Homeostasis and Inflammation in the Intestine (original) (raw)
. Author manuscript; available in PMC: 2011 Mar 19.
Abstract
The gut is home to our largest collection of microbes. The ability of the immune system to co-evolve with the microbiota during postnatal life allows the host and microbiota to coexist in a mutually beneficial relationship. Failure to achieve or maintain an equilibrium between a host and its microbiota has negative consequences for both intestinal and systemic health. In this Review, we consider the many cellular and molecular methods by which inflammatory responses are regulated to maintain intestinal homeostasis and the disease states that can ensue when this balance is lost.
Introduction
Immunology has been defined as the “science of self non-self discrimination” (Klein, 1999) with the assumption that “non-self” intrusions instigate the inflammatory response. However, the very definition of self requires further examination when we consider that we harbor microbial communities (microbiota) that contain an estimated order of magnitude more cells than our own somatic and germ cells. These communities exhibit a remarkable degree of variation within and between individuals (Grice et al., 2009; Turnbaugh et al., 2009) and provide us with traits that we have not had to evolve on our own (Turnbaugh et al., 2007). The application of culture-independent metagenomic methods has shed light on the organismal and genetic composition, and dynamic operations, of the microbial communities that integrate themselves into our various body habitats (see Hamady and Knight, 2009 for a review of the experimental and computational methods used for metagenomic analyses). These communities can harbor representatives from all three branches of life (Bacteria, Archaea and Eukarya) and their viruses. Many fundamental questions arise when considering this microbial component of our multispecies ‘self’: how has the immune system been shaped by the need to accommodate our symbionts; how does it co-evolve with a microbiota to both shape and accommodate community assembly; how does the immune system drive and tolerate the variations in microbial ecology that occur within a host’s given body habitat over time; to what extent does the continuous evolution of various microbial phylogenetic types (phylotypes) underlie this apparent tolerance; and how do the adaptive and innate immune systems retain the capacity to respond to pathogenic organisms?
The intestine represents a body habitat that vividly illustrates these issues. The distal gut of humans represents one of the most densely populated microbial ecosystems on Earth, with up to 1012 organisms packed together per milliliter or gram of luminal contents. This ecosystem is dominated by members of the Bacteria: among the 100 known phyla in this domain of life, members of the Firmicutes and the Bacteroidetes form the largest component of the distal gut community (Ley et al., 2008). Although the total number of phyla detected to date in the human gut is relatively low compared to other natural environments such as soil or the ocean, diversity at the level of species and strains is enormous. A metagenomic study of bacterial diversity in fecal samples obtained from adult monozygotic and dizygotic twin pairs and their mothers over time have revealed that (i) the communities with the greatest degree of similarity were those derived from the same individual, (ii) the degree of similarity in the gut bacterial communities of monozygotic twin pairs was not significantly different than the similarity between dizygotic twin pairs; (iii) communities are more similar within family members than between different families (i.e. family members share significantly more phylotypes than unrelated individuals), and (iv) there was not a single abundant (defined as representing >0.5% of the population) bacterial species present in all of the 154 individuals surveyed. These results emphasize that early environmental exposures are a key determinant of adult gut microbial ecology and that the hypothesis that there is a ‘core’ gut microbiota defined by abundant organismal lineages shared by all humans is likely incorrect. Shotgun sequencing of the aggregate genome (microbiome) of the fecal communities of different families revealed that different microbial communities (species assemblages) converge on the same functional state: i.e., there is a group of microbial genes represented in the guts of unrelated as well as related individuals. This ‘core’ microbiome is enriched in functions related to survival in the gut (e.g. translation, nucleotide, carbohydrate and amino acid metabolism). Genes whose proportional representation in gut communities vary among individuals comprise a ‘variable’ microbiome. Pairwise comparisons have shown that family members have functionally more similar gut microbiomes than do unrelated individuals. Thus, intra-familial and subsequent intergenerational transmission of a gut microbiome (over multiple generations), could shape the biological features of humans within a kinship, contributing to differences in the structures and operations of their innate and adaptive immune systems, and together with their H. sapiens genotypes, modulate/mediate their risks for immunopathologic states, as well as other diseases.
A second reason why the gut is such an attractive system for studying the co-evolution and co-adaptation of the immune system and microbiota is our increasing appreciation of the reciprocal nature of the regulation of the immune system and microbial community structure. The hematopoietic-derived innate and adaptive immune systems are traditionally regarded as the hardware, software, and administrators of the inflammatory response. Intestinal immune cells localize to inductive sites (Peyer’s patches, mesenteric lymph nodes, lymphoid follicles, and colonic patches) and effector sites (the epithelium and underlying lamina propria). Comparative studies of germ-free mice and their microbe-laden conventionally-raised counterparts, or of adult germ-free mice that have received gut microbial communities from conventionally-raised donors, have established that the intestinal microbiota is a key contributor to the proper structuring of these sites (Macpherson and Harris, 2004). The microbiota instructs immune cells, guides their proper assembly, and as such contributes to the proper functioning of immunologic inductive sites. Position within the mucosal barrier, and the status of the barrier (its inflammatory tone) in turn shape the development and fate choices of immune cells (see the Review by Littman et al. in this issue of Cell).
There are numerous examples of how genetically manipulated mice have revealed the role of these and other components of the immune network in accommodating the gut microbiota. Below, we highlight recent insights gleaned about these components and their interactions. Disruption of this homeostasis likely results not only in intestinal inflammatory diseases, like Crohn’s disease and ulcerative colitis, but may also contribute to “auto-immune” diseases at extra-intestinal sites, such as asthma and type 1 diabetes (T1D). We end by emphasizing how the union of metagenomic methods and gnotobiotic humanized mouse models (containing only known microorganisms) will be useful for characterizing features of human gut communities and of the immune networks that support the co-evolved homeostasis between the microbial and H. sapiens components of our multispecies ‘supraorganismal’ self.
The Mucosal Barrier and the Microbiota
The gut epithelium forms an essential element of the mucosal barrier. All five of its lineages ---goblet cell, Paneth cell, M cell, enteroendocrine cell, and absorptive enterocytes---contribute to barrier function. The gut epithelium undergoes rapid and perpetual self-renewal: this renewal is fueled by multipotential Lgr5-expressing stem cells located in the crypts of Lieberkuhn (Sato et al., 2009), and concludes with apoptosis/exfoliation of terminally differentiated cells at the tips of small intestinal villi or the villus homolog in the colon (the surface epithelial cuff). Remarkably, this process of continuous epithelial replacement occurs without disrupting the functional integrity of cell-cell junctions. Regulation of junctional integrity and paracellular permeability is especially important for immune system homeostasis as a vast diversity of microbes and food antigens hover on the luminal surface of the mucosal barrier. Pathogenic viruses, bacteria, and parasites all exploit opportunities for breaching the epithelial barrier by entering through junctions (Bergelson, 2009; O’Hara and Buret, 2008). Coxsackie B and adenovirus bind to a receptor (CAR) that co-localizes with the tight junction protein ZO-1 (Raschperger et al., 2006). Reoviruses exploit junctional proteins for entry and their spread relies on binding to JAM-A (Antar et al., 2009). Rotaviruses alter occludin localization to tight junctions by changing the levels of non-phosphorylated occludin (Beau et al., 2007). Enteropathogens, including Shigella flexneri, Clostridium difficile, and Salmonella typhimurium alter barrier function through secreted toxins or type III secretion effectors that target junctional proteins or the junction-associated cytoskeleton.
Inflammatory mediators can modulate epithelial renewal to promote host defense. IL-13, a pro-inflammatory cytokine, and CXCL10, a chemokine, accelerate turnover to drive an “epithelial escalator” that expels intestinal parasites (Cliffe et al., 2005). NF-κB, a master regulator of inflammatory response genes, functions in basal states to ensure continuous epithelial replacement and barrier integrity; under select inflammatory conditions, as in Trichuris (whipworm) infection in mice (Cliffe et al 2005), inflammatory mediators can accelerate epithelial replacement as a host defense mechanism. Mice with conditional knockout of intestinal epithelial IkB kinase-γ or both IKKα and IKKβ, lose the capacity for NF-κB activation and develop severe chronic intestinal inflammation (Nenci et al 2007). Epithelial NF-κB deficiency results in reduced expression of antimicrobial peptides and a heightened level of enterocyte apoptosis that outpaces adaptive changes in epithelial renewal; the result is a breach in the mucosal barrier and bacterial translocation (Nenci et al 2007).
