An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors (original) (raw)
. Author manuscript; available in PMC: 2010 Jul 29.
Published in final edited form as: Nat Genet. 2008 May;40(5):499–507. doi: 10.1038/ng.127
Abstract
Cancer cells possess traits reminiscent of those ascribed to normal stem cells. It is unclear, however, whether these phenotypic similarities reflect the activity of common molecular pathways. Here we analyze the enrichment patterns of gene sets associated with embryonic stem (ES) cell identity in the expression profiles of various human tumor types. Strikingly, histologically poorly differentiated tumors display preferential overexpression of genes normally enriched in ES cells, combined with underexpression of Polycomb-regulated genes. Moreover, expression of activation targets of Nanog, Oct4, Sox2 and c-Myc is observed more frequently in poorly differentiated tumors than in well-differentiated tumors. In breast cancers this ES-like signature is associated with high-grade ER-negative tumors, often of the basal-like subtype, and with poor clinical outcome. The ES signature is also present in poorly differentiated glioblastomas and bladder carcinomas. We identify a subset of ES-associated transcription regulators that are preferentially expressed in poorly differentiated tumors. Our results reveal a novel link between genes associated with ES cell identity and the histopathological traits of tumors, and support the possibility that these genes contribute to stem cell-like phenotypes displayed by many tumors.
Introduction
The apparent parallels between tumor cells and normal stem cells have generated great interest in the possible links between these two classes of cells. The hallmark traits of stem cells – self-renewal and differentiation capacity – are mirrored by the high proliferative capacity and phenotypic plasticity of tumor cells1. Moreover, tumor cells often lack the terminal differentiation traits possessed by their normal counterparts. These parallels have given rise to the hypothesis that tumors often arise from undifferentiated stem/progenitor cells, or alternatively, that cancer cells can undergo progressive de-differentiation during their development1-3. Additionally, some have proposed that cancer stem cells – a subpopulation of cancer cells possessing tumor-initiating capability – are derived from normal stem cells1,4. While certain regulators of stem cell function have been implicated in cancer pathogenesis2, a broad description of the activity of stem cell-associated regulatory networks in tumors is lacking.
The differentiation level (or grade) of human tumors is assessed routinely in the clinic, poorly differentiated tumors generally exhibiting the worst prognoses. However, this classification is based on histopathological criteria, and the underlying molecular pathways controlling tumor differentiation are poorly described. Moreover, it is not known whether a lack of histological differentiation markers in tumor cells reflects the possession of stem cell-like traits. A number of oncogenes are known to interfere with normal cell differentiation, myc being a notable example5,6, and such oncogenes could also affect tumor cell differentiation. The recent demonstration that adult fibroblasts can be reprogrammed into pluripotent ES-like cells7,8 raises the possibility that the combined expression of stem cell-associated factors and specific oncogenes could also induce a non-differentiated state in cancer cells. In fact, ectopic expression of Oct4, a central determinant of ES cell identity, is sufficient to induce tumor growth in the adult mouse9, and Polycomb complex components central to stem cell function, such as Bmi1 and Ezh2, are also oncogenic10.
These observations suggest that the regulatory networks controlling the function of stem cells may also be active in certain tumors. These networks have been the focus of much recent interest, and progress has been made particularly in the study of ES cells11. Current evidence indicates that some of the key regulators of ES cell identity – Oct4, Sox2 and Nanog – are expressed only in specific human cancer types12-15. Nevertheless, it is conceivable that different regulators – possibly paralogs of these ES cell factors – may activate stem-cell regulatory networks in other tumor types. By necessity, detection of the activity of such complex networks must rely on expression analysis of many genes in multiple cancer samples, rather than on the presence or absence of individual factors16,17.
Here, we employed recently developed gene set expression analysis methods18 to assess whether the expression signatures and regulatory networks that define human ES cell identity are also active in human tumors. Our results reveal previously undescribed links between tumor pathogenesis and the ES cell state.
Results
Gene sets reflecting ES cell identity
We wished to examine whether the regulatory networks that function in ES cells are also active in tumors. Since different subsets of the many genes involved in these networks may be active in different individual tumors, we reasoned that expression analyses of gene sets (groups of genes related through a common function, pathway or other property) could prove more revealing than single-gene analyses16,17,19. Accordingly, we set out to collect gene sets that represent the core expression signature of ES cells and reflect the activity of the regulatory pathways associated with their identity. We extracted these sets directly from published studies without modifying their contents. In order to eliminate effects of inter-species differences20,21, we used only gene sets identified in human cells.
