Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity (original) (raw)
. Author manuscript; available in PMC: 2014 Sep 12.
Abstract
Targeted genome editing technologies have enabled a broad range of research and medical applications. The Cas9 nuclease from the microbial CRISPR-Cas system is targeted to specific genomic loci by a 20-nt guide sequence, which can tolerate certain mismatches to the DNA target and thereby promote undesired off-target mutagenesis. Here, we describe an approach that combines a Cas9 nickase mutant with pairs of guide RNAs to introduce targeted double-strand breaks. Given that individual nicks in the genome are repaired with high fidelity, simultaneous nicking via appropriately offset guide RNAs effectively extends the number of specifically recognized bases in the target site. We demonstrate that paired nicking can be used to reduce off-target activity by 50–1,000 fold in cell lines and facilitate gene knockout in mouse zygotes without sacrificing on-target cleavage efficiency. This versatile strategy thus enables a wide variety of genome editing applications with higher levels of specificity.
Introduction
The ability to perturb the genome in a precise and targeted fashion is crucial for understanding genetic contributions to biological function. Genome engineering of cell lines or animal models has traditionally been accomplished through random mutagenesis or low-efficiency gene targeting. To facilitate genome editing, programmable sequence-specific DNA nuclease technologies have enabled targeted modification of endogenous genomic sequences with high efficiency, particularly in species that have proven genetically intractable (Carlson et al., 2012; Geurts et al., 2009; Takasu et al., 2010; Watanabe et al., 2012). The RNA-guided Cas9 nucleases from the microbial CRISPR (clustered regularly interspaced short palindromic repeat)-Cas systems are robust and versatile tools for stimulating targeted double-stranded DNA breaks (DSBs) in eukaryotic cells (Chang et al., 2013; Cho et al., 2013; Cong et al., 2013; Deltcheva et al., 2011; Deveau et al., 2010; Friedland et al., 2013; Gratz et al., 2013; Horvath and Barrangou, 2010; Jinek et al., 2013; Mali et al., 2013b; Wang et al., 2013), where the resulting cellular repair mechanisms (Hsu and Zhang, 2012; Perez et al., 2008; Urnov et al., 2010) – non-homologous end joining (NHEJ) or homology-directed repair (HDR) pathways – can be exploited to induce error-prone or defined alterations.
The Cas9 nuclease from Streptococcus pyogenes can be directed by a chimeric single guide RNA (sgRNA) (Jinek et al., 2012) to any genomic locus followed by a 5′-NGG protospacer-adjacent motif (PAM). A 20-nt guide sequence within the sgRNA directs Cas9 to the genomic target via Watson-Crick base pairing (Deltcheva et al., 2011; Deveau et al., 2010; Gasiunas et al., 2012; Jinek et al., 2012) and can be easily engineered to target a desired genomic locus. Recent studies of Cas9 specificity have demonstrated that although each base within the 20-nt guide sequence contributes to overall specificity, multiple mismatches between the guide RNA and its complementary target DNA sequence can be tolerated depending on the quantity, position, and base identity of mismatches (Cong et al., 2013; Fu et al., 2013; Hsu et al., 2013; Jiang et al., 2013), leading to potential off-target DSBs and indel formation. These unwanted mutations can potentially limit the utility of Cas9 for genome editing applications that require high levels of precision, such as generation of isogenic cell lines (Soldner et al., 2011) for testing causal genetic variations or in vivo and ex vivo genome editing-based therapies.
To improve the specificity of Cas9-mediated genome editing, we developed a novel strategy that combines the D10A mutant nickase (Cong et al., 2013; Gasiunas et al., 2012; Jinek et al., 2012) version of Cas9 (Cas9n) with a pair of offset sgRNAs complementary to opposite strands of the target site. While nicking of both DNA strands by a pair of Cas9 nickases leads to site-specific DSBs and NHEJ, individual nicks are predominantly repaired by the high-fidelity base excision repair pathway (BER) (Dianov and Hubscher, 2013). In a manner analogous to dimeric zinc finger nucleases (ZFNs) (Miller et al., 2007; Porteus and Baltimore, 2003; Sander et al., 2011; Wood et al., 2011) and transcription activator-like effector nucleases (TALENs) (Boch et al., 2009; Christian et al., 2010; Miller et al., 2011; Moscou and Bogdanove, 2009; Reyon et al., 2012; Sanjana et al., 2012; Wood et al., 2011; Zhang et al., 2011), where DNA cleavage relies upon the synergistic interaction of two independent specificity-encoding DNA-binding modules directing FokI nuclease monomers, this strategy minimizes off-target mutagenesis by each individual Cas9n-sgRNA complex while maintaining on-target modification rates similar to those of wild type Cas9.
