlying logic have begun to emerge. For example, SWI/SNF-independent genes containing CpG-island promoters are generally induced by a diverse range of stimuli, including multiple TLR ligands, TNFα, and serum, consistent with the absence of a nucleosome barrier at the promoter to limit activation (Ramirez-Carrozzi et al. 2009; Hargreaves and Medzhitov 2009). Conversely, SWI/SNF-dependent genes containing low CpG promoters are induced more selectively and with greater tissue-specificity, as described above for IRF3-dependent genes (Ramirez-Carrozzi et al. 2009; Hargreaves and Medzhitov 2009).
Differential displacement of the NCoR and SMRT co-repressors provides another connection between mechanistic diversity and a specific physiological response. Release of the SMRT co-repressor from its target genes was found to be catalyzed by the MEKK1 signaling pathway, whereas NCoR binding was unaffected by this pathway (Ghisletti et al. 2009). Since IFN-γ activates the MEKK1 pathway, this cytokine can lead to the selective enhancement of SMRT-associated genes, including Il12b and Ccl2, without enhancing NCoR-associated genes (Ghisletti et al. 2009). Thus, the differential response to IFN-γ may be dictated, at least in part, by the differential recruitment of NCoR and SMRT to the promoters of inducible genes during the development of responsive cell types.
Concluding Remarks
I have highlighted recent progress toward understanding the molecular mechanisms that underlie the selective activation of genes by an inflammatory stimulus. Clearly, much has been learned since studies of inflammatory gene regulation were initiated more than 25 years ago. In particular, the initial view that the selective activation of an inducible gene is dictated primarily by the combinatorial binding of a specific set of transcription factors has been replaced by models with several additional regulatory layers. From a drug discovery perspective, these additional layers of regulation suggest attractive new targets for the therapeutic modulation of distinct subsets of inducible genes. For example, inhibitors of specific H3K9 demethylases or H3K27 demethylases may selectively suppress inflammatory genes that possess H3K9 or H3K27 methylation as a barrier to activation. Similarly, inhibitors of specific kinases or methylases responsible for RelA S276 phosphorylation or K37 methylation may reduce expression of the specific subset of NF-κB target genes that require phosphorylation or methylation for their activation.
It is important to emphasize that this article focuses on only two regulatory layers that have been suggested by recent studies to be major contributors to selectivity: the use of diverse activation strategies for individual transcription factors like NF-κB and the need to overcome diverse chromatin barriers for the activation of different subsets of inducible genes. However, many other contributors to selective regulation are known to exist. For example, the numerous homodimeric and heterodimeric species that can be assembled from the five NF-κB family members have the potential to regulate distinct subsets of genes through a variety of mechanisms, including differential protein-DNA interactions and differential interactions with co-regulatory molecules (Sen and Smale 2009). Furthermore, elegant studies have revealed that NF-κB induction is subject to strict kinetic control that varies from stimulus to stimulus, raising the possibility that the duration of NF-κB induction may play an important role in defining the precise subset of genes activated in response to a stimulus (Hoffmann and Baltimore 2006). Selective regulation can also be dictated by the sequences of NF-κB recognition motifs in the promoters of inducible genes, which may influence the conformation of the DNA-bound NF-κB dimer and modulate its interaction with co-regulatory proteins (Lefstin and Yamamoto 1998; Leung et al. 2004).
In addition to the diverse mechanisms that facilitate selective gene activation by NF-κB and other transcription factors, numerous strategies have evolved for the attenuation of inflammatory gene transcription. Attenuation of an inflammatory response can be achieved, for example, by the binding of transcriptional repressors to specific target genes, by the active displacement of NF-κB and other transcription factors from their target genes, and by the differential and tightly regulated export of NF-κB complexes from the nucleus (e.g. Gilchrist et al. 2006; Natoli and Chiocca 2008; Sen and Smale 2009). A better understanding of these negative regulatory strategies may reveal additional targets for therapeutic intervention.