We compiled 13 partially overlapping gene sets, which fall into four groups (Table 1, Supplementary Table 1): (i) ES expressed genes: two sets of genes overexpressed in ES cells compared to other cells and tissues according to a multi-study compilation and meta-analysis22. (ii) Nanog, Oct4 and Sox2 (NOS) targets: four sets of genes whose promoters are bound and activated in human ES cells by each of these regulators of ES cell identity, or co-activated by all three23, and an additional set (NOS TFs) including a subset of NOS activation targets encoding transcription regulators. (iii) Polycomb targets: four sets representing genes bound by the Polycomb Repressive Complex 2 (PRC2) in human ES cells24. (iv) Myc targets: two sets of genes bound and activated by c-Myc, identified in two independent studies25,26. We hypothesized that these 13 gene sets would allow the definition of an ES-cell expression signature and, in turn, the detection of this signature in tumors.
Table 1.
Gene sets associated with human ES cell identity.
| Group | Gene Set | No. of Genes | Source | Reference |
|---|---|---|---|---|
| ES expressed | ES exp1 | 380 | Overexpressed in hES cells according to 5 or more out of 20 profiling studies | Assou et al. 2006 |
| ES exp2 | 40 | Overexpressed in hES cells according to a meta-analysis of 8 profiling studies | Assou et al. 2006 | |
| NOS targets | Nanog targets | 988 | ChIP-array of Nanog in hES cells, activated genes only | Boyer et al. 2005 |
| Oct4 targets | 290 | ChIP-array of Oct4 in hES cells, activated genes only | Boyer et al. 2005 | |
| Sox2 targets | 734 | ChIP-array of Sox2 in hES cells, activated genes only | Boyer et al. 2005 | |
| NOS targets | 179 | Overlap of above three sets | Boyer et al. 2005 | |
| NOS TFs | 37 | Transcription regulators in NOS targets set | ||
| Polycomb targets | Suz12 targets | 1040 | ChIP-array of Suz12 in hES cells | Lee et al. 2006 |
| Eed targets | 1066 | ChIP-array of Eed in hES cells | Lee et al. 2006 | |
| H3K27 bound | 1121 | ChIP-array of trimethlyated H3K27 in hES cells | Lee et al. 2006 | |
| PRC2 targets | 654 | Overlap of 3 above sets | Lee et al. 2006 | |
| Myc targets | Myc targets1 | 230 | ChIP array of c-Myc in cultured cell lines, focusing on E-box containing genes - high affinity bound subset. | Fernandez et al. 2003 |
| Myc targets2 | 775 | ChIP array of c-Myc and of Max in a Bukitt’s Lymphoma cell line, overlap set. | Li et al. 2003 |
Establishment of an ES cell gene-set enrichment pattern
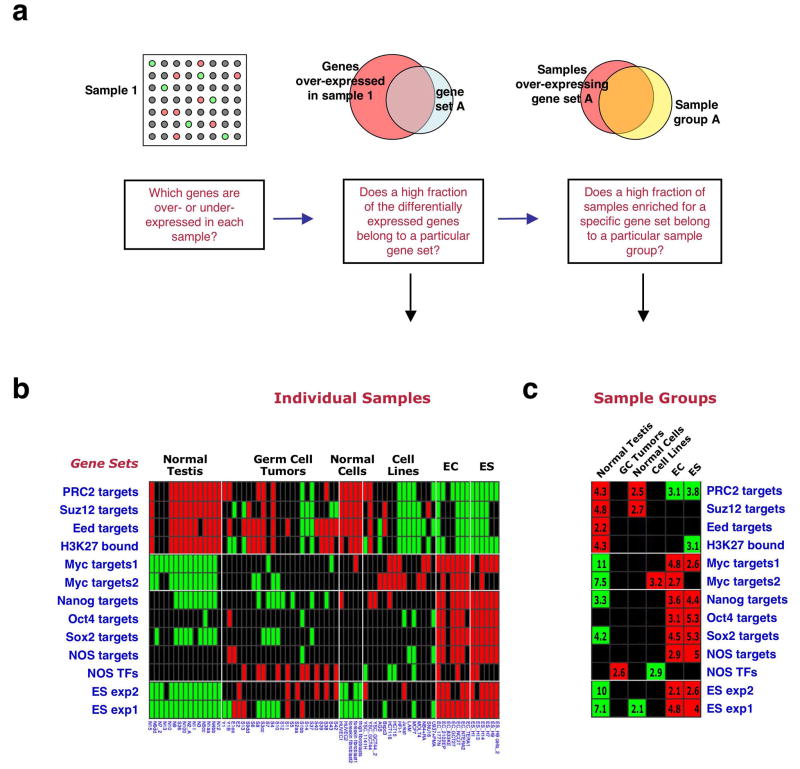
We employed a recently described computational strategy to assess the expression pattern of the above gene sets in microarray expression data18. This method tests whether the genes that are over- or underexpressed in each profiled tissue or cell line include a higher-than-randomly-expected fraction of genes from a particular gene set (Fig. 1a). A second analysis step determines whether particular sample groups (e.g., all ES cell samples) preferentially under- or overexpress particular gene sets (Fig. 1a).
