The stoichiometry of the nucleoporin 62 subcomplex of the nuclear pore in solution (original) (raw)
The stoichiometry of the nucleoporin 62 (Nup62) subcomplex of the nuclear pore complex is investigated in solution using gel filtration and analytical ultracentrifugation. The Nup58-Nup54-Nup62 complex assembles in a 1:1:1 ratio. Different stoichiometries are obtained only once the interacting, predicted, coiled-coil domains are fragmented.
Abstract
The nuclear pore complex (NPC) regulates transport between the nucleus and cytoplasm. Soluble cargo-protein complexes navigate through the pore by binding to phenylalanine-glycine (FG)-repeat proteins attached to the channel walls. The Nup62 complex contains the FG-repeat proteins Nup62, Nup54, and Nup58 and is located in the center of the NPC. The three proteins bind each other via conserved coiled-coil segments. To determine the stoichiometry of the Nup62 complex, we undertook an in vitro study using gel filtration and analytical ultracentrifugation. Our results reveal a 1:1:1 stoichiometry of the Nup62 complex, where Nup54 is central with direct binding to Nup62 and Nup58. At high protein concentration, the complex forms larger assemblies while maintaining the Nup62:Nup54:Nup58 ratio. For the homologous Nsp1 complex from Saccharomyces cerevisiae, we determine the same stoichiometry, indicating evolutionary conservation. Furthermore, we observe that eliminating one binding partner can result in the formation of complexes with noncanonical stoichiometry, presumably because unpaired coiled-coil elements tend to find a promiscuous binding partner. We suggest that these noncanonical stoichiometries observed in vitro are unlikely to be physiologically relevant.
INTRODUCTION
In the eukaryotic cell, the genetic information is stored and transcribed in the nucleus, while mRNA translation into proteins occurs in the cytoplasm. As a result, a large number and diverse set of molecules has to be transported across the double-layered membrane that is the nuclear envelope (NE). Nuclear pore complexes (NPCs) are the essential transport gates that sit in circular openings in the NE. In all eukaryotes examined, NPCs are built from multiple copies of ∼30 nucleoporins (nups; Rout et al., 2000; Cronshaw, 2002), which are arranged around a central eightfold rotational axis (Gall, 1967; Maul, 1971; Unwin and Milligan, 1982; Onischenko and Weis, 2011). The overall architecture of the NPC is conserved between eukaryotes as shown by electron microscopy (EM; Reichelt et al., 1990) and cryoEM (Akey and Radermacher, 1993; Beck et al., 2007; Frenkiel-Krispin et al., 2010). The cylindrical NPC has an outer diameter of ∼125 nm surrounding an inner pore of ∼50 nm, and a height of 60–95 nm without its asymmetrical nuclear and cytoplasmic extensions, the “nuclear basket” and “cytoplasmic filaments.” Many nups are conserved across all eukaryotes (DeGrasse et al., 2009; Neumann et al., 2010) and can be roughly categorized into three groups: membrane nups anchor the NPC to the NE, scaffold nups form the framework of the NPC, and barrier nups build the permeability barrier for selective transport (Grossman et al., 2012). In recent years, x-ray structures of individual nups and their subcomplexes have been obtained, facilitated by the modular assembly of the complex (Brohawn et al., 2009). These structures revealed common structural features between the NPC scaffold and endomembrane trafficking coats (COPI, COPII, clathrin) that appeared early in the evolution of the eukaryotic endomembrane system (Devos et al., 2004, 2006; Mans et al., 2004; Brohawn et al., 2008).
The NPC scaffold consists of four major subcomplexes. The best-characterized of these is the multimeric, ∼0.5-MDa Y complex, so named because of its characteristic shape (Lutzmann, 2002; Kampmann and Blobel, 2009; Bilokapic and Schwartz, 2012; Fernandez-Martinez et al., 2012). The Y complex consists of 7–10 members, depending on the organism, and is the main scaffolding unit of the NPC, essential for its assembly (Harel et al., 2003; Walther et al., 2003). The Ndc1 subcomplex contains membrane-bound nups that anchor the NPC scaffold into the circular openings of the NE (Onischenko et al., 2009). It binds directly to the Nup93 subcomplex (Nehrbass et al., 1996), which acts as an adaptor between the NE and the central pore facing the Nup62 subcomplex (Nsp1 subcomplex in Saccharomyces cerevisiae; Marelli, 1998; Amlacher et al., 2011).
The most central, pore-facing subcomplex is the trimeric Nup62 complex, which connects to the NPC scaffold by direct coiled-coil interaction with Nup93 (Nic96 in S. cerevisiae; Grandi et al., 1995b; Bailer et al., 2001; Schrader et al., 2008). The three Nup62 complex members—Nup62, Nup54, and Nup58—have been localized to the center of the NPC by immunoelectron microscopy (Guan et al., 1995; Hu et al., 1996). The yeast homologues Nsp1 (Carmo-Fonseca et al., 1991; Buss and Stewart, 1995), Nup57, and Nup49 are essential for cell viability (Hurt, 1988; Wente et al., 1992; Grandi et al., 1995b). Each of the three Nup62-complex members contains a coiled-coil domain that connects the binding partners (Bailer et al., 2001; Melcák et al., 2007). The coiled-coil domain is flanked N-terminally (Nup54, Nup62) or both N- and C-terminally (Nup58) by unstructured phenylalanine–glycine (FG)-repeat regions (Wente et al., 1992; Hu et al., 1996). The sequences of the coiled-coil domains are relatively well conserved, in contrast to the FG-repeat regions (Supplemental Figure S1). The FG repeats are necessary for the main function of the NPC: the selective transport through the nuclear membrane (Mohr et al., 2009). The central transport pore of the NPC is filled with protein filaments containing FG repeats, which are essential for nuclear transport (Finlay et al., 1991). In yeast, a total of ∼160 individual FG nucleoporins are located at the inner pore of the NPC (Alber et al., 2007). Small molecules (<40 kDa) can diffuse freely through the pore. Larger cargoes require active transport, facilitated by soluble nuclear transport receptors that engage in temporary binding to FG repeats (Cook et al., 2007; Chook and Süel, 2011).
