Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1 (original) (raw)
Abstract
Activation-induced cytidine deaminase (AID) is a molecule central to initiating class switch recombination, somatic hypermutation, and gene conversion of Ig genes. However, its mechanism to initiate these genetic alterations is still unclear. AID can convert cytosine to uracil on either mRNA or DNA and is involved in DNA cleavage. Although these events are expected to take place in the nucleus, overexpressed AID was found predominantly in the cytoplasm. Here, we demonstrated that AID is a nucleocytoplasmic shuttling protein with a bipartite nuclear localization signal and a nuclear export signal in its N and C termini, respectively. In addition to previously identified genetic, structural, and biochemical similarities of AID with apolipoprotein B mRNA editing catalytic polypeptide 1, an RNA editing enzyme of ApoB100 mRNA, the present finding provides another aspect to their resemblance, suggesting that both may have homologous reaction mechanisms.
The immune system has evolved specific mechanisms to defend against numerous pathogens using a limited arsenal of Ig genes. After the formation of the primary Ig repertoire by V(D)J recombination of Ig genes during the developmental process, further diversification is achieved in antigen-experienced mature IgM+ B cells by three types of genetic alterations, i.e., somatic hypermutation (SHM), gene conversion (GC), and class switch recombination (CSR). SHM and GC introduce a large number of non-templated and templated point mutations, respectively, in the Ig V region genes to raise high-affinity antibodies after selection with a limited amount of antigen. CSR takes place between two S regions that locate 5′ adjacent to each Ig heavy chain constant (CH) region gene, resulting in replacement of the most upstream Cμ gene with another downstream CH (Cγ, Cε, or Cα) gene. B cells can thus generate isotypes other than IgM, such as IgG, IgE, and IgA, without changing antigen specificity (1).
Activation-induced cytidine deaminase (AID) is expressed almost exclusively in activated B cells (2). Disruption of the AID gene in mouse and human causes the hyper-IgM phenotype by abolishing both SHM and CSR without any other signs of lymphocyte dysfunction (3, 4). Furthermore, knockout of AID in chicken B cell line DT40 also abolishes GC, which is spontaneously taking place in this cell line (5, 6). Inversely, ectopic expression of AID in non-B cells induces CSR and SHM (7–10). These results indicate that AID is a molecule central to initiating CSR, SHM, and GC, the three types of Ig gene alterations that occur in mature B lymphocytes.
Although AID is required for DNA cleavage (11), the detailed mechanism by which AID induces these genetic events is still under extensive debate. AID has the highest sequence homology with the apolipoprotein B (apoB) mRNA editing catalytic polypeptide 1 (APOBEC1), which edits a specific cytidine on mRNA of apoB100, a cholesterol carrier, converting it to mRNA of apoB 48, a triglyceride carrier. AID, like APOBEC1, conserves the catalytic motif of cytosine deaminase and has cytidine deaminase activity on cytidine (2). These structural and functional similarities between AID and APOBEC1 led to the hypothesis that AID is an RNA editing enzyme that generates mRNA for a recombinase and mutator. However, the finding of DNA deaminase activity of AID led to the proposal that AID deaminates cytosine (C) on a target DNA to generate a uracil (U)–guanine(G) base pair that triggers base excision repair activities, thereby causing DNA cleavage and subsequent DNA alterations (12–18). The additional supporting observation for this hypothesis is that uracil DNA glycosylase deficiency affects SHM base specificity and CSR efficiency (19, 20), which suggests involvement of U at some stage during the reaction.
Sequestering a protein from its target in a different intracellular compartment is one of the general strategies to regulate cellular activities that require interaction of many proteins. APOBEC1 was recently shown to shuttle between the nucleus and cytoplasm by association with importin at the N-terminal nuclear localization signal (NLS) and the nuclear export machinery at the C-terminal nuclear export signal (NES) of APOBEC1 (21). APOBEC1 associates with and carries APOBEC1 complementation factor (ACF), which recognizes apoB100 mRNA. ADAR2, the RNA editing adenosine deaminase that acts on glutamate receptor pre-mRNA, accumulates in the nucleolus but can shuttle to the nucleoplasm, where it edits the target pre-mRNA, suggesting that the ADAR2 activity can be modulated by functional sequestration from its substrate (22).