Figure 1. An ES cell gene-set enrichment pattern.
(a) Analysis method of gene set enrichment pattern18. For each sample (array) genes over- and under- expressed relative to the mean across samples are scored. The fraction of these differentially expressed genes that belong to each of the tested gene sets is then calculated, and its significance over random is estimated, producing a P value. In the second step of the analysis, the over-representation of particular sample groups among the samples enriched for each gene set is assessed. (b) Gene set enrichments in ES cells compared to other cell types. Columns represent individual samples (sample annotations on bottom), sample group names are indicated above. Rows represent individual gene sets (names indicated on left). Red – gene-set enrichment for overexpression, green - gene-set enrichment for underexpression, black – no significant enrichment. (c) Enrichment pattern across sample groups. Numbers indicate _P_-values for gene set enrichment significance within sample group, in negative log, e.g., 4 symbolizes _P_=10-4.
We first analyzed the enrichment patterns of the 13 gene sets in an expression profile dataset that included 5 human ES lines, 7 embryonic carcinoma (EC) lines, and various other normal and tumorigenic cells and tissues27. This analysis revealed that the various ES-expressed and NOS-target gene sets were, as anticipated, preferentially overexpressed in the ES and EC samples, while the Polycomb-target sets were underexpressed in these samples (Fig. 1b). The highest levels of gene set enrichment were observed for the ES exp1 set (P values ranging from P =10-28 to 10-53 in individual ES lines), which includes 380 genes shown to be overexpressed in ES cells in 5 profiling studies or more22. Myc-target gene sets showed lower enrichment levels in the ES lines. Interestingly, normal testis samples and normal cultured cells displayed an enrichment pattern opposite to that of ES cells, while the various tumor cell lines did not display a consistent gene set enrichment pattern (Fig. 1b). These findings were also evident in the sample-group analysis (Fig. 1c). The NOS TFs set was the only set enriched in the male germ-cell tumor group (Fig. 1b,c), despite the fact that only half of the genes in this small set were included in the arrays used in this study27. This interesting finding corresponds to the known activity of ES cell-associated transcription regulators in germ-cell tumors12,13.
These analyses indicated that the combined enrichment pattern of the 13 gene sets can be viewed as a gene-set based expression signature, and that a specific enrichment pattern/signature associated with ES cell identity can be defined.
Poorly differentiated breast tumors display an ES-like expression signature
We proceeded to test whether the ES-associated gene sets were enriched in human tumors. We generated a compendium of data from six published studies of breast cancer expression profiles28-33 comprising a total of 1,211 tumors. Available tumor annotations were derived from the original publications, and included tumor grade, size, estrogen receptor (ER) expression, metastasis to lymph nodes or to distant organs, and disease-associated mortality.
