Acetylation-Mediated Proteasomal Degradation of Core Histones during DNA Repair and Spermatogenesis (original) (raw)
. Author manuscript; available in PMC: 2014 May 23.
SUMMARY
Histone acetylation plays critical roles in chromatin remodeling, DNA repair, and epigenetic regulation of gene expression, but the underlying mechanisms are unclear. Proteasomes usually catalyze ATP- and polyubiquitin-dependent proteolysis. Here we show that the proteasomes containing the activator PA200 catalyze the polyubiquitin-independent degradation of histones. Most proteasomes in mammalian testes (“spermatoproteasomes”) contain a spermatid/sperm-specific α-subunit α4s/PSMA8 and/or the catalytic β-subunits of immunoproteasomes in addition to PA200. Deletion of PA200 in mice abolishes acetylation-dependent degradation of somatic core histones during DNA double-strand breaks, and delays core histone disappearance in elongated spermatids. Purified PA200 greatly promotes ATP-independent proteasomal degradation of the acetylated core histones, but not polyubiquitinated proteins. Furthermore, acetylation on histones is required for their binding to the bromodomain-like regions in PA200 and its yeast ortholog, Blm10. Thus, PA200/Blm10 specifically targets the core histones for acetylation-mediated degradation by proteasomes, providing mechanisms by which acetylation regulates histone degradation, DNA repair, and spermatogenesis.
INTRODUCTION
Proteasomes catalyze ATP- and polyubiquitin-dependent degradation of most cellular proteins and in vertebrates the generation of MHC-Class I antigenic peptides (Finley, 2009; Glickman and Ciechanover, 2002; Goldberg, 2003). The 26S proteasome consists primarily of two sub-complexes, the 20S catalytic particle and the regulatory particle at either or both ends of the 20S particle. Two major forms of the 20S “core” particle have been identified in mammals: the constitutive proteasome and the inducible variant, the immunoproteasome. The former has three catalytic subunits, β1, β2, and β5, whereas the latter has three closely related inducible subunits, β1i, β2i, and β5i. The primary regulatory particle is the 19S particle, which binds, unfolds, and translocates polyubiquitinated proteins into the 20S. However, additional proteasome activators also exist, such as the 11S complex, PA28αβ, which increases the generation of peptides appropriate for antigen presentation (Rock and Goldberg, 1999). In addition, the proteasome activator PA28γ, which is homologous to PA28α and PA28β, has been reported to promote the ubiquitin-independent degradation of certain nuclear proteins (Li et al., 2006).
The core histones, H2A, H2B, H3, and H4, form an octamer to pack DNA into the nucleosome, whereas the linker histone H1 protects internucleosomal DNA. Each octamer consists of two separate H2A–H2B dimers and a stable tetramer of two H3–H4 dimmers. The nucleosome is the unit of chromatin organization, and is critical in various cellular processes, including epigenetic regulation of gene expression, cell division, differentiation, and DNA damage response (Campos and Reinberg, 2009). During spermatogenesis, histones are largely replaced transiently by transition proteins and subsequently by protamines in postmeiotic cells (Hammoud et al., 2009; Mills et al., 1977). Histones can also be replaced at promoter regions or active gene bodies in somatic cells (Deal et al., 2010; Dion et al., 2007). However, the mechanisms underlying the replacement of these histones remain unknown (Chen and Qiu, 2012).
Lysine acetylation is extensively involved in various cellular processes, especially chromatin remodeling, DNA repair, and transcription. It influences both proteasomal degradation by either preventing or promoting polyubiquitination and lysosomal degradation of certain substrates (Chen and Qiu, 2012). Histone acetyltransferase (HAT) catalyzes the formation of the acetyllysine residue, which can be recognized by the bromodomain (BRD) in proteins, and a histone deacetylase (HDAC) removes the acetyl group (Yang and Seto, 2007). Histones are highly acetylated prior to their removal from chromatin during spermatogenesis (Gaucher et al., 2010). While the acetylation sites in histones and the enzymes that catalyze formation or removal of acetylation are widely studied, how acetylation regulates transcription, spermatogenesis, and DNA repair is still unclear (Chen and Qiu, 2012).
The proteasome activator PA200 and its ortholog in yeast, Blm10, bind to the ends of the 20S particle and stimulate the hydrolysis of small peptides and/or the unstructured tau protein in vitro (Dange et al., 2011; Schmidt et al., 2005; Ustrell et al., 2005). PA200 is present in all mammalian tissues, but is highly expressed in the testis (Ustrell et al., 2005). Deletion of PA200 markedly reduces fertility of male mice due to severe defects in spermatogenesis (Khor et al., 2006). Degradation of histones has been approached by searching for ubiquitination enzymes that catalyze their polyubiquitination, but such enzymes have not yet been identified (Liu et al., 2007). Here we show that special forms of proteasomes, which contain PA200/Blm10, specifically catalyze the acetylation-, but not polyubiquitination-, dependent degradation of the core histones during somatic DNA damage response and spermatogenesis.
RESULTS
Special Types of Proteasomes Are Predominant in Mammalian Testes
To explore the mechanisms underlying histone degradation, we first tested whether spermatogenic cells in mammals possess unusual types of proteasomes, because histones are largely lost during spermatogenesis (Mills et al., 1977). By native PAGE, we analyzed crude extracts of bovine skeletal muscle and of the seminiferous tubules from bovine testes, which primarily consist of spermatogenic cells. The proteasomes were visualized in the gels using the specific fluorogenic peptide substrate, succinyl LLVY-7-amino-4-methylcoumarin (amc). As expected, the muscle proteasomes were detected in two bands, which corresponded to doubly-capped (19S-20S-19S) and singly-capped (19S–20S) particles (Figure 1A). Proteasomes from the testis also migrated as two bands. One corresponded to 19S-20S-19S, but the other was found between the doubly- and singly-capped particles, and thus was probably intermediate in size (referred to as Lg for the large testis-specific proteasome, Figure 1A). Peptide hydrolysis by Lg appeared much stronger than that by the 19S-20S-19S, suggesting that Lg represents the predominant activity against this substrate in spermatogenic cells of bovine testes. This species also appeared to be predominant in extracts from rat and mouse testes (see Figure S1A and results shown below). When 0.02% SDS was added, a faster-migrating band (referred to as Sm for the small testis-specific proteasome) became evident in the testis sample (Figure 1A), probably because SDS activated the 20S particle by opening the gated channel for substrate entry (Smith et al., 007).
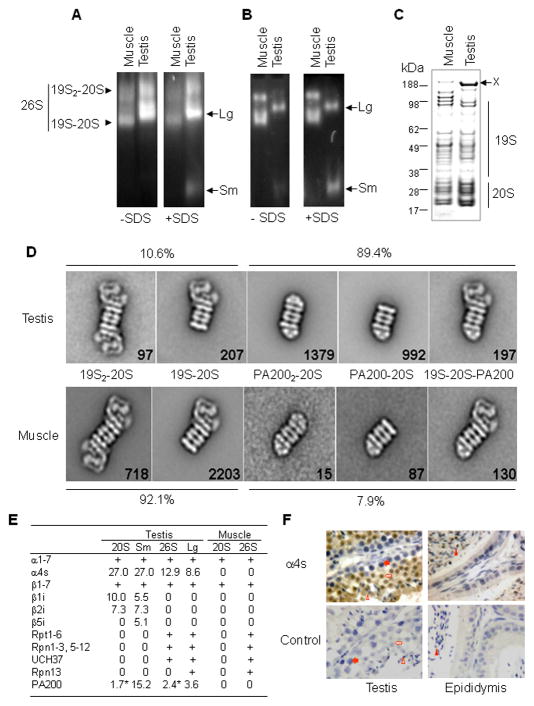
Figure 1. Identification of Two Distinct Types of Proteasomes in Mammalian Testes.