The intracellular localization of endogenous AID has not been extensively analyzed, because presently available reagents cannot visualize the endogenous AID protein by immunostaining. AID tagged by GFP revealed its predominant cytoplasmic distribution and raised the question of how cytoplasmic AID can reach its substrate in the nucleus, whether DNA or RNA (23). Here we report that AID is a nucleocytoplasmic shuttling protein with NLS and NES in its N and C termini, respectively. The finding adds another cellular biological similarity between AID and APOBEC1, suggesting their functional homology.
Materials and Methods
Retrovirus Constructs for GFP Fusion Proteins. The _Xho_I-_Not_I fragment from pEGFP-N1 (Clontech) was cloned into the _Sal_I-_Not_I site of retrovirus expression vector pFB (Stratagene), designated pFB-GFP. Human AID (hAID) and its mutants were amplified by PCR with use of Pyrobest, a high-fidelity DNA polymerase, and cloned into _Eco_RI/_Bam_HI sites of pFB-GFP. Six amino acid residues between the AID and GFP sequences, originated from a residual multiple cloning sites of pEGFP-N1, are DPPVAT. Deletion mutants were generated by PCR with internal primers that anneal where the truncations were introduced.
Cell Culture and Virus Infection. NIH 3T3 cells were maintained in DMEM (Sigma) containing 10% FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Splenocytes from AID–/– mice (3) were cultured for 2 days in RPMI medium 1640 containing 10% FCS, 50 μM 2-mercaptoethanol, 2 mM l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, 50 μg/ml lipopolysaccharide (Sigma), and 15 ng/ml mouse IL-4 (PeproTech, Boston). Splenocytes were preactivated for 2 days before infection. Cells were infected with pFB-hAID-GFP or mutant AID-GFP as described (24).
Immunofluorescence Microscopy. Cells were fixed in PLP solution (2% paraformaldehyde/10 mM NaIO4/75 mM lysine/37.5 mM PBS) for 10 min. After permeabilization with 0.5% Triton X-100 in PBS for 5 min, cells were treated with 10 μg/ml DNase-free ribonuclease (Nippon Gene, Toyama, Japan) for 120 min, and nuclei were stained with 1 μg/ml propidium iodide (PI; Sigma) for 10 min. In some experiments, cells were treated with 10 ng/ml leptomycin B (LMB), as described in the figure legends, before the fixation. LMB was provided from Dr. M. Yoshida (RIKEN Discovery Research Institute, Wako, Japan). Slides were treated with the SlowFade Antifade kit (Molecular Probes) and were analyzed under an Axiophot 2 universal microscope (Zeiss). The images were captured and processed by using an MRC-1024 laser scanning confocal imaging system (Bio-Rad).
Flow Cytometry. Cells were stained with biotinylated anti-mouse IgG1 (Pharmingen) and allophycocyanin–streptavidin (Vector). Cells were analyzed for GFP and surface IgG1 expression by FACSCalibur with cellquest software (Becton Dickinson). Dead cells were excluded from the analysis by forward scatter, side scatter, and PI gating.
Hypermutation Assay. As an artificial SHM target, a Dsred2 (Clontech) expression cassette under the control of tetracycline (tet) inducible promoter was introduced into NIH 3T3 cells with a tet-off transactivator gene (8). Dsred2-expressing cells were enriched up to 95% by G418 selection and kept with tet thereafter. wt or mutant hAID-GFP was introduced by retrovirus vector to cells, and 4 days after infection tet was removed from the culture medium. Thirteen days after tet removal, GFP+ cells were isolated by cell sorting with FACSVantage (Becton Dickinson) for DNA extraction. The purity of GFP+ cells after the sorting was 88–95%. Dsred2 coding sequence was amplified by Pyrobest and cloned into pBSKS vector. Because of the GC-rich composition of the 5′ region, only a part of the coding sequence (base number 185–660 relative to the translation start site) was determined and analyzed.
Results
AID Is a Nucleocytoplasmic Shuttling Protein. To determine the subcellular localization of AID in living cells, we have generated an expression construct for hAID tagged with GFP at the C terminus (Fig. 1). The construct was introduced into NIH 3T3 cells by a retrovirus system because this cell line is competent for both CSR and SHM when AID and appropriate artificial substrates were introduced together (7, 8). In agreement with the result in a human lymphoma cell line (23), hAID-GFP was localized primarily in the cytoplasm (Fig. 1 A Left).
