Protein Phase Separation: A New Phase in Cell Biology (original) (raw)
. Author manuscript; available in PMC: 2018 Dec 1.
Published in final edited form as: Trends Cell Biol. 2018 Mar 27;28(6):420–435. doi: 10.1016/j.tcb.2018.02.004
Abstract
Cellular compartments and organelles organize biological matter. Most well-known organelles are separated by a membrane boundary from their surrounding milieu. There are also many so-called membraneless organelles and recent studies suggest that these organelles, which are supramolecular assemblies of proteins and RNA molecules, form via protein phase separation. Recent discoveries have shed light on the molecular properties, formation, regulation, and function of membraneless organelles. A combination of techniques from cell biology, biophysics, physical chemistry, structural biology, and bioinformatics are starting to help establish the molecular principles of an emerging field, thus paving the way for exciting discoveries, including novel therapeutic approaches for the treatment of age-related disorders.
Introduction
Eukaryotic cells are composed of numerous compartments or organelles. These organelles carry out specific functions and provide spatiotemporal control over cellular materials, metabolic processes, and signaling pathways. For example, the nucleus physically separates transcription from translation; this has allowed eukaryotes to develop a complex system of posttranscriptional control, which is largely absent from prokaryotes [1]. Other examples of membrane-bound organelles include lysosomes, the endoplasmic reticulum, and synaptic vesicles. However, cells also harbor organelles that lack a delimiting membrane. These are supramolecular assemblies composed of proteins, nucleic acids, and other molecular components. They are present in the nucleus (e.g., nucleolus, nuclear speckles), as well as in the cytoplasm [e.g., stress granules (SGs), processing bodies, the centriole] [2,3]. Many of these cellular bodies were identified decades ago and numerous structural insights became available as the bodies were discovered. However, questions have remained about how these bodies form, why they form, and how their physical features contribute to biological function. These questions are starting to be answered, and recent advances in interdisciplinary approaches have fueled the emergence of insights into their organization, molecular properties, and regulation [2–4]. A growing understanding of the underlying molecular principles and the physicochemical forces that drive the formation of membraneless organelles (see Glossary) has enabled the elucidation of their diverse functions in a variety of cellular processes, including the stress response, the regulation of gene expression, and the control of signal transduction [5–8]. In the past few years, there has been increasing evidence for the involvement of membraneless organelles in age-related disorders, such as amyotrophic lateral sclerosis (ALS) [9–16]. Together, these discoveries have created a new field in cell biology, focused on understanding how organization of cellular matter into membraneless organelles contributes to function, and how their dysregulation leads to disease.
In this review, we examine the current state of the growing interest in membraneless organelles, providing insights into their biogenesis, organization, dynamics, regulation, and function. We also discuss how recent findings can give us molecular insights in age-related diseases. This could pave the way for developing novel therapeutic strategies that leverage our understanding of phase separation. Finally, we highlight the major challenges that lie ahead and questions that need to be answered quantitatively and completely in the coming years.
Membraneless Organelles Are Formed via Phase Separation
Many membraneless cytoplasmic and nuclear compartments (e.g., P bodies, SGs, the Balbiani body, germ granules, PML bodies, Cajal Bodies, nuclear speckles, and the nucleolus) have been studied for a long time. However, the forces driving their formation mostly remained enigmatic. Several early studies highlighted the dynamic nature of these assemblies [17–19]. In 2005 it was argued that Cajal bodies behave as ‘semifluid spheres suspended in semifluid nucleoplasm’ [17]. However, definitive experimental evidence for the physical nature of these assemblies was lacking. This changed in 2009, when Brangwynne, Jülicher, and Hyman showed that P granules (RNA and protein-containing bodies in embryos of Caenorhabditis elegans) have liquid-like properties and form by phase separation [20]. This is a physical process that occurs when a supersaturated solution of components spontaneously separates into two phases, a dense phase and a dilute phase, that then stably coexist. The proposed liquid-like nature of P granules was evident from their round appearance (the result of minimizing surface tension), deformability (fusion and fission events), and dynamic exchange of components. Similar observations were made 2 years later for nucleoli [21]. The liquids themselves are not ‘simple liquids’, which is a term that has specific connotations. A ‘simple liquid’, also known as a van der Waals fluid, comprises of spherical particles that interact via isotropic short-range potentials. Protein and RNA liquids are not spherical particles of uniform stickiness. Instead, they are best described as associative polymers and the liquids formed by such systems have distinctive structures that are defined by physical crosslinks that give rise to a panoply of material properties, including the possibility of spatially organized droplets where one polymer wets another [17,22–27].
Phase separation is a well-known phenomenon in polymer chemistry [28]. However, its application to biomacromolecules is a much more recent development. Some proteins, such as hemoglobin, had previously been reported to undergo phase separation at high concentration in vitro [29,30], but the significance of these observations remained unclear. Especially among crystallographers, liquid–liquid phase separation is frequently observed during crystallization trials [31]. Liquid droplet formation lowers the free energy of nucleation and thus is often a desired phenomenon in crystallization experiments [32]. However, the realization that phase separation might be the operational principle governing the formation of membraneless organelles to regulate biological functions and activities has emerged only recently. Strong support for this idea was provided by Rosen and colleagues in 2012. They showed that protein-and RNA-containing bodies could be reconstituted from purified components; they further provided evidence that these reconstituted liquid bodies can promote the nucleation of actin polymers [6]. In the years following these seminal discoveries, there has been growing appreciation that proteins and other macromolecules, such as RNAs, can form condensates that are either well-mixed or spatially organized, and switch between different material states [14,16,33]. Membraneless organelles are known more generally as biomolecular condensates, and the constituent biomolecules obey the same physical principles as other polymers (Box 1). Accumulating data underscores the variety of different phase transitions and the complex molecular and physical interactions behind these processes.
Box 1. Membraneless Organelles Can Be Liquids, Solids, or Gels.
Membraneless organelles are often referred to as liquids, but this designation also creates considerable confusion because of the mental picture this might conjure. In this regard, it is worth noting that all liquids, even so-called simple liquids made up of hard spheres, have a well-defined structure that is quantifiable in terms of pair-correlation functions. These functions show that liquids adopt ordered arrangements, rather like crystalline solids, on length scales that are of the order of magnitude of the size of a typical molecule. On longer length scales, the molecules are randomly organized, in a manner that is reminiscent of dilute gases. Aspherical molecules have spatial as well as directional order, as is the case with water and other molecular liquids, including polymeric ones. Local spatial ordering and preferred intermolecular orientations arise from hierarchies of interactions with different spatial extents and directional preferences, such as long-range electrostatics, multipolar interactions, hydrogen bonds, forces, and short-range interactions involving pi-systems.
In the world of biological phase separation, gels are often thought of as being synonymous with solids, and gelation is thought to be the process of transitioning from a liquid to solid. Gels are generated by a system-spanning network of intermolecular interactions, along which one can ‘walk across a gel by relying on the connectivity of the constitutive macromolecules. If gels have long-lived crosslinks and a high density of crosslinks, then the material properties can be consistent with those of solids. In contrast, gels with short-lived crosslinks and/or a low level of crosslinking will have material properties that are akin to those of liquids.
In reality, an organelle can be a liquid, some form of solid, a liquid–gel, a solid–gel, a crystalline–solid, a semi-crystalline– solid, or liquid–crystalline depending on the extent of spatial ordering and the directional preferences for spatial ordering. To assign an appropriate designation to a membraneless organelle, one would have to, at a minimum, measure five quantities, namely: (i) the concentrations of macromolecules within droplets to quantify density, (ii) the extent of long-range spatial order of molecules with respect to one another to quantify the intermolecular organization within the droplet, (iii) the extent of physical crosslinking amongst molecules, (iv) the interfacial tension between the droplet and its surroundings, and (v) the timescales for making and breaking bonds within droplets. Ideally, all five measurements would be performed using in vitro facsimiles of droplets and within the appropriate body in living cells to uncover the commonalities and differences between the two scenarios.
