Connections between TET proteins and aberrant DNA modification in cancer (original) (raw)
. Author manuscript; available in PMC: 2015 Oct 1.
Published in final edited form as: Trends Genet. 2014 Aug 14;30(10):464–474. doi: 10.1016/j.tig.2014.07.005
Abstract
DNA methylation has been linked to aberrant silencing of tumor suppressor genes in cancer, and an imbalance in DNA methylation/demethylation cycles is intimately implicated in the onset and progression of tumors. TET (ten-eleven translocation) proteins are Fe(II) and 2-oxoglutarate dependent dioxygenases that successively oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), thereby mediating active DNA demethylation. In this review, we focus on the pathophysiological role of TET proteins and 5hmC in cancer. We present an overview of loss-of-function mutations and abnormal expression and regulation of TET proteins in hematological malignancies and solid tumors, and discuss the potential prognostic value of assessing TET mutations and 5hmC levels in cancer patients. We also address the crosstalk between TET and two critical enzymes involved in cell metabolism, OGT and IDH. Lastly, we discuss the therapeutic potential of targeting TET proteins and aberrant DNA methylation in cancer.
Keywords: TET, 5-hydroxymethylcytosine, 5mC, 5hmC, DNA methylation, DNA demethylation, cancer, IDH, OGT
TET proteins oxidize 5-methylcytosine in DNA
DNA methylation controls diverse biological processes, including X chromosome inactivation, gene expression and genomic imprinting [1]. Dysregulated DNA methylation is frequently observed in cancer, and comprises both aberrant silencing of tumor suppressor genes due to increased DNA methylation at their promoters, as well as global DNA hypomethylation leading to decreased genome stability; both processes contribute to oncogenesis and tumor progression [2, 3]. In normal cells, DNA methylation is mediated through the coordinated actions of several DNA methyltransferases (DNMTs) which transfer a methyl group from S-adenosyl methionine (SAM) to the carbon-5 position of cytosine [4] (Figure 1). DNA methylation occurs primarily in the CpG context; replication of symmetrically methylated CpG dinucleotides leads to the production of daughter strands bearing unmethylated CpGs. The resulting hemimethylated DNA strands are normally restored to their symmetrical methylation status by the maintenance DNA methyltransferase complex (DNMT1/UHRF1), which recognizes hemimethylated CpGs [5, 6]. If this maintenance methylation does not occur, DNA becomes progressively demethylated through a “passive”, replication-dependent mechanism.
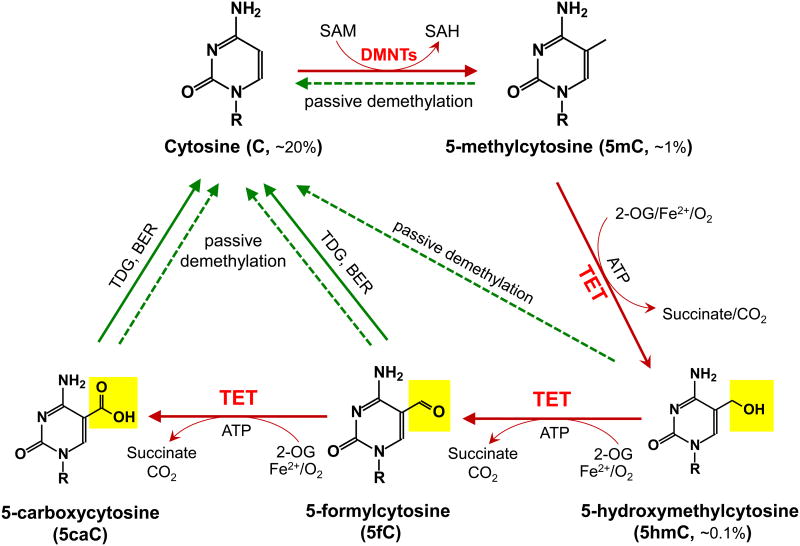
Figure 1. Schematic of major DNA methylation and demethylation pathways in mammals.
DNA methylation occurs almost exclusively as symmetrical methylation at the carbon-5 position of cytosine in the context of the dinucleotide CpG. DNMTs methylate cytosine (C, ∼20% of all bases) to yield 5-methylcytosine (5mC, ∼1% of all bases and ∼60% of all CpGs) by transferring the methyl group from S-adenosylmethionine (SAM) to cytosine. TET enzymes oxidize 5mC to 5-hydroxymethylcytosine (5hmC, ∼0.1% of all bases), 5-formylcytosine (5fC), 5-carboxylcytosine (5caC) (together, oxi-mC). Through oxi-mC production, TET proteins mediate multiple pathways of DNA demethylation, including thymine DNA glycosylase (TDG)-mediated base excision repair (BER) of 5fC:G and 5caC:G basepairs and replication-dependent passive demethylation. A recent study showed that TET proteins can oxidize thymine to 5-hydroxymethyluracil (5hmU) [143], and other studies have suggested that Activation-induced deaminase (AID)/APOBEC can mediate the deamination of 5hmC to 5hmU followed by TDG-mediated BER (reviewed in [11]). Although 5hmU:G mismatches can also be excised by TDG [144], these pathways are less well-characterized and so are not depicted here.
Recently, proteins of the Ten-Eleven Translocation (TET) family were identified as dioxygenases that utilize two key co-factors, Fe(II) and 2-oxoglutarate (2-OG), to successively oxidize the methyl group of 5mC to hydroxymethyl, formyl or carboxyl groups, thus forming the oxidised methylcytosines 5hmC, 5fC and 5caC (together termed “oxi-mC”) [7-11] (Figure 1). It is now clear that these oxi-mC intermediates facilitate DNA demethylation in at least two ways. First, they potentiate passive DNA demethylation by interfering with maintenance DNA methylation by the DNMT1/UHRF1 complex, but they also effect “active”, replication-independent DNA demethylation as discussed below (reviewed in ref [10, 11]). Second, two of the oxi-mC intermediates, 5fC and 5caC, can be excised by the DNA repair enzyme thymine-DNA glycosylase (TDG), followed by replacement with umodified cytosine through a base excision repair mechanism [8, 12-15]. TDG was originally identified as an enzyme that excised thymine from T:G mismatches, but it is also able to bind and excise 5fC and 5caC with comparable affinity and efficiency, even though these modified bases are fully base-paired with G [13, 14]. Other putative active DNA demethylation mechanisms are discussed elsewhere [10, 11, 15].
In this review, we focus on the role of TET proteins in cancer. TET proteins were named for the rare “ten-eleven translocation”, associated with both myeloid and lymphoid malignancies, that fuses the N-terminal region of the MLL gene (encoded on chromosome 10) to the C-terminal catalytic domain of TET1 (encoded on chrosome 11) [16, 17]. More recently, the gene encoding TET2 was found to be frequently mutated or deleted in a variety of hematological malignancies [18, 19]. Here, we provide an overview of TET loss-of-function mutations and aberrant TET expression or regulation in hematopoietic and solid cancers, describe the crosstalk between TET proteins and aberrant cancer metabolic pathways, and discuss the exciting possibility of targeting TET proteins with novel anti-cancer therapeutics.
Mutations in TET proteins in hematological malignancies
TET1 and TET3 are rarely mutated in hematological malignancies [20] (Box 1). By contrast, the 4q24 region of human chromosome 4 that harbours the TET2 gene recurrently undergoes microdeletions and copy number-neutral loss-of-heterozygosity (also termed uniparental disomy) in myelodysplastic syndromes (MDS) and myeloid malignancies [18, 19]. TET2 was originally identified as the relevant tumor suppressor gene in this region through the discovery of a patient with a myeloproliferative disorder who exhibited a 325 kb somatic microdeletion in 4q24 that encompassed only the TET2 gene [18]. Since then, large-scale whole-exome sequencing studies by many groups have confirmed that TET2 is one of the most frequently mutated genes in chronic myelomonocytic leukemia (CMML, ∼50%) [21-23], acute myeloid leukemia (AML, ∼20%) [24-27], and myelodysplastic syndromes (MDS, ∼20%) [18, 19, 22, 28, 29]. In many cases, deletion of TET2 in the 4q24 region is associated with a TET2 mutation on the other allele [28, 29]. Deletion of Tet2 in mouse models is also associated with dysregulated hematopoiesis (Box 2).
Box 1. Mutations of TET1 and TET3 in hematopoietic cancers.
The three members of the mammalian TET family, TET1, TET2 and TET3, are expressed in different tissues and exhibit both overlapping and distinct functions [10,11]. In early embryogenesis, TET1 and TET2 are abundantly expressed in the inner cell mass of the mouse embryo, in primordial germ cells, and in mouse embryonic stem cells (mESC) derived from the inner cell mass, and TET3 is highly expressed in the zygote [10,11]. In differentiated cell types, including neurons and cells of the hematopoietic system, the main TET family members are TET2 and TET3 [32,128]. Although mice with individual or combined genetic ablation of TET1 and/or TET2 show relatively normal embryonic and postnatal development [129,130], TET1 and TET2 have been shown to regulate mESC lineage specification [131] [132] and to exhibit distinct functions with respect to 5hmC deposition in mESC [133]; specifically, TET1 depletion diminishes 5hmC levels at transcription start sites (TSS), whereas TET2 depletion is predominantly associated with decreased 5hmC in gene bodies [133].
TET1 and TET3 are rarely mutated in hematological malignancies [20]. In a handful of rare cases of AML and ALL [16,17,134], translocation of t(10;11)(q22;q23) results in fusion of the N-terminal region of the H3K4 methyltransferase MLL1 to the DSBH domain of TET1. During disease transformation, the TET1-MLL fusion protein plays an important role in regulating the expression of transcription factors such as HOXA9, MEIS1 and PBX3 that are closely implicated in hematopoiesis [135]. TET1 is also mutated at a low frequency in T-cell acute lymphoblastic leukemia (T-ALL) [136], and TET3 is occasionally mutated in T cell lymphoma. Because all three isoforms share similar enzymatic functions and TET1 and TET3 largely remain intact in hematological malignancies, they might compensate to a greater or lesser extent for any functional defects arising from TET2 mutations during hematopoietic development. This could partially explain why TET2 loss-of-function is not by itself sufficient for disease transformation in mouse models.
Box 2. Effects of Tet2 mutations in mouse models.
The association of TET2 loss-of-function with hematopoietic dysregulation has been recapitulated in mouse models as well as during in vitro differentiation of human haematopoietic progenitors. Several groups have generated _Tet2_-deficient mouse models using targeted recombination or gene-trap techniques [45-48,137]. Although the knockout strategies were different, all the _Tet2_-deficient mice displayed similar in vivo phenotypes, exemplified by increased numbers of hematopoietic progenitor cells in the bone marrow and their skewed differentiation toward the myelomonocytic lineage [45-48,137]. Moreover, human cord blood CD34+ cells depleted of TET2 [38] or isolated from leukemia patients bearing TET2 mutations [18] showed a consistent phenotype in in vitro differentiation assays, with an increase in myeloid-lineage cells [18,38] and a decrease in erythroid-lineage cells [38,138]. 5hmC and TET2 mRNA levels both showed dynamic alterations during erythroid differentiation; moreover, regions that gained 5hmC during erythroid differentiation showed enrichment for sequence motifs recognized by erythroid transcription factors [138].
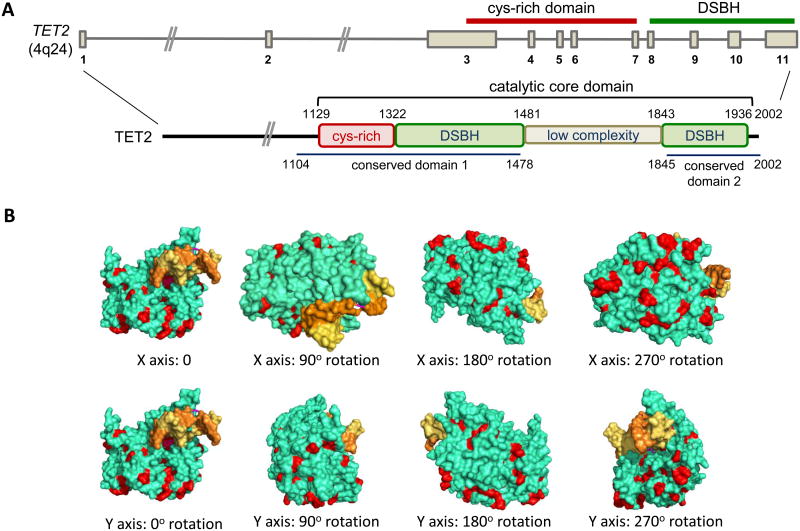
So far, >700 TET2 mutations have been identified in more than 2000 leukemia patients [26, 30, 31]. The majority of missense mutations impair the enzymatic activity of TET2, with a resultant decrease in 5hmC levels and aberrant DNA methylation [27, 32]. The missense mutations tend to be clustered in two highly conserved regions of the human TET2 protein (amino acids 1104-1478 and 1845-2002), which correspond almost exactly to the well-structured regions observed in a recently determined crystal structure of the TET2 catalytic domain [33] (Figure 2). Based on the crystal structure, many of the residues affected by the missense mutations are located on the surface of the TET2 catalytic domain [33] (Figure 2); these residues might be important for protein-protein interactions and/or may be subject to post-translational modifications (e.g., phosphorylation, ubiquitylation, SUMO'ylation, glycosylation).
Figure 2. Domain organization of TET2 and the locations of residues frequently mutated in cancer in the TET2 catalytic domain.
(A) Top, Structure of the TET2 gene on human chromosome 4q24. The regions encoding the Cys-rich domain and the catalytic double-stranded beta-helix (DSBH) domain are indicated. Bottom, The domain organization of the TET2 protein. Somatic TET2 mutations most frequently affect residues in conserved domains 1 and 2, which correspond almost exactly to the well-structured regions of the catalytic core domain. (B) The crystal structure of the catalytic core domain of TET2 ([33]; the unstructured low complexity region is not represented in the structure). Yellow and orange: the two strands of the DNA double helix; green: TET2; Red: the residues affected by selected disease-associated mutations.
Whether TET2 mutations have prognostic value for cancer patients is not yet entirely clear. A meta-analysis of a large cohort of AML patients described in eight published studies revealed a robust correlation between TET2 mutations and poor prognosis, as judged by overall survival as well as event-free survival [30]. In a smaller cohort of MDS patients, TET2 mutations significantly decreased both the time of transformation of MDS to secondary AML as well as the probability of survival [34]. However, the prognostic potential of TET2 mutations in MDS and CMML is still controversial. Based on studies of 96 and 88 patients with MDS and CMML respectively, TET2 mutations were reported to be predictive of a favorable prognosis in MDS [35] and but were negatively correlated with overall survival in CMML [36]. By contrast, results from two other groups suggested no significant correlation between TET2 mutations and overall prognosis in MDS or CMML [21, 28, 37]. Some of these discrepancies may be due to small sample sizes and could potentially be resolved by meta-analyses in which results from multiple cohorts are combined [30]. Moreover, because a subset of patients with wild type TET2 have low 5hmC [27, 32, 38], genome-wide 5hmC levels could potentially serve as a better prognostic marker than TET2 mutations [39], a hypothesis warranting further scrutiny in larger patient cohorts. Ideally, the most robust prognostic value would be provided by a combined evaluation of the mutational status of TET2 as well as other cancer-associated genes [40, 41]. TET2 mutations are also frequently observed in lymphoma, particularly in angioimmunoglastic T cell lymphomas (AITL) (∼76%) [42] and peripheral T-cell lymphoma, not otherwise specified (PTCL-NOS, 38%) [43], where they tend to be found together with mutations in RhoA as discussed below. TET2 mutations were more frequently observed in patients showing T follicular helper cells (TFH)-like features in PTCL-NOS and were associated with advanced-stage diseases and poorer clinical outcomes [43]. The prognostic value of TET2 mutations in lymphoma is still under investigation.
TET2 mutations require a second hit
Analyses of the clonal architecture and mutational allele frequency of myeloid disorders including MDS [18, 40, 41] and CMML [44] indicate that TET2 mutations often occur as an early oncogenic event and lead to clonal expansion of leukemic cells. Studies in Tet2 loss-of-function mouse models or human CD34+ cell xenograft models suggest a strong association of TET2 mutations with increased hematopoietic progenitor cell proliferation [18, 45-48]. However, deletion of TET2 by itself in mice is not sufficient to drive myeloid or lymphoid diseases. Moreover, mutations in TET2 alone were observed in older individuals with clonal hematopoiesis but no overt hematological malignancies [49]. These findings strongly imply that mutation(s) in one or more genes other than TET2 are needed for progression to a clearly malignant phenotype – the hypothesis of an initiating mutation followed by a “second hit” in cancer.
Indeed, a meta-analysis of the mutational landscape of hematological malignancies suggests that TET2 mutations are frequently observed together with mutations in a relatively small number of specific genes [40]. For instance, TET2 mutations frequently co-exist with mutations in splicing factors such as SRSF2 in MDS, CMML and mastocytosis [40, 50, 51]. Genes encoding splicing factors (e.g., SF3B1, SRSF2, U2AF1 and ZRSR2) that are involved in 3′ splice site recognition and U2 snRNP functions are frequently mutated in myeloid cancers [52-54]. Similar to TET2 mutations, mutations in these splicing factors also occur at an early stage during oncogenesis [41] [44], but have varying degrees of prognostic value. Mutation of SRSF2 is usually correlated with a poor clinical outcome [50,55]. Similarly, in MDS and CMML patients, TET2 mutations are frequently observed in conjunction with mutations in the polycomb proteins EZH2 and ASXL1 [56,57]; moreover, in mouse models, depletion of TET2 together with ASXL1 [58] or EZH2 [57] resulted in a condition resembling human MDS. EZH2 is an H3K27 methyltransferase that is the catalytic component of polycomb repressive complex 2 (PRC2); ASXL1 is the regulatory subunit of the ASXL1-BAP1 complex, a deubiquitinase for H2AK119Ub [59,60].
In PTCL and especially AITL, TET2 mutations are significantly correlated with the presence of a recurrent point mutation (G17V) in the small GTPase RHOA, which regulates cell morphology and migration [61,62]. To date, the RHOA (G17V) mutation is uniquely found in PTCL but not in other hematological malignancies, suggesting a specialized function in disease transformation in PTCL. In these T cell lymphomas, TET2 is also frequently mutated along with DNMT3A, which encodes a de novo DNA methyltransferase [63].
In summary, loss-of-function mutations in TET2, with an accompanying or independent decrease in 5hmC levels, are a common and frequently observed theme in hematological malignancies. TET2 mutations occur in early hematopoietic progenitors and are associated with clonal expansion; however, they do not by themselves result in malignant transformation. Rather, accumulating evidence from both clinical and biochemical studies points to the possibility that TET2 loss-of-function synergises with other mutations in epigenetic regulators, the pre-mRNA splicing machinery or intracellular signalling pathways to promote oncogenesis in specific hematological malignancies.
TET mutations and abnormal TET expression or regulation in solid cancers
A quick glance at the TCGA database yields the general impression that all three TET genes are mutated in solid tumors, albeit at lower frequencies than observed for TET2 in hematological malignancies. For instance, somatic mutations in all three TET proteins have been reported in colorectal cancer (CRC) [64], and TET2 mutations/deletions have been observed in clear-cell renal cell carcinoma (ccRCC, 16%) [65] as well as in metastatic castration-resistant prostate cancer but not primary prostate cancers [66]. The role of mutated TET proteins in solid cancers has not yet been firmly established. Although the bulk of the evidence suggests that TET genes are established bona fide “driver” genes whose mutations drive malignant transformation in haematopoietic malignancies and so could also be causal in solid cancers, the fact that tumors are usually genetically heterogeneous and bear many background mutations makes it important to clarify, for each mutation, whether or not it confers a true advantage in cell proliferation/survival during tumorigenesis and cancer progression. It should be possible to resolve this point by crossing the many available TET-deficient mouse models with the various available mouse models of cancer.
In addition to somatic TET mutations, TET protein expression and the global level of its dominant enzymatic product (5hmC) are markedly reduced in a wide range of solid cancers, including melanoma, prostate, breast, lung, and liver tumors [67]. Although the decrease in 5hmC might partly reflect the different rates of cell proliferation in normal and cancer cells [68], several studies have demonstrated a close correlation between decreased 5hmC levels/TET expression and robust tumor growth and metastasis [69-71], supporting the idea that TET proteins might serve as tumor suppressors in certain types of cancers (Figure 3). For instance, low expression of either TET1 alone, or both TET1 and its target TIMP2, correlated with advanced cancer stage, nodal metastases, and poor survival rate in breast cancer patients [71]. In a second scenario, the high mobility group protein HMGA2 (high mobility group AT-hook 2), which is expressed in ES cells and in several cancers but not in most normal somatic cells, was shown to inhibit TET1 and homeobox A (HOXA) expression in breast cancer cells [72]. RNAi-mediated depletion of HMGA2 increased TET1 and HOXA expression and diminished the growth and migration of cancer cell lines in mice, in a manner that correlated with increased 5hmC and decreased methylation of the TET1 and HOXA gene promoters. Moreover, when breast cancer patients were stratified according to gene expression, patients with low HMGA2 and high TET1 and HOXA expression showed better survival than patients with high HMGA2 and low TET1 and HOXA gene expression, although the expression levels of the individual genes did not predict survival [72]. Finally, TET2 expression may also be transcriptionally downregulated: for instance, high levels of DNA methylation are observed at the TET2 promoter in a number of cancers [73].
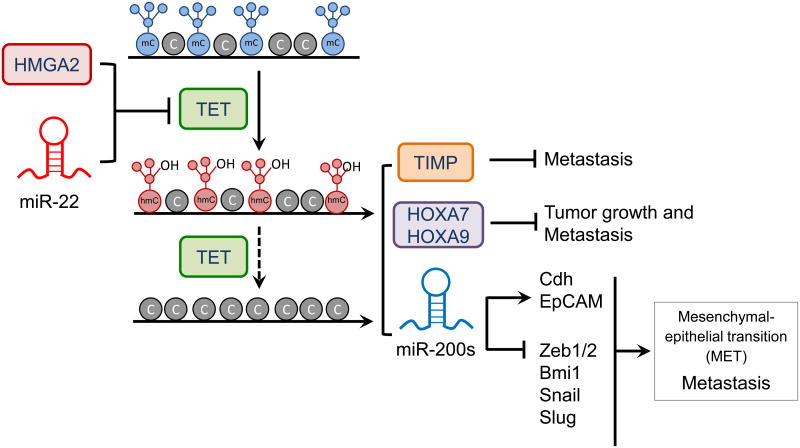
Figure 3. Regulation of TET expression by proto-oncogenic microRNAs and transcription factors in breast cancer.
Over-expression of oncogenic miR-22 or HMGA2 suppresses TET gene expression. The resulting decrease in TET protein level causes hypermethylation at promoters of genes encoding proteins (TET, the metalloproteinases TIMP, the homeobox transcription factors HOXA7 and HOXA9) or microRNAs (miR-200s) that are involved in suppressing tumor growth and metastasis. miR-200s control expression of several genes including those encoding cadherin (Cdh), the epithelial cell adhesion molecule EpCAM, the transcription factors ZEB1/2, Snail and Slug, and the polycomb group protein Bmi1.
The levels of TET proteins can be modulated by more than 30 miRNAs. In one such study, TET (especially TET2) levels were diminished by several miRNAs (including miR-125b, miR-29b, miR-29c, miR-101 and miR-7) that disrupted normal hematopoiesis and were overexpressed in AML patients harboring wild type TET2 [74]. In another study, the pro-metastatic microRNA, miR-22, suppressed TET expression in mouse mammary tumor models, and high miR-22 expression correlated with high-grade cancers and expression of genes involved in breast cancer metastasis and poor survival in human breast cancer patients [69]. The authors proposed that miR-22 directly targeted the 3′ untranslated region (3′ UTR) of TET mRNAs, leading to downregulation of TET protein expression in breast cancer cells; the resulting insufficiency of TET function led to increased promoter methylation and downregulation of the anti-metastatic microRNA, miR-200, which in in turn altered the expression of key factors associated with tumor metastasis and the mesenchymal–epithelial transition (MET) [69,75] (Figure 3). Thus both the miR-22-TET-miR-220s axis and the HMGA2-TET1-HOXA axis perturb the balance of DNA methylation/demethylation to promote breast tumor growth and metastasis (Figure 3). Similar scenarios have been reported for other types of cancers and other TET homologs: for example, decreased TET2 expression and 5hmC levels are associated with melanoma progression [70], and TET1 expression and 5hmC levels are both decreased In hepatocellular carcinoma, and this decrease is associated with poor patient outcomes [76].
Recently, it was found that TET2 protein levels can also be regulated by CXXC4, also known as IDAX [77], a negative regulator of Wnt signaling [78] that is often mutated or overexpressed in cancer. IDAX/CXXC4 originally encoded the CXXC domain of an ancestral TET2 gene, but became separated from the catalytic domain of this protein by a chromosomal inversion during evolution [77,79]. IDAX/CXXC4 recruits TET2 to genomic DNA and then downregulates TET2 through a mechanism that involves caspase activation [77]. Upregulation of IDAX/CXXC4 expression in colon carcinoma [80] might be partially responsible for the decrease in TET2 and 5hmC levels observed in colon cancer. Conversely, the downregulation of IDAX/CXXC4 frequently observed in metastatic renal cell carcinoma [81] would be expected to increase the nuclear translocation of β-catenin, thereby increasing the expression of genes associated with cell proliferation and metastasis. RINF/CXXC5, a protein closely related to IDAX/CXXC4, may have a similar function [77]. Overall, the balance between IDAX/CXXC4, RINF/CXXC5 and TET proteins may be critical in suppressing cancer transformation.
Calpain, a Ca2+-dependent cysteine protease, was recently reported to modulate TET protein stability in mESC and during neuronal differentiation [82]. Calpain is known to be upregulated in cancer cells [83], hence this might represent an additional mechanism leading to decreased TET levels and abnormal DNA methylation in cancer.
Taken together, TET proteins may be regarded as putative products of tumor suppressor genes. Both tissue-specific miRNAs (e.g., miR-22) and diverse transcriptional or post-transcriptional/post-translational regulators could modulate TET expression or function in solid tumors. Downregulation of TET expression would disrupt oxi-mC production, the normal balance of genomic DNA methylation/demethylation or both, thereby affecting downstream effectors involved in malignant transformation.
Crosstalk between TET and aberrant metabolic pathways in cancer
TET and OGT
Cancer cells typically display altered metabolism characterized by increased utilization of glucose and glutamine under aerobic conditions, a phenomenon known as the Warburg effect [84]. Increased glucose utilization funnels into increased production of the final product of the hexosamine biosynthetic pathway, UDP-N-acetylglucosamine (UDP-GlcNAc) [85], which is used by the enzyme OGT (_O_-linked β-N-acetylglucosamine transferase) to add the _O_-GlcNAc moiety to the hydroxyl groups of specific serine and threonine residues in diverse nuclear and cytosolic proteins, thus controlling their localization, stability or enzymatic activity [86] [87]. OGT also _O_-GlcNAc'ylates histones H2A, H2B and H3 [88], thus influencing gene transcription [89] and cell cycle progression [90]. Mouse ESC lacking expression of Eed and Suz12 (components of polycomb complex 2) showed decreased OGT expression and decreased levels of the _O_-GlcNAc modification [91]. Conversely, MCF-7 breast cancer cells depleted of OGT showed decreased stability of Ezh2 and reduced H3K27me3 levels [92].
Recently, several groups reported a direct interaction between the DSBH domain of TET proteins and nuclear OGT [93-98] (Figure 4). Although TET proteins can be _O_-GlcNAc'ylated by OGT [96,97], their catalytic activity appears to be unaffected [93,94,96]; instead, the interaction seems to stabilize TET protein levels without affecting mRNA processing [97], and might also modulate TET3 nuclear localization [98]. Conversely, TET proteins play an essential role in recruiting OGT to chromatin [93-95], thus enabling _O_-GlcNAc modification of histones, a process important for nucleosome assembly and entry into mitosis [90,99,100]. Several genome-wide studies have shown that TET and OGT co-localize at H3K4me3 positive, CpG-rich promoters of actively transcribed genes [93-95]. It was reported that TET2 mediates OGT modification on H2B Ser112 and is associated with highly-transcribed genes [93]; and that TET2 and TET3 recruit OGT to modify HCF1 (Host cell factor 1), an essential transcriptional regulator in the SET1/COMPASS complex, which contains the MLL methyltransferase that deposits H3K4me3 [94].
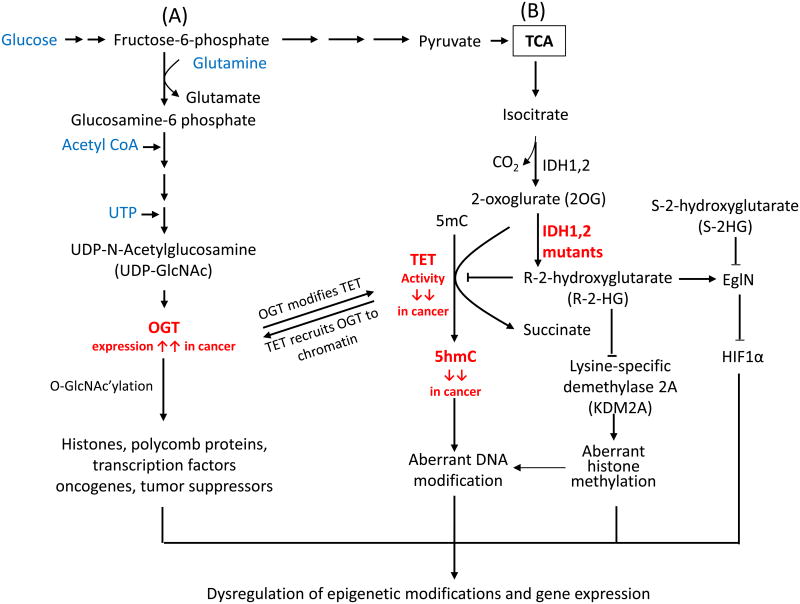
Figure 4. Crosstalk between TET and key enzymes involved in protein glycosylation and metabolism.
(A) Glucose metabolism modulates the activity of OGT. OGT (_O_-linked β-N-acetylglucosamine transferase) utilizes UDP-GlcNAc to glycosylate serine and threonine residues of diverse nuclear and cytoplasmic proteins, including enzymes involved in epigenetic regulation in cancer. TET proteins facilitate the _O_-GlcNAc modification of histones and chromatin remodeling enzymes by recruiting OGT to chromatin. Conversely, TET proteins are O-GlcNAc-modified by OGT, potentially increasing their stability and promoting TET3 nuclear localization in mESC or cultured cell lines. However, the stability of TET protein might also be regulated by other proteins, such caspase and calpain in cancer cells. (B) Isocitrate dehydrogenase (IDH) enzymes generate 2-oxoglurate (2OG), an essential co-factor for TET proteins. In contrast, mutant IDH enzymes bearing recurrent cancer-associated mutations deplete 2OG by converting it to 2-hydroxyglutarate (2HG), thus inhibiting TET enzymatic activity and decreasing the genomic levels of 5hmC. 2HG has two enantiomers, R-2HG and S-2HG. Mutant IDH enzymes only yield the oncometabolite R-2HG, which inhibits TET proteins and other dioxygenases, although it is typically less effective than S-2HG. R-2HG also promotes the enzymatic activity of EgIN prolyl-4-hydroxylases, enzymes that regulate the protein levels of the transcription factor HIF1a through hydroxylation followed by proteasomal degradation. By contrast, S-2HG can antagonize the proto-oncogenic activity induced by IDH mutants and EgIN.
These findings illustrate the potential for reciprocal crosstalk between TET proteins and pathways involved in glucose metabolism. Aberrant glucose metabolism in cancer cells may alter _O_-GlcNAc'ylation of TET proteins and therefore affect their stability; conversely, TET loss-of-function in cancer may influence the nuclear/cytoplasmic distribution of OGT, which in turn may affect the stability of tumor suppressors and oncogenes such as p53 [101], MYC [102] and β-catenin [103].
TET and IDH
Isocitrate dehydrogenases are key metabolic enzymes that function in the tricarboxylic acid (TCA) cycle; they convert isocitrate to 2-oxoglutarate (2OG, also known as α-ketoglutarate) using NADP+/NADPH as cofactors (Figure 4). 2OG is an essential cofactor for dioxygenases including TET proteins and the JmjC family of lysine demethylases [7,104]. Among the three IDH enzymes, IDH1 and IDH2 (cytosolic and mitochondrial, respectively) are frequently mutated in glioma and hematological malignancies; the observed recurrent point mutations (e.g., at residue R132 of IDH1) confer an unusual gain-of-function (reviewed in [105]). Mutant IDH1 and IDH2 convert 2OG to (R)-2-hydroxyglutarate (R-2HG), with two consequences for TETs and other dioxygenases: depletion of 2OG as well as excessive production of 2HG, an “oncometabolite” that is normally present at levels <0.1 mM but that can reach levels of 1-30 mM in cells expressing the mutant enzymes (reviewed in [105]). R-2HG inhibits the activity of TET proteins and other 2OG-dependent dioxygenases both in vitro and in vivo, although paradoxically, the R-2HG stereoisomer is considerably less effective than the S-2HG steroisomer at inhibiting the activity of TET proteins and other tested dioxygenases [106,107]. The reason for this discrepancy is not completely understood, although one group has suggested that the R-2HG stereoisomer selectively stimulates the activity of EglN prolyl hydroxylases that modify the hypoxia-inducible transcription factor HIF1α and target it for degradation [106,108]. In any case, 2HG potentiates the proliferation and impairs the differentiation of TF-1 erythroleukemia cells and 3T3-L1 cells towards the erythroid and adipocyte lineages respectively [106,109].
Recently, conditional Idh1(R132H) and Idh2(R140Q) knock-in mouse models were generated [110,111]. As anticipated, both IDH mutant mouse strains displayed high serum levels of 2HG with phenotypes resembling those of TET2 or DNMT3A deficiency, as well as age-dependent splenomegaly due to extramedullary hematopoiesis. Similar to _TET2_-deficient mice, IDH mutant mice exhibited stem/progenitor cell expansion and impaired hematopoietic differentiation, but did not develop AML. Thus as observed for TET2 deficiency, IDH mutations alone are insufficient: additional mutations are required for oncogenic progression. Indeed, transgenic mice with both Idh2(R140Q) and _Flt3_ITD (Internal tandem duplication of FMS-like tyrosine kinase 3) developed acute leukemia [111].
Notably, loss-of-function mutations of other enzymes in the TCA cycle, such as succinate dehydrogenase (SHD) and fumarate hydratase (FH), have been described in a subset of renal and uterine cancers [112,113]. These mutations, which are invariably loss-of-function, result in the accumulation of succinate and fumarate respectively [113]. Like 2HG, these endogenous metabolites inhibit the activity of 2OG-dependent dioxygenases including TET proteins and JmjC-family histone demethylases [112,113]. The impact of FH and SDH mutations on cancer growth, progression and epigenetic status requires further investigation.
Targeting DNA methylation and TET activity in cancer
The link between TET proteins and DNA methylation in cancer is not clear (Box 3). However, the data presented here indicate that the expression and activity of TET proteins are impaired in many different cancers; therefore, compounds that modulate TET activity would likely find broad use in the treatment of cancer. Among the most promising compounds are DNMT and IDH inhibitors, some of which have already been approved for use in the clinic. Potentially, compounds that increase TET expression or potentiate TET activity would also be useful. One such example is the familiar compound Vitamin C, which acts on TET proteins and a handful of other Fe(II) and 2OG-dependent dioxygenases, including those that mediate the prolyl hydroxylation of collagen [114].
Box 3. The link between TET proteins and DNA methylation.
Cancer cells display both global DNA hypomethylation and local hypermethylation of specific CpG island promoters depending on the cell type of origin [2,139,140]. The link between TET proteins and DNA methylation in cancer is not clear. One study showed that patients with low 5hmC levels (either wild type or mutant for TET2) showed global hypomethylation relative to healthy donors; the only consistently hypermethylated CpG sites were present at SP140 and AIM2 [32], as confirmed by a second group studying patients with CMML [141]. In AML, systematic analysis of a large cohort of patients revealed that TET2 and IDH1/2 mutations were mutually exclusive, supporting the idea that because mutant IDH1 inhibits TET function, TET2 and IDH1/2 act in the same leukemogenic pathways during cellular transformation [24,142]. The DNA hypermethylation reported in cancer patients bearing IDH mutations has been attributed to impaired enzymatic function of TET proteins [2,24,139]. This point is echoed in several studies of other tumor types: TF-1 erythroleukemia cells depleted of TET2 showed a phenotype similar to that of cells expressing mutant IDH [106], and in melanoma, depletion of either TET or IDH2 resulted in decreased 5hmC production [70]. Analysis of different patient cohorts and cancer subtypes, coupled with different methods of DNA methylation analysis, may yield different conclusions as to whether TET2 loss-of-function results in global hypomethylation or hypermethylation of DNA; additional insights will require analyses of DNA methylation status at individual promoters, enhancers and gene bodies and correlation with expression patterns of the associated genes. Moreover because all three TET isoforms share similar enzymatic functions and TET1 and TET3 largely remain intact in hematological malignancies, other TET proteins might compensate for functional defects arising from TET2 loss-of-function mutations during hematopoietic development.
DNA methyltransferase (DNMT) inhibitors
The DNMT inhibitors decitabine and azacitidine and their derivatives, sometimes termed “hypomethylating agents”, have been approved by the Food and Drug Administration (FDA) as a therapeutic strategy in high-risk MDS patients [115]. These compounds are cytidine analogs that bear nitrogen instead of carbon at the 5-position of the pyrimidine ring; they are randomly incorporated into genomic DNA where they bind covalently to and deplete DNMTs, thus facilitating DNA demethylation [115,116] (Figure 5). Currently, only a fraction of patients with high-risk MDS, CMML or AML respond to treatment with hypomethylating agents [115]. Potentially, one mechanism for the success of treatment with hypomethylating agents is upregulation of the TET proteins themselves: TET1 and TET3 if not expressed in the cancer cell of origin, or wild type TET2 if its expression has been downregulated due to promoter hypermethylation [73]. Indeed, 5-azacytidine treatment of aged adipose-derived mesenchymal stem cells led to upregulated TET2/3 expression and increased 5hmC levels, as well as improved proliferation and osteogenic differentiation [117].
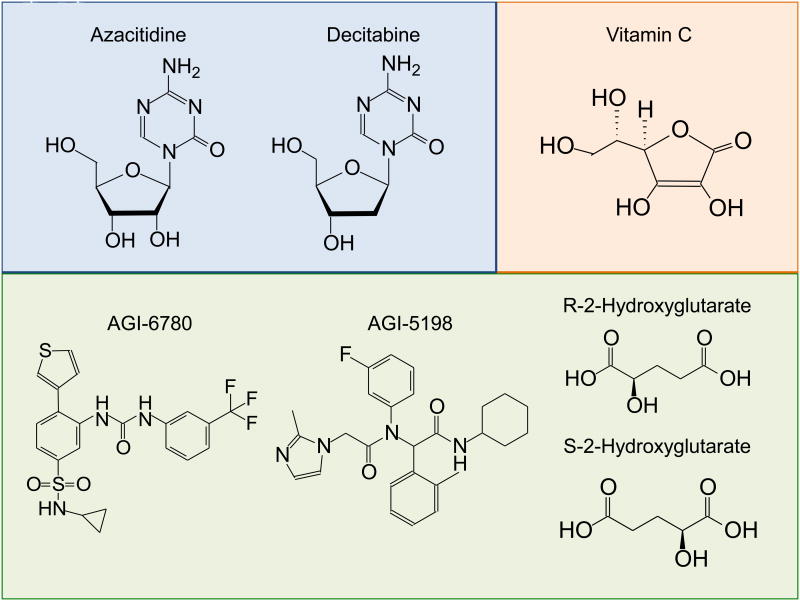
Figure 5. Structures of small molecules with potential for directly or indirectly targeting TET activity.
Shown are the chemical structures of the DNMT inhibitors azacitidine and its ribonucleoside analogue decitabine, and the structures of the IDH inhibitors AGI-6780 and -5198. Vitamin C potentiates TET activity, whereas R-2-hydroxyglutarate (R-2HG) is produced by cancer-associated mutant IDH1 and IDH2. Both R-2HG and its stereoisomer S-2HG inhibit the activity of TET proteins and other Fe(II) and 2-oxoglutarate-dependent dioxygenases, with S-2HG often being more effective.
Selective inhibitors of mutant IDH
Although the S-2HG stereoisomer is a stronger inhibitor as discussed above, the catalytic activities of TET proteins are moderately inhibited by R-2HG, the metabolic product of the recurrently-mutated IDH1 and IDH2 variants in cancer [106,108,109,113]. Therefore, preventing production of the 2HG oncometabolite is a valid therapeutic strategy. Two selective inhibitors of these mutant IDH proteins include AGI-6780 [118] and AGI-5198 [119], which induce differentiation of TF-1 erythroleukemia and TS603 glioma cells respectively (Figure 5). Further studies are needed to elucidate the effects of IDH inhibitors on TET activity.
Small molecules targeting EglN prolyl hydroxylases
The R-2HG and S-2HG enantiomers (Figure 5) promote and inhibit the activity of EglN prolyl-4-hydroxylases respectively [108,120]. EglN regulates HIF1α protein levels through proline hydroxylation followed by E3-ubiquitin ligase mediated proteasomal degradation. Because S-2HG can antagonize the increased proliferation of TF-1 cells induced by IDH mutants or depletion of TET2, cell-permeant analogs of S-2HG could potentially be exploited as anti-cancer compounds through the inhibition of pro-oncogenic EglN and enhanced degradation of HIF.
Vitamin C as a potentiator of TET protein activity
Vitamin C, also known as L-ascorbic acid, is an essential nutrient for mammals, a critical antioxidant, and an essential factor for collagen, catecholamine and carnitine biosynthesis [114,121]. In addition, Vitamin C acts as an electron donor to adjust the redox state of enzymes by recycling Fe(II), and this property facilities the enzymatic activity of many Fe(II)-2OG-dependent dioxygenases including TET proteins [114,121]. The enhancing effect of vitamin C on TET-mediated demethylation has been reported in different cellular systems, including mESC and MEF [122-125]. In mESC, vitamin C treatment caused a global increase of 5hmC levels associated with demethylation of promoters and increasing germline gene expression. In terms of DNA methylation status, vitamin C-treated ESC resemble “ground state” ESC that have been treated with GSK and MEK inhibitors (2 inhibitors, 2i), exhibiting demethylation at blastocyst-associated regions and with methylation remaining at imprinted/intracisternal A particle (IAP) regions [124,126]. The effect was specific to vitamin C over other reducing agents and antioxidants, including glutathione, NAPDH, L-cysteine and DTT [122,124], even though vitamin C – the most effective compound for recycling Fe(II) during catalysis by dioxygenases – can be substituted by other antioxidants in other physiological processes, such as the response of EglN prolyl hydroxylases to hypoxia in vitamin C-deficient mice [127]. It is worth testing whether Vitamin C might be beneficial in cancer by enhancing TET activity.
Concluding remarks
Five years after their discovery, it is well established that TET proteins are mutated or their expression levels or activities are abnormally altered in a wide range of cancers. What remains is to define their activities both in mouse models and in human cancers, determine how their dysregulation leads to disruption of the normal balance of DNA methylation and demethylation, and to find ways of manipulating their expression and activity therapeutically.
Highlights.
Loss-of-function mutations of TET proteins and decreased 5hmC are frequently observed in cancer
Numerous TET regulators influence cancer initiation and progression
There is a close reciprocal interplay between TET proteins and enzymes involved in cell metabolism
Small molecules can be exploited as modulators of TET expression and activity.
Acknowledgments
This work was supported by NIH R01 grants HD065812, AI44432 and CA151535, grant RM1-01729 from the California Institute for Regenerative Medicine and Translational Research Program Award 6187-12 from the Leukemia Society of America (to A.R.). Y.H is a Fellow of the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 2.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 4.Ooi SK, et al. Mammalian cytosine methylation at a glance. J Cell Sci. 2009;122:2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 6.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 7.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pastor WA, et al. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nature reviews: molecular cell biology. 2013 doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashimoto H, et al. Selective excision of 5-carboxylcytosine by a thymine DNA glycosylase mutant. J Mol Biol. 2013;425:971–976. doi: 10.1016/j.jmb.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, et al. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ono R, et al. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- 17.Meyer C, et al. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009;23:1490–1499. doi: 10.1038/leu.2009.33. [DOI] [PubMed] [Google Scholar]
- 18.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 19.Langemeijer SM, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 20.Abdel-Wahab O, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itzykson R, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31:2428–2436. doi: 10.1200/JCO.2012.47.3314. [DOI] [PubMed] [Google Scholar]
- 22.Tefferi A, et al. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia. 2009;23:1343–1345. doi: 10.1038/leu.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grossmann V, et al. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia. 2011;25:877–879. doi: 10.1038/leu.2011.10. [DOI] [PubMed] [Google Scholar]
- 24.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nibourel O, et al. Incidence and prognostic value of TET2 alterations in de novo acute myeloid leukemia achieving complete remission. Blood. 2010;116:1132–1135. doi: 10.1182/blood-2009-07-234484. [DOI] [PubMed] [Google Scholar]
- 26.Weissmann S, et al. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia. 2012;26:934–942. doi: 10.1038/leu.2011.326. [DOI] [PubMed] [Google Scholar]
- 27.Konstandin N, et al. Genomic 5-hydroxymethylcytosine levels correlate with TET2 mutations and a distinct global gene expression pattern in secondary acute myeloid leukemia. Leukemia. 2011;25:1649–1652. doi: 10.1038/leu.2011.134. [DOI] [PubMed] [Google Scholar]
- 28.Bejar R, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bejar R, et al. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J Clin Oncol. 2011;29:504–515. doi: 10.1200/JCO.2010.31.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu WJ, et al. Prognostic significance of TET2 mutations in adult patients with acute myeloid leukemia: a meta-analysis. Leuk Lymphoma. 2014 doi: 10.3109/10428194.2014.893308. [DOI] [PubMed] [Google Scholar]
- 31.Euba B, et al. A meta-analysis of TET2 mutations shows a distinct distribution pattern in de novo acute myeloid leukemia and chronic myelomonocytic leukemia. Leuk Lymphoma. 2012;53:1230–1233. doi: 10.3109/10428194.2011.639878. [DOI] [PubMed] [Google Scholar]
- 32.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu L, et al. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155:1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 34.Lin TL, et al. Clonal leukemic evolution in myelodysplastic syndromes with TET2 and IDH1/2 mutations. Haematologica. 2014;99:28–36. doi: 10.3324/haematol.2013.091249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosmider O, et al. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs) Blood. 2009;114:3285–3291. doi: 10.1182/blood-2009-04-215814. [DOI] [PubMed] [Google Scholar]
- 36.Kosmider O, et al. TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica. 2009;94:1676–1681. doi: 10.3324/haematol.2009.011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith AE, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116:3923–3932. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 38.Pronier E, et al. Inhibition of TET2-mediated conversion of 5-methylcytosine to 5-hydroxymethylcytosine disturbs erythroid and granulomonocytic differentiation of human hematopoietic progenitors. Blood. 2011;118:2551–2555. doi: 10.1182/blood-2010-12-324707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, et al. Decreased 5-hydroxymethylcytosine levels are associated with TET2 mutation and unfavorable overall survival in myelodysplastic syndromes. Leuk Lymphoma. 2013;54:2466–2473. doi: 10.3109/10428194.2013.778408. [DOI] [PubMed] [Google Scholar]
- 40.Haferlach T, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papaemmanuil E, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Odejide O, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123:1293–1296. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemonnier F, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120:1466–1469. doi: 10.1182/blood-2012-02-408542. [DOI] [PubMed] [Google Scholar]
- 44.Itzykson R, et al. Clonal architecture of chronic myelomonocytic leukemias. Blood. 2013;121:2186–2198. doi: 10.1182/blood-2012-06-440347. [DOI] [PubMed] [Google Scholar]
- 45.Quivoron C, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 46.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ko M, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci U S A. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Z, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011 doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Busque L, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44:1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanssens K, et al. SRSF2-P95 Hotspot Mutation is Highly Associated with Advanced Forms of Mastocytosis and Mutations in Epigenetic Regulator Genes. Haematologica. 2014 doi: 10.3324/haematol.2013.095133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meggendorfer M, et al. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML) Blood. 2012;120:3080–3088. doi: 10.1182/blood-2012-01-404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 53.Graubert TA, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2012;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Papaemmanuil E, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang SJ, et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood. 2012;119:4480–4485. doi: 10.1182/blood-2011-11-390252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gelsi-Boyer V, et al. ASXL1 mutation is associated with poor prognosis and acute transformation in chronic myelomonocytic leukaemia. Br J Haematol. 2010;151:365–375. doi: 10.1111/j.1365-2141.2010.08381.x. [DOI] [PubMed] [Google Scholar]
- 57.Muto T, et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J Exp Med. 2013;210:2627–2639. doi: 10.1084/jem.20131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abdel-Wahab O, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med. 2013;210:2641–2659. doi: 10.1084/jem.20131141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheuermann JC, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakata-Yanagimoto M, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:171–175. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 62.Palomero T, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46:166–170. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Couronne L, et al. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366:95–96. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- 64.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sato Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 66.Nickerson ML, et al. Somatic alterations contributing to metastasis of a castration-resistant prostate cancer. Hum Mutat. 2013;34:1231–1241. doi: 10.1002/humu.22346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang H, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2012 doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jin SG, et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71:7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song SJ, et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lian CG, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hsu CH, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2:568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- 72.Sun M, et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci U S A. 2013;110:9920–9925. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim YH, et al. TET2 promoter methylation in low-grade diffuse gliomas lacking IDH1/2 mutations. J Clin Pathol. 2011;64:850–852. doi: 10.1136/jclinpath-2011-200133. [DOI] [PubMed] [Google Scholar]
- 74.Cheng J, et al. An extensive network of TET2-targeting MicroRNAs regulates malignant hematopoiesis. Cell Rep. 2013;5:471–481. doi: 10.1016/j.celrep.2013.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu X, et al. Tet and TDG Mediate DNA Demethylation Essential for Mesenchymal-to-Epithelial Transition in Somatic Cell Reprogramming. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 76.Liu C, et al. Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS One. 2013;8:e62828. doi: 10.1371/journal.pone.0062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ko M, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hino S, et al. Inhibition of the Wnt signaling pathway by Idax, a novel Dvl-binding protein. Mol Cell Biol. 2001;21:330–342. doi: 10.1128/MCB.21.1.330-342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iyer LM, et al. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paez D, et al. Association of common gene variants in the WNT/beta-catenin pathway with colon cancer recurrence. Pharmacogenomics J. 2013 doi: 10.1038/tpj.2013.20. [DOI] [PubMed] [Google Scholar]
- 81.Kojima T, et al. Decreased expression of CXXC4 promotes a malignant phenotype in renal cell carcinoma by activating Wnt signaling. Oncogene. 2009;28:297–305. doi: 10.1038/onc.2008.391. [DOI] [PubMed] [Google Scholar]
- 82.Wang Y, Zhang Y. Regulation of TET protein stability by calpains. Cell Rep. 2014;6:278–284. doi: 10.1016/j.celrep.2013.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Storr SJ, et al. The calpain system and cancer. Nat Rev Cancer. 2011;11:364–374. doi: 10.1038/nrc3050. [DOI] [PubMed] [Google Scholar]
- 84.Warburg O, et al. The metabolism of tumors in the body. J Gen Physiol. 1927;7:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 86.Vosseller K, et al. Diverse regulation of protein function by O-GlcNAc: a nuclear and cytoplasmic carbohydrate post-translational modification. Curr Opin Chem Biol. 2002;6:851–857. doi: 10.1016/s1367-5931(02)00384-8. [DOI] [PubMed] [Google Scholar]
- 87.Ozcan S, et al. Modulation of transcription factor function by O-GlcNAc modification. Biochim Biophys Acta. 2010;1799:353–364. doi: 10.1016/j.bbagrm.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sakabe K, et al. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci U S A. 2010;107:19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fujiki R, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480:557–560. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fong JJ, et al. beta-N-Acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J Biol Chem. 2012;287:12195–12203. doi: 10.1074/jbc.M111.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Myers SA, et al. Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2011;108:9490–9495. doi: 10.1073/pnas.1019289108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chu CS, et al. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci U S A. 2014;111:1355–1360. doi: 10.1073/pnas.1323226111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen Q, et al. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Deplus R, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vella P, et al. Tet Proteins Connect the O-Linked N-acetylglucosamine Transferase Ogt to Chromatin in Embryonic Stem Cells. Mol Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 96.Ito R, et al. TET3-OGT interaction increases the stability and the presence of OGT in chromatin. Genes Cells. 2014;19:52–65. doi: 10.1111/gtc.12107. [DOI] [PubMed] [Google Scholar]
- 97.Shi FT, et al. Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J Biol Chem. 2013;288:20776–20784. doi: 10.1074/jbc.M113.460386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Q, et al. Differential Regulation of the Ten-Eleven Translocation (TET) Family of Dioxygenases by O-Linked beta-N-Acetylglucosamine Transferase (OGT) J Biol Chem. 2014;289:5986–5996. doi: 10.1074/jbc.M113.524140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Slawson C, et al. A mitotic GlcNAcylation/phosphorylation signaling complex alters the posttranslational state of the cytoskeletal protein vimentin. Mol Biol Cell. 2008;19:4130–4140. doi: 10.1091/mbc.E07-11-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goto H, et al. Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells. 2002;7:11–17. doi: 10.1046/j.1356-9597.2001.00498.x. [DOI] [PubMed] [Google Scholar]
- 101.Yang WH, et al. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- 102.Itkonen HM, et al. O-GlcNAc transferase integrates metabolic pathways to regulate the stability of c-MYC in human prostate cancer cells. Cancer Res. 2013;73:5277–5287. doi: 10.1158/0008-5472.CAN-13-0549. [DOI] [PubMed] [Google Scholar]
- 103.Olivier-Van Stichelen S, et al. The hexosamine biosynthetic pathway and O-GlcNAcylation drive the expression of beta-catenin and cell proliferation. Am J Physiol Endocrinol Metab. 2012;302:E417–424. doi: 10.1152/ajpendo.00390.2011. [DOI] [PubMed] [Google Scholar]
- 104.Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem Sci. 2011;36:7–18. doi: 10.1016/j.tibs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 105.Dang L, et al. IDH mutations in glioma and acute myeloid leukemia. Trends Mol Med. 2010;16:387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 106.Losman JA, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ye D, et al. R-2-hydroxyglutarate as the key effector of IDH mutations promoting oncogenesis. Cancer Cell. 2013;23:274–276. doi: 10.1016/j.ccr.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Koivunen P, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sasaki M, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kats LM, et al. Proto-Oncogenic Role of Mutant IDH2 in Leukemia Initiation and Maintenance. Cell Stem Cell. 2014;14:329–341. doi: 10.1016/j.stem.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xiao M, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Adam J, et al. Rare insights into cancer biology. Oncogene. 2013 doi: 10.1038/onc.2013.222. [DOI] [PubMed] [Google Scholar]
- 114.Englard S, Seifter S. The biochemical functions of ascorbic acid. Annu Rev Nutr. 1986;6:365–406. doi: 10.1146/annurev.nu.06.070186.002053. [DOI] [PubMed] [Google Scholar]
- 115.Yang X, et al. Targeting DNA methylation for epigenetic therapy. Trends Pharmacol Sci. 2010;31:536–546. doi: 10.1016/j.tips.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Santi DV, et al. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983;33:9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- 117.Yan X, et al. 5-azacytidine improves the osteogenic differentiation potential of aged human adipose-derived mesenchymal stem cells by DNA demethylation. PLoS One. 2014;9:e90846. doi: 10.1371/journal.pone.0090846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang F, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 119.Rohle D, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kaelin WG., Jr Cancer and altered metabolism: potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb Symp Quant Biol. 2011;76:335–345. doi: 10.1101/sqb.2011.76.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Banhegyi G, et al. Subcellular compartmentation of ascorbate and its variation in disease states. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbamcr.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 122.Yin R, et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc. 2013;135:10396–10403. doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]
- 123.Minor EA, et al. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J Biol Chem. 2013;288:13669–13674. doi: 10.1074/jbc.C113.464800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Blaschke K, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dickson KM, et al. Ascorbate-induced generation of 5-hydroxymethylcytosine is unaffected by varying levels of iron and 2-oxoglutarate. Biochem Biophys Res Commun. 2013;439:522–527. doi: 10.1016/j.bbrc.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ficz G, et al. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nytko KJ, et al. Vitamin C is dispensable for oxygen sensing in vivo. Blood. 2011;117:5485–5493. doi: 10.1182/blood-2010-09-307637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dawlaty MM, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dawlaty MM, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang Y, et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2014;111:1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lorsbach RB, et al. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 135.Huang H, et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc Natl Acad Sci U S A. 2013;110:11994–11999. doi: 10.1073/pnas.1310656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kalender Atak Z, et al. High accuracy mutation detection in leukemia on a selected panel of cancer genes. PLoS One. 2012;7:e38463. doi: 10.1371/journal.pone.0038463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kunimoto H, et al. Tet2 disruption leads to enhanced self-renewal and altered differentiation of fetal liver hematopoietic stem cells. Sci Rep. 2012;2:273. doi: 10.1038/srep00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Madzo J, et al. Hydroxymethylation at gene regulatory regions directs stem/early progenitor cell commitment during erythropoiesis. Cell Rep. 2014;6:231–244. doi: 10.1016/j.celrep.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 140.Berman BP, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yamazaki J, et al. Effects of TET2 mutations on DNA methylation in chronic myelomonocytic leukemia. Epigenetics. 2012;7:201–207. doi: 10.4161/epi.7.2.19015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Patel JP, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pfaffeneder T, et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat Chem Biol. 2014 doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- 144.Cortellino S, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]