Results
Extension of guide sequence does not improve Cas9 targeting specificity
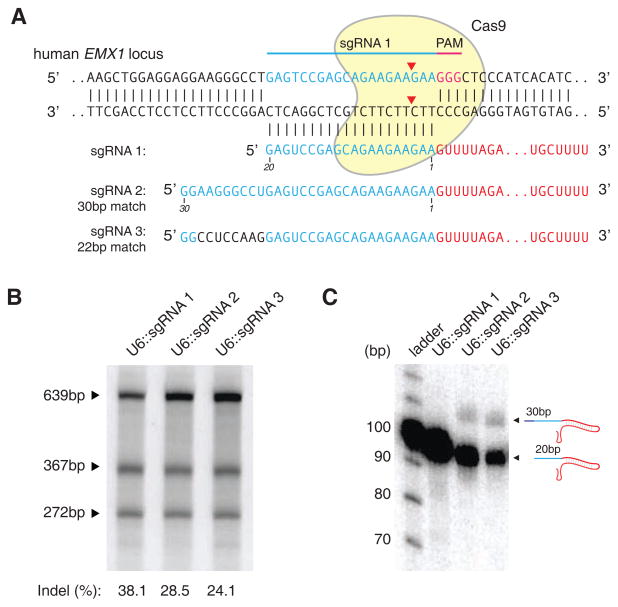
Cas9 targeting is facilitated by base-pairing between the 20-nt guide sequence within the sgRNA and the target DNA (Deltcheva et al., 2011; Deveau et al., 2010; Gasiunas et al., 2012; Jinek et al., 2012). We reasoned that cleavage specificity might be improved by increasing the length of base-pairing between the guide RNA and its target locus. To test this, we generated U6-driven expression cassettes (Hsu et al., 2013) to express three sgRNAs with 20-nt (sgRNA 1) or 30-nt guide sequences (sgRNAs 2 and 3) targeting a locus within the human EMX1 gene (Figure 1A).
Figure 1. Effect of guide sequence extension on Cas9 activity.
(A) Schematic showing Cas9 with matching or mismatching sgRNA sequences targeting the human EMX1 locus. (B) SURVEYOR assay gel showing comparable modification of target 1 by sgRNAs bearing 20- and 30-nt long guide sequences. (C) Northern blot showing that extended sgRNAs are largely reverted to 20-nt guide-length sgRNAs in HEK 293FT cells.
We and others have previously shown that while single-base mismatches between the PAM-distal region of the guide sequence and target DNA are well tolerated by Cas9, multiple mismatches in this region can significantly affect on-target activity (Fu et al., 2013; Hsu et al., 2013; Mali et al., 2013a; Pattanayak et al., 2013). To determine whether additional PAM-distal bases (21–30) could influence overall targeting specificity, we designed sgRNAs 2 and 3 to contain 10 additional bases consisting of either 9 perfectly matched or 8 mismatched bases (bases 21–28). Surprisingly, we observed that these modified sgRNAs mediated similar levels of modification at the target locus in HEK 293FT cells regardless of whether the additional bases were complementary to the genomic target (Figure 1B). Subsequent Northern blots revealed that the majority of both sgRNA 2 and 3 were processed to the same length as sgRNA 1, which contains the same 20-nt guide sequence without additional bases (Figure 1C).
Cas9 nickase generates efficient NHEJ with paired, offset guide RNAs
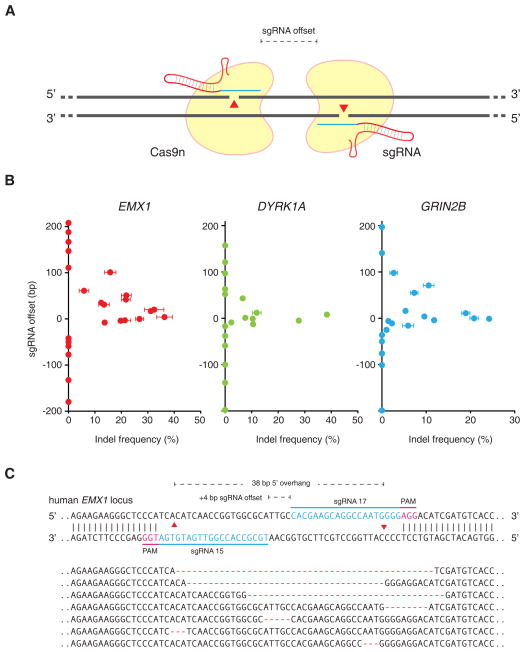
Given that extension of the guide sequence failed to improve Cas9 targeting specificity, we sought an alternative strategy for increasing the overall base-pairing length between the guide sequence and its DNA target. Cas9 enzymes contain two conserved nuclease domains, HNH and RuvC, which cleave the DNA strand complementary and non-complementary to the guide RNA, respectively. Mutations of the catalytic residues (D10A in RuvC and H840A in HNH) convert Cas9 into DNA nickases (Cong et al., 2013; Gasiunas et al., 2012; Jinek et al., 2012). As single-strand nicks are preferentially repaired by the high-fidelity BER pathway (Dianov and Hubscher, 2013), we reasoned that two Cas9 nicking enzymes directed by a pair of sgRNAs targeting opposite strands of a target locus could mediate DSBs while minimizing off-target activity (Figure 2A).
Figure 2. Double nicking facilitates efficient genome editing in human cells.
(A) Schematic illustrating DNA double-stranded breaks using a pair of Cas9 D10A nickases (Cas9n). The D10A mutation renders Cas9 able to cleave only the strand complementary to the sgRNA; a pair of sgRNA-Cas9n complexes can nick both strands simultaneously. sgRNA offset is defined as the distance between the PAM-distal (5′) ends of the guide sequence of a given sgRNA pair. (B) Efficiency of double nicking induced NHEJ as a function of the offset distance between two sgRNAs. Sequences for all sgRNAs used can be found in Table S1. (n = 3; error bars show mean ± s.e.m.) (C) Representative sequences of the human EMX1 locus targeted by Cas9n. sgRNA target sites and PAMs are indicated by blue and magenta bars respectively. Below, selected sequences showing representative indels. See also Table S1 and S2.
A number of factors may affect cooperative nicking leading to indel formation, including steric hindrance between two adjacent Cas9 molecules or Cas9-sgRNA complexes, overhang type, and sequence context, some of which may be characterized by testing multiple sgRNA pairs with distinct target sequences and offsets. To systematically assess how sgRNA offsets might affect subsequent repair and generation of indels, we first designed sets of sgRNA pairs targeted against the human EMX1 genomic locus separated by a range of offset distances from approximately -200 to 200 bp to create both 5′- and 3′-overhang products (Table S1). We then assessed the ability of each sgRNA pair, with the D10A Cas9 mutant (referred to as Cas9n; H840A Cas9 mutant is referred to as Cas9H840A), to generate indels in human HEK 293FT cells. Robust NHEJ (up to 40%) was observed for sgRNA pairs with offsets from -4 to 20 bp, with modest indels forming in pairs offset by up to 100-bp (Figure 3B, left panel). We subsequently recapitulated these findings by testing similarly offset sgRNA pairs in two other genomic loci, DYRK1A and GRIN2B (Figure 2B, right panels). Notably, across all three loci examined, only sgRNA pairs creating 5′ overhangs with less than 8bp overlap (offset greater than -8 bp) between the guide sequences were able to mediate detectable indel formation (Figure 2C).
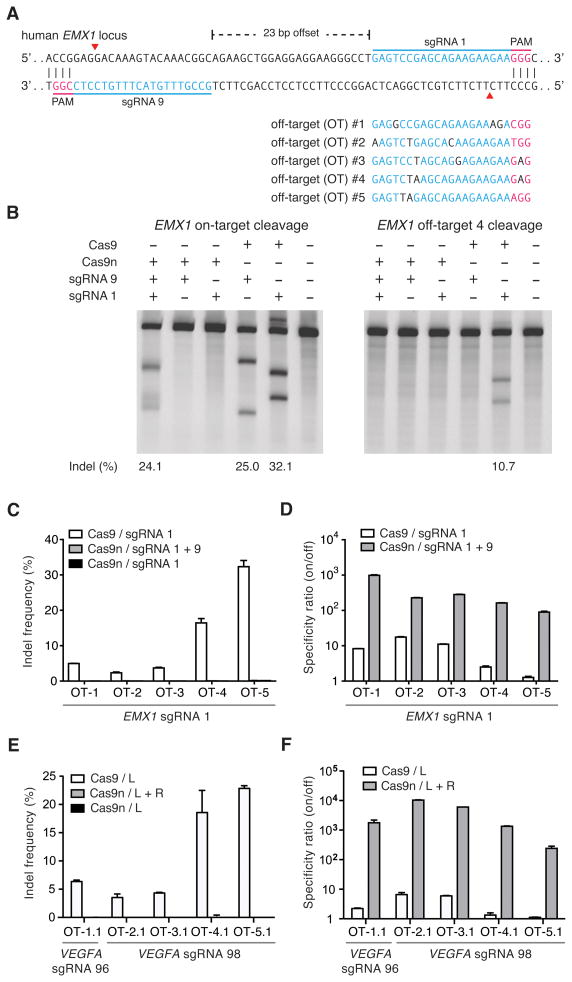
Figure 3. Double nicking facilitates efficient genome editing in human cells.
(A) Schematic illustrating DNA double strand breaks (red arrows) using Cas9 D10A nickases (Cas9n) and two sgRNAs. 5 off-target loci with sequence homology to EMX1 sgRNA 1 were selected to screen for Cas9n specificity. (B) On-target modification rate by Cas9n and a pair of sgRNAs is comparable to those mediated by wildtype Cas9 and single sgRNAs (left panel). Cas9-sgRNA1 complexes generate significant off-target mutagenesis, while no off-target locus modification is detected with Cas9n (right panel). (C) Levels of off-target modification with sgRNA 1 in HEK 293FT cells are measured by deep sequencing of five off-target loci. (D) Specificity comparison of Cas9n and wildtype Cas9 for sgRNA 1 off-target sites. Specificity ratio is calculated as on-target/off-target modification rates. (n = 3; error bars show mean ± s.e.m.) (E, F) Double nicking minimizes off-target modification at two human VEGFA loci while maintaining high specificity (on/off target modification ratio, n = 3; error bars show mean ± s.e.m.).
Importantly, each guide used in these assays is able to efficiently induce indels when paired with wildtype Cas9 (Table S1), indicating that the relative positions of the guide pairs are the most important parameters in predicting double nicking activity. Since Cas9n and Cas9H840A nick opposite strands of DNA, substitution of Cas9n with Cas9H840A with a given sgRNA pair should result in the inversion of the overhang type. For example, a pair of sgRNA that will generate a 5′ overhang with Cas9n should in principle generate the corresponding 3′ overhang instead. Therefore, sgRNA pairs that lead to the generation of a 3′ overhang with Cas9n might be used with Cas9H840A to generate a 5′ overhang. We therefore tested Cas9H840A with a broad set of sgRNA pairs but were unable to observe indel formation.
Double nicking mediates highly specific genome editing
Having established that double nicking (DN) mediates high efficiency NHEJ at levels comparable to those induced by wildtype Cas9, we next studied whether DN has improved specificity over wildtype Cas9 by measuring their off-target activities. We co-delivered Cas9n with sgRNAs 1 and 9, spaced by a 23-bp offset, to target the human EMX1 locus in HEK 293FT cells (Figure 3A). This DN configuration generated on-target indel levels similar to those generated by the wildtype Cas9 paired with each sgRNA alone (Figure 3B, left panel). Strikingly, DN did not generate detectable modification at the sgRNA 1 off-target site OT-4 by SURVEYOR assay (Figure 3B, right panel), suggesting that DN can potentially reduce the likelihood of off-target modifications.
Using deep sequencing to assess modification at 5 different sgRNA 1 off-target loci (Figure 3A), we observed significant mutagenesis at all sites with wild type Cas9 + sgRNA 1 (Figure 3C). In contrast, cleavage by Cas9n at 5 off-target sites tested was barely detectable above background sequencing error. Using the ratio of on- to off-target modification levels as a metric of specificity, we found that Cas9n with a pair of sgRNAs was able to achieve over 100-fold greater specificity relative to wild type Cas9 with one of the sgRNAs (Figure 3D). We conducted additional off-target analysis by deep sequencing for two sgRNA pairs (offsets of 16 and 20 bp) targeting the VEGFA locus, with similar results (Figure 3E). DN at these off-target loci was able to achieve 200 to over 1500-fold greater specificity than the wild-type Cas9 (Figure 3F, Table S1). Taken together, these results demonstrate that Cas9-mediated double nicking minimizes off-target mutagenesis and is suitable for genome editing with increased specificity.
Double nicking facilitates high-efficiency homology directed repair, NHEJ-mediated DNA insertion, and genomic microdeletions
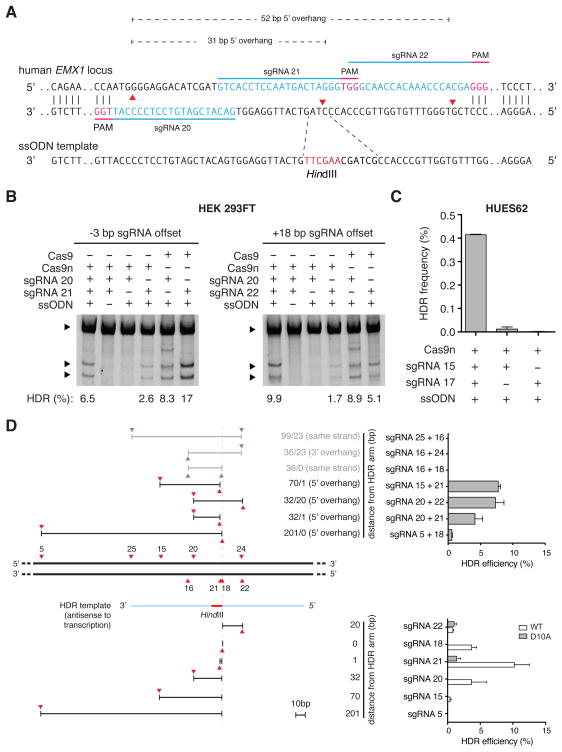
DSBs can stimulate homology directed repair (HDR) to enable highly precise editing of genomic target sites. To evaluate DN-induced HDR, we targeted the human EMX1 locus with pairs of sgRNAs offset by -3 and +18 bp (generating 31- and 52-bp 5′ overhangs), respectively, and introduced a single-stranded oligonucleotide (ssODN) bearing a _Hin_dIII restriction site as the HDR repair template (Figure 4A). Each DN sgRNA pair successfully induced HDR at frequencies higher than those of single-guide Cas9n nickases and comparable to those of wild-type Cas9 (Figure 4B). Furthermore, genome editing in embryonic stem cells or patient derived induced pluripotent stem cells represents a key opportunity for generating and studying new disease paradigms as well as developing new therapeutics. Since single nick approaches to inducing HDR in human embryonic stem cells (hESCs) have met with limited success (Hsu et al., 2013), we attempted DN in the HUES62 hES cell line and observed successful HDR (Figure 4C). To further characterize how offset sgRNA spacing affects the efficiency of HDR, we next tested a set of sgRNA pairs where the cleavage site of at least one sgRNA is situated near the site of recombination (overlapping with the HDR ssODN donor template arm). We observed that sgRNA pairs generating 5′ overhangs and having at least one nick occurring within 22bp of the homology arm are able to induce HDR at levels comparable to wildtype Cas9-mediated HDR. Double nicking HDR levels are significantly greater than single Cas9n-sgRNA nicking. In contrast, we did not observe HDR with sgRNA pairs that generated 3′-overhangs or double nicking of the same DNA strand (Figure 4D).
Figure 4. Double nicking allows insertion into the genome via HDR in human cells.
(A) Schematic illustrating HDR mediated via a single stranded oligodeoxynucleotide (ssODN) template at a DSB created by a pair of Cas9n enzymes. Successful recombination at the DSB site introduces a _Hin_dIII restriction site. (B) Restriction digest assay gel showing successful insertion of _Hin_dIII cleavage sites by double nicking-mediated HDR in HEK 293FT cells. Upper bands are unmodified template; lower bands are _Hin_dIII cleavage product. (C) Double nicking promotes HDR in the HUES62 human embryonic stem cell line. HDR frequencies are determined by deep sequencing. (n = 3; error bars show mean ± s.e.m.). (D) HDR efficiency depends on the configuration of Cas9 or Cas9n-mediated nicks. HDR is facilitated when a nick occurs near the center of the ssODN homology arm leading to a 5′-resulting overhang. HDR-compatible nicking configurations are denoted by red arrows separated by overhang regions (black lines), and non-compatible configurations are shown with brown arrows and gray lines (top panel). HDR efficiency mediated by sgRNAs 22, 18, 21, 20, 15, 5 paired with either Cas9 or Cas9n is shown for comparison (bottom panel, Table S2).
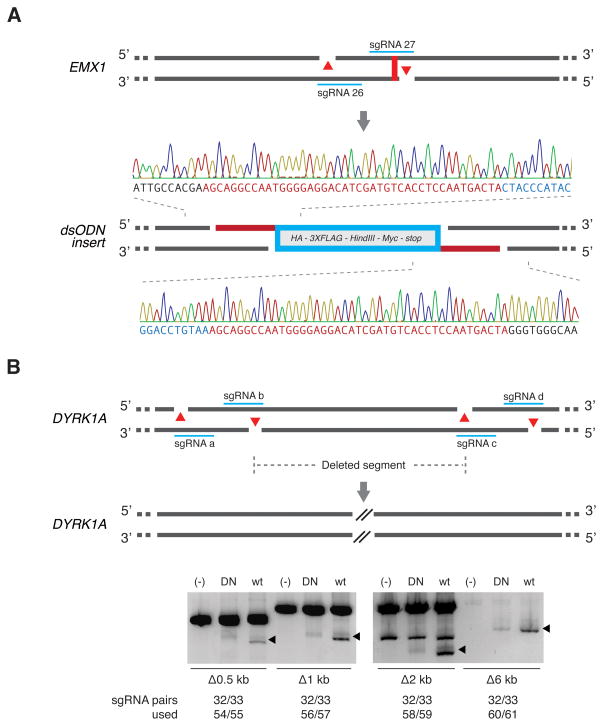
The ability to create defined overhangs could enable precise insertion of donor repair templates containing compatible overhangs via NHEJ-mediated ligation (Maresca et al., 2013). To explore this alternative strategy for transgene insertion, we targeted the EMX1 locus with Cas9n and an sgRNA pair designed to generate a 43-bp 5′-overhang near the stop codon, and supplied a double-stranded oligonucleotide (dsODN) duplex with matching overhangs (Figure 5A). The annealed dsODN insert was successfully integrated into the target with a frequency of 2.7% (1/37 screened by Sanger sequencing).
Figure 5. Multiplexed nicking facilitates non-HR mediated gene integration and genomic deletions.
(A) Schematic showing insertion of a double-stranded oligodeoxynucleotide (dsODN) donor fragment bearing overhangs complementary to 5′ overhangs created by Cas9 double nicking. The dsODN was designed to remove the native EMX1 stop codon and contains a HA tag, 3X FLAG tag, _Hind_III restriction site, Myc epitope tag, and a stop codon in frame, totaling 148 bp. Successful insertion was verified by Sanger sequencing as shown (1/37 clones screened). (B) Co-delivery of four sgRNAs with Cas9n generate long-range genomic deletions in the DYRK1A locus (from 0.5 kb up to 6 kb).
Additionally, we targeted a set of sgRNA pairs to the DYRK1A locus in HEK 293FT cells to facilitate genomic microdeletions. We generated a set of sgRNAs to mediate 0.5 kb, 1 kb, 2 kb, and 6 kb deletions (Figure 5B, Table S2: sgRNAs 32, 33, 54–61) and verified successful multiplex nicking-mediated deletion via amplicon size-based PCR screen.
Double nicking enables efficient genome modification in mouse zygotes
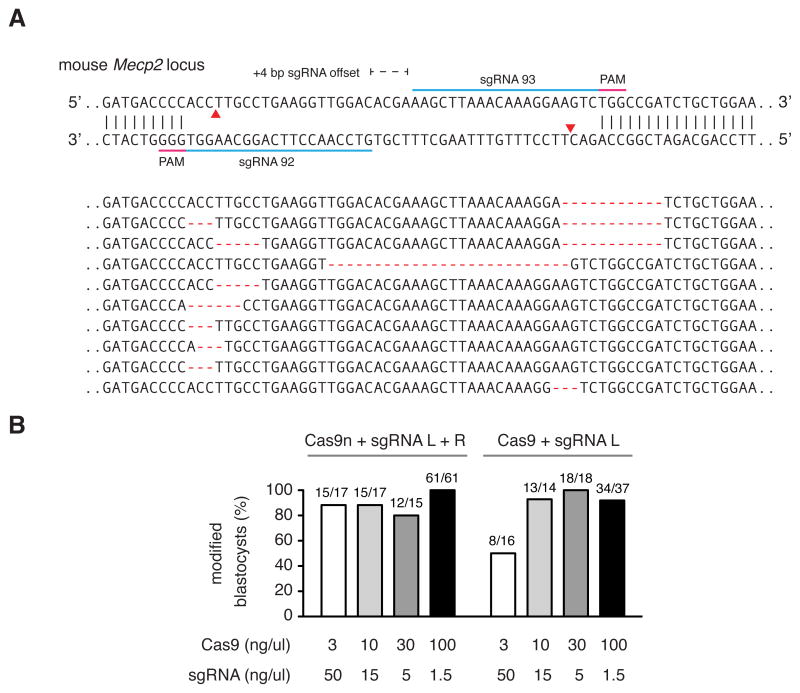
Recent work demonstrated that co-delivery of wildtype Cas9 mRNA along with multiple sgRNAs can mediate single-step generation of transgenic mice carrying multiple allelic modifications (Wang et al., 2013). Given the ability to achieve genome modification in vivo using several sgRNAs at once, we sought to assess the efficiency of multiple nicking by Cas9n in mouse zygotes. Peri-nuclear co-injection of wildtype Cas9 or Cas9n mRNA and sgRNAs into single-cell mouse zygotes allowed successful targeting of the Mecp2 locus (Figure 6A). To identify the optimal concentration of Cas9n mRNA and sgRNA for efficient gene targeting, we titrated Cas9 mRNA from 100 ng/uL to 3 ng/uL while maintaining the sgRNA levels at a 1:20 Cas9:sgRNA molar ratio. All concentrations tested for Cas9 double nicking mediated modifications in at least 80% of embryos screened, similar to those levels achieved by wildtype Cas9 (Figure 6B). Taken together, these results suggest a number of applications for double nicking-based genome editing.
Figure 6. Cas9 double nicking mediates efficient indel formation in mouse embryos.
(A) Schematic illustrating Cas9 double nicking targeting at the mouse Mecp2 locus. Representative indels are shown for mouse blastocysts co-injected with Cas9n-encoding mRNA and in vitro transcribed sgRNA pairs. (B) Efficient blastocyst modification is achieved at multiple concentrations of sgRNAs and wildtype Cas9 or Cas9n (titrating from 3 ng/uL to 100 ng/uL).
Discussion
Given the permanent nature of genomic modifications, specificity is of paramount importance to sensitive applications such as gene therapy or studies aimed at linking specific genetic variants with biological processes or disease phenotypes. Here, we have explored strategies to improve the targeting specificity of Cas9. Although simply extending the guide sequence length of sgRNA failed to improve targeting specificity, combining two appropriately offset sgRNAs with Cas9n effectively generated indels while minimizing single-stranded DNA break mutations via base excision repair. While significant off-target mutagenesis has been previously reported for Cas9 nucleases in human cells (Fu et al., 2013; Hsu et al., 2013), the DN approach could provide a generalizable solution for rapid and accurate genome editing. The characterization of spacing parameters governing successful Cas9 double nickase-mediated gene targeting reveals an effective offset window over 100-bp long, allowing for a high degree of flexibility in the selection of sgRNA pairs. Previous computational analyses have revealed an average targeting range of every 12-bp for the Streptococcus pyogenes Cas9 in the human genome based on the 5′-NGG PAM (Cong et al., 2013), which suggest that appropriate sgRNA pairs should be readily identifiable for most loci within the genome. We have additionally demonstrated DN-mediated indel frequencies comparable to wild type Cas9 modification at multiple genes and loci in both human and mouse cells, confirming the reproducibility of this strategy for high-precision genome engineering (Table S1).
The Cas9 double nicking approach is inherently similar to ZFN and TALEN-based genome editing systems, where cooperation between two hemi-nuclease domains is required to achieve double-stranded break at the target site. Systematic studies of ZFN and TALEN systems have revealed that the targeting specificity of specific ZFN and TALEN pairs can be highly dependent on the nuclease architecture (homo- or heterodimeric nucleases) or target sequence, and in some cases TALENs can be highly specific (Ding et al., 2013). Although the wildtype Cas9 system has been shown to exhibit high levels of off-target mutagenesis, the DN system is a promising solution and brings RNA-guided genome editing to similar specificity levels as ZFNs and TALENs.
Additionally, the ease and efficiency with which Cas9 can be targeted renders the DN system especially attractive. However, DNA targeting using DN will likely face similar off-target challenges as ZFNs and TALENs, where cooperative nicking at off-target sites might still occur, albeit at a significantly reduced likelihood. Given the extensive characterization of Cas9 specificity and sgRNA mutation analysis (Fu et al., 2013; Hsu et al., 2013), as well as the NHEJ-mediating sgRNA offset range identified in this study, computational approaches may be used to evaluate the likely off-target sites for a given pair of sgRNAs. To facilitate sgRNA pair selection, we developed an online web tool that identifies sgRNA combinations with optimal spacing for double nicking applications (http://www.genome-engineering.org/).
Although Cas9n has been previously shown to facilitate HDR at on-target sites (Cong et al., 2013), its efficiency is substantially lower than that of wildtype Cas9. The double nicking strategy, by comparison, maintains high on-target efficiencies while reducing off-target modifications to background levels. Nevertheless, further characterizations of DN off-target activity, particularly via whole genome sequencing and targeted deep sequencing of cells or whole organisms generated using the DN approach, are urgently needed to evaluate the utility of Cas9n DN in biotechnological or clinical applications requiring ultra-high precision genome editing. Additionally, Cas9n has been shown to induce low levels of indels at on-target sites for certain sgRNAs (Mali et al., 2013b), which may result from residual double-strand break activities and may be circumvented by further structure-function studies of Cas9 catalytic activity. As we explored these challenges, the DN strategy was shown independently to mediate HDR and NHEJ in cell lines (Mali et al., 2013a), further substantiating the potential benefit of double nicking in genome editing applications. Overall, Cas9n-mediated multiplex nicking serves as a customizable platform for highly precise and efficient targeted genome engineering and promises to broaden the range of applications in biotechnology, basic science, and medicine.
Experimental Procedures
Cell culture and transfection
Human embryonic kidney (HEK) cell line 293FT (Life Technologies) or mouse Neuro 2a (Sigma-Aldrich) cell line was maintained in Dulbecco’s modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (HyClone), 2mM GlutaMAX (Life Technologies), 100U/mL penicillin, and 100μg/mL streptomycin at 37°C with 5% CO2 incubation.
Cells were seeded onto 24-well plates (Corning) at a density of 120,000 cells/well, 24 hours prior to transfection. Cells were transfected using Lipofectamine 2000 (Life Technologies) at 80–90% confluency following the manufacturer’s recommended protocol. A total of 500ng Cas9 plasmid and 100 ng of U6-sgRNA PCR product was transfected.
Human embryonic stem cell line HUES62 (Harvard Stem Cell Institute core) was maintained in feeder-free conditions on GelTrex (Life Technologies) in mTesR medium (Stemcell Technologies) supplemented with 100ug/ml Normocin (InvivoGen). HUES62 cells were transfected with Amaxa P3 Primary Cell 4-D Nucleofector Kit (Lonza) following the manufacturer’s protocol.
SURVEYOR nuclease assay for genome modification
293FT and HUES62 cells were transfected with DNA as described above. Cells were incubated at 37°C for 72 hours post-transfection prior to genomic DNA extraction. Genomic DNA was extracted using the QuickExtract DNA Extraction Solution (Epicentre) following the manufacturer’s protocol. Briefly, pelleted cells were resuspended in QuickExtract solution and incubated at 65°C for 15 minutes, 68°C for 15 minutes, and 98°C for 10 minutes.
The genomic region flanking the CRISPR target site for each gene was PCR amplified (primers listed in Supplementary information), and products were purified using QiaQuick Spin Column (Qiagen) following the manufacturer’s protocol. 400ng total of the purified PCR products were mixed with 2μl 10X Taq DNA Polymerase PCR buffer (Enzymatics) and ultrapure water to a final volume of 20μl, and subjected to a re-annealing process to enable heteroduplex formation: 95°C for 10min, 95°C to 85°C ramping at − 2°C/s, 85°C to 25°C at − 0.25°C/s, and 25°C hold for 1 minute. After re-annealing, products were treated with SURVEYOR nuclease and SURVEYOR enhancer S (Transgenomics) following the manufacturer’s recommended protocol, and analyzed on 4–20% Novex TBE poly-acrylamide gels (Life Technologies). Gels were stained with SYBR Gold DNA stain (Life Technologies) for 30 minutes and imaged with a Gel Doc gel imaging system (Bio-rad). Quantification was based on relative band intensities. Indel percentage was determined by the formula, 100 × (1 − (1 − (b + c)/(a + b + c))1/2), where a is the integrated intensity of the undigested PCR product, and b and c are the integrated intensities of each cleavage product.
Northern blot analysis of tracrRNA expression in human cells
Northern blots were performed as previously described (Cong et al., 2013). Briefly, RNAs were extracted using the mirPremier microRNA Isolation Kit (Sigma) and heated to 95°C for 5 min before loading on 8% denaturing polyacrylamide gels (SequaGel, National Diagnostics). Afterwards, RNA was transferred to a pre-hybridized Hybond N+ membrane (GE Healthcare) and crosslinked with Stratagene UV Crosslinker (Stratagene). Probes were labeled with [gamma-32P] ATP (Perkin Elmer) with T4 polynucleotide kinase (New England Biolabs). After washing, membrane was exposed to phosphor screen for one hour and scanned with phosphorimager (Typhoon).
Deep sequencing to assess targeting specificity
HEK 293FT cells were plated and transfected as described above, 72 hours prior to genomic DNA extraction. The genomic region flanking the CRISPR target site for each gene was amplified by a fusion PCR method to attach the Illumina P5 adapters as well as unique sample-specific barcodes to the target. PCR products were purified using EconoSpin 96-well Filter Plates (Epoch Life Sciences) following the manufacturer’s recommended protocol.
Barcoded and purified DNA samples were quantified by Qubit 2.0 Fluorometer (Life Technologies) and pooled in an equimolar ratio. Sequencing libraries were then sequenced with the Illumina MiSeq Personal Sequencer (Life Technologies).
Sequencing data analysis, indel detection, and homologous recombination detection
MiSeq reads were filtered by requiring an average Phred quality (Q score) of at least 30, as well as perfect sequence matches to barcodes and amplicon forward primers. Reads from on- and off-target loci were analyzed by performing Ratcliff-Obershelp string comparison, as implemented in the Python difflib module, against loci sequences that included 30 nucleotides upstream and downstream of the target site (a total of 80 bp). The resulting edit operations were parsed, and reads were counted as indels if insertion or deletion operations were found. Analyzed target regions were discarded if part of their alignment fell outside the MiSeq read itself or if more than 5 bases were uncalled.
Negative controls for each sample provided a gauge for the inclusion or exclusion of indels as putative cutting events. For quantification of homologous recombination, reads were first processed as in the indel detection workflow, and then checked for presence of homologous recombination template CCAGGCTTGG.
Microinjection into mouse zygotes
MII-stage oocytes were collected from 8-week old superovulated BDF1 females by injecting 7.5 I.U. of PMSG (Harbor, UCLA) and hCG (Millipore). They were transferred into HTF medium supplemented with 10 mg/ml bovine serum albumin (BSA; Sigma-Aldrich) and inseminated with capacitated sperm obtained from the caudal epididymides of adult C57BL/6 male mice. Six hours after fertilization, zygotes were injected with mRNAs and sgRNAs in vitro transcribed with mMESSAGE mMACHINE T7 Kit (Life Technologies) in M2 media (Millipore) by using a Piezo impact-driven micromanipulator (Prime Tech Ltd., Ibaraki, Japan). The concentrations of Cas9 and Cas9n mRNAs and sgRNAs are described in the text and Figure 6B. After microinjection, zygotes were cultured in KSOM (Millipore) in a humidified atmosphere of 5% CO2 and 95% air at 37°C.
Genome extraction from blastocyst embryos
Following in vitro culture of embryos for 6 days, the expanded blastocysts were washed with 0.01% BSA in PBS and individually collected into 0.2 ml tubes. Five microliters of genome extraction solution (50 mM Tris-HCl, pH 8.0, 0.5% Triton-X100, 1 mg/ml Proteinase K) were added and the samples were incubated in 65°C for 3 hours followed by 95°C for 10 min. Samples were then amplified for targeted deep sequencing as described above.
Supplementary Material
01
02
Highlights.
- Cas9 nickase can facilitate targeted DNA double strand break using two guide RNAs
- Double nicking of DNA reduces off-target mutagenesis by 50–1,000 fold
- Multiplex nicking stimulates homology directed repair, microdeletion, and insertion
- Double nicking provides efficient modification of mouse zygotes
Acknowledgments
We thank Xuebing Wu and Phil Sharp for assistance with Northern blotting experiments, Joshua Weinstein and Yinqing Li for statistical consultation, and the entire Zhang Lab for their support and advice. P.D.H. is a James Mills Pierce Fellow. D.A.S. is an NSF pre-doctoral fellow. C.L. is supported by T32GM007753 from the National Institute of General Medical Sciences. This work is supported by an NIH Director’s Pioneer Award (DP1-MH100706), a NIH Transformative R01 grant (R01-DK097768) to David Altshuler, the Keck, McKnight, Damon Runyon, Searle Scholars, Klingenstein, Vallee, and Simons Foundations, Bob Metcalfe, and Jane Pauley.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Carlson DF, Tan WF, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CBA, Fahrenkrug SC. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci U S A. 2012;109:17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell research. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annual review of microbiology. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- Dianov GL, Hubscher U. Mammalian base excision repair: the forgotten archangel. Nucleic acids research. 2013;41:3483–3490. doi: 10.1093/nar/gkt076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell stem cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nature methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature biotechnology. 2013 doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui XX, Meng XD, et al. Knockout Rats via Embryo Microinjection of Zinc-Finger Nucleases. Science. 2009;325:433–433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013 doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013 doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Zhang F. Dissecting neural function using targeted genome engineering technologies. ACS Chem Neurosci. 2012;3:603–610. doi: 10.1021/cn300089k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature biotechnology. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature biotechnology. 2013a doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013b;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca M, Lin VG, Guo N, Yang Y. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome research. 2013;23:539–546. doi: 10.1101/gr.145441.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nature biotechnology. 2013 doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EE, Wang JB, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. Establishment of HIV-1 resistance in CD4(+) T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, et al. Generation of Isogenic Pluripotent Stem Cells Differing Exclusively at Two Early Onset Parkinson Point Mutations (vol 146, pg 318, 2011) Cell. 2011;146:659–659. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasu Y, Kobayashi I, Beumer K, Uchino K, Sezutsu H, Sajwan S, Carroll D, Tamura T, Zurovec M. Targeted mutagenesis in the silkworm Bombyx mori using zinc finger nuclease mRNA injection. Insect Biochem Molec. 2010;40:759–765. doi: 10.1016/j.ibmb.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Ochiai H, Sakuma T, Horch HW, Hamaguchi N, Nakamura T, Bando T, Ohuchi H, Yamamoto T, Noji S, et al. Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat Commun. 2012:3. doi: 10.1038/ncomms2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02