Conserved Organization of Centromeric Chromatin in Flies and Humans (original) (raw)
. Author manuscript; available in PMC: 2011 Oct 13.
Summary
Recent studies have highlighted the importance of centromere-specific histone H3-like (CENP-A) proteins in centromere function. We show that Drosophila CID and human CENP-A appear at metaphase as a three-dimensional structure that lacks histone H3. However, blocks of CID/CENP-A and H3 nucleosomes are linearly interspersed on extended chromatin fibers, and CID is close to H3 nucleosomes in polynucleosomal preparations. When CID is depleted by RNAi, it is replaced by H3, demonstrating flexibility of centromeric chromatin organization. Finally, contrary to models proposing that H3 and CID/CENP-A nucleosomes are replicated at different times in S phase, we show that interspersed H3 and CID/CENP-A chromatin are replicated concurrently during S phase in humans and flies. We propose that the unique structural arrangement of CID/CENP-A and H3 nucleosomes presents centromeric chromatin to the poleward face of the condensing mitotic chromosome.
Introduction
Proper segregation of duplicated sister chromatids to daughter cells during cell division is required for both cell and organism viability. The centromere/kinetochore complex is the chromosomal structure that mediates chromosome attachment to kinetochore microtubules and mitotic chromosome movement. Much is known about the proteins involved in kinetochore-mediated chromosome movement and about the role of the kinetochore in the spindle attachment checkpoint (Rieder and Khodjakov, 1997; Shah and Cleveland, 2000). Less is known about the proteins involved in assembling the kinetochore and in choosing the site for kinetochore assembly (Sullivan et al., 2001; Sullivan, 2001).
In higher eukaryotes, determination of the site of kinetochore formation (centromere identity) does not appear to depend on primary DNA sequence and instead may be regulated by an epigenetic mechanism (Karpen and Allshire, 1997; Sullivan et al., 2001). It has been suggested that the evolutionarily conserved centromere-specific histone H3-like protein CENP-A (CENtromere Protein-A) is involved in determination of the site of kinetochore formation. Several recent studies in a variety of organisms support this hypothesis, as all kinetochore proteins examined are mislocalized when CENP-A is disrupted (Blower and Karpen, 2001; Howman et al., 2000; Moore and Roth, 2001; Oegema et al., 2001). Therefore, CENP-A appears to form a link between the centromeric DNA and the protein components of the kinetochore.
CENP-A is also likely to be involved in establishing a centromere-specific chromatin structure. CENP-A was originally identified as a human autoantigen that copurified with nucleosomes from human cells (Palmer et al., 1987, 1991). Genetic evidence from S. cerevisiae suggests that histones H4 and H2A are involved in establishing the CENP-A homolog Cse4-containing centromeric nucleosome (Glowczewski et al., 2000; Meluh et al., 1998; Pinto and Winston, 2000). Overexpression of histone H3 is synthetically lethal with a Cse4 mutation, suggesting that Cse4 replaces both copies of H3 in the centromeric nucleosome (Glowczewski et al., 2000). In vitro nucleosome reconstitution using purified human CENP-A and histones H2A, H2B, and H4 has demonstrated that CENP-A nucleosomes can be homotypic, and they appear to be virtually indistinguishable from H3-containing nucleosomes (Yoda et al., 2000). The histone fold/dimerization domain of CENP-A is required for centromere targeting, and equal amounts of native CENP-A and epitope-tagged CENP-A coimmunoprecipitate from mononucleosomes (Shelby et al., 1997). These studies suggest that centromeric nucleosomes consist of CENP-A dimers and that they do not contain H3. However, no studies of the histone composition of CENP-A-containing nucleosomes in vivo, or of their physical relationship to H3-containing nucleosomes, have been reported for the larger centromeres of higher eukaryotes.
In the present study, we investigated the structure of CENP-A chromatin at fly and human metaphase kinetochores, using three-dimensional (3D) deconvolution microscopy. Next, an extended chromatin fiber technique was developed to examine the relationship between CENP-A and H3-containing nucleosomes at high resolution, in flies and humans. We also biochemically evaluated the in vivo composition of CENP-A nucleosomes and surrounding chromatin using the Drosophila CENP-A homolog (CID). Finally, extended chromatin fibers allowed us to observe the effects of CID depletion on the composition and organization of centromeric chromatin and to examine the pattern of replication in human and fly centromeric chromatin. These studies revealed conserved structural and functional features of higher eukaryotic centromeres and suggest a model for how centromeric chromatin and kinetochores are organized on metaphase chromosomes.
Results
Centromeric Chromatin Forms a Conserved Three-Dimensional Structure on Metaphase Chromosomes, which Is Separated from Inner and Outer Kinetochore Components
There has been speculation that centromeric chromatin may form a unique higher order structure; however, no detailed studies of the 3D organization of centromeric chromatin have been reported. We examined the 3D organization of centromeric chromatin in metaphase chromosomes using antibodies to Drosophila CID and human CENP-A, indirect immunofluorescence, and deconvolution microscopy (see Experimental Procedures). Three-dimensional deconvolution analysis demonstrated that CID/CENP-A chromatin in both flies and humans is present in a cylinder-like structure that extends through the depth of the chromosome (Figure 1). Z axis distortion is common to 3D reconstructions, but, in this instance, it was not sufficient to account for the depth of the observed structure (see Experimental Procedures). In addition, the presence of the CID 3D structure in Drosophila has been confirmed by EM tomography (data not shown). It has been difficult to assign a precise shape to the CENP-A/CID 3D structure, but for simplicity it will be referred to hereafter as cylindrical.
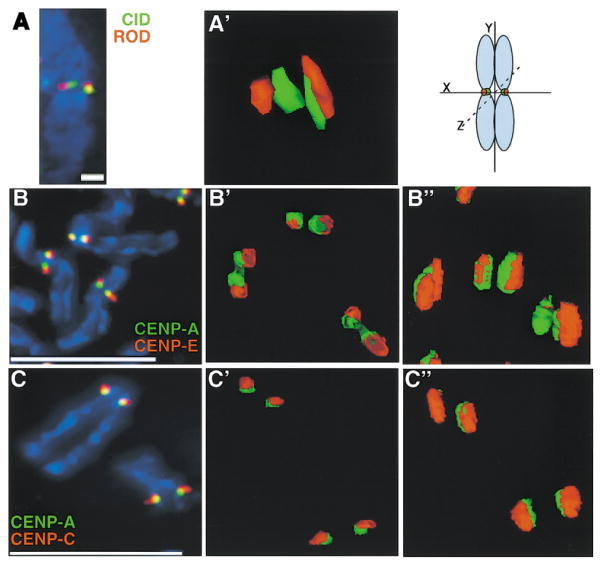
Figure 1. CENP-A Proteins form a Higher Order Structure on Metaphase Chromosomes in both Drosophila and Humans.
(A) CID (green) and ROD (Rough Deal) (red) were colocalized on three-dimensional meta-phase chromosomes from Drosophila S2 cells and showed typical double-dot staining by immunofluorescence and subsequent modeling of antibody staining.
(A′) Rotation of the chromosome 90° around the x axis showed that the double-dot staining of CID appears as a cylindrical structure that spans the depth of the chromosome, and that ROD is located next to, but minimally overlaps with, the CID cylinder. Scale bar is 5 microns.
(B and B′) CENP-E (red), a mammalian outer kinetochore protein, shows minimal overlap with CENP-A (green) by deconvolution microscopy.
(B″) Rotation of the chromosome by 35° around the x axis showed a similar cylindrical structure for CENP-A as that observed for CID at Drosophila kinetochores. Scale bar is 15 microns.
(C and C′) CENP-C (red), an inner kinetochore protein, typically shows more overlap with CENP-A (green) by 2D immunofluorescence than does CENP-E.
(C″) Rotation by 70° around the x axis showed that CENP-C also appears to be wrapped around the CENP-A structure and does not overlap significantly with CENP-A. Scale bar 15 microns. See text and Experimental Procedures for quantitation of the extent of overlap.
Previous two-dimensional (2D) immunofluorescence studies of mammalian chromosomes have suggested that CENP-A is located at the inner kinetochore plate, since it showed only partial overlap with outer kinetochore components and complete colocalization with another inner kinetochore component CENP-C (Warburton et al., 1997). We examined the 3D organization of centromeric chromatin and kinetochore components on metazoan metaphase chromosomes by simultaneously colocalizing Drosophila CID with outer kinetochore proteins (ROD [Rough Deal], ZW10 [Zeste-white 10], and human CENP-A with the outer kinetochore protein CENP-E [Cooke et al., 1997; Scaerou et al., 1999; Sunkel and Glover, 1988; Williams et al., 1998]). We also compared localization of human inner kinetochore proteins CENP-A and CENP-C; other inner kinetochore proteins in Drosophila have not yet been identified. Surprisingly, deconvolution microscopy revealed that the human inner kinetochore protein CENP-C did not completely colocalize with CENP-A and instead appeared to wrap around and partially overlap with the poleward face of CENP-A chromatin (Figure 1; see quantitation, below). Outer kinetochore proteins in flies and humans showed little overlap with CID/CENP-A and instead appeared to wrap around the CID or CENP-A/CENP-C structure (Figures 1A–1C). The domain of CENP-C antibody staining, as well as staining for outer kinetochore proteins, extended over both the top and bottom of the CENP-A structure, perhaps accounting for the overlap observed in standard 2D immunofluorescence experiments.
To examine the relationship between centromeric chromatin and the inner and outer kinetochore plates in more detail, we quantitated the extent of overlap using 3D immunofluorescence of metaphase chromosomes from human cell lines. Both GFP-tagged CENP-A and antibodies to endogenous or FLAG-tagged CENP-A were used in separate experiments on chromosomes that were prepared without formaldehyde fixation, to allow for maximum antibody accessibility. CENP-A primarily showed partial overlap (53% ± 20% of total CENP-A signal) with CENP-C antibody staining (92%, n = 35), and less frequently demonstrated complete overlap (8%, n = 3) (see Experimental Procedures). Like- wise, antibodies to CENP-E, an outer kinetochore protein, showed partial overlap (60% ± 12% of total CENP-C signal) with CENP-C antibody staining at most kinetochores (92%, n = 24), and only a few kinetochores demonstrating complete overlap (8%, n = 2). We conclude that CENP-E only partially overlaps and lies external to CENP-C and that CENP-C only partially overlaps and is located external to CENP-A. Therefore, the existing view of the spatial organization of the centromere-kinetochore complex as a purely trilaminar structure, with CENP-A and CENP-C composing the inner plate, should be modified (see Discussion).
Centromeric Chromatin in Metaphase Chromosomes Contains Histones H2A and H2B, but Not H3 or Phosphorylated H3
Cytological studies have suggested that CID-containing chromatin is completely devoid of ectopically expressed histone H3 (Ahmad and Henikoff, 2001) and that histone H3 modified by phosphorylation at serine 10 is excluded from centromeric chromatin in mammalian cells (Hendzel et al., 1997; Van Hooser et al., 1999). However, these studies did not determine if all H3, or only phosphorylated H3, is excluded from centromeric chromatin and if other native histones localize to centromeric chromatin.
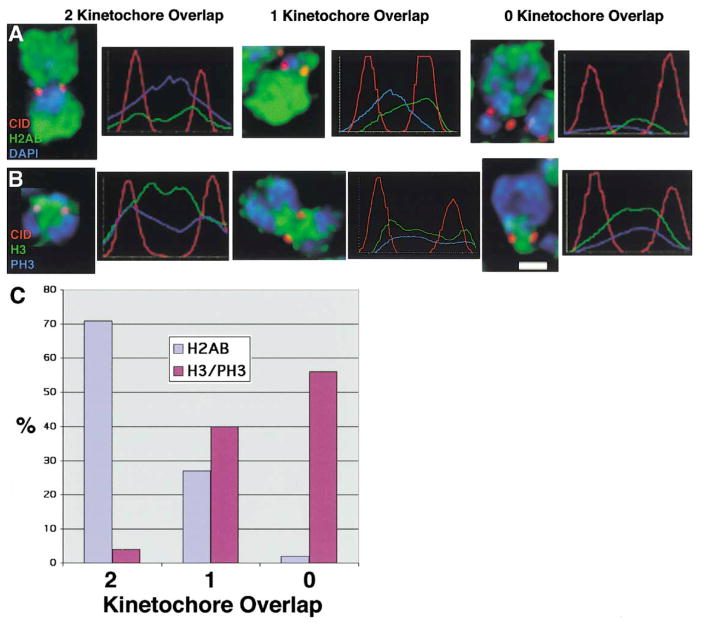
We examined the distribution of CID and core histones H2A, H2B, H3, and phosphorylated H3 (PH3), on meta-phase chromosomes, using indirect immunofluorescence and deconvolution microscopy. Distributions of these proteins were quantitated by measuring fluorescence intensities across both centromeres. CID chromatin spots colocalized extensively with histones H2AB across both centromeres (71%, n = 37), and rarely showed colocalization with only one centromere (27%, n = 14), or neither centromere (2%, n = 1) (Figures 2A and 2C). In contrast and significantly different from H2AB, H3 and PH3 overlapped minimally with CID in the area between the sister centromeres but most often was absent from both centromeres (56%, n = 32; p < 0.01) or showed significant colocalization with only one centromere (40%, n = 23), and rarely showed significant colocalization with both centromeres (4%, n = 2) (Figures 2B and 2C). Any colocalization of CID with H3/PH3 that was observed is most likely the result of variability in the cytological preparations, such as occasional distortion of a chromosome that is not completely flat against the slide. We conclude that, on metaphase chromosomes, centromeric chromatin contains histones H2A and H2B, but not significant amounts of H3 or PH3.
Figure 2. Metaphase Centromeres Contain CID and Histones H2A, H2B but Not Histone H3.
CID and core histones H2A, H2B, and H3 were localized by indirect immunofluorescence on metaphase chromosomes from S2 and Kc cells, and the fluorescence intensity was quantitated across the two sister kinetochores.
(A) CID shows three patterns of localization with respect to H2AB: colocalization with both kinetochores, colocalization with one kinetochore, or colocalization with neither kinetochore.
(B) CID shows three patterns of localization with respect to H3/PH3: colocalization with both kinetochores, colocalization with one kinetochore, or colocalization with neither kinetochore.
(C) Quantitation of the relative distribution patterns of CID localization with respect to H2AB and H3/PH3. Scale bar is 1 micron.
Centromeric Chromatin Is Composed of Interspersed Blocks of CID/CENP-A and Histone H3 Nucleosomes
Localization of proteins to condensed metaphase chromosomes provides limited resolution of highly condensed chromatin and may not accurately reflect the linear structure and organization of centromeric chromatin. A previous study of mechanically stretched meta-phase chromosomes suggested that the kinetochore is composed of repeated subunits of ACA (anticentromere antibodies recognizing CENPs-A, -B, and -C), dynein, and tubulin (Zinkowski et al., 1991). Another study using extended chromosome fibers from human cells showed that ACAs are distributed in a “beads-on-string” pattern along the fiber and colocalize with α satellite DNA (Haaf and Ward, 1994), though the spaces between CENP localizations were attributed to artifacts of the preparation. Neither study addressed the differences in histone content, the organization of centromeric chromatin, or the localization of CENP-A.
To examine the linear structure of centromeric chromatin by high-resolution immunofluorescence, extended chromatin fibers that retained DNA-associated proteins were produced from Drosophila S2 and Kc cells and stained with antibodies to CID, and histones H2AB, H3, and PH3 (see Experimental Procedures). CID antibody staining was discontinuous along chromatin fibers stretched to ~50–100 times their normal interphase lengths (average 13 ± 5 spots/fiber, n = 15), reminiscent of the repeated subunit structure reported by Zinkowski et al. (1991). In contrast, H2AB was present continuously along the same fiber (Figures 3A and 3B) and principally showed colocalization with CID (25% ± 15 of CID signal appeared as yellow, as a result of complete colocalization with H2AB, n = 10 fibers).
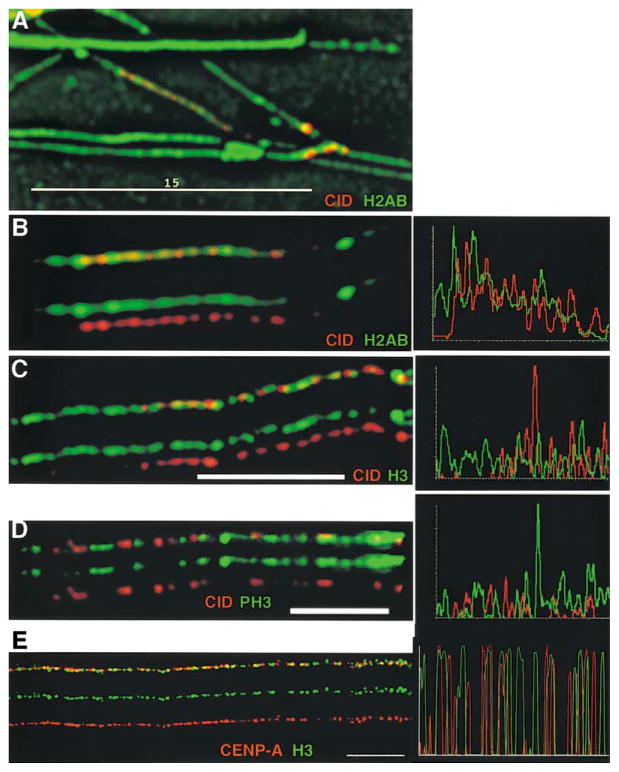
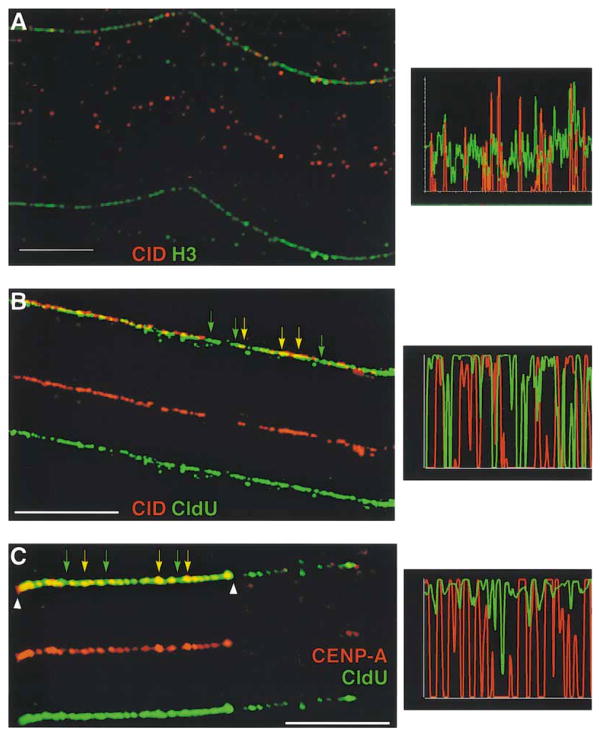
Figure 3. CID Is Colocalized with Histone H2AB on Extended Chromatin Fibers, but Is Interspersed with Histone H3.
Extended chromatin fibers were prepared from Drosophila Kc and human tissue culture cells, and CID/CENP-A and other core histones were simultaneously localized by indirect immunofluorescence. Single-color images are shown under the merged images in (B)–(D). Line graph to the right of each image shows quantitation of antibody staining along the length of a fiber.
(A) View of an entire field of chromatin fibers immunostained for H2AB (green) and CID (red). Most fibers show strong staining of H2AB antibodies continuously along their lengths, including the CID subdomains. Scale bar is 15 microns.
(B) An isolated chromatin fiber from (A) on which CID appears punctate and discontinuous, but H2AB are continuous along the fiber. (C) CID (red) and histone H3 (green) are present in close proximity along the same chromatin fiber and appear to be interspersed with one another, with very little overlap. Scale bar is 10 microns.
(D) CID (red) and histone PH3 (green) are present in close proximity along the same chromatin fiber and appear to be interspersed with one another, with some overlap. Scale bar is 5 microns.
(E) Human CENP-A (green) and H3 (red) are interspersed with infrequent overlap, similar to the organization observed in Drosophila centromeric chromatin.
Scale bar is 15 microns for (A), 5 microns for (D), and 10 microns for (C) and (E). See text and Experimental Procedures for quantitation of the extent of overlap.
Surprisingly, when fibers were stained for both H3 and CID, significant amounts of H3 were present in centromeric chromatin (Figure 3C). H3 staining appeared to alternate with CID staining and infrequently colocalized with CID staining (8% ± 7% of CID signal appeared as yellow, as a result of complete colocalization with H3, n = 10 fibers; significantly different from CID/H2AB colocalization, p < 0.01). Although centromeric chromatin assembly is likely to be a dynamic process, and may involve replacement of H3 nucleosomes with CID/CENP-A nucleosomes (Sullivan, 2001), we never observed fibers that contained only CID and not H3. CID-containing chromatin present in Drosophila centromeres extends over 200–500 kb (B.A.S. et al., unpublished data), suggesting that each block of CID or H3 chromatin occupies ~15–40 kb.
Since most fibers prepared from a randomly cycling cell population are from interphase cells, we determined if the interspersion of CID and H3 is also observed in mitotic kinetochores. We observed that blocks of the mitosis-specific PH3 modification are interspersed with CID, and infrequently colocalize with CID, as observed for unmodified H3 (Figure 3D). Therefore, we conclude that centromeric chromatin in Drosophila is composed of interspersed CID- and H3-containing chromatin during most or all stages of the cell cycle, including mitotic kinetochores.
CENP-A was also present discontinuously on fibers prepared from human cell lines, and no fibers were observed that contained CENP-A and not H3 (Figure 3E). As observed in Drosophila, other histones, such as H4, were continuously distributed along centromeric chromatin fibers (data not shown). However, similar to the pattern of CID staining, blocks of CENP-A antibody staining were interspersed with H3 staining, and infrequently overlapped with H3 (20% ± 9% overlap, n = 13 fibers) (Figure 3E). There was wide variation in the number of CENP-A antibody spots (9–70 spots/fiber; n = 25), most likely reflecting the presence of many different human centromeres, which are 2–8 times larger than Drosophila centromeres (B.A.S. et al., unpublished data). In fact, when CENP-A staining was analyzed on fibers derived from a single human chromosome, the number of spots was highly consistent (average 61 ± 10 spots/fiber, n = 10). Interestingly, human CENP-A staining was present on only one-half to two-thirds of the entire centromere-specific α satellite array (B.A.S. et al., unpublished data).
The similar organization of CENP-A/CID chromatin in humans and flies suggests that interspersed blocks of CENP-A- and H3-containing nucleosomes are an evolutionarily conserved aspect of centromere structure. How the exclusion of H3 from the CID/CENP-A cylindrical structure observed in analysis of intact metaphase chromosomes can be reconciled with interspersion on 2D fibers will be discussed below (see Discussion).
CID Mononucleosomes Contain No Histone H3, while CID Polynucleosomes Contain Histone H3
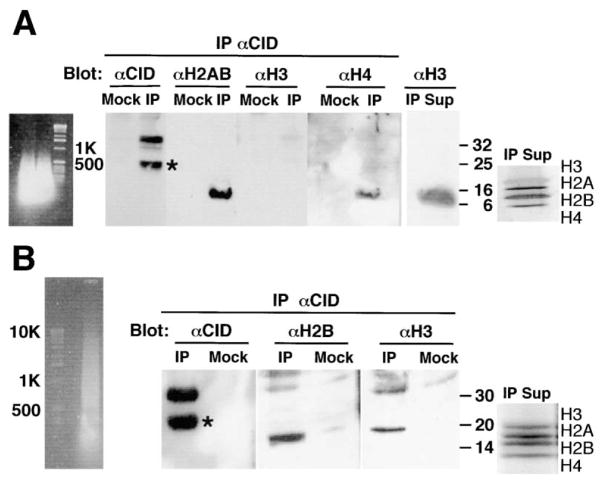
Genetic and biochemical evidence from S. cerevisiae and mammals suggest that CENP-A nucleosomes are homotypic (lack H3) (Shelby et al., 1997; Meluh et al., 1998; Yoda et al., 2000; see Introduction). However, these studies did not address the histone content of CENP-A nucleosomes or the surrounding chromatin, in vivo. To determine the histone content of CID nucleosomes in vivo, we treated S2 and Kc nuclei with extensive micrococcal nuclease digestion, immunoprecipitated nucleosomes with CID antibodies, and evaluated histone content by Western analysis (see Experimental Procedures). CID-containing nucleosomes ranging from ~100 bp to 500 bp (mono to trinucleosomes) contained CID, H2A, H2B, and H4, but not H3 (Figure 4A). The supernatant of the IP contained intact H3, and silver-stained gels of the soluble chromatin post-IP showed roughly equimolar amounts of all core histones (Figure 4A). Therefore, the inability to IP H3 within short CID chromatin is a result of complete replacement of H3 with CID, rather than H3 degradation during micrococcal nuclease digestion. We conclude that CID nucleosomes are homotypic in vivo, as predicted from previous studies, and most likely consist of two copies each of CID, H4, H2A, and H2B.
Figure 4. CID Mononucleosomes Do Not Contain Histone H3, while CID Polynucleosomes Contain Histone H3.
Soluble chromatin was prepared from Kc cells using micrococcal nuclease digestion, and CID-containing chromatin was immunoprecipitated using affinity-purified anti-CID. (A) Short polynucleosomes (primarily mono-nucleosomes) were prepared using extensive micrococcal nuclease digestion. CID and a degradation product (asterisk) were precipitated, as well as histones H2A, H2B and histone H4, while histone H3 was not coimmunoprecipitated from short CID-containing chromatin. Mock IPs did not result in the recovery of any histones. Histone H3 was found in the supernatant of the IP (IP Sup), and a silver-stained gel shows the integrity of the histones in the chromatin used in the IPs (note that after silver staining, H3 stains somewhat negatively compared to other core histones). (B) Polynucleosomes ranging from ~150 bp to 10 kb were prepared using limited micrococcal nuclease digestion. CID and a degradation product (asterisk) were immunoprecipitated in IPs, but not in mock IPs. Histone H2B as well as histone H3 was coimmunoprecipitated from long chromatin containing CID. Coomassie-stained gel shows the integrity of the histones in the chromatin used in the IPs.
To investigate the relationship between CENP-A- and H3-containing nucleosomes biochemically, we prepared polynucleosomes from Kc and S2 cells using limited micrococcal nuclease digestion, then immunoprecipitated CID, and analyzed the histone content of immunoprecipitated chromatin by Western blot. CID-containing polynucleosomes ranging from ~150 bp to ~10 kb contained CID, H2A, H2B, and H3 (Figure 4B). Coomassie staining of the IP supernatant confirmed that all of the histones were intact after micrococcal nuclease digestion. This result demonstrates that CID and H3 are present on polynucleosomes that range from 500 bp to 10 kb. Biochemical intermixing of CID and H3 in polynucleosome preparations, combined with the homotypic nature of CID and H3 mononucleosomes, confirms the separation and interspersion of CENP-A and H3 subdomains observed in centromeric chromatin fibers.
Histone H3 Replaces CID in CID-Depleted Cells
The CENP-A protein is very stable and appears to be equally partitioned to newly replicated DNA after S phase and in G2 (Shelby et al., 2000). We previously demonstrated that disruption of CID by RNA interference (RNAi) leads to decreased amounts of CID at the centromere and mitotic dysfunction (Blower and Karpen, 2001). Furthermore, the capacity to recruit outer kinetochore proteins to the kinetochore was shown to be proportional to the amount of CID. To determine if disruption of CID by RNAi results in a change in the H3:CID ratio at the centromere and in the pattern of interspersion, we simultaneously localized CID and H3 on chromatin fibers from CID RNAi-treated cells. CID staining intensity in each spot decreased markedly, and the number of CID units decreased from ~13 spots per untreated fiber to ~8 ± 1.5 (n = 18) spots per RNAi treated fiber (Figure 5A). The average distance between CID spots increased from 0.94 μm in untreated fibers to 1.87 μm in RNAi treated cells, and the regions between CID blocks were almost completely filled in by H3 (Figure 5A). We also found that CID and H3 colocalized more frequently in fibers from RNAi-treated cells. These results suggest that CID depletion increases the H3:CID ratio in centromeric chromatin and alters its structural organization. Replacement of CID by histone H3 when CID is depleted by RNAi provides evidence that the structure of centromeric chromatin is plastic, and provides a plausible explanation for the observed kinetochore dysfunction (Blower and Karpen, 2001; see Discussion). To further examine the plasticity of centromeric chromatin, we overexpressed CID and examined its localization on chromatin fibers. Consistent with a recent study in human cells (Van Hooser et al., 2001), we found that overexpressed CID is deposited throughout the entire nucleus/genome, and is nearly uninterrupted on extended chromatin fibers (data not shown).
Figure 5. H3 Replaces CID When CID Is Depleted by RNAi, and Centromeric Chromatin Replicates Synchronously in Human and Drosophila Cells.
Single-color images are shown beneath merged images. Line graph to the right of each image shows quantitation of antibody staining along a fiber.
(A) Histone H3 replaces CID when CID is depleted by RNAi. In fibers from CID RNAi-treated cells, decreased numbers of CID spots (red) are spaced further apart, and larger regions of H3 (green) are found between CID spots. Scale bar is 5 microns.
(B) Replication timing of centromeric chromatin, which contains interspersed H3 and CID nucleosomes, occurs simultaneously throughout the domain. CldU incorporation (green) on fibers shows continuous staining throughout the kinetochore chromatin (identified by CID antibodies [red]), as denoted by quantitation of signal in bar graph (right). Yellow arrows denote chromatin that contains CID and CldU. Green arrows indicate regions that lack CID staining but contain CldU staining (green), indicating that H3 chromatin adjacent to blocks of CID is replicated simultaneously with CID chromatin. Scale bar is 10 microns.
(C) Interspersed human CENP-A and H3 are replicated concurrently. Detection of replication by CldU incorporation (green) shows that both CENP-A (red) and the spaces between CENP-A staining which represent H3 (see Figure 2B) colocalize with CldU. Yellow arrows denote CENP-A chromatin that is replicated; green arrows indicate nearby regions that are devoid of CENP-A staining but show CldU staining (green). Arrowhead demonstrates that CldU staining abruptly ends at the edge of CENP-A staining and does not extend into the adjacent chromatin. Scale bar is 15 microns.
H3 and CENP-A Chromatin in the Centromere Replicate Concurrently
Recent studies in Drosophila and human cells have indicated that exclusive incorporation of CENP-A into centromeric chromatin is independent of DNA replication. Drosophila centromeres in vivo and human centromeres in cell lines are replicated asynchronously in mid to late S phase, coincident with the replication timing of bulk H3-containing chromatin (Shelby et al., 2000; Sullivan and Karpen, 2001). Furthermore, CENP-A and CID are targeted to centromeres even in the absence of replication (Ahmad and Henikoff, 2001; Shelby et al., 2000). One study failed to detect H3 within Drosophila centromeres in cell lines, and it was proposed that exclusion of H3 from centromeric nucleosomes is accomplished by replication of centromeres as isolated domains in early S phase (Ahmad and Henikoff, 2001; Henikoff et al., 2001). Our demonstration that CID and H3 are interspersed within centromeric chromatin disputes this hypothesis. However, it was important to determine if the centromeric chromatin is replicated as a single continuous domain in S phase, or if the interspersed H3 blocks are replicated at different times than nearby CID subdomains.
Asynchronously dividing cells from Drosophila and human cell cultures were labeled with the thymidine analog chlorodeoxyuridine (CldU) and extended chromatin fibers were stained with antibodies that recognized CID or CENP-A, and CldU (See Experimental Procedures). When a centromere replicated, chromatin fibers showed predominantly continuous incorporation of CldU throughout the centromeric chromatin, in both Drosophila and human cells (Figures 5B and 5C). These results indicate that H3 and CID/CENP-A subdomains are replicated concurrently in S phase and argues against special replication timing properties of H3 and CID chromatin contained within the centromere.
Interestingly, the replication timing of H3 and CENP-A in centromeric chromatin appeared to be distinct from the flanking H3-containing heterochromatin, identified by histone H3 lysine 9 methylation (data not shown; Lachner et al., 2001; Nakayama et al., 2001; Noma et al., 2001). Continuous CldU staining of interspersed H3/CENP-A domains abruptly ended at the edge of CID/CENP-A antibody staining, and vice versa (Figures 5B and 5C). This result suggests that distinct replication and/or structural boundaries exist between centromeric DNA and neighboring heterochromatic sequences, consistent with the domain organization of the centromere region observed in S. pombe and Drosophila (Blower and Karpen, 2001; Partridge et al., 2000).
Discussion
The 3D Organization and Linear Arrangement of Centromeric Chromatin Are Conserved
We have found that centromeric chromatin in both flies and humans is organized into a cylindrical 3D structure on metaphase chromosomes. This structure contains histones H2AB but appears to be devoid of H3 and PH3 (Van Hooser et al., 1999). This data suggests that the metaphase centromere is composed solely of CID/CENP-A-containing nucleosomes and that CID/CENP-A may replace all histone H3 in centromeric nucleosomes. Consistent with this data, we have found that CID mononucleosomes are homotypic in vivo and contain CID, H2A, H2B, and H4, as has been suggested by in vitro studies and the stoichiometry of CENP-A in human cells (Shelby et al., 1997; Yoda et al., 2000).
Previous 2D immunofluorescence studies in mammalian cells suggested that CENP-A and CENP-C colocalize (Warburton et al., 1997) and are therefore positioned within the same region of the inner kinetochore. CENP-A localization within subkinetochore chromatin or the inner kinetochore plates using high resolution electron microscopy has not been previously reported. The 3D deconvolution studies described here demonstrate that while CENP-A and CENP-C are indeed closely juxtaposed, they show significant nonoverlap. In cross-section, the organization we observed is consistent with human CENP-C forming a plate-like structure, as demonstrated in previous transmission EM studies (Saitoh et al., 1992). However, our data suggests that CENP-A chromatin is organized as a cylindrical structure, rather than a plate, and is predominantly located beneath the kinetochore, although some overlap with the inner plate cannot be excluded. CENP-A localization interior to CENP-C suggests that CENP-A nucleosomes are the physical foundation for kinetochore formation. CENP-A also serves as the functional foundation for kinetochore assembly, as all known kinetochore components are mislocalized in CENP-A disruptions, including CENP-C (Blower and Karpen, 2001; Howman et al., 2000; Moore and Roth, 2001; Oegema et al., 2001). CENP-C, in turn, is required for recruitment or maintenance of outer kinetochore proteins in mammals and worms. This data suggests that there is a specific order of recruitment and assembly of the kinetochore in all organisms, which mimics the 3D arrangement of the protein components.
Zinkowski et al. (1991) first proposed that the kinetochore was composed of repeats of a functional base subunit. This conclusion was based on caffeine-induced kinetochore fragmentation in the absence of DNA replication, the ability of kinetochore fragments to move along spindle microtubules, and the discontinuous appearance of CREST antibody staining on mechanically stretched metaphase kinetochores. Since many CREST sera recognize CENPs -A, -B, -C (Earnshaw and Rothfield, 1985), it was impossible to determine which of these proteins represented the fundamental base unit of the repeat. A second study in human cells showed that ACAs were distributed on extended chromatin fibers in a “beads-on-string” pattern coincident with α satellite DNA (Haaf and Ward, 1994). Gaps between ACA subunits were attributed to technical consequences of the fiber-FISH protocol. The extended fiber studies reported here suggest that these gaps actually correlate with subunits of H3 nucleosomes in flies and humans, and argue against artifacts of the fiber technique. In contrast to the exclusion of H3 from metaphase kinetochores, we have found that blocks of CID/CENP-A nucleosomes and H3 nucleosomes are interspersed on chromatin fibers and long polynucleosomal fragments. Furthermore, because CENP-A nucleosomes are arranged in a discontinuous array and CENP-A is required for the recruitment of all other kinetochore components (Blower and Karpen, 2001; Howman et al., 2000; Moore and Roth, 2001; Oegema et al., 2001), we conclude that CENP-A nucleosomes are the base subunit of the repeated centromere/kinetochore structure in diverse organisms.
Our chromatin fiber analysis indicated variation in the number of CENP-A spots, particularly on fibers from human cells. These results are particularly provocative in that they suggest that sizes of CID/CENP-A-containing regions and/or numbers of CENP-A nucleosomes may vary widely, particularly among human chromosomes (B.A.S. et al., unpublished data). CID/CENP-A-containing chromatin at Drosophila centromeres extends over 200–500 kb and over 500–1500 kb at human centromeres (B.A.S. et al., unpublished data). Our results are in agreement with recent findings indicating that the CENP-A binding domains at two human neocentromeres are 460 kb and 330 kb (Lo et al., 2001a, 2001b). In addition, we found that CENP-A antibodies stained only one-half to two-thirds of α satellite DNA regions at normal human centromeres (B.A.S. et al., unpublished data). This result suggests that the entire α satellite DNA array at a human centromere is not involved in kinetochore assembly and that α satellite DNA may have additional functions in centric regions.
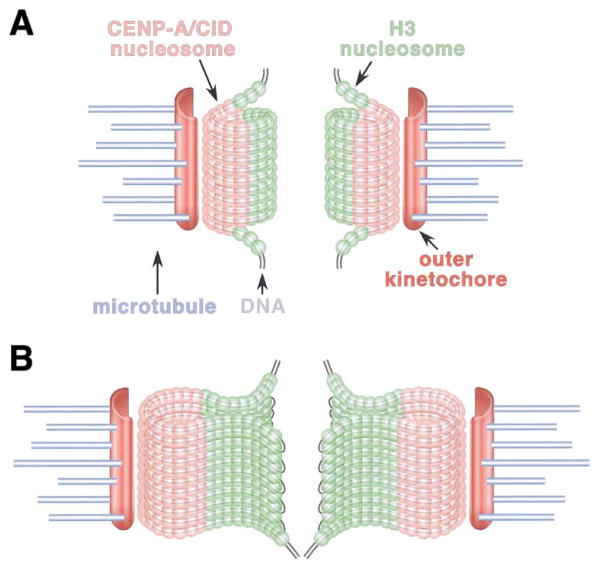
How can the linear interspersion of CID/CENP-A and H3 be reconciled with the exclusion of H3 on metaphase chromosomes? We propose that centromeric DNA may form a spiral or loop, in which blocks of CENP-A nucleosomes are oriented on the poleward faces of chromosomes and blocks of H3 nucleosomes are located toward the inner chromatid region (Figure 6). At this time, we do not know the relative proportions of CENP-A and H3 nucleosomes in the centromere and whether the number of CENP-A or H3 nucleosomes varies from block to block. Does the conserved 3D structure and organization of centromeric chromatin play a role in centromere function? We propose that the purpose of the spiral or loop structure may be to “present” centromeric chromatin to the exterior of the chromosome, where it can mediate kinetochore assembly and interactions with the spindle (Sullivan et al., 2001). If centromeric chromatin condensed along with the rest of the chromosome in a random fashion, this chromatin would most likely be hidden inside the chromatids.
Figure 6. Models for 2D and 3D Organization of Centromeric Chromatin in Drosophila and Humans.
Two models are proposed to reconcile the 2D interspersion of CENP-A and CID in centromeric chromatin fibers with the sequestration observed in three-dimensional analyses of metaphase chromosomes.
(A) Solenoid model. Blocks of CENP-A/CID and H3 nucleosomes may be arranged in a solenoid, such that CID nucleosomes are present on the poleward face of the meta-phase chromosome while H3 nucleosomes are sequestered in the region between the paired sister kinetochores. In this model, the DNA is proposed to spiral through the three dimensional cylindrical structure.
(B) Looping model. It is also possible that centromeric DNA may loop, rather than spiral, through the elliptical structure, resulting in CENP-A domains on the poleward face of the chromosome, and H3 domains located toward the region between sister centromeres.
The Role of Heterochromatin in Centromere Organization and Function
In higher eukaryotes, centromeres are uniformly embedded in large blocks of repetitive DNA that are considered highly condensed, gene-poor, and inaccessible to transcription factors. However, flanking heterochromatin behaves as a domain that is structurally and functionally distinct from CENP-A-containing centromeric chromatin in S. pombe, flies, and humans (Partridge et al., 2000). Further evidence for the existence of distinct domains within the centromere comes from our observation that chromatin immediately flanking CENP-A/CID chromatin appears to replicate at a different time than CID-containing chromatin. Discontinuity of replication has been seen in other genomic regions that are involved in epigenetic inheritance (Dalgaard and Klar, 2000, 2001). It is possible that a replication boundary between centromeric chromatin and the flanking heterochromatin is important for establishment or maintenance of the two distinct chromatin states.
We propose that flanking heterochromatin may be required to organize the higher order structure of centromeric chromatin. Flanking heterochromatin may interact with the interspersed H3 domains to produce or maintain the CENP-A cylinder (Figure 6; Sullivan, 2001). This model predicts that the interspersed H3 may display heterochromatin-like properties, such as H3 methylation at lysine 9 and HP1 binding (Lachner et al., 2001; Nakayama et al., 2001; Noma et al., 2001). Heterochromatin at the centromere may also be necessary to adopt a conformation that maintains cohesion between sister chromatids (Bernard et al., 2001; Blat and Kleckner, 1999; Tanaka et al., 1999). Furthermore, it may define the borders of the centromeric chromatin domain and prevent CENP-A chromatin from spreading into adjacent regions, as observed for neocentromere formation in flies (Maggert and Karpen, 2000, 2001).
The Evolution of Centromeric Chromatin and Kinetochore Structure
It appears that a single nucleosome containing the CENP-A homolog Cse4p is sufficient to nucleate micro-tubule interactions in S. cerevisiae (Meluh et al., 1998). The point centromeres of S. cerevisiae are likely to represent the most basic iteration of a centromere, which expanded as organisms became more complex and evolved larger chromosomes. In S. pombe, the CENP-A homolog Cnp1 is present in an apparently uninterrupted stretch of ~5–10 kb within the central core of the centromere, flanked by H3-containing chromatin (Takahashi et al., 2000). We propose that the different sizes and organizations of monocentric kinetochores, and even holocentric kinetochores, are produced by the same interspersed histone/higher order structure, present in different numbers and distributions (Sullivan et al., 2001). The holocentric kinetochores of C. elegans and other species (Pimpinelli and Goday, 1989) on the surface appear to be quite different from monocentric kinetochores but could simply represent the broadest expansion of the functional centromere base unit. In C. elegans, CENP-A (HCP-3) is present in an unusually large number of discreet foci in interphase nuclei; during mitosis these foci coalesce into the thin kinetochore ribbon present on the poleward face of each chromosome (Buchwitz et al., 1999). Thus, holocentric kinetochores appear to be organized into a 3D structure similar to humans and flies, in which centromeric chromatin is presented on the exterior face of metaphase chromosomes (Buchwitz et al., 1999; Moore and Roth, 2001; Oegema et al., 2001).
The plasticity of centromeric chromatin demonstrated here and elsewhere (reviewed in Sullivan, 2001) provides a plausible mechanism for how variations in the basic centromere unit may be established and maintained in different organisms. In our study, RNAi depletion of Drosophila CID resulted in reduced intensity and number of CID spots in fibers, an increase in the distance between spots, and an expansion of the H3 domains. Overexpression of CID resulted in continuous distribution of CID, rather than discrete arrays of CID chromatin. These results suggest that the interspersed organization of centromeric chromatin is plastic; in the absence of CID deposition, H3 chromatin is assembled on centromeric DNA, and vice versa. Reductions in the size and number of the CID blocks, and expansion of the H3 blocks, can account for the decreased recruitment of outer kinetochore proteins and increased chromosome segregation errors observed in our previous study (Blower and Karpen, 2001). In addition, variations in the amount of CENP-A in the nucleus, or the kinetics of H3 and CENP-A deposition, could be responsible for the evolution of different kinetochore sizes and interspersion patterns. The maximum extent of the centromeric chromatin could then be determined by altering the locations of specific boundary elements or the balance between the centromeric and flanking centric heterochromatin epigenetic states (see above) (Maggert and Karpen 2000, 2001).
The plasticity of centromeric chromatin organization suggests that there must be an active mechanism to maintain the balance between CID/CENP-A deposition and H3 deposition. S. pombe Mis6 and S. cerevisiae ndc10 have been demonstrated to be required for localizing or maintaining CENP-A at centromeres (Ortiz et al., 1999; Takahashi et al., 2000), although Ctf3p, the budding yeast homolog of pombe Mis6, is not required for loading Cse4p onto centromeric DNA (Measday et al., 2002). Future studies of the determinants of centromere identity must identify the proteins required for CENP-A deposition in higher eukaryotes, and for assembly and maintenance of the linear arrangement and 3D structure of centromeric chromatin reported here.
Experimental Procedures
Cloning and DNA Constructs
Histone H3 was amplified from the genomic DNA of y1 ry506 using the primers 5′-ATG GCT CGT ACC AAG-3′ and 5′-AGC ACG CTC GCC GCG-3′. The PCR product was TA cloned into pIB/V5-His (Invitrogen), which contains an in-frame V5 epitope tag. Clones were verified by sequencing. The coding region of CENP-A was amplified from ESTs (HGMP, UK) and cloned into the EcoRI site of pFLAG-CMV2 (Sigma). CENP-A coding sequence was placed 5′ to the epitope tag. GFP-CENP-A was constructed by cloning a BamHI-XhoI fragment from pUBH113 (containing human CENP-A) into the BamHI and NheI sites of pCMXGFP2 so that GFP was located 5′ to CENP-A.
Cell Culture
S2 and Kc cells were maintained as described elsewhere (Blower and Karpen, 2001). Cells expressing Drosophila V5 epitope-tagged H3 were examined using fiber preparation (see below) and compared to PH3 staining of untransfected cells. No difference was found between the localization pattern of H3-V5 and PH3 on fibers (data not shown), so we conclude these stable cell lines reflect endogenous H3 distribution. RNAi was performed as described (Blower and Karpen, 2001). Human HT1080 cells were cotransfected with 750 ng of FLAG-CENP-A or 500 ng GFP-CENP-A and a plasmid containing a gene encoding neomycin resistance. G418-resistant colonies were selected and expanded. The location of GFP-CENP-A at centromeres was tested by immunofluorescence of fixed and unfixed human nuclei and chromosomes. Localization of CENP-A-FLAG fusion protein to centromeres was confirmed by immunofluorescence using monoclonal anti-FLAG antibodies (see below). Primary human fibroblasts and HT1080 cells containing FLAG-tagged CENP-A or GFP-CENP-A were maintained in MEM (α) containing L-glutamine (Life Technologies) and supplemented with 10%–20% fetal bovine serum (Hyclone) and 1% antibiotic-antimycotic (Life Technologies). Transfections were performed using Lipofectin reagent (Life Technologies) according to the manufacturer’s instructions for adherent cells, using 0.75–2 μg of DNA and 10 μl of lipid per 4 × 106 cells.
Antibodies
Chicken anti-CID (1980 and 1981) and rabbit anti-phospho H3 were used as described (Blower and Karpen, 2001), rabbit anti-H3 (1:200 Western), rabbit anti-H2B (1:200 Western), rabbit anti-H2AB (1:200 Western) were a gift of M. Levenstein (Kadonaga Lab, UCSD). Rabbit anti-H4 (Upstate, 1:500 Western), mouse anti-V5 (Invitrogen, 1:500 immunofluorescence), and mouse anti-FLAG antibodies (Sigma; 1:200 immunofluorescence) were obtained from commercial sources. Rabbit anti-CENP-A antibodies (Valdivia et al., 1998) were used at 1:400 dilution; anti-CENP-C antibodies (du Sart et al., 1997) were used at 1:100 dilution; and rabbit anti-CENP-E (E2) antibodies (Harrington et al., 1997) were diluted at 1:500.
Cytological Preparations
S2 and Kc cells were prepared as described (Blower and Karpen, 2001), except that colcemid treatment was omitted. Extended chromatin fibers were prepared from S2 and Kc cells by centrifuging 2 × 105 cells/ml onto slides at 800 rpm for 3 min in a Cytospin 3 (Shandon, Pittsburgh, PA), and then slides were dipped into salt detergent buffer (25 mM Tris [pH 7.5], 500 mM NaCl, and 1% Triton X-100) for 10 min, slowly removed, and subsequently fixed in 4% paraformaldehyde (PFA) for 5 min. Slides were processed for immunofluorescence as described below. Unfixed human metaphase chromosomes used for 3D immunofluorescence studies were centrifuged onto glass slides as previously described (Sullivan and Schwartz, 1995). Chromatin fibers from human cells were prepared by centrifuging 104 cell/ml in 75 mM KCl onto charged microscope slides (Fisher Scientific) and lysing in salt detergent buffer supplemented with urea for 15 min before slowly removing and fixing in 4% PFA or for 20 min in cold methanol. Slides were incubated in 1× PBST (1× PBS ± 0.05% Tween-20) and blocked in 1× PBS, 1% BSA, 0.1% Triton X-100, 0.02% Na Azide for 15–30 min at room temperature before incubation with appropriate primary, then secondary, antibodies diluted in blocking buffer.
For FISH analysis after indirect immunofluorescence with CID and CENP-A antibodies, slides were again fixed in 4% formaldehyde for 15 min to crosslink antibodies to proteins and prepared for FISH as previously described (Sullivan and Warburton, 1999). Twenty-five to 300 ng of DNA probe that had been labeled by nick translation with biotin-16-dUTP (Roche) or digoxygenin-11-dUTP (Roche), or directly labeled with Spectrum Green-dUTP (Vysis) was used for each hybridization. Probe and chromosomal DNA were hybridized for 24–48 hr at 37°C and indirectly labeled probes were detected with Cy3 or fluorescein avidin.
Replication labeling on chromatin fibers from S2 and Kc cells was performed by treating unsynchronized, proliferating cultures with CldU (final concentration: 50 μM) for 2–2.5 hr. After lysis and fixation, fiber preparations were briefly washed in PBST then blocked in 1× PBS, 0.5% Triton X-100, 1% BSA, 0.02% sodium azide) for 10–20 min at room temperature. CID antibodies were used at 1.3 μg/ml. After detection with anti-chicken secondary antibodies, preparations were fixed again in 4% formaldehyde in KCM. CldU was detected using rat anti-BrdU antibodies (MAB 3424; Serotec), as described previously (Sullivan and Schwartz, 1995). Human cells were incubated with 25 μM (final concentration) CldU for 2 hr before preparing chromatin fibers. CENP-A was detected first using mouse anti-FLAG antibodies, followed by detection of CldU.
Microscopy and Image Analysis
All images were captured using an Olympus IX70 microscope and Olympus IX-HLSH100 CCD. Images were acquired using the Deltavision SoftWoRx resolve3D capture program and collected as a stack of 0.1 μm increments in the z axis; each image consisted of 1.5–2.5 μm (15–25 sections). Images were deconvolved using the conservative algorithm with 10 iterations. Deconvolved stacked images were viewed using either the Quick Projection or Volume View options. All volume renderings were also verified by imaging 1 and 4 μm fluorescent beads and performing similar rotations as used for the chromosomes. Imaging beads revealed that when viewing a z axis series a distortion occurs which stretches shapes by a factor of 1.5–1.75. This distortion does not affect the shape of the image in the x and y axes, and the net result of this distortion is that a sphere appears as a football when viewed in the z axis. All images presented are raw data, and no effort was made to correct for this optical distortion. Volume models were made using the model3D option of SoftWoRx, and all models presented were constructed directly from the data presented. Initial adjustments were made in SoftWoRx, and then the image was imported to Adobe Photoshop.
For fluorescence quantitation of CID and core histones on meta-phase chromosomes, a computer script was generated in IPLab (Vysis), and used to draw a line that passed through the center of each kinetochore, and the fluorescence intensity was measured within single optical sections. CID was judged to colocalize with core histones if the intensity graph for the core histones extended across the entire peak of the CID signal.
The overlap between CENPs -A, -C, and -E on human metaphases was quantitated with the IP lab script by drawing a line through a merge of the middle three optical sections from the deconvolved stack. Selection of middle sections was necessary to avoid false protein overlaps that result from analysis of the complete stack, where CENPs -C and -E wrap over the top and bottom of the CENP-A cylinder. CENP-A was judged to completely overlap with CENP-C, and CENP-C to completely overlap with CENP-E, if intensity curves for each antibody signal overlapped completely. The extent of partial overlap was determined by calculating the amount of overlap between intensity curves for samples that did not display complete overlap or no overlap.
For quantitation of CID/CENP-A and core histone colocalization on chromatin fibers, yellow pixels (complete colocalization) were counted and expressed as a function of the total number of CID/CENP-A pixels, using the “Select Color” and “Histogram” tools in Adobe Photoshop. Chi-squared analysis was performed on data sets using Microsoft Excel. For determining the average number of CENP-A/CID antibody spots per fiber, antibody spots were counted visually and by counting peaks on intensity graphs. For measurement of the average distance between CID spots on control and RNAi treated fibers, the distance between the adjacent spots was measured using Deltavision SoftWoRx software.
Chromatin Preparation and Immunoprecipitation
Long and short chromatin was prepared from S2 and Kc cells using a modification of a published protocol (Kanda et al., 1998). S2 or Kc cells (~109) were collected by centrifugation at 1000 × g for 5 min, and the cell pellet was resuspended in PBS, 1.5 mM MgCl2, and 1% Triton X-100. Cells were lysed by repeated vortexing during 5 min incubations on ice. Nuclei were collected by centrifugation at 1000 × g for 5 min and resuspended in Chromatin Isolation Buffer (Luger et al., 1997) supplemented with 1 mM CaCl2. Micrococcal nuclease (0.025 units) (Sigma) was added and nuclei were digested for 5 min (long chromatin) or 2 hr (short chromatin) at 37°C, and digestion was stopped by the addition of EDTA to 10 mM. Nuclei were collected by centrifugation at 20,000 × g for 10 min, resuspended in PBS, 500 mM NaCl, and 0.1% Triton X-100, and incubated on ice for ~2–8 hr. Insoluble material was removed by centrifugation at 20,000 × g for 15 min and soluble chromatin was recovered in the supernatant.
Half of the chromatin prepared from ~109 cells was used per IP. Affinity-purified chicken (~40 μg) anti-CID (20 μg 1980, 20 μg 1981) was added to soluble chromatin and incubated overnight at 4°C, 50 μg rabbit anti-chicken (Jackson ImmunoResearch) was added and incubated for 2 hr at 4°C, and chromatin was precleared by centrifugation at 20,000 × g for 15 min. The supernatant was transferred to a fresh tube and incubated at 4°C for 2 hr with 100 μl protein A beads (Sigma). Beads were collected by centrifugation at 1000 × g for 5 min and washed 6 times with PBS, 500 mM NaCl, 0.1% Triton X-100, and then suspended in 1× protein loading dye and boiled for 10 min. One quarter of each IP was separated on a 7%–18% exponential gradient SDS-PAGE gel, transferred to PVDF, and processed for western analysis using standard methods. One hundred microliters of the supernatant from each IP was analyzed by either silver or Coomassie stain and Western blot. The supernatant was extracted once with Phenol/Chloroform/Isoamylalcohol (25/24/1) and DNA was precipitated with isopropanol and electrophoresed on a 1% agarose gel.
Acknowledgments
We thank L. Cherbas for providing the Kc cell line, A. Lui for creating stable CID-expressing Kc cell lines, B. Hendrich for assistance in the construction of FLAG-CENP-A, M. Valdivia for CENP-A antibodies, A. Choo for CENP-C antibodies, H. Willard for CENP-E antibodies, J. Harrington for pUBH113, M. Levenstein (Kadonaga Lab) for H2AB and H3 antibodies, and A. Dernburg, K. Sullivan, B. Grimes, L. Pillus, and P. Heun for suggestions and advice. We also thank M. Baker (Salk DNA Sequencing Facility) for assistance with sequence analysis, and Tom Deerinck and Mark Ellisman (UCSD) for assistance with the EM tomography. Our research on eukaryotic centromere organization and function is supported by NIH grant 5R01 GM54549.
References
- Ahmad K, Henikoff S. Centromeres are specialized replication domains in heterochromatin. J Cell Biol. 2001;153:101–109. doi: 10.1083/jcb.153.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- Cooke CA, Schaar B, Yen TJ, Earnshaw WC. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma. 1997;106:446–455. doi: 10.1007/s004120050266. [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell. 2000;102:745–751. doi: 10.1016/s0092-8674(00)00063-5. [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ. A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev. 2001;15:2060–2068. doi: 10.1101/gad.200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao JI, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KH. A functional neocentromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Glowczewski L, Yang P, Kalashnikova T, Santisteban MS, Smith MM. Histone-histone interactions and centromere function. Mol Cell Biol. 2000;20:5700–5711. doi: 10.1128/mcb.20.15.5700-5711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Ward DC. Structural analysis of alpha-satellite DNA and centromere proteins using extended chromatin and chromosomes. Hum Mol Genet. 1994;3:697–709. doi: 10.1093/hmg/3.5.697. [DOI] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within peri-centromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KH. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lo AWI, Magliano DJ, Sibson MC, Kalitsis P, Craig JM, Choo KHA. A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 2001a;11:448–457. doi: 10.1101/gr.167601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AWI, Craig JM, Saffery R, Kalitsis P, Irvine DV, Earle E, Magliano DJ, Choo KHA. A 330kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 2001b;20:1–10. doi: 10.1093/emboj/20.8.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- Maggert KA, Karpen GH. Acquisition and metastability of centromere identity and function: sequence analysis of a human neocentromere. Genome Res. 2000;10:725–728. doi: 10.1101/gr.10.6.725. [DOI] [PubMed] [Google Scholar]
- Maggert K, Karpen GH. The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics. 2001;158:1615–1628. doi: 10.1093/genetics/158.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V, Hailey DW, Pot I, Givan SA, Hyland KM, Cagney G, Fields S, Davis TN, Hieter P. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 2002;16:101–113. doi: 10.1101/gad.949302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Moore LL, Roth MB. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J Cell Biol. 2001;153:1199–1208. doi: 10.1083/jcb.153.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama JI, Rice JC, Strahl BD, Allis CD, Grewal SIS. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SIS. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz J, Stemmann O, Rank S, Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Trong HL, Charbonneau H, Margolis RL. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci USA. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, Borgstrom B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- Pimpinelli S, Goday C. Unusual kinetochores and chromosome diminution in Parascaris. Trends Genet. 1989;5:310–315. doi: 10.1016/0168-9525(89)90114-5. [DOI] [PubMed] [Google Scholar]
- Pinto I, Winston F. Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J. 2000;19:1598–1612. doi: 10.1093/emboj/19.7.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Khodjakov A. Mitosis and checkpoints that control progression through mitosis in vertebrate somatic cells. Prog Cell Cycle Res. 1997;3:301–312. doi: 10.1007/978-1-4615-5371-7_24. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Tomkiel J, Cooke CA, Ratrie HD, Maurer M, Rothfield NF, Earnshaw WC. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- Scaerou F, Aguilera I, Saunders R, Kane N, Blottiere L, Karess R. The rough deal protein is a new kinetochore component required for accurate chromosome segregation in Drosophila. J Cell Sci. 1999;112:3757–3768. doi: 10.1242/jcs.112.21.3757. [DOI] [PubMed] [Google Scholar]
- Shah JV, Cleveland DW. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell. 2000;103:997–1000. doi: 10.1016/s0092-8674(00)00202-6. [DOI] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KF. A solid foundation: functional specialization of centromeric chromatin. Curr Opin Genet Dev. 2001;11:182–188. doi: 10.1016/s0959-437x(00)00177-5. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet. 1995;4:2189–2197. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- Sullivan B, Warburton P. Studying the progression of vertebrate chromosomes through mitosis by immunofluorescence and FISH. In: Bickmore W, editor. Chromosome Structural Analysis: A Practical Approach. Oxford: IRL Press; 1999. pp. 81–101. [Google Scholar]
- Sullivan B, Karpen G. Centromeric chromatin replicates in mid- to late-S phase in Drosophila. J Cell Biol. 2001;154:683–690. doi: 10.1083/jcb.200103001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Blower MD, Karpen GH. Determining centromere identity: cyclical stories and forking paths. Nat Rev Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- Sunkel CE, Glover DM. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci. 1988;89:25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Valdivia MM, Figueroa J, Iglesias C, Ortiz M. A novel centromere monospecific serum to a human autoepitope on the histone H3-like protein CENP-A. FEBS Lett. 1998;422:5–9. doi: 10.1016/s0014-5793(97)01583-4. [DOI] [PubMed] [Google Scholar]
- Van Hooser AA, Mancini MA, Allis CD, Sullivan KF, Brinkley BR. The mammalian centromere: structural domains and the attenuation of chromatin modeling. FASEB J. 1999;13(Suppl 2):S216–220. doi: 10.1096/fasebj.13.9002.s216. [DOI] [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Williams BC, Murphy TD, Goldberg ML, Karpen GH. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc Natl Acad Sci USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: a repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]