Comprehensive mapping of long range interactions reveals folding principles of the human genome (original) (raw)
. Author manuscript; available in PMC: 2010 Apr 22.
Published in final edited form as: Science. 2009 Oct 9;326(5950):289–293. doi: 10.1126/science.1181369
Abstract
We describe Hi-C, a method that probes the three-dimensional architecture of whole genomes by coupling proximity-based ligation with massively parallel sequencing. We constructed spatial proximity maps of the human genome with Hi-C at a resolution of 1Mb. These maps confirm the presence of chromosome territories and the spatial proximity of small, gene rich chromosomes. We identified an additional level of genome organization that is characterized by the spatial segregation of open and closed chromatin to form two genome-wide compartments. At the megabase scale, the chromatin conformation is consistent with a fractal globule, a knot-free conformation that enables maximally dense packing while preserving the ability to easily fold and unfold any genomic locus. The fractal globule is distinct from the more commonly used globular equilibrium model. Our results demonstrate the power of Hi-C to map the dynamic conformations of whole genomes.
The three-dimensional conformation of chromosomes is involved in compartmentalizing the nucleus and bringing widely separated functional elements into close spatial proximity (1-5). Understanding how chromosomes fold can provide insight into the complex relationships between chromatin structure, gene activity, and the functional state of the cell. Yet beyond the scale of nucleosomes, little is known about chromatin organization.
Long-range interactions between specific pairs of loci can be evaluated with Chromosome Conformation Capture (3C), using spatially constrained ligation followed by locus-specific PCR (6). Adaptations of 3C have extended the process with the use of inverse PCR (4C) (7, 8) or multiplexed ligation-mediated amplification (5C) (9). Still, these techniques require choosing a set of target loci and do not allow unbiased genome-wide analysis.
Here we report a method named Hi-C that adapts the above approach to enable purification of ligation products followed by massively parallel sequencing. Hi-C allows unbiased identification of chromatin interactions across an entire genome. Briefly: cells are crosslinked with formaldehyde; DNA is digested with a restriction enzyme that leaves a 5′-overhang; the 5′-overhang is filled, including a biotinylated residue; and the resulting blunt-end fragments are ligated under dilute conditions that favor ligation events between the cross-linked DNA fragments. The resulting DNA sample contains ligation products consisting of fragments that were originally in close spatial proximity in the nucleus, marked with biotin at the junction. A Hi-C library is created by shearing the DNA and selecting the biotin-containing fragments with streptavidin beads. The library is then analyzed using massively parallel DNA sequencing, producing a catalog of interacting fragments (Fig. 1A, SOM).
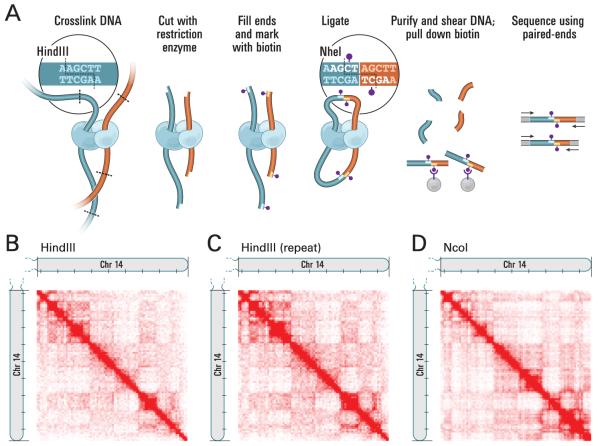
Fig. 1.
Overview of Hi-C. (A) Cells are cross-linked with formaldehyde, resulting in covalent links between spatially adjacent chromatin segments (DNA fragments: dark blue, red; Proteins, which can mediate such interactions, are shown in light blue and cyan). Chromatin is digested with a restriction enzyme (here, HindIII; restriction site: dashed line, see inset) and the resulting sticky ends are filled in with nucleotides, one of which is biotinylated (purple dot). Ligation is performed under extremely dilute conditions to create chimeric molecules; the HindIII site is lost and a NheI site is created (inset). DNA is purified and sheared. Biotinylated junctions are isolated with streptavidin beads and identified by paired-end sequencing. (B) Hi-C produces a genome-wide contact matrix. The submatrix shown here corresponds to intrachromosomal interactions on chromosome 14. Each pixel represents all interactions between a 1Mb locus and another 1Mb locus; intensity corresponds to the total number of reads (0-50). Tick marks appear every 10Mb. (C, D) We compared the original experiment to a biological repeat using the same restriction enzyme (C, range: 0-50 reads) and to results with a different restriction enzyme (D, range: 0- 100 reads, NcoI).