In mouse models, increased intestinal permeability may precede the development of non-resolving intestinal inflammation (Olson et al., 2006). Molecular defects in junctional complex proteins, such as JAM-A (Laukoetter et al., 2007), or expression of a constitutively active myosin light chain kinase, result in increased numbers of intestinal myeloid and T cells within the mucosa and more severe colitis in chemically- or T cell- induced models of inflammatory bowel disease (IBD) (Su et al., 2009). Whether tight junction abnormalities precede or follow inflammation in IBD is controversial—less subtle mucosal breaches (ulcers) occur frequently in colitis as well (Schulzke et al 2009).
Goblet cells: mucus and mucins
Mucus produced by goblet cells forms a key component of the mucosal barrier. It is a rich and consistent source of nutrients for saccharolytic bacterial members of the microbiota and a microhabitat where microbiota can embed themselves in close proximity to one another to express their nutrient sharing (syntrophic) relationships, and to avoid wash out from the continuously perfused and peristaltic gut bioreactor. Gel-forming mucins of intestinal mucus are arranged into a bilayer with a firm inner layer devoid of bacteria and a looser outer layer (Johansson et al., 2008). MUC2 is the most abundant mucin of intestinal mucus. Mice harboring MUC2 missense mutations develop chronic inflammation in their distal intestine, resembling human ulcerative colitis (UC). Misassembly of mutant MUC2 multimers leads to stress conditions in the endoplasmic reticulum (ER) and initiation of the unfolded protein response (UPR; Heazlewood et al., 2008). Goblet cell loss and muco-depletion are frequent histopathologic features in human UC (Gersemann et al., 2009). UC samples also show evidence of ER stress and accumulation of MUC2 precursors (Heazelwood et al., 2008). The interrelationship between the microbiota and barrier function is illustrated by the fact that the short chain fatty acid (SCFA) end products of microbial fermentation modulate expression of the MUC2 gene: proprionate directly increases MUC2 expression (the gene contains SCFA-responsive regulatory elements), whereas butyrate regulates expression via effects on histone acetylation and methylation plus its AP1 element (Burger-van Paassen et al., 2009).
Paneth cells: UPR and autophagy regulate intestinal inflammation
Paneth cells play a critical role in host defense through their production of zinc, lysozyme, and numerous other antimicrobial molecules. Paneth cells actively sense the microbiota and regulate their production of antimicrobial peptides via cell-autonomous MyD88-dependent activation of Toll-like receptors (TLRs), thereby limiting barrier breaches by commensal bacteria and pathogens (Vaishnava et al 2008). Several recent studies have provided insight into the importance of this cell type for intestinal homeostasis. The unfolded protein response (UPR) and autophagy play key roles in regulating Paneth cell function. Genetic alteration of the unfolded protein response gene, XBP-1 (X-box binding protein 1), a transcription factor whose activation increases protein quality control and ER expansion, or hypomorphic expression of the autophagy gene, ATG16L1, both perturb Paneth cell function. This has profound consequences for a host’s ability to maintain a productive equilibrium with its microbiota. XBP1 influences the antimicrobial activity and number of Paneth cells. Loss of XBP decreased small intestinal crypt lysozyme levels and decreased bactericidal activity against Salmonella typhimurium in a crypt homogenate. XBP also dampens the enterocyte response to pro-inflammatory stimuli, such as flagellin and TNF-α and its deletion results in basal activation of c-Jun-N-terminal kinase and NF-κB, both of which orchestrate pro-inflammatory programs (Kaser and Blumberg, 2009). There is a significant association between human IBD and several hypomorphic XBP variants (rs35873774; p-value, 1.6 × 10−5), and mice with a conditional deletion of XBP1 in intestinal epithelial cells develop spontaneous enteritis.
The UPR is activated by ER stress and ER stress induces autophagy. These two cellular homeostatic response mechanisms both play an important role in regulating inflammation. Genome wide association studies (GWAS) have identified a variant in the autophagy protein, ATG16L1, which confers increased predisposition for Crohn’s disease (CD) (Hampe et al., 2007; Rioux et al., 2007). Two recent studies in mice provide insight into how this ATG16L1 T300A variant may contribute to the multiple cellular defects intrinsic to CD. In mice hypomorphic for Atg16L1 protein expression, Paneth cells show striking defects in granule content and exocytosis (Cadwell et al., 2008). Similar results from mice with engineered deficiencies of ATG5 or ATG7 in the intestinal epithelium further support the importance of autophagy for Paneth cell function (Cadwell et al., 2008). The ATG16L1 CD variant also inhibits autophagy of the enteric pathogen, Salmonella typhimurium (Kuballa et al., 2008). In addition to the genetic association of ATG16L1 with CD, another variant of the autophagy protein, IRGM, has also been identified in association with CD (McCarroll et al., 2008).
Defensins: Cause or Effect of IBD
Compromised responses to ER stress and autophagy both affect the ability of Paneth cells to secrete antimicrobial peptides. Paneth cells, enterocytes, neutrophils, and plasma cells of the colon can express antimicrobial molecules (C-type lectins, defensins, and cathelicidins) that function in host defense and shape a host’s microbial communities. The defensins are the most widely and highly expressed antimicrobial peptides and altered defensin levels may be a factor in the pathogenesis of IBD (Ramasundara et al., 2009). Reduced Paneth cell alpha-defensin and beta-defensin levels have been observed in patients with ileal CD (Wehkamp et al., 2005) and in CD involving the colon (Fellermann et al., 2006), respectively. Both Wnt signaling and sensors of intracellular peptidoglycan cooperatively regulate alpha-defensin expression (Kobayashi et al 2005; Wehkamp et al., 2007). A mutation in NOD2 (the muramyl dipeptide recognition receptor) found in a subset of CD patients exacerbates alpha-defensin deficiency and NOD2 regulates the expression of the alpha defensin Defcr-rs10 (Kobayashi et al., 2005). Reduced expression of Tcf-4 (Wnt pathway) in ileal tissues of CD patients with ileal involvement correlates with reduced expression levels of the alpha-defensins, HD5 and HD6 (Wehkamp et al., 2007).
Members of the RegIII family of C-type lectins that bind to peptidoglycan, are expressed in Paneth cells as well as absorptive enterocytes, and are bactericidal. Reg IIIγ/HIP-PAP expression is dependent on the microbiota (Cash et al., 2006), transcriptionally regulated by MyD88-dependent signals, and the polypeptides post-translationally activated by proteolytic processing (Mukherjee et al., 2009). RegIIIγ also is regulated by a subset of NK1.1+ intestinal cells that secrete IL-22 (Sanos et al., 2009). Reg IIIγ/HIP-PAP protects the host from foodborne enteropathogens such as Listeria (Brandl et al., 2007; Sanos et al., 2009). How modulation of alpha- and/or beta-defensin or C-type lectin levels affects a host’s gut microbial community structure and affects IBD pathogenesis needs to be explored using metagenomic methods. Gnotobiotic mouse models with inducible knockout of genes encoding these anti-microbial agents, colonized with various types of microbial communities, should help address these questions.
Inflammatory Bowel Disease: Immunodeficiency or hyper-immunity
UPR and autophagy defects in CD raise questions about whether pathophysiology is rooted in immunodeficiency or dysregulated inflammation. The Paneth cell defects observed in XBP-deficient mice and the role of autophagy in both Paneth cells and in eliminating intracellular bacteria suggest immunodeficiency (Coulombe and Behr, 2009). Elevated JNK and NF-kB levels observed in the epithelium of mice with impaired ER stress responses (Kaser et al., 2008) and the high levels of active IL-1β and IL-18 in macrophages expressing Atg16L1 T300A suggest a hyper-active or dysregulated inflammatory state (Saitoh et al., 2008).
