Direct Reprogramming of Terminally Differentiated Mature B Lymphocytes To Pluripotency (original) (raw)
. Author manuscript; available in PMC: 2009 Jan 8.
Abstract
Pluripotent cells can be derived from fibroblasts by ectopic expression of defined transcription factors. A fundamental unresolved question is whether terminally differentiated cells can be reprogrammed to pluripotency. We utilized transgenic and inducible expression of four transcription factors (Oct4, Sox2, Klf4, and c-Myc) to reprogram mouse B lymphocytes. These factors were sufficient to convert non-terminally differentiated B cells to a pluripotent state. However, reprogramming of mature B cells required additional interruption with the transcriptional state maintaining B cell identity by either ectopic expression of the myeloid transcription factor CCAAT/enhancer-binding-protein-α (C/EBPα) or specific knockdown of the B cell transcription factor Pax5. Multiple iPS lines were clonally derived from both non-fully and fully differentiated B lymphocytes, which gave rise to adult chimeras with germline contribution, and to late term embryos when injected into tetraploid blastocysts. Our study provides definite proof for the direct nuclear reprogramming of terminally differentiated adult cells to pluripotency.
Introduction
Embryonic development and cellular differentiation are considered unidirectional pathways because cells undergo a progressive loss of developmental potency during cell fate specification (Gurdon, 2006). The success of somatic cell nuclear transfer (SCNT) experiments in mammalian species provided proof that the epigenetic state of adult differentiated cells is not fixed but remains pliable for reprogramming by factors present in the oocyte (Byrne et al., 2007; Jaenisch and Young, 2008; Wakayama and Yanagimachi, 2001). However, the inefficiency and ethical concerns associated with attempting to clone human somatic cells have spurred the field to search for alternative methods to achieve nuclear reprogramming (Jaenisch and Young, 2008). An important breakthrough was achieved by Yamanaka and colleagues who succeeded in directly reprogramming fibroblasts into induced pluripotent stem (iPS) cells by transduction of the four transcription factors Oct4, Sox2, Klf4 and c-Myc (Takahashi and Yamanaka, 2006). Although the initially obtained iPS cells were not normal, several groups have since advanced the direct reprogramming technique by generating iPS cells that are epigenetically and developmentally indistinguishable from embryo derived ES cells (Maherali et al., 2007; Okita et al., 2007; Wernig et al., 2007). Moreover, transgenic expression of c-Myc was found to be dispensable for reprogramming though it accelerated and enhanced the efficiency of reprogramming (Nakagawa et al., 2008; Wernig et al., 2008b). Also, the therapeutic potential of iPS cells was demonstrated in a proof of principle experiment involving transplantation and gene therapy in models of sickle cell anemia and Parkinson’s disease (Hanna et al., 2007; Wernig et al., 2008a). Finally, it has been also shown that human iPS cells can be generated by transduction of defined factors into fibroblasts (Park et al., 2008; Takahashi et al., 2007; Yu et al., 2007).
The conversion of somatic cells to a pluripotent state by SCNT or by direct in vitro reprogramming posed a number of mechanistic and technical questions. First, can terminally differentiated cells be reprogrammed to pluripotency with defined factors, or can only less differentiated cells such as somatic stem cells, undergo nuclear reprogramming to pluripotency? (Eggan et al., 2004; Hochedlinger and Jaenisch, 2002) Recently, successful reprogramming of liver cells that had activated a Cre-recombinase gene driven by a transgenic rat albumin enhancer/promoter (Postic et al., 1999), has been achieved (Aoi et al., 2008). However, as albumin gene expression marks heterogeneous cells populations in the liver in addition to hepatocytes (Matthews et al., 2004; Rountree et al., 2007), including oval cells that play an important role in liver regeneration and might serve as adult liver stem cells (Grompe, 2005; Wang et al., 2003), the question of reprogramming terminally differentiated cells remains unresolved. Moreover, it is unclear whether progressive differentiation of the donor cells affects the efficiency of in vitro reprogramming.
Development of cells along the B cell lineage allows to address these questions because sequential intrinsic genetic DNA rearrangements in the heavy and light chain immunoglobulin loci genetically mark the different consecutive stages of B cell maturation (Jung et al., 2006). Cells at the ProB stage of development initiate immunoglobulin rearrangements, a process involving the assembly of V (variable), D (diversity) and J (joining) gene segments. Assembly of the heavy chain locus (IgH) precedes that of the light chains loci (IgL) (Jung et al., 2006). In addition, the rearrangements of the IgH locus are sequential with DH to JH joining occurring on both alleles prior to VH to DHJH segment rearrangement (Papavasiliou et al., 1997). The productive assembly of VH-DHJH variable gene region indirectly signals differentiation to the next stage in which IgL chains are assembled with Igκ rearrangement generally preceding that of Igλ (Papavasiliou et al., 1997). To define the susceptibility of differentiated cells to nuclear reprogramming we used cells from this highly ordered developmental pathway that carry distinct, sequentially acquired genetic “fingerprints” that would allow accurate retrospective assessment of the developmental stage of the donor B cell nucleus that was able to generate the respective monoclonal iPS line.
In this study, we have generated iPS cells from pro and pre- B cells by transduction with the reprogramming factors Oct4, Sox2, c-Myc and Klf4 and from mature B cells by the additional over expression of C/EBPα or specific knockdown of the Pax5 transcription factor. We show that the reprogrammed cells carried the genetic rearrangements characteristic of donor non-terminally differentiated and mature terminally differentiated B cells and were able to generate adult chimeric mice and contribute to germline. Our results indicate that specific combinations of reprogramming factors can reset the genome of terminally differentiated cells to a pluripotent state.
Results
Inducible expression of reprogramming factors in the B cell lineage
Our initial strategy was to determine whether Oct4, Sox2, Klf4 and c-Myc transcription factors, which were shown to be sufficient to reprogram mouse and human fibroblast cultures (Meissner et al., 2007; Okita et al., 2007; Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007), were capable of reprogramming cells of the B cell lineage. Because of the relatively low infectivity of mouse lymphocytes with viruses, we established a system that allowed inducible transgenic expression of the four reprogramming factors in B cells. To this end, we have recently shown that doxycycline-inducible (Dox) lentiviral vectors encoding the Oct4, Sox2, c-Myc and Klf4 transcription factors are able to reprogram mouse embryonic fibroblasts (MEFs) into stable iPS cells that maintain their pluripotency after Dox withdrawal (Brambrink et al., 2008). When injected into blastocysts these cells were capable of generating postnatal chimeras which contain clonal populations of somatic cells carrying the identical proviral copies that generated the “primary” iPS cells (Wernig et al. Submitted). We reasoned that B cells derived from these chimeras, when exposed to Dox, might activate the proviral copies that induced the primary iPS cells and thus might facilitate reprogramming and the generation of “secondary” iPS cells (Figure 1A). MEFs carrying a constitutively expressed reverse tetracycline trans-activator driven by the ROSA26 promoter (R26-M2rtTA) and a knock-in of GFP into the endogenous Nanog locus (Nanog-GFP) were infected with the Dox-inducible lentiviral vectors encoding Oct4, Sox2, c-Myc and Klf4 genes (Brambrink et al., 2008). Colonies appearing after 12 days of Dox treatment (Figure 1B) were picked and propagated without Dox to establish Nanog-GFP+ iPS lines (Figure 1C and S1). The MEF derived primary iPS cells were injected into blastocysts to generate embryonic and adult chimeras (Figure 1D). Pro-B and Pre-B cells were isolated from the bone marrow and mature IgM+IgD+ B cells were purified from the spleen of 8 week old adult chimeric mice and grown in media supplemented with hematopoietic cytokines and Dox for 7 days. Chimeras derived from MEF-iPS-#1 cell line were chosen for further study as donor B cells from chimeras induced high expression levels of the 4 factors in the presence of Dox (Figure 1E).
Figure 1. Transgenic inducible expression of Oct4, Sox2, Klf4 and c-Myc in the mouse B cell lineage.
