Separase biosensor reveals that cohesin cleavage timing depends on phosphatase PP2ACdc55 regulation (original) (raw)
. Author manuscript; available in PMC: 2013 Jul 17.
Summary
In anaphase, sister chromatids separate abruptly and are then segregated by the mitotic spindle. The protease separase triggers sister separation by cleaving the Scc1/Mcd1 subunit of the cohesin ring that holds sisters together. Polo-kinase phosphorylation of Scc1 promotes its cleavage, but the underlying regulatory circuits are unclear. We developed a separase biosensor in Saccharomyces cerevisae that provides a quantitative indicator of cohesin cleavage in single cells. Separase is abruptly activated and cleaves most cohesin within one minute, after which anaphase begins. Cohesin near centromeres and telomeres is cleaved at the same rate and time. Protein phosphatase PP2ACdc55 inhibits cohesin cleavage by counteracting polo-kinase phosphorylation of Scc1. In early anaphase, the previously described separase inhibition of PP2ACdc55 promotes cohesin cleavage. Thus, separase acts directly on Scc1 and also indirectly, through inhibition of PP2ACdc55, to stimulate cohesin cleavage, providing a feedforward loop that may contribute to a robust and timely anaphase.
Introduction
During mitosis, the duplicated chromosomes are segregated into a pair of daughter nuclei. Improper control of this process can lead to errors in chromosome segregation and abnormalities in chromosome number, which are common genetic aberrations in cancer (Thompson et al., 2010). Despite the fundamental importance of chromosome segregation in biology and disease, we do not have a full quantitative understanding of the regulatory mechanisms that guide this process and ensure its accuracy.
In metaphase of mitosis, sister chromatid pairs are aligned on the metaphase plate as a result of a balance between the outward pulling forces of the spindle and the cohesive forces that hold the sister chromatids together. Sister-chromatid cohesion depends on cohesin, a 4-subunit protein complex that links the sisters as they are synthesized in S phase (Uhlmann and Nasmyth, 1998). Sister-chromatid separation is triggered in anaphase when the protease separase cleaves the Scc1/Mcd1 subunit of cohesin (or its Rec8 counterpart in meiotic cells), thereby allowing the sisters to be pulled apart by the mitotic spindle (Buonomo et al., 2000; Uhlmann et al., 1999; Uhlmann et al., 2000).
Separase is tightly regulated by its securin partner, which binds and inhibits separase activity. When all sister chromatid pairs are aligned correctly on the spindle, securin is ubiquitinated by the Anaphase-Promoting Complex (APC/C), triggering securin destruction by the proteasome and thereby promoting separase activation. In addition to its role as an inhibitor, securin is also believed to have positive roles in separase folding and localization (Agarwal and Cohen-Fix, 2002; Hornig et al., 2002); thus, securin yeast mutants display defects in separase function.
In the yeast Saccharomyces cerevisae , separase also has an important function that is independent of its catalytic activity: it promotes the initial activation of the phosphatase Cdc14. This function does not seem to be conserved in higher eukaryotes. Prior to anaphase, Cdc14 is sequestered in the nucleolus by its inhibitor Net1 and thereby rendered inactive. At anaphase onset, separase-dependent downregulation of the phosphatase PP2ACdc55 allows phosphorylation of Net1 and consequent Cdc14 release, termed the Cdc14 Early Anaphase Release, or FEAR, pathway (Queralt et al., 2006). This process depends on the Zds1 and Zds2 proteins (Queralt and Uhlmann, 2008), a pair of closely related regulators of PP2ACdc55 function (Rossio and Yoshida, 2011; Wicky et al., 2010), as well as Slk19 and Spo12 (Stegmeier et al., 2002).
Metaphase spindles exert sufficient force on the kinetochores to physically separate a DNA region surrounding sister centromeres (Pearson et al., 2001). Cleavage of cohesin alone, without Cdc14 activation, is sufficient to trigger spindle elongation and chromosome separation (Uhlmann et al., 2000). However, activation of Cdc14 by separase is required for normal spindle and chromosome behavior (Higuchi and Uhlmann, 2005). Cdc14 dephosphorylates numerous Cdk1 targets on the kinetochores and spindle (Higuchi and Uhlmann, 2005; Khmelinskii et al., 2009; Pereira and Schiebel, 2003; Woodbury and Morgan, 2007). Dephosphorylation of Cdk1 targets is also important for normal anaphase behavior in animal cells (Oliveira et al., 2010; Parry et al., 2003). Thus, the initiation of spindle elongation depends on cohesin cleavage, and dephosphorylation of Cdk1 substrates then contributes to robust spindle behavior and chromosome movement.
The polo kinase family regulates cohesin function. In vertebrates, but not in yeast, the majority of cohesin is removed from chromosome arms before metaphase by the ‘prophase pathway’, in which polo kinase phosphorylates the SA/Scc3 cohesin subunit, triggering cohesin dissociation without its cleavage by separase (Hauf et al., 2005; Waizenegger et al., 2000). In anaphase of both yeast and human cells, cleavage of the cohesin subunit Scc1 is stimulated by the phosphorylation of Scc1 by Polo, which acts primarily at two sites, one in each of the two separase cleavage motifs (Alexandru et al., 2001; Hauf et al., 2005; Hornig and Uhlmann, 2004).
Phosphorylation of chromatin-bound Scc1 by the yeast polo-related kinase Cdc5 is crucial for the timing of cohesin cleavage, particularly in securin-deficient cells, in which separase activity is less robust and the APC/C does not contribute to its activation (Alexandru et al., 2001). It seems likely that the timing of cohesin cleavage in securindeficient cells depends on mechanisms that promote an increase in Scc1 phosphorylation at the onset of anaphase. Given that Cdc5 activity rises gradually and is thought to be high before and during anaphase (Charles et al., 1998), such regulation could depend on anaphase inhibition of a phosphatase that counteracts Scc1 phosphorylation.
The phosphatase that dephosphorylates Scc1 is not known, but a prime candidate is the trimeric phosphatase PP2A. PP2A consists of a scaffold (A) subunit, a catalytic (C) subunit and one of two major regulatory subunits, B (Cdc55 in yeast) or B’ (Rts1), which are believed to confer substrate specificity and regulate phosphatase localization (Shi, 2009). As mentioned above, PP2ACdc55 activity toward Net1 is inhibited by separase in yeast, triggering the initial activation of the Cdc14 mitotic phosphatase (Queralt et al., 2006). PP2ACdc55 might also have a reciprocal inhibitory effect on separase activity, although the mechanism is unclear (Clift et al., 2009). On the other hand, PP2ARts1 has no known role in separase regulation in mitosis. It is, however, recruited by the shugoshin protein to centromeres in the first meiotic division, where it removes stimulatory phosphates from Rec8, thereby protecting it from cleavage by separase (Kitajima et al., 2006; Riedel et al., 2006).
Anaphase is a dramatic and irreversible process of the utmost importance in cell division, and it is accordingly very tightly regulated. Although the general principles of cohesin cleavage are clear, we know little about the regulatory circuits that generate robust, high-fidelity, and synchronous chromosome separation. There is evidence for at least one positive feedback loop in the cohesin cleavage system (Holt et al., 2008), but it seems likely that additional regulatory features enhance the robustness and switch-like characteristics of this system. To develop better methods for exploring this system, we generated a biosensor for separase activity in the cell, and we used this sensor to measure the kinetics and timing of cohesin cleavage relative to other anaphase events. We also used the biosensor to identify the PP2ACdc55 phosphatase as a negative regulator of cohesin cleavage, suggesting that separase-driven downregulation of PP2ACdc55 promotes cohesin phosphorylation. We speculate that this results in a coherent feedforward loop (Alon, 2007; Mangan et al., 2003) that helps reduce the impact of spurious separase activity, contributing to robust and timely cohesin cleavage.
Experimental Procedures
Strains
All strains used to measure reporter cleavage rates were derived from YGY7 (W303 matA trp1-1 ::256xLacO-TRP1 , ura3-1, leu2-3 112, his3-11, ade2-1, can1-100, GAL+). pRS303-_pCUP1_-GFP-Scc1(101–470)-LacI (pGY56), or variants encoding R180D, R268D (pGY59) or S175A, S263A (pGY65) Scc1 point mutants, were integrated at the HIS3 locus. For spindle visualization, Spc42 was tagged with mCherry. For the diploid cells with both TRP1 loci labeled with dots (Figure S4), a TetO array was inserted at the TRP1 locus of DOM90 (wild-type W303 MatA) using the two-step method described by Rohner et al (Rohner et al., 2008), and a pRS406-_pCUP1_-mCherry-TetR (pGY 90) was integrated at the URA3 locus. The Cdc5-as1 allele with the L158G gatekeeper mutation was cloned with its 5’ and 3’ UTRs into pRS306. Endogenous CDC5 was replaced with Cdc5-as1 in a two-step gene replacement.
