The tree of one percent (original) (raw)
Short abstract
As lateral gene transfer among prokaryotes and endosymbiotic gene transfer (from organelles) among eukaryotes are fundamentally not tree-like in nature, biologists need to depart from the notion that all genomes are related by a single bifurcating tree.
Abstract
Two significant evolutionary processes are fundamentally not tree-like in nature - lateral gene transfer among prokaryotes and endosymbiotic gene transfer (from organelles) among eukaryotes. To incorporate such processes into the bigger picture of early evolution, biologists need to depart from the preconceived notion that all genomes are related by a single bifurcating tree.
Evolutionary biologists like to think in terms of trees. Since Darwin, biologists have envisaged phylogeny as a tree-like process of lineage splittings. But Darwin was not concerned with the evolution of microbes, where lateral gene transfer (LGT; a distinctly non-treelike process) is an important mechanism of natural variation, as prokaryotic genome sequences attest [1-4]. Evolutionary biologists are not debating whether LGT exists. But they are debating - and heatedly so - how much LGT actually goes on in evolution. Recent estimates of the proportion of prokaryotic genes that have been affected by LGT differ 30-fold, ranging from 2% [5] to 60% [6]. Biologists are also hotly debating how LGT should influence our approach to understanding genome evolution on the one hand, and our approach to the natural classification of all living things on the other. These debates erupt most acutely over the concept of a tree of life. Here we consider how LGT and endosymbiosis bear on contemporary views of microbial evolution, most of which stem from the days before genome sequences were available.
A tree of life?
When it comes to the concept of a tree of life, there are currently two main camps. One camp, which we shall call the positivists, says that there is a tree of life, that microbial genomes are, in the main, related by a series of bifurcations, and that when we have sifted out a presumably small amount of annoying chaff (LGT), the wheat (the tree) will be there and will still our hunger for a grand and natural system [7-10]. The other camp, which we will call the microbialists, says that LGT is just as natural among prokaryotes as is point mutation, and that furthermore, it has occurred throughout microbial history. This means that even were we to agree on a grand natural classification, the process of microbial evolution underlying it would be fundamentally undepictable as a single bifurcating tree, because a substantial component of the evolutionary process - LGT - is not tree-like to begin with [1,11,12].
A recent paper by Ciccarelli et al. [9] brings these two views head-to-head. It purports to weigh in heavily for the positivists, but in doing so it inadvertently provides some of the strongest support for the microbialist camp that has been published so far. A closer look reveals why. Ciccarelli et al. [9] report an automated procedure for identifying protein families that are universally distributed among all genomes, with pipeline alignment and tree building. Their routine looked for possible cases of LGT (detected as unusual tree topologies), excluded such proteins, and reiterated the procedure until the universe of proteins had been examined. This left them with 31 presumably orthologous protein sequences present in 191 genomes each, the alignments of which were concatenated to produce a data matrix with 8,089 sites (of which only 1,212 would have remained had gapped sites been excluded). A maximum likelihood tree was inferred from this matrix, motivating a brief discussion of some important events in life's history as inferred from that tree.
Fair enough, one might say, what is there to debate? Lots. Bearing in mind that an average prokaryotic proteome represents about 3,000 protein-coding genes, the 31-protein tree of life represents only about 1% of an average prokaryotic proteome and only 0.1% of a large eukaryotic proteome. Thus, the positivists can say that there is a tree of life after all: a bit skimpier than expected, but a tree nonetheless. But the microbialists, glaring at the same data, can say that the glass is only 1% full at best, and more than 99% empty! There might be a tree there, but it is not the tree of life, it is the 'tree of one percent of life'.
Looking at the issue openly, the finding that, on average, only 0.1% to 1% of each genome fits the metaphor of a tree of life overwhelmingly supports the central pillar of the microbialist argument that a single bifurcating tree is an insufficient model to describe the microbial evolutionary process. If throwing out all non-universally distributed genes and all suspected cases of LGT in our search for the tree of life leaves us with a tree of one percent, then we should probably abandon the tree as a working hypothesis. When chemists or physicists find that a given null hypothesis can account for only 1% of their data, they immediately start searching for a better hypothesis. Not so with microbial evolution, it seems, which is rather worrying. Could it be that many biologists have their heart set on finding a tree of life, regardless of what the data actually say?