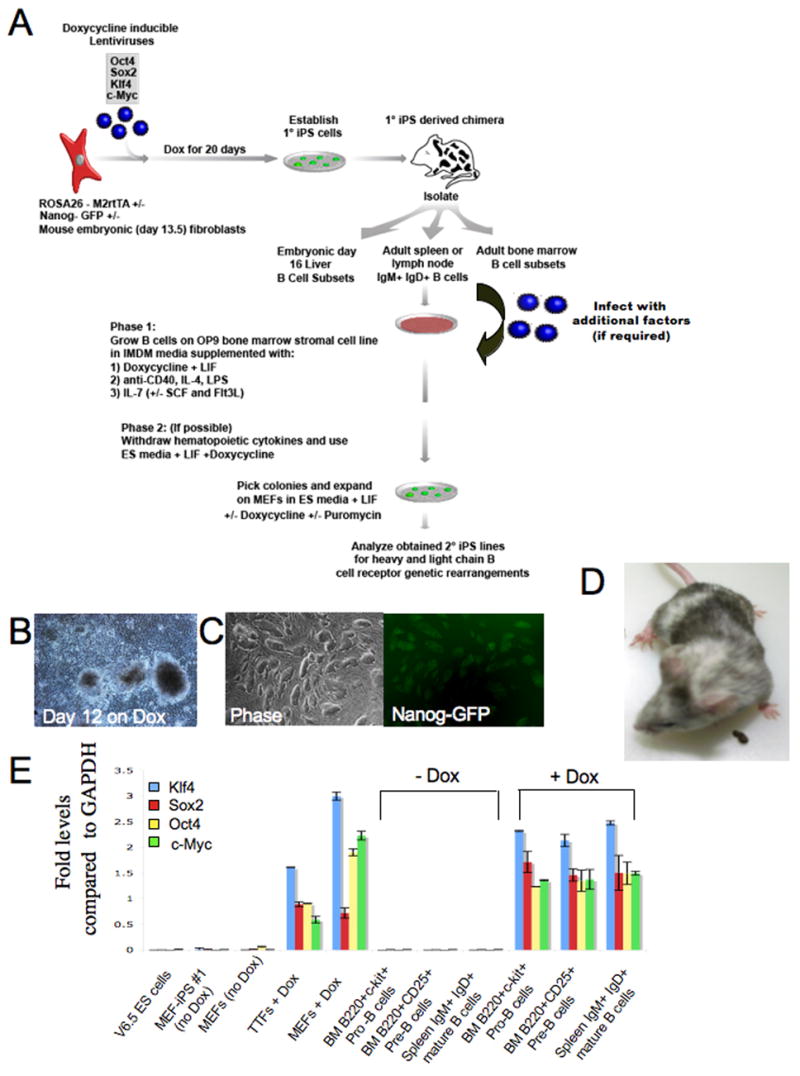
(A) Schematic drawing representing the strategy used in this study for reprogramming cells from the B cell lineage. (B) MEFs heterozygous for the ROSA26-M2rtTA and Nanog-GFP alleles were infected with Dox inducible lentiviruses encoding Oct4, Sox2, Klf4 and c-Myc. Typical morphology of colonies derived from infected MEFs cultured with Dox for 12 days. (C) ES-like morphology of Nanog-GFP+ MEF derived iPS lines (MEF-iPS#1 cell line is shown). These cell lines were injected into blastocysts to derive embryonic and adult chimeras (D). Different B cell subsets were isolated from embryonic liver or adult spleen and bone marrow tissues and used for reprogramming experiments. (E) Quantitative RT-PCR assay of viral transgene expression in MEF-iPS#1 chimera derived mouse embryonic fibroblasts (MEF) obtained at day E13.5 and 8 week old adult tail tip fibroblasts (TTF) and B cells subsets from bone marrow (BM) or spleen with (4μg/ml) or without Dox treatment for 7 days as indicated in the figure.
Reprogramming of non-terminally differentiated B cells
Initial attempts failed to reprogram bone marrow derived B cells and spleen B cells that had been cultured on irradiated feeder cells in ES media supplemented with LIF and Dox as the cells died within five days in culture (data not shown). We reasoned that addition of cytokines might be necessary to allow for an initial proliferation of the B cells that would ensure a sufficient number of cell divisions necessary to initiate epigenetic reprogramming by expression of the four factors. Therefore, we optimized culture conditions that would support immature and mature B cell growth as well as that of ES cells to ensure viability during the reprogramming process from B to iPS cells. Cells were grown on OP9 bone marrow stromal cells in media supplemented with LIF which is required for ES cell growth, with IL-7, SCF and Flt-3L which support B cell development (Milne et al., 2004), and with IL-4, anti-CD40 and LPS which are important for proliferation of mature B cells (Hayashi et al., 2005) (Figure S2). In initial experiments we detected AP positive colonies in cultures of sorted Pre- and Pro-B cell subsets derived from 8 week old adult chimera bone marrow after 14 days of Dox treatment (Figure 2A). Small flat colonies appeared 3 days after Dox induction that subsequently underwent robust expansion (Figure 2B). Around day 11 after Dox induction smooth ES-like small colonies embedded within the granulated large colonies were observed which became Nanog-GFP+ at day 14 (Figure 2B). Colonies were picked 14 days after Dox induction from 3 independent experiments and grown on MEF feeders in ES media without Dox. Within 3 passages over 90% of the picked colonies grew into homogenous ES-like Nanog-GFP+ iPS cells. In the following we will refer to these cell lines as “iB-iPS” cells (for iPS cells derived from “immature” non-fully differentiated B cells including Pre and Pro B cells)
Figure 2. Reprogramming of non-terminally differentiated B cells to pluripotency.
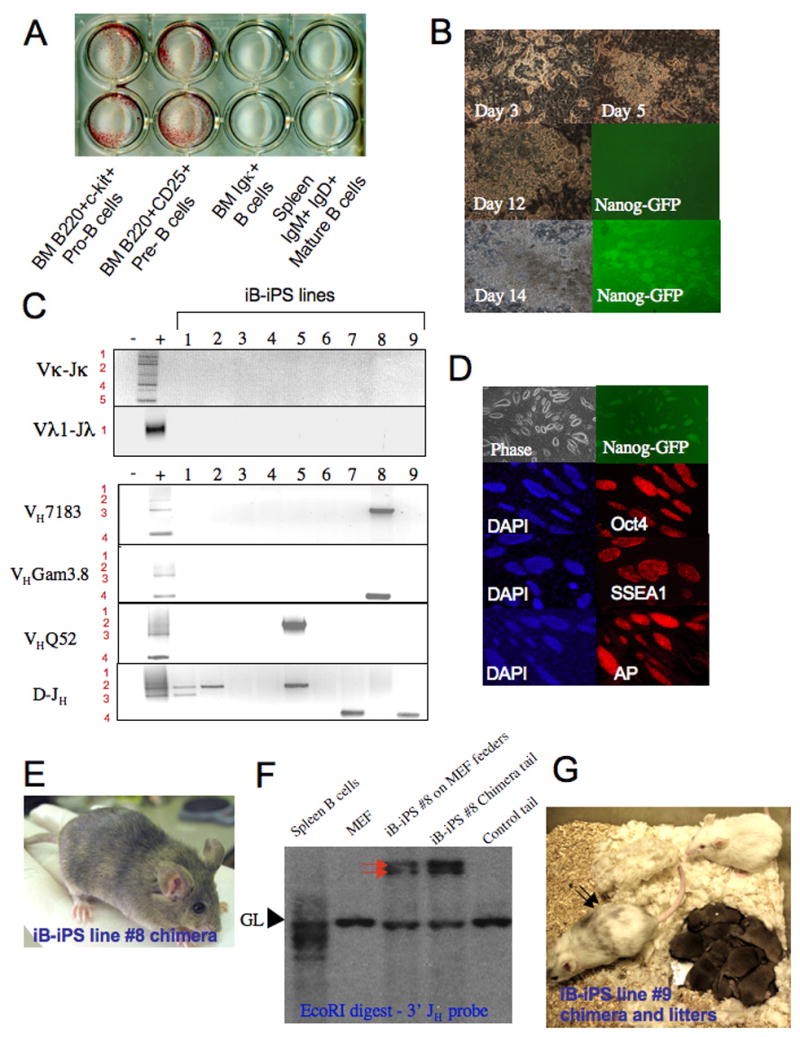
(A) 8 week old chimera derived bone marrow B cell subsets (50*10^3 cells per well) and spleen IgM+IgD+ mature B cells (250*10^3 spleen cells per well) were plated on OP9 bone marrow stroma with conditioned media and Dox. After 14 days plates were fixed and stained for AP activity. (B) Images for characteristic colonies at different time point obtained from sorted B220+CD25+ bone marrow cells. (C) PCR analysis for rearrangements in selected iB-iPS. MEFs and splenocytes were used as negative and positive controls, respectively. (D) Immunostaining for ES cell markers on a representative line iB-iPS#8. (E) A chimeric mouse from iB-iPS #8 cell line. Agouti colored hairs originate from injected iPS lines. (F) Southern blot analysis of iB-iPS line #8 grown on MEF feeders and of tail tip biopsy taken from the derived chimera. MEF and spleen B cells were used as negative and positive controls, respectively, for rearrangement detection. Red arrows indicate the two rearranged heavy chain alleles. GL indicates germline fragment representing non-rearranged configuration of the Igh locus. (G) An adult old chimeric pseudo-male mouse from iB-iPS #9 cell line was mated with Balb/C females and repeatedly achieved 100% germline transmission, as indicated by the agouti color of all litters obtained.