Cdc55, Rts1 or mCherry were subcloned together with a (GGS)x4 linker into the C-terminus of the LacI in pGY56 for construction of the phosphatase fusion reporters and the doubly labeled reporter. All Rec8 disappearing dots were identical to the Scc1 ones, with residues 110–500 of Rec8 in place of the Scc1 fragment. For the endogenous Scc3- fusion strains, Cdc55 or Rts1, along with the (GGS)x4 linker, were subcloned C-terminal of a flag3-His6 tagging vector (Holt et al., 2008). A PCR of the flag3-His6-Cdc55/Rts1 was then integrated at the C-terminus of endogenous Scc3 by homologous recombination. The pGAL-S promoter was introduced at the securin locus for conditional repression of securin expression.
Microscopy
Asynchronous cultures were grown to log phase at room temperature in synthetic complete media and plated on ConA-coated 35 mm glass bottom culture dishes. Biosensor expression was driven from the pCUP1 promoter without copper induction. For the cdc5-as1 experiment, cells were synchronized in factor alpha (10 µg per ml culture) and released into media with 15 µM CMK inhibitor or DMSO.
In a typical movie, 18 slices spanning ~6 microns were acquired every 10 s for 10 min. all movies were taken at 23°C. Microscopy was performed in the UCSF Nikon Imaging Center with a spinning disk confocal microscope comprised of a Yokogawa CSU-22 scan head, an EMCCD camera (Photometrics Cascade II or Evolve) and a Nikon TE2000 or Ti inverted microscope with a 60× / 1.2NA water immersion objective. Illumination was provided by a 50 mW 491 nm laser and 50 mW 561 nm laser.
Microscope data analysis
We developed an automated analysis procedure in MATLAB to detect and quantify the brightness of GFP-intScc1-LacI chromosomal dots. The code that implements this analysis is available for download at https://github.com/kthorn/particletrack. Dots were identified by filtering the first time point of the movie with a negative Laplacian of Gaussian filter with a standard deviation of 1 pixel. Regions above a threshold in the filtered image were taken as candidate dots, and their centroids were determined, weighted by the intensities of the initial image. These coordinates were used as initial conditions for fitting a Gaussian model to the image of the dot. For each dot, we extracted a 19×19 pixel region around it in X and Y and the full stack height in Z. We then fit a 5 model consisting of a 3D Gaussian to model the dot, a broader 3D Gaussian to model the diffuse nuclear fluorescence, and a constant background. The dot Gaussian was assumed to have cylindrical symmetry, and the standard deviation in XY was set to 1.25 pixels; the standard deviation in Z was set to 1.7 slices. These values were determined by performing unconstrained Gaussian fits to a representative selection of dots. Fixing the widths of these dots resulted in faster, more robust fits. The Gaussian used to model the diffuse nuclear background was allowed to be asymmetric in the XY plane. To account for the possibility that the dot was splitting, we also fit a second model to the image including two dot Gaussians of equal intensity. This fitting process was repeated for all time points in the movie, with the center of the nuclear Gaussian used as the center of the image for the next time point. This fitting process takes ~45–60 minutes for a 60 time point image with ~100 dots in it. A Graphical user interface (GUI) wraps this fitting process, allowing multiple movies to be queued for unattended fitting.
A second GUI takes these fit results and for each time point and decides if the one- or two-dot model is a better fit. The two-dot model is favored if it is a better fit to the data, but there is a penalty for switching models to ensure that the software does not rapidly switch between models. For each time point, the dot intensity is calculated as the amplitude of the dot Gaussian (or the sum of amplitudes in the case of the two-dot model). To find potential disappearing dots, the intensity over time is then fit to a three part function: an initial exponential decay to account for photobleaching (the time constant was determined from non-disappearing dots), followed by a linear decline (the dot disappearance) and ending at a constant value. A simulated annealing approach is used to determine the four parameters in this model: the initial and final intensities, the 6 start point of the linear decline and the total number of time-points of the linear decline, including the first and last. The latter value was used for quantification of the rate of disappearance of the dots. If the final intensity is <30% of the initial intensity, the dot is flagged as potentially disappearing and presented to the user for verification. Spindle pole bodies were tracked within each cell by using the same Laplacian of Gaussian filtering that was used to find disappearing dots. Regions above a threshold in the filtered image were identified as the spindle pole bodies, and their centroids were determined, weighted by the intensities of the initial image. This analysis was performed in each 19×19 pixel region isolated around the disappearing or splitting dot.
Statistical methods
A two-tailed T-test using MATLAB was used to test the statistical significance of biosensor rates versus the wild-type reference strain (Table S1). The statistical significance of differences between coefficients of variance (CV) was determined by drawing random samples, the same size as the parent data sets, from the combined wild type and mutant data sets. The p-value was determined as the fraction of times that these randomly chosen samples had a greater of equal difference in CV than did the difference in CV between the wild type and mutant data sets (Table S1).
In vitro separase reaction
Chromatin isolation, preparation of cell lysates containing overexpressed separase, and separase cleavage reactions were performed as described (Uhlmann et al., 1999). 7 Endogenous Scc1 was C-terminally flag-tagged, while flag tags on the fusion reporter (pGY109) and separase were N-terminal.
Kinase and phosphatase assays
GST-Scc1(1–359)-HIS8 was purified from bacteria and the GST domain was removed. Yeast HIS-Cdc5 was baculovirus-expressed and purified from insect cells. PP2ACdc55 was purified from the TAP-Cdc55 strain by one-step IgG affinity purification and TEV elution (Puig et al., 2001). For Cdc5 phosphorylation of Scc1 (Scc1-P), ~10 µg Scc1 was incubated with ~100 ng Cdc5 for 15 minutes at 30°C. For histone H1 (HH1) labeling, 40 µg HH1 (Millipore 14–155) was incubated with ~1 ng purified Clb2-Cdk1 (Loog and Morgan, 2005) for 15 min at 30°C. Substrates were purified with G-25 columns (GE 27- 5325-01). PP2ACdc55 (~0.5 µg Cdc55, and serial 3-fold dilutions) was incubated with 0.2 µg Scc1-P or 1.5 µg HH1-P for 15 min at 30°C in 20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1 % NP40. Reaction products were quantified by SDS-PAGE and PhosphorImager.
Results
A Separase Biosensor
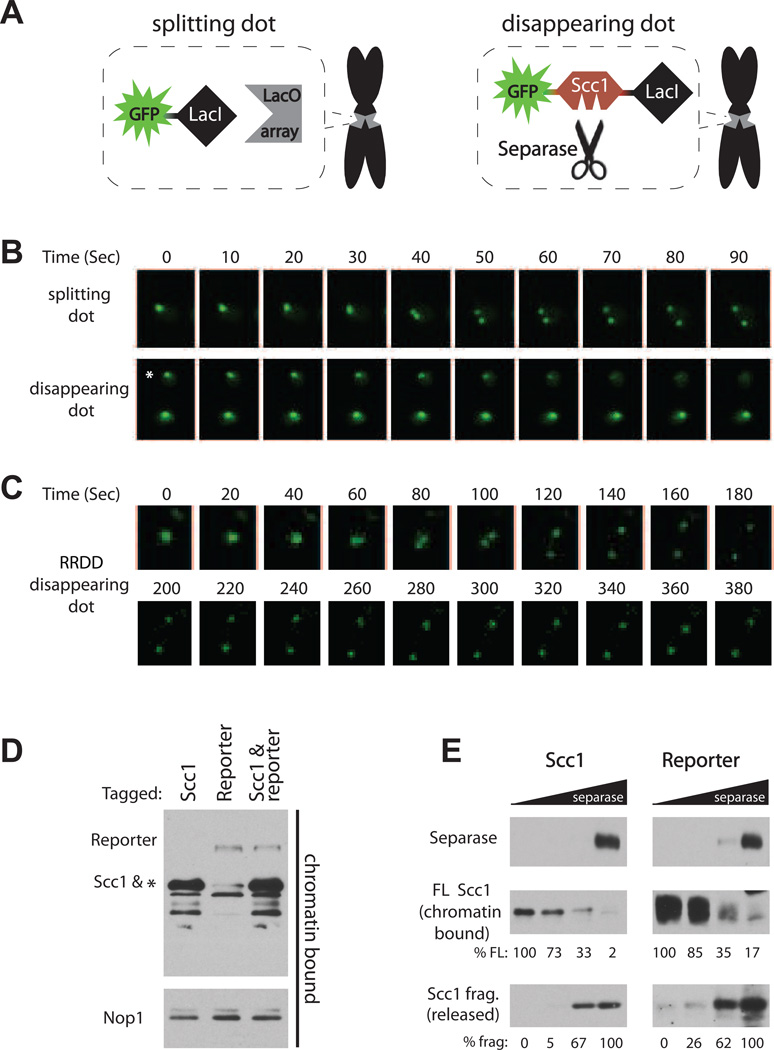
Our approach to the measurement of separase activity inside the cell was based on an established method in which a specific chromosomal locus in S. cerevisiae is tagged with a fluorescent focus (dot) by recruiting a GFP-tagged Lac repressor fusion (GFP-LacI) to an integrated Lac operator array (LacO) (Straight et al., 1997) (Figure 1A, left). In this setup, sister-chromatid separation, the endpoint of separase activity, can be visualized when the dot splits in two (Figure 1B, top; henceforth termed “splitting dot”). To visualize the dynamics of cohesin cleavage, we constructed a fusion protein with an internal fragment of Scc1 inserted between the GFP fluorophore and the LacI repressor (GFP-intScc1-LacI) (Figure 1A, right). The inserted Scc1 fragment encompasses residues 101–470, which contain both separase cleavage sites but not the terminal domains that mediate interactions with other cohesin subunits (Haering et al., 2002; Haering et al., 2004). The reporter fusion was expressed in a strain with a LacO array integrated at the TRP1 locus, 12 kb from the centromere of chromosome IV. Analysis of asynchronous cultures of this strain by video microscopy revealed that the GFP focus disappeared as the cell progressed through mitosis (Figure 1B bottom, Movie S1). Mutation of the two separase cleavage sites in the Scc1 reporter to non-cleavable residues (R180D & R268D) resulted in GFP dots that split rather than disappearing (Figure 1C), demonstrating that separase specifically cleaved the wild-type reporter. We also found that the GFP-intScc1- LacI fusion did not functionally replace endogenous Scc1 (Figure S1A), explaining why mutation to the non-cleavable form had no physiological consequence.