To unravel the composition and organization of the FG network that fills the central pore of the NPC, it is very important to gain a more detailed picture of the transport process. Questions concern the anchor points for different FG-proteins, the nature of the FG repeats (cohesive, noncohesive, etc.), and the copy numbers and stoichiometries of the individual FG-nups. For the Nup62 complex, various studies have attempted to address its composition, but the results are inconsistent. Using native Nup62 complex purifications from rat liver nuclei and subsequent size exclusion chromatography and gel electrophoresis, researchers reported Nup58-Nup54-Nup62 ratios of 1:4:4 (Finlay et al., 1991), 1:2:1 (Kita et al., 1993), and 2:4:2 (Buss and Stewart, 1995). Guan and colleagues also used purified native rat protein but applied cross-linking and scanning transmission electron microscopy (STEM) analysis to estimate a molar ratio of 1:1:1:1 (including the Nup58 splice variant Nup45; Guan et al., 1995). In Xenopus egg extracts, an equimolar ratio of purified Nup62 subcomplex was reported (Macaulay et al., 1995). Other studies reported ratios based on purifications of entire rat nuclei of Nup45:Nup58:Nup54:Nup62 equal to 2:3:2–3:1 by quantitative two-dimensional gel electrophoresis (Cronshaw, 2002). In yeast nuclei, the Nup49:Nup57:Nsp1 complex stoichiometry was measured to be 1:1:2 by quantitative immunoblotting (Rout et al., 2000); the same ratio was reported by a recent targeted proteomic measurement of the human NPC (Ori et al., 2013). Taking into account that Nup62/Nsp1 is also present in the Nup62-Nup88-Nup214 subcomplex (Nsp1-Nup82-Nup159 in yeast), two studies support an isostoichiometric occurrence of Nup62:Nup54:Nup58 within the Nup62 subcomplex (Grandi et al., 1995a; Belgareh et al., 1998). In a computational model of the yeast NPC, an Nsp1-complex stoichiometry of 1:1:1 was predicted (Alber et al., 2007). The first crystal structure of a member of the Nup62 complex was published in 2007 (Melcák et al., 2007). An 85-residue fragment of the coiled-coil region of rat Nup58 (aa 327–411) formed a tetrameric helical bundle in the crystal, consisting of a dimer of dimers. Structural differences between Nup58 crystallized in two independent crystal forms, of which only one is publicly available in the Protein Data Bank (PDB 2OSZ), were interpreted as possibly representing a sliding mechanism of Nup58 that alters the diameter of the transport channel. The ability of individual FG-nups to form oligomers was also observed for Nup62, forming linear fibers visible by EM (Buss et al., 1994), and for a 35-residue Nup54 coiled-coil fragment (aa 460–494) observable by a crystal structure (Solmaz et al., 2013). Solmaz and colleagues also reported the crystal structures of isolated coiled-coil fragments of a Nup54 (aa 346–402)–Nup62 (aa 364–419) complex and of a Nup58 (aa 327–412)–Nup54 (aa 456–494) complex (Solmaz et al., 2011). In both cases, the asymmetric unit contained a 1:2 heterotrimer. Taking all the crystallographic data together, an overall ratio of Nup58-Nup54-Nup62 of 1:2:4 was suggested; however, it was never confirmed using an independent approach.
Because the biochemically obtained Nup62 complex ratios (Finlay et al., 1991; Kita et al., 1993; Buss and Stewart, 1995; Guan et al., 1995; Rout et al., 2000; Cronshaw, 2002) are not consistent among each other and also differ substantially from the ratio deduced from the crystal structures (Melcák et al., 2007; Solmaz et al., 2011, 2013), we set out to solve this ambiguity. We determined the stoichiometry of the Nup62 complex in solution; we purified recombinant trimeric Rattus norvegicus (rn) Nup62 complex and the homologous S. cerevisiae (sc) Nsp1 complex and analyzed its molecular weight and oligomerization state by size exclusion chromatography and sedimentation equilibrium analytical ultracentrifugation.
RESULTS
Composition of the Nup58-Nup54-Nup62 complex
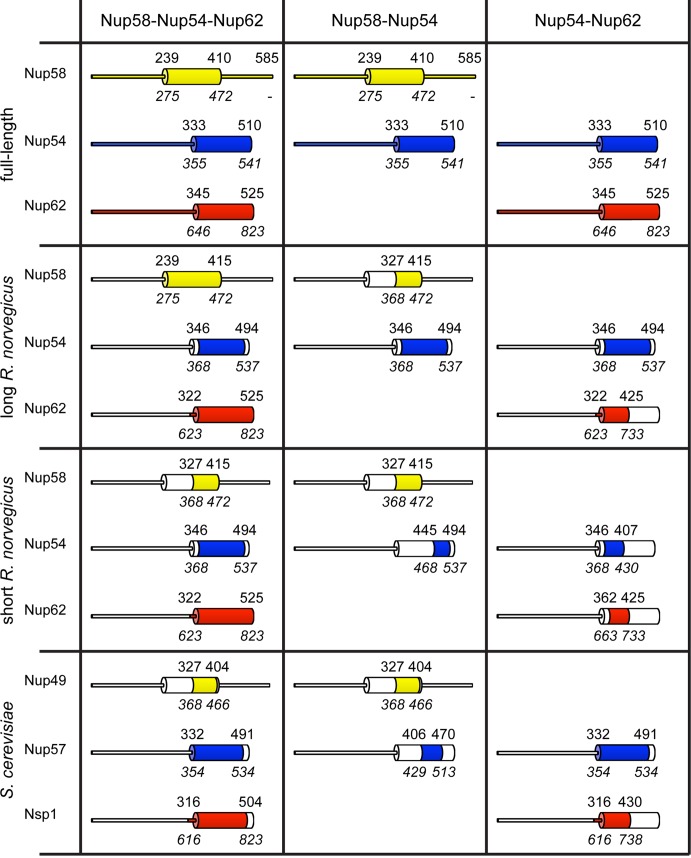
To determine the molecular composition of the rnNup58-Nup54-Nup62 complex, we coexpressed and copurified the predicted α-helical portions of the three proteins. To study the evolutionary conservation, we also studied the homologous scNup49-Nup57-Nsp1 complex (Figures 1 and S1). The “long” rnNup58-Nup54-Nup62 complex contains the entire predicted α-helical parts of Nup58 (aa 239–415), Nup54 (aa 346–494), and Nup62 (aa 322–525). N-terminally truncated Nup58 (aa 327–415) can still form a trimeric “short” rnNup58-Nup54-Nup62 complex, which exhibits higher solubility. A homologous “short” scNup49-Nup57-Nsp1 construct was designed based on the high-solubility “short” rnNup58-Nup54-Nup62 complex (Figures 1 and S1).