Genomic DNA harvested from established iB-iPS cell lines was analyzed by PCR for heavy and light chain rearrangements. Representative cell lines reprogrammed from the B220+c-Kit+ Pro-B cell subpopulation showed that some iPS lines carried DH-JH rearrangements (lines #1,2,7,9) whereas others did not show evidence for any IgH rearrangements (lines #3,4,6) (Figure 2C), as would be expected for rearrangements in the donor B cell subset at the Pro B cell stage of development. Cell lines established from the adult bone marrow derived B220+CD25+ Pre-B cells carried at least one VH-DJH rearrangement and an additional DH-JH or VH-DJH rearrangement (lines #5,8) (Figure 2C), both genetic rearrangements of the IgH locus typically observed in such B cell populations (Jung et al., 2006). Rearrangements were verified by Southern blot analysis (Figure S3). For subsequent analysis, we focused on cell lines that contained genetic evidence for IgH rearrangements, as only those can be definitively traced to cells committed to the B cell lineage. All iB-iPS cell lines stained positive for the ES markers AP, SSEA1 and Oct4 and generated differentiated teratomas (Figure 2D and S4). Furthermore, we obtained adult chimeras from several iB-iPS cell lines (Figure 2E and Table S1). Representative Southern blots of tail DNA from an iB-iPS#8 cell line derived chimera showed an identical heavy chain rearrangement pattern as the donor iB-iPS cell line (Figure 2F), thus confirming that the chimera was derived from the respective iB-iPS cell line and not from contaminating ES or MEF derived iPS cells. A chimera derived from iB-iPS line #9 produced 100% germline transmission (Figure 2G and S5). These results demonstrate that cells committed to the B cell lineage carrying DH-JH or VH-DJH rearrangements, although not fully differentiated as they lack light chain rearrangements, can be reprogrammed to a pluripotent ES-like state by the induction of the 4 transcription factors Oct4, Sox2, Klf4 and c-Myc.
Reprogramming of terminally differentiated B cells
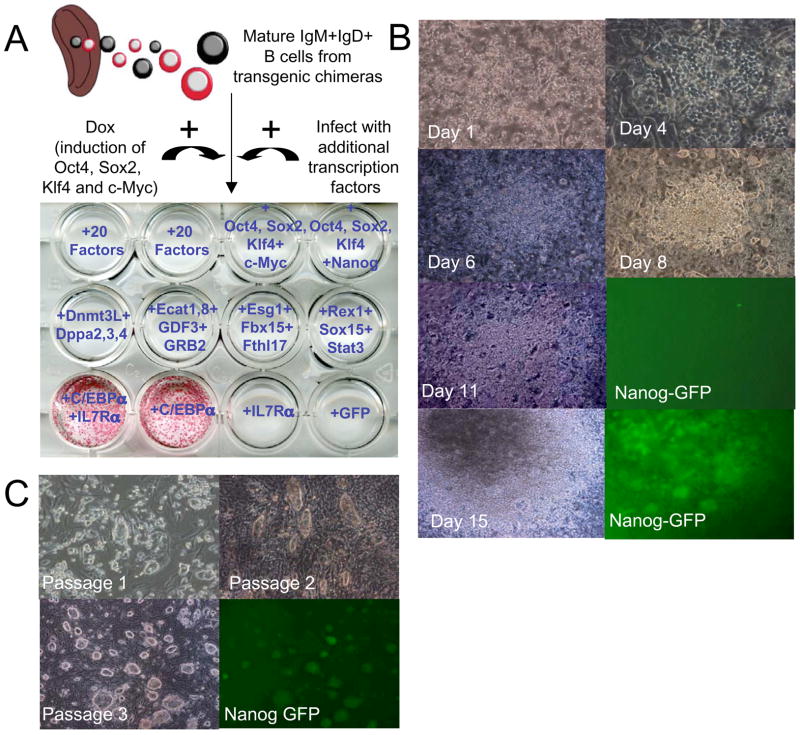
We failed to generate any reprogrammed AP+ colonies from mature spleen B cells or bone marrow derived Igκ+ cells in 3 independent experiments (Figure 2A). This was puzzling given that IgM+IgD+ mature transgenic B cells could be maintained in our culture conditions for up to 6 weeks and continued to express B cell markers (Figure S2). We tested, therefore, the hypothesis that additional pluripotency factors might be needed to achieve reprogramming of mature B cells. Dox induced IgM+IgD+ spleen B cells obtained from adult transgenic chimeras were infected with combinations of retroviruses encoding 20 different pluripotency factors, that were originally generated to screen for fibroblast reprogramming (Takahashi and Yamanaka, 2006). Yet, these experiments repeatedly yielded negative results (Figure 3A).
Figure 3. Reprogramming of terminally differentiated mature B cells to pluripotency.
IgM+IgD+ cells were isolated from spleen of a 10 week old chimera, infected for 24 hours with viral supernatants of retroviruses encoding different combinations of transcription factors and were transferred afterwards to OP9 coated wells and grown in conditioned media in the presence of Dox. “20 factors” indicate pMXs based constructs previously used to reprogram mouse fibroblasts as indicated in the Experimental Procedures section. After 14 days of Dox induction and additional viral transduction as indicated for each well, cells were stained for AP. One out of 3 independent experiments using different combinations of factors is shown. (B) Representative images of colonies at different time points after infection with C/EBPα retrovirus and Dox induction of chimeric spleen derived IgM+IgD+ mature B cell cultures. (C) Representative pictures of changes in cell morphology after passaging picked colonies at day 14 from IgM+IgD+ B cell cultures infected with C/EBPα and treated with Dox.