Molecular Determinants of Protein Phase Separation In Vitro and In Cells
Proteomic and genetic studies have identified protein components of several membraneless organelles [23,34–36]. These studies suggest that multivalency of adhesive domains and/or linear motifs is a defining feature of proteins (and perhaps RNA molecules) that drive phase transitions. Multivalency can come about in at least one of three ways: (i) folded proteins, with well-defined interaction surfaces, can form oligomers that engender multivalency of other associative patches, which participate in stereospecific interactions; (ii) folded domains can be strung together by flexible linkers to generate linear multivalent proteins; and (iii) intrinsically disordered regions (IDRs) can serve as scaffolds for multiple, distinctive short linear motifs. Of course, multivalency can also emerge by combinatorial arrangements of the three archetypes mentioned here or through emergent processes such as a structure formation within disordered regions. One feature that has attracted considerable attention is the presence of IDRs in proteins that drive phase transitions. These regions display a sequence-intrinsic preference for conformational heterogeneity (i.e., disorder) and are known as intrinsically disordered proteins/regions (IDPs/IDRs) [37]. Many IDRs have a biased amino acid composition and may be repetitive in sequence, hence, specific subsets of these IDRs are also referred to as low-complexity domains (LCDs) [23,37–39]. The formation of supramolecular assemblies enriched in IDRs/LCDs leads to membraneless organelles with a variety of different properties (Figure 1A–C). As one key example, distinct intermolecular interactions among IDRs, folded domains, and nucleic acids gives rise to a range of assembly dynamics (Box 1). For example, the Balbiani body in oocytes is a solid-like protein assembly held together by strong β-sheet interactions [36]. By contrast, many RNA-protein (RNP) granules are dynamic and liquid-like, and genetic experiments have demonstrated that IDRs can aid in their assembly [40–42]. RNA-binding proteins (RBPs) are known to have a multivalent modular domain architecture [43], which seems to be a critical factor in phase separation (Box 2). Engineered proteins containing multiple interaction domains connected by flexible linkers exhibit spontaneous liquid–liquid demixing upon interacting with their specific targets [6]. These data suggest that so-called ‘fuzzy’ interaction modes might enable a multitude of combinations amongst multivalent interaction domains and that this could be a general driver of protein phase transitions [44] (Box 3).
Figure 1.
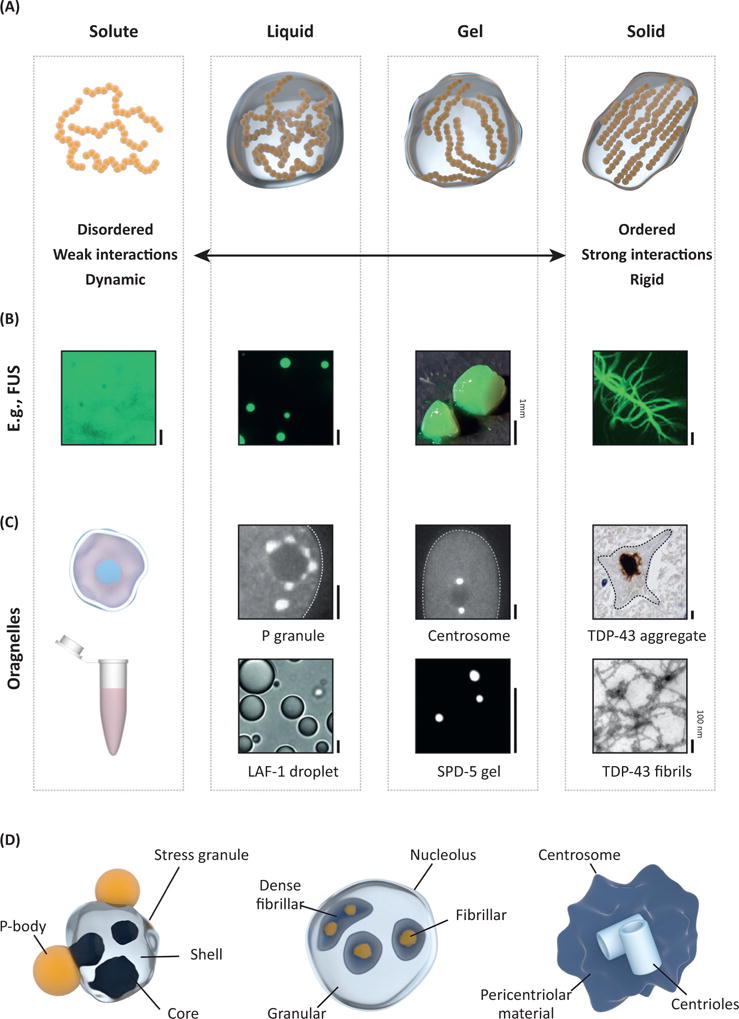
Different Characteristics of Protein Phase Transitions. (A) Material state and dynamics can vary in a wide range from liquid-like to solid states. (B) Example of the protein FUS, which can span the entire range of material states in vitro. Pictures adapted from reference [16]. (C) Examples of membraneless organelles and their reconstituted in vitro counterparts. Pictures adapted from references [48,66,72,87,142] (D) Several membraneless organelles have complex topologies with different subcompartments that may belong to different states. All scale bars 5 μm unless indicated.
Box 2. Insights from Polymer Theories and Multiscale Simulations.
Phase transitions are cooperative transitions that involve the collective effects of interacting modules from multivalent proteins. They can undergo gelation, whereby they form physically crosslinked, system-spanning networks, where the crosslinks are noncovalent interactions among associative domains/motifs. Protein polymers can also condense via a density transition, enabling the formation of a dense phase that coexists with dilute phases. The physics of gelation, or more precisely sol–gel transitions, have been deployed to understand the impact of valence of associative domains/motifs on the driving forces for phase transitions [6,143]. In contrast, the physics of density transitions explain the formation of condensed phases that are spherical in shape and display many properties that are congruent with those of liquids [20,50]. In reality, the physical principles underlying both types of transitions synergistically underlie the formation of membraneless organelles, which are better known as biomolecular condensates.
Multivalent proteins belong to a class of polymers known as associative polymers, in that they can undergo gelation aided by phase separation or gelation without phase separation. Here, valence refers to the effective numbers of adhesive domains/motifs that provide specificity in intra- as well intermolecular interactions. Recent computer simulations and adaptations of the theories of associative polymers show that multivalent proteins may be parsed into associative domains/motifs, so-called stickers, interspersed by spacers [78]. The stickers enable physical crosslinking, whereas the spacers or linkers determine whether or not gelation will be driven by phase separation. Linkers or spacers that are preferentially solvated will inhibit phase separation, whereas linkers/spacers that prefer self-associations or are indifferent about whether they interact with themselves or solvent, will enable gelation via phase separation. The key result emerging from theory, simulations, and recent experiments [87] is a clear role for intrinsically disordered regions as determinants of the nature of phase transitions, as well as the densities and organization of protein modules within droplets.
Box 3. Heterogeneity Matters for Organelle Dynamics.