Which hypotheses (if any) are we testing?
By themselves, genomes cannot tell us anything about evolution, microbial or otherwise. Evolutionary biology is about hypothesis testing: one checks to see if data from genomes provide support or not for one or the other hypothesis that was generated independently of the genome data used to test it. What ideas about early evolution that could be tested with genome data are currently discussed by specialists in the field? We consider five distinctly different views, each of which enjoys some popularity.
The rRNA tree
The first is the classical ribosomal RNA (rRNA) tree of life as constructed by Carl Woese and colleagues [13-16] from the late 1970s onwards (Figure 1a). It suggests, in its current interpretations, that the universal ancestor of all life (the progenote) was a communal collection of information-storing and information-processing entities that were not yet organized as cells. LGT is seen as the main mode of genetic novelty at the early stages of evolution, and the process of vertical inheritance arises only with the process of 'genetic annealing' from within this mixture. At this point, the emerging cellular lineages of prokaryotes and eukaryotes become refractory to LGT, and are considered to traverse a kind of 'Darwinian threshold' from the organizational state of supramolecular aggregates to the organizational state of cells. Traversing that threshold is seen as equivalent to the primary emergence, from the broth in which life arose, of the three kinds of cells that we recognize today - archaebacteria, eubacteria and eukaryotes. The classical tree [13] assumed its current shape when anciently diverged protein-coding genes suggested that the root of the universal tree lies on the bacterial branch [17,18]. This view admits that chloroplasts and mitochondria did arise via endosymbiosis, but it sees no role for mitochondria or any other kind of symbiosis in the emergence of the eukaryotic lineage, and the genetic contribution of mitochondria to eukaryotes is seen as detectable, but negligible in evolutionary or mechanistic terms [19]. The classical tree is taken by some to indicate that eukaryotes are in fact sisters of archaea at the level of the whole genome [9,16], a view that is, however, mainly founded on extrapolation from the rRNA tree to the rest of the genome without actually looking at all of the data.
Figure 1.
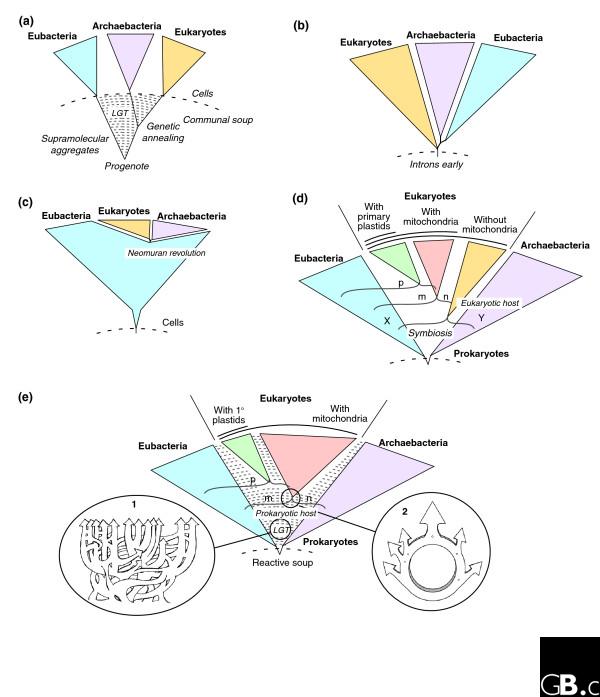
Five different current views of the general shape of microbial evolution. (a) The 'classical' tree derived from comparison of rRNA sequence and rooted with ancient paralogs. It is thought to arise from a collection of non-cellular supramolecular aggregates in the primordial soup, between which there is lateral gene transfer (LGT). A process dubbed genetic annealing gives rise to cells. In this scenario, the three domains of life - Eubacteria, Archaebacteria and Eukaryotes - branch off in that order. (b) The introns-early tree. This proposes that the ancestor of all three domains contained introns, which were lost in the Archaebacteria and Euacteria. (c) The neomuran tree. This introduces an ancestral group of organisms from which Archaeabacteria and Eukaryotes arose after the loss of the eubacterial-type cell wall in one lineage (the neomuran revolution). (d) The symbiotic tree. This proposes that the ancestor of eukaryotes originated by the endosymbiosis of one prokaryote (X) in another prokaryote host (Y), giving rise to nucleated (n) eukaryotic cells. The different groups of eukaryotes arose by subsequent separate endosymbiotic events involving various prokaryotes - the ancestors of plastids (p) and mitochondria (m) - in host cells of this lineage. (e) The prokaryote-host tree. This also incorporates endosymbiosis as the origin of mitochondria and plastids, but proposes that the endosymbiotic event that gave rise to a cell containing nucleus and mitochondria occurred in a prokaryotic host. This leads to a ring-like relationship between the ancestral organisms rather than a tree (see inset 2). This model also invokes extensive LGT throughout microbial evolution (see inset 1). See text for further details.
The introns-early tree
The introns-early (or eukaryotes-first) tree emerged when Ford Doolittle [20] suggested that the ancestral state of genes might be 'split', and that some introns in eukaryotic genes might thus be carryovers from the assembly of primordial protein-coding regions. In that case, the organizational state of eukaryotic genes (having introns) would represent the organizational state of the very first genomes [21] and the intronless prokaryotic state would be a derived condition (Figure 1b), a view that was christened 'introns-early' [22]. Doolittle has since abandoned this view [23], but it has found other proponents [24,25]. They draw upon different lines of evidence in support, and call their position 'introns-first' rather than introns-early [25]. They agree that the eubacterial root assumed for the rRNA tree is questionable and that a eukaryote root is more likely [26,27].
Some of the proponents of the introns-first hypothesis interpret various aspects of RNA processing in eukaryotes (in addition to introns), such as rRNA modification through small nucleolar RNAs (snoRNAs), as direct carryovers from the RNA world and hence as evidence for eukaryote antiquity [26,28,29]. There is no prokaryote-to-eukaryote transition in the introns-early tree, because prokaryotic genome organization is seen as a very early derivative of eukaryotic gene organization. Accordingly, the relationship of eukaryotes and prokaryotes is depicted largely as a more-or-less unresolved trichotomy [19], and the contribution of organelles or symbiosis to eukaryote evolution is admitted as existing, but negligible in terms of evolutionary significance.
The neomuran tree
The neomuran tree (Figure 1c) stems from the work of Tom Cavalier-Smith [30-32]. No theory on the relationship of prokaryotes to eukaryotes, current or otherwise, is more explicit in terms of details of mechanism [32]. In the main, it suggests that the common ancestor of all cells was a free-living eubacterium (in the most recent version of the theory, a _Chlorobium_-like anoxygenic photosynthesizer) and that eubacteria were the only organisms on Earth until about 900 million years ago. At this time, a member of the eubacteria, in recent versions an actinobacterium, lost its murein-containing cell wall and was faced with the task of reinventing a new cell wall (hence the Latin name: neo, new; murus, wall). This led to the origin of a group of rapidly evolving organisms that Cavalier-Smith calls the Neomura.
The loss of the cell wall precipitated an unprecedented process of descent with modification in this group. During a short period of time (perhaps 50 million years), the characters that are shared by archaebacteria and eukaryotes arose (for a list of those characters, see [31]). The neomuran lineage then underwent diversification into two lineages, with another long list of evolutionary changes in each. One lineage invented isoprene ether lipid synthesis and gave rise to archaebacteria. The other became phagotrophic and gave rise to the eukaryotes. In older versions of this hypothesis, some eukaryote lineages branched off before the mitochondrion was acquired; these lineages were once called the Archezoa [30]. In newer versions, the mitochondrion comes into the eukaryote lineage before any archezoan can arise. No evolutionary intermediates from the transitions of actinobacteria into neomurans, archaebacteria, and eukaryotes persist among the modern biota, which is a distressing aspect of the theory for many specialists. The neomuran theory accounts mainly for cell biological characters, but not for sequence similarity among genes.