As an alternative approach, we aimed to “sensitize” the B cells to respond to Dox dependent 4-factor induction by altering their mature B cell identity. It has been shown that over-expression of the myeloid transcription factor CCAAT/enhancer-binding protein-α (C/EBPα) is able to reprogram B cells into macrophage-like cells (Xie et al., 2004) by disrupting the function of Pax5, a transcription factor that is a master regulator of mature B cell development and function (Cobaleda et al., 2007b). We tested, therefore, whether transduction with C/EBPα would facilitate reprogramming of mature B cells. Adult spleen B cells derived from 10 week-old chimeras were transduced with a retrovirus encoding C/EBPα and/or the IL7-Rα subunit and cultured on OP9 cells in the presence of Dox to induce the four factors Oct4, Sox2, Klf4 and c-Myc. Figure 3A shows that AP positive colonies appeared after 14 days in culture in cells transduced with C/EBPα or with C/EBPα and IL7-Rα but not in cells transduced with IL7-Rα alone. Infection of transgenic B cells with C/EBPα did not result in altering the induction levels of transgenic factors upon treatment with Dox (Figure S6). After 3 days of growth on OP9, small adherent colonies were formed which continued to grow into denser granulated colonies (Figure 3B). Small round ES-like colonies appeared within the large dense granulated colonies, and Nanog-GFP+ foci were readily detected at approximately day 14 (Figure 3B and S7). Colonies isolated at day 14 were passaged on MEF feeder cells without hematopoietic cytokines or Dox and within 3 passages all lines assumed an ES-like morphology and were positive for the Nanog-GFP marker (Figure 3C). Our results suggest that transduction with C/EBPα can sensitize mature B cells to respond to the expression of Oct4, Sox2, c-Myc and Klf4 and re-express pluripotency markers. We established 120 iPS lines that were picked from independent tissue culture wells containing IgM+IgD+ B cells from adult spleen and lymph nodes at 14 days after Dox addition and C/EBPα transduction, and 9 cell lines were selected for in depth characterization (Lines 1–6 obtained from adult spleen and 7–9 from adult lymph nodes). In the following we will refer to these cell lines as “B-iPS” cells (iPS cells derived from mature “B” cells)
Next, we characterized marker expression, DNA methylation and histone marks of the B-iPS cell lines. Immunoflorescence staining showed that all B-iPS cell lines uniformly expressed ES cell markers AP, SSEA1 antigen, Oct4 and were positive for Nanog-GFP (Figure 4A). Gene expression analysis showed that B-iPS and ES cells, but not primary B cells, expressed comparable levels of Nanog, Ecat1, Rex1, Zfp296 and GDF3 genes (Figure 4B). Bisulphite sequencing was performed to determine the methylation status of Oct4 and Nanog gene promoters for iB-iPS and B-iPS cell lines. As expected, fibroblast and B cell control samples displayed extensive methylation at both promoters, whereas B-iPS and iB-iPS lines showed widespread demethylation of these regions similar to that seen in ES cells (Figure 4C). To assess the chromatin state of the cells, chromatin immunopercipitation (ChIP) and real time PCR were performed to quantify ‘active’ histone H3 lysine 4 trimethylation (H3K4me3) and ‘repressive’ histone H3 lysine 27 trimethylation (H3K27me3) methylation marks on a selected set of genes known to be bivalent in ES cells (Bernstein et al., 2006). The promoter region for the B cell transcription factor gene Pax5 displayed strong enrichment for H3K4me3 methylation in the donor mature B cells, whereas H3K27me3 methylation predominated at the silent genes Zfpm2 and Irx2 (Figure 4D). Conversely, all these genes carry equivalent enrichment for both histone modifications in B-iPS and ES cells, consistent with the notion that these bivalent domains were re-established during reprogramming (Figure 4D).
Figure 4. Marker expression and promoter methylation analysis of B-iPS lines.
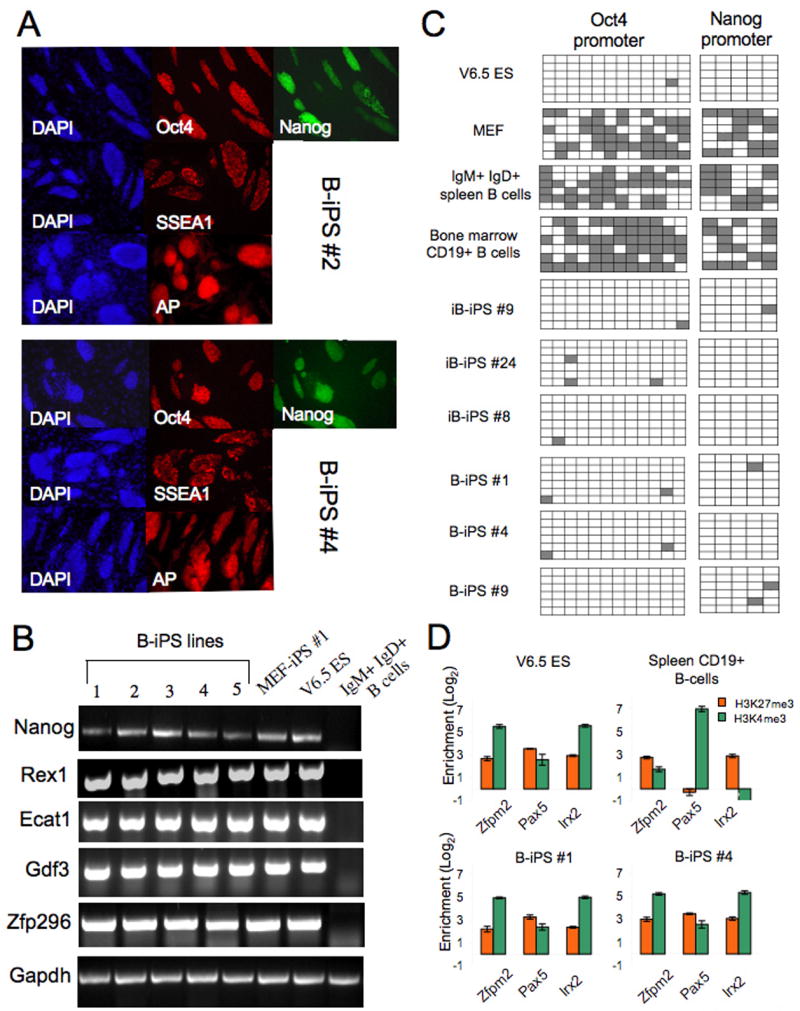
(A) Immunoflorescence staining for ES pluripotency markers Oct4, Nanog (signal obtained from GFP cassette knocked in the endogenous Nanog locus), SSEA1 and AP. (B) RT-PCR analysis of selected ES marker genes in B cells, ES cells, MEF-iPS cells and different iB-iPS and B-iPS lines. (C) Analysis of the methylation state of the Oct4 and Nanog promoters using bisulphate sequencing. Open squares indicate unmethylated and filled squares methylated CpG dinucleotides. (D) Real-time PCR after chromatin immunoprecipitation using antibodies against H3K4me3 and H3K27me3. Shown are the Log2 enrichments for several previously reported “bivalent” loci in ES cells.
Rearrangements of immunoglobulin loci in B-iPS cells confirm mature B Cell identity of the donor cells
In order to characterize the genomic rearrangements of the Ig loci in the B-iPS cells, genomic DNA from MEF depleted iPS cell lines grown on gelatin was analyzed for IgH, Igκ and Igλ rearrangements by complementary approaches that included Southern blotting, PCR and sequencing of individual PCR fragments (Alt et al., 1981; Chang et al., 1992; Cobaleda et al., 2007a; Lewis et al., 1982; Schlissel et al., 1991) (Table S2). All cell lines contained 2 heavy chain rearrangements: one was a productive in-frame V-DJ rearrangement whereas the other was either a frozen D-J rearrangement or a non-productive V-DJ rearrangement (Figure 5 A–C (lanes 1–9) and D)). These results are consistent with the well established observation that adult mature B cells in the periphery have 2 rearranged heavy chain loci (Jung et al., 2006). As predicted for mature B cells, the light chain loci had one productive in frame Igκ or Igλ light chain rearrangement (Figure 5A–C (lanes 1–9) and D–E) (Jung et al., 2006). B-iPS#9 was derived from a minor B cell subpopulation with a rearranged productive Igλ chain and kappa locus that was retained in the germline configuration (Figure 5C and E, lane 9) (Nadel et al., 1990; Oberdoerffer et al., 2003). Finally, sequences obtained from heavy and light chain rearrangements from B-iPS cell line #4 provided conclusive evidence that the donor B cell nucleus that yielded this cell line had undergone somatic hypermutation (Figure 5F), a process that occurs after antigen encounter in vivo and involves acquiring a high rate of somatic mutations at “hotspots” located throughout the DNA encoding the immunoglobulin variable region (Teng and Papavasiliou, 2007). The abundance of mostly non-silent mutations in the variable region of the productive rearrangements in this cell line (Figure 5F) shows that non-naïve B cells that have already encountered antigen in vivo are also amenable to direct reprogramming. Finally, the C/EBPα viral transgene was detected in genomic DNA from all B-iPS cell lines analyzed (Figure 5G).
Figure 5. Productive heavy and light chain rearrangements in B-iPS lines.
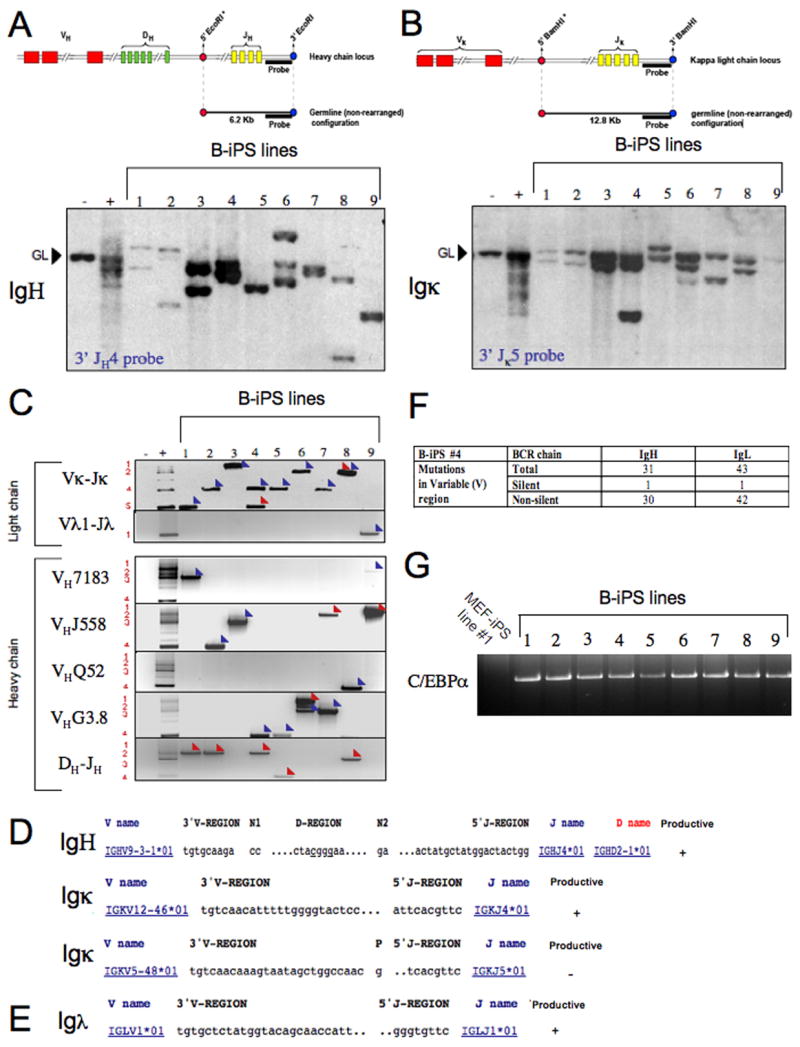
(A) Genomic DNA from B-iPS lines grown on gelatin was digested with EcoRI and analyzed for V(D)J rearrangements at the Igh locus by Southern blotting using a 3′ JH4 probe. (B) Vκ-Jκrearrangements at the Igκ locus were determined by Southern blot analysis of BamHI digested genomic DNA using a 3′Jκ5 probe. (A–B) Any rearrangement occurring in these loci abolishes the asterik marked restriction sites of the enzyme used. After rearrangements, the size of the genomic fragment bound by the probe depends on the next available 5′ restriction site in relation to the abolished asterik marked sites. MEFs served as negative control and CD19+ splenocytes were used as positive controls. GL denotes the position of germline DNA fragment. (C) PCR analysis of Vκ-Jκ and Vλ1-Jλ light chain rearrangements and of VHQ52-DJH, VH7183-DJH, VHGam3.8-DJH VHJ558-DJH and D-JH rearrangements in selected B-iPS. Rearrangements to different J segments are numbered on the left side of the panels. Detailed characterization of the different immunoglobulin gene rearrangements by cloning and sequencing was performed to confirm the presence of productive heavy and light chain rearrangements in the cell lines obtained. Blue arrows indicate “productive rearrangements” and red arrows indicate” non-productive rearrangements”. (D) Sequences from rearranged IgH and Igκ chains from B-iPS line #4 obtained by RT-PCR (Panel C, lane 4). V, D and J segments involved in IgH rearrangement are indicated, as well as V and J segments from the Igκ locus. (E) Sequencing of the Igλrearrangement (Panel C, lane 9) cloned from B-iPS line #9. (F) Summary for non-silent and silent mutations found in predicted variable region of the encoded B cell receptor as detected by sequencing productive heavy and light chain rearrangements of B-iPS line #4. (G) Transgene specific primers were designed and used to detect the presence of C/EBPα provirus in B-iPS lines.
Developmental potential of B-iPS cells
As an initial test for developmental potency we injected 8 B-iPS cell lines subcutaneously into immunodeficient (SCID) mice. Six weeks after injection, macroscopic teratomas were observed in all injected mice. Histological examination showed that the teratomas contained cell types representing all three embryonic germ layers (Figure 6A). To assess more stringently their developmental potential, individual B-iPS cell lines were injected into diploid (2N) blastocysts resulting in the generation of viable, high-contribution chimaeras from all 4 B-iPS cell lines tested (Figure 6B and Table S1). Southern blot analysis of genomic DNA isolated from B-iPS #4 and #1 derived chimeras revealed the presence of genomic fragments corresponding to rearranged Igκ alleles identical to those observed in the donor injected B-iPS cell lines (Figure 6C and S8, respectively). Importantly, B-iPS#1 cell line contributed to the germline as was evident by the derivation of offsprings carrying a constitutively expressed lentiviral transgene EGFP vector that was used for transducing B-iPS#1 cells prior to blastocyst injections (Figure 6D). Southern blot analysis confirmed segregation of the rearranged Igκ allele found in the donor B-iPS line in the GFP+ derived mice (Figure S9).
Figure 6. Developmental potential of B-iPS cell lines.
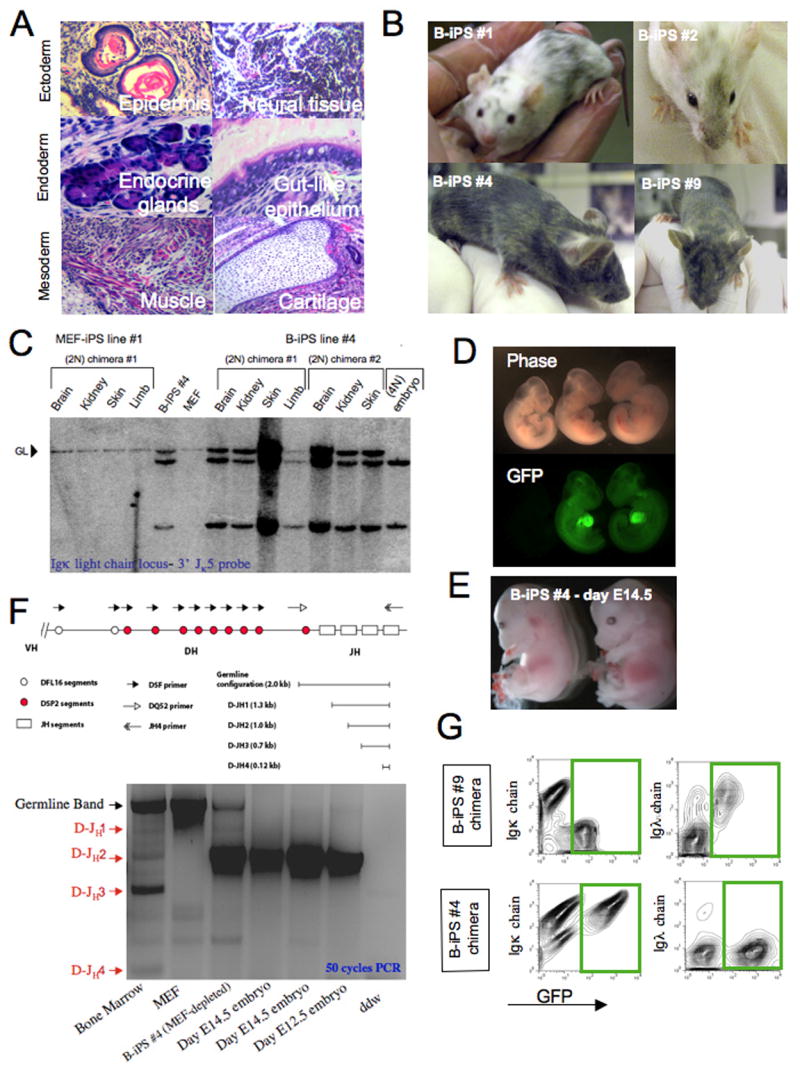
(A) Hematoxylin and eosin staining of a representative teratoma derived from B-iPS line #4. (B) Adult chimeric mice obtained from B-iPS lines in 2N Balb/C or B6D2F1 host blastocysts. (C) Southern blot analysis for detection of IgK light chain rearrangements on BamH1 digested genomic DNA (as detailed in Figure 5A) from different tissues obtained from MEF-iPS #1 derived chimeras (used as negative control) and B-iPS #4 cell line. (D) B-iPS#1 line was transduced with lentiviral ubiquitin-EGFP vector before injection in 2N blastocysts. Germline transmission was achieved from one of the chimeras, as we obtained a number of GFP+ embryos, demonstrating the transmission of constitutively expressed EGFP transgene. (E) “All B-iPS cell embryos” were generated by injection of B-iPS cell into 4N blastocysts. Live E14.5 embryos generated from B-iPS#4 cell lines. (F) Highly sensitive 50 cycle PCR analysis on 4N embryos obtained from B-iPS#4 for detection of loss of the amplicon representing germline line configuration of IgH locus in order to prove that the embryos developed entirely from iPS cells. Equimolar mix of 5′ primers DQ52 (recognizes downstream region that is always excised upon any recombination of the locus) and DSF (degenerate primer recognizing D cassettes) with 3′ JH-4 primer. This PCR could yield 4 rearrangement bands (D-JH1,2,3,4) and a germline 2 Kb band as indicated in the locus scheme. (G) Immunological analysis for detection of Igκ and Igλ surface antigen on peripheral blood B cells of chimeras obtained from constitutively GFP labeled B-iPS #4 and #9 (marked with lentiviral ubiquitin-EGFP vector before injection). Presented panels were gated on CD19+ cells and show Igκ and Igλ expression on iPS derived B cell (GFP+) and host B cells (GFP−). Green rectangles highlight B-iPS derived of GFP+ B cells.