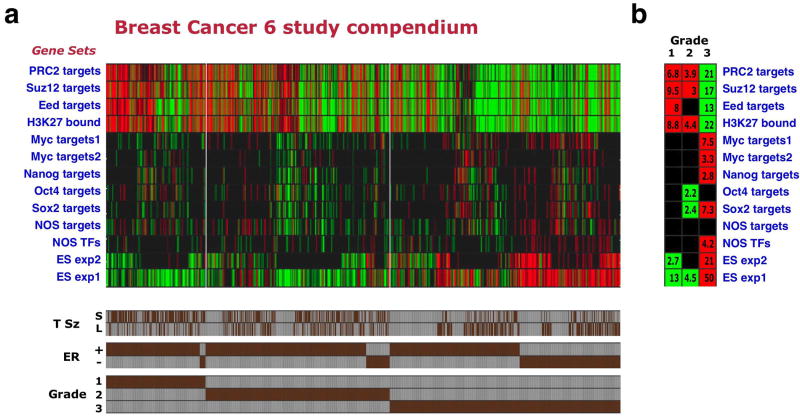
Strikingly, analysis of the enrichment patterns of the 13 gene sets across the compendium samples revealed that poorly differentiated (grade 3) breast tumors display an enrichment pattern resembling that observed in ES cells. This included underexpression of Polycomb target gene sets and overexpression of ES-expressed sets, Myc-target gene sets, and some of the NOS-target gene sets (Fig. 2a,b). Conversely, the well-differentiated, grade 1 tumors displayed an opposite pattern. The most significant enrichment levels were observed for the ES-expressed and Polycomb-target sets (_P_=10-13 to _P_=10-50 for sample group enrichments in grade 3 tumors, Fig 2b). Interestingly, the NOS TFs gene set was among those enriched in high-grade breast cancers (P =6.3×10-5). We observed this overall enrichment pattern with limited variation when analyzing the individual studies comprising the compendium separately (data not shown).
Figure 2. Poorly differentiated breast cancers display an ES-like enrichment pattern.
(a) Enrichment pattern of indicated gene sets (rows) across 1,211 breast cancer samples included in 6 profiling studies (columns). Red/green – significantly over- or underexpressed gene sets. Shown are 1,089 tumors for which both ER status and grade annotations were available. Brown bars (bottom) indicate individual tumor annotations for grade, ER status, and tumor size (T Sz), where available. S – tumor smaller than 2cm across (pathological T1), L – tumor larger than 2cm (pathological T2 or T3). (b) Gene set enrichments in the breast compendium tumors stratified by tumor grade.
These findings revealed that tumors that are defined as poorly differentiated according to purely histopathological criteria, in fact display a molecular similarity to ES cells, as reflected by the coordinated up- or down-regulation of gene sets associated with ES cell identity. This similarity suggests that the neoplastic cells within such tumors are closer in their traits to normal undifferentiated stem cells than are cells in well-differentiated tumors.
Association of the ES signature with ER-status, tumor size, and intrinsic subtype
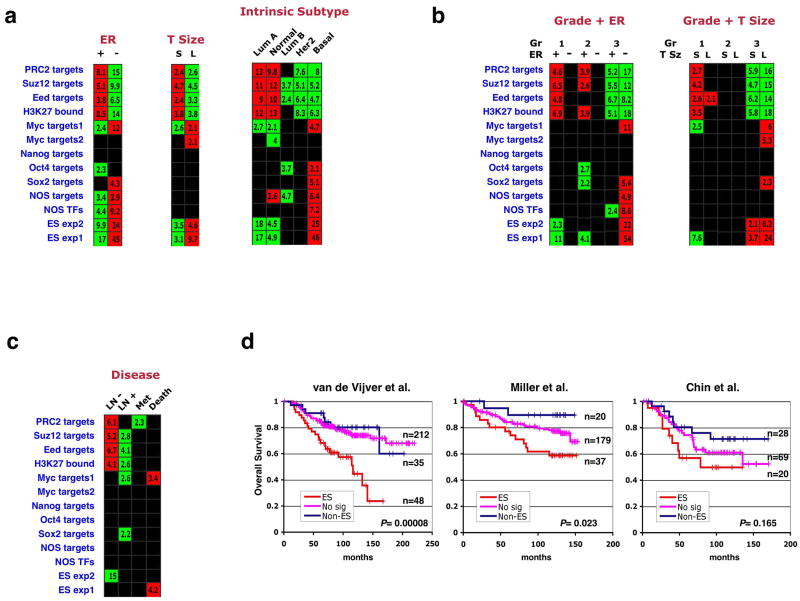
We examined whether enrichments of the 13 gene sets would be observed when the breast tumors in our compendium were stratified according to criteria other than tumor grade. Our analysis indicated that tumors lacking expression of the estrogen receptor (ER-negative) displayed an ES-like enrichment pattern when compared to the receptor-expressing (ER-positive) tumors (Fig. 2a, 3a). Most ER-negative tumors are poorly differentiated, indicating an overlap between these two categories. We therefore analyzed gene-set enrichments in the tumors stratified into six groups representing all possible combinations of these two parameters (Figure 3b). This analysis indicated that, even within the same ER status, high-grade tumors were more ES-like in their enrichment pattern than were low-grade tumors; similarly, ER-negative tumors within the same grade were more ES-like than their ER-positive counterparts (Figure 3b). The ES signature is thus independently associated with both tumor grade and ER status, with grade 3/ER-negative tumors showing the overall most significant gene-set enrichments. Subtraction of genes closely associated with ER-status did not significantly affect this enrichment pattern (Supplementary Figure 2c).
Figure 3. Association of the ES signature with ER status, tumor size, intrinsic subtype, and prognostic outcome in breast cancers.
(a) Gene set enrichments in the breast compendium tumors stratified by ER status, tumor size (T Size) or intrinsic subtype. Stratification for the latter parameter was done by employing an expression-profile based classification method35,48. S – small, L – large, Lum – luminal. (b) Samples were divided into six groups representing different combinations of tumor grade and ER-status, and enrichment of gene sets was tested across these groups (left). A similar analysis was performed for grade and tumor size (right). (c) Enrichments in tumors stratified by lymph-node metastasis absence (LN-) or presence (LN+), distant metastasis (Met), or disease-induced mortality (Death). (d) Kaplan-Meier analyses of overall survival in patients included in three of the five studies analyzed. Patients showing both overexpression of the ES exp1 set and underexpression of the PRC2 targets set were labeled as ES (red), those showing the reversed pattern were labeled as Non-ES (blue), and the remainder were labeled as No signature (No sig, pink). P values indicate significance of survival difference between the ES patient group and all other patients.
Interestingly, tumors of larger size at the time of diagnosis (more than 2 cm diameter) were also more likely to possess the ES signature compared to smaller tumors, even within a given grade (Fig. 3a,b). These results could suggest that tumors de-differentiate as they grow; alternatively, it is possible that poorly differentiated tumors are initially detected at larger sizes due to their enhanced growth rates.
Perou and colleagues have defined five “intrinsic subtypes” of breast cancer on the basis of tumor expression profiles: normal-like, luminal type A, luminal type B, HER2-like**,** and basal-like34,35. We classified the tumors in our compendium using this method and then tested whether the 13 gene sets displayed enrichments in particular subtypes. Strikingly, the basal-like tumors showed an ES-like signature with highly significant gene set enrichments (Fig. 3a), while at the other extreme, luminal type A tumors displayed the opposite pattern. Basal-like tumors are mostly grade 3 and ER-negative, possessing a high proliferation rate and a high nucleus-to-cytoplasm volume ratio36,37. Recent histological analysis revealed that these tumors often express markers of both the luminal and the myoepithelial/basal lineages present in the normal mammary duct37, suggesting that they arise from a still-unidentified, mammary stem/progenitor cell3. The association between the basal-like subtype and the ES signature suggests that, when compared with other breast cancer subtypes, these basaloid tumors may possess traits rendering them more similar to normal stem cells.
The ES signature is associated with poor prognosis
Analysis of gene-set enrichments in the breast tumors stratified according to clinical progression parameters (lymph node and distant metastasis, overall survival) revealed only weak associations of the ES-associated sets with poor outcome (Figure 3c). To assess whether presence of the ES signature is associated with poor prognosis in a more detailed manner, we performed Kaplan-Meier analyses of overall survival for patients included in five independent studies28,30,31,33,38. Tumors that displayed both overexpression of the ES exp1 set and underexpression of the PRC2 targets set were labeled as possessing the ES signature for this purpose. Patients carrying this signature displayed worse survival than the remaining patients, with significance levels varying from study to study (Figure 3d). A meta-analysis of survival data from the five studies together indicated that the mortality rate of patients carrying the ES signature was significantly higher (p<0.0001), with a hazard ratio of 2.03 (Supplementary Fig. 1). This analysis also indicated that the ES signature provides prognostic information beyond that provided by tumor grade, and is predictive of poor outcome even when patients of the basal-subtype are excluded. Thus, possession of the ES signature is indicative of aggressive tumor behavior in vivo.