Under certain circumstances, the adaptive immune system appears to compensate for immunodeficiencies such as loss of certain innate immune safeguards. Deficiency in MyD88 and/or TRIF (adaptors for the toll-like receptors) results in innate immune defects in sensing of the intestinal microbiota. Nonetheless, these mice are viable and do not develop spontaneous intestinal inflammation when raised under specified pathogen-free (SPF) conditions (Rakoff-Nahoum and Medzhitov 2008). However, when mature B cell and immunoglobulin responses are crippled by deletion of the JH gene in conjunction with MyD88 deficiency, mice cannot co-exist with their microbiota (Slack et al., 2009). T-bet−/− RAG2−/− mice, which lack adaptive immune cells (T cells, B cells, and NKT cells), develop a MyD88-independent ulcerative colitis driven by dendritic cell TNF-α over-production in response to the microbiota (Garrett et al 2007; Garrett et al 2009). Adoptive transfer of T-regulatory cells ameliorates this colitis, compensating for defects in the innate immune system that are present in T-bet−/− and RAG2−/− mice, and may explain why T-bet deficient mice have no evidence of colitis (Garrett et al 2007).
There is a well recognized increased frequency of autoimmune disorders in patients with immunodeficiency diseases: complement system deficiencies (system lupus erythematosis), common variable immunodeficiency (hemolytic anemia, rheumatoid arthritis, and thyroiditis) and T-regulatory cell abnormalities/Foxp3 mutations [X-linked IPEX syndrome (Immunodysregulation, Polyendocrinopathy, and Enteropathy)]. Certain host genetic alterations resulting in immunodeficiency may lead to changes in core features or in membership structure of microbial communities. The pathophysiology of IBD undoubtedly reflects a triad of immunodeficiency, altered microbial communities, and hyper-immunity.
M cells and enteroendocrine cells
Both M cells and enteroendocrine cells function as important sensors of intestinal luminal contents. M-cells (micro-fold cells) are present in the small intestine overlying Peyer’s patches (PP) and lymphoid follicles; they lack microvilli and the thick glycocalyx present in absorptive enterocytes, and are adept at transcytosis. M cells express toll-like receptors, α5β1 integrin, and galectin-9 on their apical surfaces, all of which facilitate M-cell sensing and transport of microbes (Kyd and Cripps, 2008). Various technical hurdles have stymied a comprehensive understanding of their functioning as sensors and details of how they relay signals to the lymphoid structures with which they interdigitate and overlie. Their importance as entry portals for a variety of infectious organisms including prions, viruses, and pathogenic bacteria and as targeted delivery sites for vaccine antigens is well recognized (Corr et al., 2008).
Enteroendocrine cells are strategically positioned to sense luminal signals, including the products of microbial metabolism, and to transduce these signals to adjacent, underlying and/or remote cell populations through secretion of various bioactive products (e.g., factors that regulate mucus secretion, modulate aspects of host energy metabolism, and regulate intestinal motility). Alterations in enteroendocrine populations are seen in mouse models of enteritis and colitis (Linden et al., 2005; O’Hara et al., 2006; Rubin et al., 2000) and in patients with IBD (El-Salhy et al., 1997). Although these associations may be casual rather than causal, enteroendocrine cells express toll-like receptors (TLR) (Bogunovic et al., 2007) and gene expression profiling of a human enteroendocrine cell line in response to dietary and microbial stimuli suggests that they may directly participate in host inflammatory responses by secreting cytokines and chemokines (Palazzo et al., 2007).
A relationship map of cell-cell regulation in the gut
The inflammatory response is a “robust” system, one that (i) responds to environmental change (ii) is regulated by negative-feedback and feed-forward controls and (iii) is modular [consisting of insulated sub-systems so that failure does not spread from one module to another] (Kitano, 2002). The presence of both adaptive and innate components provides the immune system with system safeguards as does the anatomy of the intestine at both the tissue and cellular level. At the tissue level, barriers limit exposure and promote a degree of “unawareness” (Hooper, 2009). At the cellular level, intestinal epithelial cells, myofibroblasts, stromal cells, T cells, B cells, myeloid cells, and the microbiota all participate in a network of interactions regulating the inflammatory tone of the intestine. While the cellular players continue to be characterized at ever-increasing levels of refinement and many key effector molecules are known, there are still huge gaps in our understanding of how immune cell behaviors are achieved. Many outstanding recent reviews have focused on the function of specific cell types at mucosal interfaces [DCs:(Rescigno and Di Sabatino, 2009; Strober, 2009); Macrophages: (Platt and Mowat, 2008), T cells: (van Wijk and Cheroutre, 2009); T-regulatory cells: (Barnes and Powrie, 2009; Belkaid and Tarbell, 2009; Izcue et al., 2009); Th17 cells (Korn et al., 2009); and innate T cells: (Meresse and Cerf-Bensussan, 2009)]. T-regulatory cells and the Th17 subset of CD4+ T cells are reviewed in this issue by Littman and co-workers. Here we focus on the sensing, sensor, and sensor signaling regulation of intestinal myeloid cells and briefly consider the recently recognized role of intestinal stromal cells and lymph node stromal cells in intestinal homeostasis and peripheral tolerance, respectively.
Stromal cells
Intestinal stromal cells, distinct from myofibroblasts, play a critical role in tissue regeneration and wound repair, and communicate both with the overlying epithelium and immune cells (Stappenbeck and Miyoshi, 2009). MyD88-dependent signals control the mesenchymal positioning of COX-2 expressing stromal cells necessary to maintain appropriate epithelial proliferation in response to injury (Brown et al., 2007). Intestinal stromal cells constitutively express COX-2, and produce prostaglandins (PGE(2) in a COX-2 dependent fashion; this basal prostaglandin production may shape the basal cytokine profiles of the intestine (Newberry et al 2001 2000). Caspase-1 activation in intestinal stromal cells appears to be an important mechanism by which Salmonella typhimurium initiates inflammation; this stromal cell-initiated inflammation was protective as caspase-1 deficient mice develop disseminated infection (Muller et al., 2009).
Lymph node stromal cells (LNSCs) have recently emerged as key effectors in the regulation of CD8+, but not CD4+, T cell peripheral tolerance to intestinal self-antigens (Lee et al., 2007 and Magnusson et al 2008). CD45− UEA-I+ LNSCs express Aire, a transcription factor and pro-apoptotic factor first identified in the thymus that regulates the ectopic expression of peripheral tissue antigens (Lee et al., 2007; Mathis and Benoist, 2009, Gardner JM et al 2009). While self-antigens have been detected in LNSCs (Lee et al., 2007; Nichols et al., 2008), the role of LNSCs in tolerance to microbial epitopes is unknown. Numerous viruses, parasites (Leishmania and Plasmodium), as well as prions, have been shown to target LNSCs (Mueller and Germain, 2009). Whether and/or how the microbiota shapes stromal cell representation or positioning within mesenteric and peripheral lymph nodes remains to be determined. How inflammation affects LNSC function, if at all, needs to be explored as well. Clearly, many questions remain about LNSCs functon in the induction and maintenance of tolerance (reviewed in Reynoso et al., 2009).
Dendritic cells and macrophages
The myeloid lineage’s monophagocytic cells include both dendritic cell (DC) and macrophage populations. DCs are described as the pre-eminent antigen processing and presenting cell of the immune system with the unique ability to activate both naïve and memory T cells (Steinman, 2007). This definition has stood the test of time, as the diversity and capabilities of DCs continue to unfold. DCs have been categorized into different subsets based on their functional properties and their integrin and chemokine receptor expression (Coombes and Powrie 2008). While DCs may develop from a number of distinct precursors, most DCs go through distinct maturation stages and are shaped by the local conditions of the tissues in which they reside or migrate through. There are two principal subsets of DCs: plasmacytoid (pDCs) and conventional myeloid DCs (cDCs). Key features of pDCs are their expression of toll-like receptor 7, which binds ssRNA in endosomes, and toll-like receptor 9, which binds unmethylated CpG, as well as their production of type 1 interferons. Both pDCs and cDCs localize to intestinal immune inductive and effector sites. The microbiota in combination with CD8+ T cells cooperate to regulate systemic numbers of pDCs based on observations that (i) “restricted flora’ (RF) mice have reduced pDCs numbers as compared to SPF and GF animals, and (ii) deletion of CD8 or perforin in RF and SPF mice increases pDCs numbers (Fujiwara et al., 2008). Comparisons of wild-type SPF and GF, and SPF MyD88−/− TRIF−/− mice, disclosed that the proportion and pattern of maturation marker expression of resident and migratory cDCs in lymphoid organs were quite similar; this result was to some extent unexpected, as many TLR ligands are regarded as classical activators of DC maturation programs (Wilson et al., 2008). cDCs may very well have cell-autonomous migration patterns, and it is known that a panoply of non-microbial stimuli can induce their maturation programs; for example, tissue damage or disruption of E-cadherin mediated adhesion (Jiang et al., 2007). In certain models and tissues DC distribution does appear to be influenced by the microbiota, as DCs have not been detected in the jejunum of GF pigs (Haverson L et al 2007). Intestinal DCs promote lymphocyte lineage responses that both foster co-existence with the intestinal microbiota and eradicate pathogenic organisms. They also influence the trafficking of lymphocytes in the gut by modulating integrin expression critical for intestinal homing. DCs are a nexus for linking innate and adaptive immunity and as such a central cellular node for regulation of the inflammatory response.