The generation of mice by tetraploid complementation, which involves injection of pluripotent cells in 4N host blastocysts, represents the most rigorous test for developmental potency because the resulting embryos are derived only from injected donor cells (Eggan et al., 2001). Both B-iPS lines tested (#4 and #9) were able to generate mid- and late-gestation ‘all B-iPS embryos’ after injection into 4N blastocysts (Figure 6E and S10). Sensitive PCR analysis for the detection of a 2Kb germ-line region from the B cell receptor heavy chain locus that is lost upon initiation of genetic rearrangement (Chang et al., 1992) shows that genomic DNA from B-iPS #4 cell line embryos derived by tetraploid complementation had lost the germ line band and carried only the D–J rearrangement as predicted from the repertoire in the donor nucleus (Figure 6F, compare to Figure 5C, D–J rearrangement Lane 4). This conclusion was confirmed by Southern blot analysis of genomic DNA from a day E14.5 tetraploid embryo obtained from B-iPS#4 demonstrating two rearranged Igκ locus alleles, without any evidence for a germline allele (Figure 6C).
We next tested the ability of reprogrammed mature B cells to generate monoclonal B cells in vivo as a result of the restrictions imposed by their pre-rearranged IgH and IgL loci (Hochedlinger and Jaenisch, 2002; Inoue et al., 2005; Oberdoerffer et al., 2003). To facilitate the isolation of B-iPS derived cells in chimeric mice, B-iPS lines # 4 and 9 were labeled with the GFP marker by lentiviral vector mediated transduction prior to blastocyst injection. Surface expression of Igκ and Igλ light chain proteins expressed on CD19+ cells purified from peripheral blood was evaluated by FACS staining. All GFP+ B cells in B-iPS #4 derived chimeras expressed Igκ chain, but not Igλ protein (Figure 6G), consistent with the genetic analysis that showed a functional Igκ light chain rearrangement in this cell line (Figure 5 B–C, Lane 4). In contrast, B-iPS #9 cell line derived B cells carried only a functional Igλ light chain rearrangement (Figure 6G), consistent with the analysis in Figure 5C (Lane 9). Moreover, we established pluripotent B-iPS lines from mature adult B cells by specific knockdown of Pax5 transcription factor in B cells and subsequent Dox induction of the transgenic B cells obtained from iPS derived chimeras (Figure S11). Finally, two B-iPS cell lines that were generated by direct infection of genetically unmodified mature B cells with the Oct4, Klf4, Sox2, c-Myc and C/EBPα (Figure S12). In summary, our results provide unequivocal molecular and functional proof that mature B cell donor nuclei that contain functional light and heavy chain rearrangements were reprogrammed to pluripotency.
Efficiency of reprogramming mature adult B cells to pluripotency
To estimate the efficiency of reprogramming of mature adult B cells to pluripotency, a large starting pool of CD19+ B cells isolated from the spleen of adult chimeras was infected with a C/EBPα encoding retrovirus carrying a neomycin resistance gene. After 24 hours, IgM+IgD+ B cells were plated as single cells in 96 well plates on OP9 stromal cells in cytokine conditioned medium in the presence of Dox and LIF. Five days after plating puromycin and neomycin were added to the culture medium in order to select for transgenic B cells that had also been infected with C/EBPα (Figure 7A). At day 20, wells that showed cell growth were screened by FACS for detection of Nanog-GFP+ cells (Figure S13). Wells that scored positive were expanded and Nanog-GFP iPS cells appeared within 3 passages. PCR analysis of B-iPS lines obtained confirmed that all cell lines obtained from two independent experiments originated from C/EBPα infected mature B cells that had distinct B cell receptor rearrangements (Figure 7B). Based on these data, we were able to calculate the efficiency of reprogramming by dividing the number of GFP+ wells obtained (output) by the number of C/EBPα infected transgenic B cell containing wells (puromycin and neomycin double resistant wells = input). This calculation suggested that the relative efficiency for direct reprogramming of mature B cells was approximately 1 in 30 cells (Figure 7C). We attribute the relatively high efficiency of reprogramming, compared to previously reported reprogramming efficiencies between 0.001% to 0.5% (Meissner et al., 2007; Wernig et al., 2007), to the strong Dox-mediated induction of 4 out of the 5 ectopically expressed factors that did not rely on retroviral vector infection and random proviral integrations.
Figure 7. Reprogramming efficiency of adult mature B cells into iPS cells.
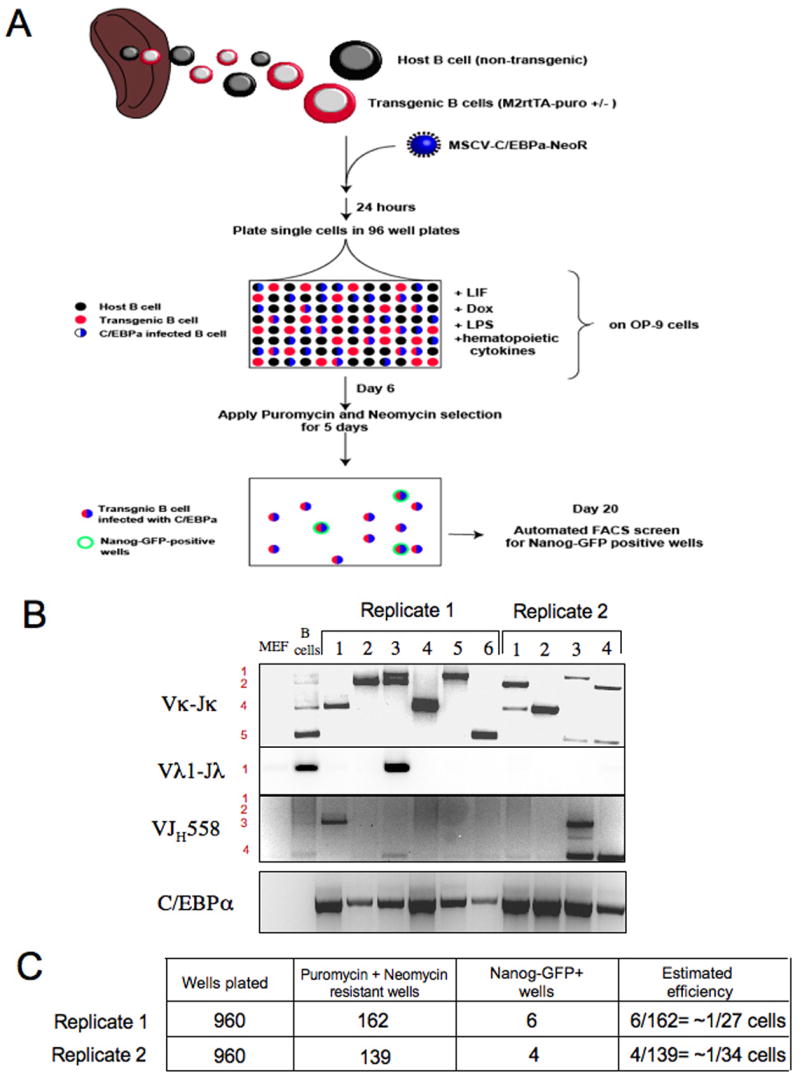
(A) Schematic representation for experiment attempting to measure reprogramming efficiency. 3*10^6 CD19+ adult B cells were infected with retrovirus encoding C/EBPα-NeoR construct and after 24 hours we sorted IgM+IgD+ mature adult B cells and plated them as single cells in 96 wells plates preplated with OP9 stromal cell line. Cells were grown in conditioned medium + Dox + LIF throughout the experiment. On day 6, culture wells were subjected to puromycin and neomycin selections for 5 days, which allowed only the growth of transgenic B cells infected with C/EBPα. On day 20, the wells containing drug resistant cells were screened for Nanog-GFP expression by FACS analysis. Wells that scored positive were subsequently passaged on MEFs in ES media and grown into iPS cell lines. (B) Established cell lines from each experiment were retrospectively confirmed to carry different B cell receptor rearrangements and to contain C/EBPα pro-viral transgenes. (C) Efficiency was determined by dividing the number of GFP+ wells by the number of wells that contained neomycin and puromycin resistant cells.
Discussion
Nuclear reprogramming, which pertains to the concept of rewiring the epigenetic state of a somatic nucleus to that of another cell type, can be achieved by different approaches. The most recently established strategy to reprogram somatic cells to pluripotency involves direct ectopic expression of defined transcription factors in somatic cells (Okita et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007). This enforced factor expression appears to initiate a sequence of stochastic events occurring over a relatively extended time period in culture that eventually leads to generation of a small fraction of cells that have acquired a stable pluripotent state (Jaenisch and Young, 2008). The transduced factors are required for an initial period of time in the reprogramming process (Brambrink et al., 2008; Stadtfeld et al., 2008) during which they may interact with endogenous pluripotency genes (Boyer et al., 2005; Loh et al., 2006) and gradually induce epigenetic changes that subsequently sustain a stable epigenetic state that is indistinguishable from that of inner cell mass derived ES cells. During this process, the de novo methyltransferases Dnmt3a and Dnmt3b also become activated and in turn methylate and silence the virally transduced factors, a process that is crucial for subsequent differentiation of the iPS cells (Brambrink et al., 2008).
Our data provide conclusive evidence that terminally differentiated mature B cells obtained from adult mice can be directly reprogrammed into ES-like cells in vitro. The donor B cell population that eventually underwent successful reprogramming had completed a complex differentiation pathway involving epigenetic and genetic changes: an initial commitment to the hematopoietic and subsequently to the B cell lineage; acquisition of productive heavy and light chain rearrangements; egression from the bone marrow to repopulate peripheral lymphoid organs in adult mice, and as observed in one of the cell lines obtained, acquisition of somatic hypermutations in variable region of B cell receptor genes in response to antigen stimuli.
Importantly, our results demonstrate that under similar induction levels of Oct4, Sox2, Klf4 and Sox2 transgenes in the B cell lineage, non-terminally differentiated and terminally differentiated B cells respond differently to these factors. We observed that robust reprogramming of fully differentiated mature B lymphocytes to pluripotency was achieved following interruption with the active transcriptional state in mature B cells. This was obtained either by overexpressing C/EBPα transcription factor, which normally plays a role in granulocyte cell fate specification (Ramji and Foka, 2002), or by knockdown of Pax5, a key transcription factor in B cells. Graf and colleagues (Xie et al., 2004) have shown that overexpression of C/EBPα converted B cells into macrophage-like cells by downregulating B cell markers through inhibition of Pax5 functions and facilitating extinction of the early B cell regulators, EBF1 and E2A transcription factors. In addition, C/EBPα induced up-regulation of components of a myeloid transcriptional network (Laiosa et al., 2006; Xie et al., 2004). Busslinger and colleagues have shown that B cells display myeloid promiscuity of gene expression following deletion of Pax5 in B lymphocytes, and that such cells can be converted into macrophage like cells when grown in the presence of myeloid cytokines (Delogu et al., 2006; Mikkola et al., 2002). These observations are relevant for understanding the mechanisms of reprogramming. We consider the following mutually non-exclusive possibilities:
- Pax5 deletion has been shown to cause loss of B cell lineage commitment and de-differentiation (Cobaleda et al., 2007a; Mikkola et al., 2002). C/EBPα may cross-antagonize key regulators of the B cell transcriptional network that maintain the mature B cell identity. This may facilitate the dedifferentiation of B cells to a less differentiated state allowing Oct4, Sox2, Klf4 and c-Myc transgene induced reprogramming. This explanation is consistent with observations that the differentiation state of the donor cells is known to influence the efficiency of reprogramming by nuclear transplantation, as neural and keratinocyte stem cells were more efficiently reprogrammed than other more differentiated cells obtained from the same lineage (Blelloch et al., 2006; Li et al., 2007).
- C/EBPα may convert mature B cells into macrophage-like cells (Xie et al., 2004) which have a different epigenetic state that possibly allows enhanced accessibility to target genes of Oct4, Sox2, Klf4, c-Myc that would facilitate the efficient induction of the endogenous auto-regulatory loop governing the pluripotent state (Boyer et al., 2005; Loh et al., 2006). Similarly, loss of Pax5 expression results in loss of inhibition of key myeloid transcription factors, and can result in reprogramming B cells into macrophage like cells (Delogu et al., 2006; Mikkola et al., 2002).
- C/EBPα over expression or Pax5 elimination may enable mature B cells to transition from a state of growing in suspension to become adherent cells in the presence of OP9 cells, which might be a rate-limiting event in their reprogramming.
- Finally, our results do not rule out the possibility that Oct4, Sox2, Klf4 or c-Myc can be sufficient at very low efficiency to reprogram mature B cells and that it is feasible that different combinations of factors may be able to reprogram B lymphocytes under different culture conditions.
The results presented in this study demonstrate that terminally differentiated cells can be directly reprogrammed to pluripotency by defined factors. At present, somatic cells have been converted to cells of other germ line lineages only by first reprogramming the cells to a pluripotent embryonic state and then by differentiation to the different cell type. Whether forced expression of other factors could “trans-differentiate” terminally differentiated cells to another cell type of a different lineage (e.g. mesoderm to endoderm or ectoderm) without first passing through a pluripotent cell stage is presently not known.
Experimental Procedures
Cell culture and viral infections
ES and established iPS cells were cultured on irradiated MEFs in DME containing 15% FCS, leukemia inhibiting factor (LIF), penicillin/streptomycin, L-glutamine, beta-mercaptoethanol and nonessential amino acids. MEFs used to derived primary iPS lines by infections with inducible lentiviruses were harvested from F1 matings between ROSA26-M2rtTA mice (Beard et al., 2006) and Nanog-GFP mice (Brambrink et al., 2008). Mouse C/EBPα cDNA was cloned into EcoRI cloning site of pLib, MSCV-Neo and pMig retroviral vectors. pMXs vectors encoding ES pluripotency genes were previously described (Takahashi and Yamanaka, 2006). Lentiviral preparation and infection with Dox inducible lentiviruses encoding Oct4, Klf4, c-Myc and Sox2 cDNA driven by the TetO/CMV promoter, were previously described (Brambrink et al., 2008). Retrovirus stocks were prepared by transient transfection of Phoenix-Eco cells using Fugene (Roche), and supernatants were harvested 48 hr later. For infection, purified B cell subsets were resuspended in IMDM with 15% FCS as well as IL-4, IL-7, Flt-3L, SCF (10ng/ml each, Peprotech), anti-CD40 (0.1μg/ml, BD-Biosciences), LPS (10ng/ml, Sigma-Aldrich) and Dox (4μg/ml). Then, 2 ml aliquots were plated onto a 24-well plate precoated with retronectin (Takara) followed by 2 ml of retrovirus supernatant to which polybrene (Sigma) was added (8 μg/ml). The plates were incubated at 37°C for 2 hours, and afterwards 1ml of viral supernatant was replaced with B cells resuspended in the cytokine conditioned media described above. Plates were centrifuged for 90 min at 900RPM and then incubated 24 hours. Infected cells were then transferred onto OP9 bone marrow stromal cells line (ATCC) in fresh cytokine and Dox supplemented media. After 14 days on Dox, colonies were picked and cultured on MEF feeder cells in ES media (without hematopoietic cytokines or Dox) and in the presence of puromycin (2 μg/ml) to eliminate any remaining OP9 cells.
V(D)J rearrangement analysis
IgH, Igκ and Igλ rearrangements were amplified by PCR using degenerate primer sets as previously described (Chang et al., 1992; Cobaleda et al., 2007a; Schlissel et al., 1991) (Table S2). To characterize individual V-DJ rearrangements, the PCR fragments were cloned in TOPO vector, and at least 5 clones corresponding to the same PCR fragment were sequenced. Sequences were analyzed with DNAPLOT search engine (http://www.dnaplot.de). V-DJ and D-J rearrangements at the Igh locus were detected by Southern blot analysis on genomic DNA of the indicated iPS lines digested with EcoRI and using a 3′JH4 probe (1.6-kb HindIII-EcoRI fragment of plasmid JH4.3) (Alt et al., 1981). Vκ-Jκ rearrangements at the Igk locus were determined by Southern blot analysis of BamHI-digested genomic DNA using a 3′Jκ5 probe (1-kb XbaI-EcoRV fragment of plasmid pBS-JκMAR) (Lewis et al., 1982).
Supplementary Material
01
Acknowledgments
We would like to thank Y. Bergman, L. Boyer and members of the Jaenisch lab for assistance. R.J. is supported by NIH grants: 5-RO1-HDO45022, 5-R37-CA084198, and 5-RO1-CA087869. J.C. is a Howard Hughes Gilliam Fellow. J.H. is a Novartis Fellow by the Helen Hay Whitney Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alt F, Rosenberg N, Lewis S, Thomas E, Baltimore D. Organization and reorganization of immunoglobulin genes in A-MULV-transformed cells: rearrangement of heavy but not light chain genes. Cell. 1981;27:381–390. doi: 10.1016/0092-8674(81)90421-9. [DOI] [PubMed] [Google Scholar]
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of Pluripotent Stem Cells from Adult Mouse Liver and Stomach Cells. Science (Science Express) 2008 doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blelloch R, Wang Z, Meissner A, Pollard S, Smith A, Jaenisch R. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem cells (Dayton, Ohio) 2006;24:2007–2013. doi: 10.1634/stemcells.2006-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T, Ruth Foreman G, Grant Welstead, Christopher J, Lengner Marius Wernig, Heikyung Suh, Rudolf Jaenisch. Sequential Expression of Pluripotency Markers during Direct Reprogramming of Mouse Somatic Cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne JA, Pedersen DA, Clepper LL, Nelson M, Sanger WG, Gokhale S, Wolf DP, Mitalipov SM. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- Chang Y, Paige CJ, Wu GE. Enumeration and characterization of DJH structures in mouse fetal liver. The EMBO journal. 1992;11:1891–1899. doi: 10.1002/j.1460-2075.1992.tb05241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007a;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nature immunology. 2007b;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- Delogu A, Schebesta A, Sun Q, Aschenbrenner K, Perlot T, Busslinger M. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24:269–281. doi: 10.1016/j.immuni.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Eggan K, Akutsu H, Loring J, Jackson-Grusby L, Klemm M, Rideout WM, 3rd, Yanagimachi R, Jaenisch R. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggan K, Baldwin K, Tackett M, Osborne J, Gogos J, Chess A, Axel R, Jaenisch R. Mice cloned from olfactory sensory neurons. Nature. 2004;428:44–49. doi: 10.1038/nature02375. [DOI] [PubMed] [Google Scholar]
- Grompe M. The origin of hepatocytes. Gastroenterology. 2005;128:2158–2160. doi: 10.1053/j.gastro.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. From nuclear transfer to nuclear reprogramming: the reversal of cell differentiation. Annual review of cell and developmental biology. 2006;22:1–22. doi: 10.1146/annurev.cellbio.22.090805.140144. [DOI] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Hayashi EA, Akira S, Nobrega A. Role of TLR in B cell development: signaling through TLR4 promotes B cell maturation and is inhibited by TLR2. J Immunol. 2005;174:6639–6647. doi: 10.4049/jimmunol.174.11.6639. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature. 2002;415:1035–1038. doi: 10.1038/nature718. [DOI] [PubMed] [Google Scholar]
- Inoue K, Wakao H, Ogonuki N, Miki H, Seino K, Nambu-Wakao R, Noda S, Miyoshi H, Koseki H, Taniguchi M, et al. Generation of cloned mice by direct nuclear transfer from natural killer T cells. Curr Biol. 2005;15:1114–1118. doi: 10.1016/j.cub.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young Stem Cells, the Molecular Circuitry of Pluripotency and Nuclear Reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annual review of immunology. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Lewis S, Rosenberg N, Alt F, Baltimore D. Continuing kappa-gene rearrangement in a cell line transformed by Abelson murine leukemia virus. Cell. 1982;30:807–816. doi: 10.1016/0092-8674(82)90285-9. [DOI] [PubMed] [Google Scholar]
- Li J, Greco V, Guasch G, Fuchs E, Mombaerts P. Mice cloned from skin cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2738–2743. doi: 10.1073/pnas.0611358104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature genetics. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Maherali N, et al. Directly Reprogrammed Fibroblasts Show Global Epigenetic Remodeling and Widespread Tissue Contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Matthews VB, Rose-John S, Yeoh GC. Genetic manipulations utilizing albumin and alpha-fetoprotein promoter/enhancers affect both hepatocytes and oval cells. Hepatology. 2004;40:759. doi: 10.1002/hep.20396. [DOI] [PubMed] [Google Scholar]
- Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nature biotechnology. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- Mikkola I, Heavey B, Horcher M, Busslinger M. Reversion of B cell commitment upon loss of Pax5 expression. Science. 2002;297:110–113. doi: 10.1126/science.1067518. [DOI] [PubMed] [Google Scholar]
- Milne CD, Fleming HE, Paige CJ. IL-7 does not prevent pro-B/pre-B cell maturation to the immature/sIgM(+) stage. European journal of immunology. 2004;34:2647–2655. doi: 10.1002/eji.200425400. [DOI] [PubMed] [Google Scholar]
- Nadel B, Cazenave PA, Sanchez P. Murine lambda gene rearrangements: the stochastic model prevails over the ordered model. The EMBO journal. 1990;9:435–440. doi: 10.1002/j.1460-2075.1990.tb08128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature biotechnology. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Novobrantseva TI, Rajewsky K. Expression of a targeted lambda 1 light chain gene is developmentally regulated and independent of Ig kappa rearrangements. The Journal of experimental medicine. 2003;197:1165–1172. doi: 10.1084/jem.20030402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, Lindner J, Cherrington AD, Magnuson MA. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. The Journal of biological chemistry. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. The Biochemical journal. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree CB, Barsky L, Ge S, Zhu J, Senadheera S, Crooks GM. A CD133-expressing murine liver oval cell population with bilineage potential. Stem cells (Dayton, Ohio) 2007;25:2419–2429. doi: 10.1634/stemcells.2007-0176. [DOI] [PubMed] [Google Scholar]
- Schlissel MS, Corcoran LM, Baltimore D. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. The Journal of experimental medicine. 1991;173:711–720. doi: 10.1084/jem.173.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining Molecular Cornerstones during Fibroblast to iPS Cell Reprogramming in Mouse. Cell Stem Cell Advance Online Publication. 2008 doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Teng G, Papavasiliou FN. Immunoglobulin somatic hypermutation. Annual review of genetics. 2007;41:107–120. doi: 10.1146/annurev.genet.41.110306.130340. [DOI] [PubMed] [Google Scholar]
- Wakayama T, Yanagimachi R. Mouse cloning with nucleus donor cells of different age and type. Molecular reproduction and development. 2001;58:376–383. doi: 10.1002/1098-2795(20010401)58:4<376::AID-MRD4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Wang X, Foster M, Al-Dhalimy M, Lagasse E, Finegold M, Grompe M. The origin and liver repopulating capacity of murine oval cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(Suppl 1):11881–11888. doi: 10.1073/pnas.1734199100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of adult rats with Parkinson’s. Proceedings of the National Academy of Sciences of the United States of America. 2008a doi: 10.1073/pnas.0801677105. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wernig MA, John P, Cassady, Rudolf Jaenisch. c-Myc Is Dispensable for Direct Reprogramming of Mouse Fibroblasts. Cell Stem Cell. 2008b;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01