Next-Generation Genomics: an Integrative Approach (original) (raw)
. Author manuscript; available in PMC: 2012 Apr 8.
Published in final edited form as: Nat Rev Genet. 2010 Jul;11(7):476–486. doi: 10.1038/nrg2795
Abstract
Integrating results from diverse experiments is an essential process in our effort to understand the logic of complex systems, such as development, homeostasis and responses to the environment. With the advent of high-throughput methods - including genome-wide association studies (GWAS), ChIP-Seq, and RNA-Seq, etc., - acquisition of genome-scale data has never been easier. Epigenetics, transcriptomics, proteomics and genomics each provide an insightful, and yet single-dimensional, view of genome function; integrative analysis promises a unified, global view. However, the large amount of information and diverse technology platforms pose multiple challenges for data access and processing. This Review discusses emerging issues and strategies related to data integration in the era of next-generation genomics.
Introduction
Driven by technological advances, recent years have witnessed a deluge of new methods for interrogating different properties of a cell on a genome-wide scale. Each offers a unique, though complementary, view of genome organization and cellular function. It is expected that integrating these datasets will provide more biological insights than using one dataset alone. Thanks to the development of next-generation DNA sequencing (NGS) technologies, the human genome has been mapped in many individuals; the challenge we now face is to understand this blueprint and to determine how errors lead to disease. The traditional approach of isolating individual genes and studying them in a model system is being is rapidly replaced by datasets generated by new high-throughput technologies, by both individual laboratories and large consortia.
Although individual datasets - including genomic, epigenomic, transcriptomic and proteomic information - are highly informative, integrating them together offers the exciting potential to answer many long-standing questions. For example: what are the functional variants of gene-distal loci identified by association studies; where are the regulatory elements; to what extent does the activity of regulatory elements contribute to disease phenotypes or to individual gene expression variation? Therefore, integrative analysis has become an essential part of experimental design in the era of next generation genomics and is no longer the preserve of bioinformaticians. However, with the diversity of the high-throughput data and the seemingly endless analyses that can be performed, data integration is posing challenges for both bench scientists and computational biologists.
In this Review, we first briefly introduce the main high-throughput approaches. We then consider the types of biological question that can be addressed through integrative analysis and insights that are starting to emerge, followed by discussion of commonly employed data integration strategies. We also consider the need for unified next-generation tools for data visualization, manipulation and analysis.
What types of genomic datasets are available?
In recent years, many high-throughput technologies have been developed to interrogate various aspects of cellular processes, including sequence and structural variation, the transcriptome, epigenome, proteome and interactome. Several recent reviews1–7 have provided in-depth discussion of various platforms, so we only briefly introduce them below. Large collaborative projects are notably involved in using and developing genome-scale techniques, as discussed in BOX 1.
Box 1. Collaborative projects and technology development.
Over the next few years, technologies such as NGS will generate a massive quantity of scientific data. Because of this, the scientific community must call for analytical tools to be developed alongside large-scale data production. For projects such as the Reference Epigenome Project, the ENCODE Project, and The Cancer Genome Atlas (TCGA), data analysis and integration are clearly defined aims.
There is a broad selection of genome-scale approaches available, some of which might be redundant or might answer a different need. For example, for techniques to map DNA methylation on a large scale, non-genome-wide approaches including reduced representation bisulfite sequencing (RRBS) and meDIP-Seq provide cheaper alternatives to full genome methylation mapping (MethlyC-seq)25. The NIH Epigenome Roadmap Consortium has undertaken the task of a comparative analysis to determine how much pertinent information is gathered from non-genome-wide approaches compared to MethylC-Seq. This comparative analysis will benefit the scientific community and could be of particular value to groups studying the role of DNA methylation across a cohort of patients, where large numbers of samples necessitates cost efficiency. It is anticipated that such collaborative projects will lead to the first epigenome-wide association studies (EWAS, epiGWAS).
Cataloging of the regulatory elements within the human and epigenomic mapping, like the sequencing of the genome itself. are not being left to individual labs. Collaborative efforts that result in a shared resource in which regulatory elements are consistently defined across the cohort of all experiments are being undertaken, for example through the Roadmap Epigenome Consortium. This project will generate the epigenomic maps for over 100 human cell types within the next several years. Similarly, the mapping of histone modifications and transcription factors in human cells by the ENCODE Consortium will provide additional insight to distal regulatory elements. Recently, several ChIP-Seq experiments for such factors and modifications have been made publicly available, giving the scientific community the opportunity to begin utilizing this resource. For model systems, Drosophila and C. elegans are being investigated by the ModENCODE Consortium 119. Currently, an effort is being made to develop a mouse ENCODE project. Maps of regulatory elements in multiple species will enable the investigation of specific questions and improve understanding of what is conserved among species.
Sequence variation data
An ultimate goal of human genetics is to map every genetic variant and link each to phenotype. Currently, two high-throughput approaches are used to catalog genetic variants: SNP genotyping arrays and re-sequencing. SNP arrays are cost-effective and this strategy has been instrumental in the identification of disease-associated genes by groups such as the International HapMap Consortium8. More recently, NGS has reduced the cost of DNA sequencing, so it is feasible to directly sequence the exomes of an individual, using methods such as Sequence Capture9, 10, or sequence individual genomes, as is being performed in the 1000 Genomes project (http://1000genomes.org). NGS can also detect copy-number variants and gene-fusion events11, 12, and in the future, NGS will likely overtake array-based detection methods, due to its superior coverage and resolution.
Transcriptomic data
NGS is also driving advances in transcriptomics2, 13. For example, RNA-Seq can detect alternative splice variants using paired-end, relatively short reads (on the Illumina and ABI platforms) or longer reads (using the Roche platform). In addition, RNA-Seq can identify transcripts arising from gene fusion events typical in cancer14 and can detect novel classes of non-coding RNAs. For example, new classes of short RNAs have been identified that originate from promoters and gene termini15 and many more large intergenic non-coding RNAs (lincRNAs) have been found16. In addition, a method that combines nuclear run-on with RNA-seq has been developed, which enables transcriptional rate in the cell to be monitored 17.
Epigenomic data
DNA methylation and covalent modifications of histone proteins have been broadly defined as epigenetic modifications18, 19 and are important for transcriptional control20–22. High-throughput technologies now allow genome-scale mapping of these modifications23–25. Several large-scale analysis techniques are available that enable the survey of DNA methylation status at nucleotide resolution throughout the genome26–30, including NGS coupled to bisulfite treatment of DNA. Chromatin immunoprecipitation followed by microarray or, more recently, by sequencing (ChIP-chip and ChIP-Seq, respectively) (see references 3 and 4 for recent reviews), can determine the genome-wide localization of histone modifications31, 32. In addition, DNase I Hypersensitivity Site footprinting coupled to genomic arrays or NGS33–37 (DHS-chip and DHS-Seq or DNase-Seq) defines regions of open chromatin structure, which can indicate potential regulatory sequences34.
Interactome data
Interactions – both physical and functional – are an important layer of information for functional genomics. ChIP-chip and ChIP-seq are able to provide genome-scale information on DNA-protein interactions and high-throughput sequencing of RNAs isolated by crosslinking and immunoprecipitation (HITS-CLIP, also known as CLIP-Seq) is emerging as an important method for understanding RNA-protein interactions38. High-throughput dissection of protein-protein interaction networks has proved a greater challenge. It is largely done via the two-hybrid system and in yeast this has been expedited by the cloning of all genes39. However, in mammalian systems we are much further away. At a lower throughput, immunoprecipitation followed by mass spectrometry is becoming more widely available40.
Technologies based on chromosomal confirmation capture (3C) provide a snapshot of long-range interactions41 between regions of DNA, which can be mediated through protein interactions. 4C42 and 5C43 provide large-scale analyses but are still limited to selected sites of interrogation (see references 44 and 45 for a comparison of methods). However, recently developed methods have demonstrated identification of long-range genomic interactions at a genomic scale through high-throughput, paired-end sequencing of the DNA fragments generated by the 3C method46–48. One method, Hi-C, maps numerous interactions in an unbiased fashion while another, ChIA-PET, identifies interactions mediated by a particular protein through a ChIP step.
In addition, high-throughput methods are being employed to define genetic and signaling pathways. For example, through large-scale RNAi screens, a number of key genes were linked to pathways regulating metastasis, apoptosis and senescence49–54, which provided new insights into cancer biology. In yeast, genetic interaction pathways are being identified through large-scale epistasis screens (E-MAPs)55, 56, and soon such approaches might be applied to other model organisms or human cells. The power of such maps was recently demonstrated by combining the information they provide with genome-wide association studies in yeast to illustrate how single mutations are mechanistically relevant to key pathways57.
Why perform integrative genomic analysis?
This broad spectrum of data provides unprecedented opportunities for investigators to address some long-standing questions related to fundamental mechanisms of genome function and disease. For example, how might particular risk-associated SNPs impact cellular function and lead to specific diseases? What functional sequences exist in the human genome? How are key developmental pathways regulated by epigenetic mechanisms? In this section we introduce some of the questions that integrative analysis is being used to answer; the methods for such integration are discussed in the following section.
Annotating functional features of the genome
A major challenge of understanding transcriptional control in higher eukaryotes is the incomplete catalog of regulatory elements, particularly long-range regulatory elements such as enhancers and insulators. As the characteristics of known regulatory elements are determined, these features can be used to identify novel elements. For example, the chromatin ‘signature’ of enhancers (Figure 1) was determined and integrative analysis of histone modifications and localization profiles of the transcriptional co-activator p300 in human cells was used to find new enhancers 58, 59. Enhancer locations were confirmed by DNase I hypersensitive site analysis and functional assay, which is an important step for validating large-scale findings.
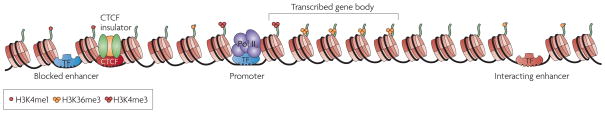
Figure 1. Annotating the genome through detecting transcription factor binding sites and histone modification states.
Promoters can be mapped by the localization of general transcription machinery and transcription factors (TF) such as RNA polymerase II (Pol II) or TAF1, or by the localization H3K4me3. The bodies of transcribed genes and noncoding RNAs are marked by H3K36me3. Enhancers can be found by distal transcription factor (TF) binding sites or by H3K4me1. This modification often coincides with H3K4me2, which has been shown to be necessary to recruit pioneering transcription factors to enhancer elements124. In addition, H3K4me1 sites overlap acetylated histone lysines, in agreement with acetylation islands outside of promoters identifying functional enhancer elements125, 126. Insulators are bound by CTCF. Nucleosomes are shown as cylinders and example histone tails are in grey. Different TFs are shown in different colours. Factors bound to the insulator include CTCF and subunits cohesion.
Although chromatin signatures define general classes of regulatory elements, their specific functions are dictated by transcription factors (TFs) that bind the elements. For the human genome, the ENCODE Consortium members and others have used genome-wide localization of key factors to define regulatory elements, such as RNA Polymerase II (RNAPII) and TAF1 for promoter elements60, CTCF for insulator elements61, STAT1 and p300 for enhancers59, 62–64, and transcriptional repressors KAP1, SUZ12, and NRSF for silencing or repressor elements24, 65, 66 (Figure 1). These results support the feasibility for genome-wide identification of _cis_-regulatory elements, but additional functional studies are necessary for specific sites of interest. However, the activities of cis-regulatory elements are often restricted to specific cell types or development stages and so a comprehensive and precise catalog of all _cis_-regulatory sequences will necessitate a thorough investigation of a multitude of TFs in various physiological conditions.
Inferring the function of genetic variants
Genome-wide association studies (GWAS) have revealed numerous SNPs that are linked to disease risk67. But one major obstacle is that if these SNPs fall within non-coding regions of the genome, our ability to assign functional roles to them is limited because functional features in the genome are still poorly defined in humans and other higher eukaryotes.
Recently, it was demonstrated that SNPs could be called from short sequenced tags acquired from Illumina sequencing during ChIP-Seq68, 69. It would be highly informative to know if TF binding sites or chromatin-marked regulatory elements (see below) contain single nucleotide variants (SNVs), which might be used to determine regulatory SNPs (rSNPs) 70–72 (Figure 2). For example, a study by Snyder and colleagues showed that SNPs found in binding regions for RNAPII and NF-kB accounted for individual variability in gene expression levels73. Studies that identify open chromatin structures have also recovered known diabetes risk-associated SNPs 74. Some algorithms that are used to find peaks of binding in ChIP-seq data have built-in SNP detection75, so identifying variants could become part of standard ChIP-Seq analysis. However, it should be noted that in all efforts to identify SNPs there is an inherent bias in mapping to the reference genome76. Therefore, additional measures should be taken to maximize mapped tags (for example, see Supplemental Methods of Reference73).
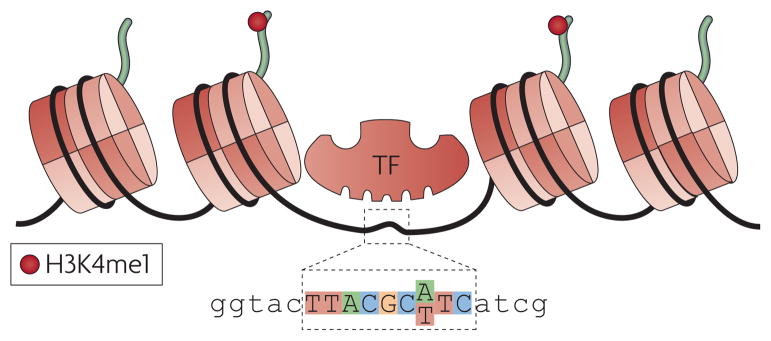
Figure 2. Identification of regulatory SNPs (rSNPs).
The sequence of a transcription factor (TF) binding site is shown with the position of an A/T polymorphism. By integrating chromatin signatures of enhancers or transcription factor binding sites with SNP data, SNPs falling with the region would be predicted as rSNPs. These could then be correlated to changes in gene expression.
Calling variants in sequence-based assays will also provide important information beyond the SNP itself as the presence of a SNP/V may enable detection of allele-specific expression. In the case of RNA-Seq, if the transcriptional output of a heterologous locus contains a variant at or near 100% frequency, it is indicative of mono-allelic expression. Allele-specific ChIP signals for transcription factors or RNAPII might offer a regulatory explanation for such allele-specific expression. For example, our group has previously demonstrated this with SNP arrays coupled with ChIP (SNP-ChIP)77. More recently, allele-specific regulatory regions in humans were identified through mapping DNase HS regions with CTCF co-localization 78. Allele-specific DNA methylation, which can now be assayed at genome-scale, can also suggest potential mechanisms for mono-allelic expression or repression, such as imprinting (see also below) 79. Therefore, integrative analysis of allelic-specific transcription factor binding, epigenomic information and large-scale phenotypic readouts such as allelic-specific RNA expression data will be key to identifying genetic or epigenetic mechanisms of gene expression. Extension of functional studies to structural variants will also be an important aim for future studies.
Understanding mechanisms of gene regulation
Because epigenetic features can control transcriptional output, and therefore traits, correlating epigenomic information and transcriptomic information can be highly informative. A classic example is genomic imprinting. Individual examples of imprinted loci – such as the H19 locus in mammals – have been studied in detail80 and illustrate the complexity of transcriptional regulation, including the combined action of insulators, enhancers, chromosome looping and epigenetic marks. Genome-scale integrative analyses will enable broader questions to be answered: how many imprinted genes are there; how many diseases does deregulation of imprinting contribute to; when does DNA methylation alter transcription factor binding and what range of factors can be affected?
Coupling histone modification data to transcriptomic data can also be valuable for the annotation of non-coding RNAs. Young and colleagues identified miRNA transcription start sites by mapping the promoter-specific modification H3K4me3 and comparing regions outside of known promoters with annotated miRNAs, conserved regions, CpG islands and histone modifications associated with transcription elongation (H3K36me3 and H3K79me2)81. Rinn and colleagues mapped the location of thousands of lincRNAs by integrating these same chromatin modifications with RNA-Seq data for expressed ncRNAs16. It is now thought that many of these large lincRNAs can influence histone modification or chromatin structure or subsequent methylation of DNA82–84.
Integration of epigenomics with genomics and transcriptomics can also provide insights into transcription-coupled RNA processing. Recently, several groups found a correlation between exon expression and levels of H3K36me385–90, and a subsequent study suggested a direct role for this modification in splicing control91. Further analysis of histone modifications in relation to splicing may provide additional insights into exon usage across genes32, 92. Integration of exon expression data with HITS-CLIP data on the interaction of splicing factors with mRNA can also help to map splicing sites precisely93. In addition, integration of data on the promoter histone modification H3K4me3 (Figure 1) with methods for capture of the 5′ ends of genes such as CAGE tags94, which can be readily adapted to NGS, will improve annotation of the transcription start sites (TSS).
In order to understand what controls the spatial organization of gene expression and how regulatory elements and proteins interact with their targets, it is useful to integrate interaction data with other datasets. For example, nuclear architecture is, at least in part, defined by how chromosomes attach to the nuclear envelope. Nuclear-membrane attached loci are typically marked by H3K9 methylation and this modification is decreased in the laminin-associated diseases Hutchinson-Gilford progeria syndrome and Facioscapulohumeral dystrophy95, 96. The nuclear-membrane attached regions often lie outside of genes, so structural variants in unannotated genomic regions may be informative to understand 3D architecture. Future studies coupling histone modification profiles, transcriptomes, structural variations and chromosomal interaction data will expedite our understanding of nuclear architecture and define new mechanisms of disease.
Approaches to an integrative analysis
Several consortia are systematically interrogating genetic variation, the transcriptome, the epigenome and the interactome on a genomic scale. Each experiment adds another dimension of data to the genome so there now are hundreds of dimensions of experimental data tethered onto the human genome (and other genomes) and this number is growing rapidly. The key to exploiting these data is integrating them. There are many ways to approach the challenge of data integration and we discuss three important – though not mutually exclusive – approaches below.
Data complexity reduction
For a growing number of sequencing based assays such as ChIP-Seq, DNase-Seq, FAIRE-Seq, RNA-Seq, or Hi-C, the result of each experiment is millions of short sequence reads, which essentially give a continuous signal of enrichment across the genome. A simple approach to reducing the complexity of this dataset from millions of data points to a more manageable hundreds or thousands of sites is to summarize each experiment as a collection of genomic regions with strong enrichment of signal. For ChIP-Seq, peak-finders discretize the genome-wide profiles into regions with enrichment and those without. Therefore, a commonly used method of data integration is to perform intersection analysis on enriched regions from different experiments. For example, Chen et al mapped a collection of 13 TFs using ChIP-Seq in mouse ES cells, used a custom peak-finder to call regions of enrichment, and observed significant co-binding of TFs97.
Although intersection analysis on discretized datasets is straightforward to perform, special attention must be paid to the underlying assumptions of data discretization. For example, blanket application of a peak finding method and set of parameters to different types of data – such as histone modifications, TF binding and open chromatin - is often ill-advised, for several reasons. Firstly, the type of experiment usually dictates a specific kind of data analysis. For instance, TFs often bind discrete, specific sites and so ChIP-Seq tags at the point of binding have a biased distribution between positive and negative strands, which can be used by peak finders to obtain excellent precision75, 98. However, this assumption is less suitable when binding or enrichment occurs contiguously across large stretches of DNA or in clusters, as is the case for certain chromatin modifications31, 99. Therefore, one must be mindful of the underlying assumptions and limitations of peak finders before applying them. Secondly, even among the same type of data, variability in data quality may necessitate calling peaks with different thresholds and/or data normalization methods. This is especially true for ChIP-Seq experiments, where variable quality of antibodies or sub-optimal ChIP conditions can lead to variable ChIP enrichment, which will require adjusting significance thresholds individually to achieve both high sensitivity and specificity.
It is important to note that the inherently noisy nature of genome-wide data means that a perfect peak finder cannot exist: in calling regions of enrichment, one can only hope to minimize, but not eliminate, false positives and false negatives. Realizing this, it is evident that we cannot simply trust peak finders blindly and that it is especially important to inspect at least some of the results by eye. Thus, if we are to perform meaningful analysis, we cannot be far removed from the original data and should follow the analysis with validation experiments.
Unsupervised integration
A more scalable method for integrating data is unsupervised learning, which approaches the data with no prior biases, knowledge, or hypotheses. To summarize a large dataset into smaller groups that can be more easily conceptualized, an unsupervised approach simply asks the question: what kinds of patterns exist in a dataset? One common assumption made by unsupervised approaches is that the interesting features of the data are the ones that occur frequently, and therefore the goal is to find common patterns. As diverse experimental methods equate frequency of genomic mapping with activity, an unsupervised analysis can treat these datasets equally and need not know the nature of the measurement. For example, Zhao and colleagues profiled 37 histone modifications in human CD4+ T cells31, 32. While the number of different possible combinations of modifications is a staggering 237 ≈ 137.4 billion, it is likely that most combinations do not exist, or occur very infrequently. To enumerate commonly occurring chromatin signatures, or other patterns, clustering can be applied. Clustering approaches are introduced in Box 2.
Box 2. Clustering.
Clustering is an integral bioinformatics tool to partition a large dataset into more easily digestible, conceptual pieces. It can be applied to a wide variety of data, but traditionally has been applied to gene expression profiles. Here, each gene is represented by a list of expression values in various cell types or conditions, and clustering identifies sets of co-expressed genes. In general, conventional clustering works well when the experimental values can be easily discretized into the clustered entities, for example RPKM-normalized expression to an associated gene.
However, for other applications, this discretization is not possible or not desired. One example is for histone modification data derived from ChIP-Seq, where the profile of experimental values over a contiguous region is informative. Conventional clustering can be applied to this data, provided that the profiles are well aligned. For example, to enumerate commonly occurring chromatin signatures in an unbiased way, conventional clustering can be applied to a subset of genomic regions such as promoters. If a pre-defined number of clusters k is expected then _k_-means clustering can be applied, otherwise hierarchical clustering can be used to offer more flexibility. Clearly, conventional clustering can be applied to a wide variety of genomic datasets, spanning genomes, epigenomes102, transcriptomes16, and interactomes120. But this method gives the best results when the set of loci examined are well-aligned, which is the case for gene definitions where excellent annotations exist. To cluster loci with poorly aligned or asymmetric chromatin signatures, or for poorly annotated loci such as gene-distal regulatory elements, our laboratory has developed an approach called ChromaSig90, 101. Given set of genomic loci, ChromaSig aligns and orients the epigenetic profiles around the loci, outputting clusters of loci that share similar profiles. Alternatively, given the genome-wide nature of epigenetic data, another clustering approach taken is to assign a cluster to every part of the genome. To accomplish this task, Jaschek et al121 employ a hidden Markov model approach to learn the most likely epigenetic states given the data.
The genome serves as a scaffold upon which high-throughput data are assembled and from a genome-centric perspective, clustering can be seen as a way of classifying genomic loci into conceptual groups with shared attributes. Clustering data from different experiments gives distinct types of conceptual groups and the first phase of data integration can be seen as enumerating the conceptual modules of each dataset. For example, clustering of RNA expression reveals co-expressed genes100, clustering of histone modifications gives loci that share similar chromatin structure90, 101, 102, protein-protein interaction clustering finds proteins in the same complex103, and genetic interaction clustering reveals members of the same or similar pathways56.
Although all modules are tethered to the genome, modules from one experiment are not linked to those from others. Thus, the next task in data integration is to connect these modules. One approach is to examine a module from one data type, for example chromatin signatures, in the context of another data type, for example DNA methylation 25, 104, 105. Alignment of data sets on a browser such as the UCSC Genome Browser 106 might be useful in this regard (Figure 3). Furthermore, the Genome Browser also contains annotations such as gene definitions, evolutionary conservation, and disease associations107. Therefore, co-clustering of new experimental data with known annotations can provide an easy bridge to hypothesis generation. In the past, when genomics consisted only of global gene expression analysis, annotation libraries such as Gene Ontology108 and the more sophisticated Gene Set Enrichment Analysis109 were developed to provide an easy way to assess the biological significance of gene hits. As datasets are now extending to include non-coding RNAs, disease-associated SNPs and regions of TF binding, it appears that “Locus Set Enrichment Analysis” will be an important part of genomics. Sets of loci that share factor binding, epigenetic modifications or disease association will provide efficient ways to form hypotheses regarding function outside of coding regions.
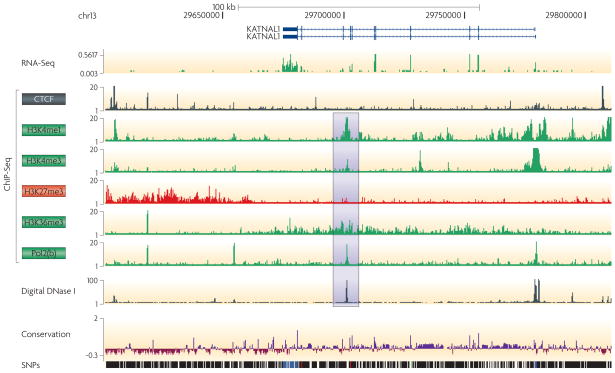
Figure 3. Data Visualization.
The UCSC Genome Browser is a tool for viewing genomic datasets. A vast amount of data is available for viewing through this browser. This example from the browser shows numerous data types, in K562 cells, from the ENCODE Consortium. A random gene was selected - KATNAL1 - that illustrates several points that can be identified by using this tool. The promoter has a typical chromatin structure (peak of H3K4me3 between the bimodal peaks of H3K4me1), is bound by Pol II, and is Dnase hypersensitive. The gene is transcribed, as indicated by RNA-Seq data, as well as H3K36me3 localization. The gene lies between two CTCF bound sites that could be tested for insulator activity. An intronic H3K4me1 peak (highlighted) predicts an enhancer element, corroborated by the DHS peak. There is a broad repressive domain of H3K27me3 downstream, which could have an open chromatin structure in another cell type.
Another approach to connecting conceptual modules involves network biology, which leverages high-throughput techniques to find relationships that connect genomic loci and conceptual groups. For example: methods to map chromosomal interactions, such as Hi-C, connect genomic loci to each other; genetic interactions from E-MAPS connect proteins to pathways; and ChIP-Seq links transcription factors to regulated genes. This second level of integration - linking different kinds of experiments - can form a knowledge base from which to extract biological insights or suggest hypotheses for further study.
As a hypothetical example, suppose we used ChIP-Seq to map a novel TF genome-wide and wanted to know the significance of its binding profile. Complicating matters, most of the binding sites are distal to promoters. Clustering reveals that a subset of binding sites share a similar chromatin environment, which suggests these sites may function similarly. Hi-C data then links this subset of binding sites with their target genes and RNA-Seq data reveals these genes are highly expressed. Finally, protein-protein and genetic interaction data reveals that some of these expressed genes belong to related but distinct protein complexes that regulate RNA splicing. Thus, data integration would allow us to efficiently propose the hypothesis that the binding of this new factor to DNA regulates the process of RNA splicing.
Often, the scope of genomic experiments performed is so diverse that it is not immediately clear how, or even if, one experiment relates to another. It is in such cases that unsupervised, data-driven approaches to integration are most useful. Unsupervised integration is a discovery tool to find correlations between two or more experiments. Novel associations lead to hypotheses of function, which can be followed up by supervised integration and by direct experimental validation (see below). In this way, high-throughput experiments are screens to identify interesting, unexpected associations. Because of the power of the approach and because the inputs required are minimal, unsupervised integration is arguably the first tool that should be applied to a new dataset, and it should be constantly run as new experiments are added to an existing dataset to find additional associations.
Supervised integration
The discovery of patterns is one output of unsupervised integration, but the patterns alone do not advance our understanding of biology or disease. Like most systems biology approaches, unsupervised integration excels at generating hypotheses. Therefore, a novel pattern is simply an observation, from which we must make and test predictions of function, often by incorporating external datasets or new experiments. This is the realm of supervised integration. Supervised integration is driven by testable hypotheses and so often relies on only a few dimensions of a full dataset.
It is important to note that the choice of data to include in supervised integration and the specific method used depend crucically on the question posed. For example, using an unsupervised clustering approach we recently observed that a set of distinct histone modifications at exons, which led to the hypothesis that these modifications mark alternatively expressed exons90. To test this hypothesis, we needed to examine these chromatin modifications in the context of expression at the exonic level and we were able to use previously published exon expression array data from the same cell type 110.
However, in most instances the impetus for supervised integration is anecdotal evidence, either through observations obtained by simply viewing genome-scale data on a browser or from previously published studies. For example, Guttman et al took advantage of previous observations that RNAPII-transcribed genes are marked by H3K4me3 at promoters and have H3K36me3 spreading into the transcribed region and searched for this chromatin signature to identify RNAPII-transcribed lincRNAs16. Thus, supervised integration starts with a prediction based on an observation and ends with a test of this prediction. This is arguably how our biological understanding is advanced most: the more predictive the hypothesis, the more biological insight gained. Therefore, observation and data integration cannot be independent from each other and there is no substitute for seeing the data with one’s own eyes. Our opinion that it is necessary to see raw data using a browser, for example, is consistent with the current trend in data visualization towards replacing traditional averaged plots with more information-rich heatmaps that simultaneously illustrate experimental profiles for thousands of loci (e.g genome-wide heatmaps of ChIP-chip data59).
As there are now tens of thousands of high-throughput experiments linked to the human genome, finding dependence relationships among the many dimensions of experimental data is essential to increasing our knowledge. In the simplest case, relationships can be discovered by correlation analysis. For example, a strong, positive correlation among the binding profiles of two transcription factors indicates that one may be dependent on another. Additionally, for genetic interactions, finding positive and negative correlations for a mutant under different conditions can allow systematic discovery of condition-dependent relationships (S. Bandyopadhyay - UCSD, personal communication).
Although informative, correlation analysis can become unwieldy as the number of datasets grows – doubling a dataset would effectively quadruple the number of computations necessary and the number of visualizations required. Luckily, machine learning techniques, notably Bayesian networks (for a primer see Needham et al111), offer a supervised approach to discover relationships among data entities. Using a probabilistic framework, Bayesian networks can find dependence relationships, for example as van Steensel et al did for a panel of chromatin modifications and chromatin-associated proteins and modifiers112. Bayesian networks can also readily integrate data from different kinds of experiments. For example, Yu et al modeled the interdependence of histone modification profiles with the binding of transcription factors, together with their relationship to gene expression113. However, it is important to note that the types of prediction that are the output by a Bayesian network critically depend on how the network is designed, which in turn depends on the question asked. For example, Jansen et al designed a Bayesian network to predict protein complexes by integrating diverse data sources including protein-protein interactions, expression and gene annotation114. In summary, Bayesian networks can find relationships among diverse kinds of data and thereby create hypotheses that can be tested experimentally.
Utilizing large-scale datasets for integrative analysis
One of the greatest challenges that comes with high-throughput technologies is the vast amount of data that they produce. The sheer amount of the data produced can be difficult to manage, especially for experiments involving next-generation DNA sequencing methods. For example, Lister et al. recently sequenced the first human methylome using bisulfite shotgun sequencing, which generated 90 Gigabases of sequence reads, representing 30X coverage of the human diploid genome25. Transferring this amount data to the NCBI public database servers took one full week. The question is: how can investigators efficiently use data of this scale for comparative analyses? This challenge can be broadly divided into two: how can bench scientists look to see how one dataset fits with others (from their own or other laboratories); and how can bioinformaticians provide better tools for integrated analyses?
For the bench scientist
In order to make strides in the era of NGS, we need tools for the bench scientist to analyze their own data in an efficient and relatively straightforward manner. We propose that a solution would be similar to an open source web browser, such as FireFox. It would have a series of “add-ons”, a core group of programmers would maintain the browser code and listen to the community for ways of updating it and, importantly, they would allow the community to build individual tools to enhance the browser’s capabilities. The ‘gatekeepers’ would ensure the tools are safe and work with the browser and users could decide which add-ons are suited to their needs. Users would also see previews and read reviews and ratings for each add-on. A tool along these lines - Galaxy 115, 116 (www.galaxy.psu.edu) - has been in development for many years and is described in Box 3, along with other popular online tools.
Box 3. Online tools for integrative analysis.
Galaxy is an online genomics analysis tool that allows users to perform a number of integrative data analyses on genomic datasets. Though not a database itself, it is directly linked into many genomic resources such as the UCSC Genome Browser. Galaxy allows users to upload data, parse it, reorder columns, and change file formats for browser compatibility. Galaxy also provides several tools for data integration. For example, it has tools for dataset intersection and union analysis, enabling users to compare their datasets with annotated genomic loci, with output directly viewable on the Genome Browser. In the process, users can create and save not just new files, but entire workflows that can be re-used and shared with others. Best of all, Galaxy provides a platform to run tools developed by the community. In the near future, tools like Galaxy will provide bench scientists a one-stop-shop for data analysis: given sequencing reads, add-ons will map these reads and call peaks, allowing for subsequent analyses.
Another popular online tool is DAVID122 (http://david.abcc.ncifcrf.gov) used for GO analysis (for a step-by-step protocol see Huang da et al.123). Therefore, using the range of tools available online, with a few clicks one can map ChIP-Seq reads at Galaxy, call peaks with CisGenome, use Galaxy’s intersection tool to find overlapped genes, and finally upload the TF-bound gene list to DAVID for GO annotation (see also Figure 4). Though not as efficient as a single tool, this method allows a significant amount of analysis to be done without the need to write new software.
It is also important to note that known and novel motif finding for peaks or promoters can be done online using CEAS (http://ceas.cbi.pku.edu.cn/) and the MEME Suite (http://meme.sdsc.edu/meme). In addition to GO annotations, understanding gene function, pathway interactions or protein-protein interaction might be of interest for key genes. A number of online tool can now assist in this [STRING (http://string-db.org/), Cytoscape (www.cytoscape.org), and MouseNET (http://mousenet.princeton.edu) are a few examples].
One potential downside of an online analytical tool, such as Galaxy, is computational load. If the majority of scientists conducting RNA- or ChIP-Seq experiments begin running Galaxy on a regular basis, will the whole system creep to a halt? Also, to prevent inefficient computation, add-ons would need to meet specific benchmarks for performance, such as time complexity and storage space as the system cannot tolerate inefficient computation. Therefore it can be argued that it may be advisable to have a stand-alone analytical system. One example of such a tool is CisGenome117 (http://www.biostat.jhsph.edu/~hji/cisgenome/), which is downloadable and compatible with several operating systems. Designed for the analysis of ChIP-chip and ChIP-Seq data, it includes a browser, file conversion tools and tools to call peaks of ChIP enrichment and to perform motif analysis. These features enable a basic workflow needed by many scientists. An example workflow using a range of tools is shown in Figure 4.
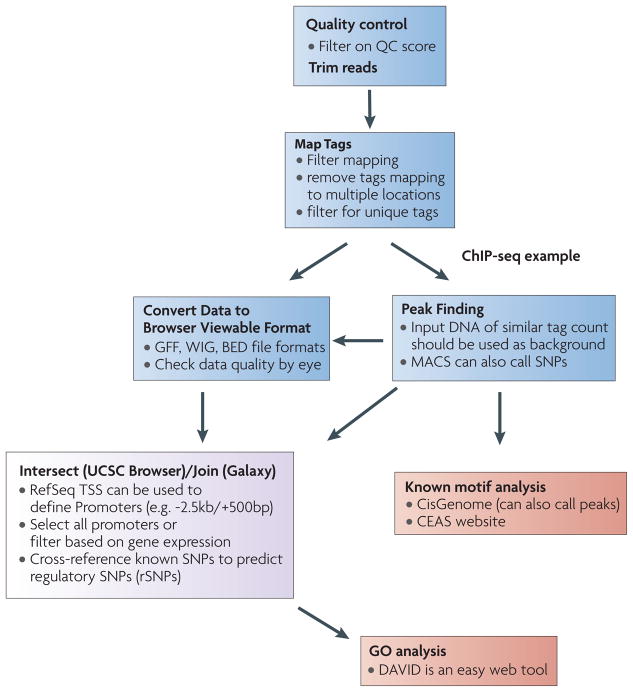
Figure 4. Flow chart for data analysis.
This example of shows a workflow for ChIP-seq data analysis that can be done by bench scientist using current resources is shown. A similar strategy could be used for other types of NGS data. Blue boxes show steps that can be performed using Galaxy. Integration or cross-sectioning of data can often be done in the UCSC browser or by joining list in Galaxy (Purple box). Downstream steps such known motif analysis and gene ontology (GO) analysis can be achieved with online or stand alone tools (Red boxes). Galaxy can also be used to establish analytical pipelines for calling SNPs that could then be integrated into sequencing-based data such as ChIP-Seq.
In the end, resources such as genome browsers are still one of our best tools. A good browser can distinguish good quality from poor quality datasets and can show trends and patterns within the data without the need for statistical measures. Such anecdotal observations can spur questions that require more sophisticated analysis. Several browsers are available, including NCBI, Ensembl and UCSC. Although the amount of data available on the UCSC browser, including many large-scale datasets106, make it very valuable, it can be slow when attempting to browse through several datasets at various locations. Other browsers such as AnnoJ (http://www.annoj.org/), which was used for visualizing the Arabidopsis and human methylomes at nucleotide resolution 25, 27, are much more dynamic. Scrolling through the genome is very rapid and tracks can be zoomed, scaled, re-ordered and removed almost instantly.
Bioinformatic hurdles
There are still a number of key issues in analyzing NGS data, several of which have been touched on in previous reviews4, 30. For example, it remains unclear how RNA-Seq data from platforms that sequence short tags will be normalized against data from longer read platforms. Also, will RNA-Seq methods be as universal as Affymetrix microarrays? Most scientists feel comfortable comparing their own and published Affymetrix platform data. It is still unclear in these early stages of data processing and normalization of RNA-Seq how relative levels of expression can be compared, especially if there is a variation in the number of reads sequenced.
To address these questions more thoroughly, it will be important to revisit data normalization. Because NGS-based assays provide a digital readout, the data is often used as is. However, different experiments are sure to provide slightly varying degrees of enrichment, possibly due to antibody differences (for ChIP-Seq or HITS-CLIP) or experimental variation. Therefore, two datasets used in a comparative analysis should first be normalized to each other. This applies to samples from different research groups, as well as samples from within a dataset. For example, if one experiment has a uniform reduction in peak height, then non-normalized peakfinding may result in calling a cell-type specific peak at a site that is actually shared. Normalization is therefore imperative in experiments examining time points of differentiation or stages of disease progression where the changes may be subtle between neighboring stages118. In this regard, we will likely benefit from the numerous normalization methodologies that have been developed for microarray analysis. However, like gene expression analysis, we are sure to find that one method does not fit all datasets and that Loess, quantile and rank order normalizations will all be useful.
Future perspectives
Data integration itself is not an end: it is designed to generate novel hypotheses and help to test them. If a hypothetical ‘Data Integrator’ existed, its most important input would not be the data to be analyzed, but a specific question to answer. Depending on the question posed, analyses of the data – from what data sources are chosen, to how normalization is performed, how controls are selected and what is precisely being calculated – can vary dramatically. A frequent misconception is that a Data Integrator is a black box that takes in data as input and outputs interesting observations (or better, papers) as output. Because unbiased integration strategies focus on a single question while supervised integration can address any number of questions, the scope of the types of analyses possible with supervised integration is much greater, and arguably endless. For this reason, it is unfeasible to automatically perform all possible integrated analyses, as if the Data Integrator were seeking both a question as well as its answer simultaneously. The choice of interesting questions must always be left to the researcher and supervised integration must be tailored to each hypothesis. It is our opinion that, while unsupervised approaches can excel at finding patterns, it will be the supervised integrative methods stemming from either unsupervised methods or simple observations that will further our understanding of biology most effectively.
The future of genomic technologies holds great promise, but for genomic data and its integration to have a more meaningful impact on our understanding of biology we must make an effort to link together all the information that is being generated. This may require a community-wide effort, akin to Wikipedia, in which information can be updated by all, but monitored for the correct citations that directly link out to Pubmed and NCBI. Each gene entry would be linked to a browser for visualizing all genomic and epigenomic information in manner similar to viewing **Gene Expression Omnibus (**GEO) profiles at NCBI. All the related information should be searchable with Google-like capabilities. That is, a search engine examines the entire text for terms and phrases and finds related information even if it does not contain the exact key words. For example, NextBio (www.nextbio.com) currently provides a similar approach when searching for genes. This integration of knowledge will make each of us a better scientist through a greater understanding of the information around us.
GLOSSARY
Nuclear run-on
An assay that directly measures the transcriptional activity of a gene by incorporation of labelled UTP into its mRNA
Histones
Small, highly conserved basic proteins, found in the chromatin of all eukaryotic cells, which associate with DNA to form a nucleosome. The N-terminal tails of histones are subject to various post-translational modifications
Two-hybrid
An assay system in which one protein is fused to an activation domain and the other to a DNA-binding domain, and both fusion proteins are expressed in cells. Expression of a reporter gene indicates that the two proteins physically interact
CAGE
(Cap analysis of gene expression). The high-throughput sequencing of concatamers of DNA tags that are derived from the initial nucleotides of 5′ mRNA
Single nucleotide variant
In addition to base substitutions covered by SNPs, SNVs also include insertions and deletions
Genomic imprinting
The epigenetic marking of a gene on the basis of parental origin, which results in monoallelic expression
Next-Generation Sequencing (NGS)
Here we define NGS as the use of sequencing platforms including Illumina/Solexa, Roche 454, ABI SOLiD, as well as newer platforms such as Helicos and Pacific Biosciences
Chromatin immunoprecipitation
A technique used to identify potential regulatory sequences by isolating soluble DNA chromatin extracts (complexes of DNA and protein) using antibodies that recognize specific DNA-binding proteins
DNase I Hypersensitivity Site footprinting
An assay that identifies regions of the genome that lack nucleosome structure and are therefore readily degraded by the enzyme DNaseI. Such regions tend to be associated with transcriptional activity. When coupled to sequencing, the ends of DNA fragments generated by treatment of chromatin with DNase I are sequenced
E-MAPs
epistatic mini-array profiles are by screening fitness of double mutants in a high-throughput manner. The results, when analyzed as a whole, can reveal both positive and negative genetic interactions between genes, and provide insights to biological pathways and protein-protein complexes in the cell
HITS-CLIP (CLIP-Seq)
A technique similar to ChIP-seq in which proteins bound to RNA -such as splicing factors - are immunoprecipitated and the RNA fragments are sequenced
MeDIP-Seq
methylated DNA is immunoprecipitated with an antibody against methylated cytocine, and then sequenced by NGS
MethylC-Seq/BS-Seq
methylated DNA is identified by shot-gun sequencing of bisulfite converted DNA, which convert unmethylated C to uracil that appears as T in sequencing reads, while leaves methylated C intact
RNA-Seq
RNA isolated from the cells are sequenced by NGS either directly, or after conversion to complement DNA (cDNA)
RRBS
reduced representation bisulfite sequencing cuts genomic DNA with restriction enzymes to enrich for CG rich regions, which are then converted via bisulfite treatment and sequenced with NGS
Sequence Capture
uses oligo microarrays or oligo-coupled beads to select for regions of the genome such as all exons (exome sequencing) for targeted sequencing
Biographies
R. David Hawkins completed his doctoral research in the laboratory of Dr. Michael Lovett at UTSW Medical Center at Dallas, where he investigated the role of transcription factors in sensory epithelia regeneration. Since 2005, he has been performing postdoctoral studies in the laboratory of Dr. Bing Ren at the Ludwig Institute for Cancer Research. His work has focused on the role of epigenetic mechanisms in human embryonic stem cell and differentiated cell fates.
Gary C. Hon completed his doctoral research in the laboratory of Bing Ren at the University of California, San Diego, USA, where he studied the chromatin modification patterns marking regulatory elements in the human genome. He is interested in investigating the epigenetic mechanisms in development and disease.
Bing Ren is Member of the Ludwig Institute for Cancer Research (LICR) and Professor of Cellular and Molecular Medicine at the University of California, San Diego School of Medicine. He directs the San Diego Epigenome Center. His lab’s research is focused on understanding the mechanisms of gene regulation in human cells. He obtained a Ph.D. degree from Harvard University in 1998, where he studied mechanisms of transcriptional repression under the guidance of Dr. Tom Maniatis. From 1998 to 2001, he continued to research mechanisms of gene regulation as a postdoctoral fellow in Dr. Richard Young’s laboratory at Whitehead Institute.
References
- 1.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farnham PJ. Insights from genomic profiling of transcription factors. Nat Rev Genet. 2009;10:605–16. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–80. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 6.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 7.Beyer A, Bandyopadhyay S, Ideker T. Integrating physical and genetic maps: from genomes to interaction networks. Nat Rev Genet. 2007;8:699–710. doi: 10.1038/nrg2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner EH, Lee C, Ng SB, Nickerson DA, Shendure J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat Methods. 2009;6:315–6. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang DY, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alkan C, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–7. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 14.Maher CA, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gingeras TR. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–32. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. Demonstrates the integration of epigenetic data with the human genome to annotate novel RNAs, where confirmed by RNA-Seq. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–8. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–73. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 23.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–9. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 24.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 25.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. In addition to providing the first human methylomes, an integrative analysis of DNA methylation, histone modifications and RNA-Seq is conducted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–9. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–36. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pomraning KR, Smith KM, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods. 2009;47:142–50. doi: 10.1016/j.ymeth.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 30.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 31.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crawford GE, et al. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat Methods. 2006;3:503–9. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–22. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabo PJ, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods. 2006;3:511–8. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 36.Dorschner MO, et al. High-throughput localization of functional elements by quantitative chromatin profiling. Nat Methods. 2004;1:219–25. doi: 10.1038/nmeth721. [DOI] [PubMed] [Google Scholar]
- 37.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–9. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walhout AJ, Vidal M. Protein interaction maps for model organisms. Nat Rev Mol Cell Biol. 2001;2:55–62. doi: 10.1038/35048107. [DOI] [PubMed] [Google Scholar]
- 40.Hutchins JR, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 328:593–9. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 42.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–54. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 43.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fullwood MJ, Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. J Cell Biochem. 2009;107:30–9. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vassetzky Y, et al. Chromosome conformation capture (from 3C to 5C) and its ChIP-based modification. Methods Mol Biol. 2009;567:171–88. doi: 10.1007/978-1-60327-414-2_12. [DOI] [PubMed] [Google Scholar]
- 46.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gobeil S, Zhu X, Doillon CJ, Green MR. A genome-wide shRNA screen identifies GAS1 as a novel melanoma metastasis suppressor gene. Genes Dev. 2008;22:2932–40. doi: 10.1101/gad.1714608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–7. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bric A, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. 2009;16:324–35. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meacham CE, Ho EE, Dubrovsky E, Gertler FB, Hemann MT. In vivo RNAi screening identifies regulators of actin dynamics as key determinants of lymphoma progression. Nat Genet. 2009;41:1133–7. doi: 10.1038/ng.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–48. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–64. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–19. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 56.Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat Methods. 2007;4:861–6. doi: 10.1038/nmeth1098. [DOI] [PubMed] [Google Scholar]
- 57.Hannum G, et al. Genome-wide association data reveal a global map of genetic interactions among protein complexes. PLoS Genet. 2009;5:e1000782. doi: 10.1371/journal.pgen.1000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 59.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–80. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–45. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hartman SE, et al. Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes Dev. 2005;19:2953–68. doi: 10.1101/gad.1371305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 64.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Geen H, et al. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 2007;3:e89. doi: 10.1371/journal.pgen.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 68.Marks H, et al. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009;19:1361–73. doi: 10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pomerantz MM, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–4. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wright JB, Brown SJ, Cole MD. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol Cell Biol. 30:1411–20. doi: 10.1128/MCB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tuupanen S, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–90. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 73.Kasowski M, et al. Variation in transcription factor binding among humans. Science. 328:232–5. doi: 10.1126/science.1183621. Demonstrates individual binding variability for Pol II and NFkB linked to SNPs and structural variants that alter individual gene expression levels, providing a functional annotation as rSNPs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gaulton KJ, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 42:255–9. doi: 10.1038/ng.530. Uses open chromatin maps to recover a type 2 diabetes associated SNP in the intron of TCF7L2. Functional assays confirm its role in enhancer activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Degner JF, et al. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics. 2009;25:3207–12. doi: 10.1093/bioinformatics/btp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maynard ND, Chen J, Stuart RK, Fan JB, Ren B. Genome-wide mapping of allele-specific protein-DNA interactions in human cells. Nat Methods. 2008;5:307–9. doi: 10.1038/nmeth.1194. [DOI] [PubMed] [Google Scholar]
- 78.McDaniell R, et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 328:235–9. doi: 10.1126/science.1184655. This study illustrates the variability of nucleotide sequences in human regulatory elements, which suggest putative regulatory SNPs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–3. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 80.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–9. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 81.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–33. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 83.Nagano T, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 84.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–81. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–41. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–5. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 88.Luco RF, et al. Regulation of alternative splicing by histone modifications. Science. 327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–54. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput Biol. 2009;5:e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luco RF, et al. Regulation of Alternative Splicing by Histone Modifications. Science. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schubeler D, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–71. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–9. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shiraki T, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci U S A. 2003;100:15776–81. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shumaker DK, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A. 2006;103:8703–8. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zeng W, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–17. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 98.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–9. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hon G, Ren B, Wang W. ChromaSig: a probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput Biol. 2008;4:e1000201. doi: 10.1371/journal.pcbi.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 103.Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci U S A. 2003;100:12123–8. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hawkins RD, Hon GC, Lee LL, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JE, Ren B. Distinct Epigenomic Landscapes of Pluripotent and Lineage-Committed Human Cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rosenbloom KR, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 38:D620–5. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Oberdoerffer S, et al. Regulation of CD45 Alternative Splicing by Heterogeneous Ribonucleoprotein, hnRNPLL. Science. 2008;321:6. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Needham CJ, Bradford JR, Bulpitt AJ, Westhead DR. Inference in Bayesian networks. Nat Biotechnol. 2006;24:51–3. doi: 10.1038/nbt0106-51. [DOI] [PubMed] [Google Scholar]
- 112.van Steensel B, et al. Bayesian network analysis of targeting interactions in chromatin. Genome Res. 2010;20:190–200. doi: 10.1101/gr.098822.109. This is an excellent example of employing supervised integration with a Bayesian network to predict interactions between chromatin-associated proteins, followed by experimental validation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yu H, Zhu S, Zhou B, Xue H, Han JD. Inferring causal relationships among different histone modifications and gene expression. Genome Res. 2008;18:1314–24. doi: 10.1101/gr.073080.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jansen R, et al. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–53. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 115.Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Using galaxy to perform large-scale interactive data analyses. Curr Protoc Bioinformatics. 2007;Chapter 10(Unit 10):5. doi: 10.1002/0471250953.bi1005s19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Blankenberg D, et al. A framework for collaborative analysis of ENCODE data: making large-scale analyses biologist-friendly. Genome Res. 2007;17:960–4. doi: 10.1101/gr.5578007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Taslim C, et al. Comparative study on ChIP-seq data: normalization and binding pattern characterization. Bioinformatics. 2009;25:2334–40. doi: 10.1093/bioinformatics/btp384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Celniker SE, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–30. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–10. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 121.Jaschek R, Tanay A. Spatial Clustering of Multivariate Genomic and Epigenomic Information. Lecture Notes in Computer Science. 2009;5541:170–183. [Google Scholar]
- 122.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 123.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 124.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–52. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roh TY, Wei G, Farrell CM, Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17:74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]