Myeloid cell sensing
Sensing of intestinal contents occurs constitutively in myeloid cells. DCs directly sense intestinal contents by extending their dendritic processes into the lumen—a carefully orchestrated cell biological feat requiring a dynamic cytoskeleton and expression of integrins and junctional proteins (Niess et al., 2005; Rescigno et al., 2001). DCs and macrophages “telesense” through epithelial intermediaries. In the sub-epithelial dome of PPs, they encounter luminal contents transcytosed by M cells. DCs and macrophages internalize not only partially degraded luminal products from absorptive enterocytes but also fragments of apoptotic epithelial cells (Huang et al., 2000). The human neonatal Fc receptor can also transport IgG/antigen complexes from the lumen across the barrier to DCs (Yoshida et al., 2004). Many of these receptors on DCs recognize microbe-associated molecular patterns, as is the case for TLRs, (discussed in detail in this issue by Akira et al), nucleotide-binding oligomerization-domain protein-like receptors (NLRs) and C-type lectin receptors, while others bind antibodies or eukaryotic cells or their components. Myeloid cells express several receptors that confer responsiveness to the classic mediators of inflammation including: vasoactive amines, complement components, vasoactive peptides, and lipid mediators (Medzhitov, 2008). They also express cytokine and chemokine receptors to gauge the inflammatory tone of the mucosal barrier and tune responses received via other receptors.
Cellular and sub-cellular sensing regulation
How is microbial sensing regulated at the cellular level in dendritic cells and macrophages? While different cellular subsets express distinct receptors, this simple statement belies the complexity intrinsic to what subsets express which receptors at certain levels. What transcriptional networks, post-transcriptional processing systems, and sub-cellular receptor targeting/localization mechanisms are responsible? Some intestinal myeloid populations under basal conditions examined ex vivo appear hypo-responsive to microbe-associated molecular patterns as compared to their counterparts isolated from lymphoid tissues. Does receptor expression alone result in this hyporesponsiveness? Pulsatile and prolonged exposure of bone marrow-derived macrophages to LPS to in vitro fails to elicit sustained pro-inflammatory cytokine expression, resulting in what is termed “macrophage LPS tolerance” (West and Heagy, 2002). Chromatin modifications regulate hyporesponsiveness. Certain pro-inflammatory TLR-induced genes have been shown to acquire specific chromatin modifications resulting in transient silencing, while other sets of genes with direct microbicidal properties appear upregulated (Foster et al., 2007). Such epigenetic mechanisms provide an essential level of regulation for inflammatory responses [reviewed in this issue by Smale]. The kinase Akt1 also contributes to regulation of LPS tolerance via its positive and negative regulation of four microRNAs which in turn regulate TLR4 expression and the transcriptional repressor SOCS1 expression (Androulidaki et al., 2009). Ligation of TLRs, pro-inflammatory cytokine receptors, and purinergic receptors (Atarashi et al 2008) would all seem to initiate signaling cascades that converge on master-regulator transcriptional factors that drive inflammatory host defense programs. However, epigenetic mechanisms appear to play a profound role in the kinetics and shape of the response that ensues.
Immunologic memory is a feature of both T and B lymphoid cells and the concept of macrophage LPS tolerance suggests that innate immune subsets may have a kind of memory as well. A host’s immunologic history, its food antigens, viral, and microbial exposures may all impact on the behavior of its sensors. How this information is stored (chromatin modification) or passed on beyond the lifetime of single cells (if at all) needs to be determined.
Sensor stimuli regulation and inflammatory stimuli
Another layer of regulation focuses on what is sensed. LPS, the TLR4 ligand, is the classic activator of the maturation program, which re-fashions DCs from sentinels to activators of naïve T cells. The gut epithelium may have the ability to inactivate certain TLR ligands. For example in zebrafish, intestinal alkaline phosphatase at the epithelial brush border dephosphorylates the lipid A moiety of LPS, which efficiently detoxifies endotoxin (Bates et al., 2007). In many Bacteroides species (common members of the mammalian distal gut microbiota (Ley et al., 2008)), the lipid A substituent of LPS lacks a 2-hydroxyl-myristoyl moiety (C14:2OH), making it a weak endotoxin that is not very immunostimulatory (Hofstad et al., 1993). Perhaps, there was selective pressure for non-stimulatory LPS, as basal not chronic activated immunity in the intestine would likely provide a more suitable/sustainable habitat.
Simultaneous engagement of multiple TLRs by products from microbial communities or an invasive pathogen may vary signal strength. TLR9 binding of DNA derived from the microbiota plays an important role in balancing T regulatory and T effector cell immune responses and in host defense for Encephalitozoon cuniculi, a micro-sporidian parasite that causes diarrheal, respiratory, and neurological diseases in immunocompromised humans (Hall et al., 2008). Another example comes from observation that components of a host’s microbiota sensed through TLR 2, 4, and 9 triggers protective T cell responses to oral infection with Toxoplasma gondii (Benson et al., 2009). TLRs and NLRs may also cross-regulate to maintain basal states in the intestine. Binding of muramyl dipeptide (MDP) to its intracellular receptor, NOD2, negatively regulates cytokine responses initiated by multiple TLRs, and in vitro studies of MDP pre-treated DCs suggest that enhanced IRF4 activity may account for the TLR signal dampening that is observed (Watanabe et al., 2008). As noted above, a subset of CD patients bears NOD2 mutations, and inadequately damped TLR signaling resulting from altered NOD2 function may contribute to an increased susceptibility to IBD.
While both mutualists/commensals, and pathogens share many similar microbe-associated molecular patterns (MAMPs), there are of course numerous pathogenic microbe-specific molecules or virulence factors—adhesive and invasive molecules, toxins, and proteases—that can either alter MAMP levels or trigger inflammation (Sansonetti and Medzhitov, 2009). Intestinal cells also appear to respond to the quorum sensing molecules which bacteria use for their communication. In vitro data suggest that bacterial quorum sensing molecules, such as N-(3-oxododecanoyl)-L-homoserine lactone and Pseudomonas quinolone signal (PQS, 2-heptyl-2-hydroxy-4-quinolone), may participate in tuning DC programs regulating T cell effector function; for example, by driving DC IL-12 production (Skindersoe et al., 2009). Bacteroides fragilis, a member of the normal human gut microbiota, expresses a number of capsular polysaccharides including one with profound immunomodulatory functions called polysaccharide A (PSA) that in mouse models of colitis lowers the pro-inflammatory cytokines IL-23 and IL-17 and increases levels of the anti-inflammatory cytokine IL-10 (Mazmanian et al., 2008). In summary, stimuli for intestinal immune sensors are broad and include nucleotides, MAMPs, bacterial polysaccharides, and even bacterial quorum sensing molecules: undoubtedly more stimuli and their sensors will be uncovered.
Sensor and Signal Transduction Pathway Regulation
Signal integration, calibration by negative and positive modulators, and cross-coupling networks regulate the set points for basal and inflammatory responses downstream of sensors. Studies of NF-kB regulation and TLR signaling have defined many elements of the signaling network map of host and microbial interaction but much territory remains uncharted.
