Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation (original) (raw)
. Author manuscript; available in PMC: 2010 Jun 1.
Published in final edited form as: Nature. 2009 Apr 30;458(7242):1180–1184. doi: 10.1038/nature07774
Abstract
Gene silencing by double-stranded RNA, denoted RNA interference, represents a new paradigm for rational drug design1. However, the transformative therapeutic potential of short interfering RNA (siRNA) has been stymied by a key obstacle—safe delivery to specified target cells in vivo2. Macrophages are particularly attractive targets for RNA interference therapy because they promote pathogenic inflammatory responses in diseases such as rheumatoid arthritis, atherosclerosis, inflammatory bowel disease and diabetes3. Here we report the engineering of β1,3-d-glucan-encapsulated siRNA particles (GeRPs) as efficient oral delivery vehicles that potently silence genes in mouse macrophages in vitro and in vivo. Oral gavage of mice with GeRPs containing as little as 20 µg kg−1 siRNA directed against tumour necrosis factor α (Tnf-α)depleted its messenger RNA in macrophages recovered from the peritoneum, spleen, liver and lung, and lowered serum Tnf-α levels. Screening with GeRPs for inflammation genes revealed that the mitogen-activated protein kinase kinase kinase kinase 4 (Map4k4) is a previously unknown mediator of cytokine expression. Importantly, silencing Map4k4 in macrophages in vivo protected mice from lipopolysaccharide-induced lethality by inhibiting Tnf-α and interleukin-1β production. This technology defines a new strategy for oral delivery of siRNA to attenuate inflammatory responses in human disease.
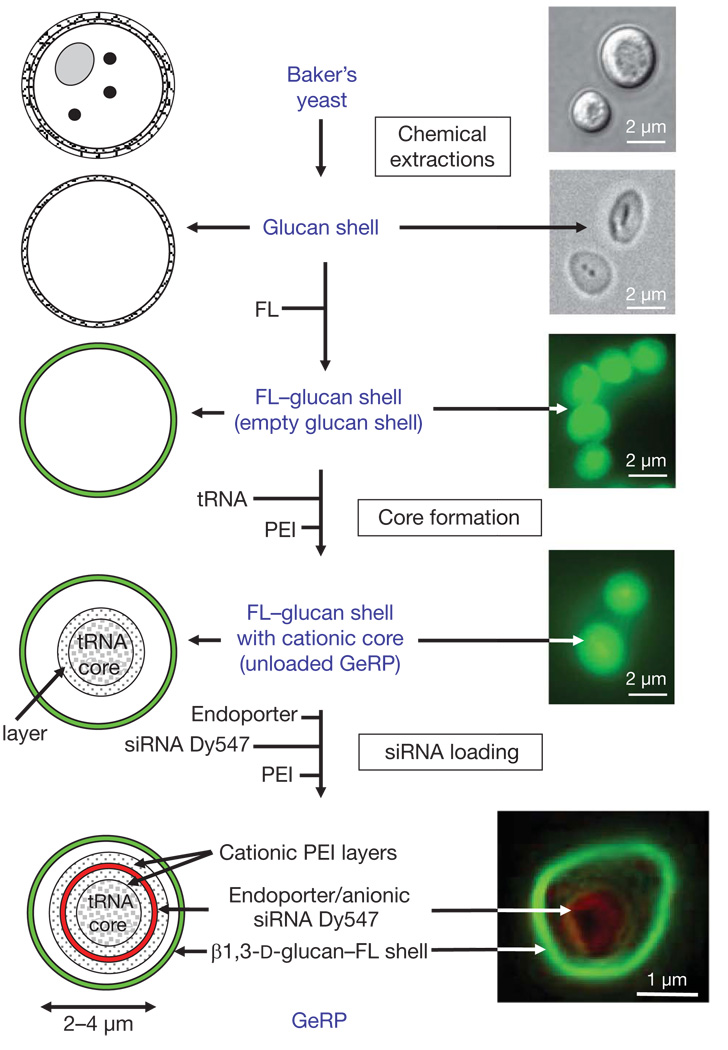
To accomplish oral delivery of siRNA to macrophages in mice, we targeted M cells in intestinal wall Peyer’s patches (Supplementary Fig. 1) to transfer micrometre-sized β1,3-d-glucan particles to the underlying gut-associated lymphatic tissue (GALT)4. Phagocytosis by macrophages and dendritic cells in the GALT occurs by means of the dectin-1 (also known as CLEC7A) receptor and perhaps other β1,3-d-glucan-receptor-mediated pathways5. Evidence suggests that GALT macrophages may traffic away from the gut and infiltrate other reticuloendothelial system tissues, such that over time some of the total body macrophages contain ingested β1,3-d-glucan particles6. We prepared hollow, porous micrometre-sized shells composed primarily of β1,3-d-glucan by treating baker’s yeast with a series of alkaline, acid and solvent extractions to remove cytoplasm and other cell wall polysaccharides7 (Fig. 1). Such empty β1,3-d-glucan shells are about 2–4 µm in diameter, and can be fluorescently labelled for tracking. The anionic siRNA within GeRPs is bound between cationic polyethylenimine (PEI) layers through electrostatic interactions in a pH-dependent manner. On phagocytosis of GeRPs by macrophages, the acidic pH in phagosomes promotes siRNA release through the porous GeRP wall.
Figure 1. Production of fluorescent GeRPs.
β1,3-d-glucan particles were purified from baker’s yeast by a series of alkaline and solvent extractions, hydrolysing outer cell wall and intracellular components and yielding purified, porous 2–4 µm, hollow β1,3-d-glucan particles (diagram of particles, left; procedure, middle; microscopy of particles, right). Empty β1,3-d-glucan particles were then labelled with fluorescein (FL, green). The cores were synthesized by absorbing yeast tRNA, PEI and Dy547-labelled siRNA (red) in a layer-by-layer format. Bottom right: confocal image of a synthesized GeRP.