We created a Hi-C library from a karyotypically normal human lymphoblastoid cell line (GM06990) and sequenced it on two lanes of an Illumina Genome Analyzer, generating 8.4 million read pairs that could be uniquely aligned to the human genome reference sequence; of these, 6.7 million corresponded to long-range contacts between segments greater than >20 Kb apart.
We constructed a genome-wide contact matrix M by dividing the genome into 1 Mb regions (‘loci’) and defining the matrix entry mij to be the number of ligation products between locus i and locus j (SOM). This matrix reflects an ensemble average of the interactions present in the original sample of cells; it can be visually represented as a heatmap, with intensity indicating contact frequency (Fig. 1B).
We tested whether Hi-C results were reproducible by repeating the experiment using the same restriction enzyme (HindIII) and using a different one (NcoI). We observed that contact matrices for these new libraries (Fig 1C, D) were extremely similar to the original contact matrix (Pearson’s r=0.990 [HindIII] and r=0.814 [NcoI]; p was negligible [<10−300] in both cases). We therefore combined the three datasets in subsequent analyses.
We first tested whether our data are consistent with known features of genome organization (1) – specifically, chromosome territories (the tendency of distant loci on the same chromosome to be near one another in space) and patterns in sub-nuclear positioning (the tendency of certain chromosome pairs to be near one another).
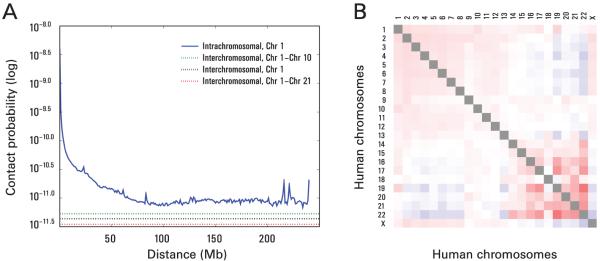
We calculated the average intrachromosomal contact probability, In (s), for pairs of loci separated by a genomic distance s (distance in base pairs along the nucleotide sequence) on chromosome n. In (s) decreases monotonically on every chromosome, suggesting polymer-like behavior in which the three-dimensional distance between loci increases with increasing genomic distance; these findings are in agreement with 3C and fluorescence in situ hybridization (FISH) (6, 10). Even at distances greater than 200 Mb, In (s) is always much greater than the average contact probability between different chromosomes (Fig. 2A). This implies the existence of chromosome territories.
Fig. 2.
The presence and organization of chromosome territories. (A) Probability of contact decreases as a function of genomic distance on chromosome 1, eventually reaching a plateau at ~90M (blue). The level of interchromosomal contact (black dashes) differs for different pairs of chromosomes; loci on chromosome 1 are most likely to interact with loci on chromosome 10 (green dashes) and least likely to interact with loci on chromosome 21 (red dashes). Interchromosomal interactions are depleted relative to intrachromosomal interactions. (B) Observed/expected number of interchromosomal contacts between all pairs of chromosomes. Red indicates enrichment, and blue indicates depletion (up to twofold). Small, gene-rich chromosomes tend to interact more with one another.
Interchromosomal contact probabilities between pairs of chromosomes (Fig. 2B) show that small, gene-rich chromosomes (chromosomes 16, 17, 19, 20, 21, 22) preferentially interact with each other. This is consistent with FISH studies showing that these chromosomes frequently co-localize in the center of the nucleus (11, 12). Interestingly, chromosome 18, which is small but gene-poor, does not interact frequently with the other small chromosomes; this agrees with FISH studies showing that chromosome 18 tends to be located near the nuclear periphery (13).
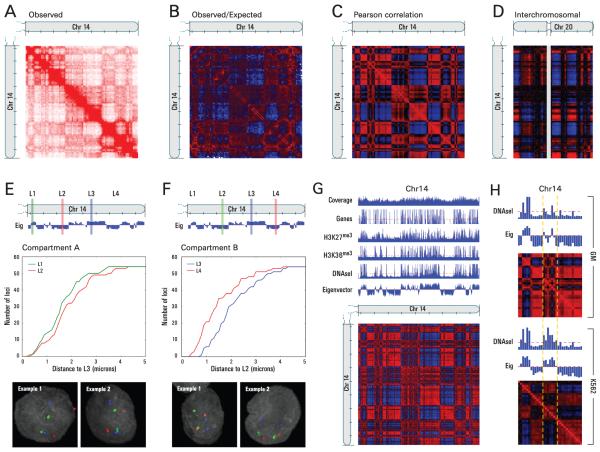
We then zoomed in on individual chromosomes to explore whether there are chromosomal regions that preferentially associate with each other. Because sequence proximity strongly influences contact probability, we defined a normalized contact matrix M* by dividing each entry in the contact matrix by the genome-wide average contact probability for loci at that genomic distance (SOM). The normalized matrix shows many large blocks of enriched and depleted interactions generating a ‘plaid’ pattern (Fig. 3B). If two loci (here 1 Mb regions) are nearby in space, we reasoned that they will share neighbors and have correlated interaction profiles. We therefore defined a correlation matrix C in which cij is the Pearson correlation between the ith row and jth column of M*. This process dramatically sharpened the plaid pattern (Fig. 3C); 71% of the resulting matrix entries represent statistically significant correlations (p ≤ 0.05).
Fig. 3.
The nucleus is segregated into two compartments corresponding to open and closed chromatin. (A) Map of chromosome 14 at a resolution of 1Mb (1 tick mark = 10Mb) exhibits substructure in the form of an intense diagonal and a constellation of large blocks (three experiments combined, range: 0-200 reads). The Observed/expected matrix (B) shows loci with either more (red) or less (blue) interactions than would be expected given their genomic distance (range: 0.2 – 5). Correlation matrix (C) illustrates the correlation (red: 1, blue: −1) between the intrachromosomal interaction profiles of every pair of 1 Mb loci along chromosome 14. The plaid pattern indicates the presence of two compartments within the chromosome. (D) Interchromosomal correlation map for chromosome 14 and chromosome 20 (red: 0.25, blue: 0.25). The unalignable region around the centromere of chromosome 20 is indicated in grey. Each compartment on chromosome 14 has a counterpart on chromosome 20 with a very similar genome-wide interaction pattern. (E,F) We designed probes for four loci (L1, L2, L3, and L4) that lie consecutively along Chromosome 14 but alternate between the two compartments (L1, L3 in A; L2, L4 in B). (E) L3 (blue) was consistently closer to L1 (green) than to L2 (red), despite the fact that L2 lies between L1 and L3 in the primary sequence of the genome. This was confirmed visually and by plotting the cumulative distribution. (F) L2 (red) was consistently closer to L4 (green) than to L3 (blue). (G) Correlation map of chromosome 14 at a resolution of 100kb. The principal component (eigenvector) correlates with the distribution of genes and with features of open chromatin. (H) A 31Mb window from the chromosome 14 is shown; the indicated region (yellow dashes) alternates between the open and closed in compartment in GM06990 (top, eigenvector and heatmap), but is predominantly open in K562 (bottom, eigenvector and heatmap). The change in compartmentalization corresponds to a shift in chromatin state (DNAseI).
The plaid pattern suggests that each chromosome can be decomposed into two sets of loci (arbitrarily labeled A and B) such that contacts within each set are enriched and contacts between sets are depleted. We partitioned each chromosome in this way using principal component analysis. For all but two chromosomes, the first principal component (PC) clearly corresponded to the plaid pattern (positive values defining one set, negative values the other) (Fig. S1). For chromosomes 4 and 5, the first PC corresponded to the two chromosome arms, but the second PC corresponded to the plaid pattern. The entries of the PC vector reflected the sharp transitions from compartment to compartment observed within the plaid heatmaps. Moreover, the plaid patterns within each chromosome were consistent across chromosomes: the labels (A and B) could be assigned on each chromosome so that sets on different chromosomes carrying the same label had correlated contact profiles, and those carrying different labels had anticorrelated contact profiles (Fig. 3D). These results imply that the entire genome can be partitioned into two spatial compartments such that greater interaction occurs within each compartment rather than across compartments.
The Hi-C data imply that regions tend be closer in space if they belong to the same compartment (A vs. B) than if they do not. We tested this using 3D-FISH, probing four loci (L1, L2, L3, and L4) on chromosome 14 that alternate between the two compartments (L1 and L3 in compartment A; L2 and L4 in compartment B) (Fig. 3E, F). 3D-FISH showed that L3 tends to be closer to L1 than to L2, despite the fact that L2 lies between L1 and L3 in the linear genome sequence (Fig. 3E). Similarly, we found that L2 is closer to L4 than to L3 (Fig. 3F). Comparable results were obtained for four consecutive loci on chromosome 22 (Fig. S2A, B). Taken together, these observations confirm the spatial compartmentalization of the genome inferred from Hi-C. More generally, a strong correlation was observed between the number of Hi-C reads mij and the three-dimensional distance between locus i and locus j as measured by FISH (Spearman’s rho=0.874, p=0.0002 [Fig. S3]), suggesting that Hi-C read count may serve as a proxy for distance.
Upon close examination of the Hi-C data, we noted that pairs of loci in compartment B showed a consistently higher interaction frequency at a given genomic distance than pairs of loci in compartment A (Fig. S4). This suggests that compartment B is more densely packed (14). The FISH data are consistent with this observation; loci in compartment B exhibited a stronger tendency for close spatial localization.
To explore whether the two spatial compartments correspond to known features of the genome, we compared the compartments identified in our 1 Mb correlation maps to known genetic and epigenetic features. Compartment A correlates strongly with the presence of genes (Spearman’s rho=0.431, p<10−137), higher expression (via genome-wide mRNA expression, Spearman’s rho=0.476, p<10−145 [Fig. S5]), and accessible chromatin (as measured by DNAseI sensitivity, Spearman’s rho=0.651, p negligible) (15, 16). Compartment A also shows enrichment for both activating (H3K36 trimethylation, Spearman’s rho=0.601, p<10−296) and repressive (H3K27 trimethylation, Spearman’s rho=0.282, p<10−56) chromatin marks (17). We repeated the above analysis at a resolution of 100 kb (Fig. 3G) and saw that while the correlation of compartment A with all other genomic and epigenetic features remained strong (Spearman’s rho>0.4, p negligible), the correlation with the sole repressive mark, H3K27 trimethylation, was dramatically attenuated (Spearman’s rho=0.046, p<10−15). On the basis of these results we concluded that compartment A is more closely associated with open, accessible, actively transcribed chromatin.
We repeated our experiment with K562 cells, an erythroleukemia cell line with an aberrant karyotype (18). We again observed two compartments; these were similar in composition to those observed in GM06990 cells (Pearson’s r=0.732, p negligible [Fig. S6]) and showed strong correlation with open and closed chromatin states as indicated by DNAseI sensitivity (Spearman’s rho=0.455, p<10−154).
The compartment patterns in K562 and GM are similar, but there are many loci in the open compartment in one cell type and the closed compartment in the other (Fig. 3H). Examining these discordant loci on karyotypically normal chromosomes in K562 (18), we observed a strong correlation between the compartment pattern in a cell type and chromatin accessibility in that same cell type (GM06990, Spearman’s rho=0.384, p=0.012; K562, Spearman’s rho=0.366, p=0.017). Thus, even in a highly rearranged genome, spatial compartmentalization correlates strongly with chromatin state.
Our results demonstrate that open and closed chromatin domains throughout the genome occupy different spatial compartments in the nucleus. These findings expand upon studies of individual loci that have observed particular instances of such interactions; both between distantly located active genes, and between distantly located inactive genes (8, 19-23).