Nuclear pore complexes possess high frequency, weakly interacting FG motifs in disordered regions, yet exhibit long recovery times in FRAP experiments [144]. What needs to be considered is that repetitive motifs or SLiMs may generate a variety of contact topologies, resulting in large numbers of iso-energetic microstates and higher entropy. This requires highly dynamic linkers (e.g., IDRs) to minimize the coupling between the binding sites and enable a multitude of arrangements. In addition, weak-affinity or nonspecific motifs may simultaneously interact with multiple target sites, or even with more binding partners via weak, short-range contacts. In contrast to the one-to-one binding model, cation–pi, pi–pi, aromatic hydrogen bonds, and van der Waals interactions are realized at different extents with alternative target sites causing uncertainty in defining contacts between the multivalent motifs. Computer simulations using a mathematical framework show that partial, heterogeneous interactions can lower the phase boundary by an order of magnitude as compared with the one-to-one binding model [145]. From this aspect, the higher-order assembly resembles an encounter complex, which facilitates productive contacts, yet enables fast reorganization of the interface. This also implies that heterogeneous systems can undergo phase transition at lower valency [145]. Different, redundant interaction patterns could be generated by large number of structural states [44]. Albeit surprising, conformational heterogeneity could also promote assembly via entropic effects, as was observed in the case of FUS [10]. Taken together, interaction and structural heterogeneity are likely the critical determinants of assembly and dynamics of membraneless organelles, and not mere multivalency of the constituent proteins. As such heterogeneity is ubiquitous to protein interactions [146], it is possible that the molecular driving forces associated with membraneless organelles are maybe not fundamentally different from traditional protein complexes and ‘lower-order’ assemblies [44].
In addition to serving as merely linkers, IDRs may also mediate ‘sticky’ interactions to promote phase transition [45]. McKnight and coworkers found that concentrated solutions of different IDRs could, over time, spontaneously form hydrogels [39], similar to existing observations made regarding FG-repeat-containing nuclear pore proteins [46]. Shortly thereafter, Taylor and coworkers discovered disease mutations in the IDRs of hnRNPA1 and hnRNPA2B1 that resulted in accelerated assembly into higher-order structures in vitro. Furthermore, these mutants promoted the spontaneous formation of SGs with dramatically reduced dynamics in living cells [33]. Subsequent work from several groups showed that the proteins containing disease-associated IDRs such as hnRNPA1 or Fused in Sarcoma (FUS) can also make liquid droplets [10,14,16,47–49]. These findings drew the attention of the entire field to the importance and functionality of IDRs in phase separation and provided an additional rationale for the abundance of protein disorder in eukaryotic proteins.
How exactly are multivalent interactions encoded in IDRs? These sequences are often enriched in uncharged polar side chains (glutamine, asparagine, glycine, serine, proline), charged amino acids (arginine, lysine, glutamic acid, aspartic acid), or aromatic residues (phenylalanine and tyrosine). Interestingly, these residues do not seem to be distributed randomly throughout the sequence, but are often found as short linear interaction motifs (SLiMs), alternating charge blocks, or degenerate repeats [50,51]. The range of sequence biases associated with IDRs that mediate phase separation indicates that there may be a range of underlying driving forces. These likely include electrostatic, dipole–dipole, pi–pi, cation–pi, hydrophobic, and hydrogen bonding interactions [9,49,50,52–56] (Figure 2A,B). Indeed, mutational studies have demonstrated that phase separation of different LCDs can be prevented by interfering with a variety of residue types [9,10,14,16,57]. Additionally, disrupting alternating charge blocks [49,53] and mutating key residues in degenerate repeats [39,46] can also perturb phase separation.
Figure 2.
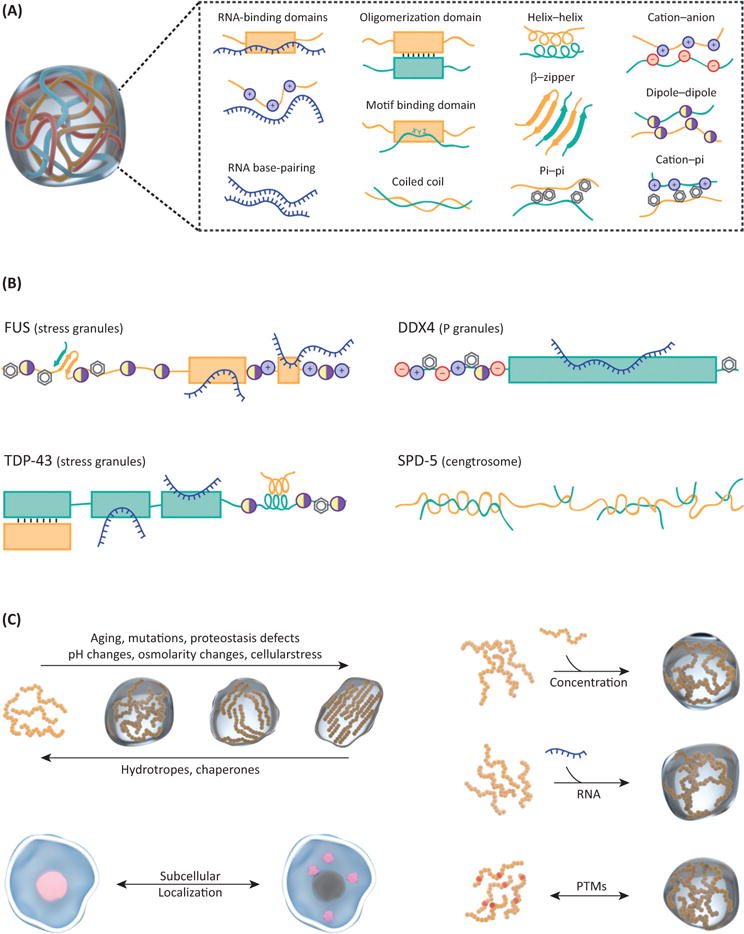
Interactions and Regulatory Mechanisms Implicated in Protein Phase Separation. (A) Overview of different kinds of contacts, which have been observed in protein phase separation. (B) Examples of phase separating proteins illustrate the importance of multivalency, highlighted by an array of interaction modules within a single protein. (C) Different mechanisms regulate the material state and nucleation of protein phase separation.
Membraneless organelles frequently contain nucleic acids, especially RNA. Moreover, the proteins associated with membraneless organelles often possess RNA-binding domains or motifs [9,23] and RNA promotes phase separation of various RBPs [9,10,14,16,58–60] (Figure 2C). Interestingly, high RNA/protein stoichiometries can inhibit phase separation as well [10,61]. RNA also regulates the nucleation and spatiotemporal distribution of membraneless organelles [62,63]. Even G-bodies, which are composed of proteins involved in glucose metabolism, require RNA for biogenesis [64]. RNA can also affect the material properties of protein droplets [48,60]. Interestingly, repetitive RNA species have recently been found to phase separate through intermolecular base-pairing interactions, once more highlighting the universality of multivalency and structural polymorphisms as drivers of phase separation [65].
Material States of Membraneless Organelles: Liquids, Hydrogels, and Aggregates
Although several proteins have been shown to phase separate in the test tube, the underlying molecular structures associated with these phase separated states in vitro or in cells remain heavily debated. The Balbiani body is dependent on stable amyloid-like interactions [36], while the pericentriolar material and postsynaptic density are mediated by interactions amongst proteins that form coiled-coils [66,67]. The picture is less clear for other assemblies. A lot of attention has been dedicated to better understand the internal structure of SGs. SGs form reversibly when cells are stressed and it is thought that the formation of SGs is a form of stress response [62]. Recent work from the Parker lab suggests that the situation in cells might be more complicated than expected, based on test tube experiments alone. It was shown that SGs contain stable cores that withstand dilution, indicating their nonliquid character. Although this does not rule out a combination of spontaneous phase separation and gelation as the route to forming SGs, the data suggest that dissolution of SGs might be a driven process to force the material out of kinetic traps [7,23] (Figure 1D). Indeed, super-resolution microscopy and crosslinking experiments have confirmed that SGs contain a labile liquid shell. These results point to a complex internal organization of SGs, a picture that, given recent work, is likely true for other membraneless organelles as well [24,25,68].