Figure 1.
Detection of separase-mediated cohesin cleavage in single cells. (A) Schematic of the experimental setup. Expression of a GFP-LacI fusion in a cell with an integrated LacO array results in a fluorescent dot that splits when sister chromatids separate (‘splitting dot’). Residues 101–470 of the cohesin subunit Scc1, including its two separase cleavage sites, were inserted between the LacI and GFP moieties, resulting in the ‘disappearing dot’. The LacO array was integrated in haploid cells at the TRP1 locus on chromosome IV, 12 kb from the centromere. (B) Representative time-lapse images from movies of cells in an asynchronous population, carrying either a splitting or disappearing dot. In the disappearing dot panel, only the top cell marked with the asterisk went through anaphase. The ‘0’ time-point was set arbitrarily. All dots shown in this study are diffraction limited. See also Movie S1. (C) Time-lapse of a representative cell with the reporter mutated in the two separase cleavage sites of Scc1 (R180D and R268D). (D) Western blot analysis of chromatin preparations from cells with flag-tagged endogenous Scc1, the GFP-intScc1-LacI reporter fusion or a combination of both in the same cell. Nop1 was used as a chromatin-bound loading control. The asterisk marks a faint cross-reactive band that co-migrates with Scc1. (E) Equal amounts of chromatin samples from (D) were subjected to an in vitro cleavage assay with a 5-fold dilution series of separase. Note that the endogenous Scc1 substrate is present in reporter reactions as well, but only the flag-tagged reporter is monitored. Due to the lower reporter abundance (see (D)), the blots for the reporter protein were exposed longer. Also, blots of the full-length (FL) substrates were exposed for shorter times than the less abundant cleavage products (frag). Quantification of the relative densities of the FL and cleavage products is shown. See also Figure S1.
We compared the timing and rate of cleavage of the fusion reporter substrate with those of endogenous Scc1. In population studies, the fusion reporter was cleaved at the same time as endogenous Scc1 in cells released from a metaphase arrest (Figure S1B) and alpha factor arrest (data not shown). We also compared the rates of reporter and Scc1 cleavage by separase in vitro . Efficient cleavage of Scc1 depends on its association with chromatin (Hornig and Uhlmann, 2004; Sun et al., 2009). We therefore extracted chromatin from cells carrying tagged variants of endogenous Scc1, the reporter fusion, or both together. We detected appreciably higher amounts of chromatin-bound endogenous Scc1, demonstrating that the amount of reporter bound at the LacO array is, as expected, far lower than the total amount of cohesin on chromatin (Figure 1D). When the chromatin was treated with separase, the fusion reporter was cleaved at the same rate as endogenous Scc1 (Figure 1E). Thus, our biosensor is a chromatin-bound protein that is cleaved specifically by separase with similar timing and efficiency to endogenous Scc1.
Quantitative Analysis of the Rate of Cohesin Cleavage in vivo
To accurately quantify the rate and timing of reporter cleavage in vivo, we developed software that tracks the fluorescent dots along the four dimensions of the acquired movies and quantifies their intensity by Gaussian fitting. This software included the capacity to quantify dots that split as they disappeared (Figure S2A, B; see Experimental Procedures), which we found to be important for mutations that uncoupled the rate of reporter cleavage from that of endogenous cohesin (see below). We acquired 10-minute movies of unperturbed asynchronous cultures, and cells in which the dot disappeared were automatically identified and manually verified. Because each movie included on average only three cells that progressed through anaphase within the ten minutes (see Movie S1), dozens of movies were required for quantification of significant numbers of cells for each strain analyzed.
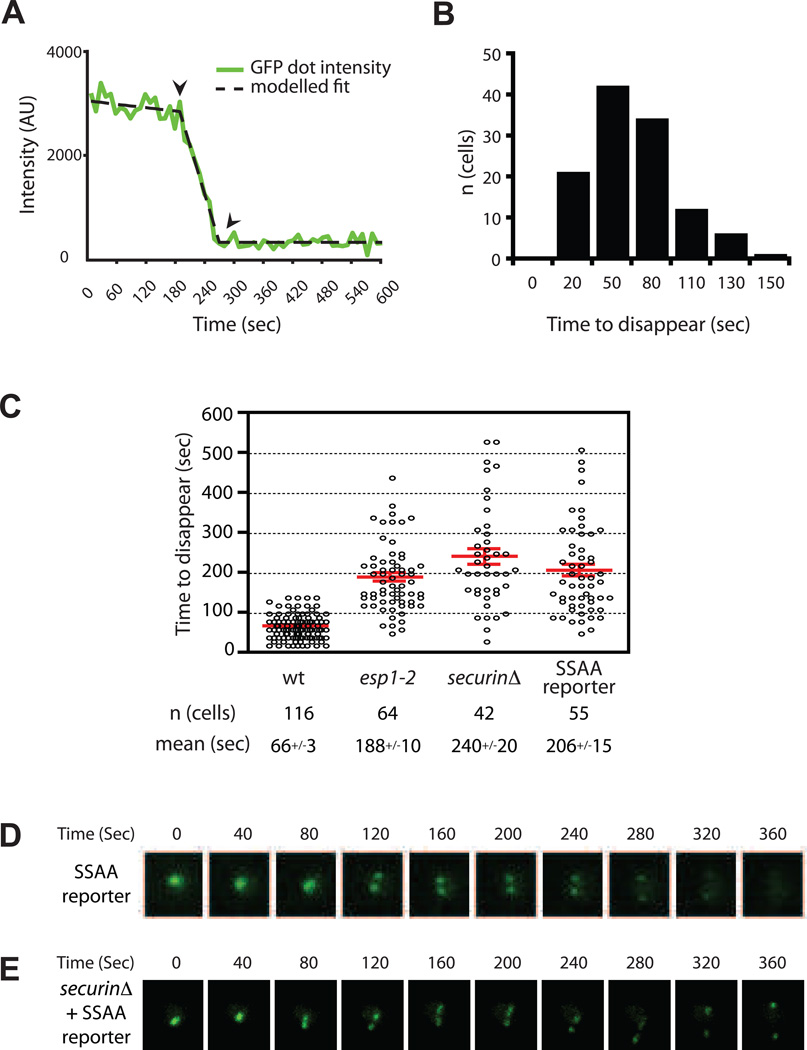
To measure the time required for reporter cleavage in single cells, the decrease in fluorescence intensity for each cell was fit to a line (Figure 2A). In wild-type cells, complete cleavage of a GFP dot required an average of 66 seconds, with a Gaussian distribution around that value (Figure 2B; Table S1). The rate of disappearance did not correlate with the initial intensity of the dot (Figure S2C), indicating that values obtained from independent experiments are comparable and confirming that the amount of substrate bound to the array is a small fraction of the total cohesin, so that variations in the amount of reporter do not significantly influence the overall rate of cleavage.
Figure 2.
Rates of cohesin cleavage in the cell. (A) The measured intensity (green) and resulting fit (black) of a representative disappearing dot. The fit is a three-part model consisting of an initial photobleaching decline, followed by a linear decrease (dot disappearance) to a baseline level. See also Figure S2 (B) The time from maximal to minimal intensity (indicated by arrowheads in (A)) was measured for 116 disappearing dots from multiple wild-type (wt) cultures, and the data plotted in a histogram. (C) The time required for dot disappearance was measured in several cells from each of the indicated strains. Each circle represents a single cell. In the SSAA reporter strain, the Scc1 fragment in the fusion reporter is mutated to alanine in the two polo phosphoacceptor sites S175 and S263 (see also Figure S3). Results with all mutants are significantly different from wt; see Table S1 for complete listing of data for all strains. (D–E) Representative time-lapse images of the SSAA reporter disappearing dot in a wild-type cell (D) or _securin_Δ cell (E). Note that 40-second time-points are shown, although 10-second intervals were analyzed. See also Movie S2.
To confirm that the biosensor is sensitive to perturbations in separase function, we analyzed mutants known to affect separase activity. We began with the temperature-sensitive esp1-2 mutant, in which separase activity is compromised even at the permissive temperature (Holt et al., 2008). At 23°C, the dots disappeared almost three times more slowly in the esp1-2 mutant (Figure 2C). Securin, the chaperonin-inhibitor of separase that is targeted for degradation by the APC/C, also has a positive role in separase folding and nuclear localization (Agarwal and Cohen-Fix, 2002; Hornig et al., 2002). Consistent with these functions, dots in cells lacking securin disappeared over three times more slowly. Finally, phosphorylation of Scc1 by the polo-like kinase Cdc5, at serine residues in the separase recognition sites (S175 and S263), is an important positive signal for its timely and efficient cleavage (Alexandru et al., 2001). When mutated at these phosphoacceptor sites (SSAA), the reporter was cleaved about threefold more slowly (Figure 2C, Movie S2). Consistent with these results, the rate of biosensor cleavage was reduced by half when the activity of the polo kinase Cdc5 was specifically inhibited (Figure S3). Thus, perturbation of either separase or the phosphorylation of its substrate leads to slower dot disappearance.
We observed that the dots of the SSAA mutant split as they disappeared (Figure 2D, Figure S2, Movie S2), which we did not detect in the other strains. Thus, the reporter in this mutant is being cleaved later than the endogenous cohesin, which is still subject to 0 phosphorylation by Cdc5. Cells lacking securin and also mutated in the phosphoacceptor sites of endogenous Scc1 are not viable (Alexandru et al., 2001). In agreement with this, dots of securinΔ cells carrying the SSAA reporter did not disappear, but instead split (Figure 2E). Dot splitting was less abrupt than in wild-type cells (compare to Fig. 1B), consistent with spindle defects observed in securinΔ cells (Holt et al., 2008).
Two residues near the N-terminus of securin are phosphorylated by Cdk1, resulting in less efficient ubiquitination of securin by the APC/C (Holt et al., 2008). The Cdc14 mitotic phosphatase, which is activated by separase in early anaphase (Queralt et al., 2006), dephosphorylates these sites, creating a positive feedback loop that might help govern separase activation (Holt et al., 2008). The securin-2A mutant, which lacks the N-terminal Cdk1 sites and is deficient in the positive feedback loop, displayed a moderate decrease in the rate of reporter cleavage (Table S1). Thus, the positive feedback loop does not have a major impact on the final rate of cohesin cleavage, and might instead be more important in the timing or reversibility of separase activation.
Cohesin is Cleaved with Similar Kinetics and Timing near the Centromere and Telomere
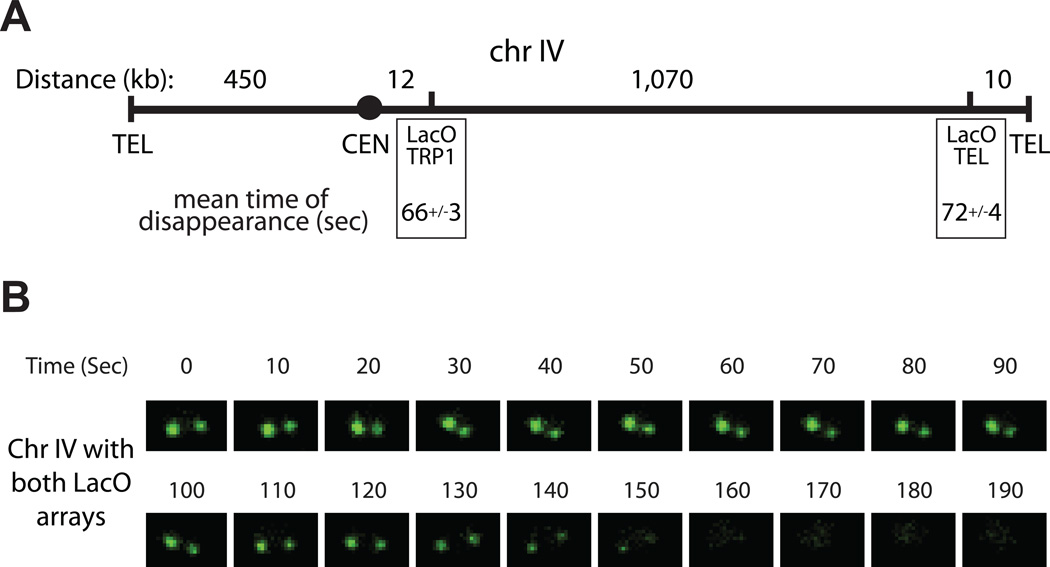
The separase biosensor allowed us to probe the spatial regulation of separase activity. In budding yeast, cohesin is distributed unevenly along the chromosomes, with a higher concentration in the 30–50 kb region around the centromere and less dense binding farther out on the chromosomal arms (Blat and Kleckner, 1999; Glynn et al., 2004). In cells with splitting dots integrated at different positions on the chromosome, dots near the centromere separate a few minutes before those at the telomere (Alexandru et al., 2001; Straight et al., 1997). It is not known whether this is a result of delayed cleavage of telomeric cohesin.
To assess cohesin cleavage at different chromosome regions, we analyzed the rate of reporter cleavage at a LacO array integrated in an intergenic locus less than 10 kb from the telomere on the long arm of chromosome IV (Figure 3A). The rate of reporter cleavage at this locus was the same as the rate measured with our standard array at the TRP1 locus, near the centromere of the same chromosome (Figure 3A). To test the relative timing of cohesin cleavage at the two loci, we analyzed cells carrying LacO arrays near both the centromere and telomere of chromosome IV. The two dots disappeared simultaneously (Figure 3B), indicating that separase cleaves cohesin at inner and outer arms with comparable rates and timing. Thus, separase is activated abruptly, both kinetically and spatially, and the delayed separation at the outer arms presumably results from the time required for spindle forces to gradually ‘unzip’ the arms.
Figure 3.
Cohesin cleavage near centromeres and telomeres occurs at the same rate and time. (A) Schematic of chromosome IV. Relevant features are labeled below (‘TEL’-Telomere, ‘CEN’-Centromere), and the distance between any two adjacent features is indicated above in kilobases. Dot disappearance rates are indicated for the ‘LacO TRP1’ array near the centromere (n=116, data same as Figure 2C) and for the ‘LacO TEL’ array near the Telomere (n=48). (B) Time-lapse images of a haploid cell with disappearing dots at both LacO arrays as shown in (A). This cell is representative of tens of cells imaged.
Anaphase Begins After Completion of Cohesin Cleavage
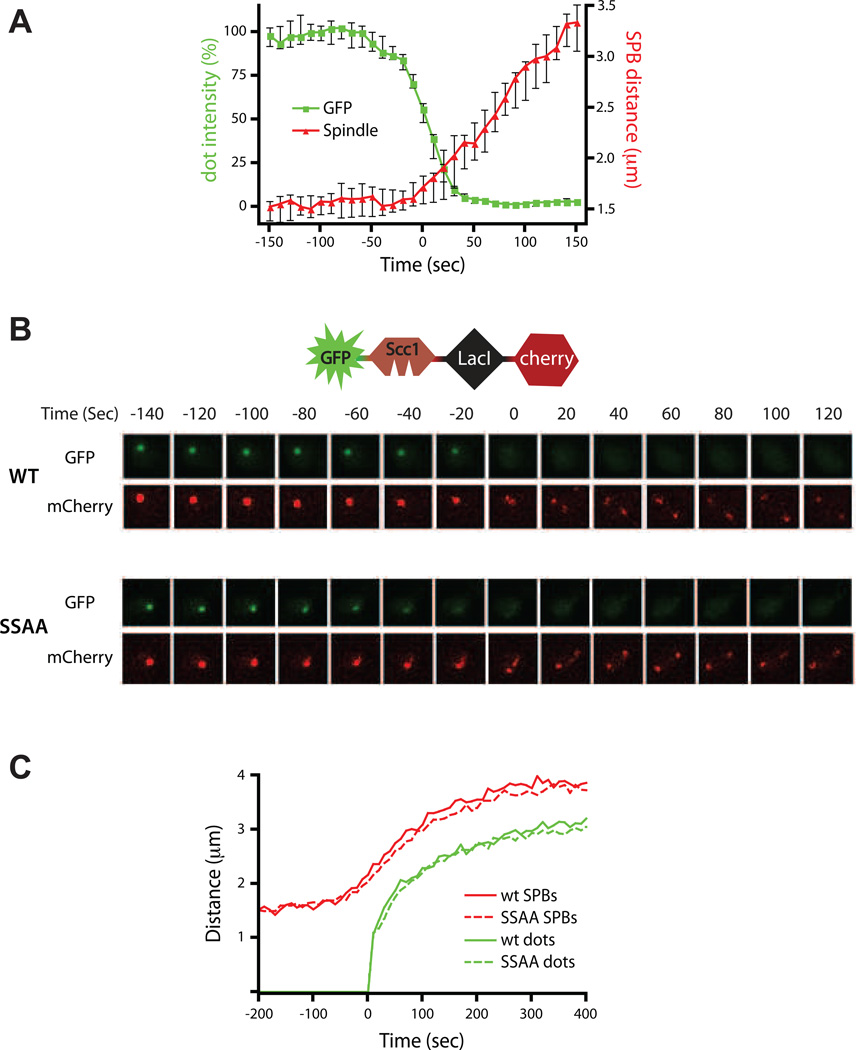
The first discernible cytological event of yeast anaphase is spindle elongation, which is likely to depend on cohesin cleavage and the unraveling of sister linkages in regions immediately surrounding the kinetochores (Pearson et al., 2001; Straight et al., 1997; Yeh et al., 2008). To assess the timing of cohesin cleavage relative to spindle elongation, we imaged the disappearing GFP dot in cells in which the spindle pole bodies were labeled with a red fluorophore (Spc42-mCherry). We found that spindle elongation was initiated just before the completion of biosensor cleavage, suggesting that chromosome segregation begins after most cohesin is cleaved (Figure 4A, Movie S3).
Figure 4.
Anaphase begins after cohesin cleavage. **(A)**GFP dot intensity (green, left axis) and 3D distance between spindle pole bodies (SPBs; red, right axis) are plotted for 24 wild-type cells carrying a disappearing dot and Spc42-mCherry. Cells were imaged every 10 seconds and plots from individual cells were centered along the time axis for disappearance of half the intensity of the dot at T=0. Plotted are the medians with 95% confidence intervals. See also Movie S3. (B) mCherry was fused to the C-terminus of the biosensor (see schematic), resulting in a green disappearing dot together with a red splitting dot in the same reporter. Time-lapse images in both color channels are shown for a representative wild-type cell (top) and a cell in which both the reporter and endogenous Scc1 carry the S175A & S263A mutations (SSAA, bottom). These data are representative of tens of cells imaged. See also Figure S4. (C) The 3D distance between the spindle pole bodies and sister chromatids (splitting dot at TRP1 locus) were measured in wild type (n=28) cells and in cells with endogenous Scc1-SSAA (n=49) with 20-second time-points. The plots from individual cells were centered on the time axis for sister separation at T=0. Plotted are the medians. The 95% confidence intervals (not shown) were all under 8% of their respective median values.
We also examined the precise relationship between reporter cleavage and sister separation by fusing the reporter protein to an additional red fluorophore that stays bound to the LacO array after cleavage by separase (GFP-intScc1-LacI-mCherry). Sisters split immediately after the disappearance of the separase biosensor (Figure 4B). The same result was seen in diploid cells carrying a GFP-intScc1-LacI disappearing dot at the TRP1 locus of one homolog of chromosome IV and an mCherry-TetR splitting dot at the same locus on the other homolog (Figure S4).
Next, we analyzed the relative timing of biosensor cleavage and sister separation in cells in which both the biosensor and the endogenous Scc1 were mutated at the two polo-kinase phosphoacceptor sites, resulting in slower cohesin cleavage. In this strain, the reporter did not split while disappearing, demonstrating that the timing and rate of reporter cleavage were coupled to those of endogenous cohesin (Figure 4B). Importantly, as in wild-type cells, sisters split after the dot had almost completely disappeared, demonstrating that the rate of dot disappearance reflects the rate of endogenous cohesin cleavage, and further suggesting that near-complete cohesin cleavage occurs before anaphase begins.
Separation of lacO arrays in our strain (at the TRP1 locus, about 12 kb from the centromere of chromosome IV) occurs one minute after the initiation of spindle elongation (Figure 4C), consistent with previous studies (Holt et al., 2008; Pearson et al., 2001; Straight et al., 1997). This is presumably because this amount of time elapses before sufficient spindle forces reach this point on the arms (Pearson et al., 2001). We observed an identical delay in SCC1-SSAA cells (Figure 4C). Thus, a reduced rate of 3 cohesin cleavage delays the initiation of anaphase but does not affect the time required from initiation of spindle elongation to separation of chromosome arms.
PP2ACdc55 Inhibits Cohesin Cleavage
The PP2A trimeric phosphatase has been implicated in regulation of cohesin cleavage by separase. The Cdc55-bound form of PP2A is particularly intriguing, as its activity is inhibited by separase during anaphase, triggering activation of the Cdc14 mitotic phosphatase (Queralt et al., 2006).
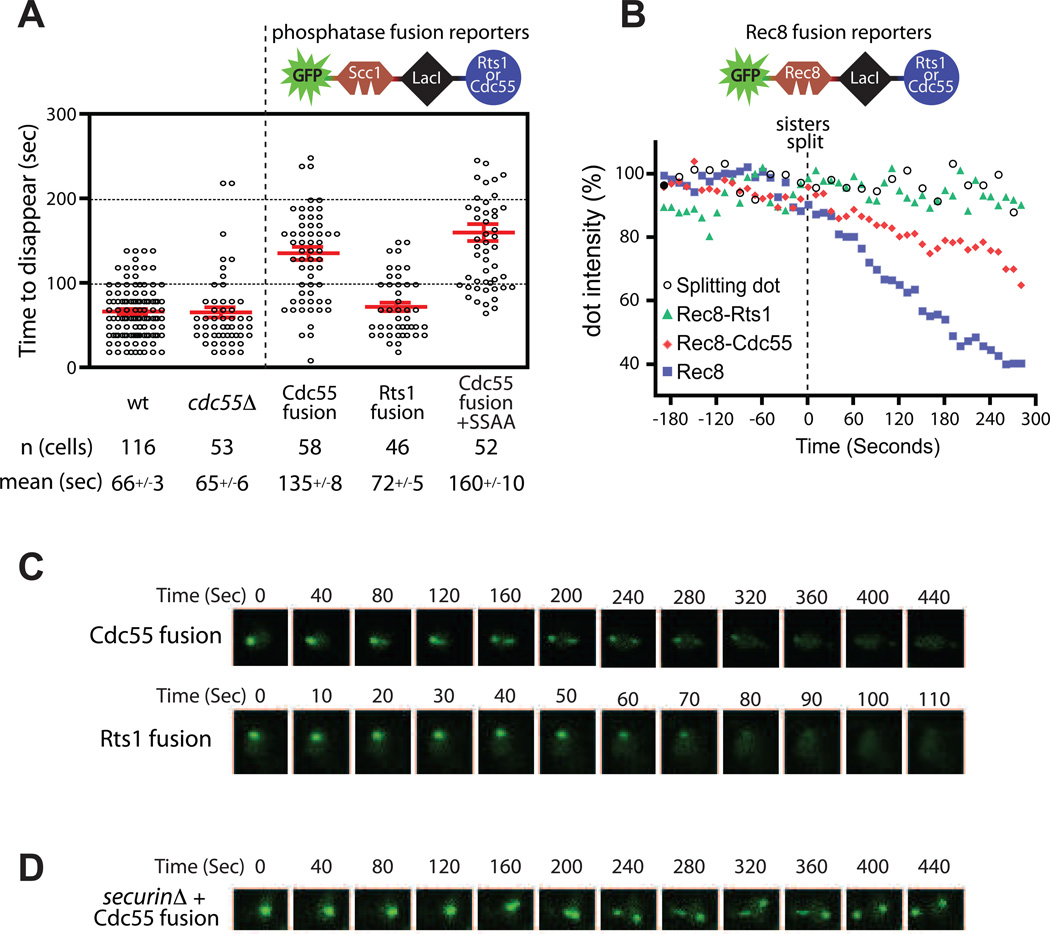
We wished to directly test the role of PP2ACdc55 in cohesin cleavage in mitotic cells. Deletion of CDC55 had no effect on the mean rate of biosensor cleavage (Figure 5A, Table S1), but this might be expected if PP2ACdc55 has an inhibitory function that is removed during anaphase (see Discussion). Interestingly, deletion of CDC55 did cause a significant increase in the variability of cohesin cleavage rates (Table S1). We further tested the role of Cdc55 by fusing it to the LacI module of our cohesin cleavage reporter (Figure 5A). As a control, we similarly fused the alternate PP2A regulator Rts1. In this approach, localization of the phosphatase is coupled to that of the disappearing dot, and it remains tethered to the DNA after separase releases the fluorescent substrate. Each of the fused regulatory subunits specifically complemented growth defects caused by its own deletion, but not those of a deletion of the other regulatory subunit, demonstrating that it successfully recruited a functional trimeric PP2A phosphatase (Figure S5A). Separase cleaved the Rts1 fusion reporter at wild-type rates (Figure 5A), suggesting that localized PP2ARts1 activity does not inhibit cleavage of the reporter. In contrast, the Cdc55 fusion was cleaved significantly more slowly than the wild-type reporter, demonstrating that 4 localized PP2ACdc55 activity can inhibit the rate of cohesin cleavage (Figure 5A, Movie S4).
Figure 5.
PP2ACdc55 inhibits cohesin cleavage. (A) The rate of cohesin cleavage in wild-type (wt) cells (data same as Figure 2C) and _cdc55_Δ cells is shown. Each of the two PP2A regulatory subunits, Rts1 or Cdc55, was fused to the LacI C-terminal module of the reporter via a flexible linker (see schematic). Rates of cohesin cleavage of the resulting fusion reporters in wild-type cells are shown. Cdc55-fusion+SSAA combines the Cdc55 fusion with two mutated Scc1 polo sites in the reporter. See also Figure S5. (B) Analysis of Rec8 disappearing dots in mitotic cells. These data show the normalized average intensities from several individual cells (n>22 for each strain), centered along the time axis at T=0 for the time of dot splitting. The Rec8 reporter construct, and the Cdc55 and Rts1 fusions, are identical to their Scc1 counterparts, with Rec8 residues 110–500 replacing the analogous region of Scc1. Rec8 disappearing dots took an average of 348 s to disappear (n=28). Splitting dots (GFP-LacI) were analyzed in parallel as a control. (CD) Time-lapse images for wild-type cells with the Cdc55 and Rts1 fusion reporters (C) and the Cdc55 fusion reporter in a _securin_Δ cell (D). Note the difference in time scales. See also Movie S4.
To further validate the activities and specificities of Rts1 and Cdc55 fusion reporters, we constructed a biosensor derived from Rec8, the meiotic counterpart of Scc1. Rec8 is phosphorylated to promote its cleavage by separase (Ishiguro et al., 2010; Katis et al., 2010), and this phosphorylation is counteracted specifically by the PP2ARts1 isoform (Kitajima et al., 2006; Riedel et al., 2006). We constructed a Rec8 reporter containing residues 110–500, which encompass all phosphorylated sites (Katis et al., 2010), and tested the capacity of Rts1 and Cdc55 fusions to affect the rate of these dots in mitotic cells.
Rec8 dots disappeared more slowly than their Scc1 counterparts, resulting in splitting before full disappearance (Figure 5B). When Rts1 was fused to the Rec8 reporter, dot disappearance was abolished. The Cdc55 fusion only partly reduced the rate of dot disappearance (Figure 5B). These results not only support a role for PP2ARts1 in Rec8 dephosphorylation but also further support the specificity of PP2ACdc55 for Scc1. The opposite specificities of the PP2A regulatory subunits toward the Scc1 and Rec8 reporters demonstrate that the reporters themselves are the targets for the phosphatase. In addition, the protection of Rec8 dots by fused Rts1 shows that the Rec8 reporter is phosphorylated in mitotic cells, and its slow rate of cleavage suggests that it is a less efficient substrate for separase than Scc1, event though Rec8 can functionally replace Scc1 in mitotic cells (Buonomo et al., 2000; Heidinger-Pauli et al., 2008). Slower Rec8 cleavage might enable more stringent regulation by dephosphorylation, a property that is essential for Rec8 protection in meiosis I. 5
PP2ACdc55 Dephosphorylates Polo-Phosphorylated Scc1
We observed that the Scc1-based Cdc55 fusion reporter, but not the Rts1 fusion, resembled the SSAA polo-phosphorylation site mutant in that the dots split while disappearing (Figure 5C, Movie S4). We therefore hypothesized that PP2ACdc55 inhibits cohesin cleavage by counteracting the stimulatory polo kinase phosphorylation of Scc1. If this were true, then a combination of the Cdc55 fusion reporter with a securin deletion would result in splitting dots, as occurs with the SSAA reporter in securinΔ cells (Figure 2E). This is indeed the case (Figure 5D). Conversely, combining the SSAA mutations and Cdc55 fusion in the same reporter did not further decrease the rate of disappearance, demonstrating that they are implicated in the same regulatory event (Figure 5A).
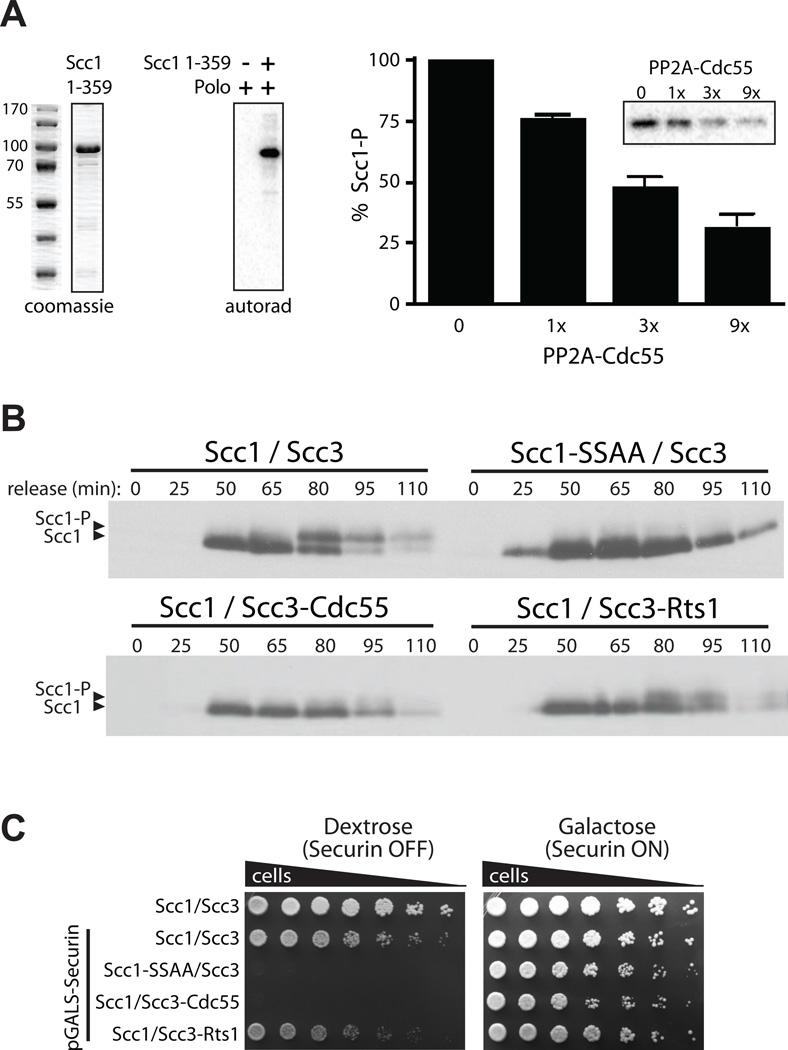
We tested whether PP2ACdc55 can remove phosphates from polo-phosphorylated Scc1 in vitro . Scc1 (residues 1–359) was purified from bacteria and phosphorylated with yeast polo kinase derived from insect cells (Figure 6A). Cdc55 was purified from yeast, and its activity was confirmed with Histone H1 (Figure S5B). PP2ACdc55 efficiently removed the polo kinase-dependent phosphates from Scc1 (Figure 6A), demonstrating that PP2ACdc55 has the capacity to counteract polo phosphorylation of Scc1.
Figure 6.
PP2ACdc55 inhibits cohesin cleavage by counteracting polo-dependent phosphorylation of Scc1. (A) Coomassie blue-stained polyacrylamide gel of a bacterially expressed Scc1 fragment encompassing residues 1–359 (left). This fragment was subjected to an in vitro kinase assay with yeast polo-kinase (Cdc5) purified from insect cells (autorad). The polo-phosphorylated Scc1 (Scc1-P) was incubated for 15 min with increasing amounts of PP2ACdc55 purified from yeast. Quantification of the input and the dephosphorylation reactions from three experiments is shown (error bars indicate SEM). A representative autoradiogram is shown in the inset. (B) Western blot analysis of endogenously-tagged Scc1-HA in cells released from α-factor arrest. Cells contained wild-type Scc1-HA and Scc3 (Scc1/Scc3), Scc1-HA mutated at the two polo sites and wild-type Scc3 (Scc1-SSAA/Scc3), or wild-type Scc1-HA with Cdc55 or Rts1 fused to Scc3 via a flexible linker (bottom panel). (C) The pGALS promoter was introduced at the endogenous securin locus in the strains in (B). Serial dilutions of the resulting strains, along with a strain carrying the endogenous securin promoter (top row), were plated on dextrose or galactose.
We next explored the ability of the PP2A regulatory subunits to dephosphorylate endogenous cohesin complexes. To this end, we fused Cdc55 or Rts1 to endogenous cohesin via a flexible tether to the Scc3 subunit, as previously described (Riedel et al., 2006). We followed Scc1 phosphorylation at the two polo sites by its mobility in SDSPAGE. Scc1 phosphorylation increased just before anaphase, resulting in a shift of a fraction of the protein that is likely the chromatin-bound subpopulation (Figure 6B) 6 (Hornig and Uhlmann, 2004). Consistent with a previous study (Alexandru et al., 2001), this shift was abolished when the two polo sites were mutated. When Cdc55 was fused to the cohesin complex, a marked loss of the phosphorylated species was observed (Figure 6B). In contrast, fusion of Rts1 had relatively little effect, providing further evidence that PP2ACdc55 specifically dephosphorylates endogenous Scc1.
We further analyzed the importance of Cdc55-dependent Scc1 dephosphorylation by combining it with securin depletion, using a strain in which the securin gene was under the control of the conditional pGALS promoter. As expected, securin shutoff in cells where endogenous Scc1 phosphorylation sites are mutated was lethal (Figure 6C) (Alexandru et al., 2001). Importantly, an identical phenotype was observed for the Cdc55 fusion. In contrast, the Rts1 fusion had only a partial growth defect in the absence of securin, once again arguing that PP2ACdc55 has greater activity toward Scc1 than PP2ARts1.
Evidence that Separase Inhibition of PP2ACdc55 is Necessary for Robust Cleavage of Cohesin
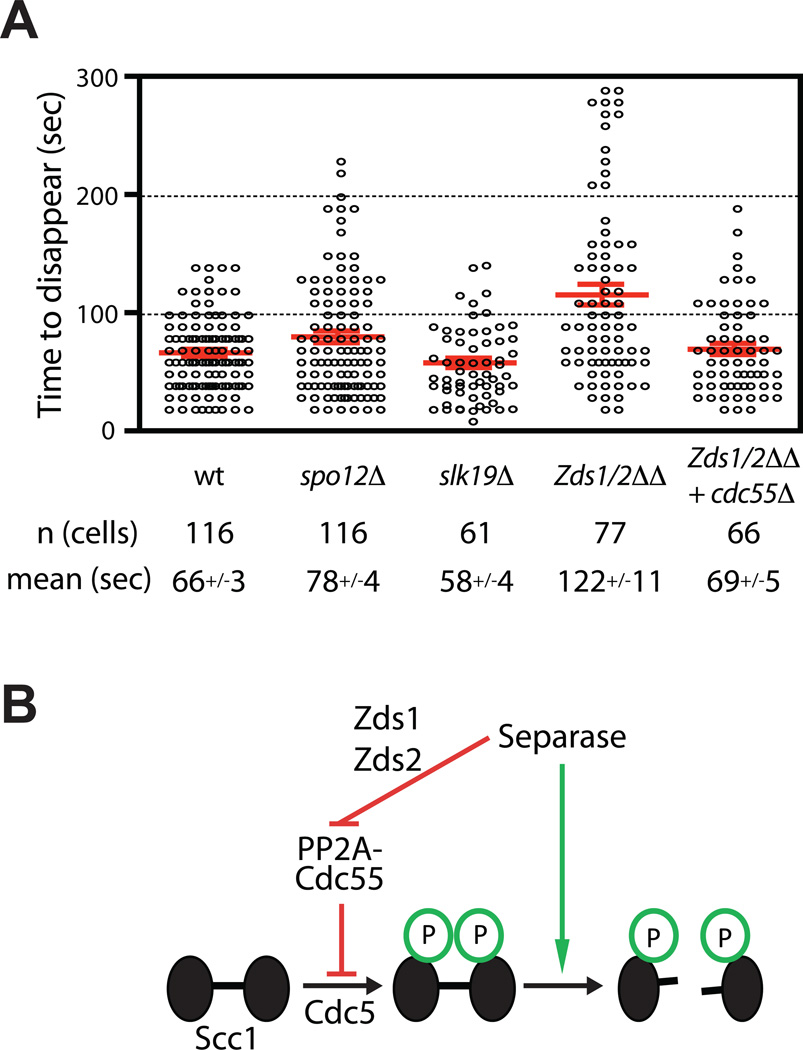
We hypothesized that PP2ACdc55 is active toward Scc1 prior to anaphase and inhibited when anaphase begins. There is a clear precedent for this regulatory mechanism: separase activation inhibits PP2ACdc55 activity toward Net1, thereby triggering the activation of Cdc14 (Queralt et al., 2006). Separase downregulation of PP2ACdc55 depends on the Zds1 and Zds2 proteins (Queralt and Uhlmann, 2008), which are stoichiometric binding partners of PP2ACdc55 that control its localization and substrate specificity (Rossio and Yoshida, 2011; Wicky et al., 2010). The release of Cdc14 also depends on the nucleolar 7 Spo12 protein and the Slk19 protein, but it is not clear whether these act upstream or downstream of PP2ACdc55 inhibition (Stegmeier et al., 2002). We reasoned that separase might also block PP2ACdc55 activity toward Scc1, resulting in increased Scc1 phosphorylation at the onset of anaphase. Consistent with this possibility, the mean rate of biosensor cleavage was half the wild-type rate in yeast lacking both Zds1 and Zds2 (Figure 7A). There was also a moderate increase in the variability of cohesin cleavage rates in the absence of Zds1 and Zds2 (Table S1). Importantly, deletion of CDC55 as well as the two ZDS genes restored a normal rate of dot disappearance, demonstrating that the effects of zds deletions are mediated by PP2ACdc55. Dot disappearance was normal in spo12Δ or slk19Δ cells, suggesting that they are specific for the Cdc14 release pathway (Table S1). Our results, together with previous evidence that PP2ACdc55 is inhibited by separase in a Zds-dependent fashion, suggest that separase promotes Scc1 phosphorylation at the onset of anaphase, thereby enhancing the rate of cohesin cleavage (Figure 7B).
Figure 7.
Separase downregulation of PP2ACdc55 is required for robust cohesin cleavage. (A) The rates of cohesin cleavage are shown for the indicated strains. For comparison, wild-type data are the same as in Figure 2C. (B) Model for PP2ACdc55 regulation of cohesin cleavage. Separase-dependent downregulation of PP2ACdc55 through the Zds1/2 proteins promotes the phosphorylation of Scc1 by Cdc5 polo kinase.
Discussion
The metaphase-anaphase transition depends on the robust timing and coordination of multiple processes. By enabling the precise measurement of the dynamics and timing of cohesin cleavage in single cells, our separase biosensor has allowed us to understand this transition in more detail. Our measurements of the timing of separase activity relative to other anaphase events are consistent with a model in which anaphase begins with the abrupt activation of separase, which targets all chromatin-bound Scc1 simultaneously 8 while at the same time inhibiting PP2ACdc55 activity toward Net1 and Scc1. Separase activation, together with increased Scc1 phosphorylation, leads to the rapid cleavage of all cohesin over a period of about one minute. The completion of Scc1 cleavage triggers the initiation of spindle elongation. Because the forces of the spindle are focused at the centromeres, the separated sister chromatids are unzipped from the centromere outwards by the mitotic spindle, whose robust elongation and stability depend on separasedependent Cdc14 activation.
We identified PP2ACdc55 as an inhibitor of mitotic cohesin cleavage in yeast, and our results demonstrate that PP2ACdc55 can counteract the stimulatory phosphorylation of Scc1 by the polo kinase Cdc5. It will be interesting to see whether this regulation is conserved in centromeric cohesin of higher eukaryotes. Vertebrate PP2ARts1 is known to protect centromeric cohesin from the prophase pathway by dephosphorylating the SA/Scc3 subunit (Hauf et al., 2005; Waizenegger et al., 2000). However, the phosphatase controlling Scc1 phosphorylation in vertebrates is not known. Given that separase-mediated inhibition of PP2ACdc55 has been demonstrated only in yeast, it is possible that Scc1 dephosphorylation is governed by distinct mechanisms in other species.
We found that dephosphorylation of Scc1 by PP2ACdc55 leads to slower cleavage of both the biosensor and endogenous cohesin by separase. The lack of a change in reporter cleavage rates in _cdc55_Δ cells is consistent with this hypothesis, as Scc1 should still be phosphorylated during anaphase in these cells. Scc1 phosphorylation might be expected to increase earlier in the cell cycle of _cdc55_Δ cells, but we detected no gross difference in the timing of Scc1 phosphorylation in a population of cdc55 Δcells released from alpha factor (data not shown). Such experiments are difficult to interpret, however, 9 because of the poor synchrony of the cell population and because _cdc55_Δ cells display defects in the timing of mitotic entry that could obscure small changes in the timing of Scc1 phosphorylation. Single cell methods with high temporal resolution will be required to allow precise measurements of Scc1 phosphorylation relative to other mitotic events.
Importantly, when PP2ACdc55 activity is rendered insensitive to separase inhibition by deletion of the ZDS genes, the rate of cohesin cleavage is reduced by half. Thus, we believe that separase activation promotes the phosphorylation of its own substrate by inhibiting the phosphatase that opposes it (Figure 7B). This regulatory relationship could result in a variation of a coherent feedforward circuit that has the potential to filter out background noise in separase activity as well as help govern the timing of cohesin cleavage (Alon, 2007; Mangan et al., 2003). Such a filter could reduce the deleterious effects of low levels of spurious separase activity – which would be especially important if this system contains positive feedback loops that amplify small signals into full separase activation.
Consistent with this possibility, we observed an increase in the variability of the rate of cohesin cleavage in cdc55 Δcells (Table S1). In these cells, the rate and timing of cohesin cleavage depends primarily on the timing of APC/C activation and securin degradation, a relatively slow process lasting about 20 minutes in human cells (Hagting et al., 2002) and about 10 minutes in yeast (D. Lu and D.O.M., unpublished data). Conversion of this gradual securin destruction process into abrupt cohesin cleavage is likely to depend on noise filters that delay cleavage until some sustained threshold level of separase activity is reached. The control of Cdc55 by separase could provide such a filter. We propose that this circuit enables the timely phosphorylation of Scc1 at anaphase onset and contributes to the remarkable coordination of anaphase events. This regulation is particularly critical in cells lacking securin, where APC/C activation does not contribute to the regulation of separase activation.