The symbiotic tree: a merger of distinct branches
At about the same time that archaebacteria and introns were being discovered, biologists were still fiercely debating the issue of whether mitochondria and chloroplasts were once free-living prokaryotes [33] or not [34]. Lynn Margulis had revived the old and controversial theories from the early 20th century regarding the endosymbiotic origin of chloroplasts and mitochondria [35,36]. Margulis's version of endosymbiotic theory was one of eukaryotes-in-pieces, and has always contained an additional partner at eukaryote origins to which no specialists other than herself have given credence: the spirochete origin of eukaryotic flagella [35-37]. Other prokaryote symbioses en route to eukaryotes involve the possible endosymbiotic origin of peroxisomes [38,39], or an endosymbiotic origin of the nucleus [40-42]. Common to those theories are a eubacterial-archaebacterial merger of some sort at the origin of eukaryotes (X and Y in Figure 1d), giving rise to a nucleated but mitochondrion-lacking cell - an archezoon [30] - followed by the origin of mitochondria.
From the viewpoint of more modern data, the spirochete origin of eukaryotic flagella can be seen as both unsupported and unnecessary [43], as can an endosymbiotic origin for peroxisomes, for which there are also no supporting data [44]. The origin of the nucleus is still debated [45].
The prokaryote-tree with LGT: a merger of ephemeral genomes
An exciting prospect predicted by all the foregoing hypotheses was that the most primitive eukaryotic lineages should lack mitochondria. That sent molecular biologists scrambling to study contemporary eukaryotes that were thought to lack mitochondria, work that unearthed findings of the most unexpected kind: all of the purportedly primitive and mitochondrion-lacking lineages were not really primitive nor did they even lack mitochondria. The mitochondria are there, it turns out, but they do not use oxygen [46,47], they are small [48], and some do not even produce ATP [49]. These 'new' members of the mitochondrial family among eukaryotic anaerobes (and some parasitic aerobes [50]) are called hydrogenosomes and mitosomes (reviewed in [51]). That pointed to the possibility that there never were any eukaryotes that lacked mitochondria; hence, the host that acquired the mitochondrion might have just been an archaeon outright (Figure 1e). Several hypotheses of this sort have been published, some of which account for the common ancestry of mitochondria and hydrogenosomes (reviewed in [52]) and some of which account for the origin of the nucleus [53].
Like the symbiotic tree, the prokaryote-host tree can accommodate LGT [54] without problems (Figure 1e, inset 1), and furthermore implies the existence of ring-like structures [55], rather than tree-like structures linking prokaryotes and eukaryotes at the level of gene content and sequence similarity (Figure 1e, inset 2). The only real difference between the symbiotic tree and the prokaryote-host tree hypotheses concerns the number of symbiotic partners involved at eukaryote origins - more than two versus two, respectively - and the existence (or nonexistence) of primitively amitochondriate eukaryotes. Both predictions are, in principle, testable with genome data, but the tests become a bit more complicated than standard phylogenetic tests, because of LGT [52].
The biggest branch is the biggest problem
For many biologists concerned with life's deeper relationships, the longest and most strongly supported branch in many current versions of the tree of life as depicted in Figure 1a or in recent papers [9,16] is also the most misleading: the central branch that implies a sister-group relationship between eukaryotes and archaebacteria [9,13]. It is misleading because at the level of genome-wide patterns of sequence similarity, eukaryotes are far more similar to eubacteria than they are to archaebacteria [56]. Put another way, eukaryotes possess more eubacteria-related genes than they possess archaebacteria-related genes [56,57]. This has escaped the attention of almost everyone, and is one of evolutionary biology's best-kept secrets, at least in circles where the rRNA tree is thought to speak for the whole genome.
An example emphasizing this point is shown in Figure 2, where the percentage amino-acid identity between eukaryotic proteins (human in this example; yeast in [56]) and their homologs in prokaryotes (when present) is depicted. Of the 5,833 human proteins that have homologs in these prokaryotes at the specified thresholds, 2,811 (48%) have homologs in eubacteria only, while 828 (14%) have homologs in archaebacteria only, and 4,788 (80%) have greater sequence identity with eubacterial homologs, whereas 877 (15%) are more similar to archaebacterial homologs (196 are ties). The proteins comprising the recent tree of life - or the tree of one percent [9] - belong almost exclusively to the informational class [57]; that is, they are involved in information storage and processing. It is well known that eukaryotic informational genes are archaea-like [55-57]. They indicate a close relationship of eukaryotes and archaebacteria, but as is clearly visible in Figure 2, they speak for only a very small minority of eukaryotic genes [56].
Figure 2.
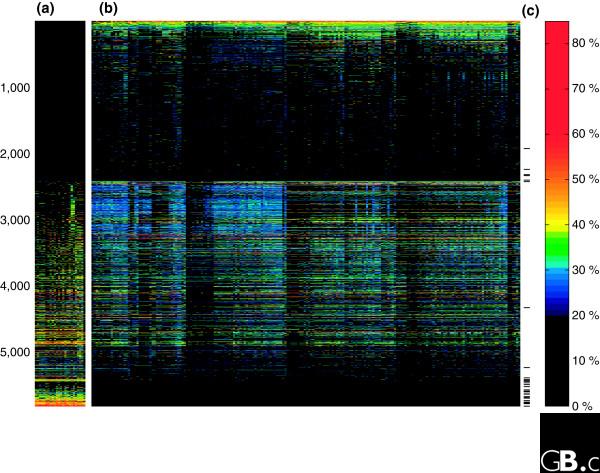
As a representative eukaryote example, the non-redundant set of human proteins (NCBI's Refseq database [70]) was compared using BLAST to a data set containing all proteins from 224 prokaryotic genomes: (a) 24 archaebacteria and (b) 200 eubacteria. In each panel, individual genomes are represented by columns and individual proteins by rows; numbers of proteins are indicated on the left and percentage amino-acid identity by the color scale shown on the right. BLAST hits with an e-value ≤ 10-20 and ≥ 20% amino-acid identity were recorded. The percent identity of the best blast hit for each human protein in each prokaryote was color coded as shown on the right and plotted with MATLAB©. The 31 proteins that were used in the recent tree of life [9] are marked with ticks in column (c). A table containing the numbers, genes, and species underlying the figure is available as additional data file 1.
Eukaryotes possess genes that they have inherited from archaebacteria and from eubacterial organelles [58]. But in plants, the acquisition of genes from cyanobacteria (plastids) has been estimated as 18% of the genome; the acquisition from mitochondria could be even greater [52]. Because such substantial gene influxes cannot be represented with bifurcating trees, they are usually just ignored.
A refreshing exception to the assumption that the tree of life is a tree to begin with is the recent paper by Rivera and Lake [55], who reported a procedure that takes LGT into account; it shows eukaryotes as the sisters of archaebacteria and eubacteria simultaneously (Figure 1e, inset 2). But Rivera and Lake [55] did not force the data onto a tree; rather, they looked to see whether the data were actually tree-like in structure, and found that a directed acyclic graph (a ring) represents the underlying evolutionary process linking prokaryotes to eukaryotes better than a tree does. They offered crisp arguments that endosymbiosis is the most likely cause for the ring-like nature of the data.
But not everyone agrees that symbiosis was important in eukaryote evolution. Some biologists, mainly from the positivist camp, categorically reject the idea that eukaryotes acquired many, or any, genes from endosymbionts, and they scorn the notion that endosymbiosis had anything to do with eukaryote origins [15,19,39]. An argument salient to that view is the sweeping claim that endosymbiosis and gene transfer from endosymbionts fails to account for the evolution of any outstanding eukaryote characters [19], such as the nucleus. A more optimistic view from the microbialist camp is that the endosymbiotic origin of mitochondria could have made a major contribution to the genetic makeup of eukaryotes [58,59]. This could account for the finding that operational genes of bacterial origin are in the majority in eukaryote genomes [52]. The origin of mitochondria could have even precipitated the origin of the nucleus via the introduction of introns into eukaryotic lineages [53]. The roles of LGT and endosymbiosis in evolution have always been controversial. Genomes attest that both processes are important [23], but neither can be handled by strictly bifurcating trees as a means to represent genome evolution.
Seeing the wood for the trees
The need to incorporate non-treelike processes into ideas about microbial evolution has long been evident [57,60-63]. But mathematicians and bioinformaticians are just now beginning to explore the biological utility of graphs that can recover and represent non-treelike process that sometimes underlie patterns of sequence similarity in molecular data and patterns of shared genes. These approaches can involve networks [64-67], rings [55], or simply tack inferred gene exchanges onto trees [4,68,69]. These newer approaches aim to recover and depict both the tree-like (vertical inheritance through common descent) and the non-treelike (LGT and endosymbiosis) mechanisms of microbial evolution. As such, they represent important advances, because both mechanisms are germane to the processes through which microbes evolve in nature.
So, are we close to having a microbial tree of life [9]? Or are we closer to rejecting a single tree as the null hypothesis for the process of microbial genome evolution [1,54]? All in all, the latter seems more likely, for if our search for the tree of life delivers the tree of one percent, then we should be searching for graphs and theories that fit the data better than a single bifurcating tree.
Additional data file
Additional data file 1 is a table containing the numbers, genes, and species on which Figure 2 is based.
Supplementary Material
Additional data file 1
A table containing the numbers, genes, and species on which Figure 2 is based
References
- Doolittle WF. If the tree of life fell, would it make a sound? In: Sapp J, editor. Microbial Phylogeny and Evolution: Concepts and Controversies. New York: Oxford University Press; 2004. pp. 119–133. [Google Scholar]
- Mirkin BG, Fenner TI, Galperin MY, Koonin EV. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 2002;12:17–25. doi: 10.1101/gr.176501. [DOI] [PubMed] [Google Scholar]
- Kunin V, Goldovsky L, Darzentas N, Ouzounis CA. The net of life: reconstructing the microbial phylogenetic network. Genome Res. 2005;15:954–959. doi: 10.1101/gr.3666505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge F, Wang LS, Kim J. The cobweb of life revealed by genome-scale estimates of horizontal gene transfer. PLoS Biol. 2005;3:e316. doi: 10.1371/journal.pbio.0030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. Interpreting the universal phylogenetic tree. Proc Natl Acad Sci USA. 2000;97:8392–8396. doi: 10.1073/pnas.97.15.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Canback B, Berg OG. Horizontal gene transfer: a critical view. Proc Natl Acad Sci USA. 2003;100:9658–9662. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Daubin V, Moran NA, Ochman H. Phylogenetics and the cohesion of bacterial genomes. Science. 2003;301:829–832. doi: 10.1126/science.1086568. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- Pal C, Papp B, Lercher MJ. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet. 2005;37:1372–1375. doi: 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria and Eukarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. The universal ancestor. Proc Natl Acad Sci USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. On the evolution of cells. Proc Natl Acad Sci USA. 2002;99:8742–8747. doi: 10.1073/pnas.132266999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NR. Time for a change. Nature. 2006;441:289. doi: 10.1038/441289a. [DOI] [PubMed] [Google Scholar]
- Iwabe N, Kuma K-I, Hasegawa M, Osawa S, Miyata T. Evolutionary relationship of archaebacteria, eubacteria and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci USA. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP, Kibak H, Dittrich P, Taiz L, Bowman EJ, Bowman BJ, Manolson MF, Poole RJ, Date T, Oshima T, et al. Evolution of the vacuolar H+-ATPase: Implications for the origin of eukaryotes. Proc Natl Acad Sci USA. 1989;86:6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Collins LJ, Penny D. Genomics and the irreducible nature of eukaryote cells. Science. 2006;312:1011–1014. doi: 10.1126/science.1121674. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. Genes in pieces: Were they ever together? Nature. 1978;272:581–582. doi: 10.1038/272581a0. [DOI] [Google Scholar]
- Doolittle WF. Revolutionary concepts in evolutionary cell biology. Trends Biochem Sci. 1980;5:147–149. doi: 10.1016/0968-0004(80)90010-9. [DOI] [Google Scholar]
- Doolittle WF. The origin and function of intervening sequences in DNA: a review. Am Nat. 1987;130:915–928. doi: 10.1086/284755. [DOI] [Google Scholar]
- Stoltzfus A, Spencer DF, Zuker M, Logsdon JM, Jr, Doolittle WF. Testing the exon theory of genes: the evidence from protein structure. Science. 1994;265:202–207. doi: 10.1126/science.8023140. [DOI] [PubMed] [Google Scholar]
- Forterre P. Thermoreduction, a hypothesis for the origin of prokaryotes. C R Acad Sci III. 1995;318:415–422. [PubMed] [Google Scholar]
- Jeffares DC, Poole AM, Penny D. Relics from the RNA world. J Mol Evol. 1998;46:18–36. doi: 10.1007/PL00006280. [DOI] [PubMed] [Google Scholar]
- Poole A, Jeffares D, Penny D. Prokaryotes, the new kids on the block. BioEssays. 1999;21:880–889. doi: 10.1002/(SICI)1521-1878(199910)21:10<880::AID-BIES11>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Forterre P, Philippe H. Where is the root of the universal tree of life? BioEssays. 1999;21:871–879. doi: 10.1002/(SICI)1521-1878(199910)21:10<871::AID-BIES10>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Penny D, Poole A. The nature of the last universal common ancestor. Curr Opin Genet Dev. 1999;9:672–677. doi: 10.1016/S0959-437X(99)00020-9. [DOI] [PubMed] [Google Scholar]
- Jeffares DC, Mourier T, Penny D. The biology of intron gain and loss. Trends Genet. 2006;22:16–22. doi: 10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The origin of eukaryote and archaebacterial cells. Ann NY Acad Sci. 1987;503:17–54. doi: 10.1111/j.1749-6632.1987.tb40596.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Cell evolution and Earth history: stasis and revolution. Philos Trans R Soc Lond B Biol Sci. 2006;361:969–1006. doi: 10.1098/rstb.2006.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L, Doolittle WF. On the prokaryotic nature of red algal chloroplasts. Proc Natl Acad Sci USA. 1975;72:2310–2314. doi: 10.1073/pnas.72.6.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. The origin of nuclei and of eukaryotic cells. Nature. 1975;256:463–468. doi: 10.1038/256463a0. [DOI] [PubMed] [Google Scholar]
- Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:225–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- Margulis L. Origin of Eukaryotic Cells. New Haven, CT: Yale University Press; 1970. [Google Scholar]
- Margulis L, Dolan MF, Guerrero R. The chimeric eukaryote: Origin of the nucleus from the karyomastigont in amitochondriate protists. Proc Natl Acad Sci USA. 2000;97:6954–6959. doi: 10.1073/pnas.97.13.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. The simultaneous symbiotic origin of mitochondria, chloroplasts and microbodies. Ann NY Acad Sci. 1987;503:55–71. doi: 10.1111/j.1749-6632.1987.tb40597.x. [DOI] [PubMed] [Google Scholar]
- de Duve C. Singularities Landmarks on the Pathways of Life. Cambridge, UK: Cambridge University Press; 2005. [Google Scholar]
- Lake JA, Rivera MC. Was the nucleus the first endosymbiont? Proc Natl Acad Sci USA. 1994;91:2880–2881. doi: 10.1073/pnas.91.8.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS. Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiike T, Hamada K, Miyata D, Shinozawa T. The origin of eukaryotes is suggested as the symbiosis of Pyrococcus into γ-proteobacteria by phylogenetic tree based on gene content. J Mol Evol. 2004;59:606–619. doi: 10.1007/s00239-004-2652-5. [DOI] [PubMed] [Google Scholar]
- Jekely G, Arendt D. Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. BioEssays. 2006;28:191–198. doi: 10.1002/bies.20369. [DOI] [PubMed] [Google Scholar]
- Gabaldon T, Snel B, van Zimmeren F, Hemrika W, Tabak H, Huynen MA. Origin and evolution of the peroxisomal proteome. Biol Direct. 2006;1:8. doi: 10.1186/1745-6150-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W. Archaebacteria (Archaea) and the origin of the eukaryotic nucleus. Curr Opin Microbiol. 2005;8:630–637. doi: 10.1016/j.mib.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Müller M. The hydrogenosome. J Gen Microbiol. 1993;139:2879–2889. doi: 10.1099/00221287-139-12-2879. [DOI] [PubMed] [Google Scholar]
- Müller M. Energy metabolism. Part I: Anaerobic protozoa. In: Marr J, editor. Molecular Medical Parasitology. London: Academic Press; 2003. pp. 125–139. [Google Scholar]
- Tovar J, Fischer A, Clark CG. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol Microbiol. 1999;32:1013–1021. doi: 10.1046/j.1365-2958.1999.01414.x. [DOI] [PubMed] [Google Scholar]
- Tovar J, León-Avila G, Sánchez LB, Sutak R, Tachezy J, van der Giezen M, Hernández M, Müller M, Lucocq JM. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature. 2003;426:172–176. doi: 10.1038/nature01945. [DOI] [PubMed] [Google Scholar]
- Williams BA, Hirt RP, Lucocq JM, Embley TM. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–869. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- van der Giezen M, Tovar J, Clark CG. Mitochondrion-derived organelles in protists and fungi. Int Rev Cytol. 2005;244:175–225. doi: 10.1016/S0074-7696(05)44005-X. [DOI] [PubMed] [Google Scholar]
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentation. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- Rivera MC, Lake JA. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature. 2004;431:152–155. doi: 10.1038/nature02848. [DOI] [PubMed] [Google Scholar]
- Esser C, Ahmadinejad N, Wiegand C, Rotte C, Sebastiani F, Gelius-Dietrich G, Henze K, Kretschmann E, Richly E, Leister D, et al. A genome phylogeny for mitochondria among alpha-pro-teobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 2004;21:1643–1660. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci USA. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Boucher Y, Nesbo CL, Douady CJ, Andersson JO, Roger AJ. How big is the iceberg of which organellar genes in nuclear genomes are but the tip? Philos Trans R Soc Lond B Biol Sci. 2003;358:39–58. doi: 10.1098/rstb.2002.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR. Ancient horizontal gene transfer. Nat Rev Genet. 2003;4:121–132. doi: 10.1038/nrg1000. [DOI] [PubMed] [Google Scholar]
- Martin W. Is something wrong with the tree of life? BioEssays. 1996;18:523–527. doi: 10.1002/bies.950180702. [DOI] [Google Scholar]
- Doolittle WF. Fun with genealogy. Proc Natl Acad Sci USA. 1997;94:12751–12753. doi: 10.1073/pnas.94.24.12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D-F, Cho G, Doolittle RF. Determining divergence times with a protein clock: update and reevaluation. Proc Natl Acad Sci USA. 1997;94:13028–13033. doi: 10.1073/pnas.94.24.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland BR, Jermiin LS, Moulton V. Improved consensus network techniques for genome-scale phylogeny. Mol Biol Evol. 2006;23:848–855. doi: 10.1093/molbev/msj061. [DOI] [PubMed] [Google Scholar]
- Holland BR, Huber KT, Moulton V, Lockhart PJ. Using consensus networks to visualize contradictory evidence for species phylogeny. Mol Biol Evol. 2004;21:1459–1461. doi: 10.1093/molbev/msh145. [DOI] [PubMed] [Google Scholar]
- Huson DH, Dezulian T, Klöpper T, Steel MA. Phylogenetic super-networks from partial trees. IEEE/ACM Trans Comput Biol Bioinform. 2004;1:151–158. doi: 10.1109/TCBB.2004.44. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Hao W, Golding GB. Patterns of bacterial gene movement. Mol Biol Evol. 2004;21:1294–1307. doi: 10.1093/molbev/msh129. [DOI] [PubMed] [Google Scholar]
- Susko E, Leigh J, Doolittle WF, Bapteste E. Visualizing and assessing phylogenetic congruence of core gene sets: a case study of the γ-proteobacteria. Mol Biol Evol. 2006;23:1019–1030. doi: 10.1093/molbev/msj113. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional data file 1
A table containing the numbers, genes, and species on which Figure 2 is based