TLR signaling is negatively regulated by at least four general mechanisms: (i) sub-cellular localization [reviewed in (Barton and Kagan, 2009)]; (ii) degradation; (iii) deubiquitination, and (iv) competition [reviewed in (Wang et al., 2009)]. A few of these regulators have an established role in controlling intestinal inflammation in mouse models while others have yet to be explored. SOCS-1, via its SH3 domain, binds and polyubiquitinates Mal (MyD88 adapter like protein), which is necessary for TLR2 and TLR4 signaling. Mice deficient in SOCS1 (except for expression in T and B cells) develop a fulminant colitis (Hanada et al., 2006). A20, first identified as a critical negative regulator of TNF-induced NF-κB activation, is also a key regulator of TLR-induced NF-κB activity (Boone et al., 2004). A20-deficient mice display severe systemic inflammation with extensive intestinal involvement (Lee et al., 2000). While A20 possess both deubiquitination and E3 ligase activity, its debuquitination of TRAF6 (a TLR down-stream effector) is what restricts TLR signaling. A20 independently regulates both NLR signaling and TLR signaling. Both in vitro and in vivo, A20 deficiency results in exaggerated MDP responses, increased RIP2 ubiquitylation, and prolonged NF-κB activation (Hitotsumatsu et al., 2008).
Other TLR negative regulators not fitting into the aforementioned categories include SIGIRR and the transcription factors aryl hydrocarbon receptor and ATF3. SIGIRR (also known as TIR8) exerts its inhibitory activity by trapping TRAF6 and IRAK1 (Wald et al., 2003). SIGIRR is highly expressed in intestinal epithelial cells and DCs: mice with genetically engineered SIGIRR deficiency exhibit increased intestinal inflammation in the dextran sulfate sodium (DSS) model of colitis (Garlanda et al., 2007). The aryl hydrocarbon receptor is a basic helix loop helix transcription factor that negatively regulates TLR4 signaling by complexing with Stat1 and NF-κB, subsequently inhibiting transcription of IL-6 (Kimura et al., 2009). Two recent studies have mapped the inflammatory response regulatory networks in innate immune cells. In macrophages, a three-component circuit consisting of an initiator, amplifier, and attenuator (NF-κB, C/EBPδ, and ATF3, respectively) discriminates between transient and persistent TLR4 signals (Litvak et al., 2009). Intestinal myeloid cells must distinguish between commensal/mutualist-derived versus pathogen-derived signals and the feed forward type 1 regulation described in Litvak et al (2009) elucidates how this process may be accomplished. Another laboratory used a combination of gene expression profiling and shRNA gene knock-down in DCs to construct a network model of DC transcriptional responses to TLR2, 4, 5, and 9 agonists (Amit et al., 2009). This landmark analysis of DC-pathogen responses provides substantial mechanistic insight into how DC achieve specific responses to diverse pathogens through a combination of core regulatory elements and fine tuners. Furthermore, the experimental approach, which coupled “observation” with “targeted perturbations”, establishes a framework that is broadly applicable for many biological systems.
Extra-intestinal effects of the gut microbiota and autoimmunity
The impact of the gut microbiota on health and disease extends beyond the gastrointestinal tract. There is a growing appreciation for the contribution of microbes to allergic and autoimmune diseases. For example, a randomized, double-blind, placebo-controlled trial reported that treatment of infants with prebiotics and probiotics reduced the cumulative incidence of atopic eczema although no characterization of microbial intestinal communities was performed (Kukkonen et al., 2007). The variable penetrance of diabetes in non-obese diabetic (NOD) mice in different animal housing facilities has been ascribed to differences in environmental microbes. A recent study suggests that the interactions between the innate immune system and intestinal microbes are a key disease-modifying factor in the development of type 1 diabetes(T1D) (Wen et al., 2008). Wen et al found that while MyD88-deficient NOD mice raised under SPF conditions were protected from T1D, germ-free MyD88-deficient NOD mice were not. Moreover, MyD88 deficiency altered the lineage composition of the gut microbiota and this altered microbiota afforded some protection from diabetes when transplanted into germ-free NOD recipients. Increasingly, alterations in the intestinal microbiota are being linked to obesity, asthma, diabetes, IBD, and other inflammatory conditions, although as noted below, much additional work is needed in this area, including assessments of the degree to which emerging associations are causally or casually related to pathogenesis.
Future directions
Next generation sequencing technology has placed mucosal immunologists and microbiologists on a pathway of discovery that should provide new insights about how microbial communities assemble during postnatal life, how their organismal and genetic composition vary during the various stages of the human lifecycle, and how their dynamic operations are shaped by and in turn shape the innate and adaptive immune systems.
Gnotobiotic mice representing both wild-type inbred strains and genetically engineered derivatives provide an opportunity to address a number of issues that cannot be readily examined in humans at the present time. Results from such analyses should help formulate hypotheses and concepts about the microbiota-immune interface that can then be tested in a more directed manner in humans. For example, the physical organization of the gut microbiota could be characterized over varying spatial scales, along the length and width of the gut and under conditions where confounding variables in humans, such as diet (a major factor determining variations in microbial community structure in the gut (Ley et al., 2008), and host genotype, can be constrained. Multicolored FISH [fluorescent in situ hybridization] (for microbial visualization) and the large collections of available antibodies and lectins (for characterizing distinct immune subsets, stromal cells, and other components of the mucosal barrier) could be used in concert to decipher the physical interactions between members of the microbiota and host. This effort could involve gnotobiotic mice harboring model ‘synthetic’ human gut microbiomes: i.e., communities of varying phylogenetic complexity, composed of sequenced and cultured members of the human gut microbiota, whose predicted genomes, proteomes, and metabolic activities are available. Since transparent zebrafish can now be reared under germ-free conditions, and the response of germ-free zebrafish to colonization with a zebrafish (or mouse) gut microbiota is very analogous to that noted in mice (Rawls et al., 2006), simplified microbial communities could be constructed that contain members engineered to express fluorescent proteins (e.g., see Rawls et al., 2007), and introduced into germ-free zebrafish engineered to contain immune cell subsets expressing distinct fluorescent proteins. The union of these methods could yield a model system that enables real time imaging of dynamic interactions between microbes, the host’s immune system and other components intimately associated with or physically remote from, the gut mucosal barrier.
Comparative metagenomic analyses are currently being conducted in patients with IBD and in suitable control populations. These analyses include 16S rRNA gene-based surveys of their gut microbiota (to determine ‘who is there), shotgun sequencing of their gut microbiomes (to characterize what genes and predicted pathways are present), characterization of their gut communities’ meta-transcriptomes and meta-proteomes (to delineate which microbial genes are expressed; Verberkmoes et al., 2009), and in some cases targeted or shotgun NMR- and mass spectrometry-based characterization of their gut microbial communities’ metabolomes. Reference control populations in these studies can consist of the individuals themselves during relapses and remissions, or family members such as discordant monozygotic co-twins with and without disease. Interpreting these vast datasets and developing hypotheses about host-microbial interactions will undoubtedly be very challenging. Such analyses should enlighten our vision of inputs or stimuli for the immune system. Bacterial metabolites and their recognition by innate immune and epithelial cells may represent new components of circuits that modulate inflammatory responses in the intestine.
Experimentally, combining gnotobiotics and metagenomics will allow for evaluation of immunoregulatory functions of complex microbial communities. Gut communities from individuals can be directly transferred to germ-free mice (Turnbaugh et al., 2009b): recipient mice could be genetically engineered so that they are sensitized for development of immunopathology because they harbor the very mutations that are also found in humans with IBD. Potentially confounding variables encountered in studies of humans, such as diet and host genotype, could be constrained by using these ‘humanized’ gnotobiotic animals. Other host phenotypes have been transferred via microbiota transplantation (e.g., Bowey et al., 2003; Turnbaugh et al., 2006; 2008). One hope from these types of experiments would be to operationally define gut microbial communities that promote or suppress inflammatory states: in this sense they could become integral parts of clinical studies as well as a platform for fulfilling Koch’s postulates where the causative “microbe” is a microbiota rather than a single pathogen. Finally, recent advances in successful engraftment of the human adaptive and innate immune systems into mice offer the tantalizing possibility of recreating a more fully humanized intestinal ecosystem populated by human immune cells and a human microbiota in gnotobiotic animals.