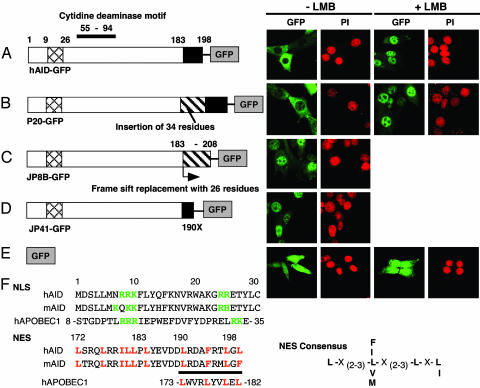
Fig. 1.
Subcellular localization of GFP-tagged AID and natural mutants. Expression vector for each GFP fusion of hAID wt (A), P20 (B), JP8B (C), JP41 (D), and GFP alone (E) was introduced by retrovirus system into NIH 3T3 cells. Confocal images of GFP signals with or without LMB treatment (10 ng/ml for 150 min) are shown together with nuclear staining with PI of the same field. The structure of each mutant is schematically illustrated (Left). Black boxes, NES; crosshatched boxes, NLS; hatched boxes, amino acid stretches caused by insertion or frame-shift. The 34- and 26-aa stretches inserted or replaced in P20 and JP8B are VTKPSTQFRRLSGPTDPQPRFEAIHSICFSLSLR and CMRLMTYETHFVLWDFDFDSNFQECHTR, respectively. (F) Amino acid sequences around NLS and NES regions of hAID, murine AID, and human APOBEC1 and NES consensus sequence are shown. Critical basic residues in NLS and conserved residues (L and F) in NES candidate are colored, and NES is underlined. Cytidine deaminase motif is indicated by a bar at the top of A. Numbers indicate residue positions.
The cytoplasmic localization may represent either dynamic but unbalanced shuttling from the nucleus or static inability in entering the nucleus. To distinguish these two cases, we treated the cells with LMB, which inhibits exportin1-dependent nuclear export (25). By this treatment, hAID-GFP changed its subcellular localization from the cytoplasm to the nucleus. After a 150-min incubation with LMB, the major GFP signal accumulated in the nucleus and only little remained in the cytoplasm (Fig. 1 A Right). The quick translocation of hAID-GFP strongly suggests that AID shuttles between the nucleus and cytoplasm and that its nuclear export pathway depends on exportin1. GFP alone, however, distributed almost evenly in the nucleus and cytoplasm, and its profile was not sensitive to LMB (Fig. 1_E_).
AID Has NES in the C-Terminal Domain. We have shown that mutant AIDs from hyper-IgM syndrome 2 patients are almost completely defective in CSR, but three mutants (P20, JP41, and JP8B) that have replaced or truncated C-terminal 17 residues retain the SHM activity (26). To examine whether these mutations have any influence on the shuttling ability of AID, GFP fusion proteins of these mutants were also introduced into NIH 3T3 cells. The GFP fusion protein of P20, which has a 34-aa insertion at residue 182 and is inactive for CSR but active for SHM (26), showed LMB-dependent nuclear accumulation, essentially the same as wt AID (Fig. 1_B_). Interestingly, JP8B carrying a frame-shift mutation at residue 183, which is almost the same position as the P20 mutation, seemed to lose the shuttling ability and accumulated in the nucleus spontaneously (Fig. 1_C_). Because an artificial truncation at residue 183 (183X), namely JP8Bdel, showed the same pattern as JP8B, the spontaneous accumulation of JP8B in the nucleus is most likely due to the lack of the C-terminal 16 residues, and not to the addition of missensed 26 amino acid residues by the frame-shift (Fig. 2_A_). Another C-terminal deletion mutant, JP41 (190X)-GFP, also accumulated in the nucleus spontaneously (Fig. 1_D_).
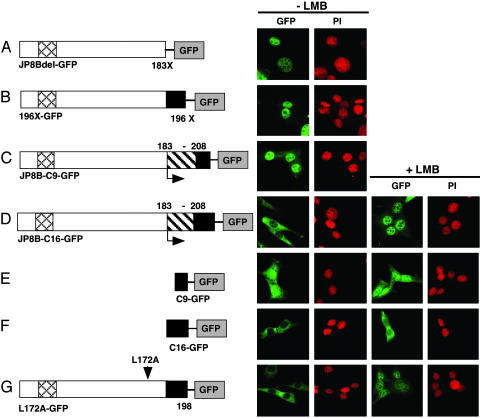
Fig. 2.
