Intersection of calorie restriction and magnesium in the suppression of genome-destabilizing RNA–DNA hybrids (original) (raw)
Abstract
Dietary calorie restriction is a broadly acting intervention that extends the lifespan of various organisms from yeast to mammals. On another front, magnesium (Mg2+) is an essential biological metal critical to fundamental cellular processes and is commonly used as both a dietary supplement and treatment for some clinical conditions. If connections exist between calorie restriction and Mg2+ is unknown. Here, we show that Mg2+, acting alone or in response to dietary calorie restriction, allows eukaryotic cells to combat genome-destabilizing and lifespan-shortening accumulations of RNA–DNA hybrids, or R-loops. In an R-loop accumulation model of Pbp1-deficient Saccharomyces cerevisiae, magnesium ions guided by cell membrane Mg2+ transporters Alr1/2 act via Mg2+-sensitive R-loop suppressors Rnh1/201 and Pif1 to restore R-loop suppression, ribosomal DNA stability and cellular lifespan. Similarly, human cells deficient in ATXN2, the human ortholog of Pbp1, exhibit nuclear R-loop accumulations repressible by Mg2+ in a process that is dependent on the TRPM7 Mg2+ transporter and the RNaseH1 R-loop suppressor. Thus, we identify Mg2+ as a biochemical signal of beneficial calorie restriction, reveal an R-loop suppressing function for human ATXN2 and propose that practical magnesium supplementation regimens can be used to combat R-loop accumulation linked to the dysfunction of disease-linked human genes.
INTRODUCTION
The longevity of various organisms is intimately linked to the lifespan of their cells (1,2). Research in the past few decades has revealed that cellular and organismal lifespan can be genetically or pharmacologically modulated opening the door for a line of thought that views aging as a malleable biological process that can be significantly, and fortunately, stalled (2,3).
Calorie restriction (CR), or a decrease in dietary glucose intake, is the most effective strategy to extend the lifespan of healthy or diseased cells (2,3). Major interconnected processes mediating beneficial effects of CR include the constriction of glycolysis, activation of the nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylase Sir2 (founding member of the conserved family of sirtuin longevity proteins), inhibition of TOR (target of rapamycin) and stabilization of the highly repetitive and thus genome destabilization prone ribosomal RNA gene repeats (rDNA) (4–11).
Living organisms typically harbor hundreds to thousands of rDNA units per cell necessary to meet its protein translation needs (10). We recently reported that, in the budding yeast Saccharomyces cerevisiae, CR can promote rDNA stability by abolishing deleterious accumulations of nucleic acid structures called R-loops (12,13). These are formed when nascent RNA competes with the DNA double helix hybridizing with one DNA strand while displacing the other. R-loops can be particularly stable if the displaced single-stranded DNA harbors so called G-quadruplex-DNA (G4DNA) sequences that lock this displaced DNA strand into a looping structure (12,14). Although many biological processes including eukaryotic gene expression can be beneficially controlled by R-loops, their aberrant accumulation compromises genome stability in different organisms from yeast to human. In fact, genes mutated in various human diseases including cancer and neurodegeneration have recently emerged as suppressors of R-loop accumulation (13,15). This highlights the importance of identifying broadly effective and practical strategies to combat deleterious R-loop buildup.
R-loops stably accumulate at G4DNA-containing sites within the intergenic regions of rDNA in yeast cells lacking the conserved RNA-binding factor Pbp1, which is orthologous to the human ATXN2 gene product mutated in some neurodegenerative diseases (12,16,17). R-loop buildup interferes with rDNA replication triggering cellular lifespan-shortening aberrant recombination within the repeats (12,18). Subjecting Pbp1 knockout (pbp1Δ) cells to a CR regimen that entails decreasing glucose concentration from 2% to 0.05% abolishes R-loop buildup restoring rDNA stability and cellular lifespan via a process that somehow requires two highly conserved R-loop suppressors, Pif1 and Rnh1/201 (RNaseH enzymes Rnh1 and Rnh201) (12). Pif1 is a helicase that suppresses R-loops by resolving G4DNA and RNA–DNA hybrids (19,20). RNaseH enzymes degrade the RNA component of hybrids and have been recently shown to counter deleterious R-loop accumulations in vivo (21,22).
Importantly, mammalian CR responses depend on highly specific diets that generally include a 20–40% decrease in daily caloric intake, conditions that are too complicated and most likely too severe for individuals already suffering from disease (2). Therefore, we aimed to gain insight into yeast responses to CR in an effort to identify more direct and practical interventions that can potentially counter R-loop accumulation in both yeast and human cells, which are separated by over a billion years of evolution.
By first investigating the potential impact of CR on common effectors of yeast Pif1 and Rnh1/201 enzymes, we uncovered a CR-induced Mg2+-driven cascade that culminates in R-loop repression. CR increases intracellular Mg2+ by boosting the expression of major Mg2+ transporters. Disrupting these transporters or decreasing environmental Mg2+ levels compromises CR-dependent repression of R-loop buildup in pbp1Δ cells. Supplementing cells directly with Mg2+ without CR targets R-loop suppressors, counters R-loop accumulation, restores rDNA stability and increases lifespan in Pbp1-deficient cells. In an effort to assess the evolutionary conservation of processes uncovered in yeast, we identified human ATXN2 as an R-loop suppressor and found that Mg2+ supplementation counters R-loop accumulation in ATXN2-deficient human cells by a process that requires analogous Mg2+ transporters and R-loop suppressors. Taken together, our work identifies Mg2+ as a biochemical signal of beneficial calorie restriction and shows that magnesium supplementation can be used to combat R-loop accumulation pertinent to various human diseases.
MATERIALS AND METHODS
Yeast strains and materials
Endogenous genes were deleted as described (23). Yeast strains and primers used are listed (Supplementary Tables S3 and S4). R-loop detection via direct pull downs employed the anti-RNA–DNA antibody from Kerafast (cat# ENH001), which exhibited lower enrichments but higher specificity compared to antibodies that we used previously in chromatin immunoprecipitation (ChIP) (12). Anti-RNAPII, anti-diAcH3K9/14, anti-TAP and anti-β-Actin antibodies were purchased from BioLegend (665004), Millipore (06-599), Sigma (P1291-500UL) and Thermo Scientific (MA1-744), respectively. Anti-GFP was a kind gift from D. Moazed (Harvard University). The Stm1 YEplac181 plasmid or empty vector control were kind gifts from F.B. Johnson (University of Pennsylvania) (24).
Yeast caloric restriction and environmental Mg2+ modulation
Media containing 0.05% glucose, instead of the standard 2% glucose, was used in YEP media for CR treatment (12,25). For yeast Mg2+ supplementation, 100 mM MgCl2 or CaCl2 was used unless otherwise indicated. For environmental Mg2+ depletion experiments, synthetic complete media was made using Yeast Nitrogen Base lacking magnesium (Formedium, CYN2801) before adding MgCl2 to a normal level of 10 mM or depleted level of 1 mM. Media was always replenished after overnight growth to ensure that cells were not glucose starved.
Magnesium concentration measurements
Exponentially growing yeast cells were harvested (5.0 × 107) in 0.75 ml of growth media and 0.25 ml of growth media containing 1 mM MagnesiumGreen (Life Technologies: M-3735) was added. Cell-MagnesiumGreen mixture was allowed to incubate for 45 min at 30°C with rotation. Cells were washed three times in growth media and allowed to incubate for an additional 30 min at 30°C with rotation. A total of 0.25 ml of cells was transferred to clear flat-bottomed 96-well plate. Absorbance was measured with a Perkin Elmer Victor3 plate reader. Relative magnesium concentration in arbitrary absorbance units was calculated using Beer's law and the following formula: [CellsMg2+] / [MediaMg2+] = AbsorbanceCells / AbsorbanceMedia, where ‘Media’ represents the growth media/magnesium-green mixture.
Yeast ChIP
ChIP analysis was performed as described with minor modifications (12,23,26). For standard ChIP assays, overnight yeast cultures (25 ml) were diluted to 50 ml with fresh media and were grown to an OD600 of 1.8 and cross-linked with 1% formaldehyde at room temperature (RT) for 30 min. The reaction was quenched with glycine at a final concentration of 125 mM for 5 min at RT. Cells were washed with cold TBS (20 mM Tris-HCl pH 7.6 and 150 mM NaCl). Cell pellets were resuspended in 400 μl of lysis buffer [50 mM HEPES-KOH pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, complete protease inhibitor (Roche)]. Cells were bead-beated with an equal volume of glass beads twice for 30 s with intermittent 2 min incubation on ice. Lysates were sonicated 3 times for 20 s at 40% amplitude with intermittent incubations on ice. Two consecutive rounds of centrifugation at 16 000 r.c.f. for 5 and 15 min clarified lysates. For non-TAP IP, 150 μl of lysate was incubated at 4°C for 2 h with 2.0 μg of antibody and further incubated with 30 μl of 50% slurry of pre-washed Protein A-Sepharose beads at 4°C for 1 h. For anti-TAP IP, 150 μl of lysate was incubated with 30 μl of 50% slurry of pre-washed IgG FastFlow beads at 4°C for 2 h. Beads were washed 3 times with lysis buffer, once with LiCl buffer (10 mM Tris-HCl pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA) and once with TE (10 mM Tris-HCl pH 8.0, 1 mM EDTA), before elution with 50 mM Tris-HCl pH 8.0, 10 mM EDTA and 1% SDS. The eluate was incubated at 65°C overnight. RNaseA was added at a final concentration of 0.2 μg/μl and incubated at 37°C for 30 min, followed by proteinase K (to 0.2 μg/μl) and glycogen (to 0.03 μg/μl) treatment for 2 h at 37°C. Dilutions for IP and input DNA were 1:2 and 1:1000, respectively. Primers are listed in Supplementary Table S4. For qPCR analyses, IP signals were divided by Input signals and normalized to CUP1 controls using the following calculations: [100 × 2 ∧ (Adjusted InputGene – Adjusted IPGene)] / [100 × 2 ∧ (Adjusted InputCUP1 – Adjusted IPCUP1)], where Adjusted Input = CTInput − log21000 and Adjusted IP = CTIP − log22. Standard deviations are shown. For EtBr-stained gel-based visualizations, relative fold enrichments were obtained according to the following calculations: [_rDNA_IP / _CUP1_IP] / [r_DNA_Input / _CUP1_Input].
Quantitative PCR (qPCR)
Quantitative real-time PCR was performed using the Bio-Rad CFX Connect Real-Time system. Ten microliters qPCR reactions contained 2X SsoAdvanced SYBR Green Supermix (Bio-Rad) or SensiFAST SYBR No-ROX qPCR (FroggaBio, BIO-98020), 200 nM for each of the forward and reverse primers and 1 μl of 10 ng/μl of genomic DNA, diluted Input or IP ChIP DNA. PCR parameters were 1 cycle of 95°C for 35 s, 60°C for 30 s, followed by 39 time of 95°C for 5 s, 60°C for 30 s and the final melt curve at 65°C to 95°C in 0.5°C increment at 5 s/step.
RNA extraction
Total RNA was prepared from logarithmically growing cells using hot phenol. Cells were resuspended in 400 μl of AE buffer [50 mM NaOAc (pH 5.3), 10 mM EDTA, in 0.1% DEPC ddH2O] and 40 μl of 10% SDS was added. RNA was extracted using 440 μl of acidic phenol (pH 4.5), vortexed and incubated at 65°C for 4 min. The samples were rapidly chilled on dry ice/EtOH bath until crystals appeared before centrifugation for 5 min at 16 000 r.c.f. at RT. The upper phase was transferred to a fresh tube, and 1 volume of phenol:chloroform 5:1 was added. The sample was then centrifuged at 16 000 rcf for 5 min at 4°C. The upper phase was transferred to a new tube before RNA precipitation. One-tenth volume of 3M NaOAc (pH 5.3) and 2.5 volumes of cold 100% EtOH were added, mixed and centrifuged at 16 000 r.c.f. for 5 min at 4°C. Upon removal of the supernatant, the RNA pellet was washed with 2.5 volumes of 80% EtOH. A total of 0.1% DEPC ddH2O was used to resuspend the pellet. A260:A280 and A260:A230 ratios from Nanodrop confirmed RNA quality. Samples were stored in −20°C until further use.
DNase-I treatment and reverse transcription (RT)
One microgram of total RNA was treated with 1 unit of DNase-I (Invitrogen) to further remove genomic DNA contaminations. Twenty microliters RT reactions were carried out using 10 mM dNTPs, 50 μM random nonamers (Sigma), 500 ng total RNA, 5X First-Strand Buffer (Invitrogen), 100 mM DTT, 40 U/μl RNaseOUT (Invitrogen) and 200 U/μl M-MLV reverse transcriptase (Invitrogen) at 23°C for 10 min, 37°C for 60 min and 70°C for 15 min. Twenty microliters minus-RT reaction without M-MLV reverse transcriptase was also set up in parallel. The RT reaction was diluted 1:4 and 4 μl were used in qPCR amplification. Linearity tests were performed and primers are listed in Supplementary Table S4.
USCE
Assays were performed as described by counting half-sectored colonies grown on media containing low adenine concentration (8,23). Cells were grown to OD600 = 0.4–0.8, sonicated briefly and spread (about 400 cells per plate) on thick plates (5 mg/l adenine). Incubation was at 30°C for 3 days, 4°C for 3 days, then RT for 1 day. Rates were obtained by dividing the number of half-sectored colonies by the total number of colonies excluding completely red colonies. For Mg2+ supplementation unequal sister chromatid exchange (USCE) experiments, cells were grown overnight under 100 mM Mg2+ conditions, re-diluted, further grown to OD600 = 0.4–0.8 and then plated on low adenine concentration media.
Yeast replicative lifespan analyses
Replicative lifespan of yeast strains was determined by micromanipulation monitoring the number of times that an initially new mother cell buds (8). Strains being compared to each other were analyzed in parallel but may be shown on separate plots for clarity as indicated in Supplementary Table S2. The Wilcoxon rank-sum statistical test was conducted.
Yeast cell culture for DNA Combing
Asynchronous cultures were grown in YPAD liquid medium in a water bath shaker at 30°C to early logarithmic phase (OD600 ∼0.20–0.30). Cells were arrested in G1 phase with 0.83 μl of 5 mg/ml alpha-factor per ml of culture volume for 75 min. An additional 0.33 μl/ml of 5 mg/ml alpha-factor was added for another 45 min. Cells were labeled with BrdU by incubating cells with 400 μg/ml for 30 min and then releasing into S-phase with 100 μg/ml pronase for 20 min before harvesting.
Agarose plug preparation for DNA combing
Cells were harvested by centrifuging 1.2 × 108 cells in pre-chilled tubes at 3000 r.p.m. for 2 min at 4°C. Cells were washed in ice cold TE50 buffer (10 mM Tris-Cl, pH 7.0, 50 mM EDTA) and resuspended in 160 μl SCE buffer (1 M sorbitol, 100 mM sodium citrate, 10 mM EDTA pH 8.0, 0.125% β-mercaptoethanol, 10 U/ml zymolase). Cells were gently mixed with 200 μl of pre-melted 1% low melting point agarose, and 110 μl of the resulting mixture was transferred into a plug mold (BioRad 170–3713) for a total of 3 plugs per sample. Once solidified, plugs were incubated in 0.5 ml SCE buffer per plug at 37°C two days, switching with fresh SCE buffer each day. Plugs were rinsed 3 × 1 ml TE50 buffer, and incubated with proteinase K solution (1 mg/ml proteinase K, 1% w/v sarkosyl, 10 mM Tris-Cl pH 7.5, 50 mM EDTA, heated to 50°C for 30 min before use) for 3 days, switching with fresh proteinase K solution each day. Plugs were removed from proteinase K solution and washed 5 times 10 min in TE50 at room temperature. Plugs were stored in 1 ml TE50 at 4°C protected from light.
Plug melting, molecular combing and immunodetection
Experiments were generally done as described (27). One plug was incubated in 150 μl of YOYO-1 solution (1 μl of YOYO-1 in 150 μl TE50) at room temperature for 30 min. Plugs were washed 3 times for 5 min with 10 ml TE50 then incubated in MES buffer (7:3 v/v MES hydrate: MES sodium salt, 50 mM, pH 5.7) for 5 min. MES buffer was removed and replaced with 2 ml fresh MES buffer, and incubated at 72°C for 10–15 min. DNA was combed onto a silanized coverslip at a constant speed of 710 μm/s and hybridized at 60°C for 90 min. Upon mounting onto glass slides, coverslips were dehydrated sequentially in 70%, 90% and anhydrous ethanol for 5 min each, and DNA was denatured in 1 M NaOH for 25 min. Coverslips were blocked in 21 μl blocking buffer (10% w/v albumin in PBS-T, 0.05% Tween-20, 2 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, pH 7.4) for 30 min, and stained with anti-BrdU solution (BrdU antibody Becton Dickson 347580 1:10 dilution in blocking buffer), anti-DNA solution (Millipore MAB3034 1:50 dilution in blocking buffer), and anti-secondary solution (Alexa Fluor 546 anti-mouse IgG1, Molecular Probes A21123, 1:50 dilution, Alexa fluor 648 anti-mouse IgG2, Molecular Probes A21241, 1:50 dilution in blocking buffer) for 1 h at 37°C in a humidity chamber, washing with PBS-T for 5 min three times between each staining. ProLong Gold was added to the cover slip and then imaged.
2D DNA electrophoresis
2D gel analysis was performed as described (12). Briefly, Bgl-II-digested gel plugs were run on a 0.4% agarose gel for 14 h at 1.32 V/cm. Lanes were excised, rotated 90 degrees and run on 1.2% agarose gel at 6.0 V/cm for 6 h. Replication arcs were analyzed following standard Southern blotting using an alpha-P32-labeled rDNA probe.
Human cells, materials and Mg2+ supplementation
HeLa parental and stable shRNA-expressing cell lines were grown in Dulbecco's Modified Eagle's Medium with 10% fetal bovine serum (FBS). Stable HeLa cell lines were generated using shAXTN2 and shCtl plasmids (Oligoengine). All cell lines were cultured in a 37°C incubator with 5% CO2 in a humidified atmosphere.
The following siRNAs were used in transient knockdown experiments involving human cell lines: siCtl. (Silencer Select), siRNaseH1 (SilencerSelect; Cat #4390826), siTRPM7 (Silencer Select #s29516), siPIF1 (ON-TARGETplus SMARTpool; cat# L-014564-01), siMAGT1 (Silencer Select #s224928) and siBRCA2 (Dharmacon; ON-Targetplus SMARTpool siRNAs L-003462-00-0005). Lipofectamine RNAiMAX (Life technologies) was used for all siRNA transfections and polyethylenimine was used for all RNaseH1 overexpression experiments in human cells.
The following antibodies were used for human cell immunofluorescence: rabbit anti-ATXN2 (Sigma cat# HPA021146), rabbit anti-RNaseH1 (Proteintech cat# 15606-1-AP), mouse anti-B23 (Sigma cat# B0556), mouse anti-UBF1 (Santa Cruz cat# sc-13125) and anti-RNA–DNA S9.6 (Kerafast). For immunoblotting, rabbit anti-ATXN2 (Novus cat# NBP-90063) was used at 1:1000.
For human supplementation experiments, media containing 10 mM MgCl2 was prepared fresh prior to every experiment and regular media was replaced with Mg2+-supplemented media or vehicle only media 24 h prior to experiments such as immunofluorescence or Western blotting.
Human knockdowns and plasmid transfections
For siRNA-dependent knockdown of TRPM7 and MAGT1, shATXN2 cells were seeded in 6-well plates and transfected with a final in-media concentration of 5 nM of siCtl, siTRPM7 or siMAGT1. Forty eight hours after post-transfection, cells were harvested and 1 × 105 cells were re-seeded onto poly-L-lysine (PLL) coated coverslips in either vehicle or 10 mM Mg2+-supplemented medium. For each treatment, at least three replicates were used for immunofluorescence and imaging analysis. Twenty four hours later, cells were fixed and subjected to S9.6 immunofluorescence as described further below.
For RNaseH overexpression analysis, HeLa stable cell lines were seeded in 6-well plates and transfected 24 h later with 0.9 μg pcDNA3-Empty or pcDNA3-RNaseH1. Forty eight hours post-transfection, cells were harvested and re-seeded onto PLL-coated coverslips, which were processed for IF 24 h later using anti-RNA–DNA S9.6 and anti-RNaseH1 to quantify RNA–DNA hybrids and confirm RNaseH1 overexpression, respectively.
For BRCA2 knockdown, 2 × 105 HeLa parental cells were plated in 6-well plates and transfected the next day with a final in-media concentration of 5 nM siControl or siBRCA2 (Dharmacon). Twenty four hours post transfection, cells were harvested and 1 × 105 cells re-seeded onto poly-L-lysine coated coverslips in either vehicle or 10 mM Mg2+-supplemented medium. For each treatment, at least three replicates were processed for subsequent IF and imaging analysis. Twenty four hours later, cells were fixed and subjected to S9.6 immunofluorescence as described below.
Human cell immunofluorescence
Cells were fixed using 1% formaldehyde for 15 min, washed 3 times with 1X PBS, permeabilized with 0.3% TritonX-100 and then washed again three times with phosphate buffered saline (PBS). Coverslips were blocked using 5% bovine serum albumin for 1 h at RT and transferred to humidified chambers for antibody incubations. Coverslips were incubated with 60 μl of primary antibody for 1 h at RT with concentrations as follows: 1:250 anti-ATXN2 (Sigma HPA021146), 1:250 anti-RNaseH1 (Proteintech, 15606-1-AP), 1:200 anti-B23 (Sigma, B0556) and 1:250 anti-UBF1 (Santa Cruz sc-13125). For all S9.6 R-loop immunofluorescence experiments, 1:500 of the antibody was used. After washing with PBS, cells were incubated with secondary antibody for 1 h in a dark chamber. Following further washing and DAPI staining, coverslips were mounted onto microscope slides using DAKO fluorescent mounting medium and then sealed with nail polish.
Microscopy
We employed a Nikon C2+ Confocal microscope coupled to NIS-elements AR software (Nikon). For yeast live cell imaging, cells were modified to endogenously express Alr1-GFP alone or endogenously express RAD52-YFP in combination with episomal Nop1-CFP, which allowed us to clearly identify cells that do not contain Rad52 foci in order to determine the percentage of cells that do actually have the foci. Cells were grown into log phase, pelleted via centrifugation, washed and resuspended in PBS before mounting onto slides or plates with a coverslip bottom for microscopy. For RNA–DNA hybrid microscopy in human cells, random fields identified by DAPI staining were captured at 100X magnification. For any given image, 7–8 2D imaging planes were acquired along the z-axis to generate 3D confocal image stacks. DAPI was used to create masks of nuclei and S9.6 intensity values for individual cells were obtained as maximum intensity planes via the NIS-elements AR (Nikon) software. Representative maximum intensity projections adjusted for background and contrast in Photoshop (Adobe) are shown.
Western blotting
For yeast, overnight cultures were diluted to 1.0 × 107 cells/ml and grown for 3 h. A total of 2.5 × 107 cells were then washed with 1 ml of cold ddH2O. Cells were spun down at 16 000 r.c.f. at 4°C for 30 s and resuspended in 500 μl cold ddH2O. Seventy five of Alkali/2-β-ME solution (prepared fresh by mixing 925 μl of 2N NaOH and 75 μl of 14.3 M 2-β-ME), was added and mixed by vortexing. The samples were incubated on ice for 10 min. Seventy five microliters of freshly prepared 50% TCA solution in ice-cold water was added, mixed well by vortexing and incubated on ice for 10 min then followed by centrifuging at 16 000 r.c.f. for 10 min at 4°C. Supernatant was discarded and the protein pellet resuspended in 40 μl of 1X SDS-loading buffer (128 mM Tris-HCl pH 8.8, 4X SDS loading buffer [200 mM Tris-HCl pH 6.8, 400 mM DTT, 8% SDS, 0.4% bromophenol blue, 40% glycerol], 14.3 M 2-β-ME). Samples were electrophoresed on an 8% acrylamide gel, and blotted with 1:1000 primary antibody.
RESULTS
Mg2+ alone or in response to CR counters R-loop buildup
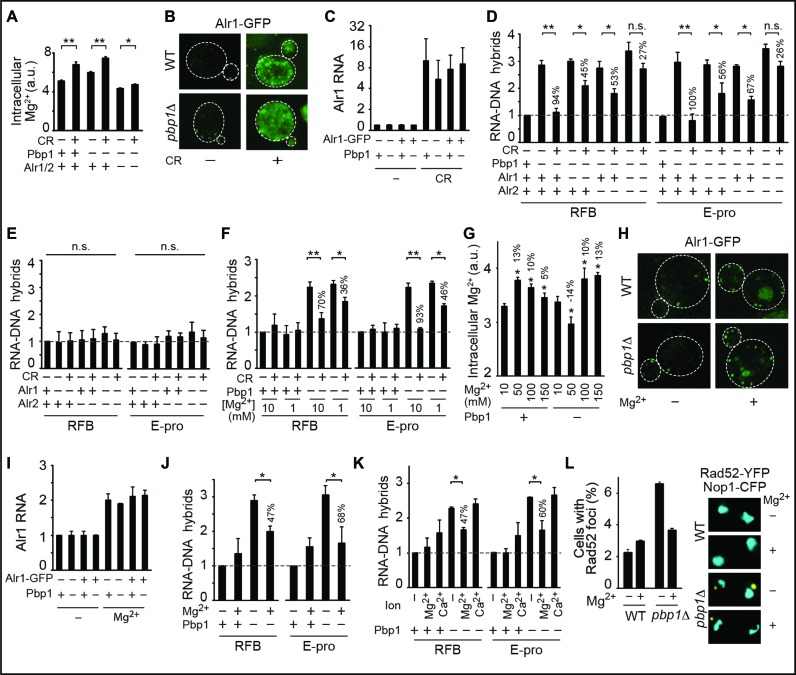
In yeast, Pbp1 binds to age-related long non-coding RNA (lncRNA) molecules at the RFB and E-pro intergenic regions of rDNA effectively limiting R-loops harboring lncRNA-DNA hybrids (10,12,28,29). Our measurements of R-loop levels at RFB and E-pro regions via anti-RNA–DNA ChIP followed by qPCR confirm that pbp1Δ cells exhibit R-loop accumulation repressible by overexpression of the RNA–DNA hybrid-resolving human RNaseH1 enzyme or by subjecting cells to CR (Supplementary Figure S1A and B) (12). CR can modulate rDNA by activating the longevity-promoting Sir2, but CR abolishes hybrid buildup in pbp1Δ and pbp1Δ sir2Δ cells (Supplementary Figure S1B) (5,7,10,12). In fact, sir2Δ cells displayed some hybrid accumulation that was also abolished by CR (Supplementary Figure S1B). Instead of using Sir2, we envisioned that CR might modulate magnesium ions (Mg2+) as both Pif1 and Rnh1/201 employ Mg2+ as a cofactor and are highly sensitive to changes in its levels (30–33). Importantly, we observed that CR increased intracellular Mg2+ levels by ∼20% in wild-type and pbp1Δ cells, where the effect was significantly compromised via deletion of the major and partly redundant Mg2+ transporters Alr1 and Alr2 (Figure 1A). CR boosted Mg2+ transporter protein levels despite down-regulating total cellular protein levels (Figure 1B, Supplementary Figure S1C). We observed similar increases in Mg2+ transporter transcript levels indicating that CR operates at the gene expression level (Figure 1C). Importantly, deletion of Alr1 and Alr2 additively compromised CR-dependent R-loop suppression in pbp1Δ cells (Figure 1D). Loss of Alr1 and/or Alr2 did not increase R-loop levels in wild-type cells (Figure 1E). In addition, we partly disrupted the ability of CR to counter R-loop buildup in pbp1Δ cells by lowering environmental Mg2+ concentration from 10 to 1 mM (Figure 1F). Our data so far reveal that CR relies on Mg2+ to abolish R-loop accumulation.
Figure 1.
Mg2+ acting downstream or independently of calorie restriction (CR) counters R-loop accumulation. (A) CR increases intracellular Mg2+ levels and this in an Alr1/2-dependent manner. Intracellular Mg2+ levels were measured using MagnesiumGreen, a chemical that fluoresces upon binding to this cation (N = 3; Mean ± SD; _t_-test, **P < 0.01, *P < 0.05). Percentage difference relative to corresponding no Mg2+ treatment cells are indicated. (B) Confocal live cell microscopy showing that CR strongly boosts Alr1-GFP protein levels. (C) Reverse transcriptase PCR coupled to quantitative PCR (RT-qPCR) reveals that CR strongly induces Alr1 transcript levels regardless of GFP tagging (N = 3; Mean±SD). (D) Anti-RNA–DNA hybrid ChIP followed by qPCR indicates that the loss of Alr1 and Alr2 additively disrupts CR-dependent repression of R-loop buildup (N = 3; Mean ± SD; _t_-test, **P < 0.01, *P < 0.05, n.s. not statistically significant). Percentage difference relative to corresponding no Mg2+ treatment cells are indicated. (E) Loss of Alr1 and/or Alr2 does not alter R-loop levels in wild-type cells as revealed by anti-RNA–DNA hybrid ChIP (N = 3; Mean ± SD; n.s. not statistically significant for One-way ANOVA). (F) Decreasing Mg2+ concentration hinders the ability of CR to suppress RNA–DNA hybrid buildup (N = 3; Mean ± SD; _t_-test, **P < 0.01, *P < 0.05). (G) Effect of different Mg2+ supplementation regimens on intracellular Mg2+ levels as measured by Magnesium Green fluorescence (N = 3; Mean ± SD; *_t_-test P < 0.05 relative to corresponding 10 mM cells). (H) Confocal microscopy showing that 100 mM Mg2+ supplementation moderately increases Alr1-GFP protein levels. (I) RT-qPCR reveals that 100 mM Mg2+ supplementation moderately induces Alr1 transcript levels regardless of GFP tagging (N = 3; Mean ± SD). (J) Moderate Mg2+ supplementation (100 mM) partly yet significantly counters RNA–DNA hybrid accumulation in pbp1Δ cells (N = 3; Mean ± SD; *_t_-test P < 0.05). (K) A total of 100 mM Mg2+, but not 100 mM Ca2+, counters RNA–DNA hybrid accumulation in pbp1Δ cells (N = 3; Mean ± SD; *_t_-test P < 0.05). (L) A total of 100 mM Mg2+ supplementation decreases Rad52-YFP focus formation in pbp1Δ cells expressing nucleolar Nop1-CFP (N = 100; Mean ± SD).
We next inquired if a direct environmental Mg2+ supplementation regimen could increase intracellular Mg2+ levels and counter R-loop buildup. We assessed the impact of low (50 mM), moderate (100 mM) and high (150 mM) Mg2+ supplementation on intracellular Mg2+ levels. Wild-type and pbp1Δ cells had similar intracellular Mg2+ levels under unsupplemented (10 mM) conditions but only moderate Mg2+ supplementation (hereafter used interchangeably with yeast Mg2+ supplementation or Mg2+) increased intracellular Mg2+ levels by ∼10% irrespective of Pbp1 presence (Figure 1G). As Mg2+ supplementation provides more of the environmental metal for uptake by cells, we did not expect supplementation to boost Mg2+ transporter expression, at least not as much as CR. Indeed, relative to CR, Mg2+ supplementation only mildly increased Mg2+ transporter transcript and protein levels without altering total cellular protein levels (compare Figure 1H,I to B,C; Supplementary Figure S1C). Differences in Alr1-GFP staining patterns are likely linked to the fact that CR is more potent than Mg2+ in terms of inducing transporter expression. Importantly, Mg2+ supplementation significantly decreased R-loop accumulation in pbp1Δ cells by 47% and 68% at RFB and E-pro, respectively (Figure 1J). This significant yet partial R-loop-suppressing effect of Mg2+ supplementation relative to that of CR is consistent with the differential increases in intracellular Mg2+ levels under these two environmental conditions (Figure 1A, D, G and J). Consistent with the sensitivity of Pif1 and RNaseH enzymes to changes in Mg2+ but not Ca2+ levels, Ca2+ supplementation failed to counter R-loop buildup (Figure 1K) (30,31). Preferential dependence of the highly conserved RNaseH enzymes on Mg2+ over Ca2+ stems from the small atomic radius and strict octahedral geometry of Mg2+ allowing this ion to support critical enzymatic reaction intermediates (Supplementary Figure S1D and E) (33). Importantly, the Mg2+ supplementation regimen used did not induce DNA damage and actually countered the elevated genome instability of pbp1Δ cells as detected by a decrease in the formation of DNA repair centers marked by the Rad52 repair and recombination protein (Figure 1L). Our work uncovers an Mg2+-dependent pathway in which CR increases intracellular Mg2+ levels in order to suppress R-loop accumulation. Importantly, this Mg2+-dependent process can be directly engaged in the absence of CR via direct Mg2+ supplementation. It is also important to note that we see changes to intracellular Mg2+ levels in wild-type and mutant cells under CR or Mg2+ supplementation conditions but the consequent benefit of countering R-loop accumulation naturally occurs in cells deficient in R-loop suppression.
R-loop suppressors are effectors of Mg2+
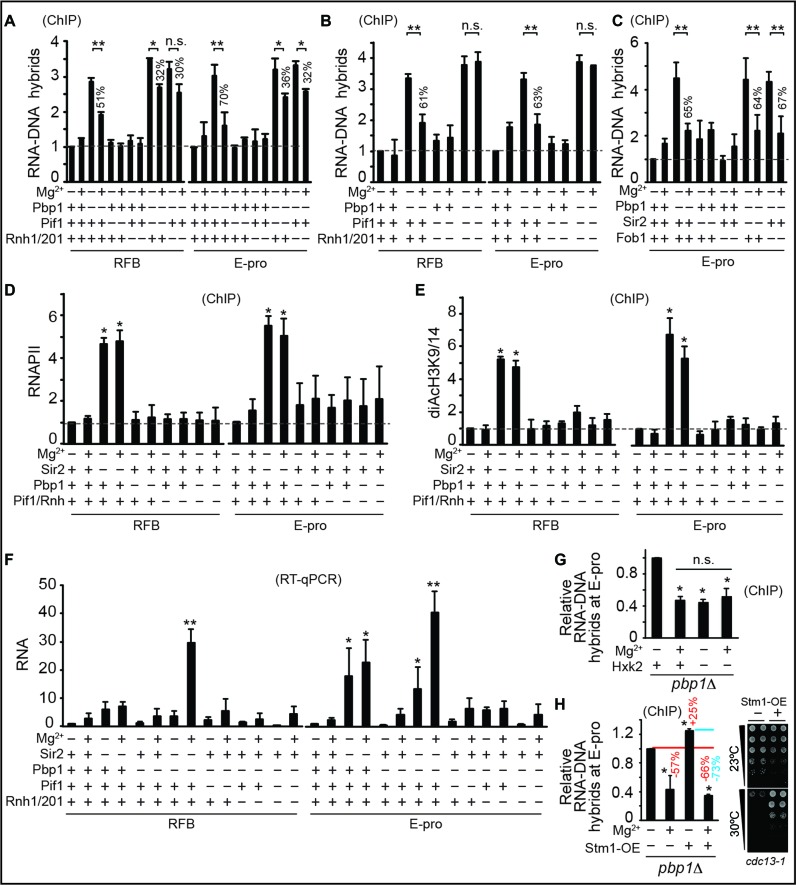
We next aimed to investigate if the Mg2+-sensitive Pif1 and Rnh1/201 do indeed fully account for R-loop suppression by Mg2+ supplementation. We observed that loss of Pif1 and Rnh1/201 additively and fully abrogated Mg2+-dependent suppression of R-loop accumulation in pbp1Δ cells but was ineffectual in wild-type cells (Figure 2A and B). Loss of Pif1 and/or Rnh1/201 did not lead to any R-loop accumulation in wild-type cells (Figure 2A and B). In addition, Mg2+-dependent repression of R-loop buildup was unaltered in cells lacking Sir2 or Fob1, which respectively mediate chromatin silencing and replication fork blocking at rDNA (Figure 2C) (10,34). Moreover, Mg2+ failed to alter enrichments of RNA Polymerase II (RNA Pol II) or the open chromatin mark diAcH3K9/14 (histone H3 acetylated at lysines 9 and 14) regardless of Pbp1 or Pif1/Rnh1/201 status (Figure 2D and E) (23). Yeast Sen1, the ortholog of human SENATAXIN, is an RNA–DNA hybrid helicase whose enzymatic function at yeast rDNA coordinates hybrid resolution and early transcriptional termination of rDNA lncRNA (17,35). In fact, decreases and increases in Sen1 activity respectively lead to increases and decreases in RNA Pol II levels at the studied loci (17,35). In contrast, Mg2+ supplementation did not alter RNA Pol II localization within our experimental conditions (Figure 2D), indicating that Sen1 is not implicated in Mg2+-dependent R-loop suppression.
Figure 2.
Mg2+ acts specifically via suppressors of R-loops to counter their accumulation. (A and B) Mg2+ supplementation requires Mg2+-sensitive Pif1 and Rnh1/201 to counter RNA–DNA hybrid accumulation. (C) Sir2 and Fob1 are dispensable for Mg2+-dependent suppression of RNA–DNA hybrid buildup. (D and E) Mg2+ supplementation does not alter the enrichment of RNA Pol II or diAcH3K9/14. Rnh symbolizes Rnh1/201. Control is sir2Δ. (F) Mg2+ does not decrease free long non-coding RNA (lncRNA) levels but can increase it especially in pbp1Δ sir2Δ cells. (G) Glycolytic constriction via hxk2Δ and Mg2+ supplementation redundantly suppress R-loop accumulation. (H) Mg2+ efficiently suppresses R-loops hyper-stabilized via Stm1 overexpression (left). Overexpressed Stm1 is functional as shown by rescue of cdc13-1 cell growth at 30°C (right). (A–H) Graphs show N = 3, Mean ± SD and _t_-test, **P < 0.01, *P < 0.05, n.s. not statistically significant.
Mg2+ also did not decrease lncRNA levels in pbp1Δ cells arguing against Mg2+-dependent R-loop repression being a reflection of decreased RNA availability for R-loop formation (Figure 2F). In fact, as expected if Mg2+-dependent R-loop resolution frees lncRNAs from hybrids, we found that Mg2+ generally increased free lncRNA levels (Figure 2F). Dependence of Mg2+ on Pif1 and Rnh1/201 cannot be explained by changes in cellular growth rates (Supplementary Figure S2A). Our findings reveal that Pif1 and Rnh1/201 additively and fully account for the ability of Mg2+ to counter R-loop buildup independently of controlling RNA Pol II, chromatin, replication, RNA turnover and overall cellular growth. Despite the total loss of R-loop suppression by Mg2+ in the absence of Pif1 and Rnh1/201, future work should still examine if additional Mg2+-responsive factors may also contribute in a more accessory role.
Next, we asked if Mg2+ supplementation overlaps with CR-induced remodeling of metabolism and if direct glycolytic constriction without environmental CR would counter R-loop accumulation redundantly with Mg2+ supplementation (4,5). Our analysis showed Mg2+ supplementation increasing expression of 60% of top CR-inducible metabolic genes in wild-type and pbp1Δ cells (Supplementary Figure S2B) (4). Half of those Mg2+-responsive genes were induced to a similar extent, while the other half was more responsive to CR. This suggests the existence of a substantial and unexpected overlap between CR and Mg2+ homeostasis. Hexokinase 2 (Hxk2) is an enzyme that phosphorylates glucose in preparation for glycolysis and hxk2Δ cells experience a glycolytic constriction effect that essentially mimics metabolic responses to environmental CR (4). Importantly, we found that hxk2Δ was redundant with Mg2+ supplementation in decreasing R-loop accumulations in pbp1Δ cells (Figure 2G) (5). We conclude that CR-induced metabolic constriction and direct Mg2+ supplementation induce overlapping processes including R-loop accumulation-suppressing Mg2+-dependent processes. Future studies in the field should assess how many of the numerous effects of CR involve or reflect increased Mg2+ levels.
We also aimed to test if we can abolish the relative R-loop-suppressing effect of Mg2+ by overexpressing the R-loop/G4DNA-stabilizing factor Stm1 (12,24). As predicted, overexpression of Stm1 in pbp1Δ cells further increased R-loop levels by ∼25% (Figure 2H). Unexpectedly, the Mg2+-dependent relative decrease in R-loop levels was 73% and 57% in pbp1Δ cells with and without Stm1 overexpression, respectively (Figure 2H) (12,24). Despite this increased relative ability of Mg2+ to repress R-loops in pbp1Δ cells overexpressing Stm1, there remained in these cells an Mg2+-resistant fraction of R-loops similar to that seen in pbp1Δ cells without Stm1 overexpression. These findings suggest that certain biochemical features of such an Mg2+-resistant fraction may underlie this resistance rather than our Mg2+ supplementation regimen reaching its maximal R-loop-suppressing capacity. Our findings also suggest that environmental CR likely triggers additional Mg2+-independent processes to fully abolish even Mg2+-resistant R-loop fractions (compare Figures 1D and 2A). Such Mg2+ independent processes are unlikely to require transcriptional silencing as CR fully suppresses hybrid buildup in pbp1Δ cells even in the absence of Sir2 (Supplementary Figure S1B). Other non-mutually exclusive possibilities are that specific chromatin modifications or that the greater increases in intracellular Mg2+ levels under environmental CR conditions may allow it to resolve all R-loops.
Mg2+-responsiveness of R-loop suppressors restores genome stability
We next wanted to test if Mg2+ physically targets R-loop suppressors to R-loop accumulating rDNA loci and decipher the impact of such a potential localization on rDNA stability and cellular longevity.
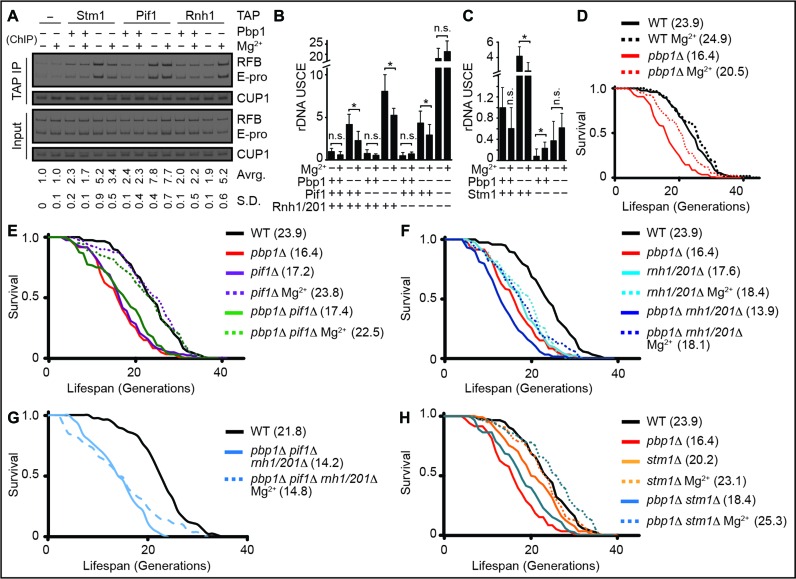
We found Pif1 and Rnh1/201 to be physically enriched at the RFB and E-pro regions of pbp1Δ cells under Mg2+ supplementation conditions (Figure 3A). Despite this increased physical targeting to R-loop accumulating loci, the levels of these R-loop-suppressing proteins were unaltered by Mg2+ (Supplementary Figure S3A). We note that Pbp1 loss was sufficient to target Pif1 to these R-loop accumulating regions whereas Rnh1 targeting additionally required Mg2+ supplementation (Figure 3A). These findings indicate that both types of R-loop-repressing enzymes are physically located at R-loop accumulating rDNA loci in the presence of Mg2+ supplementation. Differential requirements for Pif1 and Rnh1/201 responses indicate that not all R-loop suppressors are the same. More importantly, our results establish a direct physical link between R-loop suppressors and the R-loop-accumulating loci they regulate in the presence of increased Mg2+.
Figure 3.
Mg2+ increases rDNA stability and cellular lifespan by countering R-loop buildup. (A) TAP-tagged Pif1 and Rnh1 are physically located at R-loop-accumulating rDNA loci in pbp1Δ cells under Mg2+ supplementation. R-loop-stabilizing/enriched Stm1-TAP control is included and relative fold enrichments (Mean ± SD) for RFB and E-pro are shown below gels. (B) Mg2+ counters deleterious rDNA USCE in pbp1Δ cells by operating via Pif1 and Rnh1/201. (C) Mg2+ supplementation does not decrease rDNA USCE in pbp1Δ cells lacking the R-loop stabilizer Stm1. (B and C) Shown are rates (N = 5000–25 000; Mean ± SD) and _t_-test *P < 0.05 with full data and statistics in Supplementary Table S1. Not statistically significant indicated by n.s. (D–G) Mg2+ supplementation increases the replicative lifespan of cells lacking (D) Pbp1, even in the absence of (E) Pif1 or (F) Rnh1/201, (G) but not both. (H) Combining Mg2+ supplementation with stm1Δ extends the replicative lifespan of pbp1Δ cells. (D–H) Shown are replicative lifespan curves and mean lifespans (N = 80) in parentheses with full data and Wilcoxon's rank-sum test statistics in Supplementary Table S2.
R-loop buildup in pbp1Δ cells stalls rDNA replication increasing a form of genome-destabilizing aberrant recombination called USCE without disrupting global DNA replication dynamics (Figure 3B, Supplementary Figure S3B and Table S1) (8,12). Consistent with the observation that the localization of Pif1, but not Rnh1, is increased at R-loop accumulating loci in unsupplemented pbp1Δ cells, we observed that pbp1Δ pif1Δ cells, but not pbp1Δ rnh1/201Δ cells exhibited higher USCE relative to pbp1Δ cells (Figure 3B). Mg2+ significantly decreased USCE in pbp1Δ (46%), pbp1Δ pif1Δ (35%) and pbp1Δ rnh1/201Δ (32%) cells, but did not affect pbp1Δ pif1Δ rnh1/201Δ cells (Figure 3B). Mg2+ supplementation also decreased erroneous rDNA replication fork blocks typically observed in pbp1Δ cells (Supplementary Figure S3C). Thus, Mg2+ requires Pif1 and Rnh1/201 to counter R-loop-induced aberrant recombination.
We also tested if rDNA stabilization by Mg2+ is specific to R-loop repression. Stm1 promotes R-loop accumulation at rDNA in pbp1Δ cells (12). We reasoned that if Mg2+ increases rDNA stability independently of R-loop suppression, Mg2+ supplementation would still cause a similar relative decrease in rDNA instability in both pbp1Δ and pbp1Δ stm1Δ cells. Deletion of Stm1 in pbp1Δ cells increased rDNA stability and Mg2+ triggered a relative decrease in rDNA instability in pbp1Δ but not pbp1Δ stm1Δ cells (Figure 3C). Mg2+ supplementation slightly but significantly increased rDNA USCE in stm1Δ cells suggesting that some low levels of hybrids may be beneficial and that excessive hyper-suppression of hybrids may mildly increase rDNA instability (Figure 3C). Our findings indicate that Mg2+ restores rDNA stability by specifically repressing R-loop buildup.
Instability at rDNA as in pbp1Δ cells shortens replicative lifespan that is defined by the number of times that a yeast mother cell replicates its DNA and yields progeny before senescence (Figure 3D, Supplementary Table S2) (8,26,29,36). We found that Mg2+ supplementation increases the lifespan of pbp1Δ, pbp1Δ pif1Δ, pbp1Δ rnh1/201Δ, but not pbp1Δ pif1Δ rnh1/201Δ cells (Figure 3D, E, F and G; Supplementary Table S2). Failure of Mg2+ to increase the lifespan of pbp1Δ pif1Δ rnh1/201Δ cells is not due to the shorter lifespan of these cells as Mg2+ successfully increased the very short lifespan of cells lacking Npl3, another RNA-binding R-loop suppressor (Supplementary Figure S3D) (37). More importantly, by combining Mg2+ supplementation with global R-loop destabilization via stm1Δ, we extended the lifespan of pbp1Δ cells beyond that of wild-type cells (Figure 3H). Our results demonstrate that Mg2+-driven repression of DNA-destabilizing R-loop accumulation can increase or even help extend lifespan.
Mg2+-dependent repression of R-loop accumulation is evolutionarily conserved
ATXN2, the human ortholog of yeast Pbp1, physically associates to hundreds of transcripts in human cells (38). If ATXN2 counters R-loops is unknown but ATXN2 dysfunction leads to severely debilitating and often lethal neurodegenerative diseases called amyotrophic lateral sclerosis (ALS) and spinocerebellar ataxia-2 (SCA2) (39,40). In addition, we previously hypothesized that R-loop regulation may link many ALS-associated genes to each other (15).
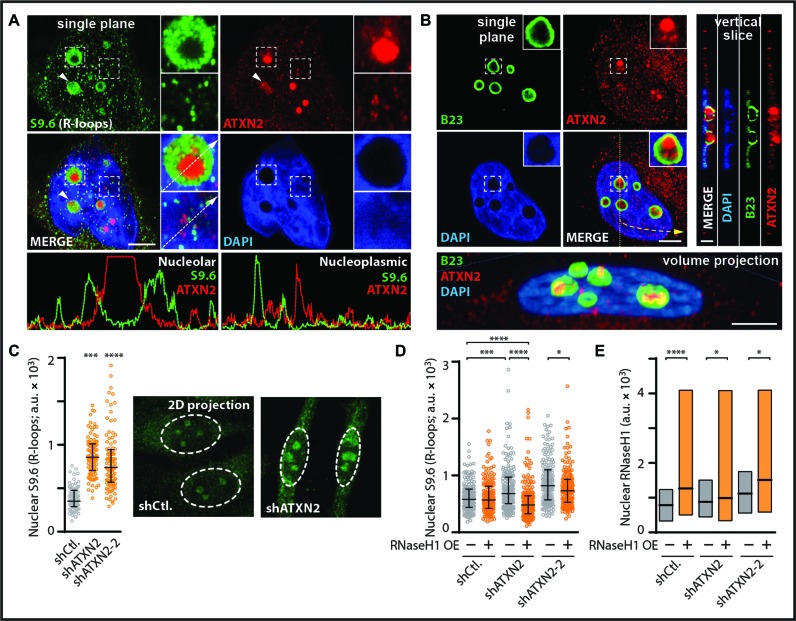
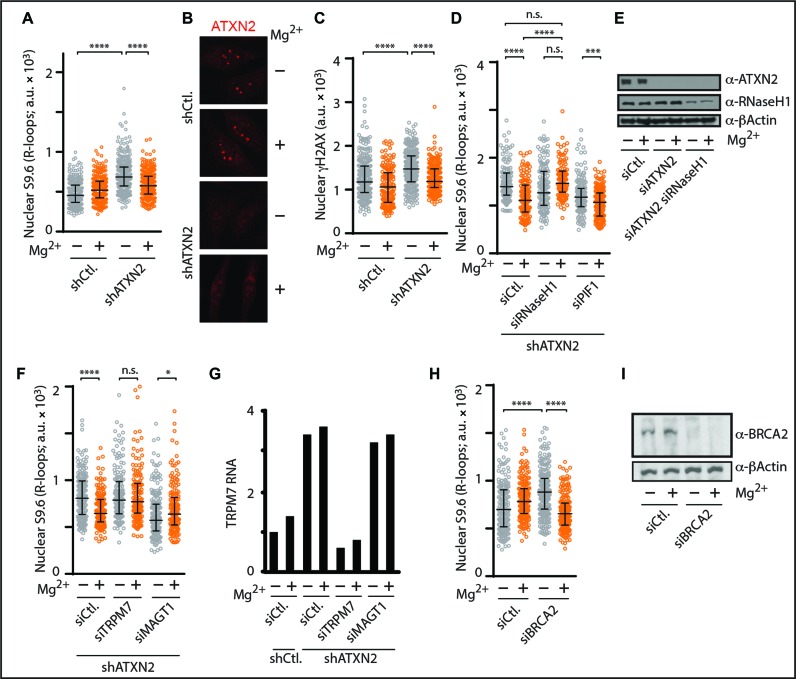
First, we aimed to study the natural localization of ATXN2 relative to R-loop signal distribution inside the cell nucleus. Immunofluorescence analysis revealed that endogenous ATXN2 localizes to hundreds of small foci in the nucleus and cytoplasm but displays a primary localization to a handful of larger nuclear foci identified as nucleoli via co-localization with nucleolar markers (Figure 4A, B, Supplementary Figure S4A and B). Co-labeling revealed an inverse correlation for ATXN2 and R-loops in the nucleoplasm and nucleoli (Figure 4A). Importantly, anti-R-loop immunofluorescence followed by single cell analysis revealed that knockdown of human ATXN2 results in a highly significant nuclear R-loop accumulation (Figure 4C, Supplementary Figure S4C). Nuclear increases were observed in both the nucleolus and nucleoplasm (Figure 4C). Over expression of the RNA–DNA hybrid suppressor human RNaseH1, but not empty vector control, significantly countered nuclear R-loop accumulation in ATXN2-deficient cells (Figure 4D and E). Thus, similar to yeast Pbp1, loss of human ATXN2 triggers nuclear R-loop accumulation.
Figure 4.
Human ATXN2 deficiency triggers nuclear R-loop accumulation. (A) Double immunofluorescence coupled to confocal microscopy reveals an inverse correlation for RNA–DNA hybrid and ATXN2 signals in HeLa cells. DAPI-stained DNA and single plane images are shown. Dashed squares outline areas shown in zoom inset images. Arrowheads highlights inverse localization of ATXN2 and R-loops even in a single nucleolus. Intensity plots below the images are for nucleolar and nucleoplasmic regions along the path of the white arrows in inset images. Bar, 5 μm. (B) Double immunofluorescence and confocal microscopy reveal that major ATXN2 foci are located within B23-outlined nucleoli of HeLa cells. DAPI-stained DNA as well as single plane, vertical slice and volume views are shown. Bar, 5 μm. (C) Short hairpin RNA (shRNA)-mediated stable knockdown of ATXN2 in HeLa cells resulted in R-loop accumulation as revealed by anti-RNA–DNA hybrid immunofluorescence and quantification of 2D images obtained by projecting confocal stacks onto a single plane. Shown are representative 2D projection images with quantification presented as a scatter plot to better capture cell population data (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001, ***P < 0.001 for the Mann–Whitney test). (D) Overexpression of human RNaseH1 (+), but not empty vector control (−), counters R-loop accumulation in shATXN2 HeLa cells as revealed by anti-RNA–DNA hybrid immunofluorescence (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001, ***P < 0.001, *P < 0.05 for the Mann–Whitney test). (E) Quantification analysis of nuclear signal from anti-RNaseH1 immunofluorescence indicating that overexpression of RNaseH1 (+), but not empty vector control (−), increases RNaseH1 protein levels in shCtl. and shATXN2 HeLa cells (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001, *P < 0.05 for the Mann–Whitney test).
We next asked if supplementing human cells with Mg2+ suppresses aberrant R-loop accumulation and promotes genome stability. Titration experiments revealed that 10 mM Mg2+ supplementation countered R-loop accumulation in ATXN2-deficient cells without altering ATXN2 knockdown efficiency or ATXN2 localization (Figure 5A and B). Mg2+ supplementation regimens higher than 10 mM did not confer increased R-loop suppression and actually exhibited elevated toxicity above 50 mM (not shown). Importantly, ATXN2 deficient cells exhibited elevated levels of the DNA damage marker γH2AX (Figure 5C). This elevated genome instability was significantly decreased upon 10 mM Mg2+ supplementation (Figure 5C). Thus, an Mg2+ supplementation regimen (10 mM) less substantial than that used in our yeast experiments (100 mM) efficiently counters R-loop accumulation and genome instability in ATXN2-deficient human cells.
Figure 5.
Mg2+ counters human R-loop accumulation by relying on RNaseH1 and the TRPM7 Mg2+ transporter. (A) Anti-RNA–DNA hybrid immunofluorescence indicates that 10 mM Mg2+ supplementation significantly decreases nuclear R-loop buildup in shATXN2 HeLa cells as shown in the scatter plot to better represent cell population data (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001 for the Mann–Whitney test). (B) Representative anti-ATXN2 immunofluorescence images from cells transiently transfected with siCtl. or siATXN2 in the presence or absence of 10 mM Mg2+ supplementation. Note how Mg2+ supplementation does not alter ATXN2 levels or localization. (C) Anti-γH2AX immunofluorescence imaging and quantitation indicate that 10 mM Mg2+ supplementation counters γH2AX accumulation in shATXN2 HeLa cells (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001 for the Mann–Whitney test). (D) Anti-RNA–DNA hybrid immunofluorescence reveals that transient knockdown of RNaseH1, but not PIF1, abolishes the R-loop-suppressing effect of 10 mM Mg2+ supplementation in shATXN2 HeLa cells (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001, ***P < 0.001 for the Mann–Whitney test; n.s. not statistically significant). (E) Immunoblots showing transient knockdown of RNaseH1 in ATXN2-deficient HeLa cells. Anti-β-Actin immunoblotting served as loading control. (F) Anti-RNA–DNA hybrid immunofluorescence reveals that transient knockdown of TRPM7, but not MAGT1, abolishes the R-loop-suppressing effect of 10 mM Mg2+ supplementation in shATXN2 HeLa cells (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001, *P < 0.05 for the Mann–Whitney test; n.s. not statistically significant). (G) RT-qPCR measuring TRPM7 RNA levels reveals that TRPM7 and MAGT1 RNA levels are unexpectedly already increased in shATXN2 HeLa cells and illustrates the efficacy of siTRPM7 in our knockdown experiments. (H) Anti-RNA–DNA hybrid immunofluorescence indicates that 10 mM Mg2+ supplementation counters nuclear R-loop buildup in siBRCA2 HeLa cells (N = 150 per condition; Mean ± Quartiles; ****P < 0.0001 for the Mann–Whitney test). (I) Immunoblots confirming the efficacy of siRNA-mediated BRCA2 knockdown. Anti-β-Actin immunoblotting served as loading control.
Based on the knowledge gained from our work in yeast, we next set out to identify the mechanistic requirements for Mg2+ to counter human R-loop accumulation. We found that RNaseH1 knockdown abolished Mg2+-dependent suppression of R-loop accumulation (Figure 5D and E). PIF1 deficiency did not compromise R-loop repression by Mg2+ despite globally decreasing R-loop levels likely due to compensation by other R-loop suppressors (Figure 5D). On another front, ALS patients from Guam, a Territory of the United States located in the northwestern Pacific Ocean, carry mutations in the TRPM7 Mg2+ transporter (41). Knockdown of TRPM7 in ATXN2-deficient cells did not alter R-loop levels but disrupted R-loop repression by Mg2+ (Figure 5F and G). Knockdown of the ALS-unrelated control MAGT1 Mg2+ transporter in ATXN2-deficient cells actually lowered R-loop levels likely due to confounding compensation by other Mg2+ transporters in the cell (Figure 5F). As a potentially similar compensation is not seen when we knockdown TRPM7, this highlights a potentially unique role for TRPM7 in Mg2+-dependent human R-loop repression (Figure 5F). We note that unlike yeast Alr1/2, transcript levels of the Mg2+ transporter TRPM7 were already elevated in ATXN2 deficient cells regardless of Mg2+ supplementation, indicating the existence of genetic interactions between ATXN2 and Mg2+ homeostasis in human cells (Figure 5F and G). Cells deficient in RNaseH1 or TRPM7 alone did not exhibit significant R-loop accumulations in our assays (not shown). We also successfully used Mg2+ to decrease R-loop buildup in cells deficient in the R-loop-repressing BRCA2 tumor suppressor indicating that Mg2+ can counter R-loop accumulations triggered by the loss of other disease-linked R-loop suppressors (Figure 5H and I) (42). We conclude that Pbp1/ATXN2 and Mg2+ supplementation acting via Mg2+ transporters, RNaseH enzymes and potentially other Mg2+-sensitive factors such as Pif1 counter deleterious R-loop accumulation in yeast and human cells evolutionary separated by over a billion years.
DISCUSSION
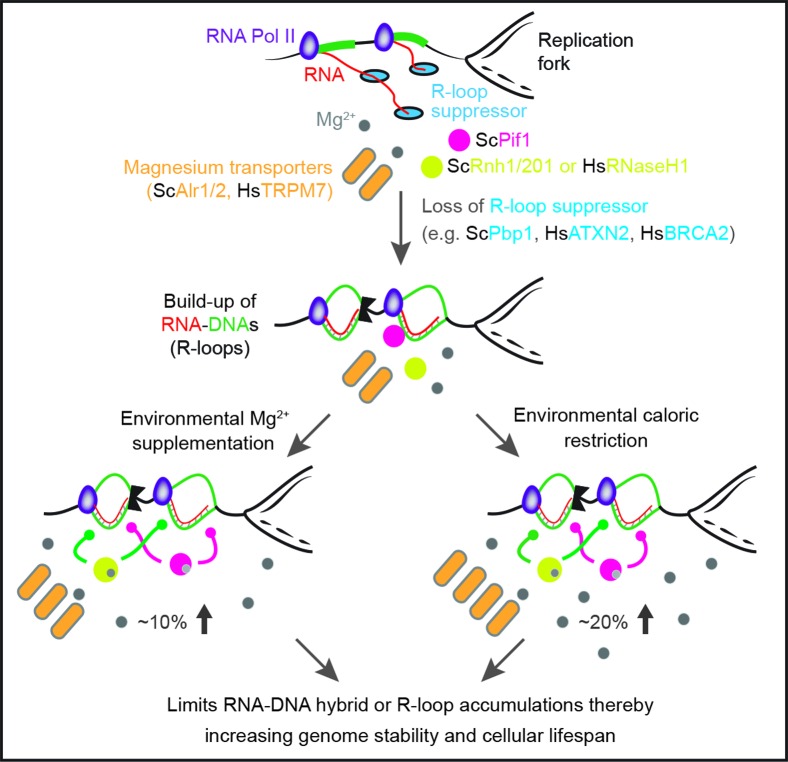
Our work uncovers a conserved Mg2+-dependent pathway that acts alone or in response to calorie restriction to counter deleterious RNA–DNA hybrids or R-loops (Figure 6). We propose that direct Mg2+ supplementation, which can engage this pathway, offers a simple and effective approach to counter genome-destabilizing R-loop buildup pertinent to different human diseases. The herein identified components of this pathway, including Mg2+ transporters and R-loop suppressors, also constitute novel targets for the development of future therapeutics focusing on the suppression of deleterious R-loop accumulation. As many cellular processes are Mg2+ sensitive, we expect that elevated intracellular Mg2+ levels, especially under calorie restriction, may modulate pathways unrelated to R-loop control (43). Thus, future work should assess the potentially broad impact of increased Mg2+ levels especially under calorie restriction. This should include a detailed characterization of the potential impact of magnesium on metabolic processes in both the nucleus and cytoplasm.
Figure 6.
Evolutionarily conserved Mg2+-dependent process alone or in response to calorie restriction suppresses genome-destabilizing RNA–DNA hybrids, or R-loops. Loss of conserved factors such as yeast Pbp1 or human ATXN2 leads to the accumulation of genome-destabilizing RNA–DNA hybrids. Environmental calorie restriction or direct and measured Mg2+ supplementation (100 mM in yeast; 10 mM in human) increases intracellular Mg2+ levels thereby engaging Mg2+-sensitive R-loop suppressors (e.g. yeast Pif1 and Rnh1/201 or human RNaseH1) allowing them to counter R-loop accumulation and restore genome stability and cellular lifespan. Sc, S. cerevisiae; Hs, H. sapiens.
Despite the potentially wide impact of Mg2+, our findings indicate that rDNA-stabilization by Mg2+ is specific to its role in R-loop suppression. First, Mg2+-dependent R-loop repression and rDNA stabilization is additively and fully abolished in the absence of Pif1 and Rnh1/201. Second, that Mg2+ but not Ca2+ counters R-loop buildup is consistent with the structure and function of these R-loop suppressing enzymes. Third, Mg2+-sensitive R-loop suppressors are directly detected at R-loop-accumulating loci under Mg2+ supplementation conditions. Fourth, Mg2+ did not alter local chromatin, RNA Pol II or lncRNA features at sites where R-loop repression is observed. Fifth, the relative stabilization of rDNA by Mg2+ is fully lost if R-loops are already independently destabilized via Stm1 deletion. In addition, consistent with our observations, a recent study identified specific roles for Mg2+ in cellular timekeeping (44). Overall, we find that Mg2+, much like hydrogen ions, can elicit specific biological responses.
We do observe similarities and differences between CR and Mg2+ supplementation in yeast. Common features include dependence on Mg2+-sensitive R-loop suppressors and increases in Mg2+ levels. Main differences are that CR more substantially increases intracellular Mg2+ levels and completely abolishes R-loop accumulation. Despite these distinctions, the level of R-loop repression that we observe correlates very well with changes in intracellular Mg2+ levels. CR in pbp1Δ cells, Mg2+ supplementation in pbp1Δ cells and CR in pbp1Δ alr1Δ alr2Δ cells triggers a 20%, 10% and 6% increase in intracellular Mg2+ levels matched by a 90–100%, 50–70% and 25% decrease in R-loop levels, respectively.
In humans, R-loop repression is emerging as a central function for various disease-linked gene products including ATXN2 (yeast Pbp1; ALS and SCA2; shown here), SETX (yeast Sen1; ALS and occulomotor apraxia type 2 (AOA2)), BRCA1/2 (breast/ovarian cancer), RNaseH2 (yeast Rnh201; Aicardi-Goutières Syndrome) and PIF1 (yeast Pif1; cellular transformation) (15,20,39,42,45–53). Mutations in the TRPM7 Mg2+ transporter are found in ALS patient populations (41). We propose that direct Mg2+ supplementation should be considered as an option to counter R-loop accumulation pertinent to several human diseases.
Taken together, our findings identify Mg2+ as a biochemical signal of beneficial calorie restriction, establish Mg2+ as an R-loop suppressing biological metal, and indicate that Mg2+ supplementation should be considered for patients suffering from various R-loop accumulation-linked diseases.
Supplementary Material
SUPPLEMENTARY DATA
Acknowledgments
The authors thank J. Greenblatt, D. Durocher, M. Ohh (Toronto, Canada) and D. Moazed (Harvard, USA) for discussions and comments on the paper.
Author Contributions: Conception, J.S.S. and K.M.; writing, K.J.A. and K.M.; text editing and westerns, J.N.Y.C.; Yeast strains, J.S.S., B.H., A.H., E.V., S.A.K.; Mg2+ measurements, J.S.S.; ChIP, J.S.S., J.N.Y.C.; USCE, J.S.S., E.V., N.X.L.; Lifespan, J.S.S., R.G., S.A.K.; Microscopy, K.J.A., J.S.S.; Growth assays, J.S.S.; RT-qPCR, J.N.Y.C; DNA combing, B.H., G.W.B.; Structure, J.E.L.
Footnotes
Present address: Jayesh S. Salvi, The Glenn Laboratories for the Biology of Aging and Department of Neurology and Neurological Sciences, Stanford University School of Medicine, Stanford, CA 94305, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research (CIHR) Vanier Doctoral Awards [to K.J.A. and J.S.S.]; Canadian Institutes of Health Research (CIHR) and Canada Research Chair [CIHR-299429 and Canada Research Chair-950230661 to K.M.]; Canadian Institutes of Health Research (CIHR) and Canadian Cancer Society Research Institute [CIHR-MOP79368 and Canadian Cancer Society Research Institute-702310 to G.W.B.]. Funding for open access charge: CIHR-299429.
Conflict of interest statement. MaRS Innovation (Toronto) did not fund this work but K.M., K.J.A., J.N.Y.C. and J.S.S. have signed a preliminary agreement with this organization and the University of Toronto to develop and commercialize intellectual property related to this study.
REFERENCES
- 1.Dillin A., Gottschling D.E., Nystrom T. The good and the bad of being connected: The integrons of aging. Curr. Opin. Cell Biol. 2014;26:107–112. doi: 10.1016/j.ceb.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szafranski K., Mekhail K. The fine line between lifespan extension and shortening in response to caloric restriction. Nucleus. 2014;5:56–65. doi: 10.4161/nucl.27929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaeberlein M., Rabinovitch P.S., Martin G.M. Healthy aging: The ultimate preventative medicine. Science. 2015;350:1191–1193. doi: 10.1126/science.aad3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin S.J., Kaeberlein M., Andalis A.A., Sturtz L.A., Defossez P.A., Culotta V.C., Fink G.R., Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 5.Lin S.J., Defossez P.A., Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 6.Kaeberlein M., Powers R.W., 3rd, Steffen K.K., Westman E.A., Hu D., Dang N., Kerr E.O., Kirkland K.T., Fields S., Kennedy B.K. Regulation of yeast replicative life span by TOR and Sch 9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 7.Imai S., Armstrong C.M., Kaeberlein M., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 8.Kaeberlein M., McVey M., Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ide S., Miyazaki T., Maki H., Kobayashi T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science. 2010;327:693–696. doi: 10.1126/science.1179044. [DOI] [PubMed] [Google Scholar]
- 10.Mekhail K., Moazed D. The nuclear envelope in genome organization, expression and stability. Nat. Rev. Mol. Cell Biol. 2010;11:317–328. doi: 10.1038/nrm2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jack C.V., Cruz C., Hull R.M., Keller M.A., Ralser M., Houseley J. Regulation of ribosomal DNA amplification by the TOR pathway. Proc. Natl. Acad. Sci. U.S.A. 2015;112:9674–9679. doi: 10.1073/pnas.1505015112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salvi J.S., Chan J.N., Szafranski K., Liu T.T., Wu J.D., Olsen J.B., Khanam N., Poon B.P., Emili A., Mekhail K. Roles for Pbp1 and caloric restriction in genome and lifespan maintenance via suppression of RNA-DNA hybrids. Dev. Cell. 2014;30:177–191. doi: 10.1016/j.devcel.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Santos-Pereira J.M., Aguilera A. R loops: New modulators of genome dynamics and function. Nat. Rev. Genet. 2015;16:583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 14.Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salvi J.S., Mekhail K. R-loops highlight the nucleus in ALS. Nucleus. 2015;6:23–29. doi: 10.1080/19491034.2015.1004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houseley J., Kotovic K., El Hage A., Tollervey D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 2007;26:4996–5006. doi: 10.1038/sj.emboj.7601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasiljeva L., Kim M., Terzi N., Soares L.M., Buratowski S. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol. Cell. 2008;29:313–323. doi: 10.1016/j.molcel.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Huertas P., Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Zhou R., Zhang J., Bochman M.L., Zakian V.A., Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. eLife. 2014;3:e02190. doi: 10.7554/eLife.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omer C.A., Faras A.J. Mechanism of release of the avian rotavirus tRNATrp primer molecule from viral DNA by ribonuclease H during reverse transcription. Cell. 1982;30:797–805. doi: 10.1016/0092-8674(82)90284-7. [DOI] [PubMed] [Google Scholar]
- 22.Lima W.F., Murray H.M., Damle S.S., Hart C.E., Hung G., De Hoyos C.L., Liang X.H., Crooke S.T. Viable RNaseH1 knockout mice show RNaseH1 is essential for R loop processing, mitochondrial and liver function. Nucleic Acids Res. 2016;44:5299–5312. doi: 10.1093/nar/gkw350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mekhail K., Seebacher J., Gygi S.P., Moazed D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 2008;456:667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith J.S., Chen Q., Yatsunyk L.A., Nicoludis J.M., Garcia M.S., Kranaster R., Balasubramanian S., Monchaud D., Teulade-Fichou M.P., Abramowitz L., et al. Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2011;18:478–485. doi: 10.1038/nsmb.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salvi J.S., Chan J.N., Pettigrew C., Liu T.T., Wu J.D., Mekhail K. Enforcement of a lifespan-sustaining distribution of Sir2 between telomeres, mating-type loci, and rDNA repeats by Rif1. Aging Cell. 2013;12:67–75. doi: 10.1111/acel.12020. [DOI] [PubMed] [Google Scholar]
- 26.Chan J.N., Poon B.P., Salvi J., Olsen J.B., Emili A., Mekhail K. Perinuclear cohibin complexes maintain replicative life span via roles at distinct silent chromatin domains. Dev. Cell. 2011;20:867–879. doi: 10.1016/j.devcel.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 27.Gallo D., Wang G., Yip C.M., Brown G.W. Analysis of replicating yeast chromosomes by DNA combing. Cold Spring Harb. Protoc. 2016;2016 doi: 10.1101/pdb.prot085118. doi:10.1101/pdb.prot085118. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi T., Ganley A.R. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- 29.Saka K., Ide S., Ganley A.R., Kobayashi T. Cellular senescence in yeast is regulated by rDNA noncoding transcription. Curr. Biol. 2013;23:1794–1798. doi: 10.1016/j.cub.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 30.Cerritelli S.M., Crouch R.J. The non-RNase H domain of Saccharomyces cerevisiae RNase H1 binds double-stranded RNA: Magnesium modulates the switch between double-stranded RNA binding and RNase H activity. RNA. 1995;1:246–259. [PMC free article] [PubMed] [Google Scholar]
- 31.Gu Y., Wang J., Li S., Kamiya K., Chen X., Zhou P. Determination of the biochemical properties of full-length human PIF1 ATPase. Prion. 2013;7:341–347. doi: 10.4161/pri.26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowotny M., Gaidamakov S.A., Crouch R.J., Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: Substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 33.Nowotny M., Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobayashi T., Heck D.J., Nomura M., Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: Requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasiljeva L., Kim M., Mutschler H., Buratowski S., Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henderson K.A., Gottschling D.E. A mother's sacrifice: What is she keeping for herself? Curr. Opin. Cell Biol. 2008;20:723–728. doi: 10.1016/j.ceb.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santos-Pereira J.M., Herrero A.B., Garcia-Rubio M.L., Marin A., Moreno S., Aguilera A. The Npl3 hnRNP prevents R-loop-mediated transcription-replication conflicts and genome instability. Genes Dev. 2013;27:2445–2458. doi: 10.1101/gad.229880.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yokoshi M., Li Q., Yamamoto M., Okada H., Suzuki Y., Kawahara Y. Direct binding of Ataxin-2 to distinct elements in 3′ UTRs promotes mRNA stability and protein expression. Mol. Cell. 2014;55:186–198. doi: 10.1016/j.molcel.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Elden A.C., Kim H.J., Hart M.P., Chen-Plotkin A.S., Johnson B.S., Fang X., Armakola M., Geser F., Greene R., Lu M.M., et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orr H.T. Cell biology of spinocerebellar ataxia. J. Cell Biol. 2012;197:167–177. doi: 10.1083/jcb.201105092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hermosura M.C., Nayakanti H., Dorovkov M.V., Calderon F.R., Ryazanov A.G., Haymer D.S., Garruto R.M. A TRPM7 variant shows altered sensitivity to magnesium that may contribute to the pathogenesis of two Guamanian neurodegenerative disorders. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11510–11515. doi: 10.1073/pnas.0505149102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhatia V., Barroso S.I., Garcia-Rubio M.L., Tumini E., Herrera-Moyano E., Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–365. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- 43.de Baaij J.H., Hoenderop J.G., Bindels R.J. Magnesium in man: implications for health and disease. Physiol. Rev. 2015;95:1–46. doi: 10.1152/physrev.00012.2014. [DOI] [PubMed] [Google Scholar]
- 44.Feeney K.A., Hansen L.L., Putker M., Olivares-Yanez C., Day J., Eades L.J., Larrondo L.F., Hoyle N.P., O'Neill J.S., van Ooijen G. Daily magnesium fluxes regulate cellular timekeeping and energy balance. Nature. 2016;532:375–379. doi: 10.1038/nature17407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R., et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skourti-Stathaki K., Proudfoot N.J., Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szafranski K., Abraham K.J., Mekhail K. Non-coding RNA in neural function, disease, and aging. Front. Genet. 2015;6:87. doi: 10.3389/fgene.2015.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice G., Patrick T., Parmar R., Taylor C.F., Aeby A., Aicardi J., Artuch R., Montalto S.A., Bacino C.A., Barroso B., et al. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. Am. J. Hum. Genet. 2007;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alzu A., Bermejo R., Begnis M., Lucca C., Piccini D., Carotenuto W., Saponaro M., Brambati A., Cocito A., Foiani M., et al. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell. 2012;151:835–846. doi: 10.1016/j.cell.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mischo H.E., Gomez-Gonzalez B., Grzechnik P., Rondon A.G., Wei W., Steinmetz L., Aguilera A., Proudfoot N.J. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moreira M.C., Klur S., Watanabe M., Nemeth A.H., Le Ber I., Moniz J.C., Tranchant C., Aubourg P., Tazir M., Schols L., et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat. Genet. 2004;36:225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- 52.Pulst S.M., Nechiporuk A., Nechiporuk T., Gispert S., Chen X.N., Lopes-Cendes I., Pearlman S., Starkman S., Orozco-Diaz G., Lunkes A., et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat. Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- 53.Sanpei K., Takano H., Igarashi S., Sato T., Oyake M., Sasaki H., Wakisaka A., Tashiro K., Ishida Y., Ikeuchi T., et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat. Genet. 1996;14:277–284. doi: 10.1038/ng1196-277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY DATA