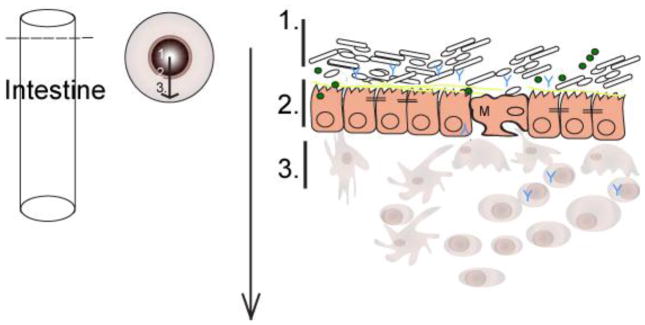
Figure 1. The gut landscape: Maintaining intestinal homeostasis.
The mucus layer, sitting atop the intestinal epithelium, is a key component of the mucosal barrier and also is both a source of nutrients and a microhabitat for bacterial members of the microbiota. The epithelial crypt-villus axis differs between the small and large intestine. The populating enterocyte populations vary as well. M cells and Paneth cells are restricted to the small intestine. Intestinal immune cells that mediate tolerance-inducing responses and participate in host defense, localize to inductive sites. These sites include Peyers patches (small intestine), lymphoid follicles, and colonic patches (large intestine) and effector sites such as the epithelium and underlying lamina propria.
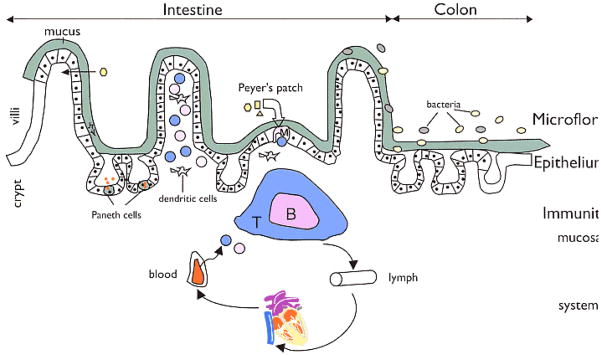
Figure 2. Regulators of host-microbial interactions in the gut.
The commensal microbiota, intestinal epithelial cells, and intestinal immune cells engage in a complex crosstalk. Epithelial cells, M cells, and dendritic cells (DCs) can directly sense and sample the intestinal contents and communicate information about the microbiota to other subsets of immune cells. The Toll-like receptors are one class of microbe-sensing molecules expressed by epithelial cells, M cells and DCs. Cytokines, chemokines, and host and microbial metabolites are key molecular mediators of intestinal homeostasis that influence responses of both host and microbe.
Acknowledgments
The authors thank the members of the Glimcher and Gordon labs for helpful discussions. Supported by NIH grants AI32412 and CA112663 (LHG), the Burroughs Wellcome Fund, and the Crohn’s and Colitis Foundation of America.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, et al. Unbiased Reconstruction of a Mammalian Transcriptional Network Mediating Pathogen Responses. Science. 2009 doi: 10.1126/science.1179050. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, Margioris AN, Tsichlis PN, Tsatsanis C. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar AA, Konopka JL, Campbell JA, Henry RA, Perdigoto AL, Carter BD, Pozzi A, Abel TW, Dermody TS. Junctional adhesion molecule-A is required for hematogenous dissemination of reovirus. Cell Host Microbe. 2009;5:59–71. doi: 10.1016/j.chom.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- Barnes MJ, Powrie F. Hybrid Treg cells: steel frames and plastic exteriors. Nat Immunol. 2009;10:563–564. doi: 10.1038/ni0609-563. [DOI] [PubMed] [Google Scholar]
- Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beau I, Cotte-Laffitte J, Amsellem R, Servin AL. A protein kinase A-dependent mechanism by which rotavirus affects the distribution and mRNA level of the functional tight junction-associated protein, occludin, in human differentiated intestinal Caco-2 cells. J Virol. 2007;81:8579–8586. doi: 10.1128/JVI.00263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Tarbell K. Regulatory T cells in the control of host-microorganism interactions. Annu Rev Immunol. 2009;27:551–589. doi: 10.1146/annurev.immunol.021908.132723. [DOI] [PubMed] [Google Scholar]
- Benson A, Pifer R, Behrendt CL, Hooper LV, Yarovinsky F. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host Microbe. 2009;6:187–196. doi: 10.1016/j.chom.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson JM. Intercellular junctional proteins as receptors and barriers to virus infection and spread. Cell Host Microbe. 2009;5:517–521. doi: 10.1016/j.chom.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Bogunovic M, Dave SH, Tilstra JS, Chang DT, Harpaz N, Xiong H, Mayer LF, Plevy SE. Enteroendocrine cells express functional Toll-like receptors. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1770–1783. doi: 10.1152/ajpgi.00249.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- Bowey E, Adlercreutz H, Rowland I. Metabolism of isoflavones and lignans by the gut microflora: a study in germ-free and human flora associated rats. Food Chem Toxicol. 2003;41:631–636. doi: 10.1016/s0278-6915(02)00324-1. [DOI] [PubMed] [Google Scholar]
- Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SL, Riehl TE, Walker MR, Geske MJ, Doherty JM, Stenson WF, Stappenbeck TS. Myd88-dependent positioning of Ptgs2-expressing stromal cells maintains colonic epithelial proliferation during injury. J Clin Invest. 2007;117:258–269. doi: 10.1172/JCI29159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger-van Paassen N, Vincent A, Puiman PJ, van der Sluis M, Bouma J, Boehm G, van Goudoever JB, van Seuningen I, Renes IB. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J. 2009;420:211–219. doi: 10.1042/BJ20082222. [DOI] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp JG, Kanther M, Semova I, Rawls JF. Patterns and scales in gastrointestinal microbial ecology. Gastroenterology. 2009;136:1989–2002. doi: 10.1053/j.gastro.2009.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon W. The Wisdom of the Body. 1. New York: Norton and Company, Inc; 1932. [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe LJ, Humphreys NE, Lane TE, Potten CS, Booth C, Grencis RK. Accelerated intestinal epithelial cell turnover: a new mechanism of parasite expulsion. Science. 2005;308:1463–1465. doi: 10.1126/science.1108661. [DOI] [PubMed] [Google Scholar]
- Corr SC, Gahan CC, Hill C. M-cells: origin, morphology and role in mucosal immunity and microbial pathogenesis. FEMS Immunol Med Microbiol. 2008;52:2–12. doi: 10.1111/j.1574-695X.2007.00359.x. [DOI] [PubMed] [Google Scholar]
- Cotran RS, Kumar Vinay, Collins Tuckey, Robbins Stanley. Robbins Pathologic Basis of Disease. 6. Philadelphia: Saunders; 1999. [Google Scholar]
- Coulombe F, Behr MA. Crohn’s disease as an immune deficiency? Lancet. 2009;374:769–770. doi: 10.1016/S0140-6736(09)61576-2. [DOI] [PubMed] [Google Scholar]
- El-Salhy M, Danielsson A, Stenling R, Grimelius L. Colonic endocrine cells in inflammatory bowel disease. J Intern Med. 1997;242:413–419. doi: 10.1046/j.1365-2796.1997.00237.x. [DOI] [PubMed] [Google Scholar]
- Fellermann K, Stange DE, Schaeffeler E, Schmalzl H, Wehkamp J, Bevins CL, Reinisch W, Teml A, Schwab M, Lichter P, et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am J Hum Genet. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- Gardner JM, Fletcher AL, Anderson MS, Turley SJ. AIRE in the thymus and beyond. Current Opinion Immunology. 2009 doi: 10.1016/j.coi.2009.08.007. epub October 13 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlanda C, Riva F, Veliz T, Polentarutti N, Pasqualini F, Radaelli E, Sironi M, Nebuloni M, Zorini EO, Scanziani E, et al. Increased susceptibility to colitis-associated cancer of mice lacking TIR8, an inhibitory member of the interleukin-1 receptor family. Cancer Res. 2007;67:6017–6021. doi: 10.1158/0008-5472.CAN-07-0560. [DOI] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Punit S, Gallini CA, Michaud M, Zhang D, Sigrist KS, Lord GM, Glickman JN, Glimcher LH. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell. 2009;16:208–219. doi: 10.1016/j.ccr.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersemann M, Becker S, Kubler I, Koslowski M, Wang G, Herrlinger KR, Griger J, Fritz P, Fellermann K, Schwab M, et al. Differences in goblet cell differentiation between Crohn’s disease and ulcerative colitis. Differentiation. 2009;77:84–94. doi: 10.1016/j.diff.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA NISC Comparative Sequencing Program. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–2. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, Grigg ME, Berzofsky JA, Belkaid Y. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Research. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- Hanada T, Kobayashi T, Chinen T, Saeki K, Takaki H, Koga K, Minoda Y, Sanada T, Yoshioka T, Mimata H, et al. IFNgamma-dependent, spontaneous development of colorectal carcinomas in SOCS1-deficient mice. J Exp Med. 2006;203:1391–1397. doi: 10.1084/jem.20060436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverson K, Rehakova Z, Sinkora J, Sver L, Bailey M. Immune development in jejunal mucosa after colonization with selected commensal gut bacteria: a study in germ-free pigs. Vet Immunol Immunopathol. 2007;119:243–253. doi: 10.1016/j.vetimm.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, Thornton DJ, Png CW, Crockford TL, Cornall RJ, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson PM, Cosgrove GP, Vandivier RW. State of the art. Apoptosis and cell homeostasis in chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2006;3:512–516. doi: 10.1513/pats.200603-072MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitotsumatsu O, Ahmad RC, Tavares R, Wang M, Philpott D, Turer EE, Lee BL, Shiffin N, Advincula R, Malynn BA, et al. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstad T, Skaug N, Sveen K. Stimulation of B lymphocytes by lipopolysaccharides from anaerobic bacteria. Clin Infect Dis. 1993;16(Suppl 4):S200–202. doi: 10.1093/clinids/16.supplement_4.s200. [DOI] [PubMed] [Google Scholar]
- Hooper LV. Do symbiotic bacteria subvert host immunity? Nat Rev Microbiol. 2009;7:367–374. doi: 10.1038/nrmicro2114. [DOI] [PubMed] [Google Scholar]
- Huang FP, Platt N, Wykes M, Major JR, Powell TJ, Jenkins CD, MacPherson GG. A discrete subpopulation of dendritic cells transports apoptotic intestinal epithelial cells to T cell areas of mesenteric lymph nodes. J Exp Med. 2000;191:435–444. doi: 10.1084/jem.191.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Annu Rev Immunol. 2009;27:313–338. doi: 10.1146/annurev.immunol.021908.132657. [DOI] [PubMed] [Google Scholar]
- Jiang A, Bloom O, Ono S, Cui W, Unternaehrer J, Jiang S, Whitney JA, Connolly J, Banchereau J, Mellman I. Disruption of E-cadherin-mediated adhesion induces a functionally distinct pathway of dendritic cell maturation. Immunity. 2007;27:610–624. doi: 10.1016/j.immuni.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Blumberg RS. Endoplasmic reticulum stress in the intestinal epithelium and inflammatory bowel disease. Semin Immunol. 2009;21:156–163. doi: 10.1016/j.smim.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Naka T, Nakahama T, Chinen I, Masuda K, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J Exp Med. 2009;206:2027–2035. doi: 10.1084/jem.20090560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- Klein J. Self-nonself discrimination, histocompatibility, and the concept of immunology. Immunogenetics. 1999;50:116–123. doi: 10.1007/s002510050587. [DOI] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS One. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukkonen K, Savilahti E, Haahtela T, Juntunen-Backman K, Korpela R, Poussa T, Tuure T, Kuitunen M. Probotics and prebiotic galacto-oligosaccharides in the prevention of allergic diseases: a randomized, double-bind, placebo-controlled trial. J Allergy Clin Immunol. 2007;119:192–8. doi: 10.1016/j.jaci.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Kyd JM, Cripps AW. Functional differences between M cells and enterocytes in sampling luminal antigens. Vaccine. 2008;26:6221–6224. doi: 10.1016/j.vaccine.2008.09.061. [DOI] [PubMed] [Google Scholar]
- Laukoetter MG, Nava P, Lee WY, Severson EA, Capaldo CT, Babbin BA, Williams IR, Koval M, Peatman E, Campbell JA, et al. JAM-A regulates permeability and inflammation in the intestine in vivo. J Exp Med. 2007;204:3067–3076. doi: 10.1084/jem.20071416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Epardaud M, Sun J, Becker JE, Cheng AC, Yonekura AR, Heath JK, Turley SJ. Peripheral antigen display by lymph node stroma promotes T cell tolerance to intestinal self. Nat Immunol. 2007;8:181–190. doi: 10.1038/ni1427. [DOI] [PubMed] [Google Scholar]
- Ley R, Peterson DA, Gordon JI. Ecological and evolutionary forces that shape microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden DR, Foley KF, McQuoid C, Simpson J, Sharkey KA, Mawe GM. Serotonin transporter function and expression are reduced in mice with TNBS-induced colitis. Neurogastroenterol Motil. 2005;17:565–574. doi: 10.1111/j.1365-2982.2005.00673.x. [DOI] [PubMed] [Google Scholar]
- Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy KA, Lampano AE, Nykter M, Shmulevich I, Aderem A. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol. 2009;10:437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- Magnusson FC, Liblau RS, von Boehmer H, Pittet MJ, Lee JW, Turley SJ, Khazaie K. Direct presentation of antigen by lymph node stromal cells protects against CD8 T-cell-mediated intestinal autoimmunity. Gastroenterology. 2008;134:1028–1037. doi: 10.1053/j.gastro.2008.01.070. [DOI] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host & Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D, Benoist C. Aire. Annu Rev Immunol. 2009;27:287–312. doi: 10.1146/annurev.immunol.25.022106.141532. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- Meresse B, Cerf-Bensussan N. Innate T cell responses in human gut. Semin Immunol. 2009;21:121–129. doi: 10.1016/j.smim.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Mueller SN, Germain RN. Stromal cell contributions to the homeostasis and functionality of the immune system. Nat Rev Immunol. 2009;9:618–629. doi: 10.1038/nri2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Partch CL, Lehotzky RE, Whitham CV, Chu H, Bevins CL, Gardner KH, Hooper LV. Regulation of C-type lectin antimicrobial activity by a flexible N-terminal prosegment. J Biol Chem. 2009;284:4881–4888. doi: 10.1074/jbc.M808077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller AJ, Hoffmann C, Galle M, Van Den Broeke A, Heikenwalder M, Falter L, Misselwitz B, Kremer M, Beyaert R, Hardt WD. The S. Typhimurium effector SopE induces caspase-1 activation in stromal cells to initiate gut inflammation. Cell Host Microbe. 2009;6:125–136. doi: 10.1016/j.chom.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, Huth M, Nikolaev A, Neufert C, Madison B, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- Newberry RD, McDonough JS, Stenson WF, Lorenz RG. Spontaneous and continuous cyclooxygenase-2-dependent prostaglandin E2 production by stromal cells in the murine small intestine lamina propria: directing the tone of the intestinal immune response. J Immunol. 2001;166:4465–4472. doi: 10.4049/jimmunol.166.7.4465. [DOI] [PubMed] [Google Scholar]
- Newberry RD, Stenson WF, Lorenz RG. Cyclooxygenase-2-dependent arachidonic acid metabolites are essential modulators of the intestinal immune response to dietary antigen. Nat Med. 1999;5:900–906. doi: 10.1038/11341. [DOI] [PubMed] [Google Scholar]
- Nichols LA, Chen Y, Colella TA, Bennett CL, Clausen BE, Engelhard VH. Deletional self-tolerance to a melanocyte/melanoma antigen derived from tyrosinase is mediated by a radio-resistant cell in peripheral and mesenteric lymph nodes. J Immunol. 2007;179:993–1003. doi: 10.4049/jimmunol.179.2.993. [DOI] [PubMed] [Google Scholar]
- Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- O’Hara JR, Buret AG. Mechanisms of intestinal tight junctional disruption during infection. Front Biosci. 2008;13:7008–7021. doi: 10.2741/3206. [DOI] [PubMed] [Google Scholar]
- O’Hara JR, Skinn AC, MacNaughton WK, Sherman PM, Sharkey KA. Consequences of Citrobacter rodentium infection on enteroendocrine cells and the enteric nervous system in the mouse colon. Cell Microbiol. 2006;8:646–660. doi: 10.1111/j.1462-5822.2005.00657.x. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Fukushima K, Naito H, Funayama Y, Unno M, Takahashi K, Kitayama T, Matsuno S, Ohtani H, Takasawa S, et al. Increased expression of HIP/PAP and regenerating gene III in human inflammatory bowel disease and a murine bacterial reconstitution model. Inflamm Bowel Dis. 2003;9:162–170. doi: 10.1097/00054725-200305000-00003. [DOI] [PubMed] [Google Scholar]
- Olson TS, Reuter BK, Scott KG, Morris MA, Wang XM, Hancock LN, Burcin TL, Cohn SM, Ernst PB, Cominelli F, et al. The primary defect in experimental ileitis originates from a nonhematopoietic source. J Exp Med. 2006;203:541–552. doi: 10.1084/jem.20050407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo M, Balsari A, Rossini A, Selleri S, Calcaterra C, Gariboldi S, Zanobbio L, Arnaboldi F, Shirai YF, Serrao G, et al. Activation of enteroendocrine cells via TLRs induces hormone, chemokine, and defensin secretion. J Immunol. 2007;178:4296–4303. doi: 10.4049/jimmunol.178.7.4296. [DOI] [PubMed] [Google Scholar]
- Platt AM, Mowat AM. Mucosal macrophages and the regulation of immune responses in the intestine. Immunol Lett. 2008;119:22–31. doi: 10.1016/j.imlet.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Medzhitov R. Innate immune recognition of the indigenous microbial flora. Mucosal Immunol. 2008;1(Suppl 1):S10–14. doi: 10.1038/mi.2008.49. [DOI] [PubMed] [Google Scholar]
- Ramasundara M, Leach ST, Lemberg DA, Day AS. Defensins and inflammation: the role of defensins in inflammatory bowel disease. J Gastroenterol Hepatol. 2009;24:202–208. doi: 10.1111/j.1440-1746.2008.05772.x. [DOI] [PubMed] [Google Scholar]
- Raschperger E, Thyberg J, Pettersson S, Philipson L, Fuxe J, Pettersson RF. The coxsackie- and adenovirus receptor (CAR) is an in vivo marker for epithelial tight junctions, with a potential role in regulating permeability and tissue homeostasis. Exp Cell Res. 2006;312:1566–1580. doi: 10.1016/j.yexcr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–33. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Goodman AL, Trent CM, Gordon JI. In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc Natl Acad Sci USA. 2007;104:7622–7627. doi: 10.1073/pnas.0702386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescigno M, Di Sabatino A. Dendritic cells in intestinal homeostasis and disease. J Clin Invest. 2009;119:2441–2450. doi: 10.1172/JCI39134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- Reynoso ED, Lee JW, Turley SJ. Peripheral tolerance induction by lymph node stroma. Adv Exp Med Biol. 2009;633:113–127. doi: 10.1007/978-0-387-79311-5_10. [DOI] [PubMed] [Google Scholar]
- Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin DC, Zhang H, Qian P, Lorenz RG, Hutton K, Peters MG. Altered enteroendocrine cell expression in T cell receptor alpha chain knock-out mice. Microsc Res Tech. 2000;51:112–120. doi: 10.1002/1097-0029(20001015)51:2<112::AID-JEMT2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Sala C, Grainger DC, Cole ST. Dissecting regulatory networks in host-pathogen interaction using chIP-on-chip technology. Cell Host Microbe. 2009;5:430–437. doi: 10.1016/j.chom.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, Diefenbach A. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti PJ, Medzhitov R. Learning tolerance while fighting ignorance. Cell. 2009;138:416–420. doi: 10.1016/j.cell.2009.07.024. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Schulzke JD, Ploeger S, Amasheh M, Fromm A, Zeissig S, Troeger H, Richter J, Bojarski C, Schumann M, Fromm M. Epithelial tight junctions in intestinal inflammation. Ann N Y Acad Sci. 2009;1165:294–300. doi: 10.1111/j.1749-6632.2009.04062.x. [DOI] [PubMed] [Google Scholar]
- Skindersoe ME, Zeuthen LH, Brix S, Fink LN, Lazenby J, Whittall C, Williams P, Diggle SP, Froekiaer H, Cooley M, et al. Pseudomonas aeruginosa quorum-sensing signal molecules interfere with dendritic cell-induced T-cell proliferation. FEMS Immunol Med Microbiol. 2009;55:335–345. doi: 10.1111/j.1574-695X.2008.00533.x. [DOI] [PubMed] [Google Scholar]
- Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, Lawson MA, Geuking MB, Beutler B, Tedder TF, Hardt WD, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325:617–620. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Angenent LT, Gordon JI. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat Immunol. 2004;5:569–573. doi: 10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Miyoshi H. The role of stromal stem cells in tissue regeneration and wound repair. Science. 2009;324:1666–1669. doi: 10.1126/science.1172687. [DOI] [PubMed] [Google Scholar]
- Steinman RM. Lasker Basic Medical Research Award. Dendritic cells: versatile controllers of the immune system. Nat Medicine. 2007;13:1155–9. doi: 10.1038/nm1643. [DOI] [PubMed] [Google Scholar]
- Strober W. The multifaceted influece of the mucosal microflora on mucosal dendritic cell responses. Immunity. 2009;31:377–388. doi: 10.1016/j.immuni.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Su L, Shen L, Clayburgh DR, Nalle SC, Sullivan EA, Meddings JB, Abraham C, Turner JR. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology. 2009;136:551–563. doi: 10.1053/j.gastro.2008.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host & Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel B, Duncan A, Ley RE, Sogin ML, Jones J, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. Science Translational Med 1. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk F, Cheroutre H. Intestinal T cells: facing the mucosal immune dilemma with synergy and diversity. Semin Immunol. 2009;21:130–138. doi: 10.1016/j.smim.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, Lefsrud MG, Apajalahti J, Tysk C, Hettich RL, Jansson JK. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;1:179–89. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- Wald D, Qin J, Zhao Z, Qian Y, Naramura M, Tian L, Towne J, Sims JE, Stark GR, Li X. SIGIRR, a negative regulator of Toll-like receptor-interleukin 1 receptor signaling. Nat Immunol. 2003;4:920–927. doi: 10.1038/ni968. [DOI] [PubMed] [Google Scholar]
- Wang J, Hu Y, Deng WW, Sun B. Negative regulation of Toll-like receptor signaling pathway. Microbes Infect. 2009;11:321–327. doi: 10.1016/j.micinf.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Asano N, Murray PJ, Ozato K, Tailor P, Fuss IJ, Kitani A, Strober W. Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J Clin Invest. 2008;118:545–559. doi: 10.1172/JCI33145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, Shen B, Schaeffeler E, Schwab M, Linzmeier R, et al. Reduced Paneth cell alpha-defensins in ileal Crohn’s disease. Proc Natl Acad Sci U S A. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehkamp J, Wang G, Kubler I, Nuding S, Gregorieff A, Schnabel A, Kays RJ, Fellermann K, Burk O, Schwab M, et al. The Paneth cell alpha-defensin deficiency of ileal Crohn’s disease is linked to Wnt/Tcf-4. J Immunol. 2007;179:3109–3118. doi: 10.4049/jimmunol.179.5.3109. [DOI] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MA, Heagy W. Endotoxin tolerance: A review. Crit Care Med. 2002;30:S64–S73. [PubMed] [Google Scholar]
- Wilson NS, Young LJ, Kupresanin F, Naik SH, Vremec D, Heath WR, Akira S, Shortman K, Boyle J, Maraskovsky E, et al. Normal proportion and expression of maturation markers in migratory dendritic cells in the absence of germs or Toll-like receptor signaling. Immunol Cell Biol. 2008;86:200–205. doi: 10.1038/sj.icb.7100125. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Claypool SM, Wagner JS, Mizoguchi E, Mizoguchi A, Roopenian DC, Lencer WI, Blumberg RS. Human neonatal Fc receptor mediates transport of IgG into luminal secretions for delivery of antigens to mucosal dendritic cells. Immunity. 2004;20:769–783. doi: 10.1016/j.immuni.2004.05.007. [DOI] [PubMed] [Google Scholar]