Missense mutations in several SG proteins cause neurodegenerative disorders such as ALS, and both mutant and wild type proteins are found aggregated in neurons [69–72]. While SGs are dynamic assemblies, these aggregates may have a fibrillar architecture [73,74]. FUS and hnRNPA1 are examples of such SG proteins, with long, low-complexity IDRs that are mutated in ALS patients [75,33]. Initial studies from McKnight and coworkers found that these disordered domains can form reversible hydrogels, and this is dependent on labile kinked β sheets [39,76]. Interestingly, repeated cycles of gelling/dissolution of hydrogels promoted a transition toward irreversible gels [14,15]. A different mechanism is suggested by studies of liquid droplets formed by full-length FUS and hnRNPA1, or their LCDs [14,16]. In such liquid droplets, the LCDs seem to retain their tendency to be disordered, which is similar to their monomeric state [10,56]. However, for many droplets formed by low-complexity IDRs, especially in the case of constructs that contain the full-length protein, an eventual maturation into fibrillar solid aggregates occurs. Interestingly, the rate of maturation is enhanced by ALS-causing mutations [14,16] (Figure 2C). Both the labile-to-stable gel [15] and liquid-to-solid [14,16] transitions could explain the pathological conversion of SGs to aggregates in ALS [77], but their exact relation to both cellular SGs and aggregates remains undetermined.
How Is Specificity Generated and Maintained?
Interestingly, many proteins reside in multiple distinct membraneless organelles [9,23,69]. As these proteins are significantly enriched in multivalent proteins, the question inevitably arises as to what determines the specificity and ensures the integrity of these assemblies: how is fusion of distinct membraneless organelles prevented? How are distinct subcompartments within membraneless organelles maintained (e.g., in the nucleolus; Figure 1D)? How can a multiphase system such as the nucleolus be assembled and controlled? Recent work has suggested that differences in surface tension of protein droplets could mediate the formation of such multiphase droplets [24]. The key components of two subnucleolar compartments can phase separate independently, but the resulting droplets have different surface tensions. When mixed together, these droplets do not fuse but arrange in a droplet-within-a-droplet topology, which appears strikingly similar to the nucleolus. This example hints at a more general principle that could underlie multiphase behavior in other membraneless organelles.
Besides physical properties, specificity of granule assembly may derive from the specificity of direct protein–protein interactions. IDPs and proteins undergoing phase separation are enriched in SLiMs and degenerate repeats, which can serve as primary protein-binding modules [51] (Figure 2A,B). Specificity may be related to additional features of IDPs, such as the number and spacing of repetitive binding motifs (multivalency), their post-translational modifications (PTMs), or the dynamics of the intervening linkers [44,78]. Nonspecific electrostatic interactions, especially with RNA, could be critical to nucleate droplet assembly, and different IDRs respond differently to changing ionic strength [10,49]. Although SLiMs possess some degree of specificity, the multiplicative effects of multiple SLiMs may determine the material properties and composition of a given assembly.
Additionally, several key proteins in membraneless organelles possess folded dimerization or oligomerization domains. For example, G3BP1 contains a folded dimerization domain sufficient for SG targeting [79]. Other examples include components of PML and Cajal bodies, and nuclear speckles [80–82]. TDP-43 can phase separate by dimerizing via a transient alpha helix in its LCD [11], and can multimerize through its folded N terminal domain [83] (Figure 2B). This suggests that for some proteins, phase separation may occur via two distinct mechanisms that are physically coupled by the protein structure. A convincing demonstration for such a mechanism of assembly has come from elegant work by the Brangwynne lab [84]. Shin et al. used a plant-derived light-inducible protein oligomerization domain to provide an optogenetic route for driving protein– protein interactions. Fusion of this oligomerization domain to LCDs known to drive phase separation yielded synthetic proteins that formed liquid droplets in cells upon light stimulation. These so-called ‘optodroplets’ indicate that the combination of specific oligomerization domains with LCDs indeed is a potent mechanism to mediate specific cellular phase transitions. In addition to oligomerization domains, coiled-coils and β-zippers could provide the requisite multiplicative sticky interactions that are needed to drive the formation of membraneless organelles [39,66,67,76] (Figure 2B). Interestingly, labile β-zipper regions, as the ones driving FUS gelation, have recently been found to be enriched in numerous disordered proteins [85].
It has also become clear that there is a clear distinction between structural components of membraneless organelles and client proteins, which only target the compartment [36,66]. However, the exact molecular characteristics discriminating between these behaviors currently remain unknown. Preference of client proteins for certain assemblies could simply be mediated by the physical restrictions introduced by the constituent components of the compartment. The array of interactions in a protein droplet/gel creates a network with a specific mesh size. This mesh size could act as a diffusion barrier by allowing free diffusion of small molecules below the mesh size through the network, while limiting the entry of larger ones [49,86,87]. Additionally, membraneless organelles could be anchored in space, hereby preventing diffusion and fusion events. Aggresomes, for example, are perinuclear misfolded protein deposits kept in place by a cytoskeletal cage, which prevents their diffusion through the cell [88].
Spatiotemporal Regulation
Given the rapidly expanding range of proteins that are being shown to undergo spontaneous phase separate in the test tube, it remains puzzling how cells can exercise precise control over this process. For example, several RBPs appear fully soluble at cellular concentrations well above their in vitro saturation concentration [14,16], yet their transition to a different phase only occurs under specific conditions. Put simply, how is the cell able to avoid spontaneous and uncontrollable phase separation? This question is closely related to the above-mentioned ideas surrounding the origins of specificity of phase transitions.
Work from different labs has shown that both serine and tyrosine phosphorylation can control phase separation [12,52,89,90], and the same holds true for arginine methylation [49,91] and sumoylation [80]. Importantly, the activity of the dual specificity kinase DYRK3, which partitions into SG, was shown to be necessary for SG dissolution [92], suggesting that there might be specific cellular switches controlling these processes. Interestingly, proteins prone to phase separation, seem to be enriched in residues that are targeted by PTMs [38]. Indeed, PTMs can dramatically alter the charge or other properties of these IDRs/LCDs, hence, modifying the sequence-intrinsic driving forces to phase separate [49,52] (Figure 2C).
Another way for the cell to control phase transitions is by controlling the cellular concentrations and intracellular distribution (i.e., diffusivities of proteins that mediate phase separation) (Figure 2C). The cellular concentrations of hnRNPA1 are higher than the in vitro saturation concentration, yet these molecules remain soluble in the nucleus for reasons that remain unclear [14]. Blocking nuclear import of hnRNPA1, which leads to its accumulation in the cytoplasm, leads to the spontaneous formation of SGs [14]. Interestingly, nuclear transport factors are themselves components of SGs, suggesting that nucleocytoplasmic transport processes might control phase separation in multiple, albeit unknown ways [69].
Since RNA is involved in enabling the formation of multiple membraneless organelles, availability of specific RNA species may also regulate phase separation in time and space (Figure 2C). For example, expression of the noncoding NEAT-1 RNA is essential for para-speckle formation [93]. Also, polysome disassembly upon cellular stress results in the cytoplasmic availability of free mRNA, which subsequently nucleates SGs. Inhibiting poly-some disassembly prevents SG formation, even when the stress response is activated [18]. Moreover, the canonical stress-granule marker poly(A)-binding protein (PAB1) undergoes phase separation in response to heat stress, which leads to the release of its bound RNA [54]. This suggests a complex relationship between translational responses and SG formation.
Disease, Pathology, and Aging
Several key proteins in neurodegenerative disorders are components of membraneless organelles. Hence, misregulation in the formation, maintenance, or clearance of these assemblies may provide a stepping-stone for pathological aggregation [70,71]. Indeed, spontaneous maturation of dynamic protein droplets and hydrogels to solid aggregates has been observed over the course of hours in the test tube and in cells [14–16,24,84]. This conversion indicates that the dynamic assemblies may be metastable or inherently unstable, and specific cellular processes keep them from solidifying (Figure 2C). The fact that these liquid-to-solid transitions are accelerated by disease mutations further highlights the significance of phase transition to pathology [11,14–16]. These disease mutations seem to target β-zippers in IDRs, which makes them more prone to fold into stable amyloid structures [14,33,94]. Yet, it is important to note that there is currently no direct evidence that pathological protein aggregates in patient brains result from solidification of SGs or other membraneless organelles.
Disease mutations might also affect phase separation through the generation of aberrant protein and RNA species. Repeat expansion disorders prove especially interesting in this regard. Several of these disorders involve the formation of repeat RNA foci, which trap RBPs, resulting in their loss of function [95]. Interestingly, such repeat RNAs can themselves phase separate through multivalent base pairing, mimicking the foci observed in patients [65]. Additionally, several of these expanded repeat RNAs have been found to be translated, generating peptide repeats [96]. Translation of ALS-causing GGGGCC repeat expansions, for example, produces different dipeptide repeats [97–99]. Two of them, namely glycine– arginine and proline–arginine dipeptide repeats, localize to different membraneless organelles, including SGs [9,12,13,57,100]. SGs positive for these pathogenic peptides were less dynamic, and moreover, recruited aggregation-prone proteins such as TDP-43 [9,13].
SG srequire autophagy for clearance [101]. Interestingly, mutations in autophagy genesare the cause of various diseases, including ALS [102] and the efficacy of autophagy is also known to decrease with age [103]. Besides autophagy, chaperones are also involved in both maintaining SG fluidity and their clearance [104,105]. These observations suggest that the inability of the cell to tightly control these assemblies may lead to pathological aggregation. Nuclear transport is also known to deteriorate with aging [106] and is being increasingly implicated in protein aggregation diseases [69,107–112].
Mitochondrial dysfunction is a cornerstone of aging and neurodegeneration [113], potentially causing a reduction in ATP levels that could affect the regulation of membraneless organelles. Numerous SG proteins contain ATPase domains, and lowering cellular ATP levels decreases the dynamic character of these organelles [23]. Additionally, there is evidence that cellular ATP can act as a chemical hydrotrope, directly preventing phase separation and aggregation. This feature of ATP is independent of its role in providing energy for active cellular processes [114]. Hence, defects in mitochondrial respiration may promote protein aggregation in aging and disease, either through an overall reduction in cellular ATP levels, or by the impairment of ATP-dependent processes that maintain the liquidity of membraneless organelles.
Given the importance of PTMs in the regulation of phase separation, it is interesting to note that several pathological protein aggregates show specific PTM signatures. For example, Tau phosphorylation is a hallmark of pathology in Alzheimer’s disease [115], and interestingly, tau phosphorylation promotes aggregation and phase separation in vitro [116].
Although protein aggregation and phase transitions are mostly studied in the context of neurodegenerative disorders, they are implicated in a wide variety of pathological conditions, including viral infections and cancer. Several of the key proteins associated with neurodegeneration are also implicated in different types of cancer [117]. For example, the LCD of FUS, which is involved in SG targeting and aggregation in ALS, has been shown to undergo oncogenic fusion events in liposarcomas [118]. Indeed, cancer-related fusion proteins are often enriched in disordered low-complexity domains, indicating this may be a common mechanism [119,120]. Indeed, the transcriptional activation potential of FUS LCD, as well as its human homologs EWSR1 and TAF15, implicated together in a family of cancers, is highly correlated with their in vitro hydrogel binding capability and ability to recruit the C-terminal domain of polymerase II to such hydrogels [90]. The mechanism through which FUS LCD and its homologs mediate transcriptional activation remains unclear, but is thought to involve phase separation [5,10,121,122]. Additionally, increased SG and paraspeckle formation have been linked to a poor prognosis for cancer survival [123–125]. Lastly, aggregation of the tumor suppressor p53, resulting in its loss of function, is a major mechanism in cancer [126], and compounds preventing its aggregation have been successful in preclinical animal models [127].
SGs have also been implicated in the antiviral stress response [128] and viruses have evolved numerous ways of interfering with SG assembly. Moreover, some viruses, such as flaviviruses, including Zika, even hijack SG proteins to aid in their replication [128,129]. Although still in its infancy, we would argue that since protein aggregation and phase separation are implicated in numerous human pathological conditions, a better understanding of these processes will help us develop novel therapeutic strategies in the wider field of human medicine.
Road Toward Novel Therapy?
As outlined in the foregoing discussion, protein phase separation is suspected to be intimately linked to pathological protein aggregation and disease. The ultimate vindication of our understanding of its pathological importance would be a demonstrated ability to harness this information and devise new ways of treatment. Cellular phase transitions can be targeted by different chemicals interfering with hydrophobic [7,57,130] or polar [65] interactions. However, such general approaches would be expected to target a wide range of membraneless organelles in the cell [7,57,65] and may therefore be poorly situated as therapeutic options.
Antisense oligonucleotides (ASOs) offer a likely suitable selective approach to specifically knockdown (KD) key players in these aberrant phase transitions. Although ASOs targeting pathological proteins were successful in different mouse models [131], their application is limited to nonessential proteins. In case of essential proteins, ASOs could target nonessential functional partners, which regulate phase transition. The feasibility of this approach was illustrated in the case of the essential protein TDP-43 in ALS models. Ataxin-2 was previously identified as an ALS disease modifier in animal models and in humans [132]. Subsequently, it has been shown that Ataxin-2 directly recruits TDP-43 to SGs, providing a putative mechanism for how it may promote TDP-43 aggregation [133]. Unlike TDP-43 KD, Ataxin-2 KD is well tolerated in mice. Compellingly, KD of Ataxin-2 in an ALS mouse model reduced the number of TDP-43 aggregates in the spinal cord of the affected mice and dramatically extended survival [133]. Similarly, KD of SG protein Tia-1, which is known to interact with Tau, was also shown to prevent Tau pathology and toxicity in neuronal culture and rodent models [134,135]. These observations convincingly demonstrate that targeting phase transitions through ASO technology could be a viable strategy to halt pathological aggregation in TDP-43 and Tau proteinopathies, and possibly in other protein aggregation diseases.
Lastly, given that protein aggregation and phase separation are tightly controlled by the cell’s protein degradation and chaperone machinery [101,104,105,136], ongoing efforts are focused on finding drugs that upregulate these pathways [137], or on the generation of potent engineered disaggregases which could antagonize pathological phase transitions [138,139]. Unraveling the complex regulation of protein phase separation will be key in identifying new pathways, which could be targeted to correct pathological phase transitions.
Concluding Remarks
In recent years it has become clear that numerous cellular organelles are formed through the process of phase separation. Although these organelles have been studied for decades (or in the case of the nucleolus over a century) their dynamic nature and its relevance to their formation, function, and physiopathology has only recently come to light. Leveraging prior insights from polymer chemistry has dramatically advanced our understanding of membraneless organelles in this rapidly progressing field of cell biology, and inspired new approaches to further explore their underlying biophysics. Compellingly, these recent findings are already opening novel avenues to target aberrant protein phase transitions in human disease. Additionally, understanding the relationship between sequence and the resulting material state may also lead to novel synthetic biomaterials [140,141].
We must be fully aware, though, that we are far from completely understanding the complex biology behind membraneless organelles and their functional roles (Box 4). To this end, we have compiled a list of, in our opinion, the key outstanding questions that remain unanswered (see Outstanding Questions). Addressing these questions will be of pivotal importance for gaining further insight into protein phase separation. Developing novel molecular biological and cell biological tools will be essential for this endeavor. At the moment, purification and high-resolution structural studies of membraneless organelles present a bottleneck, especially in a cellular context. Only by generating tools that can specifically target individual granules we will be able to repurpose them for new disease treatments, and translate our basic biological knowledge to the bedside. Protein phase separation has not yet given us all its secrets, and an exciting future lies ahead of us.
Box 4. What Is the Function of Membraneless Organelles?
Numerous proteins have seemingly evolved the ability to drive the formation of or be recruited to membraneless organelles. Yet why do cells need such compartments? What is their biochemical function? Surprisingly, these questions remain largely unanswered.
Compartmentalization in different forms and scales is widely used by organisms. Our stomach is a well-defined organ, which serves one main purpose (i.e., digesting food through acid hydrolysis). Obviously, such a chemical reaction is best carried out if the body has a means to concentrate both food and the acid into one singular compartment. Additionally, this compartmentalization protects other organs from exposure to acid. Similarly, our cells have evolved a strikingly similar mechanism. Lysosomes create an acidic compartment to degrade cellular waste. Through their membrane-barrier lysosomes can both concentrate the reaction components and at the same time protect the rest of the cell from its harmful effects. Another example of organ–organelle parallels includes fat tissue and lipid droplets, which store energy under the form of lipids for later use. Additionally, bodies and cells can amplify signals from the environment by compartmentalizing signal reception: our eyes focus incoming light onto our retina, which concentrates the light receptors. On the subcellular level, neurons also concentrate their receptors in distinct substructures, namely the synapses.
From these analogies four main functions arise for compartmentalization, being: (i) concentration of (bio)chemical reactions, (ii) sequestering harmful components, (iii) storage of biomolecules, and (iv) signal amplification. Interestingly, all these functions exist in the realm of membraneless organelles. First, concentration of cytoskeleton components through phase separation promotes their nucleation into filaments [66,147], and similarly, splicing is controlled by multivalent assembly of splicing factors on mRNA [148]. Second, although protein aggregates in disease are considered harmful, accumulating evidence suggests that they could be an initial rescue mechanism of the cell to sequester the more toxic protein oligomers [149,150]. Third, numerous assemblies function as storage granules, as they sequester proteins and other biomolecules under times of stress or quiescence for later reuse [64,151]. Fourth, by concentrating receptors and signaling molecules, a cell can amplify certain signaling pathways. In light of this, different membrane receptors achieve exactly this through protein phase separation [6,8].
Although we have not fully unraveled the complex function of phase separation in the cell, these examples give us a glimpse into why cells could clearly benefit from the formation of membraneless organelles.
Highlights.
Phase separation is known to play a role in a variety of cellular processes, including formation of classical membraneless organelles, signaling complexes, the cytoskeleton, and numerous other supramolecular assemblies.
The concept of phase separation provides a new framework for our understanding of the functional role of sequence degeneracy (low-complexity) and protein disorder.
Accumulating evidence points to a key role for phase transitions in human diseases associated with protein aggregation, and to the misregulation of membraneless organelles in disease.
Understanding the physical principles and molecular interactions behind protein phase separation could inspire novel biomaterials.
Outstanding Questions.
What are the exact biological functions of phase separation? Why did cells evolve membraneless organelles? What makes liquid/gel assemblies functionally different from canonical protein complexes?
We have a basic understanding of the physical force(s) driving phase separation, yet deeper insights into the interactions at the atomic level will be pivotal in better understanding these phases.
What are the essential and nonessential components of different membraneless organelles, and what are their sequence and structural properties?
Despite a few examples, we know surprisingly little about how cells spatiotemporally regulate phase separation. This will be key in understanding how biology regulates physics. Which regulatory pathways are involved?
A predictive framework on how specificity of membraneless organelle composition is generated is currently completely lacking. Which principles target proteins and RNAs to specific phases and what prevents the coalescence of distinct membraneless organelles?
While progress is being made in determining the internal structure of test tube granules, we mostly lack the tools to pursue this question in living cells. How can we investigate the internal organization of membraneless organelles in living systems?
What are the differences between physiological and pathological assemblies? Which factors drive this conversion in disease?
How do disease mutations and aging specifically affect phase separation of membraneless organelle components? What are the associated molecular events?
Why do diseases associated with protein aggregation display such profound cell type specificity? What makes (specific) neurons especially sensitive to perturbations in proteostasis?
Can we harness our growing understanding of biological phase separation to develop novel therapeutic treatment options? Can we devise ways to specifically target membraneless organelles or interactions within them?
Acknowledgments
This manuscript is based on discussions held at the Phase Transitions in Biology and Disease meeting held in Leuven, May 2nd–3th, 2017, and the seminal discoveries of Dr Anthony A. Hyman, Dr Michael K. Rosen, and others. The authors would like to thank Dr Roy Parker, Dr J. Paul Taylor, Dr Clifford Brangwynne, and Dr Steve McKnight for helpful discussions during the preparation of this manuscript. The authors would like to thank Dr Alex Holehouse and Dr Rohit V. Pappu for contributing text and providing helpful comments. The two anonymous reviewers are gratefully acknowledged for their thorough review and critique of the manuscript. M.F. acknowledges financial support of GINOP-2.3.2-15-2016-00044, and the Hungarian Academy of Sciences. S.B and L.V.D.B are supported by KU Leuven (‘Opening the future’ and C1), the Fund for Scientific Research Flanders (FWO-Vlaanderen), the ‘Agency for Innovation by Science and Technology in Flanders’ (IWT-Vlaanderen) and the ALS Liga (Belgium). S.B. acknowledges a long-term fellowship from EMBO. P.T. is supported by Odysseus grant G.0029.12 from FWO-Vlaanderen. J.S. is supported by a grant from the European Research Council under the European Union’s Horizon 2020 Framework Programme ERC Grant agreement 647458 (MANGO). N.L.F. acknowledges support by NIGMS R01GM118530.
Glossary
Gel
defined by a system- or droplet-spanning network formed by the constituent macromolecules. The crosslinks are either covalent bonds (chemical gel) or noncovalent bonds (physical gel). The material properties of gels vary in a broad range dictated by the lifetimes of crosslinks, the extents of crosslinking, and the crosslinking patterns. Associative polymers can form physical gels. Water-soluble polymers form hydrogels
Intrinsically disordered protein/region
proteins containing regions that adopt multiple structures or exhibit a fast conformational exchange in their native state
Liquid
one of the four fundamental states of matter. Characterized by a definite volume, but no fixed shape. Liquids minimize their surface area (to reduce surface tension), which often leads to the formation of spherical droplets. If two droplets fuse, they also adopt a spherical shape. In liquids, local spatial ordering (i.e., preferred intermolecular distances and orientations) does not exceed the dimensions of a few molecules, beyond which the molecules are randomly organized. This leads to fast reorganization of liquid structure and also enables an exchange of components with the surroundings
Liquid–crystalline
a material state, which shares properties with both liquids and crystals. The constituent molecules are oriented in a crystal-like manner, yet can flow, similarly to liquids. Ordering is considerable on a molecular scale, at least into one direction. Liquid crystals frequently undergo phase transition in response to temperature changes
Liquid–liquid demixing
two liquids coexist as separate phases instead of a mixed solution (see phase separation)
Low-complexity domain (LCD)
a protein segment, which is enriched in or composed of only a few amino acids. These often follow simple patterns, like tandem repeats, and are associated with fast evolutionary rates
Material state
there are four material states or phases in which matter can occur: gas, liquid, solid, and plasma
Membraneless organelle
a non-membrane-bound cellular compartment. Membraneless organelles are usually composed of protein and nucleic acids assemblies and sample a broad range of material states
Phase separation
phase separation reflects a demixing transition, in which a homogenous and well-mixed solution rearranges itself such that distinct regions of space are occupied by a distinct concentration of species. In the simplest case of a binary mixture (polymer and solution), phase separation yields a high concentration region and a low concentration region. In Flory-Huggins solution theory, the free energy of mixing associated with a binary mixture includes a single interdispersing term (χ). If this term is favorable (χ < 0), the components will form a homogenous well-mixed solution. If the interdispersing term is unfavorable (χ > 0) demixing will occur and a two-phase solution will appear
Phase transition
while phase separation refers to a demixing of an initial homogeneous solution, phase transition describes the switch in phase of a molecule (e.g., from liquid to solid)
Solid
although both liquids and solids are termed as condensed matter, they differ in the range of long-range organization of their components and dynamics. Material states of membraneless organelles can also be crystalline, semi-crystalline, or liquid–crystalline depending on the extent of spatial ordering and the directional preferences for spatial ordering
References
- 1.Glisovic T, et al. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitrea DM, Kriwacki RW. Phase separation in biology; functional organization of a higher order. Cell Commun Signal. 2016;14:1. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uversky VN. Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol. 2017;44:18–30. doi: 10.1016/j.sbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Hyman AA, et al. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 5.Boulay G, et al. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell. 2017;171:163–178. doi: 10.1016/j.cell.2017.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li P, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler JR, et al. Distinct stages in stress granule assembly and disassembly. Elife. 2016;5:e18413. doi: 10.7554/eLife.18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su X, et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352:595–599. doi: 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeynaems S, et al. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell. 2017;65:1044–1055. doi: 10.1016/j.molcel.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burke KA, et al. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conicella AE, et al. ALS mutations disrupt phase separation mediated by alpha-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 2016;24:1537–1549. doi: 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon I, et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee KH, et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167:774–788. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molliex A, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami T, et al. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel A, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 17.Handwerger KE, et al. Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Mol Biol Cell. 2005;16:202–211. doi: 10.1091/mbc.E04-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kedersha N, et al. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrei MA, et al. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brangwynne CP, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 21.Brangwynne CP, et al. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trcek T, et al. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat Commun. 2015;6:7962. doi: 10.1038/ncomms8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain S, et al. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feric M, et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fei J, et al. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J Cell Sci. 2017;130:4180–4192. doi: 10.1242/jcs.206854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang JT, et al. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife. 2014;3:e04591. doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitt JN, et al. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol. 2000;219:315–333. doi: 10.1006/dbio.2000.9607. [DOI] [PubMed] [Google Scholar]
- 28.Flory PJ. Thermodynamics of high polymer solutions. J Chem Phys. 1942;10:10. [Google Scholar]
- 29.Broide ML, et al. Binary-liquid phase separation of lens protein solutions. Proc Natl Acad Sci U S A. 1991;88:5660–5664. doi: 10.1073/pnas.88.13.5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galkin O, et al. Liquid-liquid separation in solutions of normal and sickle cell hemoglobin. Proc Natl Acad Sci U S A. 2002;99:8479–8483. doi: 10.1073/pnas.122055299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dumetz AC, et al. Protein phase behavior in aqueous solutions: crystallization, liquid-liquid phase separation, gels, and aggregates. Biophys J. 2008;94:570–583. doi: 10.1529/biophysj.107.116152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ten Wolde PR, Frenkel D. Enhancement of protein crystal nucleation by critical density fluctuations. Science. 1997;277:1975–1978. doi: 10.1126/science.277.5334.1975. [DOI] [PubMed] [Google Scholar]
- 33.Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen JS, et al. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 35.Fong KW, et al. Whole-genome screening identifies proteins localized to distinct nuclear bodies. J Cell Biol. 2013;203:149–164. doi: 10.1083/jcb.201303145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boke E, et al. Amyloid-like self-assembly of a cellular compartment. Cell. 2016;166:637–650. doi: 10.1016/j.cell.2016.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 38.Varadi M, et al. Functional advantages of conserved intrinsic disorder in RNA-binding proteins. PLoS One. 2015;10:e0139731. doi: 10.1371/journal.pone.0139731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilks N, et al. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Decker CJ, et al. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reijns MAM, et al. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lunde BM, et al. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu H, Fuxreiter M. The structure and dynamics of higher-order assemblies: amyloids, signalosomes, and granules. Cell. 2016;165:1055–1066. doi: 10.1016/j.cell.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banjade S, et al. Conserved interdomain linker promotes phase separation of the multivalent adaptor protein Nck. Proc Natl Acad Sci U S A. 2015;112:E6426–E6435. doi: 10.1073/pnas.1508778112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frey S, et al. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- 47.Lin Y, et al. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elbaum-Garfinkle S, et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nott TJ, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brangwynne CP, et al. Polymer physics of intracellular phase transitions. Nat Phys. 2015;11:899–904. [Google Scholar]
- 51.Tompa P, et al. A million peptide motifs for the molecular biologist. Mol Cell. 2014;55:161–169. doi: 10.1016/j.molcel.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 52.Monahan Z, et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36:2951–2967. doi: 10.15252/embj.201696394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pak CW, et al. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riback JA, et al. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell. 2017;168:1028–1040. doi: 10.1016/j.cell.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vernon RM, et al. Pi-pi contacts are an overlooked protein feature relevant to phase separation. Elife. 2018;7:e31486. doi: 10.7554/eLife.31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brady JP, et al. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc Natl Acad Sci U S A. 2017;114:E8194–E8203. doi: 10.1073/pnas.1706197114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin Y, et al. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell. 2016;167:789–802. doi: 10.1016/j.cell.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz JC, et al. RNA seeds higher-order assembly of FUS protein. Cell Rep. 2013;5:918–925. doi: 10.1016/j.celrep.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X, et al. RNA stores tau reversibly in complex coacervates. PLoS Biol. 2017;15:e2002183. doi: 10.1371/journal.pbio.2002183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H, et al. RNA controls polyQ protein phase transitions. Mol Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Banerjee PR, et al. Reentrant phase transition drives dynamic substructure formation in ribonucleoprotein droplets. Angew Chem Int Ed Engl. 2017;56:11354–11359. doi: 10.1002/anie.201703191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kedersha N, et al. Stress granules and cell signaling: more than just a passing phase. Trends Biochem Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berry J, et al. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci U S A. 2015;112:E5237–E5245. doi: 10.1073/pnas.1509317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin M, et al. Glycolytic enzymes coalesce in G bodies under hypoxic stress. Cell Rep. 2017;20:895–908. doi: 10.1016/j.celrep.2017.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–247. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woodruff JB, et al. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell. 2017;169:1066–1077. doi: 10.1016/j.cell.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 67.Zeng M, et al. Phase transition in postsynaptic densities underlies formation of synaptic complexes and synaptic plasticity. Cell. 2016;166:1163–1175. doi: 10.1016/j.cell.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.West JA, et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J Cell Biol. 2016;214:817–830. doi: 10.1083/jcb.201601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boeynaems S, et al. Inside out: the role of nucleocytoplasmic transport in ALS and FTLD. Acta Neuropathol. 2016;132:159–173. doi: 10.1007/s00401-016-1586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramaswami M, et al. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li YR, et al. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 73.Lin WL, Dickson DW. Ultrastructural localization of TDP-43 in filamentous neuronal inclusions in various neurodegenerative diseases. Acta Neuropathol. 2008;116:205–213. doi: 10.1007/s00401-008-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kao PF, et al. Detection of TDP-43 oligomers in frontotemporal lobar degeneration-TDP. Ann Neurol. 2015;78:211–221. doi: 10.1002/ana.24431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murray DT, et al. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017;171:615–627. doi: 10.1016/j.cell.2017.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu-Yesucevitz L, et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harmon TS, et al. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife. 2017;6:e30294. doi: 10.7554/eLife.30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tourriere H, et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Shen TH, et al. The mechanisms of PML-nuclear body formation. Mol Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell. 2000;11:4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marzahn MR, et al. Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 2016;35:1254–1275. doi: 10.15252/embj.201593169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Afroz T, et al. Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat Commun. 2017;8:45. doi: 10.1038/s41467-017-00062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shin Y, et al. Spatiotemporal control of intracellular phase transitions using light-activated optodroplets. Cell. 2017;168:159–171. doi: 10.1016/j.cell.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hughes MP, et al. Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks. Science. 2018;359:698–701. doi: 10.1126/science.aan6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Updike DL, et al. P granules extend the nuclear pore complex environment in the C. elegans germ line. J Cell Biol. 2011;192:939–948. doi: 10.1083/jcb.201010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wei MT, et al. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat Chem. 2017;9:1118–1125. doi: 10.1038/nchem.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnston JA, et al. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aumiller WM, Jr, Keating CD. Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat Chem. 2016;8:129–137. doi: 10.1038/nchem.2414. [DOI] [PubMed] [Google Scholar]
- 90.Kwon I, et al. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ryan VH, et al. Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol Cell. 2018;69:465–479. doi: 10.1016/j.molcel.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wippich F, et al. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 93.Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thompson MJ, et al. The 3D profile method for identifying fibril-forming segments of proteins. Proc Natl Acad Sci U S A. 2006;103:4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wojciechowska M, Krzyzosiak WJ. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum Mol Genet. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cleary JD, Ranum LP. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum Mol Genet. 2013;22:R45–R51. doi: 10.1093/hmg/ddt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mori K, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 98.Ash PE, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zu T, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yamakawa M, et al. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum Mol Genet. 2015;24:1630–1645. doi: 10.1093/hmg/ddu576. [DOI] [PubMed] [Google Scholar]
- 101.Buchan JR, et al. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Renton AE, et al. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rubinsztein DC, et al. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 104.Ganassi M, et al. A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol Cell. 2016;63:796–810. doi: 10.1016/j.molcel.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 105.Mateju D, et al. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 2017;36:1669–1687. doi: 10.15252/embj.201695957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.D’Angelo MA, et al. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boeynaems S, et al. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci Rep. 2016;6:20877. doi: 10.1038/srep20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang K, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jovicic A, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Freibaum BD, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Grima JC, et al. Mutant huntingtin disrupts the nuclear pore complex. Neuron. 2017;94:93–107. doi: 10.1016/j.neuron.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gasset-Rosa F, et al. Polyglutamine-expanded hunting-tin exacerbates age-related disruption of nuclear integrity and nucleocytoplasmic transport. Neuron. 2017;94:48–57. doi: 10.1016/j.neuron.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 114.Patel A, et al. ATP as a biological hydrotrope. Science. 2017;356:753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- 115.Grundke-Iqbal I, et al. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci U S A. 1986;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ambadipudi S, et al. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat Commun. 2017;8:275. doi: 10.1038/s41467-017-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Boeynaems S, et al. Phasing in on the cell cycle. Cell Div. 2018;13:1. doi: 10.1186/s13008-018-0034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rabbitts TH, et al. Fusion of the dominant-negative transcription regulator chop with a novel gene Fus by translocation T(12-16) in malignant liposarcoma. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 119.Hegyi H, et al. Intrinsic structural disorder confers cellular viability on oncogenic fusion proteins. PLoS Comput Biol. 2009;5:e1000552. doi: 10.1371/journal.pcbi.1000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Latysheva NS, et al. Molecular principles of gene fusion mediated rewiring of protein interaction networks in cancer. Mol Cell. 2016;63:579–592. doi: 10.1016/j.molcel.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Toretsky JA, Wright PE. Assemblages: functional units formed by cellular phase separation. J Cell Biol. 2014;206:579–588. doi: 10.1083/jcb.201404124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Janke AM, et al. Lysines in the RNA polymerase II C-terminal domain contribute to TAF15 fibril recruitment. Biochemistry. 2017 doi: 10.1021/acs.biochem.7b00310. Published online September 25, 2017. http://dx.doi.org/10.1021/acs.biochem.7b00310. [DOI] [PMC free article] [PubMed]
- 123.Grabocka E, Bar-Sagi D. Mutant KRAS enhances tumor cell fitness by upregulating stress granules. Cell. 2016;167:1803–1813. doi: 10.1016/j.cell.2016.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Somasekharan SP, et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J Cell Biol. 2015;208:913–929. doi: 10.1083/jcb.201411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Adriaens C, et al. p53 induces formation of NEAT1 IncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22:861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 126.Xu J, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol. 2011;7:285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
- 127.Soragni A, et al. A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell. 2016;29:90–103. doi: 10.1016/j.ccell.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.White JP, Lloyd RE. Regulation of stress granules in virus systems. Trends Microbiol. 2012;20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hou S, et al. Zika virus hijacks stress granule proteins and modulates the host stress response. J Virol. 2017 doi: 10.1128/JVI.00474-17. Published online June 7, 2017. http://dx.doi.org/10.1128/JVI.00474-17. [DOI] [PMC free article] [PubMed]
- 130.Kroschwald S, et al. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife. 2015;4:e06807. doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schoch KM, Miller TM. Antisense oligonucleotides: translation from mouse models to human neurodegenerative diseases. Neuron. 2017;94:1056–1070. doi: 10.1016/j.neuron.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Becker LA, et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544:367–371. doi: 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vanderweyde T, et al. Interaction of tau with the RNA-binding protein TIA1 regulates tau pathophysiology and toxicity. Cell Rep. 2016;15:1455–1466. doi: 10.1016/j.celrep.2016.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Apicco DJ, et al. Reducing the RNA binding protein TIA1 protects against tau-mediated neurodegeneration in vivo. Nat Neurosci. 2018;21:72–80. doi: 10.1038/s41593-017-0022-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guo LL, et al. A cellular system that degrades misfolded proteins and protects against neurodegeneration. Mol Cell. 2014;55:15–30. doi: 10.1016/j.molcel.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Barmada SJ, et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10:677–685. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yasuda K, et al. FUS inclusions disrupt RNA localization by sequestering kinesin-1 and inhibiting microtubule detyrosination. J Cell Biol. 2017;216:1015–1034. doi: 10.1083/jcb.201608022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jackrel ME, et al. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell. 2014;156:170–182. doi: 10.1016/j.cell.2013.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Quiroz FG, Chilkoti A. Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat Mater. 2015;14:1164–1171. doi: 10.1038/nmat4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rauscher S, Pomes R. The liquid structure of elastin. Elife. 2017;6:e26526. doi: 10.7554/eLife.26526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Johnson BS, et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Banani SF, et al. Compositional control of phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schmidt HB, Gorlich D. Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. Elife. 2015;4:04251. doi: 10.7554/eLife.04251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tuu Szabo B, et al. Simulations of higher-order protein assemblies using a fuzzy framework. bioRxiv. 2018 Published online January 15, 2018. http://dx.doi.org/10.1101/248062.
- 146.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 147.Hernandez-Vega A, et al. Local nucleation of microtubule bundles through tubulin concentration into a condensed tau phase. Cell Rep. 2017;20:2304–2312. doi: 10.1016/j.celrep.2017.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gueroussov S, et al. Regulatory expansion in mammals of multivalent hnRNP assemblies that globally control alternative splicing. Cell. 2017;170:324–339. doi: 10.1016/j.cell.2017.06.037. [DOI] [PubMed] [Google Scholar]
- 149.Arrasate M, et al. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 150.Cremers CM, et al. Polyphosphate: a conserved modifier of amyloidogenic processes. Mol Cell. 2016;63:768–780. doi: 10.1016/j.molcel.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wallace EW, et al. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell. 2015;162:1286–1298. doi: 10.1016/j.cell.2015.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]