NES activity of the C-terminal 16 amino acid residues of hAID. Confocal images of GFP signals with or without LMB (10 ng/ml for 150 min) are shown together with PI staining of the same field: GFP fusion proteins of JP8Bdel (A), 196X (B), JP8B-C9 (C), JP8B-C16 (D), C9 (F), C16 (E), and L172A (G). C16 and C9 indicate C-terminal peptides of 16 and 9 amino acid residues, respectively. The structure of each mutant is shown (Left). Arrowhead, point mutation; horizontal arrow, replacement; X, termination; black box, NES; crosshatched box, NLS.
Exportin1 recognizes a leucine-rich NES on a target protein and exports it from the nucleus (27). The localization profiles of hyper-IgM syndrome 2 AID mutants indicate the presence of a NES in the C-terminal region of AID. We checked the amino acid composition of this region to see whether it matches the well documented NES motif. We found repeats of hydrophobic residues (leucine, valine, and phenylalanine) in every third to fourth interval with LXL (or LXF in mice) at the C-terminal end, which appeared to fit the NES motif (Fig. 1_F_) (28). To confirm the existence of NES, we constructed artificial mutants of the C-terminal domain. The deletion of C-terminal three and six residues (196X and 193X, respectively) resulted in the nuclear dominant pattern of GFP signal (Fig. 2_B_ and data not shown), suggesting the three residues (LXL) at the C terminus of AID are the essential part of the NES. We then examined whether the C-terminal region is sufficient for the nuclear export. Two strategies were taken: addition of the C-terminal 16 residues back to the JP8B or to the N terminus of GFP. The addition of the 16 residues restored cytoplasmic localization to both JP8B (JP8B-C16) and GFP (C16-GFP), whereas the C-terminal nine residues did not (Fig. 2 C_–_F). The restored cytoplasmic proteins responded to LMB, although the degree of nuclear accumulation was much higher for JP8B-C16 than for C16-GFP.
There is another leucine-rich NES candidate at residues 172–183 (Fig. 1_F_). Despite the marked similarity to the NES consensus, point mutations of these leucines did not give any indications of the NES activity (Fig. 2_G_ and data not shown). Taking all of these observations together, we conclude that AID has a functional NES within the C-terminal 16 residues 183–198.
AID Has a Bipartite NLS in the N-Terminal Domain. AID was shown to shuttle between the cytoplasm and nucleus, but how does it enter the nucleus? As described above, JP8B-C16 responded to LMB much more quickly and completely than did C16-GFP. Because GFP itself has no specific localization signal, it is more likely that JP8B has a specific signal for active import to the nucleus. From an alignment with other APOBEC1-related sequences (21, 29), it appeared that AID has a putative bipartite NLS, two clusters of basic residues, at residues 8–25 (Fig. 1_F_). To analyze the putative NLS activity, we constructed N-terminal region mutants of AID tagged with GFP. A mutant with deletion of residues 2–26, ΔN26hAID-GFP lost the capacity to enter the nucleus, as shown by blocking nuclear export with the LMB treatment (Fig. 3_A_). The result is consistent with the notion that the putative NLS region is indeed functional.
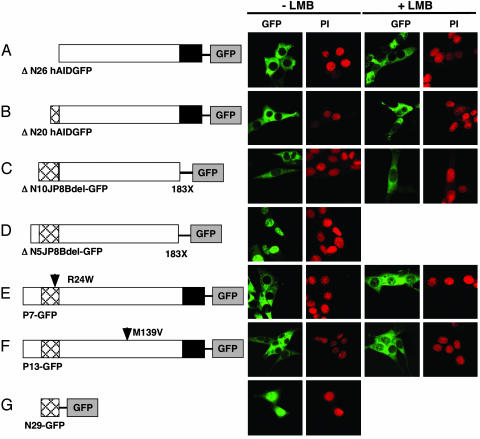
Fig. 3.
N-terminal NLS activity of hAID. Subcellular localizations of N-terminal truncation mutants ΔN26hAID (A), ΔN20hAID (B), ΔN10JP8Bdel (C), and ΔN5JP8Bdel (D) and point mutated natural mutants P7 (E) and P13 (F) as well as GFP protein fused with the N-terminal peptides (G) are shown with PI staining. The structure of each mutant is shown (Left). Arrowhead, point mutation; X, termination; black box, NES; crosshatched box, NLS. LMB treatments were done at 10 ng/ml for 150 min.