FIGURE 1:
Protein complexes used in this study. Definitions of all Nup58-Nup54-Nup62, Nup58-Nup54, and Nup54-Nup62 complexes used in our study. Narrow tubes represent FG-repeat regions; broad cylinders represent α-helical, predicted coiled-coil domains. Numbers indicate residue positions in the R. norvegicus proteins. Numbers in italics indicate protein residues in the S. cerevisiae homologues. Regions present in the particular complex are colored. The scNup49-Nup57-T4 Lysozyme complex contains, in addition to the residues of the scNup49-Nup57 complex, T4 Lysozyme N-terminally fused to Nup57.
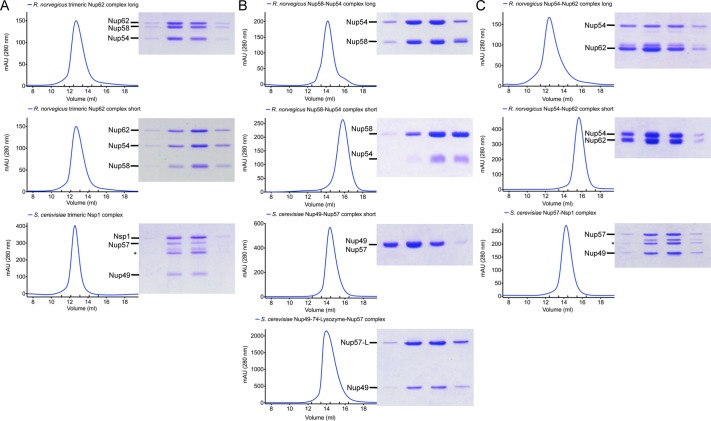
All three heterotrimeric complexes were analyzed by gel filtration (Figure 2A). They elute as single peaks from a gel-filtration column, without any significant signs of complex disassembly during the experiment. Based on visual observation of the peak fractions on Coomassie blue–stained SDS–PAGE gels, all samples appear homogenous in composition and have a protein ratio of 1:1:1 (Figure 2A; scNup57 is partially proteolyzed).
FIGURE 2:
Final purity of protein complexes used in AUC experiments. Size exclusion chromatograms and SDS–PAGE gels of protein complexes used in AUC experiments. Proteins were purified as described in Materials and Methods. Final purity as obtained after gel filtration with S200 10/300 column of (A) Nup58-Nup54-Nup62 complexes, (B) Nup58-Nup54 complexes, and (C) Nup54-Nup62 complexes, shown by size exclusion chromatograms and SDS–PAGE gels. Nup57 autodegradation fragments are indicated with asterisks.
For accurate mass determination, we assayed the three trimeric complexes by sedimentation equilibrium analytical ultracentrifugation (SE-AUC). Fitted data with residuals for all samples are shown in Figure S2. The long rnNup58-Nup54-Nup62 complex was fitted about equally well with monomer-dimer (M-D), monomer-trimer (M-Tri), monomer-tetramer (M-Tetr), and also higher monomer-oligomer models. Fitting to a single-species model resulted in systematic errors, indicating the 1:1:1 complex can form higher-order oligomers. The molecular weights calculated from AUC data for the long rnNup58-Nup54-Nup62 complex were 59.1 kDa (M-D), 54.7 kDa (M-Tri), and 63.2 kDa (M-Tetr) and fit well to the expected molecular weight of 63.8 kDa of a 1:1:1 composition (Table 1). The short rnNup58-Nup54-Nup62 complex was successfully fitted with monomer-dimer and monomer-trimer models, again not with a single-species model. The calculated molecular weights were 61.3 kDa (M-D) and 55.0 kDa (M-Tri), respectively. These values correspond reasonably well to a 1:1:1 ratio with an expected molecular weight of 53.8 kDa. The dissociation constants for the monomer-dimer model, calculated with UltraScan II, of the long and short rnNup58-Nup54-Nup62 complexes were 52.0 and 49.3 μM, respectively.
TABLE 1:
Results of SE-AUC experiments.
| Complex | Expected MW in kDa (various protein ratios)a | Calculated MW in kDab | Fitting modelc | Rotor speedsd | SD of residuals | ||
|---|---|---|---|---|---|---|---|
| R. norvegicus trimeric Nup62 long | 63.8 (1:1:1) | — | — | 59.1 | M-D | 1–4 | 6.5711E-03 |
| 54.7 | M-Tri | 1–4 | 5.6120E-03 | ||||
| 63.2 | M-Tetr | 1–4 | 5.4685E-03 | ||||
| R. norvegicus trimeric Nup62 short | 53.8 (1:1:1) | — | — | 61.3 | M-D | 1–4 | 8.5899E-03 |
| 55.0 | M-Tri | 1–4 | 7.8944E-03 | ||||
| S. cerevisiae trimeric Nsp1 | 56.9 (1:1:1) | — | — | 51.7 | Single | 1–4 | 6.1343E-03 |
| 54.7 (1:1:1) | |||||||
| R. norvegicus Nup58-Nup54 long | 30.1 (1:1) | 42.8 (2:1) | 47.6 (1:2) | 53.2 | Single | 2–4 | 7.1562E-03 |
| R. norvegicus Nup58-Nup54 short | 18.6 (1:1) | 31.3 (2:1) | 24.6 (1:2) | 29.9 | Single | 2–4 | 5.6959E-03 |
| S. cerevisiae Nup49-Nup57 | 22.2 (1:1) | 34.7 (2:1) | 32.0 (1:2) | 32.3 | Single | 2–4 | 6.5178E-03 |
| S. cerevisiae Nup49-T4-L-Nup57 | 40.7 (1:1) | 53.2 (2:1) | 68.9 (1:2) | 48.3 | Single | 2–4 | 6.3640E-03 |
| R. norvegicus Nup54-Nup62 long | 31.0 (1:1) | 49.9 (2:1) | 43.2 (1:2) | 48.0 | M-Tri | 1–4 | 8.5302E-03 |
| 48.3 | M-Tetr | 1–4 | 7.8707E-03 | ||||
| R. norvegicus Nup54-Nup62 short | 16.4 (1:1) | 25.2 (2:1) | 24.1 (1:2) | 26.0 | Single | 2–4 | 5.2776E-03 |
| S. cerevisiae Nup57-Nsp1 | 35.8 (1:1) | 56.7 (2:1) | 50.7 (1:2) | 27.0 | Single | 2–4 | 6.2001 E-03 |
| 33.6 (1:1) | 54.5 (2:1) | 46.3 (1:2) |
The short scNup49-Nup57-Nsp1 complex was successfully fitted to a single-species model. The expected molecular weight of a 1:1:1 complex is 56.9 kDa, reasonably well matched by the experimentally determined molecular weight of 51.7 kDa (Table 1). As mentioned above, scNup57 degrades partially as visible on SDS–PAGE (Figure 2A). The smaller, more abundant of the two degradation fragments was identified via mass spectrometry to have a molecular weight of 18.4 kDa, 2.2 kDa shorter than nondegraded scNup57 (20.6 kDa), thereby reducing the complex weight to 54.7 kDa. Although AUC cannot resolve the size difference between the nondegraded and the partially fragmented scNup49-Nup57-Nsp1 complex, it does not impact the main result of this analysis. We show that the investigated rnNup62/scNsp1 complexes are organized in a 1:1:1 ratio, with rnNup58-Nup54-Nup62 exhibiting a modest tendency to former higher-order oligomers. This difference in oligomerization behavior might explain why the size exclusion peak of the yeast complex appears sharper than the peaks of the rat complexes (Figure 2A).
Binary subcomplexes reveal direct interactions within the Nup62/Nsp1 complex
To find how the three Nup62/Nsp1 components directly interact, we coexpressed and copurified all three binary combinations and tested for complex formation. rnNup54 can form a stable complex both with rnNup62 alone as well as with rnNup58 alone (Figure 2, B and C). On the other hand, rnNup58 and rnNup62 do not form a stable complex. The same behavior was seen when we assayed the yeast homologues (unpublished data). Thus we conclude that Nup54 in metazoa and Nup57 in yeast are in the center of the complex bridging Nup62 (scNsp1) and Nup58 (scNup49).
Composition of an incomplete Nup58-Nup54 complex
To identify the molecular ratio of the binary Nup58-Nup54 subcomplex, we eliminated Nup62 from the trimeric Nup58-Nup54-Nup62 complex (Figure 1). We also tested the published Nup58-Nup54 crystal construct (Solmaz et al., 2011). Both constructs were purified and both eluted in a single peak in gel filtration (Figure 2B). We further designed a minimal scNup49-Nup57 construct, stable on a size exclusion column. Interestingly, although the ends of the rnNup58/scNup49 fragment are very similar between the crystal structure and our minimal yeast construct, the rnNup54/scNup57 fragments are shifted (Figures 1 and S1). When aligned, the rnNup54 and the scNup57 fragments share a common core of only 25 residues. The minimal yeast constructs extends N-terminally by 39 additional residues, while the rat construct extends by 24 residues C-terminally (Figures 1 and S1). Purification of a yeast complex analogous to the published rnNup58-Nup54 crystal structure failed due to poor protein expression.
We again performed SE- AUC experiments with the three binary rnNup58-Nup54 and scNup49-Nup57 complexes. Fitted data with residuals for the samples are shown in Figure S2. Data of all three complexes fit a single-species model. The calculated molecular weight of the long rnNup58-Nup54 complex was 53.2 kDa. A 1:1 stoichiometry (30.1 kDa) was expected, assuming the binary complex is organized like the trimeric complex but without Nup62. However, the calculated mass is closest to a Nup58:Nup54 ratio of 1:2, suggesting that Nup54 can substitute for the missing Nup62. In contrast, the crystal Nup58-Nup 54 construct has a measured molecular weight of 29.9 kDa, which fits best with a Nup58:Nup54 ratio of 2:1 (31.3 kDa), rather than 1:2, as seen in the published crystal structure (PDB code 3T98). This 2:1 ratio is also supported by the relative band intensities on SDS–PAGE gels (Figure 2B). Thus, depending on how the fragments are chosen, different Nup58:Nup54 ratios can be obtained in vitro. Finally, the yeast Nup49-Nup57 complex is experimentally determined as 32.3 kDa, which could be explained by either a 2:1 (34.7 kDa) or a 1:2 stoichiometry (32.0 kDa) (Table 1). To increase the weight differences and thus better distinguish between the two stoichiometries, we fused T4 Lysozyme to the N terminus of scNup57. In this configuration, the complex size is most consistent with a 2:1 ratio (scNup49:scNup57), the same composition we also observed for the rat Nup58-Nup54 crystal construct. We conclude that, in absence of Nup62/scNsp1, the two remaining proteins Nup58/scNup49 and Nup54/scNup57 still form a trimeric complex, where Nup62/scNsp1 is substituted by either of the two remaining components, depending on the sequence context.
Composition of an incomplete Nup54-Nup62 complex
To analyze the molecular composition of an incomplete Nup54-Nup62 complex, we first eliminated Nup58 from the short rnNup58-Nup54-Nup62 complex. Although we were able to copurify the binary construct, the amounts were not sufficient for AUC analysis (Figure S3). However, we had previously determined a minimal scNup57-Nsp1 construct containing the full α-helical portion of scNup57 and a C-terminally truncated scNsp1 compared with scNsp1 in the scNup49-Nup57-Nsp1 construct (Figure 1). We translated this construct to rat (Figure S1) and expressed both complexes and obtained them in sufficient purity and amount for AUC analysis (Figure 2C). In addition, we used a construct equal to the rnNup54-Nup62 crystal construct previously used by Solmaz et al. (2011; Figure 1).
We performed SE-AUC experiments with these three subcomplexes. Fitted data with residuals for all samples are shown in Figure S2. The long rnNup54-Nup62 complex was successfully fitted to monomer-trimer and monomer-tetramer models. The resulting molecular weights were 48.0 kDa (M-Tri) and 48.3 kDa (M-Tetr). This matches a Nup54:Nup62 ratio of either 2:1 (49.9 kDa) or 1:2 (43.1 kDa) but not of 1:1 (31.0 kDa) (Table 1). Band intensities on SDS–PAGE gels support a Nup54:Nup62 ratio of 1:2 (Figure 2C). The crystal construct was successfully fitted with the single-species model. A molecular weight of 26.0 kDa was calculated, also indicating a Nup54:Nup62 ratio of 2:1 (25.2 kDa) or 1:2 (24.1 kDa) but not a 1:1 ratio (16.4 kDa). The composition of the rnNup54-Nup62 subcomplex is, in analogy to the rnNup58-Nup54 subcomplex, different from the composition of the three-protein complex. The rnNup54 surface involved in binding of the missing protein rnNup58 might be occupied by an additional copy of rnNup54 or, more likely, rnNup62. The yeast homologue of the long rnNup54-Nup62 complex was fitted to a single-species model and has a calculated molecular weight of 27.0 kDa. Different from the rat complex, this is indicating a 1:1 ratio with 35.8 kDa or 33.6 kDa (see degradation issue of Nup57, discussed above) rather than a 2:1 or 1:2 ratio. Taken together, incomplete rnNup54-Nup62 and scNup57-Nsp1 complexes exhibit different behavior in compensating for the missing rnNup58/scNup49 component.
Oligomerization state of the Nup58-Nup54-Nup62 complex
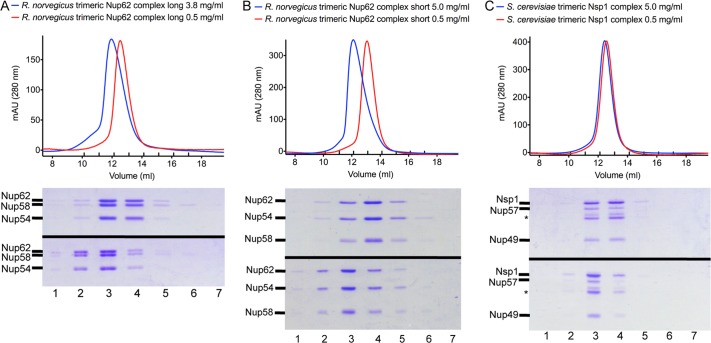
We noticed that the N-terminal residues 322–361 of rnNup62 resulted in higher-order oligomerization of the rnNup62-Nup54-Nup58 complex (unpublished data). To further investigate this behavior, we analyzed the concentration-dependent oligomerization state of long and short rnNup58-Nup54-Nup62 complexes by gel filtration and SDS–PAGE (Figure 3). For each complex, a low protein concentration of 0.5 mg/ml and a high protein concentration of 3.8 and 5.0 mg/ml, respectively, were compared.
FIGURE 3:
Concentration-dependent oligomerization of Nup58-Nup54-Nup62 complexes. Size exclusion chromatograms and SDS–PAGE gels of a high (5 mg/ml or 3.8 mg/ml) and a low (0.5 mg/ml) concentration of (A) the long rnNup58-Nup54-Nup62 complex, (B) the short rnNup58-Nup54-Nup62 complex, and (C) the scNup49-Nup57-Nsp1 complex. scNup57 autodegradation fragments are indicated by asterisks. Experiments were performed as described in Materials and Methods.
Both complexes from rat showed a significant concentration-dependent change in their elution profile when analyzed on a Superdex S200 10/300 size exclusion column. The peak of the short complex shifted up by 1.0 ml (13.1–12.1 ml), and the peak of the long complex shifted up by 0.6 ml (12.6–12.0 ml; Figure 3, A and B). In both cases, the protein ratios do not change as a result of the molecular weight increase. These results suggest that the rat complex undergoes a concentration-dependent change in oligomerization state, consistent with the results of the SE-AUC data analysis.
We also examined the concentration-dependent behavior of the scNup49-Nup57-Nsp1 complex. In contrast to the rat complex, it displayed only an insignificant change of the peak elution volume between the two concentrations tested (12.6–12.4 ml; Figure 3C).
DISCUSSION
In this study, we examined the oligomerization state of the Nup62 complex. We find that the Nup62 complex is organized in a 1:1:1 stoichiometry, as shown by our gel filtration, SDS–PAGE, and SE-AUC data. This ratio is also observed for the homologous Nsp1 complex from S. cerevisiae, separated by an estimated ∼1.5 billion years of evolution (Hedges et al., 2004). Therefore the 1:1:1 stoichiometry appears to be a common, well-conserved design principle of the Nup62/Nsp1 complex (Figure 4). Fittingly, the amino acid sequences of the α-helical portions of Nup58, Nup54, and Nup62 are each relatively well conserved between distant eukaryotic species, more so, for example, than the large class of scaffold nucleoporins with stacked α-helical architecture (Figure S1). This observation suggests that the specific interactions between the three complex members are important. It possibly also indicates interaction sites with other proteins. Nup93 is a known candidate, but there might be others yet to be identified.
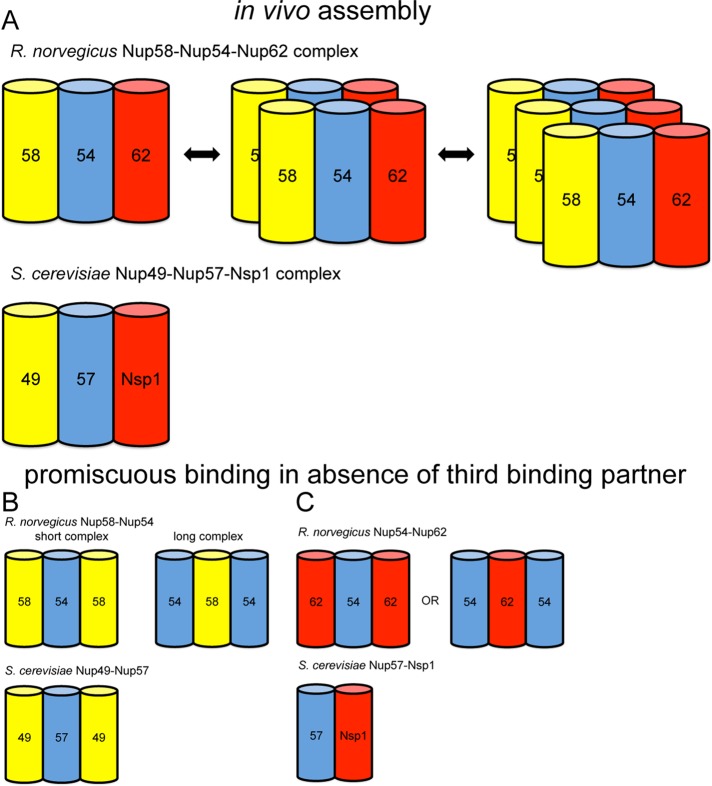
FIGURE 4:
Models of protein subcomplexes of the Nup62/Nsp1 complex. Complex models are based on size exclusion chromatography and AUC data. Cylinders symbolize predicted coiled-coil portions of nucleoporins. Double arrows indicate dynamic exchange between oligomeric states. Cartoon representations of oligomerization behavior and composition of (A) the trimeric complexes, (B) the rnNup54-Nup58/scNup49-Nup57 two-component complexes, and (C) the rnNup54-Nup62/scNup57-Nsp1 two-component complexes. We suggest that complexes depicted in (A) likely represent the complex ensemble in vivo, whereas complexes depicted in (B) and (C) represent assemblies, which, likely artificially, occur in vitro in the absence of the third binding partner.
In a series of recent publications, the rat Nup62 complex was analyzed using a crystallographic approach (Melcák et al., 2007; Solmaz et al., 2011, 2013). Using rather small, ∼4- to 10-kDa coiled-coil fragments of dimeric subassemblies, the authors hypothesized that the in vivo composition of the Nup58-Nup54-Nup62 complex would be 1:2:4, inconsistent with our data. Coiled-coil regions in proteins are, by and large, interaction domains (Lupas and Gruber, 2005), and understanding the specificity of these represents an intense research field (Grigoryan and Keating, 2008). It is well known that the use of fragmented coiled-coil portions, taken out of their natural context, can yield noncognate assemblies. We tested this by using six incomplete binary assemblies of Nup62 components. These tests reveal that rnNup54/scNup57 is the central helical element in the trimeric assembly, with direct interaction with the two remaining proteins, rnNup62/scNsp1 and rnNup58/scNup49, which do not directly interact in our experiments. We further observe that, if we take one binding partner out, the now-unoccupied binding site on the remaining protein does not necessarily stay vacant but is often bound promiscuously. Consequently, the resulting incomplete complex might have a 1:1 stoichiometry, but can also generate 1:2 or 2:1 complexes, depending on the sequence context. The fact that we do not see any of these noncanonical interactions occurring when we copurify the trimeric rnNup62/scNsp1 complex strongly supports that the physiologically relevant stoichiometry is 1:1:1.
One prominent aspect of the hypothesis that the Nup62/Nsp1 complex might assemble in various stoichiometries is that it could potentially explain opening and closing of the NPC as suggested by EM and tomographic studies (Kiseleva et al., 1998; Beck et al., 2004). The fact that we observe a uniform stoichiometry of the Nup62/Nsp1 complex in solution neither supports nor challenges the observation that NPCs might dilate. Most current models consider the Nup62/Nsp1 complex anchored to the main NPC scaffold via Nup93/Nic96. The main scaffold itself is largely composed of the latticework generated by β-propellers and the different types of stacked α-helical domains, which are prominently represented in the Nup93/Nic96 complex and the Y complex. Because some of these scaffold proteins have intrinsic flexibility, observed structural dynamics within the assembled NPC can easily be explained by this characteristic alone. It is also important to ask what role a postulated dilation mechanism might play in the NPC transport function. At this point, it is still an open question whether the NPC scaffold needs to be dynamically arranged or whether it can simply function as a rigid entity once it is assembled. Because transport of most substrates through the wide central channel occurs simultaneously in both directions, flexibility of the scaffold is not an obvious necessity.
Another interesting consequence of a stable 1:1:1 stoichiometry of the Nup62/Nsp1 complex is that it supports the notion that the distribution of the various FG repeat–containing Nups within the NPC is organized and not arbitrary. FG repeats can be classified in different categories, and their behavior is not uniform (Patel et al., 2007; Frey and Görlich, 2009; Yamada et al., 2010). That the stoichiometry of the Nup62/Nsp1 complex is highly conserved suggests that the FG network has a rather specific composition whose functional significance we have yet to fully understand. In support of a discrete composition of the Nup62/Nsp1 complex, it was recently shown that hydrogels made up of various Nup62-Nup54-Nup58 combinations exhibit substantially different transport characteristics (Labokha et al., 2013)
In conclusion, our analysis suggests that the universally conserved trimeric Nup62/Nsp1 complex arranges in a 1:1:1 stoichiometry. It will now be interesting to see where exactly this complex is anchored to the NPC scaffold. The organization of the different FG-nups within the transport channel is still largely unclear but needs to be understood to arrive at an accurate description of facilitated transport through the NPC.
MATERIALS AND METHODS
Construct design
DNA coding for coiled-coil portions of R. norvegicus Nup58, Nup54, and Nup62 and S. cerevisiae homologues Nup49, Nup57, and Nsp1 was cloned into modified pET-Duet plasmids containing either two or three expression cassettes. In all cases, the first cassette contained an N-terminal 6×His tag. In addition, the S. cerevisiae Nup49-Nup57-Nsp1 construct contained a cleavable T4 Lysozyme fused to the N terminus of Nsp1; the S. cerevisiae Nup49-Nup57-T4 Lysozyme construct contained T4 Lysozyme fused to the N-terminus of Nup57; and the S. cerevisiae Nup57-Nsp1 construct contained a C-terminal 6×Arg tag. All tags are cleavable by thrombin or 3C protease. The two Nup49-Nup57-Nsp1 constructs used for N-terminal truncation studies of Nsp1 contained T4 Lysozyme fused to the N-terminus of Nup49 (see Table S2).
Expression and purification of protein complexes
Proteins were coexpressed in Escherichia coli BL21 (DE3) RIL in Luria–Bertani medium and induced with 200 μM isopropyl-β-d-1-thiogalactopyranoside at 18°C overnight. Cells were resuspended in solubilization buffer (50 mM sodium-phosphate, pH 8.0, 500 mM NaCl, 30 mM imidazole, pH 8.0, and 5 mM β-mercaptoethanol) and lysed using a homogenizer (Constant Systems, Kennesaw, GA). Protein complexes were bound to Ni-affinity resin (GE Healthcare, Waukesha, WI) and eluted with 250 mM imidazole (pH 8.0). After dialysis at 4°C against purification buffer (10 mM Tris/HCl, pH 8.0, 150 mM NaCl, 0.1 mM EDTA and 1 mM dithiothreitol [DTT]), samples were subjected to ion-exchange chromatography followed by proteolytic removal of tags and size exclusion chromatography. These steps were all performed at 4°C.
Analysis of concentration-dependent protein oligomerization behavior
Purified R. norvegicus protein complexes containing Nup58, Nup54, Nup62, or their S. cerevisiae homologues were each concentrated to 0.5 and 5.0 mg/ml. The long R. norvegicus Nup58-Nup54-Nup62 complex could maximally be concentrated to 3.8 mg/ml. The two concentrations of each complex were analyzed on a Superdex 200 10/300 GL (GE Healthcare) size exclusion column, and elution volumes were compared. The peak fractions were analyzed by SDS–PAGE.
SE-AUC experiments
Purified protein samples in purification buffer (10 mM Tris/HCl, pH 8.0, 150 mM NaCl, 0,1 mM EDTA, and 1 mM DTT) were concentrated to an absorption of 0.4 at 280 nm, centrifuged for 10 min at 10,000 × g to remove particulates, and filled into two sample chambers of an Epon 6-Channel Centerpiece (Beckman Coulter, Brea, CA). The AUC was performed in an Optima XL-I analytical ultracentrifuge with an An-Ti-50 Rotor (Beckman Coulter). The rotor and samples were incubated in the centrifuge for 1 h at 20°C before the start of centrifugation. Each AUC experiment was performed at 20°C with four rotor speeds: 10,000, 15,000, 20,000, and 25,000 rpm, starting with the lowest. At equilibrium, sample cells were scanned five times at 280 nm, with three replicas of each scan.
AUC data analysis
Data analysis of SE data was performed with UltraScan II (Demeler, 2013). Rotor speeds were included in analysis if, after removal of data points <0.1 or >0.9 AU 280 nm, remaining data contained an optical density gradient ≥0.4 AU 280 nm and a channel radius fraction of at least 0.07 cm. The partial specific volume at 20°C (vbar (20°C)) and the molar extinction coefficient at 280 nm ε(280nm) were calculated using UltraScan II, based on the protein sequence (Table S1). Data were fitted to multiple models; the most appropriate model was chosen based on visual inspection of the residual maps and best statistical parameters. Fit results contain the calculated molecular weight and statistical parameters (Table 1).
Supplementary Material
Supplemental Materials
Acknowledgments
We thank D. Pheasant for assistance with the analytical ultracentrifuge, E. Spooner for mass spectrometry analysis, and S. Bilokapic for general experimental advice. This work was supported by National Institutes of Health grant GM077537 (T.U.S.) and a Pew Scholar Award (T.U.S.) and used the MIT Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (NSF-0070319). A.U. was supported by the German Academic Exchange Service.
Abbreviations used:
FG
phenylalanine–glycine
NPC
nuclear pore complex
Nup
nucleoporin
Footnotes
REFERENCES
- Akey CW, Radermacher M. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J Cell Biol. 1993;122:1–19. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- Bailer SM, Balduf C, Hurt E. The Nsp1p carboxy-terminal domain is organized into functionally distinct coiled-coil regions required for assembly of nucleoporin subcomplexes and nucleocytoplasmic transport. Mol Cell Biol. 2001;21:7944–7955. doi: 10.1128/MCB.21.23.7944-7955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Förster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- Beck M, Lucˇic´ V, Förster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- Belgareh N, Snay-Hodge C, Pasteau F, Dagher S, Cole CN, Doye V. Functional characterization of a Nup159p-containing nuclear pore subcomplex. Mol Biol Cell. 1998;9:3475–3492. doi: 10.1091/mbc.9.12.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S, Schwartz TU. 3D ultrastructure of the nuclear pore complex. Curr Opin Cell Biol. 2012;24:86–91. doi: 10.1016/j.ceb.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Partridge JR, Whittle JRR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss F, Kent H, Stewart M, Bailer SM, Hanover JA. Role of different domains in the self-association of rat nucleoporin p62. J Cell Sci. 1994;107:631–638. doi: 10.1242/jcs.107.2.631. [DOI] [PubMed] [Google Scholar]
- Buss F, Stewart M. Macromolecular interactions in the nucleoporin p62 complex of rat nuclear pores: binding of nucleoporin p54 to the rod domain of p62. J Cell Biol. 1995;128:251–261. doi: 10.1083/jcb.128.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Kern H, Hurt EC. Human nucleoporin p62 and the essential yeast nuclear pore protein NSP1 show sequence homology and a similar domain organization. Eur J Cell Biol. 1991;55:17–30. [PubMed] [Google Scholar]
- Chook YM, Süel KE. Nuclear import by karyopherin-βs: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Cronshaw JM. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics. 2009;8:2119–2130. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler B. UltraScan II. A Comprehensive Data Analysis Software Package for Analytical Ultracentrifugation Experiments. San Antonio: Department of Biochemistry, University of Texas Health Science Center.; 2013. www.ultrascan.uthscsa.edu. [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci USA. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Martinez J, et al. Structure-function mapping of a heptameric module in the nuclear pore complex. J Cell Biol. 2012;196:419–434. doi: 10.1083/jcb.201109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J Cell Biol. 1991;114:169–183. doi: 10.1083/jcb.114.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkiel-Krispin D, Maco B, Aebi U, Medalia O. Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture. J Mol Biol. 2010;395:578–586. doi: 10.1016/j.jmb.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Frey S, Görlich D. FG/FxFG as well as GLFG repeats form a selective permeability barrier with self-healing properties. EMBO J. 2009;28:2554–2567. doi: 10.1038/emboj.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Octagonal nuclear pores. J Cell Biol. 1967;32:391–399. doi: 10.1083/jcb.32.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Emig S, Weise C, Hucho F, Pohl T, Hurt EC. A novel nuclear pore protein Nup82p which specifically binds to a fraction of Nsp1p. J Cell Biol. 1995a;130:1263–1273. doi: 10.1083/jcb.130.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO J. 1995b;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan G, Keating AE. Structural specificity in coiled-coil interactions. Curr Opin Struct Biol. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman E, Medalia O, Zwerger M. Functional architecture of the nuclear pore complex. Annu Rev Biophys. 2012;41:557–584. doi: 10.1146/annurev-biophys-050511-102328. [DOI] [PubMed] [Google Scholar]
- Guan T, Müller S, Klier G, Panté N, Blevitt JM, Haner M, Paschal B, Aebi U, Gerace L. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol Biol Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Hedges S, Blair JE, Venturi ML, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt EC. A novel nucleoskeletal-like protein located at the nuclear periphery is required for the life cycle of Saccharomyces cerevisiae. EMBO J. 1988;7:4323–4334. doi: 10.1002/j.1460-2075.1988.tb03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann M, Blobel G. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol. 2009;16:782–788. doi: 10.1038/nsmb.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E, Goldberg MW, Allen TD, Akey CW. Nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNA export. J Cell Sci. 1998;111:223–236. doi: 10.1242/jcs.111.2.223. [DOI] [PubMed] [Google Scholar]
- Kita K, Omata S, Horigome T. Purification and characterization of a nuclear pore glycoprotein complex containing p62. J Biochem. 1993;113:377–382. doi: 10.1093/oxfordjournals.jbchem.a124054. [DOI] [PubMed] [Google Scholar]
- Labokha AA, Gradmann S, Frey S, Hülsmann BB, Urlaub H, Baldus M, Görlich D. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 2013;32:204–218. doi: 10.1038/emboj.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas AN, Gruber M. The structure of α-helical coiled coils. Adv Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- Lutzmann M. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270:254–262. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- Marelli M. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J Cell Biol. 1998;143:1813–1830. doi: 10.1083/jcb.143.7.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul GG. On the octagonality of the nuclear pore complex. J Cell Biol. 1971;51:558–563. doi: 10.1083/jcb.51.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcák I, Hoelz A, Blobel G. Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science. 2007;315:1729–1732. doi: 10.1126/science.1135730. [DOI] [PubMed] [Google Scholar]
- Mohr D, Frey S, Fischer T, Güttler T, Görlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009;28:2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U, Rout MP, Maguire S, Blobel G, Wozniak RW. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J Cell Biol. 1996;133:1153–1162. doi: 10.1083/jcb.133.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann N, Lundin D, Poole AM. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS One. 2010;5:e13241. doi: 10.1371/journal.pone.0013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E, Stanton LH, Madrid AS, Kieselbach T, Weis K. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J Cell Biol. 2009;185:475–491. doi: 10.1083/jcb.200810030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E, Weis K. Nuclear pore complex—a coat specifically tailored for the nuclear envelope. Curr Opin Cell Biol. 2011;23:293–301. doi: 10.1016/j.ceb.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori A, et al. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol Syst Biol. 2013;9:648. doi: 10.1038/msb.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- Reichelt R, Holzenburg A, Buhle EL, Jarnik M, Engel A, Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader N, Stelter P, Flemming D, Kunze R, Hurt E, Vetter IR. Structural basis of the Nic96 subcomplex organization in the nuclear pore channel. Mol Cell. 2008;29:46–55. doi: 10.1016/j.molcel.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Solmaz SR, Blobel G, Melcák I. Ring cycle for dilating and constricting the nuclear pore. Proc Natl Acad Sci USA. 2013;110:5858–5863. doi: 10.1073/pnas.1302655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solmaz SR, Chauhan R, Blobel G, Melcák I. Molecular architecture of the transport channel of the nuclear pore complex. Cell. 2011;147:590–602. doi: 10.1016/j.cell.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin PN, Milligan RA. A large particle associated with the perimeter of the nuclear pore complex. J Cell Biol. 1982;93:63–75. doi: 10.1083/jcb.93.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Wente SR, Rout MP, Blobel G. A new family of yeast nuclear pore complex proteins. J Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada J, et al. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol Cell Proteomics. 2010;9:2205–2224. doi: 10.1074/mcp.M000035-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials