Formins and Microtubules (original) (raw)
. Author manuscript; available in PMC: 2011 Feb 1.
Published in final edited form as: Biochim Biophys Acta. 2009 Jul 23;1803(2):164–173. doi: 10.1016/j.bbamcr.2009.07.006
Abstract
Formins have recently been recognized as prominent regulators of the microtubule (MT) cytoskeleton where they modulate the dynamics of selected MTs in interphase and mitosis. The association of formins with the MT cytoskeleton and their action on MT dynamics are relatively unexplored areas, yet growing evidence supports a direct role in their regulation of MT stability independent of their activity on actin. Formins regulate MT stability alone or in combination with accessory MT binding proteins that have previously been implicated in the stabilization of MTs downstream of polarity cues. As actin and MT arrays are typically remodeled downstream of signaling pathways that orchestrate cell shape and division, formins are emerging as excellent candidates for coordinating the responses of the cytoskeletal in diverse regulated and homeostatic processes.
1. Regulation of MT dynamics and MT stabilizing proteins
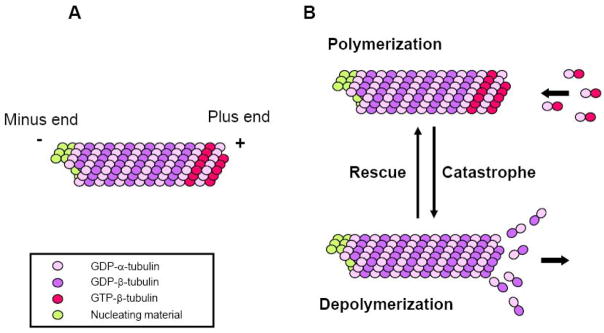
MTs are polarized polymers assembled by the lateral interaction of 12–15 protofilaments. Each protofilament is composed of α/β tubulin heterodimers that are held together by non-covalent bonds [1–4] (Fig. 1A). Because the α/β tubulin heterodimer polymerizes in a head to tail fashion and is intrinsically a polarized molecule, the resulting microtubule is a polarized polymer with α-tubulins exposed at one end (the “minus” end) and β-tubulins at the other (the “plus” end) [4]. Tubulin heterodimers can add onto or dissociate from either end of a microtubule, but there is greater tendency for addition to occur at the faster growing “plus” end. In most cells, the minus end is typically anchored to nucleating factors, such as the γ–tubulin complex, so most dynamics occur at the plus end.
Figure 1. Microtubule structure and dynamic instability.
(A) A MT is a polarized polymer of α/β tubulin heterodimers. During polymerization the GTP bound to the β tubulin subunit is hydrolyzed (GTP is also bound to the α-subunit, but this is not exchangeable or hydrolyzed). GTP-bound subunits decorate the plus end while GDP-bound subunits form the core of the MT. The minus end of the MTs is usually anchored to nucleating material in cells and does not exchange subunits. (B) MT dynamic instability. In most cells MTs alternate between phases of polymerization and depolymerization at their plus ends. Transitions from shrinkage to growth are known as rescues while the opposite reaction is know as catastrophe. Most MT dynamics in vitro and in vivo can be described by the rates of growth and shrinkage and the frequencies of rescues and catastrophes.
MT dynamics are characterized by phases of slow growth and rapid shrinkage with infrequent transitions, known as catastrophe and rescue, between these two states (Fig. 1B). This behavior, termed dynamic instability, was originally described using purified tubulin assembled in vitro [5], but is also characteristic of MTs in vivo. The β-subunit of tubulin binds GTP and this is hydrolyzed upon assembly into MTs (Fig. 1). It is thought that the maintenance of a small cap of GTP-containing subunits at the end of the MT promotes growth, whereas the exposure of a GDP-containing subunit promotes shrinkage. MT dynamics in vitro and in vivo are also dependent on regulatory proteins such as microtubule associated proteins (MAPs) and members of the kinesin family of molecular motors. Conventional MAPs, such as tau and MAP2, contribute to the stability of MTs by their ability to bind adjacent tubulin subunits within a MT and may contribute to higher ordered MT arrays by bundling MTs [6]. XMAP215 and its homologs are unconventional MAPs in that they bind tubulin heterodimers and promote MT polymerization by enhancing tubulin heterodimer association with growing MT plus ends [7–9]. Since they remain associated with MTs through multiple rounds of subunit addition, they have been referred to as MT polymerases. Members of the “KinI” class of kinesins generally do not possess motor activity to translocate along MTs, but instead act as MT depolymerases to rapidly diassasemble MTs [10–12]. Another class of MT interacting proteins are plus end tracking proteins or +TIPs that preferentially interact with the plus end of growing MTs and often mediate MT plus end attachment to cortical or intracellular sites in response to signal transduction pathways that regulate MT stabilization and positioning. We refer the readers elsewhere for most recent advances on these proteins [13].
MT dynamic instability has been proposed as a general mechanism by which MT plus ends probe the cellular environment in search of contacts with target sites or organelles distributed throughout the cell (the “search and capture” model) [14]. For example, during mitosis MT capture has been implicated in kinetochore attachment of chromosomes to spindle MTs and this is essential for spindle assembly. In migrating cells, attachment of MT plus ends to cortical sites is responsible for the selective stabilization of a subset of MTs oriented towards the wound edge and for the reorientation of the centrosome to the direction of migration.
Here, we summarize the literature on the biochemical activities of formins toward MTs, the localization of formins to the MT cytoskeleton and their role in modulating the dynamics and functions of MT arrays in cellular processes.
2. Biochemical studies of MT–formin interaction
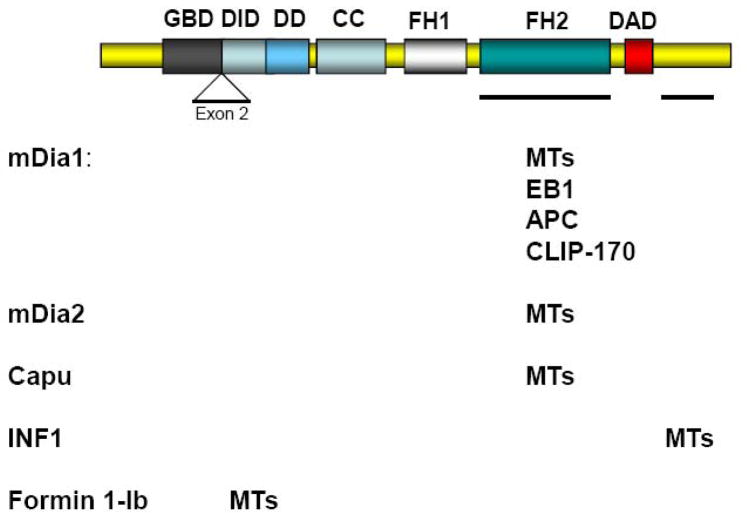
The recognition that formins may regulate MTs initially came from in vivo studies (see below). Only recently have biochemical studies begun to explore how formins interact with MTs and affect their dynamic properties. An initial indication that formins might directly interact with MTs to alter their dynamics came from a study showing that a constitutively active form of mDia2 lacking its regulatory domains colocalized with a subset of MTs in vivo and could bind to MTs when synthesized in vitro [15]. Subsequent studies have shown that at least four of the mammalian formins (mDia1, mDia2, Formin1-Ib and INF1) and one of the Drosophila formins, Cappuccino (Capu), can interact directly with MTs [16–19]. The MT binding site for mDia1, mDia2 and Capu appears to be within the FH2 domain, the same domain required for actin filament elongation (Fig. 2). These data raise the possibility that the MT and the actin cytoskeletons may be mutually exclusive for binding to these formins, although this remains to be tested. In addition, given that the FH2 domain is the most widely conserved domain among formins it is likely that other formin family members will be found to interact with and affect the dynamics of MTs through this domain. Interestingly, an FH2 containing fragment of INF2, a mammalian formin with a unique biochemical ability to accelerate actin filament depolymerization in addition to promoting actin filament assembly [138, 139], has been recently found to bind to MTs with at least as high affinity as mDia (Ramabhadran and Higgs, personal communication).
Figure 2. MT binding regions on formins.
The diagram represents the linear organization of domains in an idealized diaphanous-related formin. The domains known to interact with MTs or MT-interacting proteins for each of the formins are indicated. Note that most formins function as dimers and that Formin 1-Ib does not have a GBD or a DAD domain. GBD, GTPase binding domain; DID, DAD-interacting domain; DD, dimerization domain; CC, coiled-coil domain; FH1, formin homology 1; FH2, formin homology 2 domain; DAD, diaphanous autoregulatory domain.
Whereas the FH2 domain appears to be generally involved in linking diaphanous related formins (DRFs) to MTs, other regions have been implicated in binding MTs in other formins. Formin1 has a number of splice variants and an alternatively spliced N-terminal domain encoded by exon-2 (isoform Ib) was found to be necessary and sufficient for localizing Formin 1-Ib to MTs in vitro and in vivo [16]. Unlike mDia, the MT associated sequence did not appear to induce any gross change in MT organization or in cell shape when overexpressed, indicating that it may primarily act as targeting signal rather then a MT regulating motif [16]. With the exception of isoform IV and V, the exon-2 encoded peptide is shared by all the remaining Formin1 isoforms, indicating a conserved function [16]. Exon-2 does not exhibit homology to other formins or MT binding domains [16]. Interestingly, immuno-gold localization of isoform IV, which lacks exon-2, showed that this formin also localized along MTs in primary fibroblasts, suggesting that multiple MT targeting motifs are present in this class of proteins [20].
MT binding through a previously uncharacterized domain has also been reported for INF1, a relatively unexplored formin with the FH1 and FH2 domains near the N-terminus and a C-terminal region consisting of a unique polypeptide sequence [19]. INF1 was shown to directly associate with MTs via a novel bipartite MT binding domain in the C-terminal region whereas no binding to MTs was detected with an isolated FH2 domain of INF1 [19].
To date, the only formin-MT interaction characterized in any detail has been that of mDia2. The FH1FH2 fragment of mDia2 binds to MTs with an estimated Kd of 6.1 μM and a stoichiometry at saturation of 1:4.7 (mDia2:tubulin). These results suggest that FH1FH2mDia2 binds along the MT lattice, a prediction confirmed by localizing the protein on MTs in vitro [18].
The effect of formin-MT interactions on MT dynamics has also begun to be explored with in vitro experiments. The FH1FH2 fragment mDia2 stabilizes MTs against cold- and dilution-induced depolymerization, suggesting that it reduces tubulin subunit dissociation from MTs. Similar results have been obtained with the FH1FH2 domain of mDia1 (Bartolini & Gundersen, unpublished observation). Analysis of the dynamics of MTs grown off axonemal seeds, revealed that FH1FH2mDia2 slowed the rate of MT assembly by about 30% and the disassembly rate by 50%, although some MTs stopped depolymerizing completely [18]. Given that traditional MAPs, such as tau and MAP2, stimulate the rate of polymerization, this suggests that mDia1 and mDia2 may represent a unique class of MT regulators. The effects of other formins on in vitro MT dynamics have not been reported as yet.
In addition to their effects on MT dynamics, several studies show that formins can bundle MTs in vitro or bundle MTs with actin filaments. At high concentrations, FH1FH2mDia2 bundles MTs in vitro [18]. This bundling activity does not seem to be required to stabilize MTs from depolymerization [18]. An FH2 fragment of Capu was reported to cause crosslinking of MTs with actin filaments in vitro [17]. Since Capu interacts with both MTs and actin filaments via its FH2 domain, this activity may reflect the dimeric nature of Capu or the formation of Capu oligomers.
3. MT stabilization in migrating cells
Most MTs in interphase cultured cells exhibit dynamic instability and short half-lives (5–10 min). Yet, it has long been known that a subset of MTs do not undergo dynamic instability and have much longer half-lives (> 1hr) [21, 22]. These long-lived MTs (hereafter referred to as “stable MTs”), also show enhanced resistance to MT depolymerizing agents and accumulate post-translational modifications, such as detyrosination and acetylation, as a result of their longevity [23–26]. Stable MTs are found in polarizing cells (usually oriented along the axis of developing polarity) and generally increase in numbers in differentiated cells [27–30]. The posttranslational detyrosination and/or acetylation of tubulin comprising stable MTs enhances the interaction of kinesin-1 with MTs [31–33] allowing for preferential transport of certain cargoes such as vimentin filaments [34, 35], recycling transferrin receptors [36], and JIP1 [33].
The molecular basis for the selective formation of stable MTs among an array of dynamic MTs has remained an enduring problem in cell biology. A distinguishing property of stable detyrosinated MTs (termed “Glu MTs” after the new C-terminal glutamate residue of α-tubulin exposed by detyrosination) is that they do not add or lose subunits at their growing plus ends indicating that their ends are distinct from dynamic MTs [21, 22]. Indeed, a study employing a permeabilized cell model showed that the resistance of these stable MTs to either incorporate or lose subunits was lost if the stable MTs were first experimentally severed [37]. These studies suggest that stable Glu MTs may arise by a capping activity working on the ends of the MTs. Traditional MAPs, such as MAP2 and tau, tend to enhance the stability of the entire array of MTs within the cell (or cell compartment) and do not cap MTs, suggesting that a new type of MAP must be involved.
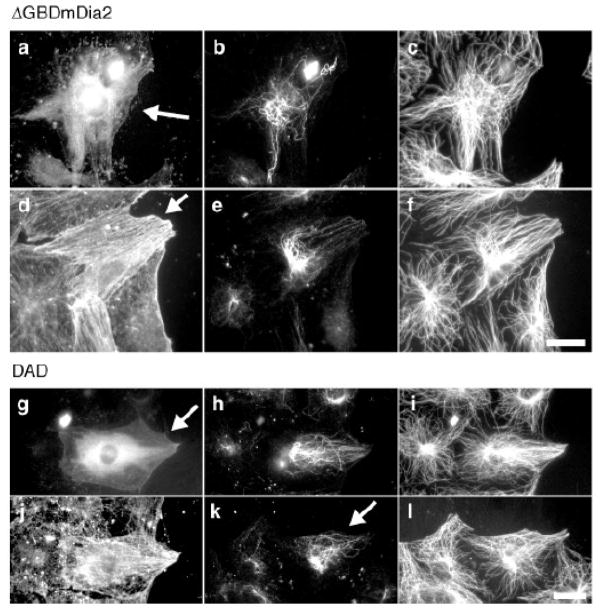
Studies of the regulation of stable MTs in migrating fibroblasts and other cells have recently lead to the idea that the mDia class of formins plays a key role in formation of stable MTs. In wounded monolayers of migrating fibroblasts, stable Glu MTs selectively form in the leading edge of the cell before migration and hence are oriented in the direction that the cell will migrate [38]. Treatment of starved fibroblasts with serum or lysophosphatidic acid (LPA), which was identified as the active, stable MT inducing factor in serum, induces the formation of stable MTs within 5 minutes suggesting that a signal transduction pathway regulates the formation of these stable MTs [39, 40]. Based upon the known activation of Rho GTPase downstream of the LPA receptor [41, 42], Rho was tested and found to be necessary and sufficient for the induction of stable Glu MTs in fibroblasts [40]. mDia was subsequently identified as the Rho effector mediating Rho stabilization of MTs in fibroblasts by using a Rho effector domain screen in which a series of Rho effector domain mutants with known effector binding profiles [43] were screened for their ability to stimulate Glu MTs [15]. Activation of endogenous mDia by expressing the Dia autoregulatory domain (DAD), stimulates Glu MT formation even in cells where Rho is inhibited showing that mDia is sufficient to induce MT stability and that other Rho effectors are not necessary (Fig. 3). Recent studies using RNAi approaches have found that mDia1 is essential for generating stable Glu MTs in a number of different cell types, including fibroblasts [44, 45], glioma cells [46] and endodermal cells [47].
Figure 3. Active mDia induces the formation of Glu MTs in fibroblasts.
a–f, Constitutively actin ΔGBDmDia2–MycGFP was microinjected into nuclei of serum-starved NIH3T3 cells, and after 3–4 h, cells were fixed and immunostained for stable (Glu) and dynamic (Tyr) MTs and actin. One cell is shown in a–c, another in d–f (arrows denote expressing cells; GFP fluorescence is not shown for cell in d–f). a, GFP; b, e, Glu MTs; c, f, Tyr MTs; d, actin. The Glu MTs in the mDia2-expressing cell in (e) are oriented towards the wound edge, whereas those in the expressing cell in (b) are not. g–l, DAD-mycGFP was microinjected into nuclei of serum-starved NIH3T3 cells, and after 3 h cells were fixed and immunostained for Glu and Tyr MTs and actin. One cell is shown in g–j, another in k, l (arrows denote expressing cells; GFP fluorescence is not shown for cell in k, l). g, GFP; h, k, Glu MTs; i, l, Tyr MTs; j, actin. The Glu MTs in these cells were oriented towards the wound edge as seen in (e). Scale bars, 20 μm. Reproduced by permission of the authors [15].
How does mDia contribute to MT stability in vivo? Early studies showed that the MT stabilizing activity of mDia towards MTs in cells resides within its FH2 domain [15, 48]. This raised the possibility that mDia may mediate its effects on MTs through the well-characterized ability of the FH2 domain to nucleate and elongate actin filaments [49]. However, a recent study showed that FH2 domain fragments of mDia2 with point mutations that block mDia2’s ability to stimulate actin polymerization in vitro and in vivo, still induce stable Glu MT formation in starved fibroblasts [18]. As mentioned earlier, biochemical studies now support the idea that mDia 1 and 2 have independent activity to stabilize MTs in vitro [18].
Although mDia has intrinsic stabilization activity toward MTs, it is likely that in cells, mDia works in conjunction with other proteins to generate capped and long-lived MTs. In the biochemical study referred to above, the FH1FH2 fragment of mDia2 only infrequently blocked depolymerization of MTs upon dilution-induced disassembly, suggesting that additional regions of the molecule or additional proteins may be needed to generate fully capped and stabilized MTs. In fibroblasts, mDia1 binds directly to EB1 and adenomatous polyposis coli protein (APC), two MT +TIPs, and appears to function upstream or perhaps in concert with these factors [48]. EB1 and APC also bind directly to each other and domain binding studies with mDia2 indicate that it is possible that the three proteins may form a complex to stabilize MTs [48].
mDia may also regulate the activity of kinases that contribute to MT stability. LPA treatment of starved fibroblasts stimulates the PKC dependent phosphorylation of GSK3β leading to the inhibition of its activity [50]. Notably, mDia1 was recently shown to be necessary for the LPA stimulated phosphorylation of GSK3β through a pathway involving novel isoforms of PKC (nPKCs) [45]. GSK3β is a known negative regulator of MT stability and in neurons it seems to phosphorylate MAPs leading to decreased MT stability [51–53]. In non-neuronal cells, the targets of GSK3β phosphorylation are unknown, but APC is a prominent substrate raising the possibility that GSK3β may regulate MT stability through this protein. The constitutive phosphorylation of MT stabilizing proteins by GSK3β may explain the observation that acute treatment of cells with protein phosphatase inhibitors selectively destabilizes stable MTs [54].
The question of whether or not mDia1 may also contribute to regulating the properties of dynamic MTs has been raised in a number of studies. Breast carcinoma cells form broad lamellipodia and lamella in response to growth factor stimulation [55] [56]. MTs normally penetrate the broad lamella, but in the absence of mDia1, they fail to [56]. Disruption of Memo, an effector of the ErbB2 receptor, or Rho GTPase yields a similar phenotype and measurements of MT dynamics in Memo depleted cells showed that MT dynamics were altered with increased catastrophes and decreased rescues. Whether formation of stabilized MTs was affected was not tested, yet these results suggest that mDia1 may be important to maintain growing MTs in breast carcinoma cells.
While virtually all studies in migrating cells (and neurons and dividing cells, see below) consistently find that the mDia family of formins promotes MT stability, there is one report that mDia1 has the opposite effect. In osteoclasts, which are large multinucleated cells that make adhesive contacts with the substratum known as podosomes, mDia1 or Rho activation, inhibits the formation of stabilized, acetylated MTs [57]. Interestingly, in osteoclasts mDia1 interacted with HDAC6, a tubulin and cortactin deacetylase. The same study confirmed the importance of Rho in promoting stable MTs in fibroblasts, indicating that the results in osteoclasts are cell type specific. These results emphasize that cellular context is likely to be important for how formins affect MTs.
Although mDia1 is both necessary and sufficient in generating stable MTs in several cell types, it was recently reported that full length INF1 and its C-terminally located MT binding sequence alone were sufficient to induce MT bundling and formation of acetylated MTs following overexpression in cultured cells [19]. Notably, a significant reduction in the levels of acetylated MTs (but not Glu MTs) was observed when INF1 expression was silenced by siRNA [19] suggesting that this formin may have a necessary role in those cell types in which acetylated MTs are required for maintenance or differentiation.
4. MT stabilization in neurons
Stable MTs are abundant in axonal processes and are required for proper axon specification and outgrowth and for neuromuscular junction formation [58–60]. A recent study used local activation of photoactivatable taxol to show that increasing MT stability alone was sufficient to enhance axon formation and outgrowth from one of the undifferentiated minor neurites of hippocampal cells [61]. Using overexpression of dominant negative and constitutively active constructs as well as siRNA knockdowns, Rho and mDia1 [62] and mDia2 [63] have been shown to play a role in axon elongation in cultured cerebellar granular cells. These studies attributed the axon promoting effects of mDia to Rac GTP levels [62] or the binding of mDia to the PAX6 transcription factor, which is involved in process outgrowth [63]. Given the mobilization of the actin cytoskeleton and formation of stable MTs during axon outgrowth, it is important to determine whether the capabilities of formins to regulate these processes are important for axon outgrowth.
A study of the Drosophila neuromuscular junction (NMJ), showed that Diaphanous, an ortholog of the mammalian mDia, but not Capu, is involved in the growth of the NMJ and functions downstream of the tyrosine receptor phosphatase Dlar and the Rho GEF trio [60]. A loss of function Diaphanous mutant showed reduced bouton number but increased size and disruption of actin foci and stable MTs within the NMJ. Measurements of MT growth using GFP-EB1 revealed that growth rates were increased in the Diaphanous mutant, suggesting that the formin may play a significant role in regulating MT stability in the NMJ.
5. MT stabilization in cell division
Formins are well-established regulators of cytokinesis where they are thought to contribute to the assembly of the cytokinetic ring [64]. However, there is growing evidence that they may also contribute to the assembly or function of the mitotic spindle through their effects on MTs. The initial observation that suggested that formins may affect the mitotic spindle was the localization of mDia1 to kinetochore and astral MTs following detergent extraction [65]. This spindle localization was independent of RhoA binding or the integrity of the actin cytoskeleton [65]. Point mutations within the N-terminal Dia-inhibitory domain (DID) and the adjacent coiled-coil domain (CC) inhibited the association of GFP-tagged mDia1 to the spindle, suggesting a role for this region in the association of mDia1 to spindle MTs [65]. Interestingly, the homologous region in Fus1 and Cdc12, two formin-related proteins implicated in the localization of actin structures during mating and cytokinesis respectively in S. pombe, co-localizes with γ-tubulin in the spindle pole body in growing yeast cells [66], suggesting that tubulin targeting sequences present in related formins may be evolutionarily conserved.
The same N-terminal region in mDia1 was also identified in a two hybrid screen as an interacting site for PKD2 [67], a cation channel that when mutated accounts for ~ 15% of type 2 forms of autosomal dominant polycystic kidney disease [68–70]. PKD2 co-localized with mDia1 to the mitotic spindle and down-regulation of mDia1 by RNAi resulted in the loss of PKD2 specific signal from the spindle of dividing HeLa cells. As mDia1 depletion also significantly decreased intracellular Ca2+ release, the authors speculate that PKD2-mDia1 interaction may be critical for Ca2+ release during mitosis. Whether this process affects MT dynamics or the function of the spindle is yet to be tested.
mDia1 has also been implicated in early mitotic spindle organization where it appears to function downstream of the Rho-GEF Lfc and Rho. Lfc associates with MTs both in interphase and mitosis [71, 72]. Inactivation of Lfc by siRNA, Rho inhibition by C3 treatment and injection of an anti-mDia1 antibody into prophase Rat-2 cells all induced mitotic arrest and a disorganized mitotic spindle [72]. A detailed analysis revealed that the spindle was rotated about the z-axis and that kinetochores failed to attach properly to spindle MTs [72]. mDia1 has not been specifically localized to kinetochores, so it is possible that the lack of attachment of kinetochores to spindle MTs may be an indirect effect of mDia1 on tethering of astral MTs to the cortex. Interfering with mDia1 did not affect spindle function in all cell types (for example, no effect was seen in HeLa cells). While this may reflect cell-type-dependent differences in the requirement for mDia1 or mDia3 (see below), it clearly raises the need for further studies on the role of formins in early spindle function.
Another formin, mDia3 has been specifically localized at kinetochores and been implicated in the proper assembly of the metaphase spindle and the congression of chromosomes to the metaphase plate [73]. mDia3 is regulated by Cdc42 and this lead to its identification as the likely downstream effector mediating Cdc42 effects on spindle assembly early in mitosis. In HeLa cells, mDia3 localization to kinetochores is Cdc42-dependent and dominant negative Cdc42 or mDia3 (but not mDia1) knockdown both caused inhibition of prometaphase progression as revealed by chromosome misalignment and maintenance of Mad2 localization to kinetochores of mis-aligned chromosomes [73]. Inhibition of mDia3 ultimately resulted in chromosome missegregation and aneuploidy [73]. Importantly, Y. Mao and colleagues have found that mDia3 knockdown in HeLa cells reduced stabilization of kinetochore MTs and this accounted for the inappropriate chromosome attachment to spindle MTs (Y. Mao, Columbia University, unpublished observations).
These studies point to a role for MT stabilization by mDia3 in spindle assembly and suggest that the activity of the mDia family of formins toward MTs is active in both interphase and mitotic cells. It will be interesting to determine whether EB1 and APC, which contribute to MT stabilization by mDia1 in interphase cells, also contribute to MT capture by mDia3 at kinetochores during mitosis. Both EB1 and APC localize to kinetochores in a MT dependent manner [74–76] and chromosome segregation defects have been reported in mammalian cultured cells carrying a truncated version of APC [77, 78] or in cells depleted of APC or EB1 expression [76, 79, 80]. More recently, a study combining imaging, immunodepletion and in vitro reconstitution experiments in Xenopus egg extracts has uncovered a role for APC and EB1 in mediating kinetochore attachment to spindle MTs in cooperation with the mitotic checkpoint kinase BubR1 [81].
Aside from their well known role in the regulation of cytokinesis through the assembly of the actin-based cleavage furrow, formins may also contribute to cytokinesis through their action on MTs. In cultured mammalian cells all three mDia isoforms localize during cytokinesis to the central spindle, a structure composed of interdigitating stable MTs [82, 83]. A recent study has found that mDia2 silencing by siRNA, but not that of mDia1 or mDia3, resulted in an increased number of binucleated NIH3T3 cells and that the block in cytokinesis was largely caused by inhibition of cleavage furrow ingression due to the induction of actin contraction at ectopic sites on the cell cortex [83]. The defect in cytokinesis completion was not rescued by an mDia2 mutant deficient in actin nucleation, strongly arguing that mDia2 contributes to actin assembly necessary for the cytokinetic ring [83]. This study does not formally rule out the possibility that formins may also position the furrow through their action on MTs. Also, whereas this study concluded that only mDia2 was involved in cytokinesis in NIH3T3 cells an earlier report showed that mDia1 was also important for cytokinesis in NIH3T3 cells (82), suggesting that more than one formin may be involved in this process. Independent observations of furrow positioning have reported the existence of another population of stable MTs extending past chromosomes, that are believed to stimulate cleavage furrow formation by interacting with the cell cortex and/or by delivering signals to the cell cortex [84]. The nature of the MT-stabilizing or MT-delivered signals has not yet been identified but it is interesting to note that mDia2 also localizes to the equatorial region where this class of MTs is predicted to be stabilized [83].
6. MT stabilization in virus infection
Formins are also critical mediators of viral infections through their action on MT dynamics. Analysis of the mechanism of infection of human herpes virus 8 (HHV-8 also known as Kaposi’s sarcoma-associated herpesvirus) in cultured human fibroblasts has shown that Rho GTPases and their effector hDia2 (ortholog of mDia3) are regulators of MT-dependent trafficking of HHV-8 capsids towards the nucleus where the viral DNA replication occurs [85]. In this study, a transient reorganization of the MT cytoskeleton into bundles and upregulation in the levels of acetylated MTs was observed early upon viral entry [85]. The induction in MT hyperacetylation by HHV-8 was comparable to induction by LPA and dependent on RhoA activation [85]. In addition, co-precipitation analyses of hDia2 with Rho-GTP as early as 1 minute post infection demonstrated that HHV-8 increases the formation of Rho-hDia2 complexes [85]. These observations identified hDia2 as the RhoA effector in the regulation of MT dynamics by HVV-8 infection and suggested a functional parallelism with the MT stabilization pathway that is activated in migrating cells downstream of LPA signaling [15, 40, 45, 48, 86].
In migrating fibroblasts, localized stabilization of MTs by Rho-mDia was shown to be dependent on integrin signaling and on the activation of FAK, suggesting the existence of an “adhesion checkpoint” for MT stability [86]. In this system, FAK was necessary for the activation of mDia by Rho, and appeared to regulate the formation and localization of lipid raft domains to mediate mDia activation [86]. Interestingly, HHV-8 also utilizes α3β1 integrin molecules to infect adherent cells through the binding of its virion envelope glycoprotein gB [87] and induction of FAK phosphorylation was shown to be induced upon infection [88]. HHV-8 entry is significantly reduced in FAK-null fibroblasts [89] and in the presence of inhibitors of the cellular tyrosine kinases, Src and PI3-kinases, all molecules known to be activated downstream of FAK [88, 90, 91]. A recent study conducted in human primary endothelial cells has also underscored an essential role for lipid rafts in the post-binding and entry stages of infection of HVV-8 and in the HVV-8 induction of MT stability by virus entry [92].
Vaccinia virus-induced cell motility is another example where mDia is a key effector in viral modulation of MT dynamics. In the life cycle of the virus, inhibition of RhoA was shown to be necessary for virus morphogenesis and induction of host-cell motility [93]. The viral protein F11L was shown to be responsible for RhoA inhibition by directly competing with ROCK for RhoA interaction [93]. Loss of F11L expression by siRNA resulted in the accumulation of immature intracellular enveloped virus and in inefficient virus spreading from cell to cell [93]. In a follow-up study, vaccinia virus infection was found to be marked by a significant increase in the dynamics of MTs of the infected cells at 8–10 hours post-infection, resulting in loss of stability of MTs largely concentrated at the cell periphery [94]. A combination of imaging and loss of function analyses confirmed that this induction of MT dynamics was dependent on Rho inhibition in virus infected cells and that mDia1 depletion mirrored the loss of MT stability observed upon infection [94]. By analogy with the Rho-mDia dependent stabilization of a subset of MTs in migrating cells, these results suggest that F11L may be acting as a negative regulator of this process to allow a successful infection cycle. Also, a marked disorganization of cell adhesion has been reported during infection [94], suggesting that the same Integrin-Rho-mDia dependent “cell adhesion checkpoint” described in fibroblasts needs to be shut down for efficient viral infection.
7. Centrosome reorientation in migrating cells and T-cells
The centrosome is reoriented along the axis of polarity in migrating cells, in T cells interacting with their targets and possibly in neurons during axon outgrowth [95]. The reorientation of the centrosome biases MTs nucleated at the centrosome to align with the axis of polarity and this in turn allows MT directed transport to contribute to the establishment and maintenance of cell polarity. Studies have implicated the GTPase Cdc42, the Par polarity proteins and the MT motor dynein in centrosome orientation [95–97]. Recently, the possibility that formins also contribute to centrosome orientation in some types of migrating cells [46, 98] and in T cells [99] has been raised. In glioma cells and in mouse embryo fibroblasts (MEFs) siRNA depletion of mDia1 prevents both the formation of stable Glu MTs and the orientation of the centrosome to a position between the nucleus and the leading edge [46, 98]. In contrast, siRNA depletion of mDia1 has no detectable effect on centrosome orientation in NIH3T3 cells (Andrés-Delgado L., Bartolini F., Gundersen G.G., unpublished observations). This result suggests that in NIH3T3 cells the formation of stable Glu MTs and centrosome reorientation are separately regulated during polarization. This is consistent with the observation that in NIH3T3 cells centrosome reorientation occurs when formation of stable Glu MTs is blocked experimentally [100, 45, 48]. Centrosome reorientation in migrating fibroblasts was recently shown to require the concerted activity of both the actin and MT cytoskeletons [101]. Actin retrograde flow moved the nucleus away from the leading edge while MTs, dynein and Par proteins were required to maintain the centrosome in the cell center as the nucleus moved rearward [101]. Thus, it is unclear whether in glioma cells or MEFs mDia1 contributes to centrosome reorientation through its effects on MTs, or whether its actin activity might be involved. The possibility that formins contribute to actin retrograde flow or nuclear movement has not been explored.
When T cells engage their targets, an “immunological synapse” of clustered T cell receptors (TCRs), signaling molecules and actin forms at the site of interaction. T cells also reorient their centrosome to a position between the nucleus and immune synapse and this reorientation is important for prolonged TCR signaling as well as for lytic functions of cytotoxic T cells and natural killer cells [102]. The centrosome is oriented by movement toward the immunological synapse in a process that involves MTs and dynein [103, 104]. Two formins, mDia1 and FMNL1, appear to be essential for the reorientation of the centrosome in T cells [99]. Both proteins localize in a “cloud” around the centrosome, suggesting they may play a direct role in centrosome positioning. It will be interesting to see whether these formins act directly on the centrosome, centrosome MTs and/or the actin cytoskeleton to reorient the centrosome in T cells and other cells.
8. Formin-mediated cross –talk between actin and MTs in Yeast
In both budding (S.cerevisiae) and fission (S.pombe) yeast there is no evidence that formins directly regulate MTs. Yet, there is abundant evidence that formins mediate cross-talk between the actin and MT cytoskeletons in these organisms. Interestingly in budding yeast, formins indirectly affect the cytoplasmic MT array through their action on actin cables, whereas in fission yeast, MTs appear to affect the localization of actin cables by controlling the localization of formins.
The budding yeast formin Bni1 was the first formin shown to stimulate actin polymerization and accumulated evidence suggests that Bni1 and a second formin Bnr1 are responsible for formation of actin cables originating from the bud tip and neck, respectively [105–108]. These formins play an important role in cytokinesis and in directing Myo2 and Myo4 (type V myosins) cargos toward the bud for polarized cell growth. The actin cables generated by these formins orient cytoplasmic MTs toward the bud and in doing so, orient the nucleus and spindle along the mother-bud axis [109]. The mechanism responsible for the MT orientation along the actin cables has been established through a series of elegant studies. Myo4 binds to the adaptor protein Kar9 which in turn binds to Bim1, the homolog of the mammalian +TIP, EB1 [110–114]. MTs reaching the bud undergo a controlled shrinkage while remaining attached thus drawing the nucleus toward the bud [115]. Kip3, a plus-end kinesin, which is also able to depolymerize MTs, may contribute to this controlled shrinkage of the MT [116–118]. It is still unclear what anchors MTs to the cortex to resist the pulling force exerted by the shrinking MTs.
The fission yeast formin, for3, plays an analogous role in actin cable formation but functions during interphase at the two ends of the cell where growth occurs [119, 120]. MTs are involved in for3 function by delivering the +TIPs tea1 (a kelch-repeat protein) and tea4 (an SH3-domain protein) to cell tips at sites where they recruit the formin for3 [121]. Deletion of for3 disrupts cable formation and delocalizes actin patches. MT dynamics are unchanged in for3 deleted cells, but MT organization is altered and there is an increased number of MT bundles, an effect that may be the result of increased cell rounding in the for3Δ cells [119, 120]. Overexpression of a truncated for3 protein, lacking presumed regulatory domains, causes MTs to redistribute to the periphery in large bundles [120]. In mammalian cells, this phenotype is observed with a number of overexpressed MAPs leaving open the possibility that for3 may more directly affect MTs in fission yeast.
9. Formin-mediated cross –talk between actin and MTs in higher eukaryotes
In contrast to the situation in yeast, in mammalian and insect cells, formins directly regulate both actin filaments and MTs. When the Rho GTPases that regulate formins become activated in mammalian and insect cells, it follows that both cytoskeletal systems will respond simultaneously. Although the responses will be temporally related, they need not be spatially coupled. This seems to be the case in the formation of actin filaments and stable MTs in mammalian fibroblasts by mDia [18]. Nonetheless, there is also evidence that some formins can cross-link actin filaments and MTs, and in these cases, formin activation in cells may orchestrate the formation of complex, mixed cytoskeletal systems.
The first observation of a role of formins in orchestrating MTs and actin filaments in mammalian cells came from a study on the effects of an active mDia1 fragment, containing only the FH1 and FH2 regions, on cell shape [122]. Expression of this active mDia1 fragment in HeLa cells not only induced actin bundles and cell elongation, but also resulted in the alignment of MTs in parallel to the actin bundles and the long axis of the cell [122]. Aligned MTs and actin filaments frequently terminated in mDia1-enriched patches near focal adhesions and drug treatments showed that MTs but not actin filaments were responsible for the change in cell shape [122]. Interestingly, mutations of three lysines in the C-terminal region of the FH2 domain of the mDia1 construct inhibited both cell elongation and MT alignment, suggesting that the coalignment of MTs and actin is dependent on an intact FH2 region [122]. Coalignment of actin filaments and MTs was also reported in the case of FHOD1, another diaphanous-related formin that was originally shown to bind to Rac1 and induce the formation of bundled actin fibers to which the formin localizes [123–125]. Expression of constitutively active FHOD1 induced longitudinal alignment of actin fibers and MTs in Hela cells but not NIH3T3. This phenotype required the integrity of both the FH1 and FH2 regions suggesting that actin filament-MT coalignment required the actin nucleation activity of FHOD1 [125]. Both MTs and actin bundles were required for FHOD1-induced cell elongation, suggesting that in the case of FHOD1 MT coordination with actin fibers is a necessary condition to bring about changes in cell shape [125]. MT and actin co-alignment was also observed in HeLa cells overexpressing INF1[19]. Whether these phenotypes reflect actual cross-linking of actin and MTs mediated by the formin is unclear: in some cases (FHOD1) the formin is localized on one of the filaments, whereas in other cases (mDia1 and INF1), there is no significant accumulation of the formin along the filaments. Also, it is unclear why coalignment is observed in some cells and not others.
Capu, the Drosophila homolog of mammalian formin 2, has been proposed to directly mediate the crosstalk between the MT and the actin cytoskeletons in developing fly oocytes [17]. Mutations in Capu and the actin nucleator Spire were previously known to result in the premature onset of cytoplasmic streaming in Drosophila oocytes, a process known to be dependent on both microfilaments and MTs [126]. Notably, Capu, the actin nucleator spire and Rho, which binds Spire and Capu [17], all accumulated in the region where MTs and microfilaments overlap in the oocyte [17]. The isolation of a weak allele of Capu with a mutation in the FH2 domain (_capu_2F) that does not affect its actin nucleation activity, indicated that this region may be necessary for the coordination between actin filaments and MTs that is required in ooplasmic streaming [17]. Interestingly, in vitro MTs/microfilaments crosslinking assays using purified components showed that the FH2 domain of Capu can crosslink F-actin and MTs and that this activity is abrogated by the _capu_2F mutation in the FH2 domain, suggesting that the same mutation may be working by inhibiting crosslinking activity also in vivo [17]. The same study provided evidence that Spire can modulate the crosslinking activity of Capu in vitro and that this regulation is itself dependent on Rho1, leading to a model in which Rho1, Spire and Capu are all functionally coordinated to orchestrate the temporally regulated crosstalk between the actin and MT cytoskletons during germline development in Drosophila [17]. A recent paper has challenged this model by providing evidence for a role of Spire and Capu in oocyte streaming through their coordinated ability to form an isotropic actin mesh in the ooplasm that regulates the polarized arrangement of MTs rather than through a direct role on MTs [127]. According to the authors, loss of the actin mesh by mutations in either Capu or Spire would only indirectly affect the polarization of the MT network by interfering with the speed of kinesin-dependent organelle trafficking, a process known to determine the rate and the extent of ooplasmic flow [127].
A type of formin mediating crosstalk in mammalian cells that is more similar to that observed in fission yeast is observed during macrophage phagocytosis. Macrophages have at least two pathways for phagocytosis. One is activated by immunoglobulin binding to surface Fc receptor and is dependent on Rac and Cdc42 signaling and actin polymerization. The other is activated by complement binding to complement receptor and is dependent on Rho signaling, MTs and to a lesser extent, actin [128, 129]. During phagocytosis, mDia1 accumulates around the forming phagocytic cup and is necessary to stimulate actin polymerization at this site [130, 131]. A recent study revealed that the MT +TIP, CLIP-170, was important for efficient phagocytosis of complement-coated particles and for localizing actin and mDia1 near phagocytic sites [132]. CLIP-170 interacted directly with the FH2 domain of mDia1 in vitro without affecting mDia1’s ability to stimulate actin polymerization. During phagocytosis, the interaction between CLIP-170 and mDia1 was reduced, suggesting that CLIP-170 may contribute to mDia1 localization, but not activity. Interestingly, the Cdc42 and Rac effector IQGAP1 has also been implicated in localizing mDia1 (through a direct interaction with the DID domain of mDia1) during phagocytosis and in the leading edge of migrating cells [133]. IQGAP1 forms a complex with CLIP-170 [134], raising the possibility that at least during phagocytosis mDia1, CLIP-170 and IQGAP1 function together.
Concluding remarks
Growing evidence supports a role for higher eukaryotic formins in regulating the dynamics of the MT cytoskeleton independently of their role in actin assembly. Both FH2 and unique domains outside of the actin nucleation region have been mapped as discrete regions that interact with MTs. Initial studies show that formins affect MTs in a manner that is different from conventional MAPs and instead of conveying stability by enhancing MT growth, enhance MT stability by reducing subunit disassembly. Formins regulate MT stability in both interphase and mitotic cells and seem to mediate the effects of Rho GTPases on MTs. In addition, formins orchestrate complex actin and MT arrays, suggesting that formins may function in many cellular processes requiring cytoskeletal coordination.
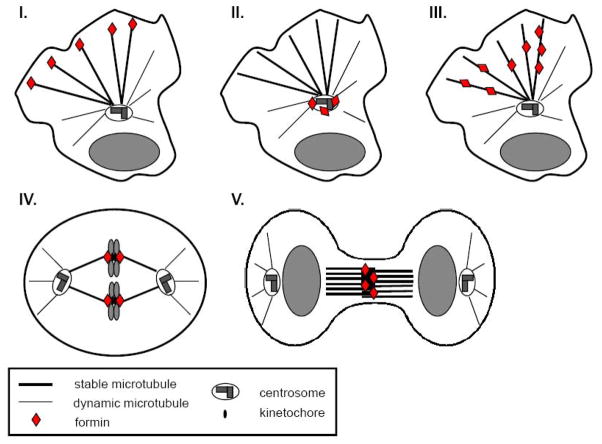
Figure 4. Schematic indicating cellular sites of formin action on MT arrays.
In migratory cells formins can regulate MTs at their plus ends (I), in proximity of the centrosome (II), or through binding along MTs (III). In cell division formins act on MT plus ends at kinetochore during mitosis (IV) and may act on central spindle MTs during cytokinesis (V).
Table 1.
Association of formins with the MT cytoskeleton and their action on MT arrays in vivo.
| Formin | Rho GTPase | MT effect | MT localization | Cellular process | Refs |
|---|---|---|---|---|---|
| mDia1 | Rho | stabilization | Glu MT plus ends | cell migration | [15,44,45,48] |
| alignment with actin | unknown | cell elongation | [122] | ||
| centrosome orientation | centrosome | cell migration, T cell-target interaction | [46,99] | ||
| spindle position | midbody, spindle MTs | mitosis, cytokinesis | [65,72,82,83] | ||
| stabilization | unknown | virus infection | [94] | ||
| mDia2 | Rho, Cdc42 | stabilization * | interphase MTs | cell migration | [15,18,45,48] |
| unknown | midbody | cytokinesis | [82,83] | ||
| stabilization | unknown | virus infection | [85] | ||
| mDia3 | Cdc42 | stabilization | kinetochore | mitosis | [73] |
| FHOD1 | Rac1 | alignment with actin * | unknown | cell elongation | [125] |
| INF1 | unknown | stabilization | interphase MTs | unknown | [19] |
| alignment with actin * | interphase MTs | unknown | [19] | ||
| Formin-1b | unknown | unknown | interphase MTs | unknown | [16] |
| Cappuccino | Rho | alignment with actin | unknown | cytoplasmic streaming in oocytes | [17] |
| Bni1 | Rho1,3 | orientation | unknown | nucleus/spindle orientation | [135–137] |
| For3 | Rho3 | orientation | unknown | polarized cell growth | [119,120] |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Downing KH, Nogales E. Tubulin and microtubule structure. Curr Opin Cell Biol. 1998;10(1):16–22. doi: 10.1016/s0955-0674(98)80082-3. [DOI] [PubMed] [Google Scholar]
- 2.Downing KH, Nogales E. New insights into microtubule structure and function from the atomic model of tubulin. Eur Biophys J. 1998;27(5):431–6. doi: 10.1007/s002490050153. [DOI] [PubMed] [Google Scholar]
- 3.Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391(6663):199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 4.Nogales E, et al. High-resolution model of the microtubule. Cell. 1999;96(1):79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- 5.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312(5991):237–42. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 6.Cassimeris L, Spittle C. Regulation of microtubule-associated proteins. Int Rev Cytol. 2001;210:163–226. doi: 10.1016/s0074-7696(01)10006-9. [DOI] [PubMed] [Google Scholar]
- 7.Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19(1):31–5. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Al-Bassam J, et al. Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure. 2007;15(3):355–62. doi: 10.1016/j.str.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Brouhard GJ, et al. XMAP215 is a processive microtubule polymerase. Cell. 2008;132(1):79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moores CA, et al. A mechanism for microtubule depolymerization by KinI kinesins. Mol Cell. 2002;9(4):903–9. doi: 10.1016/s1097-2765(02)00503-8. [DOI] [PubMed] [Google Scholar]
- 11.Moores CA, et al. Regulation of KinI kinesin ATPase activity by binding to the microtubule lattice. J Cell Biol. 2003;163(5):963–71. doi: 10.1083/jcb.200304034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shipley K, et al. Structure of a kinesin microtubule depolymerization machine. Embo J. 2004;23(7):1422–32. doi: 10.1038/sj.emboj.7600165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9(4):309–22. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 14.Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45(3):329–42. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- 15.Palazzo AF, et al. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat Cell Biol. 2001;3(8):723–9. doi: 10.1038/35087035. [DOI] [PubMed] [Google Scholar]
- 16.Zhou F, Leder P, Martin SS. Formin-1 protein associates with microtubules through a peptide domain encoded by exon-2. Exp Cell Res. 2006;312(7):1119–26. doi: 10.1016/j.yexcr.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Rosales-Nieves AE, et al. Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat Cell Biol. 2006;8(4):367–76. doi: 10.1038/ncb1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartolini F, et al. The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J Cell Biol. 2008;181(3):523–36. doi: 10.1083/jcb.200709029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young KG, et al. INF1 is a novel microtubule-associated formin. Mol Biol Cell. 2008;19(12):5168–80. doi: 10.1091/mbc.E08-05-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dettenhofer M, Zhou F, Leder P. Formin 1-isoform IV deficient cells exhibit defects in cell spreading and focal adhesion formation. PLoS ONE. 2008;3(6):e2497. doi: 10.1371/journal.pone.0002497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulze E, Kirschner M. Dynamic and stable populations of microtubules in cells. J Cell Biol. 1987;104(2):277–88. doi: 10.1083/jcb.104.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webster DR, et al. Differential turnover of tyrosinated and detyrosinated microtubules. Proc Natl Acad Sci U S A. 1987;84(24):9040–4. doi: 10.1073/pnas.84.24.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gundersen GG, Kalnoski MH, Bulinski JC. Distinct populations of microtubules: tyrosinated and nontyrosinated alpha tubulin are distributed differently in vivo. Cell. 1984;38(3):779–89. doi: 10.1016/0092-8674(84)90273-3. [DOI] [PubMed] [Google Scholar]
- 24.Gundersen GG, Khawaja S, Bulinski JC. Postpolymerization detyrosination of alpha-tubulin: a mechanism for subcellular differentiation of microtubules. J Cell Biol. 1987;105(1):251–64. doi: 10.1083/jcb.105.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kreis TE. Microtubules containing detyrosinated tubulin are less dynamic. Embo J. 1987;6(9):2597–606. doi: 10.1002/j.1460-2075.1987.tb02550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol. 1987;104(2):289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gundersen GG, Bulinski JC. Microtubule arrays in differentiated cells contain elevated levels of a post-translationally modified form of tubulin. Eur J Cell Biol. 1986;42(2):288–94. [PubMed] [Google Scholar]
- 28.Gundersen GG, Khawaja S, Bulinski JC. Generation of a stable, posttranslationally modified microtubule array is an early event in myogenic differentiation. J Cell Biol. 1989;109(5):2275–88. doi: 10.1083/jcb.109.5.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bulinski JC, Gundersen GG. Stabilization of post-translational modification of microtubules during cellular morphogenesis. Bioessays. 1991;13(6):285–93. doi: 10.1002/bies.950130605. [DOI] [PubMed] [Google Scholar]
- 30.Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4(12):938–47. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- 31.Liao G, Gundersen GG. Kinesin is a candidate for cross-bridging microtubules and intermediate filaments. Selective binding of kinesin to detyrosinated tubulin and vimentin. J Biol Chem. 1998;273(16):9797–803. doi: 10.1074/jbc.273.16.9797. [DOI] [PubMed] [Google Scholar]
- 32.Dunn S, et al. Differential trafficking of Kif5c on tyrosinated and detyrosinated microtubules in live cells. J Cell Sci. 2008;121(Pt 7):1085–95. doi: 10.1242/jcs.026492. [DOI] [PubMed] [Google Scholar]
- 33.Reed NA, et al. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16(21):2166–72. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Gurland G, Gundersen GG. Stable, detyrosinated microtubules function to localize vimentin intermediate filaments in fibroblasts. J Cell Biol. 1995;131(5):1275–90. doi: 10.1083/jcb.131.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreitzer G, Liao G, Gundersen GG. Detyrosination of tubulin regulates the interaction of intermediate filaments with microtubules in vivo via a kinesin-dependent mechanism. Mol Biol Cell. 1999;10(4):1105–18. doi: 10.1091/mbc.10.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin SX, Gundersen GG, Maxfield FR. Export from pericentriolar endocytic recycling compartment to cell surface depends on stable, detyrosinated (glu) microtubules and kinesin. Mol Biol Cell. 2002;13(1):96–109. doi: 10.1091/mbc.01-05-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Infante AS, et al. Detyrosinated (Glu) microtubules are stabilized by an ATP-sensitive plus-end cap. J Cell Sci. 2000;113( Pt 22):3907–19. doi: 10.1242/jcs.113.22.3907. [DOI] [PubMed] [Google Scholar]
- 38.Gundersen GG, Bulinski JC. Selective stabilization of microtubules oriented toward the direction of cell migration. Proc Natl Acad Sci U S A. 1988;85(16):5946–50. doi: 10.1073/pnas.85.16.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagasaki T, Gundersen GG. Depletion of lysophosphatidic acid triggers a loss of oriented detyrosinated microtubules in motile fibroblasts. J Cell Sci. 1996;109( Pt 10):2461–9. doi: 10.1242/jcs.109.10.2461. [DOI] [PubMed] [Google Scholar]
- 40.Cook TA, Nagasaki T, Gundersen GG. Rho guanosine triphosphatase mediates the selective stabilization of microtubules induced by lysophosphatidic acid. J Cell Biol. 1998;141(1):175–85. doi: 10.1083/jcb.141.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ridley AJ, Hall A. Signal transduction pathways regulating Rho-mediated stress fibre formation: requirement for a tyrosine kinase. Embo J. 1994;13(11):2600–10. doi: 10.1002/j.1460-2075.1994.tb06550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kranenburg O, et al. Activation of RhoA by lysophosphatidic acid and Galpha12/13 subunits in neuronal cells: induction of neurite retraction. Mol Biol Cell. 1999;10(6):1851–7. doi: 10.1091/mbc.10.6.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sahai E, Alberts AS, Treisman R. RhoA effector mutants reveal distinct effector pathways for cytoskeletal reorganization, SRF activation and transformation. Embo J. 1998;17(5):1350–61. doi: 10.1093/emboj/17.5.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goulimari P, et al. Galpha12/13 is essential for directed cell migration and localized Rho-Dia1 function. J Biol Chem. 2005;280(51):42242–51. doi: 10.1074/jbc.M508690200. [DOI] [PubMed] [Google Scholar]
- 45.Eng CH, Huckaba TM, Gundersen GG. The formin mDia regulates GSK3beta through novel PKCs to promote microtubule stabilization but not MTOC reorientation in migrating fibroblasts. Mol Biol Cell. 2006;17(12):5004–16. doi: 10.1091/mbc.E05-10-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamana N, et al. The Rho-mDia1 pathway regulates cell polarity and focal adhesion turnover in migrating cells through mobilizing Apc and c-Src. Mol Cell Biol. 2006;26(18):6844–58. doi: 10.1128/MCB.00283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kodama A, et al. ACF7: an essential integrator of microtubule dynamics. Cell. 2003;115(3):343–54. doi: 10.1016/s0092-8674(03)00813-4. [DOI] [PubMed] [Google Scholar]
- 48.Wen Y, et al. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6(9):820–30. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- 49.issue, S.X.a.Y.f.t.s., 2009.
- 50.Fang X, et al. Lysophosphatidic acid is a bioactive mediator in ovarian cancer. Biochim Biophys Acta. 2002;1582(1–3):257–64. doi: 10.1016/s1388-1981(02)00179-8. [DOI] [PubMed] [Google Scholar]
- 51.Shi SH, et al. APC and GSK-3beta are involved in mPar3 targeting to the nascent axon and establishment of neuronal polarity. Curr Biol. 2004;14(22):2025–32. doi: 10.1016/j.cub.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 52.Jiang H, et al. Both the establishment and the maintenance of neuronal polarity require active mechanisms: critical roles of GSK-3beta and its upstream regulators. Cell. 2005;120(1):123–35. doi: 10.1016/j.cell.2004.12.033. [DOI] [PubMed] [Google Scholar]
- 53.Yoshimura T, et al. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell. 2005;120(1):137–49. doi: 10.1016/j.cell.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 54.Gurland G, Gundersen GG. Protein phosphatase inhibitors induce the selective breakdown of stable microtubules in fibroblasts and epithelial cells. Proc Natl Acad Sci U S A. 1993;90(19):8827–31. doi: 10.1073/pnas.90.19.8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Condeelis JS, et al. Lamellipodia in invasion. Semin Cancer Biol. 2001;11(2):119–28. doi: 10.1006/scbi.2000.0363. [DOI] [PubMed] [Google Scholar]
- 56.Zaoui K, et al. Memo-RhoA-mDia1 signaling controls microtubules, the actin network, and adhesion site formation in migrating cells. J Cell Biol. 2008;183(3):401–8. doi: 10.1083/jcb.200805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Destaing O, et al. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. J Cell Sci. 2005;118(Pt 13):2901–11. doi: 10.1242/jcs.02425. [DOI] [PubMed] [Google Scholar]
- 58.Arimura N, Kaibuchi K. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat Rev Neurosci. 2007;8(3):194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- 59.Roos J, et al. Drosophila Futsch regulates synaptic microtubule organization and is necessary for synaptic growth. Neuron. 2000;26(2):371–82. doi: 10.1016/s0896-6273(00)81170-8. [DOI] [PubMed] [Google Scholar]
- 60.Pawson C, Eaton BA, Davis GW. Formin-dependent synaptic growth: evidence that Dlar signals via Diaphanous to modulate synaptic actin and dynamic pioneer microtubules. J Neurosci. 2008;28(44):11111–23. doi: 10.1523/JNEUROSCI.0833-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Witte H, Neukirchen D, Bradke F. Microtubule stabilization specifies initial neuronal polarization. J Cell Biol. 2008;180(3):619–32. doi: 10.1083/jcb.200707042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arakawa Y, et al. Control of axon elongation via an SDF-1alpha/Rho/mDia pathway in cultured cerebellar granule neurons. J Cell Biol. 2003;161(2):381–91. doi: 10.1083/jcb.200210149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tominaga T, et al. The Rho GTPase effector protein, mDia, inhibits the DNA binding ability of the transcription factor Pax6 and changes the pattern of neurite extension in cerebellar granule cells through its binding to Pax6. J Biol Chem. 2002;277(49):47686–91. doi: 10.1074/jbc.M207539200. [DOI] [PubMed] [Google Scholar]
- 64.issue, Z.a.W.f.t.s., 2009.
- 65.Kato T, et al. Localization of a mammalian homolog of diaphanous, mDia1, to the mitotic spindle in HeLa cells. J Cell Sci. 2001;114(Pt 4):775–84. doi: 10.1242/jcs.114.4.775. [DOI] [PubMed] [Google Scholar]
- 66.Petersen J, et al. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J Cell Biol. 1998;141(5):1217–28. doi: 10.1083/jcb.141.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rundle DR, Gorbsky G, Tsiokas L. PKD2 interacts and co-localizes with mDia1 to mitotic spindles of dividing cells: role of mDia1 IN PKD2 localization to mitotic spindles. J Biol Chem. 2004;279(28):29728–39. doi: 10.1074/jbc.M400544200. [DOI] [PubMed] [Google Scholar]
- 68.Kimberling WJ, et al. Autosomal dominant polycystic kidney disease: localization of the second gene to chromosome 4q13–q23. Genomics. 1993;18(3):467–72. doi: 10.1016/s0888-7543(11)80001-7. [DOI] [PubMed] [Google Scholar]
- 69.Peters DJ, et al. Chromosome 4 localization of a second gene for autosomal dominant polycystic kidney disease. Nat Genet. 1993;5(4):359–62. doi: 10.1038/ng1293-359. [DOI] [PubMed] [Google Scholar]
- 70.Mochizuki T, et al. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science. 1996;272(5266):1339–42. doi: 10.1126/science.272.5266.1339. [DOI] [PubMed] [Google Scholar]
- 71.Benais-Pont G, et al. Identification of a tight junction-associated guanine nucleotide exchange factor that activates Rho and regulates paracellular permeability. J Cell Biol. 2003;160(5):729–40. doi: 10.1083/jcb.200211047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bakal CJ, et al. The Rho GTP exchange factor Lfc promotes spindle assembly in early mitosis. Proc Natl Acad Sci U S A. 2005;102(27):9529–34. doi: 10.1073/pnas.0504190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yasuda S, et al. Cdc42 and mDia3 regulate microtubule attachment to kinetochores. Nature. 2004;428(6984):767–71. doi: 10.1038/nature02452. [DOI] [PubMed] [Google Scholar]
- 74.Su LK, et al. APC binds to the novel protein EB1. Cancer Res. 1995;55(14):2972–7. [PubMed] [Google Scholar]
- 75.Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001;1(1):55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 76.Kaplan KB, et al. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nat Cell Biol. 2001;3(4):429–32. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- 77.Green RA, Kaplan KB. Chromosome instability in colorectal tumor cells is associated with defects in microtubule plus-end attachments caused by a dominant mutation in APC. J Cell Biol. 2003;163(5):949–61. doi: 10.1083/jcb.200307070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tighe A, V, Johnson L, Taylor SS. Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J Cell Sci. 2004;117(Pt 26):6339–53. doi: 10.1242/jcs.01556. [DOI] [PubMed] [Google Scholar]
- 79.Green RA, Wollman R, Kaplan KB. APC and EB1 function together in mitosis to regulate spindle dynamics and chromosome alignment. Mol Biol Cell. 2005;16(10):4609–22. doi: 10.1091/mbc.E05-03-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Draviam VM, et al. Misorientation and reduced stretching of aligned sister kinetochores promote chromosome missegregation in EB1- or APC–depleted cells. Embo J. 2006;25(12):2814–27. doi: 10.1038/sj.emboj.7601168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang J, Ahmad S, Mao Y. BubR1 and APC/EB1 cooperate to maintain metaphase chromosome alignment. J Cell Biol. 2007;178(5):773–84. doi: 10.1083/jcb.200702138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tominaga T, et al. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol Cell. 2000;5(1):13–25. doi: 10.1016/s1097-2765(00)80399-8. [DOI] [PubMed] [Google Scholar]
- 83.Watanabe S, et al. mDia2 Induces the Actin Scaffold for the Contractile Ring and Stabilizes its Position during Cytokinesis in NIH 3T3 Cells. Mol Biol Cell. 2008 doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Canman JC, et al. Determining the position of the cell division plane. Nature. 2003;424(6952):1074–8. doi: 10.1038/nature01860. [DOI] [PubMed] [Google Scholar]
- 85.Naranatt PP, et al. Kaposi’s sarcoma-associated herpesvirus modulates microtubule dynamics via RhoA-GTP-diaphanous 2 signaling and utilizes the dynein motors to deliver its DNA to the nucleus. J Virol. 2005;79(2):1191–206. doi: 10.1128/JVI.79.2.1191-1206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Palazzo AF, et al. Localized stabilization of microtubules by integrin- and FAK-facilitated Rho signaling. Science. 2004;303(5659):836–9. doi: 10.1126/science.1091325. [DOI] [PubMed] [Google Scholar]
- 87.Akula SM, et al. Integrin alpha3beta1 (CD 49c/29) is a cellular receptor for Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell. 2002;108(3):407–19. doi: 10.1016/s0092-8674(02)00628-1. [DOI] [PubMed] [Google Scholar]
- 88.Sharma-Walia N, et al. Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 envelope glycoprotein gB induces the integrin-dependent focal adhesion kinase-Src-phosphatidylinositol 3-kinase-rho GTPase signal pathways and cytoskeletal rearrangements. J Virol. 2004;78(8):4207–23. doi: 10.1128/JVI.78.8.4207-4223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Krishnan HH, et al. Focal adhesion kinase is critical for entry of Kaposi’s sarcoma-associated herpesvirus into target cells. J Virol. 2006;80(3):1167–80. doi: 10.1128/JVI.80.3.1167-1180.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Naranatt PP, et al. Kaposi’s sarcoma-associated herpesvirus induces the phosphatidylinositol 3-kinase-PKC-zeta-MEK-ERK signaling pathway in target cells early during infection: implications for infectivity. J Virol. 2003;77(2):1524–39. doi: 10.1128/JVI.77.2.1524-1539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sharma-Walia N, et al. ERK1/2 and MEK1/2 induced by Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) early during infection of target cells are essential for expression of viral genes and for establishment of infection. J Virol. 2005;79(16):10308–29. doi: 10.1128/JVI.79.16.10308-10329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Raghu H, et al. Lipid rafts of primary endothelial cells are essential for Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8-induced phosphatidylinositol 3-kinase and RhoA-GTPases critical for microtubule dynamics and nuclear delivery of viral DNA but dispensable for binding and entry. J Virol. 2007;81(15):7941–59. doi: 10.1128/JVI.02848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Valderrama F, et al. Vaccinia virus-induced cell motility requires F11L-mediated inhibition of RhoA signaling. Science. 2006;311(5759):377–81. doi: 10.1126/science.1122411. [DOI] [PubMed] [Google Scholar]
- 94.Arakawa Y, Cordeiro JV, Way M. F11L-mediated inhibition of RhoA-mDia signaling stimulates microtubule dynamics during vaccinia virus infection. Cell Host Microbe. 2007;1(3):213–26. doi: 10.1016/j.chom.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 95.Li R, Gundersen GG. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat Rev Mol Cell Biol. 2008;9(11):860–73. doi: 10.1038/nrm2522. [DOI] [PubMed] [Google Scholar]
- 96.Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106(4):489–98. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- 97.Etienne-Manneville S, Hall A. Cell polarity: Par6, aPKC and cytoskeletal crosstalk. Curr Opin Cell Biol. 2003;15(1):67–72. doi: 10.1016/s0955-0674(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 98.Goulimari P, et al. LARG and mDia1 link Galpha12/13 to cell polarity and microtubule dynamics. Mol Biol Cell. 2008;19(1):30–40. doi: 10.1091/mbc.E06-11-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gomez TS, et al. Formins regulate the actin-related protein 2/3 complex-independent polarization of the centrosome to the immunological synapse. Immunity. 2007;26(2):177–90. doi: 10.1016/j.immuni.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Palazzo AF, et al. Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr Biol. 2001;11(19):1536–41. doi: 10.1016/s0960-9822(01)00475-4. [DOI] [PubMed] [Google Scholar]
- 101.Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121(3):451–63. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 102.Sancho D, et al. Regulation of microtubule-organizing center orientation and actomyosin cytoskeleton rearrangement during immune interactions. Immunol Rev. 2002;189:84–97. doi: 10.1034/j.1600-065x.2002.18908.x. [DOI] [PubMed] [Google Scholar]
- 103.Kuhn JR, Poenie M. Dynamic polarization of the microtubule cytoskeleton during CTL-mediated killing. Immunity. 2002;16(1):111–21. doi: 10.1016/s1074-7613(02)00262-5. [DOI] [PubMed] [Google Scholar]
- 104.Combs J, et al. Recruitment of dynein to the Jurkat immunological synapse. Proc Natl Acad Sci U S A. 2006;103(40):14883–8. doi: 10.1073/pnas.0600914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Evangelista M, et al. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol. 2002;4(1):32–41. doi: 10.1038/ncb718. [DOI] [PubMed] [Google Scholar]
- 106.Sagot I, et al. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4(8):626–31. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- 107.Sagot I, Klee SK, Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat Cell Biol. 2002;4(1):42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- 108.Pruyne D, et al. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297(5581):612–5. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- 109.Pruyne D, et al. Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol. 2004;20:559–91. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- 110.Yin H, et al. Myosin V orientates the mitotic spindle in yeast. Nature. 2000;406(6799):1013–5. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
- 111.Schott D, Huffaker T, Bretscher A. Microfilaments and microtubules: the news from yeast. Curr Opin Microbiol. 2002;5(6):564–74. doi: 10.1016/s1369-5274(02)00369-7. [DOI] [PubMed] [Google Scholar]
- 112.Hwang E, et al. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J Cell Biol. 2003;161(3):483–8. doi: 10.1083/jcb.200302030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Korinek WS, et al. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 2000;287(5461):2257–9. doi: 10.1126/science.287.5461.2257. [DOI] [PubMed] [Google Scholar]
- 114.Bloom K. It’s a kar9ochore to capture microtubules. Nat Cell Biol. 2000;2(6):E96–8. doi: 10.1038/35014089. [DOI] [PubMed] [Google Scholar]
- 115.Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149(4):863–74. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.DeZwaan TM, et al. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J Cell Biol. 1997;138(5):1023–40. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee L, et al. Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J Cell Biol. 1999;144(5):947–61. doi: 10.1083/jcb.144.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gupta ML, Jr, et al. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8(9):913–23. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- 119.Feierbach B, Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr Biol. 2001;11(21):1656–65. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- 120.Nakano K, et al. The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J Cell Sci. 2002;115(Pt 23):4629–39. doi: 10.1242/jcs.00150. [DOI] [PubMed] [Google Scholar]
- 121.Martin SG, et al. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev Cell. 2005;8(4):479–91. doi: 10.1016/j.devcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 122.Ishizaki T, et al. Coordination of microtubules and the actin cytoskeleton by the Rho effector mDia1. Nat Cell Biol. 2001;3(1):8–14. doi: 10.1038/35050598. [DOI] [PubMed] [Google Scholar]
- 123.Westendorf JJ. The formin/diaphanous-related protein, FHOS, interacts with Rac1 and activates transcription from the serum response element. J Biol Chem. 2001;276(49):46453–9. doi: 10.1074/jbc.M105162200. [DOI] [PubMed] [Google Scholar]
- 124.Gasteier JE, et al. Activation of the Rac-binding partner FHOD1 induces actin stress fibers via a ROCK-dependent mechanism. J Biol Chem. 2003;278(40):38902–12. doi: 10.1074/jbc.M306229200. [DOI] [PubMed] [Google Scholar]
- 125.Gasteier JE, et al. FHOD1 coordinates actin filament and microtubule alignment to mediate cell elongation. Exp Cell Res. 2005;306(1):192–202. doi: 10.1016/j.yexcr.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 126.Theurkauf WE. Premature microtubule-dependent cytoplasmic streaming in cappuccino and spire mutant oocytes. Science. 1994;265(5181):2093–6. doi: 10.1126/science.8091233. [DOI] [PubMed] [Google Scholar]
- 127.Dahlgaard K, et al. Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev Cell. 2007;13(4):539–53. doi: 10.1016/j.devcel.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Castellano F, Chavrier P, Caron E. Actin dynamics during phagocytosis. Semin Immunol. 2001;13(6):347–55. doi: 10.1006/smim.2001.0331. [DOI] [PubMed] [Google Scholar]
- 129.Caron E, Hall A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 1998;282(5394):1717–21. doi: 10.1126/science.282.5394.1717. [DOI] [PubMed] [Google Scholar]
- 130.Colucci-Guyon E, et al. A role for mammalian diaphanous-related formins in complement receptor (CR3)-mediated phagocytosis in macrophages. Curr Biol. 2005;15(22):2007–12. doi: 10.1016/j.cub.2005.09.051. [DOI] [PubMed] [Google Scholar]
- 131.Brandt DT, et al. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178(2):193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lewkowicz E, et al. The microtubule-binding protein CLIP-170 coordinates mDia1 and actin reorganization during CR3-mediated phagocytosis. J Cell Biol. 2008;183(7):1287–98. doi: 10.1083/jcb.200807023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Brandt DT, Grosse R. Get to grips: steering local actin dynamics with IQGAPs. EMBO Rep. 2007;8(11):1019–23. doi: 10.1038/sj.embor.7401089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fukata M, et al. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell. 2002;109(7):873–85. doi: 10.1016/s0092-8674(02)00800-0. [DOI] [PubMed] [Google Scholar]
- 135.Lee L, et al. Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J Cell Biol. 1999;144(5):947–61. doi: 10.1083/jcb.144.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Miller RK, Matheos D, Rose MD. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J Cell Biol. 1999;144(5):963–75. doi: 10.1083/jcb.144.5.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fujiwara T, et al. Bni1p regulates microtubule-dependent nuclear migration through the actin cytoskeleton in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(12):8016–27. doi: 10.1128/mcb.19.12.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chhabra ES, Higgs HN. INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem. 2006;281(36):26754–67. doi: 10.1074/jbc.M604666200. [DOI] [PubMed] [Google Scholar]
- 139.Chhabra ES, Ramabhadran V, Gerber SA, Higgs HN. INF2 is an endoplasmic reticulum-associated formin protein. J Cell Sci. 2009;122(9):1430–40. doi: 10.1242/jcs.040691. [DOI] [PMC free article] [PubMed] [Google Scholar]