It is appropriate to conclude by considering the likely impact of these diverse contributors to selective gene transcription on the long-term goal of defining complete gene regulation networks in response to an inflammatory stimulus. More specifically, a major goal is to connect each transcription factor that is directly activated by a stimulus first to its primary response target genes, and then to secondary target genes activated by an increasingly complex mixture of transcription factors that become expressed and activated as the response continues. Considerable progress has been reported toward the elucidation of inflammatory gene regulation networks through detailed transcriptome analyses, often coupled to bioinformatic analyses of transcription factor recognition motifs in the promoters of inducible genes and genome-wide ChIP analyses of transcription factor binding sites (Amit et al. 2009; Litvak et al. 2009, and references therein). These studies have already provided meaningful new insights. However, the difficulty of using computational and genomics approaches to rigorously define gene regulation networks is greatly increased by evidence that target gene activation is dictated, not only by a specific set of transcription factors, but also by the need to remove diverse chromatin barriers and the need for signal-dependent activation and association of diverse co-regulatory molecules. Because of this diversity, it will likely be necessary to incorporate chromatin properties of target genes into computational strategies for defining gene regulation networks. It may also be necessary to consider co-regulatory requirements for transcription factor function at each target gene, as well as the signal transduction pathways needed to activate each co-regulatory function. The successful integration of emerging mechanistic insights with bioinformatic and genomics strategies should move the field closer to the goal of elucidating definitive inflammatory gene regulation networks.
Acknowledgments
I would like to thank Sankar Ghosh, Alexander Hoffmann, Ruslan Medzhitov, Gioacchino Natoli, Simona Saccani, and Peter Tontonoz for helpful discussions and critical comments on this article. Work in our laboratory on gene regulation in response to inflammatory stimuli is funded by NIH grants 5R01AI073868, 5R01GM086372, and 5R01CA127279.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Amir-Zilberstein L, Ainbinder E, Toube L, Yamaguchi Y, Handa H, Dikstein R. Differential regulation of NF-kappaB by elongation factors is determined by core promoter type. Mol Cell Biol. 2007;27:5246–5259. doi: 10.1128/MCB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, Schubert LA, Birditt B, Shay T, Goren A, Zhang X, Smith Z, Deering R, McDonald RC, Cabili M, Bernstein BE, Rinn JL, Meissner A, Root DE, Hacohen N, Regev A. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E, Thanos D. Virus infection induces NF-κB-dependent interchromosomal associations mediating monoallelic IFN-b gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- Arnosti DN, Kulkarni MM. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem. 2005;94:890–898. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- Beutler BA. TLRs and innate immunity. Blood. 2009;113:1399–1407. doi: 10.1182/blood-2008-07-019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B, Cerami A. Cachectin and tumour necrosis factor as two sides of the same biological coin. Nature. 1986;320:584–588. doi: 10.1038/320584a0. [DOI] [PubMed] [Google Scholar]
- Bohmann D, Bos TJ, Admon A, Nishimura T, Vogt PK, Tjian R. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science. 1987;238:1386–1392. doi: 10.1126/science.2825349. [DOI] [PubMed] [Google Scholar]
- Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, Natoli G. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kappaB-dependent gene activity. EMBO J. 2006;25:798–810. doi: 10.1038/sj.emboj.7600977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd KE, Wells J, Gutman J, Bartley SM, Farnham PJ. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey MF, Peterson CL, Smale ST. Transcriptional regulation in eukaryotes: Concepts, strategies, and techniques. Cold Spring Harbor Laboratory Press; NY: 2009. [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Collart MA, Belin D, Vassalli JD, de Kossodo S, Vassalli P. Gamma interferon enhances macrophage transcription of the tumor necrosis factor/cachectin, interleukin 1, and urokinase genes, which are controlled by short-lived repressors. J Exp Med. 1986;164:2112–2118. doi: 10.1084/jem.164.6.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- de Laat W, Klous P, Kooren J, Noordermeer D, Palstra RJ, Simonis M, Splinter E, Grosveld F. Three-dimensional organization of gene expression in erythroid cells. Curr Top Dev Biol. 2008;82:117–139. doi: 10.1016/S0070-2153(07)00005-1. [DOI] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, Bucci G, Caganova M, Notarbartolo S, Casola S, Testa G, Sung WK, Wei CL, Natoli G. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009 doi: 10.1038/emboj.2009.271. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Jimi E, Zhong H, Hayden MS, Ghosh S. Epigenetic regulation of NF-kB dependent gene expression. Genes Dev. 2008;22:1159–1173. doi: 10.1101/gad.1657408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S, Vaidya S, O’Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- Dynan WS, Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Ea CK, Baltimore D. Regulation of NF-κB activity through lysine monomethylation of p65. Proc Natl Acad Sci. 2009;106:18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke DR, Ng SY, Shastry BS, Roeder RG. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980;19:717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- Foster SL, Medzhitov R. Gene-specific control of the TLR-induced inflammatory response. Clin Immunol. 2009;130:7–15. doi: 10.1016/j.clim.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev GP. On the structural organization of operon and the regulation of RNA synthesis in animal cells. J Theor Biol. 1969;25:473–490. doi: 10.1016/s0022-5193(69)80034-2. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Gierer A. Molecular models and combinatorial principles in cell differentiation and morphogenesis. Cold Spr Harb Symp Quant Biol. 1973;38:951–961. doi: 10.1101/sqb.1974.038.01.097. [DOI] [PubMed] [Google Scholar]
- Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, Kennedy K, Hai T, Bolouri H, Aderem A. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Baltimore D. Circuitry of NF-kappaB signaling. Immunol Reviews. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe. 2008;3:352–363. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Annu Rev Immunol. 2009;27:313–338. doi: 10.1146/annurev.immunol.021908.132657. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002;115(Pt 4):689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- Karin M, Lawrence T, Nizet V. Innate immunity gone awry: Linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Kayama H, Ramirez-Carrozzi VR, Yamamoto M, Mizutani T, Kuwata H, Iba H, Matsumoto M, Honda K, Smale ST, Takeda K. Class-specific regulation of pro-inflammatory genes by MyD88 pathways and IkappaBzeta. J Biol Chem. 2008;283:12468–12477. doi: 10.1074/jbc.M709965200. [DOI] [PubMed] [Google Scholar]
- Kovesdi I, Reichel R, Nevins JR. Identification of a cellular transcription factor involved in E1A trans-activation. Cell. 1986;45:219–228. doi: 10.1016/0092-8674(86)90386-7. [DOI] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Lai D, Wan M, Wu J, Preston-Hurlburt P, Kushwah R, Grundstrom T, Imbalzano AN, Chi T. Induction of TLR4-target genes entails calcium/calmodulin-dependent regulation of chromatin remodeling. Proc Natl Acad Sci. 2009;106:1169–1174. doi: 10.1073/pnas.0811274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping gene expression in activated and resting primary macrophages by IL-10. J Immunol. 2002;169:2253–2263. doi: 10.4049/jimmunol.169.5.2253. [DOI] [PubMed] [Google Scholar]
- Lau LF, Nathans D. Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells: coordinate regulation with c-fos or c-myc. Proc Natl Acad Sci USA. 1987;84:1182–1186. doi: 10.1073/pnas.84.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Lee W, Haslinger A, Karin M, Tjian R. Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature. 1987;325:368–372. doi: 10.1038/325368a0. [DOI] [PubMed] [Google Scholar]
- Lefevre P, Witham J, Lacroix CE, Cockerill PN, Bonifer C. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol Cell. 2008;32:129–139. doi: 10.1016/j.molcel.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- Leung TH, Hoffmann A, Baltimore D. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Li Y, Reddy MA, Miao F, Shanmugam N, Yee JK, Hawkins D, Ren B, Natarajan R. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem. 2008;283:26771–26781. doi: 10.1074/jbc.M802800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford JR, Smith GT, Chi Y, Deshaies RJ. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy KA, Lampano AE, Nykter M, Shmulevich I, Aderem A. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol. 2009;10:437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Thanos D. Modifying gene expression programs by altering core promoter chromatin architecture. Cell. 2002;110:261–271. doi: 10.1016/s0092-8674(02)00822-x. [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- Milanesi G, Brody EN, Grau O, Geiduschek EP. Transcriptions of the bacteriophage T4 template in vitro: separation of “delayed early” from “immediate early” transcription. Proc Natl Acad Sci USA. 1970;66:181–188. doi: 10.1073/pnas.66.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- Motoyama M, Yamazaki S, Eto-Kimura A, Takeshige K, Muta T. Positive and negative regulation of nuclear factor-kappaB-mediated transcription by IkappaB-zeta, an inducible nuclear protein. J Biol Chem. 2005;280:7444–7451. doi: 10.1074/jbc.M412738200. [DOI] [PubMed] [Google Scholar]
- Munshi N, Agalioti T, Lomvardas S, Merika M, Chen G, Thanos D. Coordination of a transcriptional switch by HMBI(Y) acetylation. Science. 2001;293:1133–1136. doi: 10.1126/science.293.5532.1133. [DOI] [PubMed] [Google Scholar]
- Muratani M, Kung C, Shokat KM, Tansey WP. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell. 2005;120:887–899. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Nathan C. Points of control in inflammation. Nature. 2002;420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- Natoli G, Chiocca S. Nuclear ubiquitin ligases, NF-kappaB degradation, and the control of inflammation. Sci Signal. 2008;1:pe1. doi: 10.1126/stke.11pe1. [DOI] [PubMed] [Google Scholar]
- Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D, Maniatis T, Harrison SC. Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-β enhancer. EMBO J. 2004;23:4384–4393. doi: 10.1038/sj.emboj.7600453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-β enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- Prywes R, Roeder RG. Inducible binding of a factor to the c-fos enhancer. Cell. 1986;47:777–784. doi: 10.1016/0092-8674(86)90520-9. [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauscher FJ, Cohen DR, Curran T, Bos TJ, Vogt PK, Bohmann D, Tjian R, Franza BR. Fos-associated protein p39 is the product of the jun proto-oncogene. Science. 1988;240:1010–1016. doi: 10.1126/science.3130660. [DOI] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G. Two waves of nuclear factor kappaB recruitment to target promoters. J Exp Med. 2001;193:1351–1359. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Natoli G. Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev. 2002;16:2219–2224. doi: 10.1101/gad.232502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariban E, Imamura K, Luebbers R, Kufe D. Transcriptional and posttranscriptional regulation of tumor necrosis factor gene expression in human monocytes. J Clin Invest. 1988;81:1506–1510. doi: 10.1172/JCI113482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savinova OV, Hoffmann A, Ghosh G. The Nfkb1 and Nfkb2 proteins p105 and p100 function as the core of high-molecular-weight heterogeneous complexes. Mol Cell. 2009;34:591–602. doi: 10.1016/j.molcel.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R, Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappa B by a posttranslational mechanism. Cell. 1986;47:921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- Sen R, Smale ST. Selectivity of the NF-kappaB response. Cold Spr Harb Perspect Biol. 2009 doi: 10.1101/cshperspect.a000257. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JP, Utz PJ, Durand DB, Toole JJ, Emmel EA, Crabtree GR. Identification of a putative regulator of early T cell activation genes. Science. 1988;241:202–205. doi: 10.1126/science.3260404. [DOI] [PubMed] [Google Scholar]
- Siebenlist U, Durand DB, Bressler P, Holbrook NJ, Norris CA, Kamoun M, Kant JA, Crabtree GR. Promoter region of interleukin-2 gene undergoes chromatin structure changes and confers inducibility on chloramphenicol acetyltransferase gene during activation of T cells. Mol Cell Biol. 1986;6:3042–3049. doi: 10.1128/mcb.6.9.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46:567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- van Essen D, Engist B, Natoli G, Saccani S. Two modes of transcriptional activation at native promoters by NF-kappaB p65. PLoS Biol. 2009;7:e73. doi: 10.1371/journal.pbio.1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang X, Hussain S, Zheng Y, Sanjabi S, Ouaaz F, Beg AA. Distinct roles of different NF-kappa B subunits in regulating inflammatory and T cell stimulatory gene expression in dendritic cells. J Immunol. 2007;178:6777–6788. doi: 10.4049/jimmunol.178.11.6777. [DOI] [PubMed] [Google Scholar]
- Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Plevy SE, Smale ST. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11:665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Groudine M. Chromosomal subunits in active genes and an altered conformation. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wu C, Bingham PM, Livak KJ, Holmgren R, Elgin SC. The chromatin structure of specific genes I: Evidence for higher order domains of defined DNA sequence. Cell. 1979;16:797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. Transcriptional competence and the active marking of tissue-specific enhancers in embryonic and induced pluripotent stem cells. Genes Dev. 2009 doi: 10.1101/gad.1861209. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto KR, Alberts BM. Steroid receptors: elements for modulation of eukaryotic transcription. Annu Rev Biochem. 1976;45:721–746. doi: 10.1146/annurev.bi.45.070176.003445. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Yamazaki S, Uematsu S, Sato S, Hemmi H, Hoshino K, Kaisho T, Kuwata H, Takeuchi O, Takeshige K, Saitoh T, Yamaoka S, Yamamoto N, Yamamoto S, Muta T, Takeda K, Akira S. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature. 2004;430:218–222. doi: 10.1038/nature02738. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Takeda K. Role of nuclear IkappaB proteins in the regulation of host immune responses. J Infect Chemother. 2008;14:265–269. doi: 10.1007/s10156-008-0619-y. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Watts J, Xu J, Wandzioch E, Smale ST, Sekiya T. Pioneer factors, genetic competence, and inductive signaling: programming liver and pancreas progenitors from the endoderm. Cold Spr Harb Symp Quant Biol. 2008;73:119–126. doi: 10.1101/sqb.2008.73.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the co-activator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- Zhong H, May MJ, Jimi E, Ghosh S. Phosphorylation of nuclear NF-κB governs its association with either HDAC-1 or CBP/p300: a mechanism for regulating the transcriptional activity of NF-κB. Mol Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- Zinn K, DiMaio D, Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983;34:865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]