It has recently been proposed that Zds1 and Zds2 are stoichiometric subunits of the PP2ACdc55 complex that help regulate its localization and substrate specificity (Rossio and Yoshida, 2011; Wicky et al., 2010). Nuclear levels of PP2ACdc55 decline slightly in late mitosis, and deletion of both ZDS genes results in the accumulation of Cdc55 in the nucleus (Rossio and Yoshida, 2011). Thus, the Zds proteins appear to promote exclusion of PP2ACdc55 from the nucleus in late mitosis. Forcing Cdc55 into the nucleus inhibits Cdc14 release (Rossio and Yoshida, 2011), just as we found that forcing Cdc55 localization on cohesin slows cohesin cleavage. These results are consistent with the idea that separase normally inhibits the function of PP2ACdc55 by promoting its nuclear exclusion in a _ZDS_-dependent manner. An alternative possibility is that separase and the Zds proteins are not linked in the same pathway, but rather that an increased nuclear concentration of PP2ACdc55 in the zds1/2 mutant simply promotes Scc1 (and Net1) dephosphorylation, leading to the slower dot disappearance we observed and enhanced Cdc14 release (Queralt and Uhlmann, 2008). We believe the direct linkage model is more likely, based on the previously observed physical interaction between Zds1 and separase (Queralt and Uhlmann, 2008). Also, separase overexpression cannot trigger the release of Cdc14 in the absence of the Zds proteins, suggesting that they are necessary for separase induced Cdc14 release (Queralt and Uhlmann, 2008). Further biochemical studies will be required to clarify the regulatory connections between separase and the Zds proteins. In 1 any case, the inhibitory effects of ZDS gene deletion on cohesin cleavage clearly support our model that PP2ACdc55 counteracts polo phosphorylation of Scc1.
The specificity of the regulatory PP2A subunits towards the Scc1 and Rec8 cohesin subunits is striking. These two regulatory subunits have very distinct folds; while Rts1/B’ has a HEAT-repeat elongated superhelical structure (Cho and Xu, 2007; Xu et al., 2006), Cdc55/B is a 7-bladed WD40 propeller (Xu et al., 2008). Interestingly, substrate specificity appears to be conferred, at least in part, after the regulatory subunit is recruited to the phosphatase, as demonstrated by our fusion proteins. This suggests a role for the regulatory subunit at the level of substrate orientation or catalysis. Indeed, the Rts1/B’ regulatory subunit makes direct interactions with three surface features of the catalytic subunit in close proximity to the active site (Cho and Xu, 2007; Xu et al., 2006), while Cdc55/B makes a single interaction (Xu et al., 2008).
Our study provides a detailed description of the process of cohesin cleavage. We observed an abrupt onset of separase activation, followed by a constant rate of cohesin cleavage that was completed in approximately one minute. Separase biosensors near the centromere and the telomere were cleaved with similar timing and rates. Interestingly, the cell appears to contain small amounts of separase (under 50 molecules per cell (Ghaemmaghami et al., 2003), and it is remarkable that this small enzyme population can process all of the established cohesin along the sister chromatids (likely several hundred complexes per cell), as well as cleave at least one additional substrate (Slk19) and control Cdc14 release, in such a synchronous and abrupt manner. It is also intriguing that a prolonged securin degradation process culminates in a relatively sharp activation of separase and cleavage of cohesin. We propose that separase-mediated regulation of 2 PP2ACdc55 activity towards Scc1 contributes to the observed abruptness of cohesin cleavage. We also suspect that future work will lead to the discovery of additional regulatory interactions that enhance the robust and switch-like properties of this fundamental process.
Supplementary Material
01
02
03
04
05
Highlights.
- Cohesin is cleaved abruptly in one minute, just before sister-chromatid separation.
- Cohesin is cleaved near centromeres and telomeres with the same rate and timing.
- PP2ACdc55 counteracts polo-phosphorylation of cohesin to inhibit cleavage.
- Separase inhibition of PP2ACdc55 may promote cleavage via a feedforward loop.
Acknowledgements
We thank the members of the Morgan lab for helpful discussions and comments on the manuscript, and F. Uhlmann, S. Gasser and J. Taunton for reagents. GY performed all the experiments with guidance from DOM; KT developed the software for quantitative analysis of microscopy. This work was supported by a postdoctoral fellowship 119209 from the American Cancer Society (GY) and a grant (GM094173) from the National 8 Institute of General Medical Sciences (DOM). Microscopy data for this study were acquired at the Nikon Imaging Center at UCSF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal R, Cohen-Fix O. Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes Dev. 2002;16:1371–1382. doi: 10.1101/gad.971402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S cerevisiae. Curr Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- Cho US, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–57. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- Clift D, Bizzari F, Marston AL. Shugoshin prevents cohesin cleavage by PP2A(Cdc55)-dependent inhibition of separase. Genes Dev. 2009;23:766–780. doi: 10.1101/gad.507509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Lowe J. Structure and stability of cohesin's Smc1-kleisin interaction. Mol Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Hagting A, Den Elzen N, Vodermaier HC, Waizenegger IC, Peters JM, Pines J. Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC/C switches from activation by Cdc20 to Cdh1. J Cell Biol. 2002;157:1125–1137. doi: 10.1083/jcb.200111001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Unal E, Guacci V, Koshland D. The kleisin subunit of cohesin dictates damage-induced cohesion. Mol Cell. 2008;31:47–56. doi: 10.1016/j.molcel.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Higuchi T, Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Krutchinsky AN, Morgan DO. Positive feedback sharpens the anaphase switch. Nature. 2008;454:353–357. doi: 10.1038/nature07050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig NC, Knowles PP, McDonald NQ, Uhlmann F. The dual mechanism of separase regulation by securin. Curr Biol. 2002;12:973–982. doi: 10.1016/s0960-9822(02)00847-3. [DOI] [PubMed] [Google Scholar]
- Hornig NC, Uhlmann F. Preferential cleavage of chromatin-bound cohesin after targeted phosphorylation by Polo-like kinase. EMBO J. 2004;23:3144–3153. doi: 10.1038/sj.emboj.7600303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro T, Tanaka K, Sakuno T, Watanabe Y. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol. 2010;12:500–506. doi: 10.1038/ncb2052. [DOI] [PubMed] [Google Scholar]
- Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell. 2010;18:397–409. doi: 10.1016/j.devcel.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A, Roostalu J, Roque H, Antony C, Schiebel E. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev Cell. 2009;17:244–256. doi: 10.1016/j.devcel.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- Mangan S, Zaslaver A, Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J Mol Biol. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- Oliveira RA, Hamilton RS, Pauli A, Davis I, Nasmyth K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat Cell Biol. 2010;12:185–192. doi: 10.1038/ncb2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DH, Hickson GR, O'Farrell PH. Cyclin B destruction triggers changes in kinetochore behavior essential for successful anaphase. Curr Biol. 2003;13:647–653. doi: 10.1016/s0960-9822(03)00242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Queralt E, Lehane C, Novak B, Uhlmann F. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell. 2006;125:719–732. doi: 10.1016/j.cell.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Queralt E, Uhlmann F. Separase cooperates with Zds1 and Zds2 to activate Cdc14 phosphatase in early anaphase. J Cell Biol. 2008;182:873–883. doi: 10.1083/jcb.200801054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- Rohner S, Gasser SM, Meister P. Modules for cloning-free chromatin tagging in Saccharomyces cerevisae. Yeast. 2008;25:235–239. doi: 10.1002/yea.1580. [DOI] [PubMed] [Google Scholar]
- Rossio V, Yoshida S. Spatial regulation of Cdc55-PP2A by Zds1/Zds2 controls mitotic entry and mitotic exit in budding yeast. J Cell Biol. 2011;193:445–454. doi: 10.1083/jcb.201101134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Sun Y, Kucej M, Fan HY, Yu H, Sun QY, Zou H. Separase is recruited to mitotic chromosomes to dissolve sister chromatid cohesion in a DNA-dependent manner. Cell. 2009;137:123–132. doi: 10.1016/j.cell.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–R295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Wicky S, Tjandra H, Schieltz D, Yates J, 3rd, Kellogg DR. The Zds proteins control entry into mitosis and target protein phosphatase 2A to the Cdc25 phosphatase. Mol Biol Cell. 2010;22:20–32. doi: 10.1091/mbc.E10-06-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury EL, Morgan DO. Cdk and APC activities limit the spindlestabilizing function of Fin1 to anaphase. Nat Cell Biol. 2007;9:106–112. doi: 10.1038/ncb1523. [DOI] [PubMed] [Google Scholar]
- Xu Y, Chen Y, Zhang P, Jeffrey PD, Shi Y. Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation. Mol Cell. 2008;31:873–885. doi: 10.1016/j.molcel.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xing Y, Chen Y, Chao Y, Lin Z, Fan E, Yu JW, Strack S, Jeffrey PD, Shi Y. Structure of the protein phosphatase 2A holoenzyme. Cell. 2006;127:1239–1251. doi: 10.1016/j.cell.2006.11.033. [DOI] [PubMed] [Google Scholar]
- Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05