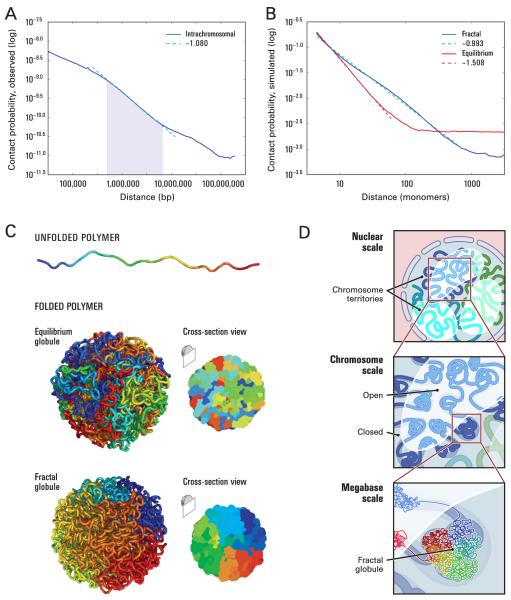
Finally, we sought to explore the internal structure of the open and closed chromatin domains that correspond to the compartments seen in the plaid correlation maps. We closely examined the average behavior of intrachromosomal contact probability as a function of genomic distance, calculating the genome-wide distribution I(s). When plotted on log log axes, I(s) exhibits a prominent power law scaling between ~500 kb and ~7 Mb, where contact probability scales as s−1 (Fig. 4A). This range corresponds to the known size of open and closed chromatin domains.
Fig. 4.
The local packing of chromatin is consistent with the behavior of a fractal globule. (A) Contact probability as a function of genomic distance, averaged across the genome (blue) shows a power law scaling between 500kb and 7Mb (shaded region) with a slope of −1.08 (fit shown in cyan). (B) Simulation results for contact probability as a function of distance (1 monomer~6 nucleosomes~1200 bp, SOM) for equilibrium (red) and fractal (blue) globules. The slope for a fractal globule is very nearly −1 (cyan), confirming our prediction (SOM). The slope for an equilibrium globule is −3/2, matching prior theoretical expectations. The slope for the fractal globule closely resembles the slope we observed in the genome. (C) Top: An unfolded polymer chain, 4000 monomers (4.8 Mb) long. Coloration corresponds to distance from one endpoint, ranging from blue to cyan, green, yellow, orange, and red. Middle: An equilibrium globule. The structure is highly entangled; loci that are nearby along the contour (similar color) need not be nearby in 3D. Bottom: A fractal globule. Nearby loci along the contour tend to be nearby in 3D, leading to monochromatic blocks both on the surface and in cross-section. The structure lacks knots. (D) Genome architecture at three scales. Top: Two compartments, corresponding to open and closed chromatin, spatially partition the genome. Chromosomes (blue, cyan, green) occupy distinct territories. Middle: Individual chromosomes weave back-and-forth between the open and closed chromatin compartments. Bottom: At the scale of single megabases, the chromosome consists of a series of fractal globules.
Power-law dependencies can arise from polymer-like behavior (24). Various authors have proposed that chromosomal regions can be modeled as an ‘equilibrium globule’ – a compact, densely knotted configuration originally used to describe a polymer in a poor solvent at equilibrium (25, 26). (Historically, this specific model has often been referred to simply as a ‘globule’; some authors have used the term ‘equilibrium globule’ to distinguish it from other globular states [See below].) Grosberg et al. proposed an alternative model, theorizing that polymers, including interphase DNA, can self-organize into a long-lived, non-equilibrium conformation that they described as a ‘fractal globule’ (27, 28). This highly compact state is formed by an unentangled polymer when it crumples into a series of small globules in a ‘beads-on-a-string’ configuration. These beads serve as monomers in subsequent rounds of spontaneous crumpling until only a single globule-of-globules-of-globules remains. The resulting structure resembles a Peano curve, a continuous fractal trajectory that densely fills three-dimensional space without crossing itself (29). Fractal globules are an attractive structure for chromatin segments because they lack knots (30) and would facilitate unfolding and refolding, e.g. during gene activation, gene repression, or the cell cycle. In a fractal globule, contiguous regions of the genome tend to form spatial sectors whose size corresponds to the length of the original region (Fig. 4C). In contrast, an equilibrium globule is highly knotted and lacks such sectors; instead, linear and spatial positions are largely decorrelated after at most a few megabases (Fig. 4C). The fractal globule has not previously been observed (28).