(A) The proteasomes in bovine testes migrate differently from those in muscle on native PAGE. Proteasomes in crude tissue extracts were detected by incubating the gel with LLVY-amc in the absence or presence of 0.02% SDS, and visualized under UV light. Two unusual bands were labeled as Lg and Sm, respectively.
(B) Proteasomes purified from seminiferous tubules of the testis are found primarily as large (Lg) and small (Sm) testis-specific particles. Proteasomes were purified using chromatography and glycerol gradient fractionation. In order to enrich the testis-specific proteasomes, the fractions with typical 26S proteasomes were not collected, and results were analyzed as in (A).
(C) Analysis of proteasomes by SDS-PAGE and Coomassie blue staining. The letter X indicates a 200 kDa protein.
(D) The majority of proteasomes purified from the testis contain one or two small particles. The purified proteasomes were analyzed by electron microscopy, and class averages of different types of proteasomes from the testis and muscle were shown. Numbers in each class average indicate the total number of proteasomes in the classes.
(E) Mass spectrometric analysis of testis proteasomes. The testis proteasomes from 4 bands in Figure S1E were applied for mass spectrometry, and the percentile coverage of amino acids for each protein was shown. The 20S and the 26S proteasomes from skeletal muscle were included as references. The detection of regular proteasome subunits was indicated by “+”, whose coverage of amino acids was 3.6–51.4%. An asterisk indicates that immunoblotting could not confirm the detection by mass spectrometry.
(F) α4s protein is specifically present in spermatids and sperm as shown by immunohistochemical assays. The antigen–antibody complexes on mouse tissue paraffin sections were stained in brown, and nuclei were in blue. In control, purified IgG was used as the primary antibody. The filled arrow (spermatocyte), the open arrow (round spermatid), the open triangle (elongated spermatid), and the filled triangle (sperm) point to the corresponding cells.
See also Figures S1 and S2.
The proteasomes were then purified by ion exchange chromatography from seminiferous tubules of the testis by excluding the typical 26S particles. The purified preparations contained primarily large (Lg) and small (Sm) forms of testis proteasomes (Figure 1B). The Sm forms of the proteasome migrated more slowly than typical 20S particles (Figure S1). On SDS-PAGE gels (Figure 1C), the distribution patterns of proteasomal subunits from the testis were generally similar to those from muscle, but the levels of all 20S subunits appeared greater than those of 19S subunits. In addition, a major band (indicated as X) at approximately 200 kDa was found in the proteasomes purified from the testis. Following negative staining electron microscopy (EM), all proteasomes in side views were selected for single particle classification and averaging analysis. Five different types of proteasomes were found in both the testis and muscle (Figure 1D). The first two types were typical 26S proteasomes with one or both ends of the 20S particle capped with the 19S particle. The other three types had at the ends of the 20S one or two smaller structures, which resemble PA200 or its yeast ortholog Blm10 (Ortega et al., 2005; Schmidt et al., 2005), but differ from PA28-containing complexes (Cascio et al., 2002) (Figure S1). About 90% of proteasomes from the testis contained this small structure resembling PA200. In comparison, only ~8% of the proteasomes from muscle contained such a component (Figure 1D). Thus, both Lg and Sm proteasomes from the testis (i.e., spermatoproteasomes) appear to contain PA200.
Subunits of Testis-Specific Proteasomes
To further analyze the subunits of the proteasomes from spermatogenic cells, we collected fractions from the glycerol gradient into two pools (a and b in Figure S1D), which were then run on native PAGE (Figures S1E and S1F). The individual bands shown in Figure S1E were cut out and subjected to mass spectrometric analysis. In addition to the typical subunits found in muscle 20S particles (α1–7 and β1–7), three immunoproteasomal catalytic subunits (β1i, β2i, and β5i) were detected in either the testis 20S or the small testis proteasome (Sm) (Figure 1E). PA200 was detected in all 4 samples from the testis (Figure 1E), but its presence in the 20S particle and the doubly-capped 26S proteasome was not confirmed by immunoblotting (Figures S1F). In contrast, neither immunoproteasome subunits nor PA200 was detectable in the muscle 20S or 26S proteasomes by this approach (Figure 1E). Surprisingly, a novel subunit with 82% identity to α4/PSMA7, which is referred to as α4s, was detected in all testis proteasomes, and was expressed specifically in spermatids and sperm (Figures 1F and S2). Thus, PA200, α4s, and the catalytic subunits of the immunoproteasome seem to be subunits of the testis-specific proteasomes.
The testis consists of multiple cell types, including spermatogonia, spermatocytes, spermatids/sperm, sertoli cells, and leydig cells (Kotaja et al., 2004). When levels of proteins were analyzed in isolated sperm and various testis cell lines, α4s and β2i were detected only in sperm, and β1i was in sertoli cells and spermatocytes, whereas β5i, PA200, 19S subunits (such as Rpt2, Rpt4, and Rpn7), and PA28α were expressed in all these cell types (Figure 2A). β1i, β2i, and β5i were abundant in the testis and spleen, but not in C2C12 myoblast cells (Figure 2A). As demonstrated by glycerol gradient ultracentrifugation of GC-2spd spermatocyte or testis extracts, PA200, β1i, and β5i (like Rpt4, β1, β2, and β5) were primarily present in the fractions with proteasomal activity. In contrast, PA28α was mostly in the low molecular weight fractions that lacked activity (Figure S3).
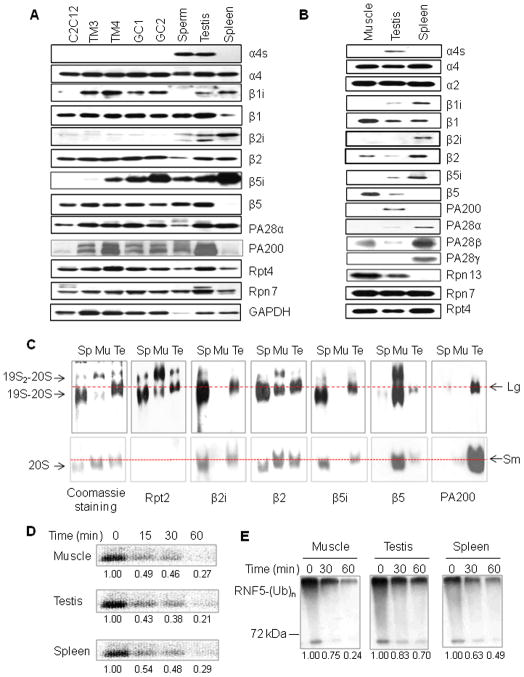
Figure 2. Testis–Specific Proteasomes Possess Distinct Subunits and Activities.
(A) Proteasome subunits in various cell lines derived from testes. Extracts of mouse spleen, the testis, sperm, testis cell lines (including spermatogonium GC1-spg, spermatocyte GC-2spd, leydig cell TM3, and sertoli cell TM4), and a muscle-related cell line (C2C12) were subjected for SDS-PAGE, and proteasomal subunits were analyzed by immunoblotting.
(B) Analysis of the purified proteasomes by immunoblotting following SDS-PAGE. To purify all forms of the proteasomes from testes, almost all the eluted fractions with proteasome activity from each step were collected and used in the next purification step.
(C) Subunits for the testis-specific proteasomes. The proteasomes purified from the testis (Te), skeletal muscle (Mu), and spleen (Sp) were separated on native PAGE, and stained with Coomassie blue or analyzed by immunoblotting. Lg and Sm indicate the positions for the large and small testis-specific proteasomes, respectively.
(D) Spermatoproteasomes degrade denatured core histones in a rate similar to other proteasomes. The [125I]-labeled calf core histones at 3.75 μM were incubated with proteasomes (0.4 μg/ml) for indicated periods of time, separated by 15% SDS-PAGE, and analyzed with PhosphoImager. Their relative levels were shown under the bands. Similar results were obtained from at least three independent experiments.
(E) Spermatoproteasomes are not efficient in degrading ubiquitinated RNF5. The polyubiquinated species of RNF5 [RNF5-(Ub)n] prepared in vitro as in Figure S4F were incubated with proteasomes (0.4 μg/ml) for indicated periods of time. Ubiquitin conjugates were analyzed by immunoblotting with an anti-ubiquitin antibody. Similar results were obtained from at least three independent experiments.
See also Figures S3 and S4.
To confirm the presence of the above subunits in testis proteasomes, we purified all forms of the proteasomes from the spermatogenic cells of testes. When analyzed by immunoblotting following SDS-PAGE, α4s, β1i, β5i, and PA200, but not β2i, were detected in the purified testis proteasomes. The 11S proteasome activators PA28α and PA28β were present, but only at low levels (Figure 2B). When analyzed by non-denaturing PAGE, the 26S proteasomes from muscle were present mainly as doubly-capped with appreciable singly-capped 26S complexes, and those from spleen (immunoproteasomes) were primarily found as singly-capped with some doubly-capped structures (Figure 2C). In contrast, most proteasomes purified from the testis contained PA200, and appeared in a band between the two 26S bands (i.e., the large testis-specific proteasomes) and a small amount migrated with the doubly-capped 26S particles (Figure 2C). The presence of the 19S complexes in these structures was confirmed by immunoblotting against Rpt2. β2i and β5i were present in the proteasomes from the testis and spleen, but not in those from muscle, which contained instead β2 and β5 (Figure 2C). Thus, both large (19S-20S-PA200) and small (20S-PA200 or PA200-20S-PA200) testis-specific proteasomes appear distinct in containing PA200, the catalytic subunits of the immunoproteasome, and the novel α-subunit (α4s).
Spermatoproteasomes Are Not Efficient in Degrading Ubiquitinated Proteins
The 20S catalytic particle contains three catalytic sites with different specificities (Rock and Goldberg, 1999). Despite the similar catalytic subunits in the particles from testis and spleen, the testis proteasomes purified as in Figure 1B differed from other species in their peptidase activities (Figures S4A–S4C). To examine the capacity of the spermatoproteasomes to degrade protein substrates, we first utilized the purified casein, which has little tertiary structure and thus can be digested by proteasomes without ubiquitination (Kisselev et al., 1998). Proteasomes from muscle appeared about twice as active as those from testis or spleen in hydrolyzing β-casein (P<0.01; Figure S4D). As expected, bortezomib (Velcade), a specific proteasome inhibitor at 50 nM almost completely inhibited the chymotrypsin-like activities of all three preparations (Figure S4E), but reduced the degradation of β-casein by about 50% (Figure S4D). Because most histones are eliminated during spermatogenesis, we also utilized the [125I]-labeled denatured core histones in the degradation assay, but found that all these three types of proteasomes degraded denatured core histones at similar rates (Figure 2D).
Because the physiological substrates of the 26S proteasome are usually polyubiquitinated proteins, we compared the ability of these proteasome preparations to degrade ubiquitinated RNF5, a ubiquitin ligase. RNF5 promotes formation of the K48-linked ubiquitin chains and proteasomal degradation of its substrates (Zhong et al., 2009). When FLAG-tagged RNF5 was purified from 293T cells and further auto-ubiquitinated in vitro (Figure S4F), the polyubiquitinated species of RNF5 were then incubated with proteasomes. The testis-specific proteasomes were much less efficient in degrading polyubiquitinated RNF5 than those from muscle or spleen (Figure 2E). Since there was no any detectable release or any increase in the levels of free RNF5 in the degradation assay (data not shown and Figure S4G), the reduced levels in polyubiquitinated RNF5 must be caused by degradation, instead of deubiquitination. Similar results were obtained, when polyubiquitinated RNF5 was replaced with polyubiquitinated Nrdp1, another ubiquitin ligase (Qiu and Goldberg, 2002) (Figures S4H and S4I). Thus, the testis-specific proteasomes degrade unstructured casein and denatured core histones in vitro in the rates comparable to those from muscle or spleen, but are much less efficient in degrading polyubiquitinated proteins.
PA200-Deficient Mouse Testes Are Defective in Core Histone Replacement
To determine whether spermatoproteasomes might degrade histones differently in vivo, we tested whether PA200 was required for histone replacement during spermatogenesis using PA200-deficient mice. As reported previously (Khor et al., 2006), deletion of PA200 increased markedly apoptosis in testes and reduced male fertility, but did not cause any other apparent phenotypic changes in the mice (Figure 3A and data not shown). Sperm differentiation in mice proceeds through 16 distinct steps. The core histones, H2B and H3, disappeared at the early stage of elongated spermatids (step 9 of spermatogenesis) in wild-type mice. However, in PA200-knockout mice, both H2B and H3 remained detectable at the end of the elongation stage of the spermatids (step 11), though these histones were lost in the elongated spermatids with fully condensed chromatin (e.g., steps 15–16) (Figures 3B and S5A–C). In contrast, PA200-deficiency did not retard the disappearance of the linker histone H1 in elongated spermatids, while reducing its relative levels in most diploid cells in testes (Figures 3B and S5D).
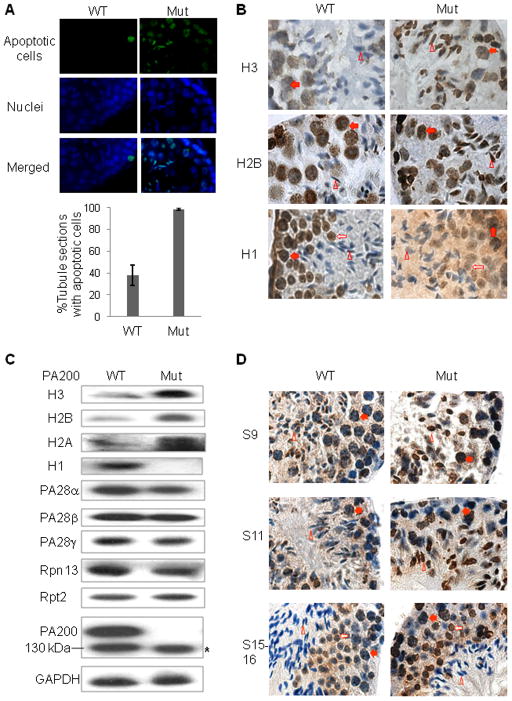
Figure 3. Deletion of PA200 in Mice Retards Disappearance of Core Histones in Elongated Spermatids.
(A) Deletion of PA200 increases the rate of apoptosis in testis. Apoptotic cells in testis paraffin sections of the 15-week-old wild-type or PA200-deficient mice were detected by fluorometric tunnel assay (green). The nucleus was stained by DAPI (4′,6-diamidino-2-phenylindole, blue). Only few apoptotic cells (usually <5) were detected in each apoptosis-positive tubule section from wild-type mice, but much more (mostly >5) from the PA200-deficient mice. Data are represented as mean +/− SEM.
(B) Deletion of PA200 leads to accumulation of core histones in elongated spermatids. Histones in testis paraffin sections of the 15-week-old wild-type or PA200-deficient mice were detected by immunohistochemistry (brown), and nuclei were stained with haematoxylin (blue). The steps of spermatogenesis were 11 for both H2B and H3, and 13–14 for H1. The filled arrow (spermatocyte), the open arrow (round spermatid), and the open triangle (elongated spermatid) point to the corresponding cells.
(C) PA200 deficiency increases the levels of the core histones in soluble testis extracts. Testis homogenates from the wild-type or PA200-deficient mice were prepared in regular buffer, and were analyzed by immunoblotting following SDS-PAGE. The asterisk denotes a polypeptide, which did not complex with the 20S particle in Figure S5A.
(D) Deletion of PA200 elevates the levels of H4K16ac in round and elongating spermatids. Testis paraffin sections were prepared and stained, and cells were labeled as in (B), but the primary antibody was anti-H4K16ac (Millipore #07-329).
See also Figure S5.
Histones are usually packed in chromatin, and can be extracted under high-salt or acidic conditions. We reasoned that they should be released from chromatin before degradation. Elongated spermatids at steps 9–11 are just a very small fraction of all the cells in the testis, and thus changes in the levels of total histones would not be expected to be detectable by western blot analysis. Therefore, we analyzed histones extracted by the regular buffer, and found that deletion of PA200 markedly increased the levels of the core histones H2A, H2B, and H3 in the soluble testis extracts, and dramatically decreased the levels of H1 (Figure 3C). It is known that H4 is acetylated at K16 (H4K16ac) prior to the removal of the core histones in elongating spermatids (Lu et al., 2010). It is noteworthy that deletion of PA200 elevated the levels of H4K16ac in round and elongating spermatids (Figure 3D). Thus, PA200 appears to promote the selective loss of the core histones (and especially the acetylated species) in elongated spermatids.
PA200/Blm10 Promotes Acetylation-Associated Degradation of Core Histones during Somatic DNA Double-Strand Breaks
Since histone acetylation happens prior to histone removal in elongated spermatids and at the sites near DNA double-strand breaks (DSBs) (Gaucher et al., 2010; Murr et al., 2006), we wondered whether acetylation promotes histone degradation in response to DSBs. In GC-2spd cells, DSBs induced by γ-irradiation, as marked by phosphorylation of histone H2AX (γ-H2AX) (Huen et al., 2007), had little effect on the levels of the core histones. However, in the presence of the histone deacetylase (HDAC) inhibitor, trichostatin A (TSA), which markedly increased the levels of acetylated H4K16 (H4K16ac), γ-irradiation led to a dramatic decrease in the levels of non-acetylated H2B and H4 at 20 min or 60 min post-irradiation (Figure 4A). At 120 min post-irradiation, the levels of H2B and H4 bounced back markedly, probably because the cells had recovered from the damage. Accordingly, treatment of GC-2spd cells with TSA in conjunction with the DNA-damaging agent, methyl methanesulfonate (MMS), also led to a dramatic decrease in the levels of H2B and H4 (Figure 4B). Similarly, in wild-type mouse embryonic fibroblast (MEF) cells, the joint treatment of γ-irradiation and TSA led to a marked decrease in the levels of H2B and H4 at 20 min or 60 min post-irradiation (Figures 4C and S6A). In contrast, in the PA200-deficient MEF cells, this joint treatment had almost no effect on the levels of the core histones (Figures 4C and S6B). Since the core histones are semi-conservatively replicated during DNA replication (Zhu and Reinberg, 2011), this dramatic loss of the core histones within only 20 min after irradiation (compared to the ~24 h doubling time for the MEF cells) must result from accelerated degradation of these proteins rather than from reduced transcription or translation during DNA replication. Thus, PA200 is apparently required for the acetylation-associated degradation of the core histones in response to DNA double-strand breaks.
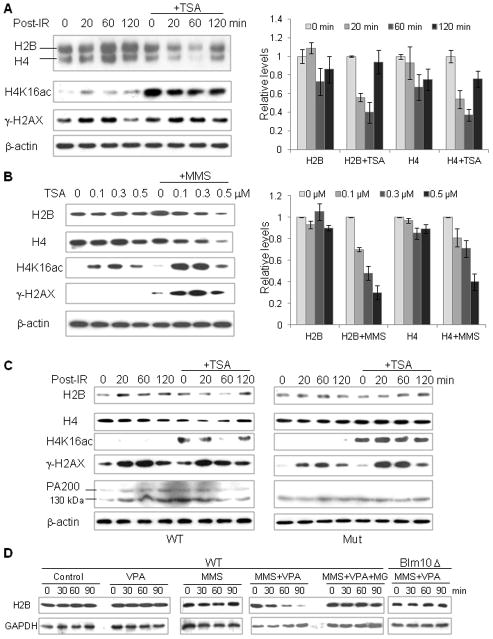
Figure 4. PA200/Blm10 Is Required for Acetylation-Associated Degradation of Core Histones during Somatic DNA Damage.
(A) Joint treatment of irradiation with trichostatin A reduces the levels of the core histones. GC-2spd cells were treated with or without TSA (0.3 μM), irradiated by a 60Co gamma irradiator for 15 min (1 Gy/min), and then incubated for the time periods as indicated. The levels of the histones and the loading control, β-actin, were analyzed by immunoblotting following lysis of cells by SDS sample buffer. The levels of H2B and H4 were quantified by densitometry (normalized to the loading control), and data are represented as mean +/− SEM.
(B) Treatment with TSA and MMS decreases the levels of the core histones. GC-2spd cells were treated with 25 μg/ml cycloheximide and TSA at the concentrations as indicated in the absence or the presence of 0.004% (0.472 mM) MMS for 4 h. The levels of histones and the loading control, β-actin, were analyzed as in (A).
(C) Treatment with both irradiation and TSA reduces the levels of the core histones in wild-type, but not in PA200-deficient, MEF cells. Wild-type (WT) or PA200-deficient (Mut) MEF cells were treated and analyzed as in (A). A probably non-specific 130-kDa band was recognized by the anti-PA200 antiserum.
(D) Acetylation- and Blm10-dependent degradation of the core histones in diploid yeast treated with MMS. Wild-type or the Blm10-deficient diploid budding yeast was treated with MMS and/or VPA in the absence or presence of MG132 (10 μM) for the time indicated. The levels of H2B and GAPDH were analyzed by western blot following lysis of yeast by SDS sample buffer.
See also Figure S6.
To further confirm these results, we sought to explore the role of the PA200 ortholog in S. cerevisiae, Blm10, in the histone degradation in response to DSBs. In diploid yeast, the joint treatment with MMS and the HDAC inhibitor, valproic acid (VPA), also dramatically reduced the levels of the core histone, H2B. Treatment with the proteasome inhibitor, MG132, or deletion of Blm10 blocked this reduction (Figures 4D and S6C). Thus, DNA double-strand breaks promote the acetylation-associated proteasomal degradation of the core histones, and PA200/Blm10 is required for this process in both yeast and mammalian somatic cells.
PA200/Blm10 Binds Acetylated Core Histones via Bromodomain-Like Regions
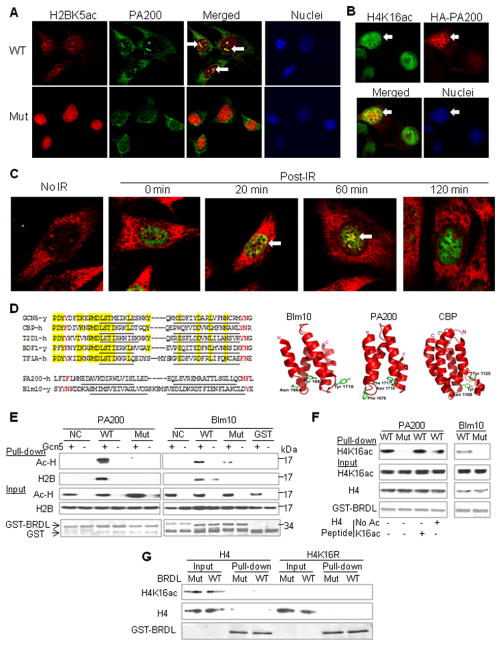
To test whether PA200 directly modulates the levels of the acetylated histones, cellular localization of PA200 was examined. In the wild-type MEF cells, an antiserum against PA200 recognized punctuate structures in the nuclei, where acetylated H2B (H2BK5ac) was also present. The staining of acetylated H2B in the PA200-deficient cells was much stronger than in the wild-type cells, a further indication that PA200 promotes the acetylation-dependent degradation of the core histones (Figure 5A). Since the PA200 antiserum showed some non-specific staining, especially in the cytosol (as seen in the PA200-deficient cells in Figure 5A), we transfected COS-7 cells with HA-tagged PA200 and demonstrated that HA-PA200 partially co-localized with acetylated H4 (H4K16ac) (Figure 5B). These results suggest that PA200 co-localizes with acetylated core histones in the nuclei. Furthermore, we examined whether PA200 is recruited to DNA damage loci following γ-irradiation. In the wild-type MEF cells treated with TSA, γ-H2AX was induced by γ-irradiation, and co-localized with PA200 in nuclei at 20 min and 60 min post-irradiation (Figure 5C), when the core histones were degraded as shown in Figure 4C. These findings raised the possibility that acetylation serves as a marker on the core histones and is directly recognized by PA200/Blm10 at the loci of DNA double-strand breaks.
Figure 5. PA200/Blm10 Binds Acetyllysine Residue in Core Histones via BRD-Like Regions.
(A) Co-localization of PA200 with H2BK5ac. In the wild-type mouse embryonic fibroblast (MEF) cells, PA200 was visualized with an antiserum against PA200 from rabbit (green), and H2BK5ac was detected by a specific antiserum from mouse (red), while nuclei were stained with DAPI. One of co-localization loci in each cell was indicated by an arrow. At least 20 cells were analyzed, and similar results were obtained for almost all the cells. The PA200-deficient MEF cells (Mut) served as controls for the specificity of the anti-PA200 antiserum.
(B) Partial co-localization of transfected HA-PA200 with H4K16ac. COS-7 cells were transfected with the N-terminally HA-tagged PA200, and immunofluorescence staining was carried out using anti-HA (mouse) and anti-H4K16ac (rabbit). A representative of about 20 cells transfected with HA-PA200 was indicated by an arrow.
(C) PA200 is recruited to DNA damage loci following γ-irradiation. The wild-type MEF cells were pre-treated with 0.3 μM of TSA for 2 h, irradiated by a 60Co gamma irradiator for 15 min (1 Gy/min), and then incubated for 0, 20, 60, or 120 min. PA200 was visualized with an antiserum against PA200 from rabbit (red), and γ-H2AX was detected by a specific monoclonal antibody from mouse (green). Cells with co-localized PA200 and γ-H2AX (yellow) were indicated by arrows. In control, the cells were not irradiated before immunostaining. The results were representative of more than 16 cells for each treatment.
(D) BRDL regions in PA200/Blm10. Left panels, alignment of the regions containing critical hydrophobic residues in the BRD-like (BRDL) regions of PA200/Blm10 with those in known BRDs from yeast Gcn5, human CBP, human T2D1, yeast BDF1, and human TF1A. α-Helices were underlined, the highly conserved residues were shaded in yellow, and the potential acetyllysine- recognizing residues were in red. Right panel, 3D structure of BRDL regions in yeast Blm10 and human PA200 in comparison with the BRD in human CBP (Dhalluin et al., 1999).
(E) BRD-like regions specifically bind acetyl-lysine on core histones in vitro. GST-fused BRDL regions from PA200/Blm10 (WT) and their mutants (N1716T/F1717S in PA200 and Y1663H/N1664D in Blm10) (Mut) were expressed and purified from E. coli, and incubated with the histones, which were acetylated by Gcn5 HAT domain. Non-relevant regions of Blm10 (aa1980–2073) and PA200 (aa1296–1377) served as negative controls (NC). Following a pull-down assay using GSH-beads, acetylated histones (Ac-H) and H2B were analyzed by western blot with an anti-acetyllysine antibody and an anti-H2B antibody, respectively. GST fusion proteins were stained by Coomassie blue.
(F) H4K16ac derived in HeLa cells binds BRD-like regions. Acetylated histones were purified from HeLa cells. The H4 peptide (aa1–21) with or without acetylation at K16 was included as indicated at ~200-fold of histone molecules. GST pulldown experiments were carried out as in (E), and H4K16ac was analyzed with a specific anti-H4K16ac antibody.
(G) Acetylation at K16 is not sufficient for bacterially-expressed histone H4 to bind the BRDL region. The bacterially-expressed wild-type H4 or H4K16R was acetylated by TIP60, and incubated with the GST-BRDL of PA200 or its mutant for a GST pulldown assay.
See also Figure S7.
Therefore, we searched for acetylation-recognizing regions in PA200/Blm10. The acetyllysine-binding bromodomain (BRD) usually comprises a left-handed bundle of four α helices with two adjacent hydrophobic loops (ZA and BC loops), where acetyl-Lys is anchored to an Asn residue (Filippakopoulos et al., 2012) (Figure 5D). Based on its crystal structure (Sadre-Bazzaz et al., 2010), Blm10 contains a BRD-like (BRDL) region at aa1648-1732, which forms 4 similar α-helices with critical hydrophobic residues (Tyr1663Asn1664/ Tyr1710) in the adjacent loops. Human PA200 is predicted to have a similar region of aa1650-1738 with Phe1676/ Asn1716 Phe1717. Unlike typical BRDs, which share many other conserved residues (Filippakopoulos et al., 2012), the BRDL regions in PA200 and Blm10 do not share any significant sequence homology with known BRDs (Figure 5D).
To test whether these regions in PA200/Blm10 bind acetylated histones, we expressed and purified this domain as a GST-fusion protein. Histones were isolated from rabbit thymus, and were further acetylated by the HAT domain of Gcn5 in vitro. The acetyl-histones could be specifically pulled down with the BRDL region aa1648-1732, but not a non-relevant region, of Blm10. Furthermore, the mutations at this region (Y1663H/N1664D) abolished the association (Figure 5E). Similarly, the BRDL region aa1650-1738 of human PA200, but not its corresponding mutant form (N1716T/F1717S) or a non-relevant region of PA200, bound acetyl-histones directly (Figures 5E and S7A-B). Although the nature of the acetylated histones pulled down with the BRDL regions remained to be further characterized, an anti-H2B antibody recognized a band from acetylated histones at ~14-kDa, suggesting that acetylation on H2B was required for binding the BRDL regions (Figure 5E).
As shown above, acetylation of histone H4 at K16 (H4K16ac) was involved in histone degradation. To further validate these results, we purified the naturally-acetylated histones from HeLa cells. After incubating them with GST-BRDL of PA200 or Blm10, H4K16ac could be specifically pulled down by the WT BRDL region, but not its mutant (Figure 5F). However, the H4 peptide with K16ac could not compete with the full-length histone in this assay (Figure 5F), hinting that additional posttranslational modifications might be required. Bacteria lack most posttranslational modifications of proteins in mammalian cells. Therefore, we expressed and purified the full-length histone H4 from bacteria, and acetylated it at K16 in vitro using the HAT, TIP60, but the BRDL region of PA200 could not bind this type of acetylated H4 (Figure 5G). Accordingly, the BRDL regions of PA200 and Blm10 also could not bind the N-terminal histone peptides (including H4K16ac [aa1-21]), which were only modified by acetylation at a single Lys residue (data not shown). Thus, the BRD-like regions of Blm10 and PA200 can bind acetyl-histones in vitro, but additional posttranslational modifications of histones might be required to assist the binding.
PA200/Blm10 Targets Acetyl-Histones for Proteasomal Degradation
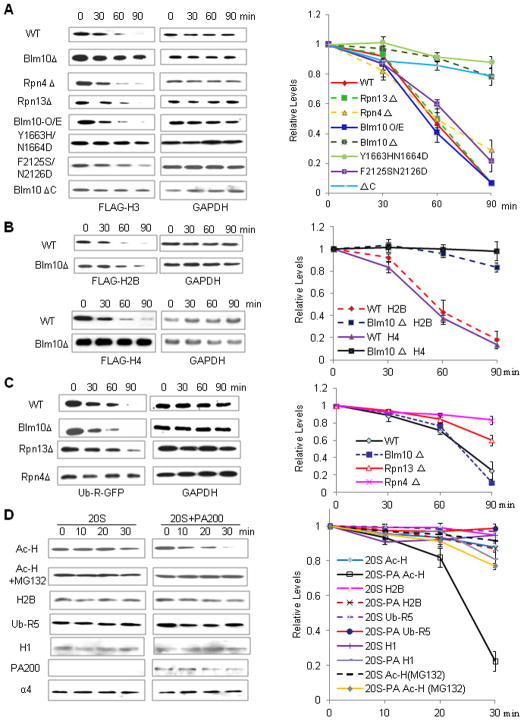
Excess histones in a cell can cause genome instability (Singh et al., 2009). Delayed disappearance of histones in elongated spermatids or release of histones from DSB sites should resemble ectopic overexpression of histones, which in yeast leads to an accumulation of excess histones (Singh et al., 2009). Therefore, the FLAG-tagged core histones were inducibly overexpressed in budding yeast to examine the mechanisms for the Blm10/PA200-mediated degradation of the core histones. Unlike deletion of the 19S subunits, Rpn4 and Rpn13, deficiency of Blm10 stabilized the ectopically-expressed histone H3 (Figures 6A and S7C–D). Overexpression of the HA-tagged Blm10 by itself did not promote FLAG-H3 degradation (Figure 6A), suggesting that the endogenous Blm10 was sufficient for the destruction of the excess histones. Furthermore, mutations (Y1663H/N1664D) or deletion (Blm10 ΔC) of the BRD-like region in Blm10 stabilized FLAG-H3 (Figure 6A). Similarly, deletion of Blm10 also stabilized the ectopically-expressed FLAG-tagged histones H2B and H4 (Figure 6B). However, unlike Rpn4 or Rpn13, Blm10 was not required for the ubiquitin-dependent degradation of the N-end rule substrate, ubiquitin-R-GFP (Green fluorescent protein) (Figure 6C).
Figure 6. PA200/Blm10-Containing Proteasomes Selectively Degrade Acetylated Core Histones.
(A) Blm10 targets the ectopically expressed H3 for degradation via BRD-like regions. Wild-type BY4741 (WT) or mutant yeast carrying the pHHF1-Gal-10/1-FLAG-HHT1 plasmids encoding the galactose-inducible FLAG-tagged H3 was used to perform a histone degradation assay, analyzed by immunoblotting, and quantified by densitometry. The relative levels of histones were obtained by normalizing to the loading control (GAPDH).
(B) Blm10 deficiency stabilizes ectopically expressed H2B and H4. Wild-type BY4741 (WT) or mutant yeast carrying the inducible H2B or H4 was constructed and analyzed as in (A).
(C) Blm10 deficiency does not stabilize ectopically expressed Ub-R-GFP. Wild-type or mutant yeast carrying the galactose-inducible C-terminally His-tagged Ub-R-GFP was used to perform a degradation assay as in (A).
(D) PA200 specifically stimulates degradation of acetylated core histones. Purified 20S particle was incubated with the acetylated histones (Ac-H), unmodified histones (monitored by H2B), or poly-ubiquitinated RNF5 (Ub-R5) in the absence or presence of the purified PA200 for the time as indicated. The levels of the substrates were analyzed by immunoblotting and quantified by densitometry (normalized to the proteasome subunit α4).
See also Figure S7.
To exclude the possibility that PA200/Blm10 is indirectly involved in histone degradation (e.g., in proteasome assembly), we purified PA200 from bovine testes by traditional chromatography (Figure S7E). The purified PA200 greatly promoted degradation of the acetylated core histones by the 20S particles in the absence of ATP (Figure 6D). Since the proteasome inhibitor MG132 blocked this degradation, the observed effect should not be caused simply by deacetylation. In contrast, PA200 stimulated degradation of unmodified H2B only slightly, and had almost no effect on degradation of the polyubiquitinated RNF5 (Figure 6D). Thus, PA200/Blm10 appears to target the acetylated histones for proteasomal degradation in a polyubiquitination-independent manner.
DISCUSSION
PA200 Is Essential for Programmed Degradation of Core Histones during Spermatogenesis
In mammals, the proteasomes bearing one or two PA200 in the regulatory particles and β1i, β2i, β5i, and/or α4s in the 20S particle are testis-specific. Unlike the other alternative 20S subunits (Murata et al., 2007), α4s is in the outer α-ring and thus lacks catalytic activity, and might preferentially interact with regulatory complexes such as PA200. Possibly, these unique properties might allow the development of drugs against certain testis tumors or even male contraceptives that specifically target sperm development by blocking the proteasomes essential for spermatogenesis.
An excess of histones blocks transcription, leads to enhanced DNA-damage sensitivity, and triggers chromosome aggregation or loss (Singh et al., 2009). Delayed disappearance of the core histones in elongated spermatids in PA200-deficient mice also led to accumulation of the core histones and induced apoptosis (Figure 3). Thus, our findings provide a mechanism for the observation that deletion of PA200 leads to malformed spermatids or sperm in mouse testes, and thereby dramatically reduces fertility in male mice (Khor et al., 2006).
Acetylation and PA200 Mediate Polyubiquitin-Independent Proteasomal Degradation of Core Histones
The core histones are removed from the vicinity of double-strand DNA breaks in yeast (Tsukuda et al., 2005), and PA200 accumulates on chromatin during DNA repair (Blickwedehl et al., 2008). We show that the elevated acetylation (induced by HDAC inhibitors) enhanced core histone degradation upon DNA damage in both yeast and mammalian somatic cells. Phosphorylation of histones by Rad53 is required for degradation of excess histones in yeast (Singh et al., 2009), and monoubiquitination of histones catalyzed by the ubiquitin ligase, RNF8, promotes the replacement of histones by protamines during spermiogenesis (Huen et al., 2007; Lu et al., 2010). This study demonstrates that the BRD-like regions in PA200/Blm10 bind the acetylated histones, but other uncharacterized protein modifications are probably also required. Many BRDs bind the combined marks, and the flanking modifications, such as acetylation and phosphorylation, can strongly influence the binding to the acetylated site (Filippakopoulos et al., 2012). Thus, acetylation mediates the proteasomal degradation of the core histones, presumably with the assistance of other posttranslational modifications.
Numerous HDAC inhibitors are under investigation in clinical trials as anticancer agents, especially in conjunction with other treatments such as chemotherapy and radiation therapy (Shabason et al., 2011). We demonstrate here that HDAC inhibitors potentiate the acetylation-mediated histone degradation induced by radiation or the DNA-damaging agent MMS, providing a mechanism that may contribute to their clinical applications.
A Model for Core Histone Degradation during Spermatogenesis and Somatic DNA Repair
As proposed in Figure 7A, during spermatogenesis, the core histones in postmeiotic spermatids 1) undergo acetylation and probably other uncharacterized posttranslational modifications, 2) are recognized by the BRD-like region in PA200, 3) and are degraded by spermatoproteasomes, 4) leading to their replacement by the transition proteins, which are eventually replaced by protamines. In response to DNA double strand breaks (Figure 7B), the core histones near the damage sites on DNA in somatic cells are, at least partly, also acetylated and degraded by the PA200/Blm10-containing proteasomes so that DNA repair proteins can reach the damage sites. In addition, it is possible that other co-factors might function together with PA200/Blm10 to fulfill this task and promote histone degradation in vivo.
Figure 7. Models for Acetylation-Mediated Degradation of Core Histones.
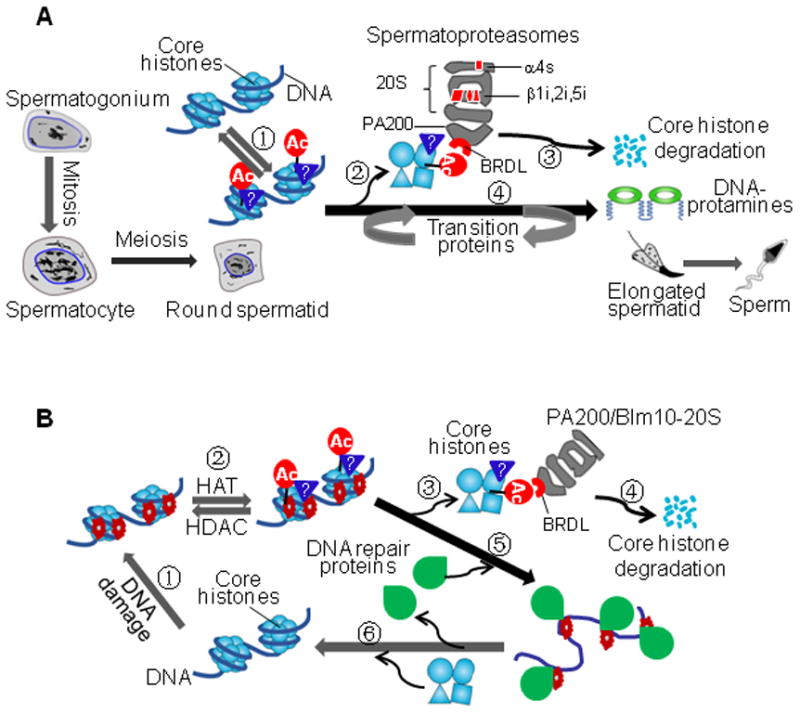
(A) Core histone degradation by spermatoproteasomes during spermatogenesis. During elongation of haploid spermatids, the BRD-like (BRDL) region in PA200 recognizes the core histones with acetylation and other uncharacterized posttranslational modifications, dissociates the histones from the nucleosome, and leads to cleavage of the core histones into small peptides. Meanwhile, transition proteins are recruited into the chromatin, and are eventually replaced by protamines.
(B) Coupling of core histone degradation with somatic DNA repair. DNA double-strand breaks trigger acetylation and other uncharacterized posttranslational modifications on the core histones. Targeting and release of core histones would allow DNA repair proteins to fix the damaged DNA. Meanwhile, the acetylated core histones are, at least partly, degraded by the PA200/Blm10-containing proteasomes. Following repair of the damaged DNA, the newly synthesized core histones join the DNA to form new nucleosomes.
About 4% of the histones on the male haploid genome are retained in mature sperm in nucleosomes, which contain either the standard histones or variants (Hammoud et al., 2009). Epigenetic information encoded in these retained nucleosomes is passed onto the next generation, while nearly all the other core histones are destroyed. The removal of these core histones near the sites of DNA damage seems to be critical for efficient repair of the damaged DNA. Thus, this acetylation-mediated degradation of the core histones may help assure accurate passing of the epigenetic information in sperm to next generations and maintaining the genetic information after somatic DNA damage.
Histone acetylation plays critical roles in epigenetic regulation of gene expression. We demonstrate that acetylation on the lysine residue can also serve as a signal for proteasomal degradation. Almost all known BRDs, which recognize the acetyllysine residue, share a moderate sequence homology (Filippakopoulos et al., 2012). Although the BRDL regions of PA200/Blm10 structurally resemble BRD, they share almost no sequence homology with any known BRDs. Our findings may be important in identifying other new acetyllysine-binding proteins bearing BRD-like regions. It is attractive to speculate that some non-histone proteins may also be targeted for acetylation-mediated degradation, if their acetylation can be recognized by PA200/Blm10.
EXPERIMENTAL PROCEDURES
Details of proteasome purification, recombinant protein production, mutant mouse and yeast generation, regular or electron microscopy, GST pull-down, protein degradation assays, and immuno-fluorescent or -histochemical analysis are available in the Extended Experimental Procedures. Briefly, purification of proteasomes and PA200 from bovine or rabbit tissues was carried out as described (Qiu et al., 2006; Ustrell et al., 2005) with some modifications. IP, western blot, histone purification, and proteasome activity analysis were performed according to established protocols or their adaptations.
Supplementary Material
01
02
HIGHLIGHTS.
- Most proteasomes in mammalian testes are “spermatoproteasomes” that contain PA200
- Deletion of PA200 in mice delays core histone disappearance in elongated spermatids
- PA200/Blm10 promotes degradation of acetyl-histones during somatic DNA damage
- Binding of histones to bromodomain-like regions in PA200/Blm10 depends on acetylation
Acknowledgments
We thank D. Finley for critical comments on the manuscript, Y. Cong, D. Finley, and O. J. Rando for reagents, T. Walz for providing the original EM images of Blm10 and PA28, H. Matsumoto and C. Xiong for technical assistance, as well as O. Gozani, D. Patel, H. Wang, and Z. Wang for assistance in the acetyl-peptide binding assay. T.C. thanks K. Tanaka for support and encouragement. This work was supported by grants from the Ministry of Science and Technology of China (2012CB910300), the National Natural Science Foundation of China (30525033 and 30730024), and the Fundamental Research Funds for the Central Universities of China to X.-B. Q., a National Basic Research Program of China (2012CB518700) to C.H. L., grants-in-aid for Scientific Research on Priority Areas, MEX, Japan to T.C., and a grant from NIGMS (5R01 GM51923) to A.L.G.
Footnotes
Supplemental Information includes 7 figures and Extended Experimental Procedures, which can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blickwedehl J, Agarwal M, Seong C, Pandita RK, Melendy T, Sung P, Pandita TK, Bangia N. Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability. Proc Natl Acad Sci U S A. 2008;105:16165–16170. doi: 10.1073/pnas.0803145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Cascio P, Call M, Petre BM, Walz T, Goldberg AL. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J. 2002;21:2636–2645. doi: 10.1093/emboj/21.11.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YS, Qiu XB. Transcription-coupled replacement of histones: degradation or recycling? J Genet Genomics. 2012;39:575–580. doi: 10.1016/j.jgg.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Dange T, Smith D, Noy T, Rommel PC, Jurzitza L, Cordero RJ, Legendre A, Finley D, Goldberg AL, Schmidt M. Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism. J Biol Chem. 2011;286:42830–42839. doi: 10.1074/jbc.M111.300178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert J, Barsyte-Lovejoy D, Felletar I, Volkmer R, Müller S, Pawson T, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher J, Reynoird N, Montellier E, Boussouar F, Rousseaux S, Khochbin S. From meiosis to postmeiotic events: the secrets of histone disappearance. FEBS J. 2010;277:599–604. doi: 10.1111/j.1742-4658.2009.07504.x. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor B, Bredemeyer AL, Huang CY, Turnbull IR, Evans R, Maggi LB, Jr, White JM, Walker LM, Carnes K, Hess RA, et al. Proteasome activator PA200 is required for normal spermatogenesis. Mol Cell Biol. 2006;26:2999–3007. doi: 10.1128/MCB.26.8.2999-3007.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev AF, Akopian TN, Goldberg AL. Range of sizes of peptide products generated during degradation of different proteins by archaeal proteasomes. J Biol Chem. 1998;273:1982–1989. doi: 10.1074/jbc.273.4.1982. [DOI] [PubMed] [Google Scholar]
- Kotaja N, Kimmins S, Brancorsini S, Hentsch D, Vonesch JL, Davidson I, Parvinen M, Sassone-Corsi P. Preparation, isolation and characterization of stage-specific spermatogenic cells for cellular and molecular analysis. Nat Methods. 2004;1:249–254. doi: 10.1038/nmeth1204-249. [DOI] [PubMed] [Google Scholar]
- Li X, Lonard DM, Jung SY, Malovannaya A, Feng Q, Qin J, Tsai SY, Tsai MJ, O’Malley BW. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell. 2006;124:381–392. doi: 10.1016/j.cell.2005.11.037. [DOI] [PubMed] [Google Scholar]
- Liu Z, Miao D, Xia Q, Hermo L, Wing SS. Regulated expression of the ubiquitin protein ligase, E3(Histone)/LASU1/Mule/ARF-BP1/HUWE1, during spermatogenesis. Dev Dyn. 2007;236:2889–2898. doi: 10.1002/dvdy.21302. [DOI] [PubMed] [Google Scholar]
- Lu LY, Wu J, Ye L, Gavrilina GB, Saunders TL, Yu X. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell. 2010;18:371–384. doi: 10.1016/j.devcel.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills NC, Van NT, Means AR. Histones of rat testis chromatin during early postnatal development and their interactions with DNA. Biol Reprod. 1977;17:760–768. doi: 10.1095/biolreprod17.5.760. [DOI] [PubMed] [Google Scholar]
- Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Ortega J, Heymann JB, Kajava AV, Ustrell V, Rechsteiner M, Steven AC. The axial channel of the 20S proteasome opens upon binding of the PA200 activator. J Mol Biol. 2005;346:1221–1227. doi: 10.1016/j.jmb.2004.12.049. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Goldberg AL. Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc Natl Acad Sci U S A. 2002;99:14843–14848. doi: 10.1073/pnas.232580999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- Sadre-Bazzaz K, Whitby FG, Robinson H, Formosa T, Hill CP. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol Cell. 2010;37:728–735. doi: 10.1016/j.molcel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Walz T, Finley D. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat Struct Mol Biol. 2005;12:294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- Shabason JE, Tofilon PJ, Camphausen K. Grand rounds at the National Institutes of Health: HDAC inhibitors as radiation modifiers, from bench to clinic. J Cell Mol Med. 2011;15:2735–2744. doi: 10.1111/j.1582-4934.2011.01296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11:925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustrell V, Pratt G, Gorbea C, Rechsteiner M. Purification and assay of proteasome activator PA200. Methods Enzymol. 2005;398:321–329. doi: 10.1016/S0076-6879(05)98026-9. [DOI] [PubMed] [Google Scholar]
- Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Zhu B, Reinberg D. Epigenetic inheritance: uncontested? Cell Res. 2011;21:435–441. doi: 10.1038/cr.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02