NLS activity is not restored by adding back the N-terminal 6 or 16 residues, as shown with ΔN20hA ID-GFP and ΔN10JP8Bdel-GFP (Fig. 3 B and C). On the other hand, a mutant with deletion of residues 2–5, ΔN5-JP8Bdel-GFP, is transported into the nucleus (Fig. 3_D_). Consistently, a point mutation at residue arginine 24, namely P7, showed impaired nuclear transport (Fig. 1_E_). P13 with one point mutation at residue methionine 139 also appeared to be LMB-insensitive. Both P7 and P13 are functionally null mutants for both CSR and SHM (26). However, P7 has a significant deaminase activity in Escherichia coli, whereas P13 has no such activity, although its deaminase motif is intact (data not shown), suggesting that protein conformational change is associated with the P13 mutation. To examine whether the bipartite NLS of AID is active for an unrelated protein, we added the N-terminal 29 residues of AID to the N terminus of GFP. The nuclear GFP level was increased by the addition of NLS (Fig. 3_G_ vs. Fig. 1_E_). These results indicate that AID has a functional bipartite NLS in the N terminus, which works efficiently in a correctly folded AID.
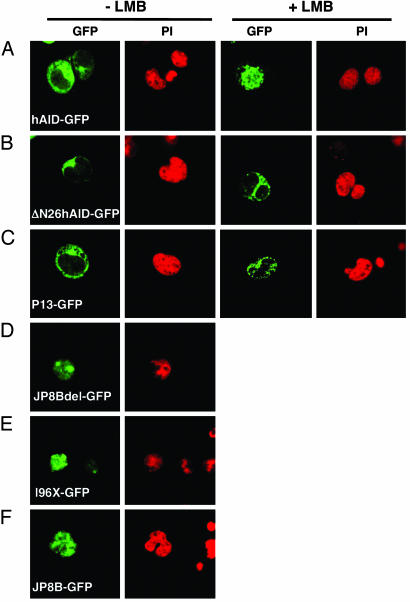
Nucleocytoplasmic Shuttling of AID in B Lymphocytes. To examine whether the subcellular localization of AID is regulated in the same manner in B lymphocytes as in fibroblasts, we infected AID–/– spleen B lymphocytes with wt and mutant hAID-GFP-expressing retroviruses. wt hAID-GFP was localized in the cytoplasm and translocated into the nucleus upon the LMB treatment, whereas ΔN26hAID-GFP and P13-GFP were localized in the cytoplasm regardless of the LMB treatment (Fig. 4 A–C). Mutants lacking the C-terminal NES (JP8Bdel, 196X, and JP8B) accumulated in the nucleus without LMB (Fig. 4 D–F). Therefore, we conclude that the localization of AID is regulated similarly in B lymphocytes and in fibroblasts.
Fig. 4.
B lymphocytes show intracellular AID-GFP patterns similar to those of fibroblasts. Selected wt (A) and mutant AID-GFP representative for normal, disturbed import (B and C) or export (D_–_F) were introduced by retrovirus vector to preactivated AID–/– B lymphocytes. Photos were taken 3 days after infection. LMB treatments were done at 10 ng/ml for 150 min.
Relevance of the Localization Signals to SHM and CSR. Analyses of P20, JP41, and JP8B mutants have shown that the C-terminal 17 amino acid residues of AID are critical for CSR but not for SHM and GC (26, 30). Because this C-terminal region well overlaps with the NES, we examined SHM activity of GFP fusion of NES deleted mutants, for which we used NIH 3T3 cells carrying a Dsred2 reporter gene driven by the tet inducible promoter. When AID is expressed in NIH 3T3, SHM accumulates on the actively transcribed reporter gene (8). As shown in the Table 1, all GFP fusion proteins with NES deletion exhibited SHM activity with higher frequencies than wt AID. In addition, it is indicated that the N-terminal five residues are dispensable for SHM because ΔN5-JP8Bdel-GFP is active in SHM. The result suggests that the efficient export from the nucleus is not critical for induction of SHM.
Table 1. Mutation frequency induced by mutant AID-GFP fusion protein.
| Clone mutated/total | Mutation (del. or ins.) | Total bases | Frequency per 104 | P values versus* | ||
|---|---|---|---|---|---|---|
| Sequence | None | hAID | ||||
| 196X | 7/10 | 16 (1) | 4,760 | 33.6 | <0.001 | <0.001 |
| 193X | 5/10 | 12 (0) | 4,760 | 25.2 | <0.001 | 0.006 |
| JP8Bdel | 8/10 | 20 (3) | 4,730 | 42.2 | <0.001 | <0.001 |
| ΔN5JP8Bdel | 7/10 | 20 (1) | 4,714 | 42.4 | <0.001 | <0.001 |
| hAID | 7/33 | 12 (2) | 15,679 | 7.7 | 0.005 | |
| None | 1/27 | 1 (0) | 12,851 | 0.8 |
To determine whether the most C-terminal three residues (which are essential residues as NES but are dispensable for SHM) are relevant to CSR activity, we introduced 196X-GFP into lipopolysaccharide- and IL-4 stimulated spleen B cells of AID–/– mice (3). On day 3, CSR efficiency in 196X-GFP-infected cells was slightly higher than background (0.35% IgG+) but very low (3–6% IgG+) compared with wtAID-GFP (38–47% IgG+). We conclude that the three residues at the C terminus are critical for effective induction of CSR.
Because it is likely that AID edits its target (RNA or DNA) in the nucleus (29), the defect of NLS may result in the total loss of the AID activity. In fact, a point mutation at residue arginine 24, namely P7, causes a complete loss of nuclear localization and function (26). Similarly, the NLS deletion mutants (Δ2–10, Δ11–20, and Δ2–20) of mouse AID completely lost their CSR and SHM activities together with nuclear localization activity (unpublished data). We conclude that the NLS region is critical for the AID function for CSR and SHM.
Discussion
AID is thought to deaminate C on either RNA or DNA to initiate the Ig gene alteration reactions (1, 18). Because not only DNA editing but also RNA editing are thought to take place in the nucleus (29), AID is likely to function in the nucleus. In support of this view, AID fused with the hormone binding domain of the human estrogen receptor (AID-ER) quickly migrates to the nuclear fraction in concomitance with CSR induction after addition of the estrogen analogue that activates AID-ER by dissociating it from hsp90 (24). However, the AID-GFP fusion protein was previously demonstrated to localize in the cytoplasm (23). Here we have shown that AID has NLS and NES and can thereby dynamically shuttle between the nucleus and cytoplasm. The predominant cytoplasmic localization pattern of AID-GFP represents the biased balance to the export from the nucleus, because the blockade of a nuclear export pathway evokes its rapid and robust accumulation in the nucleus. The predominance of the nuclear export does not seem to be specific to the cell type, because the fibroblasts, B lymphocytes, and human embryonic kidney cell line showed essentially the same pattern (Figs. 1 A and 4_A_ and data not shown). The nuclear localization of AID appears to be essential to its function, because all of the NLS mutations tested so far lost their activities for CSR and SHM in mammalian cells.
The role of NES signal may be 2-fold. First, the amount of AID in the nucleus at the steady state must be tightly controlled to maintain a minimal level, because excessive amounts of nuclear AID may induce unregulated SHM not only in Ig genes (Table 1) but also in non-Ig genes (31). Second, according to the RNA editing hypothesis, edited mRNA in the AID–cofactor complex should be transported to the cytoplasm for translation. In case of APOBEC1, the majority of edited ApoB100 mRNA are exported to the cytoplasm as a complex associated with APOBEC1 and ACF to avoid non-sense mediated decay (21).
Several possibilities can be raised to explain why NES mutants showed higher mutator activities on the artificial substrate. One may speculate that AID requires specific cofactors not only for CSR but also for SHM. Cofactors for CSR and SHM may compete for association with the AID molecule. Because association of the CSR specific cofactor requires the C-terminal 17 residues of AID (26, 30), which overlaps with NES, the C-terminal NES mutants can associate only with the SHM cofactor, giving rise to enhanced SHM activities. Because P20 lost CSR activity specifically despite normal nuclear export, the presence of NES is not sufficient for CSR activity, indicating that interaction of AID with the CSR-specific cofactor may require a larger region than NES. Another possibility, which does not exclude the possiblity mentioned above, is that longer retention of AID in the nucleus may increase the chance to meet its substrate, either pre-mRNA or DNA.
Like AID, APOBEC1 shuttles between the nucleus and cytoplasm and has both bipartite NLS and NES (21). Overexpressed APOBEC1 localizes in the nucleus and cytoplasm, and the preference is variable among cell types (21, 32, 33). ACF expressed alone localizes almost exclusively in the cytoplasm (21), but ACF is imported into the nucleus when coexpressed with APOBEC1 in a single cell. The NES activity of APOBEC1 is also sensitive to LMB. These observations suggest that a complex formation of APOBEC1 and ACF is essential not only for target recognition but also for modulation of the intracellular localization. Moreover, glycerol gradient sedimentation analysis revealed that the active nuclear editing complex of APOBEC1 and ACF differs from that of the inactive cytoplasmic complex (34).
AID and APOBEC1 are known to have genetic and structural homology that indicates their evolutionary relationship, namely chromosomal locus proximity and conservation of essential motifs (2, 29, 35). Both AID and APOBEC1 are known to function as a complex containing the homodimer and another protein for recognition of target (26, 34, 36, 37). apoB mRNA edited by APOBEC1 has to be translated to manifest its physiological function. Likewise, DNA cleavage activity of AID depends on de novo protein synthesis (ref. 24 and unpublished data). The present cellular biological findings that both APOBEC1 and AID are nuclear-cytoplasmic shuttling proteins containing NLS and NES at their N- and C-terminal regions, respectively, strengthens their similarity. Such extensive similarities between AID and APOBEC1 favor the hypothesis that they share functional homology, i.e., RNA editing.
Acknowledgments
We thank Dr. M. Yoshida for providing LMB, Ms. Y. Sasaki for excellent technical assistance, Drs. S. Fagarasan and K. Kinoshita for critical reading of the manuscript, and Ms. K. Saito for preparation of the manuscript. This work was supported by a Center of Excellence Grant from the Ministry of Education, Science, Sports, and Culture of Japan.
Abbreviations: AID, activation-induced cytidine deaminase; wt, wild type; SHM, somatic hypermutation; CSR, class switch recombination; APOBEC1, apolipoprotein B mRNA editing catalytic polypeptide 1; GC, gene conversion; NES, nuclear export signal; NLS, nuclear localization signal; ACF, APOBEC1 complementation factor; hAID, human AID; LMB, leptomycin B; PI, propidium iodide; tet, tetracycline.
References
- 1.Honjo, T., Kinoshita, K. & Muramatsu, M. (2002) Annu. Rev. Immunol. 20**,** 165–196. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu, M., Sankaranand, V. S., Anant, S., Sugai, M., Kinoshita, K., Davidson, N. O. & Honjo, T. (1999) J. Biol. Chem. 274**,** 18470–18476. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu, M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y. & Honjo, T. (2000) Cell 102**,** 553–563. [DOI] [PubMed] [Google Scholar]
- 4.Revy, P., Muto, T., Levy, Y., Geissmann, F., Plebani, A., Sanal, O., Catalan, N., Forveille, M., Dufourcq-Labelouse, R., Gennery, A., et al. (2000) Cell 102**,** 565–575. [DOI] [PubMed] [Google Scholar]
- 5.Arakawa, H., Hauschild, J. & Buerstedde, J. M. (2002) Science 295**,** 1301–1306. [DOI] [PubMed] [Google Scholar]
- 6.Harris, R. S., Sale, J. E., Petersen-Mahrt, S. K. & Neuberger, M. S. (2002) Curr. Biol. 12**,** 435–438. [DOI] [PubMed] [Google Scholar]
- 7.Okazaki, I. M., Kinoshita, K., Muramatsu, M., Yoshikawa, K. & Honjo, T. (2002) Nature 416**,** 340–345. [DOI] [PubMed] [Google Scholar]
- 8.Yoshikawa, K., Okazaki, I. M., Eto, T., Kinoshita, K., Muramatsu, M., Nagaoka, H. & Honjo, T. (2002) Science 296**,** 2033–2036. [DOI] [PubMed] [Google Scholar]
- 9.Martin, A. & Scharff, M. D. (2002) Proc. Natl. Acad. Sci. USA 99**,** 12304–12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin, A., Bardwell, P. D., Woo, C. J., Fan, M., Shulman, M. J. & Scharff, M. D. (2002) Nature 415**,** 802–806. [DOI] [PubMed] [Google Scholar]
- 11.Petersen, S., Casellas, R., Reina-San-Martin, B., Chen, H. T., Difilippantonio, M. J., Wilson, P. C., Hanitsch, L., Celeste, A., Muramatsu, M., Pilch, D. R., et al. (2001) Nature 414**,** 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickerson, S. K., Market, E., Besmer, E. & Papavasiliou, F. N. (2003) J. Exp. Med. 197**,** 1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen-Mahrt, S. K. & Neuberger, M. S. (2003) J. Biol. Chem. 278**,** 19583–19586. [DOI] [PubMed] [Google Scholar]
- 14.Ramiro, A. R., Stavropoulos, P., Jankovic, M. & Nussenzweig, M. C. (2003) Nat. Immunol. 4**,** 452–456. [DOI] [PubMed] [Google Scholar]
- 15.Bransteitter, R., Pham, P., Scharff, M. D. & Goodman, M. F. (2003) Proc. Natl. Acad. Sci. USA 100**,** 4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhuri, J., Tian, M., Khuong, C., Chua, K., Pinaud, E. & Alt, F. W. (2003) Nature 422**,** 726–730. [DOI] [PubMed] [Google Scholar]
- 17.Sohail, A., Klapacz, J., Samaranayake, M., Ullah, A. & Bhagwat, A. S. (2003) Nucleic Acids Res. 31**,** 2990–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersen-Mahrt, S. K., Harris, R. S. & Neuberger, M. S. (2002) Nature 418**,** 99–103. [DOI] [PubMed] [Google Scholar]
- 19.Rada, C., Williams, G. T., Nilsen, H., Barnes, D. E., Lindahl, T. & Neuberger, M. S. (2002) Curr. Biol. 12**,** 1748–1755. [DOI] [PubMed] [Google Scholar]
- 20.Di Noia, J. & Neuberger, M. S. (2002) Nature 419**,** 43–48. [DOI] [PubMed] [Google Scholar]
- 21.Chester, A., Somasekaram, A., Tzimina, M., Jarmuz, A., Gisbourne, J., O'Keefe, R., Scott, J. & Navaratnam, N. (2003) EMBO J. 22**,** 3971–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sansam, C. L., Wells, K. S. & Emeson, R. B. (2003) Proc. Natl. Acad. Sci. USA 100**,** 14018–14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rada, C., Jarvis, J. M. & Milstein, C. (2002) Proc. Natl. Acad. Sci. USA 99**,** 7003–7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doi, T., Kinoshita, K., Ikegawa, M., Muramatsu, M. & Honjo, T. (2003) Proc. Natl. Acad. Sci. USA 100**,** 2634–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishi, K., Yoshida, M., Fujiwara, D., Nishikawa, M., Horinouchi, S. & Beppu, T. (1994) J. Biol. Chem. 269**,** 6320–6324. [PubMed] [Google Scholar]
- 26.Ta, V. T., Nagaoka, H., Catalan, N., Durandy, A., Fischer, A., Imai, K., Nonoyama, S., Tashiro, J., Ikegawa, M., Ito, S., et al. (2003) Nat. Immunol. 4**,** 843–848. [DOI] [PubMed] [Google Scholar]
- 27.Ullman, K. S., Powers, M. A. & Forbes, D. J. (1997) Cell 90**,** 967–970. [DOI] [PubMed] [Google Scholar]
- 28.Bogerd, H. P., Fridell, R. A., Benson, R. E., Hua, J. & Cullen, B. R. (1996) Mol. Cell. Biol. 16**,** 4207–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wedekind, J. E., Dance, G. S., Sowden, M. P. & Smith, H. C. (2003) Trends Genet. 19**,** 207–216. [DOI] [PubMed] [Google Scholar]
- 30.Barreto, V., Reina-San-Martin, B., Ramiro, A. R., McBride, K. M. & Nussenzweig, M. C. (2003) Mol. Cell 12**,** 501–508. [DOI] [PubMed] [Google Scholar]
- 31.Okazaki, I. M., Hiai, H., Kakazu, N., Yamada, S., Muramatsu, M., Kinoshita, K. & Honjo, T. (2003) J. Exp. Med. 197**,** 1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eto, T., Kinoshita, K., Yoshikawa, K., Muramatsu, M. & Honjo, T. (2003) Proc. Natl. Acad. Sci. USA 100**,** 12895–12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, Y. & Smith, H. C. (1997) Proc. Natl. Acad. Sci. USA 94**,** 13075–13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sowden, M. P., Ballatori, N., Jensen, K. L., Reed, L. H. & Smith, H. C. (2002) J. Cell Sci. 115**,** 1027–1039. [DOI] [PubMed] [Google Scholar]
- 35.Muto, T., Muramatsu, M., Taniwaki, M., Kinoshita, K. & Honjo, T. (2000) Genomics 68**,** 85–88. [DOI] [PubMed] [Google Scholar]
- 36.Lau, P. P., Zhu, H. J., Baldini, A., Charnsangavej, C. & Chan, L. (1994) Proc. Natl. Acad. Sci. USA 91**,** 8522–8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehta, A., Kinter, M. T., Sherman, N. E. & Driscoll, D. M. (2000) Mol. Cell. Biol. 20**,** 1846–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]