The ‘equilibrium globule’ and ‘fractal globule’ models make very different predictions concerning the scaling of contact probability with genomic distance s. The equilibrium globule model predicts that contact probability will scale as s−3/2, which we do not observe in our data. We analytically derived the contact probability for a fractal globule and found that it decays as s−1 (SOM); this corresponds closely with the prominent scaling we observed (−1.08).
The equilibrium and fractal globule models also make differing predictions about the three-dimensional distance between pairs of loci (s1/2 for an equilibrium globule, s1/3 for a fractal globule). While three-dimensional distance is not directly measured by Hi-C, we note that a recent paper using 3D-FISH reported an s1/3 scaling for genomic distances between 500kb and 2Mb (26).
We used Monte Carlo simulations to construct ensembles of fractal globules and equilibrium globules (500 each). The properties of the ensembles matched the theoretically-derived scalings for contact probability (fractal: s−1, equilibrium: s−3/2) and three dimensional distance (fractal: s1/3, equilibrium: s1/2). These simulations also illustrated the lack of entanglements [measured using the knot-theoretic Alexander polynomial (31)] and the formation of spatial sectors within a fractal globule (Fig. 4B).
We conclude that at the scale of several megabases, the data are consistent with a fractal globule model for chromatin organization. Of course, we cannot rule out the possibility that other forms of regular organization might lead to similar findings.
We focused here on interactions at relatively large scales (37). Hi-C can also be used to construct comprehensive, genome-wide interaction maps at finer scales by increasing the number of reads. This should enable the mapping of specific long-range interactions between enhancers, silencers, and insulators (32-34). To increase the resolution by a factor of n, one must increase the number of reads by a factor of n2. As the cost of sequencing falls, detecting finer interactions should become increasingly feasible. In addition, one can focus on subsets of the genome by using chromatin immunoprecipitation or hybrid capture (35, 36).
Supplementary Material
Supplementary Figures
Supplementary text
Acknowledgments
Supported by the Fannie and John Hertz Foundation Graduate Fellowship, the National Defense Science and Engineering Graduate Fellowship, the National Science Foundation Graduate Fellowship, the National Space Biomedical Research Institute, and Grant Number T32 HG002295 from the National Human Genome Research Institute (E.L.), a fellowship from the American Society of Hematology (T.R), Award Number R01HL06544 from the National Heart, Lung, And Blood Institute and R37DK44746 from the National Institute of Diabetes and Digestive and Kidney Diseases (M.G.), NIH grant U54HG004592 (J.S), i2b2 (Informatics for Integrating Biology & the Bedside) and the NIH-supported Center for Biomedical Computing at Brigham and Women’s Hospital (L.M.), Grant Number HG003143 from the National Human Genome Research Institute and a Keck Foundation distinguished young scholar award (J.D.). We thank J. Goldy, K. Lee, S. Vong, and M. Weaver for assistance with DNaseI experiments, A. Kosmrlj for discussions and 16 code; A .P. Aiden, X. R. Bao, M. Brenner, D. Galas, W. Gosper, A. Jaffer, A. Melnikov, A. Miele, G. Giannoukos, C. Nusbaum, A.J.M. Walhout, L. Wood, and K. Zeldovich for discussions; and L. Gaffney and B. Wong for help with visualization. We also acknowledge the ENCODE chromatin group at Broad Institute and Massachusetts General Hospital.
Footnotes
REFERENCES
- 1.Cremer T, Cremer C. Nat Rev Genet. 2001 Apr;2:292. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 2.Sexton T, Schober H, Fraser P, Gasser SM. Nat Struct Mol Biol. 2007 Nov 5;14:1049. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 3.Dekker J. Science. 2008 Mar 28;319:1793. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misteli T. Cell. 2007 Feb 23;128:787. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 5.Kosak ST, Groudine M. Genes Dev. 2004 Jun 15;18:1371. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- 6.Dekker J, Rippe K, Dekker M, Kleckner N. Science. 2002 Feb 15;295:1306. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Z, et al. Nat Genet. 2006 Nov;38:1341. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 8.Simonis M, et al. Nat Genet. 2006 Nov;38:1348. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 9.Dostie J, et al. Genome Res. 2006 Oct;16:1299. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokota H, van den Engh G, Hearst JE, Sachs RK, Trask BJ. J Cell Biol. 1995 Sep;130:1239. doi: 10.1083/jcb.130.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyle S, et al. Hum Mol Genet. 2001 Feb 1;10:211. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- 12.Tanabe H, Habermann FA, Solovei I, Cremer M, Cremer T. Mutat Res. 2002 Jul 25;504:37. doi: 10.1016/s0027-5107(02)00077-5. [DOI] [PubMed] [Google Scholar]
- 13.Croft JA, et al. J Cell Biol. 1999 Jun 14;145:1119. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekker J. J Biol Chem. 2008 Dec 12;283:34532. doi: 10.1074/jbc.M806479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabo PJ, et al. Nat Methods. 2006 Jul;3:511. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 16.Hesselberth JR, et al. Nat Methods. 2009 Apr;6:283. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikkelsen TS, et al. Nature. 2007 Aug 2;448:553. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naumann S, Reutzel D, Speicher M, Decker HJ. Leuk Res. 2001 Apr;25:313. doi: 10.1016/s0145-2126(00)00125-9. [DOI] [PubMed] [Google Scholar]
- 19.Osborne CS, et al. Nat Genet. 2004 Oct;36:1065. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 20.Brown JM, et al. J Cell Biol. 2008 Sep 22;182:1083. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dernburg AF, et al. Cell. 1996 May 31;85:745. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- 22.Shopland LS, et al. J Cell Biol. 2006 Jul 3;174:27. doi: 10.1083/jcb.200603083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraser P, Bickmore W. Nature. 2007 May 24;447:413. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 24.Gennes P. G. d. Scaling concepts in polymer physics. Cornell University Press; Ithaca, N.Y.: 1979. p. 324. [Google Scholar]
- 25.Münkel C, Langowski J. Physical Review E. 1998;57:5888. [Google Scholar]
- 26.Mateos-Langerak J, et al. Proc Natl Acad Sci U S A. 2009 Mar 10;106:3812. doi: 10.1073/pnas.0809501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grosberg AY, Nechaev SK, Shakhnovich EI. J. Phys. France. 1988;49:2095. [Google Scholar]
- 28.Grosberg A, Rabin Y, Havlin S, Neer A. EPL (Europhysics Letters) 1993;373 [Google Scholar]
- 29.Mandelbrot BB. The fractal geometry of nature. W.H. Freeman; New York: 1983. p. 468. Updated and augm. [Google Scholar]
- 30.Vasilyev OA, Nechaev SK. Theoretical and Mathematical Physics. 2003;134:142. [Google Scholar]
- 31.Kolesov G, Virnau P, Kardar M, Mirny LA. Nucleic Acids Res. 2007 Jul;35:W425. doi: 10.1093/nar/gkm312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blackwood EM, Kadonaga JT. Science. 1998 Jul 3;281:60. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 33.Bell AC, West AG, Felsenfeld G. Science. 2001 Jan 19;291:447. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 34.Phillips JE, Corces VG. Cell. 2009 Jun 26;137:1194. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuo MH, Allis CD. Methods. 1999 Nov;19:425. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 36.Gnirke A, et al. Nat Biotechnol. 2009 Feb;27:182. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hi-C Sequence data has been deposited in the Genbank database (http://www.ncbi.nlm.nih.gov/Genbank/), Accession no. ###. Expression results are available at GEO (http://www.ncbi.nlm.nih.gov/geo/), Accession no. ###. ChIP-Seq data is available at the UCSC Browser (http://genome.ucsc.edu/). Additional visualizations are available at http://hic.umassmed.edu.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary text