The Drosophila Cyclin D–Cdk4 complex promotes cellular growth (original) (raw)
Abstract
Mammalian cyclin D–Cdk4 complexes have been characterized as growth factor-responsive cell cycle regulators. Their levels rise upon growth factor stimulation, and they can phosphorylate and thus neutralize Retinoblastoma (Rb) family proteins to promote an E2F-dependent transcriptional program and S-phase entry. Here we characterize the in vivo function of Drosophila Cyclin D (CycD). We find that Drosophila CycD–Cdk4 does not act as a direct G1/S-phase regulator, but instead promotes cellular growth (accumulation of mass). The cellular response to CycD–Cdk4-driven growth varied according to cell type. In undifferentiated proliferating wing imaginal cells, CycD–Cdk4 caused accelerated cell division (hyperplasia) without affecting cell cycle phasing or cell size. In endoreplicating salivary gland cells, CycD–Cdk4 caused excessive DNA replication and cell enlargement (hypertrophy). In differentiating eyes, CycD–Cdk4 caused cell enlargement (hypertrophy) in post-mitotic cells. Interaction tests with a Drosophila Rb homolog, RBF, indicate that CycD–Cdk4 can counteract the cell cycle suppressive effects of RBF, but that its growth promoting activity is mediated at least in part via other targets.
Keywords: cell cycle/cell growth/Cyclin D–Cdk4/Drosophila/Rb
Introduction
Cell proliferation normally requires not only DNA replication and mitosis, but also a doubling of cell mass—growth—with each division cycle. Most of the well characterized cell cycle regulators act specifically to promote DNA replication or mitosis and have little direct role in controlling cytoplasmic growth. However, several genes that act early in the mammalian cell cycle remain ambiguous with respect to whether they regulate cell cycle progression, cellular growth or both. These genes, the D-type cyclin CycD1, its target pRb and the specific Cyclin D (CycD) inhibitor p16_INK4A_, were discovered as oncogenes and tumor suppressors (Friend et al., 1988; Motokura et al., 1991; Withers et al., 1991; Serrano et al., 1993).
Molecular studies have focused on the CycD-dependent kinases as specific promoters of the G1–S transition (Sherr, 1996; Sherr and Roberts, 1999). The prevailing view holds that CycD–Cdk complexes promote G1 progression by phosphorylating and thereby neutralizing ‘pocket proteins’ of the Retinoblastoma family including pRb, p107 and p130. In the active hypo-phosphorylated state these pocket proteins bind transcription factors, notably E2Fs, inhibiting their function and in some cases converting them into repressors of transcription (Beijersbergen and Bernards, 1996). Upon phosphorylation by CycD–Cdk4 and then CycE–Cdk2 the pocket proteins release the E2Fs, allowing them to promote transcription of a large set of cell cycle genes including enzymes for nucleotide synthesis and G1 cyclins such as CycE (Dyson, 1998; Harbour et al., 1999). This mobilization of gene expression is thought to be the event that makes DNA replication and cell cycle progression inexorable.
Much correlative and functional data support this paradigm. Binding of the pocket proteins to the various E2Fs has been extensively characterized, and is highly correlated with the phosphorylation state of the pocket proteins and the position of cells within the cell cycle (Beijersbergen and Bernards, 1996; Dyson, 1998; Harbour et al., 1999). Overexpressed E2F can trigger S-phases in quiescent, serum-starved cells, whereas overexpressed pRb generally inhibits cell cycle progression. Forced expression of CycD1 or loss of pRb can also shorten the G1 period (Jiang et al., 1993; Quelle et al., 1993; Resnitzky et al., 1994; Herrera et al., 1996), and overexpressed CycD1–Cdk4 can suppress a pRb-mediated G1 arrest (Connell-Crowley et al., 1997; Leng et al., 1997). Even more compelling are experiments demonstrating that CycD1 is dispensable in cells lacking pRb function (Lukas et al., 1994), and that CycD1 can be substantially complemented by its downstream target, cyclin E, in vivo in the mouse (Geng et al., 1999). However, overexpressed CycD1 is not sufficient to trigger cell cycle entry in quiescent, serum-starved cells (Kato et al., 1994; Matsushime et al., 1994), and gene knockouts show that murine CycD1, CycD2 and Cdk4 are not required for proliferation of most types of cells in vivo (Fantl et al., 1995; Sicinski et al., 1995, 1996; Rane et al., 1999; Tsutsui et al., 1999).
In cultured mammalian cells, expression of CycD1 and its kinase partner Cdk4 are induced by serum growth factors (Matsushime et al., 1991, 1994), and so are thought to act as ‘growth factor sensors’ that link extracellular cues to the cell cycle machinery (Sherr, 1996; Sherr and Roberts, 1999). Elegant studies of the yeast G1 cyclin CLN3 suggest how such a mechanism might work (Polymenis and Schmidt, 1997). CLN3 levels are positively coupled to overall rates of cellular metabolism via short upstream open reading frames that selectively inhibit its translation when growth conditions are poor and ribosomes are scarce. Since CLN3 protein is highly unstable, it only accumulates to the threshold levels required to trigger S-phase when rates of cellular biosynthesis (and growth) are high. Thus CLN3 acts as a ‘growth sensor’. Several characteristics of the D-type cyclins have prompted the suggestion that they too may couple rates of biosynthesis to cell cycle progression (Rosenwald, 1996; Polymenis and Schmidt, 1999). Like CLN3, the D-type cyclins act early in the cell cycle to trigger the expression of later-acting genes, they are highly unstable, and they are regulated post-transcriptionally (Rosenwald et al., 1993; Sewing et al., 1993; Muise-Helmericks et al., 1998).
Although the ‘growth sensor’ model accounts for much experimental data, it falls short of explaining how CycD1 can promote hyperplastic growth (Motokura et al., 1991; Withers et al., 1991; Wang et al., 1994; Robles et al., 1996). This is because inducing cell cycle progression is not in itself sufficient to promote cellular growth. In fact, accelerating the cell cycle without a corresponding increase in cellular growth results in cell diminution and cell death (Neufeld et al., 1998). Thus the question arises: does cyclin D promote cellular growth in addition to cell cycle progression? Perhaps. Indeed, studies of Cyclin D–Cdk function in transgenic and knockout mice and plants have revealed deregulation of growth more often than specific defects in cell cycle progression (see Discussion).
Here we address these issues in the fruit fly, Drosophila melanogaster. Drosophila has one known D-type cyclin homolog (Finley et al., 1996) and one known cyclin D-specific kinase subunit, Cdk4 (previously called Cdk4/6; Sauer et al., 1996). Drosophila CycD interacts with Drosophila Cdk4 in two-hybrid assays (Sauer et al., 1996) and in vivo (Meyer et al., 2000), and is capable of rescuing G1 cyclin deficiency in yeast (Finley et al., 1996). CycD is expressed in Drosophila embryos and imaginal discs during developmental stages when cell proliferation is widespread, but its function has not been examined in depth (Finley et al., 1996). The studies described here and in the accompanying paper (Meyer et al., 2000) suggest that the primary function of CycD–Cdk4 in Drosophila is to stimulate cellular growth.
Results
Drosophila CycD–Cdk4 function is not limited to G1–S progression
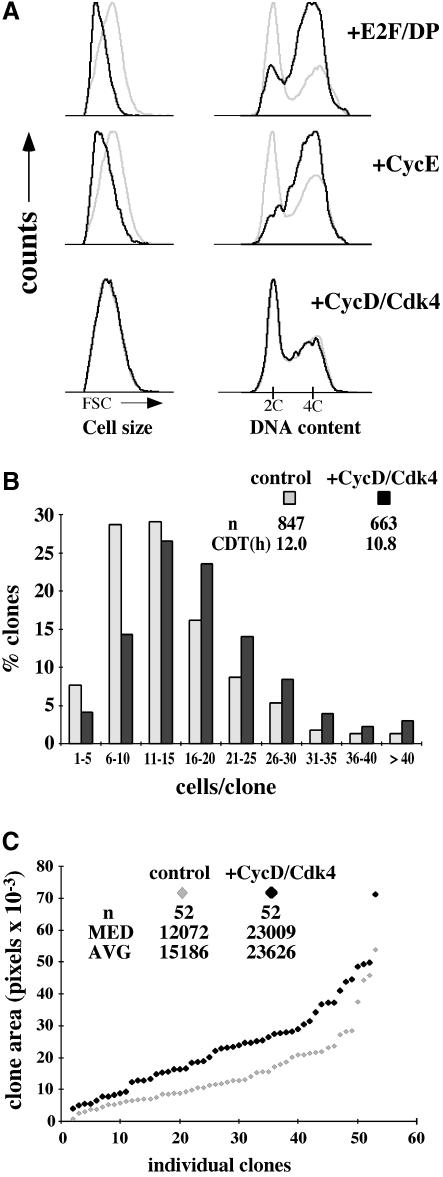
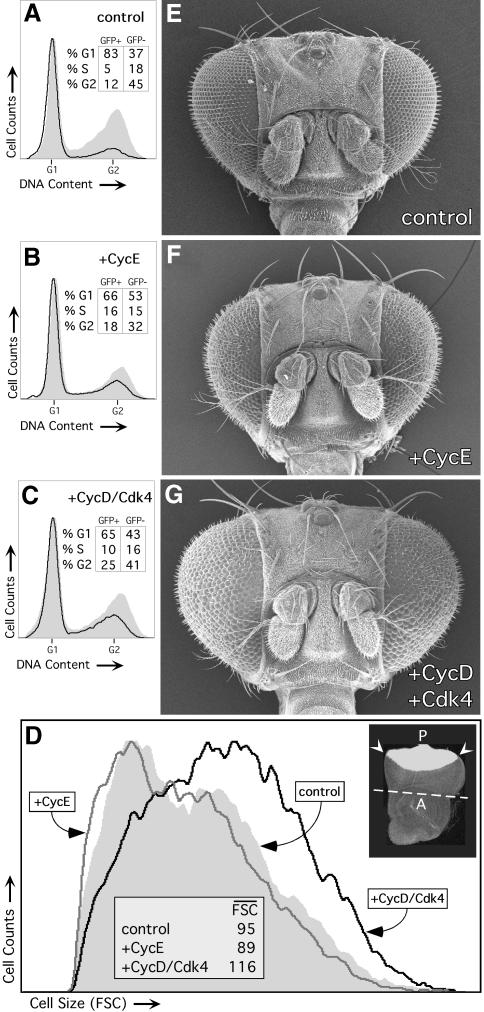
To assess the function of Drosophila Cyclin D, we first studied the effects of overexpressing it in actively proliferating wing imaginal disc cells. Cell clones overexpressing CycD and/or Cdk4 and marked with co-expressed green fluorescent protein (GFP) were induced in developing wings using the flip-out Gal4 system (Pignoni and Zipursky, 1997; Neufeld et al., 1998). Tissues were dissociated and analyzed by fluorescence activated cell sorting (FACS) 48 h post-induction (h.p.i.), to determine the proportion of cells in each phase of the cell cycle, and to assess effects on cell size (Figure 1A). In these and other experiments we observed that expressing either CycD or Cdk4 alone had no detectable effects (Figures 2 and 5 and Table I; Datar, 2000). Hence we simultaneously expressed both CycD and Cdk4. CycD–Cdk4-overexpressing cells were compared with wild-type, non-expressing cells from the same discs. In parallel experiments, overexpressed Drosophila E2F/DP or cyclin E truncated the G1 phase and decreased the cell size (Figure 1A). However, we were surprised to find no changes in cell cycle phasing or cell size in CycD–Cdk4-overexpressing cells (Figure 1A). These data differ markedly from the effects of cyclin D1 or D2 overexpression demonstrated in cultured vertebrate cells (Jiang et al., 1993; Quelle et al., 1993; Resnitzky et al., 1994; Resnitzky and Reed, 1995) and suggest that the Drosophila CycD–Cdk4 complex does not promote G1–S transitions directly in vivo.
Fig. 1. Effects of overexpressed CycD–Cdk4 in imaginal wing disc cells. (A) Overexpression clones were induced 48 h after egg deposition (AED) using the flip-out Gal4 technique and analyzed by FACS at 96 h after AED. Light traces represent GFP-negative (wild-type) cells, and dark traces represent GFP-positive (experimental) cells. Relative cell size is shown on the left by forward light scatter (FSC), and cell cycle profile (DNA) is on the right. (B) Clones were induced at 72 h AED and analyzed after fixation at 115.5 h AED. The distribution of number of cells per clone as a percentage of the total number of clones for each genotype is shown. Number of clones scored (n) and median cell doubling time (CDT) are indicated, P <0.002. Control cells expressed GFP alone. (C) Clones were induced at 48 h AED. Discs were fixed at 115.5 h AED. Fifty-two random clones of each genotype were analyzed and plotted individually in order of increasing size. Values for median and average area per clone are indicated, P <0.002. All controls were done in parallel.
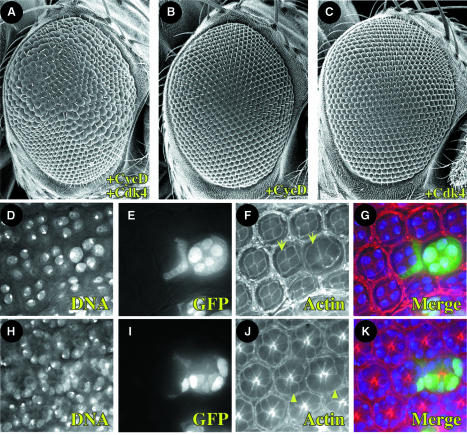
Fig. 2. Clonal expression of CycD–Cdk4 promotes cellular hypertrophy in the eye. (A–C) Overexpression clones were induced during embryonic development (12–16 h AED), and adult eyes were analyzed 3 days after eclosion. Lateral view SEMs at 200×. Anterior is to the right, dorsal is up. Genotypes are (A) hs-FLP; UAS-CycD UAS-Cdk4; Act>Cd2>Gal4 UAS-GFP; (B) hs-FLP; UAS-CycD; Act>Cd2>Gal4 UAS-GFP; (C) hs-FLP; UAS-Cdk4; Act>Cd2>Gal4 UAS-GFP. (D–K) Clones overexpressing CycD–Cdk4 were induced at 72 h AED, and pupal discs were fixed 96 h later at 48 h after puparium formation (APF). Optical sections are of the same eye field at superficial (cone cell level, D–G) and deep (photoreceptor level, H–K) layers. GFP marks cells overexpressing CycD–Cdk4 (E, I, G and K), DAPI marks nuclei (D, H, G and K) and rhodamine-conjugated phalloidin marks cell membranes (F, J, G and K). Photoreceptor cells (arrowhead) and cone cells (arrow) are labeled accordingly. Compare analogous cell types with or without GFP (+/– CycD–Cdk4).
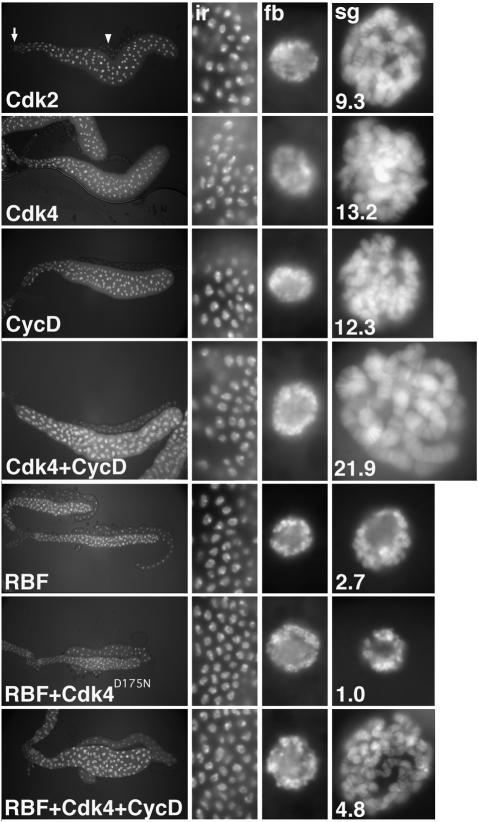
Fig. 5. Opposing effects of CycD–Cdk4 and RBF on salivary gland growth. Salivary glands were dissected from larvae expressing either Cdk2, Cdk4, CycD, Cdk4 + CycD, RBF, RBF + Cdk4D175N-myc or RBF + Cdk4 + CycD under F4-GAL4/UAS control, at third instar wandering stage. Panels from left to right show DNA labeling in complete salivary glands, and at a higher but constant magnification in imaginal ring cells (ir), fat body cell nuclei (fb), and salivary gland cell nuclei (sg). The DNA signal ratio of expressing salivary gland nuclei and control, non-expressing fat body nuclei averaged from at least three different glands as described by Weiss et al. (1998) is indicated by the white numbers in the right panels. Cdk2 expression, shown for a control, was indistinguishable from wild type (Weiss et al., 1998; data not shown).
Table I. Eye phenotypes after manipulation of CDK activity.
| UAS | GAL4 | ||
|---|---|---|---|
| eyeless (ey) | GMR | sevenless (sev) | |
| Cdk2 | o | o | o |
| Cdk4 | o | o | o |
| Cdk4D175N | o | o | o |
| CycD | o | o | o |
| CycD + Cdk2 | o | o | o |
| CycD + Cdk4 | o | +++ | +++ |
| RBF | – – – | o | o |
| RBF + Cdk4 | – – – | o | o |
| RBF + CycD | – | o | o |
| RBF + Cdk4 + CycD | – | ++ | + |
| CycD + Cdk4D175N | o | + | ++ |
| RBF + Cdk4D175N | – – – | o | o |
| RBF + CycD + Cdk4D175N | – – | (+) | + |
CycD–Cdk4 accelerates cell division in the developing wing
To assess the effect of CycD–Cdk4 on rates of cell growth and proliferation, we evaluated CycD–Cdk4-expressing cell clones in situ in wing imaginal discs. Although overexpressed CycD–Cdk4 did not alter cell cycle phasing (Figure 1A), it did affect the proliferative rate of cells in the developing wing (Figure 1B). At 44 h.p.i. we noted a significant increase in the number of cells in CycD–Cdk4-expressing clones, which had a median of 16 cells, compared with 12 cells in controls (Figure 1B). This corresponded to a 10% decrease in the cell doubling time, from 12 h in controls to 10.8 h in cells overexpressing CycD–Cdk4. Since cells overexpressing CycD–Cdk4 were not reduced in size (Figure 1A), it follows that they must acquire more mass than control cells over the same developmental period. Consistent with this we observed that CycD–Cdk4-overexpressing clones encompassed 150% of the area of controls at 44 h.p.i., and 180% of the area of controls at 67 h.p.i. (Figures 1C and 4C). We conclude that CycD–Cdk4 can stimulate cellular and clonal growth. In the context of the developing wing this increased growth results in faster cell division without detectable changes in cell size.
Fig. 4. Opposing effects of CycD–Cdk4 and RBF in wing imaginal discs. (A) Flip-out Gal4 clones expressing GFP and the genes indicated were induced at 72 h AED and fixed at 115.5 h AED. Distribution of clone size for the genotypes given is shown. Cell doubling times (CDTs) are indicated. Total number of clones analyzed (n): control = 161; CycD–Cdk4 = 158; RBF = 184; CycD–Cdk4 + RBF = 144. Comparison of cell number/clone for CycD–Cdk4 + RBF with each of the other three genotypes, P <0.003. (**B**) Clones were induced at 48 h AED and analyzed by FACS at 96 h AED. Light trace represents GFP-negative (wild-type) cells, and dark trace represents GFP-positive (experimental) cells. Ratio of mean cell size (GFP+/GFP–) is indicated at top right. (**C**) Clones were induced at either 48 or 72 h AED and fixed at 115 or 116 h AED, respectively. Comparison of clone areas between genotypes at each time-point, _P_ <0.05, except between control and CycD–Cdk4 + RBF, _P_ >0.6. Median cell number/clone was established for 44 h clones of each genotype: control, 18; CycD–Cdk4, 25; RBF, 8; CycD–Cdk4 + RBF, 14, P <0.04 for comparison of cell number/clone between all genotypes, including control and CycD–Cdk4 + RBF. This number divided into the average clone area gave a relative cell size, shown in parentheses in (C). CycD–Cdk4 + RBF-expressing clones encompassed as great an area as control clones with significantly fewer cells per clone; thus, CycD–Cdk4 + RBF cells are larger than wild type.
CycD–Cdk4 promotes hypertrophy in post-mitotic cells in the eye
We next asked what effect CycD–Cdk4 overexpression would have on the development of adult tissues. Adult wings containing CycD–Cdk4-expressing clones showed no gross abnormalities in overall shape, veination, bristle or trichome patterning (not shown). Optical sections through pupal wings failed to reveal significant changes in cell morphology or cell density within clones overexpressing CycD–Cdk4 (Datar, 2000).
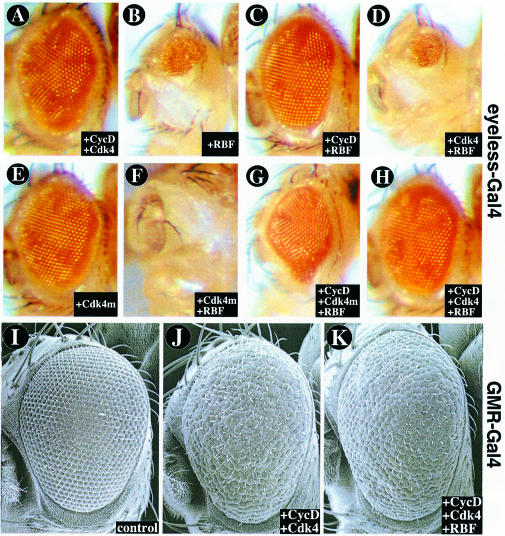
However, we observed a striking enlargement of ommatidiae in adult eyes that clonally overexpressed CycD–Cdk4 (Figure 2A). This phenotype required expression of both CycD and Cdk4 (Figure 2B and C) and was clearly mosaic. Optical sections of pupal eyes confirmed that overexpressed CycD–Cdk4 led to increased cell size (hypertrophy) and that this effect was cell autonomous (Figure 2D–K). We observed a mild disorganization in the regular arrangement of ommatidia, but no specific defects in cell differentiation or overall patterning. Hypertrophy occurred in several cell types and structures, including primary pigment and cone cells (Figure 2D–G), photoreceptors (Figure 2H–K) and interommatidial bristles (not shown). In ommatidiae overexpressing CycD–Cdk4, we often observed five cone cells instead of the normal four (Figure 2D–G), suggesting that an extra cell division occurred.
We also induced CycD–Cdk4 in the eye using the GMR-Gal4 and sev-Gal4 drivers, which are expressed starting late in eye development posterior to the morphogenetic furrow (MF), as cells enter their ultimate or penultimate cell cycle and begin to differentiate (Figure 3D; Freeman, 1996). Using these drivers to induce CycD–Cdk4 expression we found that all ommatidiae were enlarged, as was the entire eye, which bulged out of the head in an ominous fashion (Figure 3G and Table I). Light and transmission electron microscopy of pupal and adult eyes from GMR-Gal4, UAS-CycD, UAS-Cdk4 flies revealed enlarged photoreceptor cell bodies and rhabdomeres, and excessive accumulations of actin (not shown), suggesting that CycD–Cdk4 increased cellular protein content as well as cell volume. Driving expression of two copies of CycD–Cdk4 with a single copy of GMR-Gal4 led to even larger ommatidiae and eyes, indicating dose dependence (not shown). We did not observe significant eye overgrowth when CycD was expressed alone or when it was co-expressed with a kinase-impaired mutant form of Cdk4 (_Cdk4_D175N; Meyer et al., 2000; Table I). Thus, the bulging eye phenotype appeared to require phosphorylation of CycD–Cdk4 substrates. In comparison, CycE (Figure 3F) or E2F (not shown) did not cause eye overgrowth when expressed under GMR-Gal4 control, even though both factors promote ectopic DNA replication and cell proliferation in the eye (Figure 3B; Richardson et al., 1995; Asano et al., 1996; Du et al., 1996b).
Fig. 3. GMR-Gal4-driven expression of CycD–Cdk4, but not CycE, causes hypertrophy of the eye. A–D show FACS analyses of eye discs in which GMR-Gal4 was used to co-express UAS-GFP with the genes indicated in differentiating cells posterior to the morphogenetic furrow (MF; see inset in D). (A–C) Expressing GFP-positive, posterior cells are denoted by black traces, and GFP-negative, anterior cells are denoted by gray shaded plots. Both CycE (B) and CycD–Cdk4 (C) increase S and G2 in cells posterior to the MF (dark traces). (D) Cell size of GFP-positive posterior cells measured by FSC. Mean FSC values are listed. Right inset shows GFP expression in a control eye–antennal disc. The division between the posterior (P) eye region and the anterior (A) antennal region is marked (white line), and the MF is indicated (arrowheads). FACS was performed on the eye region only. (E–G) show SEM images of GMR-Gal4/CyO (control, E); GMR-Gal4/UAS-CycE (+CycE, F); and GMR-Gal4 UAS-CycD UAS-Cdk4/CyO (+CycD–Cdk4, G) females. Genotypes used in (A–D) were the same except for the inclusion of UAS–GFP.
To assess these phenotypes further we performed FACS on eye discs in which UAS-GFP was co-expressed with either UAS-CycD–Cdk4 or UAS-CycE under GMR-Gal4 control (Figure 3A–D). FACS analysis showed that overexpression of either CycD–Cdk4 or CycE significantly increased the fraction of S- and G2-phase cells posterior to the MF (GFP+ cells, Figure 3B and C). Normally these cells are nearly all arrested in G1 (Figure 3A), and thus both cyclins appeared to perturb the normal program of cell cycle exit at differentiation. The effects of CycD–Cdk4 and CycE on cell size, however, were dramatically different. CycD–Cdk4 greatly increased the size of posterior eye cells, whereas CycE caused a slight decrease in cell size (Figure 3D). This cell size effect was not phase specific: even G1 cells overexpressing CycD–Cdk4 were much larger than controls (not shown). These results are consistent with our findings in wing cells in that CycD–Cdk4 appeared to promote both cell cycle progression and cellular growth, whereas CycE affected only cell cycle progression. In addition, neither CycE nor CycD–Cdk4 completely prevented the G1 arrest that accompanies cell differentiation, but only CycD–Cdk4 appeared to continue promoting growth in post-mitotic cells, leading to cellular hypertrophy.
Interactions between CycD–Cdk4 and RBF
Many studies have indicated that the major substrates for vertebrate D-type cyclin–Cdk complexes are the Rb family proteins (for reviews see Beijersbergen and Bernards, 1996; Sherr, 1996; Dyson, 1998). Previously we showed that a Drosophila homolog of Rb, RBF (Du et al., 1996a; Du and Dyson, 1999), could inhibit cell division when overexpressed in the wing imaginal disc. RBF did not immediately block cell growth, and as a result there was a large increase in cell size (Neufeld et al., 1998). To test interactions between CycD–Cdk4 and RBF we co-expressed all three genes together in wings (Figure 4), salivary glands (Figure 5) or eyes (Figure 6).
Fig. 6. CycD–Cdk4 and RBF interactions in the eye. (A–H) show light micrographs of eyes overexpressing the genes indicated under ey-Gal4 control, which activates UAS target genes throughout the eye beginning early in development. Using the ey-Gal4 driver, overexpressed RBF suppresses eye growth (B), and this effect is counteracted by CycD (C) or CycD–Cdk4 (H). The catalytically inactive variant Cdk4D175N-myc (Cdk4m) acts synergistically with RBF in suppressing eye growth (F and G). (I–K) show SEM images of eyes overexpressing the genes indicated under GMR-Gal4 control, which activates its UAS targets late in eye development at the onset of cell differentiation. Using GMR-Gal4, CycD–Cdk4 causes overgrowth (J), but this effect is not neutralized by co-expressed RBF (K).
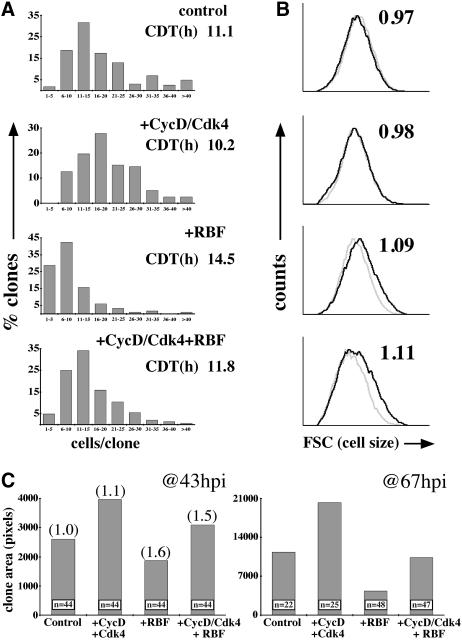
Using the flip-out Gal4 method in the wing, we found that cells co-expressing CycD–Cdk4 + RBF cycled more rapidly than cells expressing RBF alone, and thus that CycD–Cdk4 attenuated the inhibitory effects of RBF on cell cycle progression (Figure 4A). Interestingly, although cell clones co-expressing CycD–Cdk4 + RBF had fewer cells than wild-type controls, they encompassed substantially more area (Figure 4C; 43 h.p.i.). Flow cytometry (Figure 4B) and in situ cell size measurements (Figure 4C) confirmed that the increased mass of these clones was due to increased cell size. The large size of cells co-expressing CycD–Cdk4 + RBF, despite a nearly normal division rate, suggests that CycD–Cdk4 promoted extra growth even whilst RBF slowed cell cycle progression. A logical inference, corroborated below, is that CycD–Cdk4 promotes growth via targets other than RBF. This effect was less evident at a later time-point (67 h.p.i.), perhaps because cell cycle suppression by RBF eventually throttled even the growth of CycD–Cdk4-expressing clones (Figure 4C).
We also performed RBF–CycD interaction tests in the larval salivary gland, a differentiated tissue in which cell growth is accomplished by cycles of DNA endoreplication. To express UAS-linked target genes, we used the F4-Gal4 driver (Weiss et al., 1998), which commences its expression late in embryogenesis after cell proliferation in the salivary primordium is complete and stays active throughout the larval stages. Results obtained in the salivary glands (Figure 5) were consistent with those described above for the wing. F4-Gal4-driven expression of CycD–Cdk4 resulted in oversized salivary glands with nuclei containing excessively endoreduplicated polytene chromosomes (Figure 5). Co-expression of cyclin D with the kinase impaired variant Cdk4D175N or Cdk2 did not induce this size increase, suggesting that the protein kinase activity of cyclin D–Cdk4 complexes is required for the stimulation of salivary gland DNA endoreplication and growth (not shown). Although expression of Cdk4D175N alone had little effect on salivary growth, overexpressed RBF strongly inhibited both growth and DNA endoreplication. Even stronger growth suppression was observed when RBF was co-expressed with Cdk4D175N. As in wings and eyes, the growth-inhibitory effects of RBF were attenuated by simultaneous co-expression of CycD–Cdk4 (Figure 5).
Similar results were obtained when RBF and CycD–Cdk4 were co-expressed in the eye using ey-Gal4, which is expressed throughout the eye primordium beginning very early in its development. Expression of RBF under ey-Gal4 control caused a dramatic reduction of the adult eye (Figure 6B). This loss of eye tissue was suppressed virtually completely when CycD–Cdk4 was co-expressed with RBF (Figure 6H and Table I), providing further evidence that CycD–Cdk4 can functionally inactivate RBF. Interestingly, suppression of RBF was also observed when CycD was expressed without the Cdk4 kinase subunit (Figure 6C) or to a lesser extent when CycD was co-expressed with the kinase-impaired variant Cdk4D175N (Figure 6G and Table I). This suggests that CycD may suppress RBF function by utilizing an endogenous Cdk, or in a kinase-independent fashion.
Although ectopic RBF suppressed growth in developing wings, salivary glands and eyes, our previous analysis (Neufeld et al., 1998) suggested that growth inhibition by RBF was secondary to its effects on cell cycle progression. The tests described above are consistent with this interpretation, since in all cases both cell cycle progression (DNA replication) and growth were affected coincidentally. However, these tests cannot rule out the possibility that RBF inhibits cellular growth directly. Therefore we used the late acting eye specific drivers, GMR-Gal4 and sev-Gal4, to test whether RBF could suppress growth in non-cycling cells. Expression of RBF alone using these drivers had little effect on eye size (Table I; Du et al., 1996a), suggesting that RBF cannot suppress the post-mitotic growth that normally occurs in the eye. Moreover, co-expressed RBF did not substantially suppress eye overgrowth caused by GMR-Gal4- or sev-Gal4-driven CycD–Cdk4 (Figure 6J and K and Table I). Given this, a parsimonious interpretation of all these interaction tests (Figures 4–6) is that whereas CycD–Cdk4 can promote cellular growth in both proliferating and non-cycling cells, RBF can suppress growth only in cells that are undergoing cycles of DNA replication. In this case, growth suppression by ectopic RBF most likely stems from its ability to inhibit cell cycle progression.
Loss of RBF does not accelerate growth
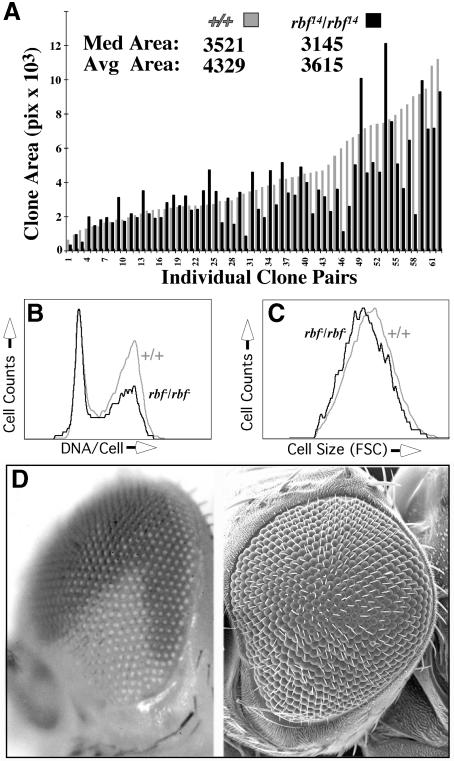
To further test RBF function we used FLP/FRT-mediated mitotic recombination to generate cell clones homozygous for a null allele of rbf (_rbf_14; Du and Dyson, 1999). We measured areas of _rbf_14/14 cell clones in wing discs, and compared these with the areas of their rbf+/+ sister clones (‘twin spots’). Cell clones mutant for rbf were slightly smaller than their wild-type twin spots, though the difference was not statistically significant (Figure 7A). Thus, loss of RBF did not confer a growth advantage. FACS analysis of _rbf_14/14 cells showed a reduced cell size and, surprisingly, an increased G1 population (Figure 7B and C). Microscopic examination revealed many pyknotic nuclei associated with _rbf_14/14 mutant clones, suggesting elevated levels of apoptosis. Reduced cell size and poor viability have also been observed in hyperproliferative cell clones overexpressing E2F/DP (Figure 1A; Neufeld et al., 1998), and similar characteristics have been described for mouse embryo fibroblasts lacking pRB, p107 or p130 (Herrera et al., 1996; LeCouter et al., 1998a,b). One explanation for these phenotypes is that they arise when cell division rates outpace rates of cell growth (Neufeld et al., 1998). This may also be the case for _rbf_14/14 cells.
Fig. 7. Loss of RBF does not promote overgrowth in the wing or the eye. (A) rbf_14/rbf_14 and +/+ sister clones (twin spots) were induced by FLP/FRT-mediated mitotic recombination at 48 h AED, and scored in wing imaginal discs at 115 h AED. The areas of both mutant and wild-type clones were measured; rbf_14/rbf_14 mutant clone areas are displayed as black bars, and are paired with their +/+ wild-type sister clones (gray bars). Data are arrayed according to size of the sister clone. Values for median and average clone areas are indicated. P >0.1. Genotype: w; FRT18A 2_π_myc/FRT18A rbf_14; hs-FLP/+._ (B) FACS analysis of rbf_14/rbf_14 cells (black trace) in wing discs, showing an increase in the G1 cell population relative to control cells (gray trace) from the same discs. Genotype: w; FRT18A P[w+ub-GFPnls]/FRT18A rbf_1_4; hs-FLP/+. (C) FACS analysis of rbf_14/rbf_14 cells (black trace) in wing discs, showing a decreased cell size as measured by FSC relative to internal control cells (gray trace). Genotype as in (B). (D) An eye containing a large rbf_14/rbf_14 clone (white area) generated using the Minute technique. An SEM of the same eye is shown to the right; note that the rbf_14/rbf_14 clone does not show enlarged ommatidia. Approximately 100 such eyes were examined. Genotype: w; FRT18A P[w+ub-GFPnls] M(1)15D_RpS52/FRT18A rbf_14; hs-FLP/+.
Finally, we generated large _rbf_14/14 clones in the eye using the Minute technique (see Materials and methods). Although these clones populated large fractions of the eye (Figure 7D) they did not exhibit the hypertrophic characteristics noted when CycD–Cdk4 was overexpressed (Figures 2 and 3). Instead, _rbf_14/14 clones in the adult eye exhibited slight to moderate hypoplasia and mild defects such as missing or duplicated inter-ommatidial bristles and fused ommatidiae (Figure 7D). Similar phenotypes have been noted in eyes overexpressing E2F (Du et al., 1996a) or cyclin E (Richardson et al., 1995), both of which promote extra cell division in the eye without increasing growth (Figure 3). These results are consistent with the interpretation that RBF functions to inhibit cell cycle progression, rather than to inhibit cellular growth directly. If this is the case, stimulation of cellular growth by CycD–Cdk4 must be, at least in part, independent of RBF.
Discussion
Drosophila CycD–Cdk4 promotes cellular growth
Here and in Meyer et al. (2000), we have used several sensitive assays to probe the in vivo function of Drosophila CycD–Cdk4. We find that Drosophila CycD–Cdk4 does not have a specialized function in promoting G1 progression, but instead promotes accumulation of cellular mass, i.e. growth (Figure 8). Stimulation of growth by ectopic Drosophila CycD–Cdk4 was clearly cell autonomous, and occurred in many different cell types. Interestingly, the cellular response to excess growth caused by CycD–Cdk4 varied. In proliferating epithelial cells of the wing disc, ectopic CycD–Cdk4 led to a more rapid cell cycle and more cells (hyperplasia), but had little effect on G1/S/G2 phasing or cell size. In endoreplicative cells in the salivary gland, ectopic CycD–Cdk4 caused excessive DNA endoreplication and increased the cell size (hypertrophy). In neuronal cells of the differentiating eye, overexpressed CycD–Cdk4 caused unscheduled cell proliferation, but also promoted growth in post-mitotic cells, leading to cellular hypertrophy. Although ectopic CycD–Cdk4 did promote extra cell cycles in a number of contexts, it did not appear to block developmentally programmed cell differentiation, or the exit from the cell cycle that normally accompanies it.
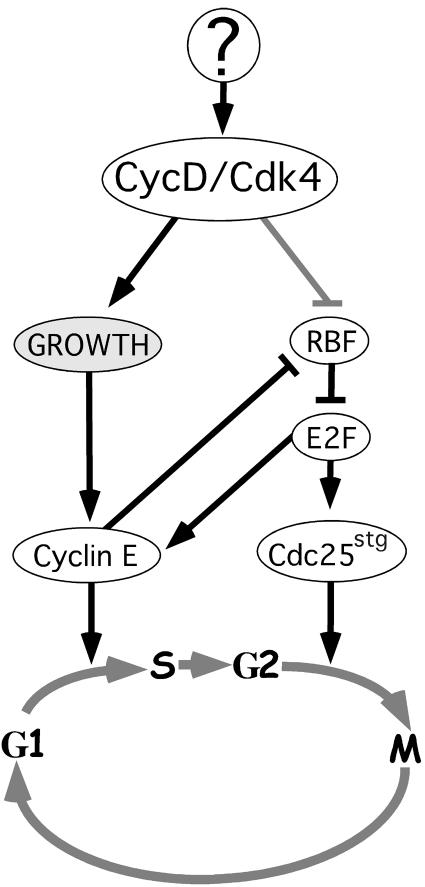
Fig. 8. Model for CycD–Cdk4 function based on data presented here and in Neufeld et al. (1998) and Prober and Edgar (2000).
The effects of CycD–Cdk4 differed profoundly from those of its presumed targets, RBF, E2F and CycE (Knoblich et al., 1994; Richardson et al., 1995; Asano et al., 1996; Du et al., 1996b; Neufeld et al., 1998; Weiss et al., 1998). In numerous assays, RBF, E2F and CycE behaved as specific cell cycle regulators which affected cellular growth only indirectly, via their effects on the cell cycle. It is also noteworthy that the cellular function we suggest for Drosophila CycD–Cdk4 differs from that proposed for CLN3, which is sometimes thought of as a yeast analog of cyclin D. Translational control and rapid protein turnover allow CLN3 levels to track the cell’s biosynthetic rate, making it in effect a ‘growth sensor’ (Polymenis and Schmidt, 1997). Drosophila CycD however, rather than measuring growth, actually stimulates it. In light of this it is interesting to note that our studies using other growth drivers in the fly wing (dMyc and Ras1) indicate that cyclin E levels are growth responsive and controlled post-transcriptionally (Prober and Edgar, 2000). Thus cyclin E, rather than cyclin D, may be the ‘growth sensor’ that links biosynthetic activity to G1/S progression in Drosophila (Figure 8).
While our findings differ from those in cultured mammalian cells, they are remarkably consistent with data from in vivo studies of the murine D-type cyclins. Just as overexpressing CycD–Cdk4 in the fly wing or eye caused hyperplasia, targeted overexpression of CycD1 in mice can promote epidermal, mammary and thymic hyperplasia (Wang et al., 1994; Robles et al., 1996; Rodriguez-Puebla et al., 1999), and mutational activation of Cdk4 can cause pancreatic hyperplasia (Rane et al., 1999). Likewise, overexpressed cyclin D3 can promote endomitosis in platelets (Zimmet et al., 1997), much as overexpressed Drosophila CycD–Cdk4 drives extra DNA endoreplication in salivary gland cells. Overexpression of a D-type cyclin in plants has also been shown to accelerate growth (Cockcroft et al., 2000), suggesting that the ability of Cdk–cyclin complexes to stimulate biosynthesis may be widely conserved. These parallels are also evident in loss-of-function studies. CycD1 knockout mice are smaller than their littermates and display hypoplasia of the retina and mammary epithelium, tissues that normally express CycD1 at high levels (Fantl et al., 1995; Sicinski et al., 1995). Mice lacking CycD2 display hypoplastic growth phenotypes (Sicinski et al., 1996). Cdk4 mutant mice, like Cdk4 mutant Drosophila, are also small (Rane et al., 1999; Tsutsui et al., 1999).
Although it is not yet clear whether these growth effects in other systems are cell autonomous, studies of mouse embryo fibroblasts (MEFs) suggest that some are. Proliferating MEFs lacking CycD1 or Cdk4 have normal G1/S/G2 profiles and so, like our studies in flies, cast doubt upon a direct role for CycD–Cdk4 in G1–S progression. However, CycD1 or Cdk4 mutant MEFs are delayed in activating DNA replication upon serum stimulation (Fantl et al., 1995; Rane et al., 1999; Tsutsui et al., 1999), an effect that could be attributed to impaired growth. MEFs from mice lacking p16_INK4A_, a specific inhibitor of CycD-dependent kinases, also exhibit accelerated growth (Serrano et al., 1993). Taken together, all of these observations fall into place to support the view that one cellular function of cyclin D–Cdk complexes is to stimulate cellular growth.
Does CycD–Cdk4 act through RBF?
D-type cyclins are believed to function primarily by suppressing the function of the ‘pocket’ proteins, pRb, p107 and p130 (Beijersbergen and Bernards, 1996). We describe numerous genetic interactions between CycD–Cdk4 and RBF that indicate in vivo cross-regulation of these gene products. CycD–Cdk4 counteracted the growth-suppressive effects of RBF in wing and eye imaginal discs (Figures 4 and 6), and also in the endoreplicating salivary gland (Figure 5). As in vertebrates, these interactions might reflect direct phosphorylation and neutralization of RBF by CycD–Cdk4 kinase (see also Meyer et al., 2000). However, these interactions could also be indirect consequences of the effects of CycD–Cdk4 on cellular growth. For instance, growth stimulation by CycD–Cdk4 may increase the activity of CycE–Cdk2, which is also a potent suppressor of RBF activity (Figure 8; Du et al., 1996a; Neufeld et al., 1998;Meyer et al., 2000).
Although the CycD–RBF interactions we describe confirm current paradigms to some extent, they also highlight the differences between CycD–Cdk4 and RBF function. For instance, RBF’s ability to suppress growth was limited to cells actively undergoing DNA replication, such as proliferating cells in the imaginal discs (Figures 4 and 6) and endoreplicating cells in the salivary gland (Figure 5). In the post-mitotic eye, where CycD–Cdk4 is a potent stimulator of growth, RBF is virtually inert (Du et al., 1996a; Figure 6K). Moreover, loss of RBF did not enhance growth in either proliferating or post-mitotic cells, and thus did not phenocopy gain of CycD–Cdk4 function. Indeed, all the data on RBF suggest that it functions specifically to suppress cell cycle progression (see also Du et al., 1996a; Neufeld et al., 1998; Du and Dyson, 1999), and that its growth-suppressive effects are secondary consequences of this function. CycD–Cdk4, in contrast, promoted growth in both proliferating and post-mitotic cells. An important implication of our analysis is, therefore, that Drosophila CycD–Cdk4 must promote growth via targets other than RBF. These growth-regulatory targets remain unknown, but the requirement for a catalytically active Cdk4 subunit suggests that they are phosphorylation substrates. Potential candidates include pocket proteins other than RBF, or factors that affect biosynthesis directly such as regulators of protein synthesis or turnover. Consistently, recent reports describe non-Rb targets for mammalian CycD1 (Hirai and Sherr, 1996; Zwijsen et al., 1997), and indicate that the transforming activity of CycD1 does not require Rb binding (Zwicker et al., 1999).
How CycD–Cdk4 fits into the hierarchy of genes that regulate growth in Drosophila is unclear, and will remain obscure until mutations in the cycD gene are identified. It is tempting to speculate that CycD–Cdk4 might mediate the growth effects of the WNT, BMP and EGF sig naling pathways, which orchestrate patterned growth in Drosophila and throughout the animal kingdom. However, there are presently scant data to support this hypothesis in flies (but see Diehl et al., 1998). Recent work has revealed that insulin receptor/Pi3K/AKT signaling is also an important growth control pathway in Drosophila, and may respond to nutritional conditions (Böhni et al., 1999; Verdu et al., 1999; Weinkove et al., 1999). Like CycD–Cdk4, genes in this pathway appear to be growth specific and to have little role in tissue patterning or differentiation. But there are clear distinctions between CycD–Cdk4-driven growth and Pi3K-driven growth. In the wing for instance, CycD–Cdk4 accelerates growth in a balanced fashion such that cell cycle phasing and cell size remain normal, whereas Pi3K-driven growth is characterized by a truncated G1 phase and increased cell size (Weinkove et al., 1999). The origin of these differences is presently unclear. One plausible explanation is that CycD–Cdk4 can promote G2–M progression, perhaps by repressing RBF and activating Cdc25/Stg (Figure 8; Neufeld et al., 1998), whereas Pi3K cannot. In support of this idea it has recently been shown that human Cdk4 activity is required for the G2–M transition in HeLa cells (Gabrielli et al., 1999).
There is an abundance of correlative evidence associating cyclin D with diverse cancers (Wang et al., 1994). It is telling that nearly all cases are associated with elevated levels of cyclin D function as a result of amplification or translocation of the cyclin D locus, or inactivation of the cyclin D-specific inhibitor, p16INK4A. When we increased cyclin D–Cdk4 activity in Drosophila, we induced overgrowth as well. In thinking about how the human D-type cyclins are involved in the progression of cancer, it may prove fruitful to consider them, not as a regulators of G1 progression, but as promoters of cellular growth.
Materials and methods
Fly stocks
We identified a Drosophila cyclin D cDNA clone, LD05713 (Berkeley Drosophila Genome Project/HHMI EST Project, accession No. AA246773), which in addition to the 452 amino acids previously identified (Finley et al., 1996), encodes 25 or 29 N-terminal amino acids within its putative coding region (two potential initiator methionines are at position 1 or 5, in bold type): M A D I M D L L C S E I I V Y E S D P S L Y R L N K R Q Q. This full-length cDNA includes the pRb-binding motif, LXCXE (underlined) and is 39% identical to human cyclins D1 and D2 at the amino acid level. A 2.1 kb _Not_I–_Kpn_I fragment of LD05713 was ligated into pUAST (Brand and Perrimon, 1993). UAS-CycD transgenics (chromosomes II and III) were generated by P-element-mediated transformation.
Transgenes (chromosome)
UAS-E2F (III) (Neufeld et al., 1998); UAS-DP (III) (gift from N.Dyson); UAS-CycE (III) (Lane et al., 1996); UAS-RBF (III) (Du et al., 1996a; Neufeld et al., 1998); actin 5c>CD2>Gal4 (III) (Pignoni and Zipursky, 1997); UAS-GFP(S65T)nls (II, III) (Neufeld et al., 1998); GMR-Gal4 (II) (Freeman, 1996); ey-Gal4 (gift from Barry Dickson); UAS-Cdk4 (II) (Meyer et al., 2000); UAS-Cdk4D175N (Meyer et al., 2000); Ub-GFP(S65T)nls #27 (I) (gift from Welcome Bender).
Proliferation and growth rate measurements
Larvae from 2 h egg collections were transferred to yeasted vials, 50 larvae/vial, 24 h after egg deposition (AED). Overexpression clones were induced at 37°C for 20 min or 34.2°C for 30 min at either 48 or 72 h AED and were evaluated at 115 h AED for cell count or clone size as in Neufeld et al. (1998). When counting cells/clone for cell division rates, ideal clone density was 5–10 clones/disc. Clones were imaged on a Bio-Rad MRC-600 or a Leica TCSSP confocal microscope. Cell doubling times were derived using the formula (log2/log_n_)h, where n = median number of cells per clone and h = age of clone. P values were calculated using a two tailed student’s _t_-test. Controls were done in parallel, and each experiment was scored blind and performed at least twice. Mitotic clones were induced using the FLP/FRT technique (Xu and Rubin, 1993), imaged on a Leica TCSSP confocal microscope at 20× magnification, and clone areas measured using the histogram function of Adobe Photoshop. FACS analysis was performed as in Neufeld et al. (1998).
Histology
Larval imaginal discs were treated as in Neufeld et al. (1998). Pupal discs were dissected 48 h after puparium formation (APF) and stained as described (Hariharan et al., 1995). Images were captured on a Deltavision SA/30 microscope (Applied Precision). Apoptosis was analyzed as in Neufeld et al. (1998) or by observing pyknotic nuclei in the basal plane of the disc epithelium. Electron microscopy was performed on a JEOL JSM5800 scanning electron microscope as described (Kimmel et al., 1990).
Supplementary material
Supplementary material to this paper is available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Wei Du and Russ Finley for sharing flies, reagents and unpublished data, Chuck Sherr for discussing data prior to publication; Kristy Seidel for statistical analysis; the FHCRC Biotechnology, Flow Cytometry, Electron Microscopy and Image Analysis Facilities; Tom Neufeld, Leslie Saucedo and Jon Cooper for advice and discussion; and members of the Edgar lab for comments on the manuscript. S.A.D. was supported by the Medical Scientist Training Program at the University of Washington and NIH Training Grant T32 HD07183. B.A.E. is a Rita Allen Scholar and is supported by NIH R01 GM51186.
References
- Asano M., Nevins,J.R. and Wharton,R.P. (1996) Ectopic E2F expression induces S-phase and apoptosis in Drosophila imaginal discs. Genes Dev., 10, 1422–1432. [DOI] [PubMed] [Google Scholar]
- Beijersbergen R.L. and Bernards,R. (1996) Cell cycle regulation by the retinoblastoma family of growth inhibitory proteins. Biochim. Biophys. Acta, 1287, 103–120. [DOI] [PubMed] [Google Scholar]
- Böhni R., Riesgo-Escovar,J., Oldham,S., Brogiolo,W., Stocker,H., Andruss,B.F., Beckingham,K. and Hafen,E. (1999) Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell, 97, 865–875. [DOI] [PubMed] [Google Scholar]
- Brand A.H. and Perrimon,N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Cockcroft C.E., den Boer,B.G., Healy,J.M. and Murray,J.A. (2000) Cyclin D control of growth rate in plants. Nature, 405, 575–579. [DOI] [PubMed] [Google Scholar]
- Connell-Crowley L., Harper,J.W. and Goodrich,D.W. (1997) Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol. Biol. Cell, 8, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datar S.A. (2000) Developmental regulation of growth and cell cycle progression in Drosophila melanogaster: a larval growth arrest screen and molecular and genetic analysis of the Cyclin D/Cdk4 complex. In Molecular Cell Biology. University of Washington, Seattle, WA, p. 147. [Google Scholar]
- Diehl J.A., Cheng,M., Roussel,M.F. and Sherr,C.J. (1998) Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev., 12, 3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W. and Dyson,N. (1999) The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. EMBO J., 18, 916–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Vidal,M., Xie,J.-E. and Dyson,N. (1996a) RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev., 10, 1206–1218. [DOI] [PubMed] [Google Scholar]
- Du W., Xie,J.E. and Dyson,N. (1996b) Ectopic expression of dE2F and dDP induces cell proliferation and death in the Drosophila eye. EMBO J., 15, 3684–3692. [PMC free article] [PubMed] [Google Scholar]
- Dyson N. (1998) The regulation of E2F by pRB-family proteins. Genes Dev., 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- Fantl V., Stamp,G., Andrews,A., Rosewell,I. and Dickson,C. (1995) Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev., 9, 2364–2372. [DOI] [PubMed] [Google Scholar]
- Finley R.L. Jr, Thomas,B.J., Zipursky,S.L. and Brent,R. (1996) Isolation of Drosophila cyclin D, a protein expressed in the morphogenetic furrow before entry into S phase. Proc. Natl Acad. Sci. USA, 93, 3011–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. (1996) Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell, 87, 651–660. [DOI] [PubMed] [Google Scholar]
- Friend S.H., Dryja,T.P. and Weinberg,R.A. (1988) Oncogenes and tumor-suppressing genes. N. Engl. J. Med., 318, 618–622. [DOI] [PubMed] [Google Scholar]
- Gabrielli B.G., Sarcevic,B., Sinnamon,J., Walker,G., Castellano,M., Wang,X.Q. and Ellem,K.A. (1999) A cyclin D–Cdk4 activity required for G2 phase cell cycle progression is inhibited in ultraviolet radiation-induced G2 phase delay. J. Biol. Chem., 274, 13961–13969. [DOI] [PubMed] [Google Scholar]
- Geng Y., Whoriskey,W., Park,M.Y., Bronson,R.T., Medema,R.H., Li,T., Weinberg,R.A. and Sicinski,P. (1999) Rescue of cyclin D1 deficiency by knockin cyclin E. Cell, 97, 767–777. [DOI] [PubMed] [Google Scholar]
- Harbour J.W., Luo,R.X., Dei Santi,A., Postigo,A.A. and Dean,D.C. (1999) Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell, 98, 859–869. [DOI] [PubMed] [Google Scholar]
- Hariharan I.K., Hu,K.Q., Asha,H., Quintanilla,A., Ezzell,R.M. and Settleman,J. (1995) Characterization of rho GTPase family homologues in Drosophila melanogaster: overexpressing Rho1 in retinal cells causes a late developmental defect. EMBO J., 14, 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera R.E., Sah,V.P., Williams,B.O., Mäkelä,T.P., Weinberg,R.A. and Jacks,T. (1996) Altered cell cycle kinetics, gene expression and G1 restriction point regulation in _Rb_-deficient fibroblasts. Mol. Cell. Biol., 16, 2402–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H. and Sherr,C.J. (1996) Interaction of D-type cyclins with a novel myb-like transcription factor, DMP1. Mol. Cell. Biol., 16, 6457–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Kahn,S.M., Zhou,P., Zhang,Y.J., Cacace,A.M., Infante,A.S., Doi,S., Santella,R.M. and Weinstein,I.B. (1993) Overexpression of cyclin D1 in rat fibroblasts causes abnormalities in growth control, cell cycle progression and gene expression. Oncogene, 8, 3447–3457. [PubMed] [Google Scholar]
- Kato J.Y., Matsuoka,M., Strom,D.K. and Sherr,C.J. (1994) Regulation of cyclin D-dependent kinase 4 (cdk4) by cdk4-activating kinase. Mol. Cell. Biol., 14, 2713–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel B.E., Heberlein,U. and Rubin,G.M. (1990) The homeo domain protein Rough is expressed in a subset of cells in the developing Drosophila eye where it can specify photoreceptor cell subtype. Genes Dev., 4, 712–727. [DOI] [PubMed] [Google Scholar]
- Knoblich J.A., Sauer,K., Jones,L., Richardson,H., Saint,R. and Lehner,C.F. (1994) Cyclin E controls S-phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell, 77, 107–120. [DOI] [PubMed] [Google Scholar]
- Lane M.E., Sauer,K., Wallace,K., Jan,Y.N., Lehner,C.F. and Vaessin,H. (1996) Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell, 87, 1225–1235. [DOI] [PubMed] [Google Scholar]
- LeCouter J.E., Kablar,B., Hardy,W.R., Ying,C., Megeney,L.A., May,L.L. and Rudnicki,M.A. (1998a) Strain-dependent myeloid hyperplasia, growth deficiency and accelerated cell cycle in mice lacking the Rb-related p107 gene. Mol. Cell. Biol., 18, 7455–7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCouter J.E., Kablar,B., Whyte,P.F., Ying,C. and Rudnicki,M.A. (1998b) Strain-dependent embryonic lethality in mice lacking the retinoblastoma-related p130 gene. Development, 125, 4669–4679. [DOI] [PubMed] [Google Scholar]
- Leng X., Connel-Crowley,L., Goodrich,D. and Harper,J.W. (1997) S-phase entry upon ectopic expression of G1 cyclin-dependent kinases in the absence of retinoblastoma protein phosphorylation. Curr. Biol., 7, 709–712. [DOI] [PubMed] [Google Scholar]
- Lukas J., Muller,H., Bartkova,J., Spitkovsky,D., Kjerulff,A.A., Jansen-Durr,P., Strauss,M. and Bartek,J. (1994) DNA tumor virus oncoproteins and retinoblastoma gene mutations share the ability to relieve the cell’s requirement for cyclin D1 function in G1. J. Cell Biol., 125, 625–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushime H., Roussel,M.F., Ashmun,R.A. and Sherr,C.J. (1991) Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell, 65, 701–713. [DOI] [PubMed] [Google Scholar]
- Matsushime H., Quelle,D.E., Shurtleff,S.A., Shibuya,M., Sherr,C.J. and Kato,J.Y. (1994) D-type cyclin-dependent kinase activity in mammalian cells. Mol. Cell. Biol., 14, 2066–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C.A., Jacobs,H.W., Datar,S.A., Du,W., Edgar,B.A. and Lehner,C.F. (2000) Drosophila Cdk4 is required for normal growth and dispensable for cell cycle progression. EMBO J., 19, 4533–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motokura T., Bloom,T., Kim,H.G., Jüppner,H., Ruderman,J.V., Kronenberg,H.M. and Arnold,A. (1991) A novel cyclin encoded by a bcl1-linked candidate oncogene. Nature, 350, 512–515. [DOI] [PubMed] [Google Scholar]
- Muise-Helmericks R.C., Grimes,H.L., Bellacosa,A., Malstrom,S.E., Tschilis,P.N. and Rosen,N. (1998) Cyclin D expression is controlled post-transcriptionally via a phosphatidylinositol 3-kinase/AKT-dependent pathway. J. Biol. Chem., 273, 29864–29872. [DOI] [PubMed] [Google Scholar]
- Neufeld T.P., de la Cruz,A.F.A., Johnston,L.A. and Edgar,B.A. (1998) Coordination of growth and cell division in the Drosophila wing. Cell, 93, 1183–1193. [DOI] [PubMed] [Google Scholar]
- Pignoni F. and Zipursky,S. (1997) Induction of Drosophila eye development by decapentalegic. Development, 124, 271–278. [DOI] [PubMed] [Google Scholar]
- Polymenis M. and Schmidt,E.V. (1997) Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev., 11, 2522–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenis M. and Schmidt,E.V. (1999) Coordination of cell growth with cell division. Curr. Opin. Genet. Dev., 9, 76–80. [DOI] [PubMed] [Google Scholar]
- Prober D.A. and Edgar,B.A. (2000) Ras promotes cellular growth in the Drosophila wing. Cell, 100, 435–446. [DOI] [PubMed] [Google Scholar]
- Quelle D.E., Ashmun,R.A., Shurtleff,S.A., Kato,J.-y., Bar-Sagi,D., Rousel,M.F. and Sherr,C.J. (1993) Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev., 7, 1559–1571. [DOI] [PubMed] [Google Scholar]
- Rane S.G., Dubus,P., Mettus,R.V., Galbreath,E.J., Boden,G., Reddy,E.P. and Barbacid,M. (1999) Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in β-islet cell hyperplasia. Nature Genet., 22, 44–52. [DOI] [PubMed] [Google Scholar]
- Resnitzky D. and Reed,S.I. (1995) Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol. Cell. Biol., 15, 3463–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnitzky D., Gossen,M., Bujard,H. and Reed,S.I. (1994) Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol., 14, 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson H., O’Keefe,L.V., Marty,T. and Saint,R. (1995) Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development, 121, 3371–3379. [DOI] [PubMed] [Google Scholar]
- Robles A.I., Larcher,F., Whalin,R.B., Murillas,R., Richie,E., Gimenez-Conti,I.B., Jorcano,J.L. and Conti,C.J. (1996) Expression of cyclin D1 in epithelial tissues of transgenic mice results in epidermal hyperproliferation and severe thymic hyperplasia. Proc. Natl Acad. Sci. USA, 93, 7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Puebla M.L., LaCava,M. and Conti,C.J. (1999) Cyclin D1 overexpression in mouse epidermis increases cyclin-dependent kinase activity and cell proliferation in vivo but does not affect skin tumor development. Cell Growth Differ., 10, 467–472. [PubMed] [Google Scholar]
- Rosenwald I.B. (1996) Deregulation of protein synthesis as a mechanism of neoplastic transformation. BioEssays, 18, 243–250. [DOI] [PubMed] [Google Scholar]
- Rosenwald I.B., Lazaris-Karatzas,A., Sonenberg,N. and Schmidt,E.V. (1993) Elevated levels of cyclin D1 protein in response to increased expression of eukaryotic initiation factor 4E. Mol. Cell. Biol., 13, 7358–7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K., Weigmann,K., Sigrist,S. and Lehner,C.F. (1996) Novel members of the cdc2-related kinase family in Drosophila: cdk4/6, cdk5, PFTAIRE and PITSLRE kinase. Mol. Biol. Cell, 7, 1759–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Hannon,G.J. and Beach,D. (1993) A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature, 366, 704–707. [DOI] [PubMed] [Google Scholar]
- Sewing A., Burger,C., Brusselbach,S., Schalk,C., Lucibello,F.C. and Muller,R. (1993) Human cyclin D1 encodes a labile nuclear protein whose synthesis is directly induced by growth factors and suppressed by cyclic AMP. J. Cell Sci., 104, 545–555. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. (1996) Cancer cell cycles. Science, 274, 1672–1677. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. and Roberts,J.M. (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev., 13, 1501–1512. [DOI] [PubMed] [Google Scholar]
- Sicinski P., Donaher,J.L., Parker,S.B., Li,T., Fazeli Am Gardner,H., Haslam,S.Z., Bronson,R.T., Elledge,S.J. and Weinberg,R.A. (1995) Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell, 82, 621–630. [DOI] [PubMed] [Google Scholar]
- Sicinski P. et al. (1996) Cyclin D2 is a FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature, 384, 470–474. [DOI] [PubMed] [Google Scholar]
- Tsutsui T., Hesabi,B., Moons,D.S., Pandolfi,P.P., Hansel,K.S., Koff,A. and Kiyokawa,H. (1999) Targeted disruption of CDK4 delays cell cycle entry with enhanced p27 (Kip1) activity. Mol. Cell. Biol., 19, 7011–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu J., Burtovich,M.A., Wilder,E.L. and Birnbaum,M.J. (1999) Cell autonomous regulation of cell and organ growth in Drosophila by AKT/PKB. Nature Cell Biol., 1, 500–513. [DOI] [PubMed] [Google Scholar]
- Wang T.C., Cardiff,R.D., Zukerberg,L., Lees,E., Arnold,A. and Schmidt,E.V. (1994) Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature, 369, 669–671. [DOI] [PubMed] [Google Scholar]
- Weinkove D., Neufeld,T.P., Twardzik,T., Waterfield,M.D. and Leevers,S.J. (1999) Regulation of imaginal disc cell size, cell number and organ size by Drosophila class IA phosphoinositide 3-kinase and its adaptor. Curr. Biol., 9, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Weiss A., Herzig,A., Jacobs,H. and Lehner,C.F. (1998) Continuous Cyclin E expression inhibits progression through endoreduplication cycles in Drosophila. Curr. Biol., 8, 239–242. [DOI] [PubMed] [Google Scholar]
- Withers D.A., Harvey,R.C., Faust,J.B., Melnyk,O., Carey,K. and Meeker,T.C. (1991) Characterization of a candidate bcl-1 gene. Mol. Cell. Biol., 11, 4846–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T. and Rubin,G.M. (1993) Analysis of genetic mosaics in developing and adult Drosophila tissues. Development, 117, 1223–1237. [DOI] [PubMed] [Google Scholar]
- Zimmet J.M., Ladd,D., Jackson,C.W., Stenberg,P.E. and Ravid,K. (1997) A role for cyclin D3 in the endomitotic cell cycle. Mol. Cell. Biol., 17, 7248–7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwicker J., Brusselbach,S., Jooss,K.U., Sewing,A., Behn,M., Lucibello,F.C. and Muller,R. (1999) Functional domains in cyclin D1: pRb-kinase activity is not essential for transformation. Oncogene, 18, 19–25. [DOI] [PubMed] [Google Scholar]
- Zwijsen R.M., Wientjens,E., Klompmaker,R., van der Sman,J., Bernards,R. and Michalides,R.J. (1997) CDK-independent activation of estrogen receptor by cyclin D1. Cell, 88, 405–415. [DOI] [PubMed] [Google Scholar]