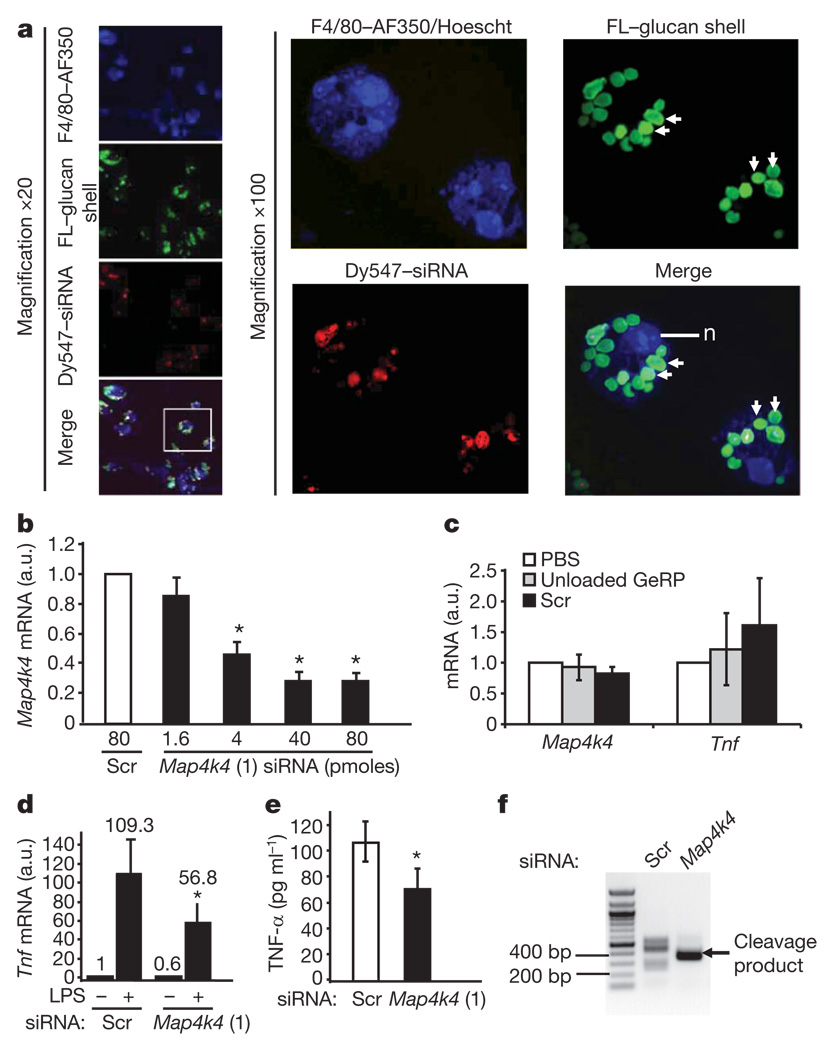
Preliminary experiments using two siRNA oligonucleotides against Tnf-α within GeRPs showed extensive phagocytosis of GeRPs by the primary macrophages and gene silencing (Supplementary Fig. 2c, d). We then screened in peritoneal exudate cell macrophages (PECs) for candidate intracellular signalling proteins that might control Tnf-α expression. One of these was the Map4k4, a germinal centre protein kinase that facilitates Tnf-α signalling itself8–10. β1,3-d-glucan shells derivatized with a green fluorescein probe were loaded with scrambled or_Map4k4_ siRNA (oligo 1 in Supplementary Table 1) coupled to a red Dy547 fluorescent probe and incubated with PECs (10:1 GeRP-to-cell ratio) for 12 h. Confocal microscopy showed that about 90% of the macrophages had internalized at least one fluorescein–GeRP (Fig. 2a, left panels), whereas most cells had internalized multiple fluorescein–GeRPs (Fig. 2a, right panels show two cells at × 100 magnification). Remarkably, a 70–80% knockdown of Map4k4 mRNA was achieved in 106 PECs with as little as 40 pmoles siRNA within GeRPs (Fig. 2b), whereas PBS, unloaded GeRPs or those containing scrambled siRNA had no effect (Fig. 2c).
Figure 2. In vitro treatment of GeRPs containing Map4k4 siRNA silence Map4k4 expression and inhibit LPS-induced Tnf-α production in macrophages.
a, Confocal images of cultured PECs treated with siRNA–GeRPs (red/green). PECs were stained for F4/80 (blue) and nuclei (n) (Hoescht; blue). Arrows indicate GeRPs. b, Map4k4 mRNA expression from PECs treated with increasing Map4k4 siRNA concentrations. a.u., arbitrary units. c, Map4k4 and Tnf mRNA expression from PECs treated with PBS, unloaded GeRPs or scrambled (Scr) GeRPs. d, Tnf mRNA expression from PECs treated with Scr- or _Map4k4_-siRNA-containing GeRPs, ±6 h LPS (1 µg ml−1). e, Tnf-α secretion from PECs treated with Scr or Map4k4 GeRPs. Statistical significance was determined by ANOVA and Tukey post test. *P < 0.05. Results are mean ± s.e.m. f, PCR amplification of 5′ RACE products produces a band (arrow) reflecting the predicted RNAi-mediated mRNA cleavage product in _Map4k4_- but not Scr-siRNA-treated PECs.
Lipopolysaccharide (LPS), a major structural component of the outer membrane of Gram-negative bacteria, activates monocytes and macrophages to produce inflammatory cytokines, such as Tnf-α and interleukin (Il)-1β11. PECs (106) were incubated with 107 GeRPs containing 40 pmoles of scrambled or Map4k4 siRNA for 48 h, and then treated with saline or LPS for an additional 6 h. _Tnf_mRNA levels were decreased by 40% in unstimulated cells treated with _Map4k4_-siRNA-containing GeRPs compared to GeRPs containing scrambled siRNA (Fig. 2d). Map4k4 silencing inhibited the LPS-induced Tnf expression by 50% (Fig. 2d) or more (Supplementary Fig. 2b). Map4k4 silencing in PECs also resulted in an average 30% decrease of LPS-induced Tnf-α protein secreted into the medium (Fig. 2e). GeRPs with scrambled siRNA, unloaded GeRPs (GeRPs containing transfer RNA/PEI cores, but no siRNA) or phosphate buffered saline (PBS) failed to affect Tnf expression (Fig. 2c) or secretion (Supplementary Fig. 3), or expression of interferon response genes (Supplementary Fig. 12).
To determine whether depletion of Map4k4 mRNA in the GeRP-treated macrophages, as shown in Fig. 2b, is mediated by an RNA interference (RNAi)-dependent gene-silencing mechanism, we performed rapid amplification of 5′ complementary DNA ends (5′ RACE) analysis to identify the cleavage sites of Map4k4 mRNA12. After treatment of PECs with GeRPs containing Map4k4 siRNA, but not scrambled siRNA, a unique 5′ RACE product could be detected on agarose gels (Fig. 2f). Sequence analysis of the cloned polymerase chain reaction (PCR) products revealed that 94 out of 100 products were derived from mRNA cleaved at the predicted cut site (ACTA/TGGC; not shown).
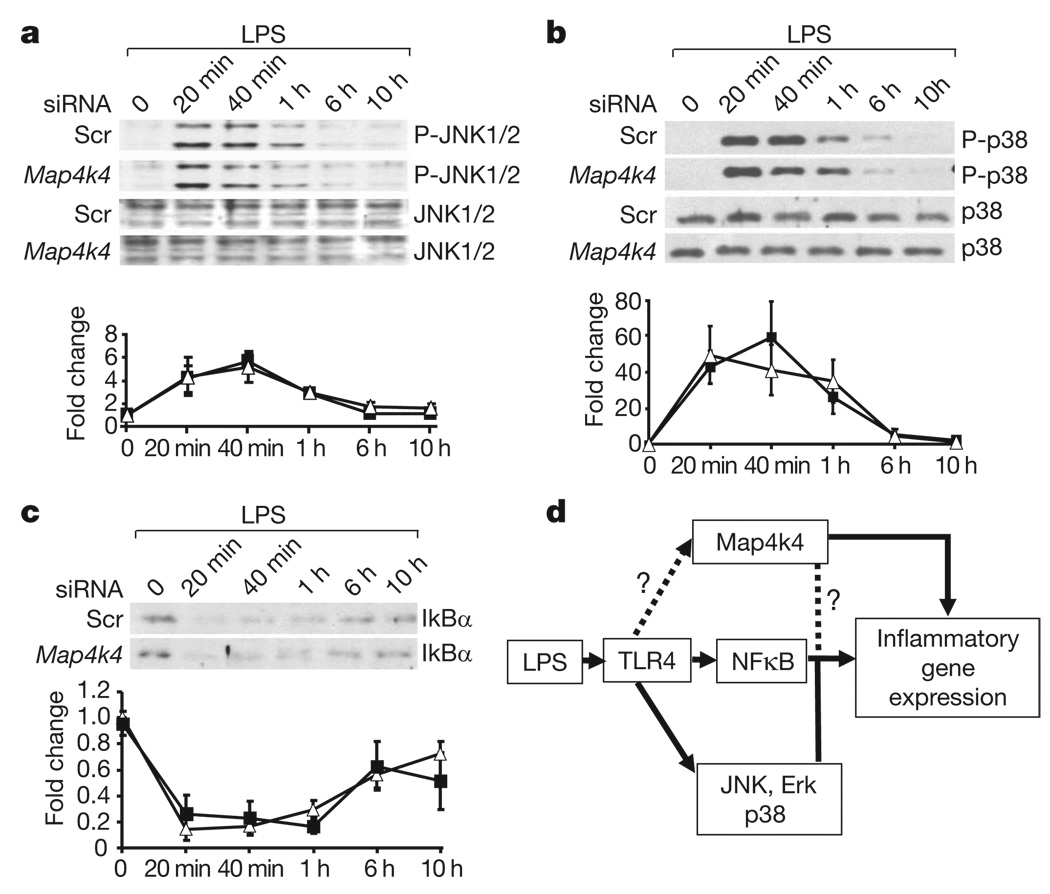
cJun N-terminal kinases 1 and 2 (JNK1 (also known as Mapk8) and JNK2 (Mapk9)), extracellular signal-related kinase 1 (Erk1) and 2 (Erk2) (also known as Mapk3 and Mapk1, respectively), p38 (Mapk14) mitogen-activated protein kinase (MAPK) and nuclear factor κB (NFκB) pathways regulate Tnf-α production in macrophages 13,14. Interestingly, we found that Map4k4 defines a proinflammatory pathway that activates Tnf-α expression independently of the JNK1/2, p38 and NFκB pathways (Fig. 3) or Erk1/2 (not shown). These data demonstrate that Map4k4 is a new target for suppression of Tnf-α expression in LPS-induced macrophage inflammatory responses.
Figure 3. Map4k4 silencing fails to affect LPS activation of JNK, p38 and NFκB signalling pathways.
PECs treated with GeRPs containing scrambled (Scr) or Map4k4 (oligo 1) siRNA, and stimulated with 1 µg ml−1 LPS for the indicated amounts of time were analysed for phospho- (P-) and total JNK1/2 (a) and p38 MAPK (b), and for total IκBα (c). Representative blots are shown from three experiments. Immunoblot signals are expressed in arbitrary units (a.u.) and data represent mean ± s.e.m. (n = 3). Filled squares, Scr siRNA; open triangles, Map4k4 siRNA. d, Schematic diagram of potential Map4k4 signalling to modulate the expression of inflammatory genes such as Tnf and Il1b.
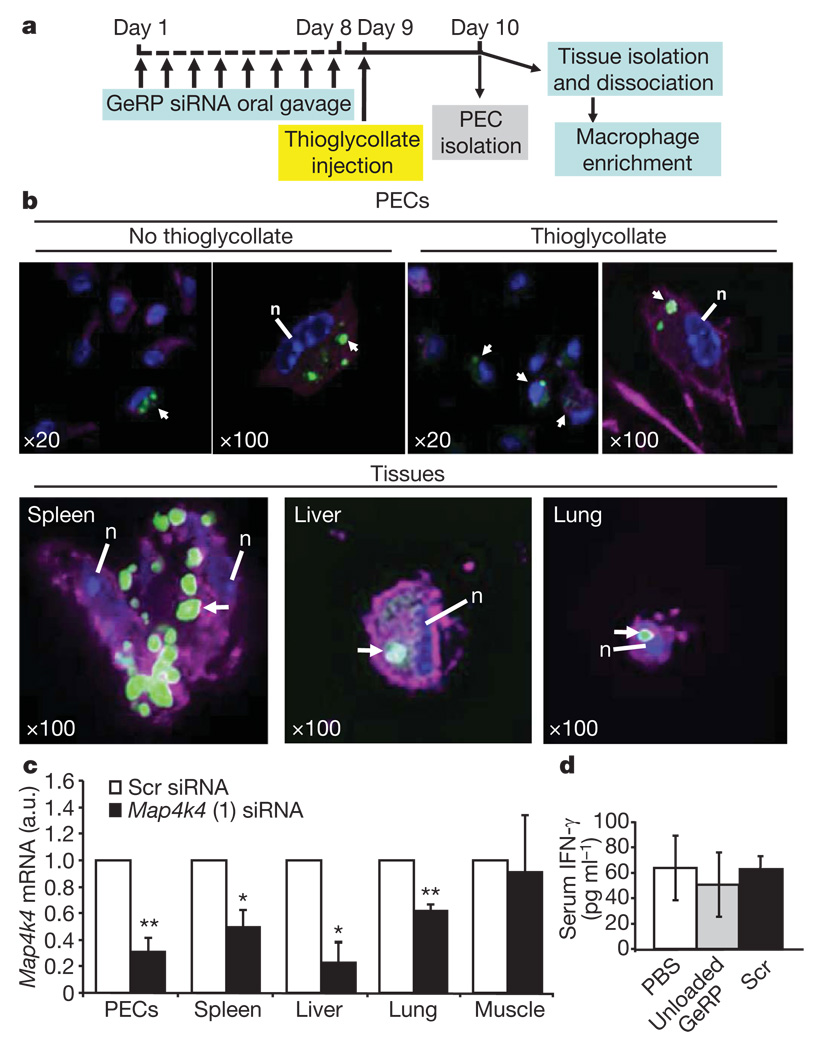
To evaluate GeRPs for oral delivery of siRNA-mediated gene silencing in vivo, mice were given Dy547-conjugated scrambled or Map4k4 siRNA (20 µg kg−1) contained within fluorescein–GeRPs (4 × 109 fluorescein–GeRPs per kg or about 108 GeRPs in 200 µl per mouse) by oral gavage once daily for 8 consecutive days (Fig. 4a). Staining the PECs recovered from these mice with the macrophage-specific antibody F4/80 (Emr1)–AF633 (magenta) revealed that the fluorescein–GeRPs containing Dy547–siRNA were taken up by GALT macrophages, which then migrate out into peripheral tissues (Fig. 4b). Co-localization of AlexaFluor633, fluorescein–GeRPs and Dy547 fluorescent signals in adherent macrophages was readily observed. Notably, Map4k4 mRNA expression was inhibited by 70% in PECs isolated from mice orally gavaged with _Map4k4_-siRNA-containing GeRPs compared to scrambled-siRNA-containing GeRPs (Fig. 4c). This level of knockdown is much greater than the relatively lower number of macrophages that could be observed to contain GeRPs in Fig. 4b, indicating possible degradation of the GeRPs and loss of detectable signal even though siRNA-mediated knockdown persists.
Figure 4. Orally administered GeRPs containing Map4k4 siRNA attenuate Map4k4 mRNA expression in PECs and macrophages from spleen, lung and liver.
a, Time line of GeRP (scrambled (Scr) or Map4k4, oligo 1) administration and PEC/tissue isolation. b, Confocal microscopy of PECs and tissue macrophages containing GeRPs (green). Macrophages were stained with F4/80–AF633 (magenta). Arrows indicate cells containing GeRPs. Nuclei (n) were stained with 4,6-diamidino-2-phenylindole (DAPI, blue). Magnification is indicated in the panels. c, Map4k4 mRNA expression in PECs and adherent cells from tissues. d, Serum INF-γ levels from mice gavaged with PBS, unloaded GeRPs or GeRPs containing 20 µg kg−1 of Scr siRNA (n = 5). Statistical significance was determined by ANOVA and Tukey post test. **P < 0.01 and *P < 0.05. Results are mean ± s.e.m.
Map4k4 silencing was also analysed in macrophages that had migrated to other tissues on day 10 of the protocol. Significant depletions of about 50%, 80% and 40% in Map4k4 mRNA levels were observed in macrophage-enriched cells isolated from spleen, liver and lung tissues, respectively, in mice treated with _Map4k4_-siRNA-containing GeRPs compared to scrambled-siRNA-containing GeRPs (Fig. 4c). In a parallel experiment, we could identify fluorescein–GeRP-containing macrophages isolated from spleen, liver and lung tissues of mice orally gavaged with fluorescently labelled GeRPs (Fig. 4b, bottom panel). Only a small proportion of macrophages enriched from these tissues contained GeRPs when examined by confocal microscopy (not shown). Tissue sections analysed by fluorescence microscopy also revealed infiltration of spleen, liver and lung with macrophages containing fluorescein–glucan shells (Supplementary Fig. 4b). Consistent with the lack of gene silencing in cells from skeletal muscle (Fig. 4c), GeRP-containing macrophages were rare in this tissue (Supplementary Fig. 4b). Taken together, these data indicate that macrophages in the GALT internalize orally absorbed GeRPs, undergo siRNA-mediated gene silencing and migrate into tissues throughout the body.
We confirmed that gene silencing by orally delivered GeRPs can be mediated by multiple siRNAs by using a different Map4k4 siRNA or either of two Tnf siRNA oligonucleotides previously found to be effective on macrophages in vitro (Supplementary Figs 2a, c and d and 5). We also found efficacy of _Map4k4_-siRNA-containing GeRPs to silence Map4k4 expression in macrophages after delivery by intraperitoneal (i.p.) injection (Supplementary Fig. 6c). Gene-specific silencing following oral delivery of GeRPs containing siRNA directed against fatty acid binding protein 4, which had no effect on Map4k4 or Tnf expression, was also documented (data not shown). Importantly, oral gavage of GeRPs containing either siRNA or no siRNA (unloaded GeRPs) did not change interferon-γ levels in serum (Fig. 4d), consistent with results in vitro (Supplementary Fig. 12). Serum levels of the liver enzymes, aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were within normal ranges (AST <255 IU l−1; ALT <77 IU l−1; Supplementary Fig. 13). Initial experiments indicate that the gene silencing with unmodified siRNA lasted about 8 days after the termination of oral GeRPs administration (not shown). Thus, efficient knockdown of three genes with five different siRNA sequences using orally delivered GeRPs has been achieved (Fig. 4c, Supplementary Fig. 5, and data not shown).
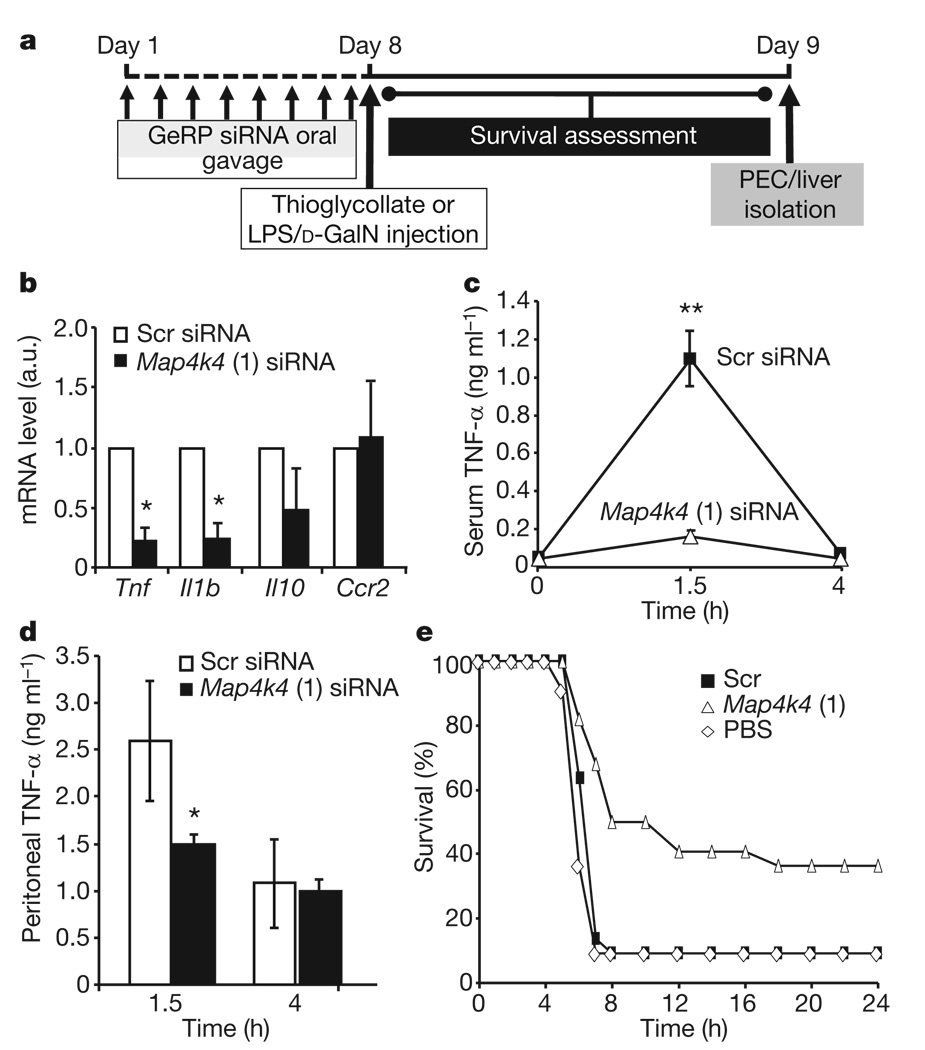
Notably, an 80% decrease in Tnf expression in PECs was observed in mice orally gavaged with GeRPs containing Map4k4 siRNA versus scrambled siRNA (Fig. 5b). This was accompanied by an equally marked 80% knockdown of the inflammatory cytokine Il-1β, but not Il-10 nor chemokine receptor 2 (Ccr2), known to be downregulated by LPS15. We found no effect of oral gavage of empty β1,3-d-glucan shells (Supplementary Fig. 9), β1,3-d-glucan shells containing tRNA and PEI, or GeRPs containing scrambled siRNA on serum Tnf-α levels before LPS treatment compared to PBS administration (Supplementary Fig. 11). Importantly, Tnf siRNA does not silence Il-1β, showing specificity of this broad anti-inflammatory response to Map4k4 knockdown (Supplementary Fig. 7).
Figure 5. Map4k4 silencing by oral gavage with GeRPs inhibits LPS-induced Tnf-α production in vivo.
a, Time line of siRNA and LPS/d-GalN administration. b, Tnf, Il1b, Il10 and Ccr2 mRNA expression in unstimulated PECs from mice gavaged with Scr- and _Map4k4_-siRNA-containing GeRPs (n = 3). Statistical significance was determined by a two-tailed Student’s _t_-test. c, d, Serum (c) and peritoneal (d) Tnf-α levels in siRNA-treated mice 1.5 h and 4 h after LPS/d-GalN administation (n = 10). Statistical significance was determined by ANOVA and Tukey post test; **P < 0.001 and *P < 0.05. e, Survival percentage of mice gavaged with PBS or Scr- and _Map4k4_-siRNA-containing GeRPs, followed by LPS/d-GalN administration (PBS, n = 11; Scr and Map4k4, n = 22). Statistical significance was determined by Kaplan–Meier analysis and Mantel–Cox testing (P < 0.01; see Supplementary Table 3). Results are the mean of three independent experiments. All results are mean ± s.e.m.
Consistent with previous reports16, we found circulating Tnf-α levels are strongly increased 1.5 h after LPS/d-galactosamine (d-GalN) injection into mice and then decreased to basal levels after 4 h. Oral delivery of GeRPs containing Map4k4 siRNA blocked the increase in serum Tnf-α protein (Fig. 5c) and Tnf-α levels in peritoneal fluid 1.5 h after LPS/d-GalN injection (Fig. 5d). Similarly, serum Tnf-α levels in response to LPS were decreased in mice orally gavaged with GeRPs containing other Map4k4 or Tnf siRNA oligonucleotides (Supplementary Fig. 8a), but not with scrambled siRNA or unloaded GeRPs (Supplementary Fig. 11a). These data demonstrate downregulation of the Tnf-α response to an inflammatory stimulus through oral delivery of _Map4k4_-siRNA-containing GeRPs.
The lethality observed in LPS/d-GalN-challenged animals is attributed to inflammatory cytokine toxicity and can be mimicked by administration of Tnf-α and Il-1β, which synergize with each other17. We therefore tested whether _Map4k4_-siRNA-containing GeRPs could protect against this toxicity. Figure 5e shows that 90% of the control mice orally gavaged with scrambled-siRNA-containing GeRPs before LPS/d-GalN injection died between 4 h and 8 h after LPS/d-GalN injection, whereas 50% of mice treated with _Map4k4_-siRNA-containing GeRPs survived for 8 h and 40% survived the LPS challenge long term (Supplementary Tables 2 and 3). Similarly, protection was obtained with the alternate Map4k4 siRNA (oligo 2) and two Tnf siRNA species (Supplementary Fig. 8b). Hepatocyte apoptosis in response to LPS injection18 was also attenuated by orally delivered _Map4k4_-siRNA-containing GeRPs (Supplementary Fig. 10), whereas serum insulin and glucose levels were unaffected (Supplementary Fig. 14). Thus, oral gavage of _Map4k4_-siRNA-containing GeRPs significantly protects mice from LPS/d-GalN-induced lethality through inhibition of Tnf-α and Il-1β production in macrophages.
The in vivo potency of 20 µg siRNA per kg in GeRPs to mediate gene silencing is 5 to 250 times greater than that in previous studies reporting systemic delivery. Intravenous injection of siRNA formulations requires doses ranging from 125 µg kg−1 to 50 mg kg−1 in mice19–24 and 1 mg kg−1 in nonhuman primates25. For attenuation of LPS-induced lethality in mice by i.p. injection, 1.2 mg Tnf siRNA per kg was required26. The high potency of orally delivered siRNA within GeRPs (20 µg kg−1) is even more surprising because unmodified siRNA was used in our studies. This high potency is probably due to protection of siRNA against nuclease degradation by PEI within GeRPs, low nonspecific binding of GeRPs en route to the gut, and high efficiency of GeRP uptake by phagocytic cells in the GALT. Furthermore, the siRNA loading capacity within β1,3-d-glucan shells is far greater than we used here, and can potentially deliver combinations of siRNA, DNA, proteins and small molecules. We also cannot rule out the possibility that other cell types are also targeted by oral delivery of GeRPs.
Injectable anti-TNF-α protein therapeutics are successful commercial products for the treatment of rheumatoid arthritis, ankylosing spondylitis, Crohn’s disease and psoriasis27. Macrophage-mediated pathogenesis is also well characterized in mouse models of obesity-associated insulin resistance28 and atherosclerosis29, whereas such autoimmune diseases as type 1 diabetes involve the deleterious actions of inflammatory cytokines30. Further development of GeRP-mediated delivery of siRNA to attenuate inflammation for these and other human maladies will be a major focus of our future studies.
METHODS SUMMARY
Preparation of GeRPs
Small interfering RNA was incorporated into the interior of porous, hollow β1,3-d-glucan shells purified from baker’s yeast by a layer-by-layer synthesis strategy to make GeRPs (Fig. 1). In brief, carrier tRNA was absorbed into β1,3-d-glucan shells and cationic complexes formed by PEI trapping7. Negatively charged siRNA was absorbed onto encapsulated positively charged complexes and coated with a PEI layer to produce multi-layered GeRP formulations.
Cell culture and GeRP treatment
PECs were isolated from ten-week-old C57BL6/J male mice 1–5 days following i.p. injection with thioglycollate broth. PECs were incubated for 48 h with GeRPs or fluorescein–GeRPs at a 10:1 particle-to-cell ratio.
GeRP administration
Ten-week-old C57BL6/J male mice were injected i.p. daily for 3 days, or gavaged daily for 8 days, with 2 × 109 or 4 × 109 GeRPs per kg (200 µl), respectively, containing 20 µg kg−1 scrambled or Map4k4 siRNA.
5′ RACE and nested PCR
Messenger RNA was purified from PECs treated for 48 h with GeRPs loaded with scrambled or Map4k4 siRNA. The GeneRacer Kit (Invitrogen) was used to produce 5′ RACE products, followed by nested PCR. GeneRacer primers and gene-specific primers were used. Products were cloned into the pCR4.1 TOPO vector for sequencing.
LPS lethality test
Ten-week-old C57BL6/J male mice were treated with GeRPs containing scrambled or Map4k4 siRNA, and then i.p. injected with 25 mg of d-galactosamine and 0.25 µg Escherichia coli LPS. Animals were monitored for survival over 24 h. Blood and peritoneal fluid were collected 1.5 h and 4 h after LPS/d-GalN injection for secreted Tnf-α measurements.
All procedures involving animals were approved by the Institutional Animal Care and Use Committee at University of Massachusetts Medical School.
METHODS
Preparation of β1,3-d-glucan shells
β1,3-d-glucan shells were prepared as described previously7. Baker’s yeast (AB Mauri Food Inc.) was alkaline-and-acid-extracted and dried at 20–25 °C following alcohol and acetone extractions. β1,3-d-glucan shells are 2–4 µm hollow, porous microspheres consisting primarily of β1,3-d-glucan and typically contain 5 × 1011 particles per g7.
Fluorescein labelling of β1,3-d-glucan shells
β1,3-d-glucan shells (1 g) were washed with sodium carbonate buffer (0.1 M, pH 9.2) and resuspended in 0.1 l carbonate buffer. 5-(4,6-dichlorotriazinyl) aminofluorescein (Invitrogen; 1 mg ml−1 in DMSO) was added to the buffered β1,3-d-glucan shell suspension (10% v/v) and mixed at 20–25 °C in the dark for 16 h. Tris buffer (2 mM) was added, incubated for 15 min and β1,3-d-glucan shells washed with sterile pyrogen-free water until the colour was removed. The β1,3-d-glucan shells were then dehydrated with absolute ethanol and acetone and dried in the dark at 20–25 °C.
Preparation of GeRPs
For empty GeRPs, dry β1,3-d-glucan shells were mixed with a volume of yeast RNA (Sigma; 10 mg ml−1 in 50 mM Tris HCl, pH8, 2 mM EDTA and 0.15 M NaCl (TEN)) to minimally swell the shells, and were incubated for 2 h at room temperature to allow for RNA absorption as described previously7. Neutral PEI (Aldrich; 25 kDa branched PEI; 2 mg ml−1 in TEN, pH7) was added in excess to form β1,3-d-glucan -shell-encapsulated RNA complexes, and these were resuspended by homogenization and incubated for at least 1 h. The β1,3-d-glucan-shell-encapsulated cationic complexes were sterilized in 70% ethanol, washed and resuspended in sterile pyrogen-free saline, counted by a haematocytometer (200×), diluted to 1 × 109 shells per ml in saline and stored at −20 °C.
For loading of siRNA into GeRPs for in vitro experiments, 1 × 107 empty GeRPs were diluted with sterile saline and incubated with 1 nmole Endo-Porter (Gene Tools) for 1 h at 20–25 °C. Next, 40 pmoles of siRNA was added and incubated at 20–25 °C for 2 h (all of the in vitro experiments contained 40 pmoles siRNA with the exception of the in vitro dose response, which loaded 1.6, 4, 40 or 80 pmoles of siRNA). Then, PEI (5 µg in saline) was added while vortexing and incubated at 20–25 °C for 20 min to trap the siRNA in the GeRPs. The PEI was quenched by the addition of 600 µl of complete DMEM. siRNA-loaded GeRPs were added to cells in culture at a concentration of 1 × 107 GeRPs per 1 × 106 cells. For loading of siRNA into GeRPs for in vivo experiments, the same procedure was followed with an adjusted ratio of 40 pmoles of siRNA in 1 × 108 empty GeRPs per dose. Empty GeRPs were incubated with 1 nmole Endo-Porter for 1 h at 20–25 °C. Next, 40 pmoles of siRNA was added, incubated at 20–25 °C for 2 h and trapped with PEI (50 µg) for 20 min at 20–25 °C. The siRNA-loaded GeRPs were then washed, resuspended in saline and sonicated to ensure homogeneity of the GeRP preparation. For each experiment, GeRP batches were aliquoted into tubes for daily dosing, flash-frozen in liquid nitrogen and stored at −20 °C. The final oral dose of GeRPs in 200 µl contains 40 pmole siRNA in 1 × 108 GeRPs.
Tissue macrophage isolation
C57BL6/J male mice (ten-week-old) were administered 1 × 108 GeRPs by daily oral gavage for 8 days. On day 9, mice were i.p. injected with thioglycollate. Cells from spleen, liver, lung, muscle and PECs were isolated on day 10. Tissues were cut into small pieces, washed with PBS and digested at 37 °C for 30 min with agitation using 5 mg ml−1 collagenase. Digested tissues were filtered through 70-µm pore nylon mesh, centrifuged, and the cells were plated in plastic dishes for 2–3 h in medium (DMEM plus 10% fetal bovine serum). The cells were washed with PBS to remove non-adherent cells, and adherent cells were used for real-time PCR and microscopy.
For confocal experiments, mice were gavaged with a single 2 × 108 dose of GeRPs, and tissue macrophages isolated after 24 h as described previously.
Microscopy
Fixed cells from in vitro and in vivo experiments were incubated with F4/80 primary antibody (AbD Serotec) followed by AlexaFluor350 or 633 secondary antibody (Invitrogen). Nuclei were stained with Hoescht 33342 or DAPI as denoted in the figure legends.
Isolation of RNA and real-time PCR
RNA isolation was performed according to the Trizol reagent protocol (Invitrogen). Complementary DNA was synthesized from 1 µg of total RNA using iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s instructions. For real-time PCR, synthesized cDNA forward and reverse primers along with the iQ SYBR Green Supermix were run on the MyIQ real-time PCR system (Bio-Rad). Sequences of the primers used were designed with Primer Bank (Supplementary Table 4). The ribosomal mRNA 36B4 was used as an internal loading control, as its expression did not change over a 24 h period with the addition of LPS, TNF-α or siRNA against the genes used in this study.
ELISA assay
Tnf-α levels in the PEC supernatant, plasma and peritoneal fluids and interferon-γ levels in plasma were measured using mouse ELISA kits (Pierce) as recommended by the manufacturer.
AST and ALT measurement
To test for liver toxicity, levels of ALT and AST activity in serum were measured using a commercial kit (Fisher Scientific) according to the manufacturer’s instructions.
Histology and TUNEL assay
Tissue sections were stained with F4/80–AlexaFluor405 antibody (AbD-Serotec) and were haematoxylin-stained. TUNEL assay was performed on liver sections from mice challenged with LPS/d-GalN according to the manufacturer’s instructions (Upstate). TUNEL images were obtained using a Zeiss Axiovert 200 inverted microscope equipped with a Zeiss AxioCam HR CCD camera with 1,300 × 1,030 pixels basic resolution and a Zeiss Plan NeoFluar 20×/0.50 Ph2 (DIC II) objective. Zeiss Aprochromat 100×/1.40 Oil (440780).
Tissue and cell images were obtained with a Solamere CSU10 Spinning Disk confocal system mounted on a Nikon TE2000-E2 inverted microscope. Images were taken with a multi-immersion ×20 objective with a NA = 0.75, oil:W.D. = 0.35 mm, or a ×100 Plan Apo VC objective NA = 1.4, oil:W.D. = 0.13 mm.
Supplementary Material
supplementary
Acknowledgements
We appreciate critical reading of the manuscript and suggestions by C. Mello, V. Ambros, G. Hannon, S. Corvera and J. Sullivan. We thank P. Zamore for advice on methods. We also appreciate the technical help of P. Furcinitti, A. Burkhart and A. Goller. This work was supported by The University of Massachusetts Diabetes and Endocrinology Center (DK 32520), including its Genomics, Bioinformatics and Imaging Cores, the Diabetes Genome Anatomy Project (DK 60837), Commonwealth Medicine and NIH grant DK 30898.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions G.R.O., M.A. and M.P.C. initially conceptualized the study. M.A., G.J.T., M.W., M.C. and S.M.N. performed experiments, and all authors participated in designing experiments, and analysing and interpreting data. M.A. and G.J.T. contributed equally to this work. G.R.O. and E.S. provided the β1,3-d-glucan-shell-encapsulated cationic materials and they with M.A. and G.J.T. developed the GeRP formulations used in these studies. M.A., G.R.O. and M.P.C. wrote the manuscript.
Author Information The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Grimm D, Kay MA. Therapeutic application of RNAi: is mRNA targeting finally ready for prime time? J. Clin. Invest. 2007;117:3633–3641. doi: 10.1172/JCI34129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duffield JS. The inflammatory macrophage: a story of Jekyll and Hyde. Clin. Sci. (Lond.) 2003;104:27–38. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 4.Beier R, Gebert A. Kinetics of particle uptake in the domes of Peyer’s patches. Am. J. Physiol. 1998;275:G130–G137. doi: 10.1152/ajpgi.1998.275.1.G130. [DOI] [PubMed] [Google Scholar]
- 5.Herre J, Gordon S, Brown GD. Dectin-1 and its role in the recognition of β-glucans by macrophages. Mol. Immunol. 2004;40:869–876. doi: 10.1016/j.molimm.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Vazquez-Torres A, et al. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature. 1999;401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 7.Soto ER, Ostroff GR. Characterization of multilayered nanoparticles encapsulated in yeast cell wall particles for DNA delivery. Bioconjug. Chem. 2008;19:840–848. doi: 10.1021/bc700329p. [DOI] [PubMed] [Google Scholar]
- 8.Tang X, et al. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPARγ, adipogenesis, and insulin-responsive hexose transport. Proc. Natl Acad. Sci. USA. 2006;103:2087–2092. doi: 10.1073/pnas.0507660103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tesz GJ, et al. Tumor necrosis factor α (TNFα) stimulates Map4k4 expression through TNFα receptor 1 signaling to c-Jun and activating transcription factor 2. J. Biol. Chem. 2007;282:19302–19312. doi: 10.1074/jbc.M700665200. [DOI] [PubMed] [Google Scholar]
- 10.Bouzakri K, Zierath JR. MAP4K4 gene silencing in human skeletal muscle prevents tumor necrosis factor-α-induced insulin resistance. J. Biol. Chem. 2007;282:7783–7789. doi: 10.1074/jbc.M608602200. [DOI] [PubMed] [Google Scholar]
- 11.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Frank-Kamenetsky M, et al. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc. Natl Acad. Sci. USA. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barthel R, et al. Regulation of tumor necrosis factor alpha gene expression by mycobacteria involves the assembly of a unique enhanceosome dependent on the coactivator proteins CBP/p300. Mol. Cell. Biol. 2003;23:526–533. doi: 10.1128/MCB.23.2.526-533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsytsykova AV, et al. Post-induction, stimulus-specific regulation of tumor necrosis factor mRNA expression. J. Biol. Chem. 2007;282:11629–11638. doi: 10.1074/jbc.M611418200. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Y, Yang Y, Warr G, Bravo R. LPS down-regulates the expression of chemokine receptor CCR2 in mice and abolishes macrophage infiltration in acute inflammation. J. Leukoc. Biol. 1999;65:265–269. doi: 10.1002/jlb.65.2.265. [DOI] [PubMed] [Google Scholar]
- 16.Endo Y, et al. Enhancement by galactosamine of lipopolysaccharide (LPS)-induced tumour necrosis factor production and lethality: its suppression by LPS pretreatment. Br. J. Pharmacol. 1999;128:5–12. doi: 10.1038/sj.bjp.0702747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okusawa S, Gelfand JA, Ikejima T, Connolly RJ, Dinarello CA. Interleukin 1 induces a shock-like state in rabbits. Synergism with tumor necrosis factor and the effect of cyclooxygenase inhibition. J. Clin. Invest. 1988;81:1162–1172. doi: 10.1172/JCI113431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silverstein R. D-galactosamine lethality model: scope and limitations. J. Endotoxin Res. 2004;10:147–162. doi: 10.1179/096805104225004879. [DOI] [PubMed] [Google Scholar]
- 19.Filleur S, et al. SiRNA-mediated inhibition of vascular endothelial growth factor severely limits tumor resistance to antiangiogenic thrombospondin-1 and slows tumor vascularization and growth. Cancer Res. 2003;63:3919–3922. [PubMed] [Google Scholar]
- 20.McCaffrey AP, et al. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 21.Peer D, Zhu P, Carman CV, Lieberman J, Shimaoka M. Selective gene silencing in activated leukocytes by targeting siRNAs to the integrin lymphocyte function-associated antigen-1. Proc. Natl Acad. Sci. USA. 2007;104:4095–4100. doi: 10.1073/pnas.0608491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song E, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nature Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 23.Soutschek J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 24.Wesche-Soldato DE, et al. In vivo delivery of caspase-8 or Fas siRNA improves the survival of septic mice. Blood. 2005;106:2295–2301. doi: 10.1182/blood-2004-10-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmermann TS, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 26.Sorensen DR, Leirdal M, Sioud M. Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J. Mol. Biol. 2003;327:761–766. doi: 10.1016/s0022-2836(03)00181-5. [DOI] [PubMed] [Google Scholar]
- 27.Shealy DJ, Visvanathan S. Anti-TNF antibodies: lessons from the past, roadmap for the future. Handb. Exp. Pharmacol. 2008;181:101–129. doi: 10.1007/978-3-540-73259-4_5. [DOI] [PubMed] [Google Scholar]
- 28.Ferrante AW., Jr Obesity-induced inflammation: a metabolic dialogue in the language of inflammation. J. Intern. Med. 2007;262:408–414. doi: 10.1111/j.1365-2796.2007.01852.x. [DOI] [PubMed] [Google Scholar]
- 29.Hansson GK, Libby P. The immune response in atherosclerosis: a double-edged sword. Nature Rev. Immunol. 2006;6:508–519. doi: 10.1038/nri1882. [DOI] [PubMed] [Google Scholar]
- 30.Shoda LK, et al. A comprehensive review of interventions in the NOD mouse and implications for translation. Immunity. 2005;23:115–126. doi: 10.1016/j.immuni.2005.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary