Nkx2.2 repressor complex regulates islet β-cell specification and prevents β-to-α-cell reprogramming (original) (raw)
In order to determine how cell fates are determined and maintained, Sussel and colleagues studied the endogenous regulatory pathway by which β-cell identity is maintained. They show that pancreatic β cells can be reprogrammed by a mutation in a single transcription factor, Nkx2.2, leading to repression of Arx, an α-cell-specific factor. This paper provides novel insight into an in vivo link between cell-specific DNA methylation events, gene regulation, and cellular reprogramming.
Keywords: islet development, β-cell reprogramming, Nkx2.2, DNMT3a, transcriptional repression, DNA methylation
Abstract
Regulation of cell differentiation programs requires complex interactions between transcriptional and epigenetic networks. Elucidating the principal molecular events responsible for the establishment and maintenance of cell fate identities will provide important insights into how cell lineages are specified and maintained and will improve our ability to recapitulate cell differentiation events in vitro. In this study, we demonstrate that Nkx2.2 is part of a large repression complex in pancreatic β cells that includes DNMT3a, Grg3, and HDAC1. Mutation of the endogenous Nkx2.2 tinman (TN) domain in mice abolishes the interaction between Nkx2.2 and Grg3 and disrupts β-cell specification. Furthermore, we demonstrate that Nkx2.2 preferentially recruits Grg3 and HDAC1 to the methylated Aristaless homeobox gene (Arx) promoter in β cells. The Nkx2.2 TN mutation results in ectopic expression of Arx in β cells, causing β-to-α-cell transdifferentiation. A corresponding β-cell-specific deletion of DNMT3a is also sufficient to cause Arx-dependent β-to-α-cell reprogramming. Notably, subsequent removal of Arx in the β cells of Nkx2.2TNmut/TNmut mutant mice reverts the β-to-α-cell conversion, indicating that the repressor activities of Nkx2.2 on the methylated Arx promoter in β cells are the primary regulatory events required for maintaining β-cell identity.
In the developing embryo, cell fates are achieved through the regulated activation and repression of transcription factors, which act as intermediaries in response to the secretion of inductive signals. The acquisition of cell fate is accomplished by a series of differentiation steps that were once thought to be irreversible. More recently, however, studies in multiple systems have demonstrated that dedifferentiation and transdifferentiation are mechanisms commonly used during developmental programs and in paradigms of regeneration (Tsonis et al. 2004; Mirsky et al. 2008; Jopling et al. 2010, 2011). The ability to reprogram somatic cells into pluripotent cells by the expression of transcription factors has also demonstrated the feasibility of direct reprogramming that does not require an intermediate dedifferentiation step (Takahashi and Yamanaka 2006; Boland et al. 2009). Following this breakthrough, there have been many examples of tissue reprogramming, including the transdifferentiation of fibroblasts into functional cardiomyocytes or multilineage blood progenitors (Ieda et al. 2010; Selvaraj et al. 2010; Szabo et al. 2010).
Similar to most other developmental systems, lineage determination and cell specification in the developing pancreas involve an organized series of events consisting of appropriately timed extrinsic signaling that regulates hierarchies of transcriptional networks (Gittes 2009). In addition, recent studies have exploited the known islet transcriptional networks to transdifferentiate nonpancreatic or exocrine tissue into endocrine cells. Ectopic expression of different combinations of pancreatic transcription factors—including Pdx1, Ngn3, NeuroD, MafA, and _Nkx6.1_—promotes liver-to-pancreatic β-cell reprogramming (Chan et al. 2003; Kojima et al. 2003; Song et al. 2007; Delisle et al. 2009; Nagaya et al. 2009; Gefen-Halevi et al. 2010). Similarly, ectopic expression of a defined set of transcription factors in adult pancreatic acinar cells results in an acinar-to-β-cell conversion (Zhou et al. 2008). Recent studies have also demonstrated a previously unappreciated plasticity between the different islet cell types in embryonic and adult pancreas. Misexpression of the α-cell-specific transcription factor Aristaless homeobox gene (Arx) in fetal β cells is sufficient to cause a β-to-α-cell conversion (Collombat et al. 2007). Furthermore, Thorel et al. (2010) have shown that mice with a >90% reduction in β cells are able to replenish the lost β cells through the up-regulation of β-cell factors in α cells to induce α-to-β-cell transdifferentiation. Most recently, epigenetic manipulations have also demonstrated the importance of DNA methylation and histone modifications in maintaining β-cell identity and driving islet cell differentiation (Haumaitre et al. 2009; Dhawan et al. 2011). These groundbreaking studies have underscored the potential of using reprogramming methods to generate novel sources of islet cells.
The homeodomain transcription factor Nkx2.2 is required for cell fate decisions in the pancreatic islet and cell patterning in the ventral neural tube (Sussel et al. 1998; Briscoe et al. 1999). It has previously been shown that Nkx2.2 acts as a transcriptional repressor to regulate ventral neural patterning through its interaction with the corepressor Groucho-4 (Grg4) (Muhr et al. 2001). This interaction is mediated by a motif called the tinman (TN) domain, which shares sequence homology with the core region of the engrailed homology-1 domain in the transcriptional repressor Engrailed and through which Grg/TLE proteins interact (Lints et al. 1993; Smith and Jaynes 1996; Jimenez et al. 1997). In the developing pancreas, Nkx2.2 appears to function as either a transcriptional repressor or an activator, depending on the temporal- or cell-specific environment (Watada et al. 2000; Cissell et al. 2003; Raum et al. 2006; Doyle and Sussel 2007; Doyle et al. 2007; Anderson et al. 2009).
To further investigate the regulatory role of Nkx2.2 in the developing pancreas and its dependence on Grg interactions, we generated mice containing amino acid substitutions in the highly conserved TN domain of the endogenous Nkx2.2 gene (Nkx2.2TNmut/TNmut). In this study, we demonstrate that the TN domain is required for the in vivo interaction between Nkx2.2 and Grg3 and that the interaction with Grg proteins is required for two independent stages of development. At embryonic day 13.5 (E13.5), during the initiation of the secondary transition, the Nkx2.2 TN domain is necessary for the β- versus ɛ-cell fate decision. Subsequently, after β cells have formed, the Nkx2.2 TN domain is essential for maintaining β-cell identity through the repression of the α-cell determinant Arx. In this analysis, we determined that although Nkx2.2 is present in both α and β cells, it preferentially binds the promoter of the α-cell-specific factor Arx in β cells when the promoter is in a hypermethylated state. Furthermore, we demonstrate that Nkx2.2 is present in a complex with the de novo methyltransferase Dnmt3a and repressor cofactor Grg3. In the β-cell context, Nkx2.2 preferentially recruits Grg3 and HDAC1 to the Arx promoter to repress Arx transcription. Disruption of the interaction between Nkx2.2 and Grgs in mice leads to ectopic expression of Arx in β cells and is sufficient to cause β-to-α-cell reprogramming during embryonic development and in the adult. We further show that by removing Arx specifically in the Nkx2.2TNmut/TNmut β cells, the β-to-α-cell conversion can be prevented. These combined in vitro and in vivo studies elucidate how cell-specific DNA modifications and transcriptional regulatory events come together to establish and maintain islet β-cell identity.
Results
Mutation of the Nkx2.2 TN domain disrupts the in vivo interaction between Nkx2.2 and the Grg3 corepressor and leads to diabetes in adult Nkx2.2TNmut/TNmut mice
In the pancreas, Grg2 and Grg3 are coexpressed with Nkx2.2 (Doyle et al. 2007; Hoffman et al. 2008), and Grg3 interacts with Nkx2.2 via the conserved TN domain (Doyle et al. 2007). To determine whether the physical interaction between Nkx2.2 and Grg proteins is necessary for appropriate pancreatic islet development and/or function, we generated mice that express a mutant allele of Nkx2.2 containing amino acid substitutions within the core sequence of the TN domain (Nkx2.2TNmut/TNmut) (Fig. 1A). Presence of the mutant allele was monitored using allele-specific primers (Fig. 1B; Materials and Methods). Heterozygous Nkx2.2TNmut/+ and homozygous Nkx2.2TNmut/TNmut mice display grossly normal pancreatic morphology and histology at birth (Fig. 1C,D; data not shown). Importantly, mutation of the TN domain does not appear to affect Nkx2.2 mRNA or protein stability in Nkx2.2TNmut/+ and Nkx2.2TNmut/TNmut mice (Fig. 1E,F; data not shown); however, we confirmed that mutation of the TN domain is sufficient to abolish the endogenous Nkx2.2–Grg3 interaction (Fig. 1G). The Nkx2.2TNmut/TNmut mice survive postnatally, but the male mice begin to display overt hyperglycemia by 3.5 wk of age (Supplemental Fig. 1A,B). By 6 wk, both male and female Nkx2.2TNmut/TNmut mice are severely diabetic in fed and fasted conditions, with compromised fertility (Fig. 2A; Supplemental Fig. 1C). The Nkx2.2TNmut/TNmut mice do not survive beyond 8 wk of age. Histological analysis of the 6-wk-old Nkx2.2TNmut/TNmut pancreas indicates that total islet cell numbers are unchanged, but the mutant islets are smaller and more disorganized than in wild-type littermate controls (Fig. 2B,C,F). Consistent with the phenotype of the Nkx2.2−/− mice, which form fewer α and no β cells, the Nkx2.2TNmut/TNmut islets have fewer insulin-producing β cells (Fig. 2D). Surprisingly, however, the number of glucagon-producing α cells appears to be increased (Fig. 2E). The Nkx2.2TNmut/TNmut mice do not display any apparent defects in δ-, pancreatic polypeptide (PP), or acinar cell populations (data not shown).
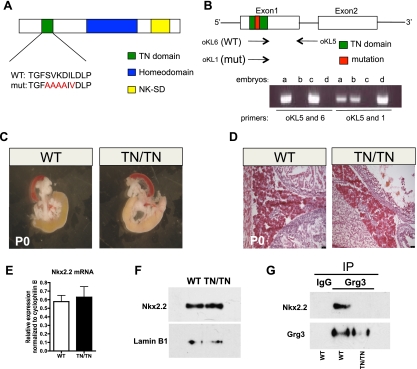
Figure 1.
The Nkx2.2 TN domain mutation disrupts the Nkx2.2–Grg3 interaction. (A) Recombinase-mediated cassette exchange (RMCE) was used to knock-in amino acid substitutions into the Nkx2.2 TN domain. The mutations are shown in red. (B) Allele-specific primers were used to distinguish between wild-type and mutant alleles when genotyping. (C) At birth, Nkx2.2TNmut/TNmut pancreata exhibit normal size and morphology compared with wild-type mice (n = 3). (D) H&E-stained section of wild-type and mutant pancreas showing normal endocrine and acinar morphology (n = 3). (E) Quantitative PCR demonstrates that Nkx2.2 transcript levels from whole embryonic pancreas (E18.5) are unchanged in the mutant (n = 5). (F) Protein levels of Nkx2.2 are unchanged in P0 pups. Lamin B1 was used as a loading control (n = 5). (G) Coimmunoprecipitation of Grg3 and Nkx2.2 from whole pancreas at birth demonstrates a loss of interaction between the two proteins in the Nkx2.2TNmut/TNmut mice (n = 3).
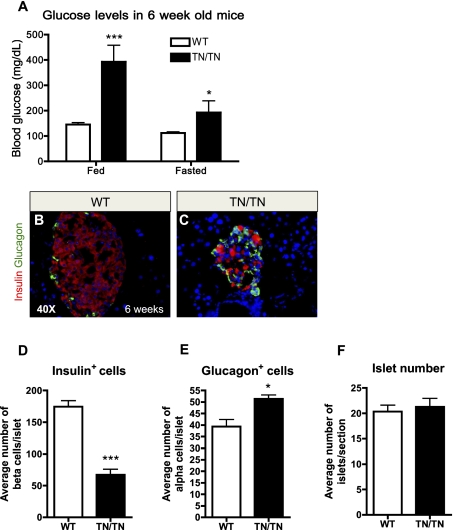
Figure 2.
Nkx2.2TNmut/TNmut mice develop diabetes. (A) Nkx2.2TNmut/TNmut mice exhibit hyperglycemia during fed and fasted states (n = 5). (B,C) Wild-type and mutant islet at 6 wk old immunostained for insulin and glucagon. By 6 wk of age, the Nkx2.2TNmut/TNmut mice contain smaller islets with fewer β cells and more α cells (n = 3). (D_–_F) Quantification of β cells and α cells at 4 wk of age demonstrates a significant loss of β cells and a significant increase in α cells, while no change in total islet number was observed (n = 3). (*) P < 0.05; (***) P < 0.001.
Nkx2.2 TN domain is required for appropriate islet cell development
Nkx2.2 is required for the primary induction of most endocrine cell lineages; in Nkx2.2−/− embryos, ghrelin-expressing cells form at the expense of all β cells and the majority of α and PP cells (Sussel et al. 1998). To determine whether the disrupted islet structure and abnormal islet cell ratios observed in the Nkx2.2TNmut/TNmut adult mice are caused by pancreatic defects manifested during embryonic development, we assessed endocrine cell formation in Nkx2.2TNmut/TNmut embryos. Unlike the Nkx2.2−/− embryos, which already display endocrine cell differentiation phenotypes by E9.5, the Nkx2.2TNmut/TNmut embryos display no apparent defects in α- and β-cell numbers at E12.5 (Fig. 3A–C). In agreement with the cell quantification, there are no detectable differences in glucagon or insulin gene expression between the Nkx2.2TNmut/TNmut embryos and their wild-type littermates (Fig. 3L). By E13.5, however, at the onset of the secondary transition, there is a reduction in β-cell numbers and insulin expression, with a corresponding increase in ɛ-cell numbers and ghrelin expression (Fig. 3L; Supplemental Fig. 1D–F,I,J). At E15.5, during the peak of endocrine cell differentiation, insulin-producing β-cell numbers remain significantly decreased, with a corresponding reduction in insulin (ins1 and ins2) gene expression (Fig. 3D,E,H,L; data not shown). The reduction in β-cell numbers does not appear to be due to apoptosis, as indicated by the absence of caspase3 expression in the insulin-producing cell populations or elsewhere in the pancreas (Supplemental Fig. 2A–D). Furthermore, ɛ-cell numbers and ghrelin gene expression are increased concomitantly with the loss of β cells, suggesting that, similar to the Nkx2.2−/− allele, ɛ cells are forming at the expense of the β-cell population (Fig. 3F–H,L). These phenotypes persist through the end of gestation (Fig. 3I–L).
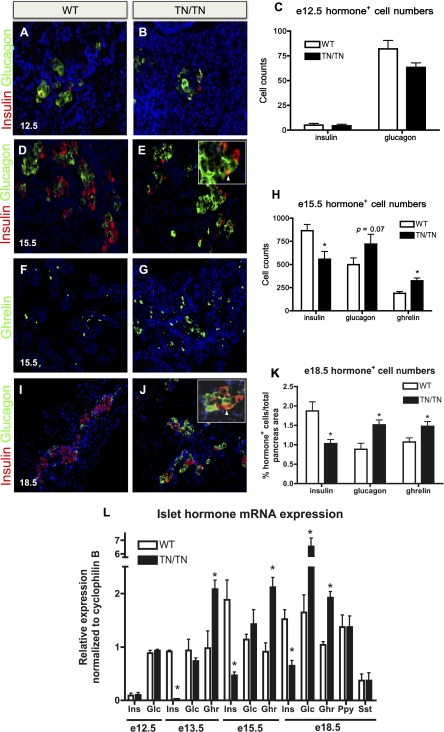
Figure 3.
Nkx2.2TNmut/TNmut mice display decreased insulin expression and increased glucagon and ghrelin expression. (A–L) Characterization of insulin-, glucagon-, and ghrelin-expressing cells in Nkx2.2TNmut/TNmut mice (n = 3–5). (A_–_C,L) Hormone-positive cell numbers and expression were unaltered at E12.5. (D_–_H,L) By E15.5, Nkx2.2TNmut/TNmut mice display increased ɛ-cell numbers, slightly elevated α-cell numbers, and decreased numbers of β cells. (I–L) By E18.5, the glucagon population is significantly increased, while the insulin and ghrelin populations remain decreased and increased, respectively. (*) P < 0.05. Arrowheads indicate glucagon/insulin costaining in E and J.
Surprisingly, in contrast to the Nkx2.2−/− phenotype, there is a significant increase in both α-cell numbers and glucagon gene expression in the Nkx2.2TNmut/TNmut pancreata by E18.5 (Fig. 3I–L). The increase in α cells does not appear to be caused by excessive replication, since we do not detect proliferating glucagon-positive cells before or during expansion of the α-cell population (Supplemental Fig. 2E–H). Although there is an increase in the numbers of α and ɛ single-hormone-positive cells, this is not associated with an increase in the glucagon+ ghrelin+ coexpressing population (data not shown). In contrast, we do observe an increase in the number of cells that coexpress insulin and glucagon, despite the total reduction in β-cell numbers (Fig. 3E,J, insets). Throughout gestation, PP and somatostatin levels remain unaffected in Nkx2.2TNmut/TNmut pancreata, and there are no obvious changes in exocrine tissue morphology, acinar cell numbers, or exocrine enzyme gene expression (Fig. 3L; data not shown).
Islet cell-specific transcription factor expression is disrupted in Nkx2.2TNmut/TNmut pancreas
Interestingly, the Nkx2.2TNmut/TNmut phenotype differs substantially from that observed in the Nkx2.2−/− embryos. While the less severe β-cell and ɛ-cell phenotypes could be attributed to mutations in the TN domain, having a more moderate reduction in Nkx2.2 function than the null allele, the resulting increase in α-cell numbers is more difficult to interpret. To begin to characterize the underlying molecular changes that may be contributing to the Nkx2.2TNmut/TNmut endocrine cell phenotype, we assessed the expression of several pancreatic developmental markers at E14.5, when alterations in insulin and ghrelin cell populations are evident. Similar to Nkx2.2−/− mice, expression of Pdx1 is unchanged at E14.5 and we see a reduction in Ngn3 expression, although there is no apparent reduction in the endocrine progenitor population (Fig. 4A; Anderson et al. 2009; data not shown). In contrast to the Nkx2.2−/− mice, Nkx6.1 expression is not significantly down-regulated at E14.5 (Fig. 4A).
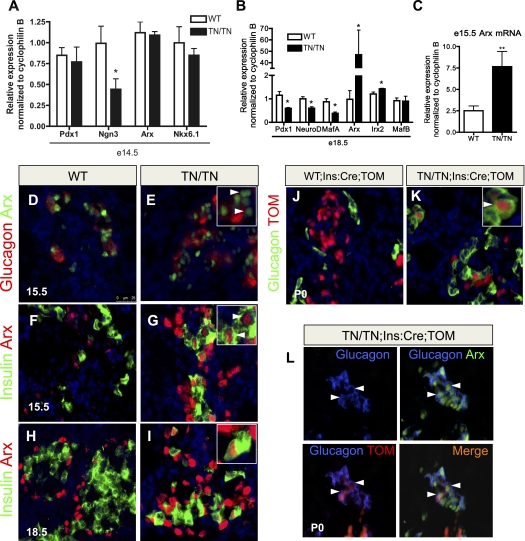
Figure 4.
Mutations in the Nkx2.2 TN domain cause Arx misexpression in β cells and β-to-α-cell reprogramming. (A) Islet cell-specific transcription factor expression at E14.5 (n = 5). (B) By E18.5, β-cell factors are reduced, while α-cell factors are increased, reflecting the changes associated with these cell types (n = 5). (C) Nkx2.2TNmut/TNmut mice exhibit a threefold to fourfold increase in Arx expression at E15.5, prior to the changes in glucagon expression and α-cell number (n = 5). (D,E) There are increased numbers of Arx+ cells and more Arx+/glucagon− cells in Nkx2.2TNmut/TNmut mice at E15.5, when the increase in Arx expression is first observed. (F–I) Between E15.5 and continuing through gestation, Arx is misexpressed in the β-cell population of Nkx2.2TNmut/TNmut mice. (J,K) Using the β-cell-specific Ins:Cre and the Rosa26:tomato reporter, β-cell-derived glucagon-positive cells are frequently detected in Nkx2.2TNmut/TNmut mice. (L) β-Cell-derived α cells express Arx (n = 3). (*) P < 0.05; (**) P < 0.01. Arrowheads in E indicate Arx+/glucagon− cells. Arrowheads in G, I, and K indicate copositive cells. Arrowheads in L indicate triple-positive cells.
By E18.5, altered expression of the islet lineage-specific factors corresponded well with the observed changes in islet cell ratios caused by the TN domain mutation. Consistent with the reduction in β-cell numbers, Pdx1, NeuroD, and MafA are significantly decreased by E18.5 (Fig. 4B). Alternatively, the α-cell markers Arx and Irx2 are significantly elevated by E18.5, with increases in Arx expression being close to 70-fold over wild-type levels (Fig. 4B). MafB expression does not appear changed; however, this may be attributed to the fact that MafB is expressed in both α- and β-cell populations during embryogenesis; the loss of _MafB_-expressing β cells would be compensated by the increase in _MafB_-expressing α cells. By 4 wk of age, MafB is expressed in the α cells in both wild-type and Nkx2.2TNmut/TNmut mice (Supplemental Fig. 3).
Interestingly, although the general changes in expression levels of the α- and β-cell-specific genes corresponded well with the observed alterations in the α- and β-cell numbers in the Nkx2.2TNmut/TNmut embryos, we could detect a threefold to fourfold increase in the expression of the α-cell transcription factor Arx at E15.5, prior to a significant increase in the number of glucagon+ cells (Fig. 4C). To identify the cellular source of increased Arx expression, we performed immunofluorescence analysis to correlate Arx expression with the different pancreatic populations. At E15.5, we observe a large increase in the number of Arx-expressing cells that do not coexpress glucagon in the Nkx2.2TNmut/TNmut (Fig. 4D,E). Surprisingly, at the same stage of development, we begin to observe a number of insulin-producing cells that ectopically express Arx (Fig. 4F,G). By the end of gestation, the Nkx2.2TNmut/TNmut pancreas contains Arx+, insulin+ β cells and Arx+, glucagon+ α cells, whereas pancreata from wild-type littermates show mutually exclusive expression of Arx and insulin. (Fig. 4H,I,L).
Mutation of the Nkx2.2 TN domain results in a β-to-α-cell conversion due to the ectopic expression of Arx in β cells
Previous studies have demonstrated that misexpression of Arx in embryonic β cells is sufficient to convert β cells into α cells (Collombat et al. 2007). Since we observe elevated Arx expression and Arx+ insulin-producing cells at E15.5, prior to a significant increase in α-cell numbers, we postulated that mutation of the Nkx2.2 TN domain may cause aberrant derepression of the Arx gene in β cells to trigger their transdifferentiation into α cells. To test this hypothesis, we generated Nkx2.2TNmut/TNmut mice carrying the Ins:Cre transgene (Herrera 2000) and either the Rosa26:tomato or Rosa26:LacZ reporter alleles (Soriano 1999; Madisen et al. 2010) to allow us to lineage trace the mutant β cells. While Rosa26 reporter expression is restricted to the insulin-expressing population in control Nkx2.2+/+;Ins:Cre;Rosa26:LacZ/tomato islets (Fig. 4J; Supplemental Fig. 4A,B), we detect Rosa26:tomato reporter expression in >2% of the glucagon-expressing cells of the Nkx2.2TNmut/TNmut ;Ins:Cre; Rosa26:LacZ/tomato islets at postnatal day 0 (P0) (Fig. 4K,L; Supplemental Fig. 4C,J). Many of the glucagon and tomato double-positive cells coexpress Arx, supporting the idea that Arx is up-regulated in the β-cell-derived glucagon-expressing population that appears in the absence of functional Nkx2.2 (Fig. 4L). Consistent with the discovery that β cells are reprogrammed to an α-cell fate in the Nkx2.2TNmut/TNmut mice, expression of a number of β-cell transcription factors—including Pdx1, MafA, and Nkx6.1—can be detected in a small population of cells that are in the process of converting to glucagon-expressing α cells (Supplemental Fig. 4D–I). The insulin- and glucagon-coexpressing population present in the Nkx2.2TNmut/TNmut pancreas may also represent this transitioning population (Fig. 3E,J). It should be noted that costaining of tomato with ghrelin cannot be detected, indicating, in contrast to the α-cell lineage, the observed increase in ɛ cells is not due to β-cell reprogramming (Supplemental Fig. 4K,L).
Since the Nkx2.2TNmut/TNmut pups are not born with a metabolic phenotype but continue to lose β cells and become increasingly hyperglycemic with age, we wished to determine whether the β-to-α-cell conversion continues to occur in postnatal animals. For this analysis, we generated Nkx2.2TNmut/TNmut mice carrying the Pdx1:CreER transgene (Gu et al. 2002) and the Rosa26:tomato reporter allele. To avoid outcomes that could be associated with a hyperglycemic environment, we induced 3-wk-old mice with tamoxifen prior to the onset of overt hyperglycemia (Supplemental Fig. 1). Five days following tamoxifen induction, we can detect tomato reporter expression in glucagon-producing α cells in the Nkx2.2TNmut/TNmut; Pdx1:CreER; Rosa26:tomato islets (n = 3), indicating that β-to-α-cell reprogramming continues to occur postnatally (Supplemental Fig. 5A–D). Glucagon expression and tomato reporter expression were mutually exclusive in control Nkx2.2TNmut/+; Pdx1:CreER; Rosa26:tomato littermates receiving tamoxifen. Nkx2.2TNmut/TNmut; Pdx1:CreER; Rosa26:tomato islets that did not receive tamoxifen were devoid of tomato reporter expression, as expected (Supplemental Fig. 5E,F).
Nkx2.2 directly binds and represses the methylated Arx promoter in β cells
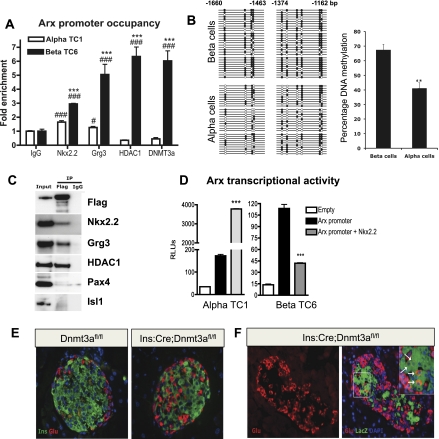
Given the dramatic increase in Arx expression in the Nkx2.2TNmut/TNmut β cells, it is possible that disruption of the interaction between Nkx2.2 and Grg3 interferes with repression of the Arx promoter in β cells. To determine whether Arx is a direct target of Nkx2.2, we performed in silico analysis of the Arx promoter to identify a highly conserved Nkx2.2-binding site located ∼1.4 kb upstream of the transcriptional start site (Supplemental Fig. 6A; Hill et al. 2011). Chromatin immunoprecipitation (ChIP) of this region with Nkx2.2 antibody in islet cell lines demonstrated that endogenous Nkx2.2 normally occupies this region of the Arx promoter in both α and β cells, although binding is significantly enriched in βTC6 cells (Fig. 5A). Furthermore, Grg3 and HDAC1 are preferentially recruited to the Arx promoter in β cells, supporting the idea that Nkx2.2 may function to recruit Grg3 to repress Arx expression in the β-cell population. Further analysis of the Arx promoter revealed the presence of CpG islands surrounding the conserved Nkx2.2-binding site, which, if methylated, could also contribute to the mechanism of Arx repression in β cells. Accordingly, bisulfite analysis for the Nkx2.2-binding region of the Arx promoter identified differential methylation patterns in β versus α cells; the Arx promoter is hypermethylated in β cells (Fig. 5B). Since the de novo DNA methyltransferase Dnmt3a has been shown to be important for establishing methylation patterns during development and promotes site-specific methylation through its interaction with transcription factors (Hervouet et al. 2009), we investigated whether Dnmt3a was also differentially present on the Arx promoter. Consistent with a role for Dnmt3a in the cell-specific regulation of Arx promoter methylation, Dnmt3a occupies the Arx promoter specifically in β cells (Fig. 5A). Furthermore, coimmunoprecipitation analysis demonstrates that Flag-tagged Dnmt3a can form a protein complex with Nkx2.2, but not other β-cell factors such as Pax4 and Isl1 (Fig. 5C). Grg3 and HDAC1 are also part of this complex, suggesting that Dnmt3a and Nkx2.2 cooperate to facilitate recruitment of a repressor complex to the Arx promoter in β cells. Interestingly, the Nkx2.2 TN domain appears to be dispensable for a direct interaction between Nkx2.2 and Dnmt3a (Supplemental Fig. 6B), suggesting that the two factors interact through another Nkx2.2 domain and/or through other proteins within the complex.
Figure 5.
Nkx2.2 directly represses Arx in βTC6 cells but not αTC1 cells. (A) Nkx2.2, Grg3, HDAC1, and Dnmt3a are highly enriched on the Arx promoter in βTC6 cells (n = 3). (B) Bisulfite sequencing analysis and quantification for the Nkx2.2-binding region of Arx locus (−1660 to −1463 base pairs [bp] and −1374 to −1162) in isolated β and α cells. (C) Coimmunoprecipitation analysis for the interaction of Dnmt3a with Nkx2.2, Grg3, HDAC1, Pax4, and Isl1 using immunoprecipitation with Flag tag antibody on Min6 cells transfected with Flag-Dnmt3a construct, and Western blotting with Flag tag, Nkx2.2, Grg3, HDAC1, Pax4, and Isl1 antibodies. (D) Nkx2.2 induces the expression of a 2.5-kb region of the Arx promoter fused to luciferase in αTC1 cells. Nkx2.2 represses the Arx promoter activity 3.5-fold in βTC6 cells (n = 4). (E) Immunostaining of representative pancreatic sections from 3-mo-old Dnmt3afl/fl (control) and Ins:Cre:Dnmt3afl/fl littermates for insulin and glucagon. (F) Representative pancreatic section from a 3-mo-old Ins:Cre:Dnmt3afl/fl:Rosa26:LacZ animal showing immunostaining for glucagon in the left panel and an overlay of glucagon with β-galactosidase activity (LacZ), in the right panel. The inset marks a magnified representative area, showing a number of LacZ and glucagon double-labeled cells (white arrows). (***) P < 0.001 versus α TC; (#) P < 0.05; (###) P < 0.001 versus IgG in A. (**) P < 0.01 versus β cells in B. (***) P < 0.001 versus Arx promoter in D.
To investigate whether Nkx2.2 specifically represses the Arx promoter in β cells, we subcloned a 2.5-kb region of the Arx promoter located upstream of the transcriptional start site into the pGL3 basic promoter-less luciferase reporter vector. Cotransfection of pGL3-Arx and Nkx2.2 into αTC1 cell lines resulted in a significant induction of Arx transcriptional activity, whereas cotransfection of pGL3-Arx and Nkx2.2 into βTC6 cell lines elicited a 65% repression of transcription (Fig. 5D). These data suggest that Nkx2.2 represses Arx activity in β cells through its interaction with Dnmt3a and Grg3 and preferential recruitment of HDAC1 to the Arx promoter in β cells. These findings are consistent with the in vivo results, suggesting that the Nkx2.2 TN domain is essential for the recruitment of Grg3 and maintaining repression of Arx in the β-cell population.
β-Cell-specific deletion of Dnmt3a also results in a β-to-α-cell conversion
Given the enrichment of Dnmt3a on the Arx promoter in β cells and its presence in the Nkx2.2, Grg3, and HDAC1 repressor complex, we wished to determine whether Dnmt3a is critical for proper islet cell type maintenance, similar to Nkx2.2. Interestingly, mice with a β-cell-specific deletion of Dnmt3a (Ins:Cre; Dnmt3afl/fl) phenocopy the Nkx2.2TNmut/TNmut mice; α cells are increased with a corresponding loss of β cells (Fig. 5E). Furthermore, genetic lineage tracing in Ins:Cre;Dnmt3afl/fl;Rosa26:LacZ mice demonstrates that the β-cell loss associated with deletion of Dnmt3a results in β-to-α-cell transdifferentiation, similar to the Nkx2.2TNmut/TNmut mice (Fig. 5F).
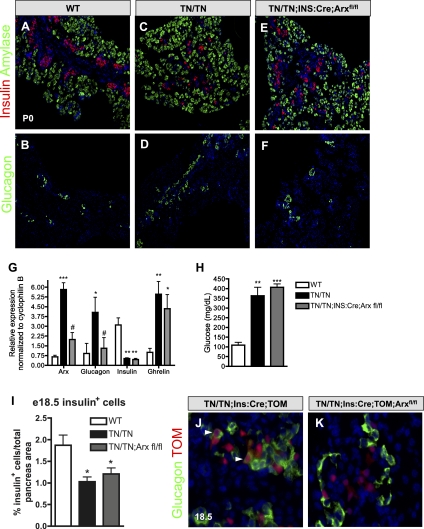
Removal of Arx from Nkx2.2TNmut/TNmut β cells is sufficient to prevent β-cell reprogramming, but fails to restore β-cell maturation and function
To determine whether inhibiting the ectopic expression of Arx in β cells is sufficient to prevent the Nkx2.2TNmut/TNmut β-to-α-cell reprogramming phenotype, we crossed Ins:Cre; Arxfl/fl mice (Herrera 2000; Marsh et al. 2009) into the Nkx2.2TNmut/TNmut background to generate mice that would remove the ectopically expressed Arx specifically from the Nkx2.2TNmut/TNmut β cells. Wild-type mice do not normally express Arx in β cells, and, accordingly, Ins:Cre; Arxfl/fl mice develop normal islets and are indistinguishable from wild-type mice (Supplemental Fig. 7A). Alternatively, deletion of Arx from the β cells of Nkx2.2TNmut/TNmut mice returned Arx to wild-type expression levels, confirming that the elevated Arx expression was due primarily to its ectopic expression in β cells (Fig. 6G). Furthermore, removal of Arx from Nkx2.2TNmut/TNmut β cells normalized both glucagon mRNA levels and α-cell numbers (Fig. 6B,D,F,G). Lineage tracing of the β cells in the Nkx2.2TNmut/TNmut;Ins:Cre; Arxfl/fl mice did not identify any β-cell-derived glucagon-expressing cells (Fig. 6J,K), suggesting that the deletion of Arx is sufficient to prevent the β-to-α-cell transdifferentiation. On the other hand, the β-cell deletion of Arx is not sufficient to restore β-cell numbers or insulin expression at E18.5, and adult Nkx2.2TNmut/TNmut; Ins:Cre;Arxfl/fl mice remained hyperglycemic (Fig. 6A,C,E,G–I). This suggests that the TN domain regulates additional Nkx2.2 targets, which are responsible for β-cell formation, maintenance, and function. Similarly, elevated ghrelin levels are unaffected by the removal of Arx from the Nkx2.2TNmut/TNmut mutant β cells, demonstrating that _Arx_-independent Nkx2.2 TN domain functions are also required for the ɛ- versus β-cell fate choice during embryogenesis (Fig. 6G).
Figure 6.
β-cell-specific loss of Arx in the Nkx2.2TNmut/TNmut mice rescues glucagon and Arx expression. (A–F) Characterization of insulin- and glucagon hormone-positive cells (n = 3). (G) Transcript levels of hormones in wild-type, Nkx2.2TNmut/TNmut, and Nkx2.2TNmut/TNmut;Ins:Cre;Arxfl/fl mice (n = 5). (H) Fed blood glucose levels in 7-wk-old male and female mice (n = 5). (I) Quantification of β cells as a percentage of total pancreas area in wild-type, Nkx2.2TNmut/TNmut, and Nkx2.2TNmut/TNmut;Ins:Cre;Arxfl/fl mice. (J,K) Using the β-cell-specific Ins:Cre and the Rosa26:tomato reporter, β-cell-derived ghrelin-positive cells are not detected in Nkx2.2TNmut/TNmut mutant mice (n = 3). (*) P < 0.05; (**) P < 0.01; (***)P < 0.001 versus wild type. (#) P < 0.05 versus Nkx2.2TNmut/TNmut.
To investigate whether the Ins:Cre; Dnmt3afl/fl phenotype is also dependent on Arx, we tested whether simultaneous depletion of Dnmt3a and Arx could rescue the β-to-α-cell transdifferentiation process. siRNAs targeting Dnmt3a, Arx, or a combination of the two genes were transfected into Min6 cells. Similar to the Ins:Cre; Dnmt3afl/fl mice, knockdown of Dnmt3a in Min6 cells resulted in β-to-α-cell reprogramming: Expression of insulin, Pdx1, and Pax4 was reduced, with a concomitant up-regulation of glucagon, Arx, and MafB. Combined siRNA knockdown of Arx with Dnmt3a was able to prevent the corresponding alterations in α- and β-cell marker expression, indicating that, similar to Nkx2.2, Dnmt3a-mediated reprogramming is dependent on Arx misexpression (Supplemental Fig. 7B).
Discussion
Transcriptional repression is widely appreciated for its role in determining lineage and cell specification, as repressive mechanisms are known to be key regulators of cellular commitment in many tissues and organisms (Gray et al. 1995; Gray and Levine 1996; Laslo et al. 2006; Jepsen et al. 2007; Rajasekhar and Begemann 2007; Li and Davidson 2009; Nishi et al. 2009). Nkx2.2 has been shown to act as a transcriptional repressor to regulate ventral neural patterning through its interaction with the corepressor Grg4 (Muhr et al. 2001). The ability of Nkx2.2 to act as a transcriptional repressor in the developing pancreas has also been shown (Doyle et al. 2007). Grg2 and Grg3, rather than Grg4, are coexpressed with Nkx2.2 in the pancreas and can also interact with Nkx2.2 through the groucho interaction TN domain (Doyle et al. 2007; Hoffman et al. 2008). In this study, we mutated the Nkx2.2 TN domain to determine the role of Grg-mediated Nkx2.2 repressor activity in the developing pancreas. We demonstrate that mutation of the Nkx2.2 TN domain abolishes the interaction between Nkx2.2 and Grg3 protein in vivo, and this is sufficient to cause a shift from the β-cell to the ɛ-cell lineage. Furthermore, many of the remaining β cells are reprogrammed into α cells due to the β-cell-specific derepression of the α-cell regulator Arx. Remarkably, it appears that Arx is the primary downstream target of Nkx2.2 in the maintenance of β-cell fate; removal of Arx from the Nkx2.2TNmut/TNmut β cells appears to completely inhibit the β-to-α-cell conversion.
The phenotypes associated with Nkx2.2TNmut/TNmut suggest that TN domain-mediated functions are required in two distinct processes within the pancreas: primary islet cell fate decisions and maintenance of β-cell identity. Initially, the TN domain appears to influence the competency of the Ngn3+ endocrine progenitor to differentiate into ɛ cells versus β cells, similar to what we observed in the Nkx2.2−/−-null mice. A proposed defect in cell fate choice regulation is supported by the observation that the corresponding alterations of β-cell and ɛ-cell ratios occurs at the onset of the secondary transition, which marks the first major wave of β-cell differentiation. Furthermore, we demonstrated that the increase in ghrelin-producing cells is not due to reprogramming of the β-cell population, since we did not observe ghrelin cells that were derived from the lineage-labeled β-cell population (Supplemental Fig. 4L). Although the Nkx2.2TNmut/TNmut and Nkx2.2−/− mice both display an increase in ɛ-cell numbers, coupled with a corresponding decrease in β cells, the phenotype is much more dramatic in the Nkx2.2−/− mice, which have a complete absence of β cells at all developmental stages. The less severe phenotype associated with the Nkx2.2TNmut/TNmut mice could indicate that the interaction between Nkx2.2 and Grgs is not completely disrupted, and residual activity is sufficient to allow for the formation of a small number of β cells. Alternatively, it is possible that other domains of the Nkx2.2 protein are responsible for β-cell formation during the primary transition stage of pancreas development, which would be consistent with the absence of a β-cell phenotype in the Nkx2.2TNmut/TNmut mice prior to E12.5 (Fig. 3A–C). A third possibility is that additional Nkx2.2 protein domains can compensate for the loss of TN domain activity to allow the formation of a small number of β cells throughout development. These possibilities are not mutually exclusive and will be explored in future studies to gain a more complete understanding of the role of Grg-mediated regulation of Nkx2.2 function in β-cell lineage decisions.
The Nkx2.2 TN domain also appears to play a distinct role in maintaining the β cells that are able to form in the Nkx2.2TNmut/TNmut mice. We show that by E18.5, there is an additional reduction in β cells that corresponds to an unexpected increase in α-cell numbers. Unlike the early changes we observed in β-cell versus ɛ-cell ratios, we do not observe a significant increase in α-cell numbers until the end of gestation. The observation that α-cell genes are becoming ectopically expressed in insulin-producing populations, combined with the β-cell lineage-tracing analysis, indicates that many of the α cells are derived from insulin-producing cells. This implies that Nkx2.2 is functioning within the β cell to maintain its cellular identity and prevent transdifferentiation into the α-cell fate. Interestingly, in the Nkx2.2TNmut/TNmut mice, we do not observe immediate and simultaneous reprogramming of all β cells in response to dysregulation of Arx. Instead, the transdifferentiation event occurs gradually over time and continues in the postnatal animal. This would suggest that, in addition to the activation of α-cell lineage inducers, such as Arx, a number of additional (perhaps more resistant) cellular events, such as global chromatin remodeling, must occur to allow reprogramming to be initiated. Further epigenetic analyses and gene expression profiling of FACs-purified β-cell populations derived from the Nkx2.2TNmut/TNmut mice during intermediate stages of transdifferentiation may elucidate the β-cell-determining features that must be overcome to allow β-to-α-cell reprogramming.
Interestingly, the Nkx2.2TNmut/TNmut mice also differ quite significantly from the Nkx2.2−/− phenotype with regards to PP- and α-cell formation. Unlike the Nkx2.2−/− mice, which display an ∼50% reduction in PP cells, there is no apparent change in PP-cell numbers or PPY expression in the Nkx2.2TNmut/TNmut mice, suggesting that PP-cell formation is not regulated by TN domain functions. Even more puzzling was the unexpected α-cell phenotype associated with the Nkx2.2TNmut/TNmut mice. We previously demonstrated that the Pdx1:Nkx2.2hdEnR repressor derivative was sufficient to fully rescue α-cell numbers in the Nkx2.2−/− mice (Doyle et al. 2007), suggesting that Nkx2.2 repressor function is sufficient for α-cell formation. However, removal of the Nkx2.2 TN repressor domain does not appear to negatively affect the α-cell population at any stage of development, and instead we observe an increase in α-cell numbers. This may indicate that Grg-dependent repression is dispensable for the α-cell lineage specification or perhaps that other regions of the Nkx2.2 protein can compensate for the loss of this interaction and/or provide an alternative mechanism of repressive activity. We are currently exploring the possibility that the conserved NK2-SD domain, the defining domain of the Nkx2 family, can contribute to Nkx2.2 repressor activity in vivo, as has been suggested previously (Watada et al. 2000). The data presented here now give us a greater understanding of a previously unanticipated role for the Nkx2.2 TN domain in maintaining β-cell identity through its repression of the Arx promoter in β cells. This function would not be revealed in Nkx2.2−/− mice, since there are no β cells available to reprogram. This unexpected result not only begins to elucidate the distinct functions of Nkx2.2 in different islet cell types and at different stages of development, but also emphasizes the usefulness of generating hypomorphic and/or domain-specific alleles to understand the nuances of a protein's function in vivo.
Our study also re-emphasizes the importance of maintaining the appropriate regulation of Arx expression in the different islet cell types. Previous studies have demonstrated that Arx is essential for specifying the α- and PP-cell lineages (Collombat et al. 2003, 2005, 2007). Disruption of Arx leads to reduced numbers of α cells coupled with a corresponding increase in β and δ cells (Collombat et al. 2003). Alternatively, misexpression of Arx in β cells is sufficient to convert β cells to α or PP cells (Collombat et al. 2007). Interestingly, although we recapitulate the β-to-α-cell reprogramming phenomenon with the derepression of Arx in β cells, we are unable to detect significant changes in PP expression. It is possible that the increased levels of Arx caused by the Nkx2.2 TN mutation are sufficient for the α-cell fate but are not sufficient for conversion to a PP-cell fate. This is consistent with studies showing that protein expression levels and dosing requirements of transcription factors are important for controlling islet cell development and islet cell fate choices (Fujitani et al. 2006; Wang et al. 2009). Alternatively, removal of the Nkx2.2 TN domain likely affects additional downstream effectors that may function to counter the Arx regulation of the PP fate.
Differential expression of transcription factors can allow for tissue- and cell-specific gene regulation; however, when a factor is expressed in more than one cell type, higher-order regulation will be required to control cell- and/or promoter-specific regulation. Differential promoter regulation can be achieved by cell-specific cofactor interactions or at the level of DNA accessibility or both. In the case of Nkx2.2, we demonstrated that it preferentially binds the Arx promoter in β cells, although Nkx2.2 is expressed and functions in both α and β cells. The differential binding ability of Nkx2.2 may be influenced by the methylation state of the Arx promoter that surrounds the Nkx2.2 consensus binding site and is possibly facilitated by the DNA modifications induced by Dnmt3a. A cooperative role between these factors for maintaining β-cell-specific Arx repression and β-cell identity is supported by the similar reprogramming phenotypes displayed by the β-cell-specific deletion of Dnmt3a and the Nkx2.2 TN mutant mice. Therefore, it is possible that differential DNA methylation and the presence of Dnmt3a on the Arx promoter provide a cell-specific platform for Nkx2.2 binding on the Arx promoter in β cells. However, Dnmt3a is also expressed in both α and β cells, suggesting that its preferential recruitment to the Arx promoter in β cells must also rely on a β-cell-specific factor. Nkx6.1 is a possible candidate for mediating β-cell-specific recruitment of Dnmt3a to the Arx promoter, and we are able to demonstrate that Nkx6.1 is also present on the Arx promoter in β cells (Supplemental Fig. 6). Further studies will be necessary to determine the sequence of events that occur on the Arx promoter with regards to protein recruitment and DNA methylation to initiate and maintain transcriptional repression of Arx in β cells.
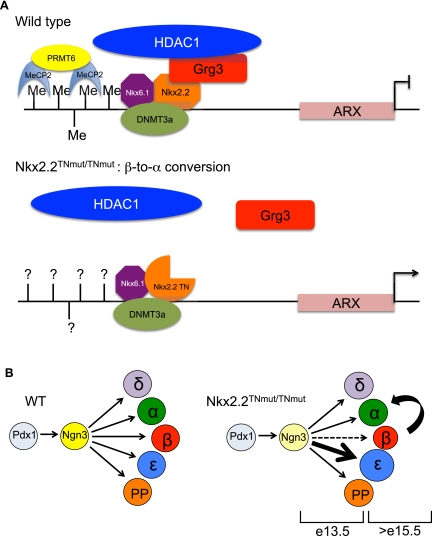
Since we can also detect Nkx2.2 bound to the Arx promoter in α cells, albeit at lower levels, it remains possible that Nkx2.2-mediated repression of Arx in β cells also depends on a combinatorial effect between methyl-binding proteins and Nkx2.2. Dhawan et al. (2011) demonstrated that the methyl-binding protein MeCP2 is also bound to the methylated Arx promoter in β cells. Methyl-binding proteins have been shown to tether histone deacetylase complexes, such as NuRD, to methylated DNA (Zhang et al. 1999). Furthermore, recruitment of these repressor complexes can be facilitated by sequence-specific transcription factors (Kehle et al. 1998; Liang et al. 2008; Fu et al. 2011). Since Nkx2.2 preferentially recruits Grg3 and HDAC1 to the Arx promoter in β cells, it is possible that Nkx2.2 cooperates with methyl-binding proteins to recruit histone deacetylases and achieve repression of Arx. Since it appears that the initiation and maintenance of Arx expression in β cells is crucial for maintaining β-cell identity, it is likely that a combination of these mechanisms is necessary to cooperatively repress Arx and retain β-cell integrity (schematized in Fig. 7A). Additionally, given the small but significant recruitment of Nkx2.2 and Grg3 to the Arx promoter in α cells (Fig. 5A) and the Nkx2.2-mediated induction of Arx in α cells (Fig. 5D), we cannot rule out the possibility that Nkx2.2 and Grg3 may be facilitating the activation of Arx in α cells. While Grgs are generally described as corepressors, a recent study demonstrated that Grg3 is able to function as both an activator and a repressor during adipogenesis (Villanueva et al. 2011).
Lineage-specific gene expression patterns rely on a complex interplay between transcription factors, cofactors, and DNA accessibility. This process becomes much more complicated when a single factor has differential functions across development and within unique cell types. Our analysis highlights the complex regulation of lineage commitment within the endocrine pancreas and draws attention to the need to dissect out the cell-specific functions of each regulatory factor to fully understand lineage specification and maintenance. The phenotype of mice carrying a disruption of the Nkx2.2 TN domain has revealed that the Grg-mediated repressor activity of Nkx2.2 controls two stages of development (schematized in Fig. 7B): (1) Grg-mediated Nkx2.2 repression is not required for the early commitment of β versus α cells, but is required for the correct balance of β and ɛ cells that form at the beginning of the secondary transition. (2) The functions mediated by this interaction are also critical for the maintenance of β-cell identity by preventing ectopic expression of Arx in β cells. This emphasizes the importance of understanding the spatial and temporal regulation of cofactors in lineage commitment decisions and underscores the potential importance of epigenetic and transcription factor interactions to further regulate promoter occupancy. Our studies, coupled with those of Dhawan et al. (2011), suggest that Nkx6.1, Nkx2.2, Dnmt3a, and MeCP2 converge on methylated DNA of the Arx promoter specifically in β cells to recruit histone modifiers (HDAC1 and PRMT6, respectively), in order to silence Arx expression (schematized in Fig. 7). Further understanding of the precise molecular events of islet cell-specific transcriptional networks on individual promoters will aid in formulating effective protocols to induce and maintain appropriate endocrine lineage commitment from alternative cell sources, such as embryonic stem cells or induced pluripotent cells, for the treatment of diabetes.
Figure 7.
Model of Nkx2.2 regulation of the Arx promoter in β cells. (A) Our data, in combination with the study by Dhawan et al. (2011), support a model whereby a repressor complex comprised of Nkx2.2, Grg3, and Dnmt3a preferentially recruits HDAC1 to the Arx promoter to silence Arx expression specifically in the pancreatic β cells to prevent β-to-α-cell conversion. We can also detect the presence of Nkx6.1 bound to this promoter region, which possibly confers cell-specific recruitment of the Nkx2.2/Dnmt3a complex to the Arx promoter. Since Nkx2.2TNmut/TNmut mice no longer retain a Nkx2.2/Grg3 interaction, Grg3 and HDAC1 are no longer recruited to the Arx promoter, allowing ectopic expression of Arx in β cells. It is not yet known whether Nkx2.2TNmut/TNmut mice affect Dnmt3a recruitment and subsequent methylation of the Arx promoter. (B) Grg3-mediated Nkx2.2 repression controls two developmental stages of pancreas development. We first observe aberrations in the proper specification of β and ɛ cells at E13.5 in the Nkx2.2TNmut/TNmut mice, when an increased number of ɛ cells are forming at the expense of β cells. The remaining β cells then go on to transdifferentiate to α cells. This transdifferentiation is evident by E18.5 and continues to occur postnatally.
Materials and methods
Generation of Nkx2.2TNmut mice
The Nkx2.2 TN domain mutant (Nkx2.2TNmut) mice were generated using the two-step recombination-mediated cassette exchange (RMCE) technology (Chen et al. 2011) to insert amino acid substitutions in the endogenous Nkx2.2 locus. To do so, we first engineered an Nkx2.2 loxed cassette acceptor (Nkx2.2LCA) allele using the strategy and plasmids described in Chen et al. (2011) (L Arnes, K Leclerc, JM Friel, SB Hipkens, MA Magnuson, and L Sussel, in prep.). Second, to generate the Nkx2.2TNmut allele, we then made a basal exchange Nkx2.2 targeting vector containing a 5.1-kb genomic (129-S6) DNA fragment encompassing the two Nkx2.2 coding exons. PCR-mediated site-directed mutation of the TN domain was performed using the QuikChange Site-Directed Mutagenesis kit (Stratagene) to replace the conserved TN domain core SVKD12–15 amino acids with four alanine residues and the invariant leucine (L17) with valine, an equally hydrophobic amino acid. We also introduced a SalI restriction site for genotyping purposes. All mutations were verified by DNA sequencing. The Nkx2.2TNmut exchange vector was used to perform RMCE using mouse embryonic stem cells containing the Nkx2.2LCA allele. A dual, positive-negative selection strategy was used to identify clones that had undergone cassette exchange (Chen et al. 2011), which were then confirmed by DNA PCR. Positive clones were further analyzed by Southern blot analysis for the presence of the novel SalI restriction site. Nkx2.2TNmut/+ embryonic stem cells were injected into blastocysts obtained from the mating of C57Bl/6 mice, then transferred to pseudopregnant C57Bl/6 mice. Male chimeras were bred to C57Bl/6J female mice, and agouti offspring were PCR-genotyped using Nkx2.2 allele-specific primers that distinguish between the wild-type and Nkx2.2TNmut allele (Fig. 1B): oKL6 (FWD, wild type), GTCAAGGACATCTTGGACCTTCCG; oKL1 (FWD, Nkx2.2TNmut), TTTGCAGCAGCGGCAATC; and oKL5 (REV, wild type and Nkx2.2TNmut), ATCCGGGATGGGACTTGGACATTA. The FRT-flanked hygromycin cassette was subsequently removed by mating to FlpE mice (Rodriguez et al. 2000). All procedures to generate the Nkx2.2TNmut mice were approved by the Vanderbilt University Animal Care and Use Committee.
Animal maintenance
All mice were maintained on a Swiss Black (Taconic) background and housed and treated according to the Columbia University Institutional Animal Care and Use Committee approval protocol. Genotyping of Nkx2.2TNmut mice and embryos was performed using Nkx2.2 allele-specific primers (described above). Genotyping of Arxflox/flox mice, Ins:Cre mice, Rosa26:Tomato mice, Rosa26:LacZ mice, and Pdx:CreER mice was performed as previously described (Herrera 2000; Gu et al. 2002; Marsh et al. 2009; Madisen et al. 2010).
Immunohistochemistry
Tissues were fixed in 4% PFA for 4 h or overnight, washed in cold PBS, incubated in 30% sucrose, and cryopreserved. Immunofluorescence was performed on 7 μM sections. See Supplemental Table 1 for a list of antibodies used. Fluorescent images were obtained with a Nikon Eclipse 80i microscope, Q-image camera, and ImagePro software (Media Cybernetics). Confocal images were captured using a Zeiss LSM 510 upright (pinhole = 1). For quantification purposes, stained cells were counted manually on every fourth section (E12.5 and eE13.5) and every eighth section (E15.5) throughout the entire pancreas for the wild type (n = 3) and mutant (n = 3). For animals at E18.5, the number of stained cells, which were counted on every eighth section, was divided by total pancreas area. All values are expressed as mean ± SEM. Statistical analysis was performed using a two-tailed Student's unpaired _t_-test. Results were considered significant when P < 0.05.
RNA analysis
Total RNA was isolated from whole pancreatic tissue from the respective embryonic ages (RNeasy, Qiagen), and cDNA was prepared using the SuperScript III kit (Invitrogen) and random hexamer primers. Quantitative PCR was performed using 200 ng of cDNA, PCR master mix (Eurogentec), and TaqMan probes (ABI Assays on Demand). See Supplemental Table 2 for a list of primers used. All genes were normalized to Cyclophilin B and were quantified with ABI prism software. An n ≥ 4 was obtained for all mouse populations. All values are expressed as mean ± SEM. Statistical analysis was performed using a two-tailed Student's unpaired _t_-test. Results were considered significant when P < 0.05.
ChIP
α-TC1 and β-TC6 cells were grown to ∼80% confluency in a 15-cm culture dish and were formaldehyde-cross-linked according to the ChIP-IT Express kit (Active Motif). Cross-linked chromatin was fragmented by sonication using a Diagnode BioRupter (8 min to 30 sec on/off). Three micrograms of mouse α-Nkx2.2 (Developmental Studies Hybridoma Bank [DSHB]), rabbit α-TLE3 (Santa Cruz Biotechnologies), rabbit α-HDAC1 (Santa Cruz Biotechnologies), rabbit α-Nkx6.1 (Beta Cell Biology Consortium), or mouse α-IgG (AbCam) was added to the sheared chromatin. The antibody/chromatin complexes were left to rotate end to end overnight at 4°C. Antibody/chromatin complexes were pulled down using ChIP-grade protein G magnetic beads (Cell Signaling). Chromatin was washed, eluted, and reverse-cross-linked, followed by protease treatment. Chromatin fragments were then analyzed by quantitative PCR using SYBR Green fluorescence with the following primers: Arx (FWD), TCCTCCACCATTTGAGGGTA; and (REV), GCAACTTGAGGGGGTACAGA. All values are expressed as mean ± SEM. Statistical analysis was performed using a two-tailed Student's unpaired _t_-test. Results were considered significant when P < 0.05.
Coimmunoprecipitation and Western
Nuclear extraction was performed on excised pancreata from P0 mouse pups using the Nuclear Complex Co-IP kit (Active Motif). Nuclear extract was pooled from five wild-type or five mutant animals. One-hundred-fifty micrograms of nuclear extract and 5 μg of rabbit anti-TLE3 (Santa Cruz Biotechnologies), rabbit α-Dnmt3a (Santa Cruz Biotechnologies), mouse α-Nkx2.2 (DSHB), or rabbit anti-IgG (Chemicon) were incubated overnight at 4°C, rotating end to end. Protein/antibody complexes were pulled down using protein G DynaBeads (Invitrogen). Protein/antibody complexes were washed, eluted, and run on a NuPAGE Novex 10% Bis-Tris gel (Invitrogen). Proteins were transferred onto a nitrocellulose membrane (GE Healthcare) and probed using mouse anti-Nkx2 or rabbit α-Dnmt3a. For immunoprecipitation control, the membrane was stripped, washed, and reprobed with the antibody that was used for the initial immunoprecipitation.
Luciferase assays
To generate the _Arx_-Luciferase reporter, a 2.5-kb fragment of the Arx genomic region upstream of the transcriptional start site was PCR-amplified from mouse tail genomic DNA using the following primers: oRS18 (FWD), ACGCGTTGCACCTCCCCCTTTACAGTGGGTGATTA; and oRS19 (REV), AGATCTTGGGCTCAGAGAGCGGACCT. MluI and BglII sites were incorporated into the 5′ and 3′ ends, respectively, to facilitate cloning into the pGL3 basic promoter-less luciferase reporter vector using MluI and BglII cloning sites. The resulting plasmid was verified by DNA sequencing. Luciferase assays were performed in αTC1 and βTC6 cell lines using the Dual-Luciferase Reporter Assay System (Promega) as described previously (Hill et al. 2010).
Tamoxifen treatment in Pdx:CreER;R26R:Tomato mice
Pdx:CreER was induced with a 5-mg intraperitoneal injection of tamoxifen (Sigma) in 3-wk-old mice every 48 h for 4 d. On the fifth day, pancreata were harvested and fixed in paraformaldehyde, as described above, and immunostained for indicated hormones.
In vitro siRNA-mediated knockdown
Knockdown of Dnmt3a, Arx, and Dnmt3a+Arx in Min6 cells was performed by transfection of specific-targeting small inhibitory RNAs or scrambled controls (Dharmacon Research, Inc.) using Lipofectamine-2000 (Invitrogen), according to the manufacturer's instructions, in OPTI-MEM medium. Min6 cells were transfected with appropriate siRNAs or scrambled controls every 3 d (average transfection efficiency, 65%–80%), and samples were harvested at 4 d post-transfection, as indicated. RNA was isolated from cells using TRI Reagent (MRC) and treated with DNase (Ambion) according to the manufacturers' instructions. One microgram of RNA was used for preparation of single-stranded cDNA using SuperScript III reverse transcriptase (Invitrogen) by the oligodT priming method. Real-time RT–PCRs were performed using the Fast SYBR Green Master Mix (Applied Biosystems) and the 7900HT Fast Real-Time PCR equipment (Applied Biosystems). The expression levels of each transcript were normalized to the housekeeping gene Cyclophilin. Each real-time PCR experiment shown is a representative from at least three independent experiments. Primers for RT–PCR are listed in Supplemental Table 2.
Acknowledgments
We thank Catherine Lee May (CHOP) and Jeffrey Golden (CHOP) for providing the _Arx_-floxed mice, and Kanako Miyabayashi (Kyushu University, Fukuoka, Japan) for providing Arx antibody. We thank Jonathon Hill for identifying the conserved Nkx2.2-binding site within the Arx promoter. We thank Drs. Lori Zeltser and Stephanie Padilla for critical reading of the manuscript, Ms. Kathy D. Shelton for assembling the gene targeting vector, and the staff of the Vanderbilt Transgenic/ES Cell Shared Resource for their expert performance of the blastocyst microinjection experiments. This work was supported by NIH grants DK0272504 (to L.S.), DK082590 (to L.S. and J.P.), DK042502 (to M.A.M.), DK072473 (to M.A.M.), and DK068763 (to A.B.). Additional support was provided by the Columbia University DERC (P30 DK63608) and CTSA grants.
Footnotes
Supplemental material is available for this article.
References
- Anderson KR, Torres CA, Solomon K, Becker TC, Newgard CB, Wright CV, Hagman J, Sussel L 2009. Cooperative transcriptional regulation of the essential pancreatic islet gene NeuroD1 (β2) by Nkx2.2 and neurogenin 3. J Biol Chem 284: 31236–31248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK 2009. Adult mice generated from induced pluripotent stem cells. Nature 461: 91–94 [DOI] [PubMed] [Google Scholar]
- Briscoe J, Sussel L, Serup P, Hartigan-O'Connor D, Jessell TM, Rubenstein JL, Ericson J 1999. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature 398: 622–627 [DOI] [PubMed] [Google Scholar]
- Chan L, Fujimiya M, Kojima H 2003. In vivo gene therapy for diabetes mellitus. Trends Mol Med 9: 430–435 [DOI] [PubMed] [Google Scholar]
- Chen SX, Osipovich AB, Ustione A, Potter LA, Hipkens S, Gangula R, Yuan W, Piston DW, Magnuson MA 2011. Quantification of factors influencing fluorescent protein expression using RMCE to generate an allelic series in the ROSA26 locus in mice. Dis Model Mech 4: 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cissell MA, Zhao L, Sussel L, Henderson E, Stein R 2003. Transcription factor occupancy of the insulin gene in vivo. Evidence for direct regulation by Nkx2.2. J Biol Chem 278: 751–756 [DOI] [PubMed] [Google Scholar]
- Collombat P, Mansouri A, Hecksher-Sorensen J, Serup P, Krull J, Gradwohl G, Gruss P 2003. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev 17: 2591–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Hecksher-Sorensen J, Broccoli V, Krull J, Ponte I, Mundiger T, Smith J, Gruss P, Serup P, Mansouri A 2005. The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the α- and β-cell lineages in the mouse endocrine pancreas. Development 132: 2969–2980 [DOI] [PubMed] [Google Scholar]
- Collombat P, Hecksher-Sorensen J, Krull J, Berger J, Riedel D, Herrera PL, Serup P, Mansouri A 2007. Embryonic endocrine pancreas and mature β cells acquire α and PP cell phenotypes upon Arx misexpression. J Clin Invest 117: 961–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delisle JC, Martignat L, Dubreil L, Sai P, Bach JM, Louzier V, Bosch S 2009. Pdx-1 or Pdx-1-VP16 protein transduction induces β-cell gene expression in liver-stem WB cells. BMC Res Notes 2: 3 doi: 10.1186/1756-0500-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan S, Georgia S, Tschen SI, Fan G, Bhushan A 2011. Pancreatic β cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell 20: 419–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MJ, Sussel L 2007. Nkx2.2 regulates β-cell function in the mature islet. Diabetes 56: 1999–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MJ, Loomis ZL, Sussel L 2007. Nkx2.2-repressor activity is sufficient to specify α-cells and a small number of β-cells in the pancreatic islet. Development 134: 515–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Qin L, He T, Qin J, Hong J, Wong J, Liao L, Xu J 2011. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res 21: 275–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Boyer DF, Gannon M, Kawaguchi Y, Ray M, Shiota M, Stein RW, Magnuson MA, Wright CV 2006. Targeted deletion of a _cis_-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes Dev 20: 253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefen-Halevi S, Rachmut IH, Molakandov K, Berneman D, Mor E, Meivar-Levy I, Ferber S 2010. NKX6.1 promotes PDX-1-induced liver to pancreatic β-cells reprogramming. Cell Reprogram 12: 655–664 [DOI] [PubMed] [Google Scholar]
- Gittes GK 2009. Developmental biology of the pancreas: a comprehensive review. Dev Biol 326: 4–35 [DOI] [PubMed] [Google Scholar]
- Gray S, Levine M 1996. Transcriptional repression in development. Curr Opin Cell Biol 8: 358–364 [DOI] [PubMed] [Google Scholar]
- Gray S, Cai H, Barolo S, Levine M 1995. Transcriptional repression in the Drosophila embryo. Philos Trans R Soc Lond B Biol Sci 349: 257–262 [DOI] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA 2002. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129: 2447–2457 [DOI] [PubMed] [Google Scholar]
- Haumaitre C, Lenoir O, Scharfmann R 2009. Directing cell differentiation with small-molecule histone deacetylase inhibitors: the example of promoting pancreatic endocrine cells. Cell Cycle 8: 536–544 [DOI] [PubMed] [Google Scholar]
- Herrera PL 2000. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development 127: 2317–2322 [DOI] [PubMed] [Google Scholar]
- Hervouet E, Vallette FM, Cartron PF 2009. Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics 4: 487–499 [DOI] [PubMed] [Google Scholar]
- Hill JT, Chao CS, Anderson KR, Kaufman F, Johnson CW, Sussel L 2010. Nkx2.2 activates the ghrelin promoter in pancreatic islet cells. Mol Endocrinol 24: 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JT, Anderson KR, Mastracci TL, Kaestner KH, Sussel L 2011. Novel computational analysis of protein binding microarray data identifies direct targets of Nkx2.2 in the pancreas. BMC Bioinformatics 12: 62 doi: 10.1186/1471-2105-12-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BG, Zavaglia B, Beach M, Helgason CD 2008. Expression of Groucho/TLE proteins during pancreas development. BMC Dev Biol 8: 81 doi: 10.1186/1471-213X-8-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D 2010. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, Glass CK, Hermanson O, Rosenfeld MG 2007. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 450: 415–419 [DOI] [PubMed] [Google Scholar]
- Jimenez G, Paroush Z, Ish-Horowicz D 1997. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev 11: 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Belmonte JC 2010. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464: 606–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Boue S, Izpisua Belmonte JC 2011. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat Rev Mol Cell Biol 12: 79–89 [DOI] [PubMed] [Google Scholar]
- Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, Muller J 1998. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282: 1897–1900 [DOI] [PubMed] [Google Scholar]
- Kojima H, Fujimiya M, Matsumura K, Younan P, Imaeda H, Maeda M, Chan L 2003. NeuroD-βcellulin gene therapy induces islet neogenesis in the liver and reverses diabetes in mice. Nat Med 9: 596–603 [DOI] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H 2006. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 126: 755–766 [DOI] [PubMed] [Google Scholar]
- Li E, Davidson EH 2009. Building developmental gene regulatory networks. Birth Defects Res C Embryo Today 87: 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, et al. 2008. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol 10: 731–739 [DOI] [PubMed] [Google Scholar]
- Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP 1993. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119: 419–431 [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13: 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh E, Fulp C, Gomez E, Nasrallah I, Minarcik J, Sudi J, Christian SL, Mancini G, Labosky P, Dobyns W, et al. 2009. Targeted loss of Arx results in a developmental epilepsy mouse model and recapitulates the human phenotype in heterozygous females. Brain 132: 1563–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsky R, Woodhoo A, Parkinson DB, Arthur-Farraj P, Bhaskaran A, Jessen KR 2008. Novel signals controlling embryonic Schwann cell development, myelination and dedifferentiation. J Peripher Nerv Syst 13: 122–135 [DOI] [PubMed] [Google Scholar]
- Muhr J, Andersson E, Persson M, Jessell TM, Ericson J 2001. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell 104: 861–873 [DOI] [PubMed] [Google Scholar]
- Nagaya M, Katsuta H, Kaneto H, Bonner-Weir S, Weir GC 2009. Adult mouse intrahepatic biliary epithelial cells induced in vitro to become insulin-producing cells. J Endocrinol 201: 37–47 [DOI] [PubMed] [Google Scholar]
- Nishi Y, Ji H, Wong WH, McMahon AP, Vokes SA 2009. Modeling the spatio-temporal network that drives patterning in the vertebrate central nervous system. Biochim Biophys Acta 1789: 299–305 [DOI] [PubMed] [Google Scholar]
- Rajasekhar VK, Begemann M 2007. Concise review: roles of polycomb group proteins in development and disease: a stem cell perspective. Stem Cells 25: 2498–2510 [DOI] [PubMed] [Google Scholar]
- Raum JC, Gerrish K, Artner I, Henderson E, Guo M, Sussel L, Schisler JC, Newgard CB, Stein R 2006. FoxA2, Nkx2.2, and PDX-1 regulate islet β-cell-specific mafA expression through conserved sequences located between base pairs −8118 and −7750 upstream from the transcription start site. Mol Cell Biol 26: 5735–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet 25: 139–140 [DOI] [PubMed] [Google Scholar]
- Selvaraj V, Plane JM, Williams AJ, Deng W 2010. Switching cell fate: the remarkable rise of induced pluripotent stem cells and lineage reprogramming technologies. Trends Biotechnol 28: 214–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ST, Jaynes JB 1996. A conserved region of engrailed, shared among all en-, gsc-, Nk1-, Nk2- and msh-class homeoproteins, mediates active transcriptional repression in vivo. Development 122: 3141–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YD, Lee EJ, Yashar P, Pfaff LE, Kim SY, Jameson JL 2007. Islet cell differentiation in liver by combinatorial expression of transcription factors neurogenin-3, β2, and RIPE3b1. Biochem Biophys Res Commun 354: 334–339 [DOI] [PubMed] [Google Scholar]
- Soriano P 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21: 70–71 [DOI] [PubMed] [Google Scholar]
- Sussel L, Kalamaras J, Hartigan-O'Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, German MS 1998. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic β cells. Development 125: 2213–2221 [DOI] [PubMed] [Google Scholar]
- Szabo E, Rampalli S, Risueno RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M 2010. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 468: 521–526 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, Herrera PL 2010. Conversion of adult pancreatic α-cells to β-cells after extreme β-cell loss. Nature 464: 1149–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsonis PA, Madhavan M, Tancous EE, Del Rio-Tsonis K 2004. A newt's eye view of lens regeneration. Int J Dev Biol 48: 975–980 [DOI] [PubMed] [Google Scholar]
- Villanueva CJ, Waki H, Godio C, Nielsen R, Chou WL, Vargas L, Wroblewski K, Schmedt C, Chao LC, Boyadjian R, et al. 2011. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab 13: 413–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Jensen JN, Seymour PA, Hsu W, Dor Y, Sander M, Magnuson MA, Serup P, Gu G 2009. Sustained Neurog3 expression in hormone-expressing islet cells is required for endocrine maturation and function. Proc Natl Acad Sci 106: 9715–9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watada H, Mirmira RG, Kalamaras J, German MS 2000. Intramolecular control of transcriptional activity by the NK2-specific domain in NK-2 homeodomain proteins. Proc Natl Acad Sci 97: 9443–9448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev 13: 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA 2008. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature 455: 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]