The ES signature is not a direct reflection of proliferation rates
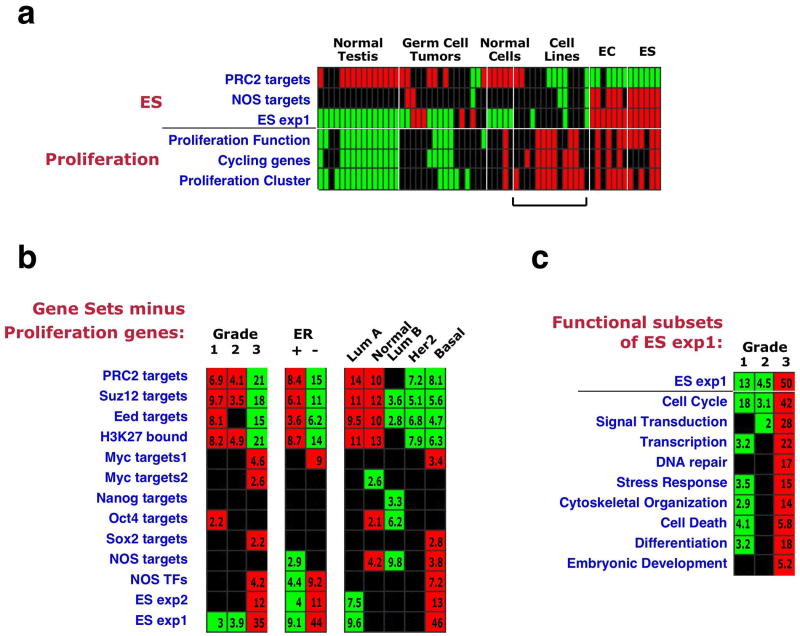
Breast cancer grade is determined using metrics for duct formation, nuclear atypia, and mitotic index; poorly differentiated tumors therefore often contain more proliferating cells than do well-differentiated tumors. Since ES cells are characterized by a high proliferation rate39, we wished to assess the contribution of proliferation-related genes to the observed gene set enrichments. We collected three different sets of proliferation genes (Supplementary Table 3): genes functionally involved in proliferation (based on Gene Ontology (GO)), genes displaying cell-cycle stage-specific expression40, and genes belonging to a “Proliferation Cluster” defined in human breast tumor expression data35. We first examined the enrichment pattern of these sets in ES cells relative to other cell types. Interestingly, the proliferation-associated sets were overexpressed in ES and EC samples, and, also, in most cultured tumor cell lines (Fig. 4a). This enrichment pattern contrasted with that displayed by the ES gene sets, which were not overexpressed in the tumor cell lines despite the rapid proliferation of the latter (Fig. 4a). This finding indicates that the ES gene sets specifically reflect an ES-like phenotype, rather than a general state of rapid proliferation
Figure 4. Contribution of proliferation-associated genes to the ES signature.
(a) Enrichment patterns in individual samples of ES cells and other cell types27 of the indicated ES gene sets (as in Fig. 1), and of three Proliferation gene sets: Proliferation Function - genes functionally associated with cell proliferation (compiled from several GO categories), Cycling Genes – genes showing cell-cycle stage-specific expression40, Proliferation Cluster – defined in tumor expression data35. Bar indicates difference in enrichment pattern between ES gene sets and proliferation gene sets in cultured tumor cell lines. (b) Gene set enrichment patterns across grade, ER-status and intrinsic subtypes after subtraction of the Proliferation Function genes from all gene sets. Subtraction of the two other proliferation sets is shown in Supplementary Fig. 2. (c) Enrichment pattern across grade of different subsets of the ES exp1 gene set, based on their cellular function (GO).
Next, we generated three amended versions of our original gene sets, from which genes included in each of the proliferation sets were removed. Analysis of the enrichment patterns of the modified gene sets revealed that most of the sets still displayed highly significant enrichments (Fig. 4b, Supplementary Fig. 2a,b), indicating that the ES signature can be detected in high grade, ER-negative and basal tumors even without the influence of proliferation-associated genes. However, for some gene sets, including the Nanog targets, Oct4 targets and Sox2 targets, the association with tumor grade was dependent to a greater extent on the inclusion of proliferation genes (Fig. 4b, Supplementary Fig. 2a,b).
Lastly, we extracted subsets of genes from the ES exp1 gene set, each associated with a specific cellular function, according to GO annotations (Fig. 4c). Strikingly, in addition to the Cell Cycle subset, all other functional subsets tested were significantly overexpressed in high-grade cancers as well (Fig. 4c). These combined results indicate that while proliferation-related genes are an inherent part of the ES signature, many other genes, involved in a variety of other, distinct cellular functions, also contribute to this signature and to its enrichment in specific tumors.
The ES signature appears in poorly differentiated cancers of various types
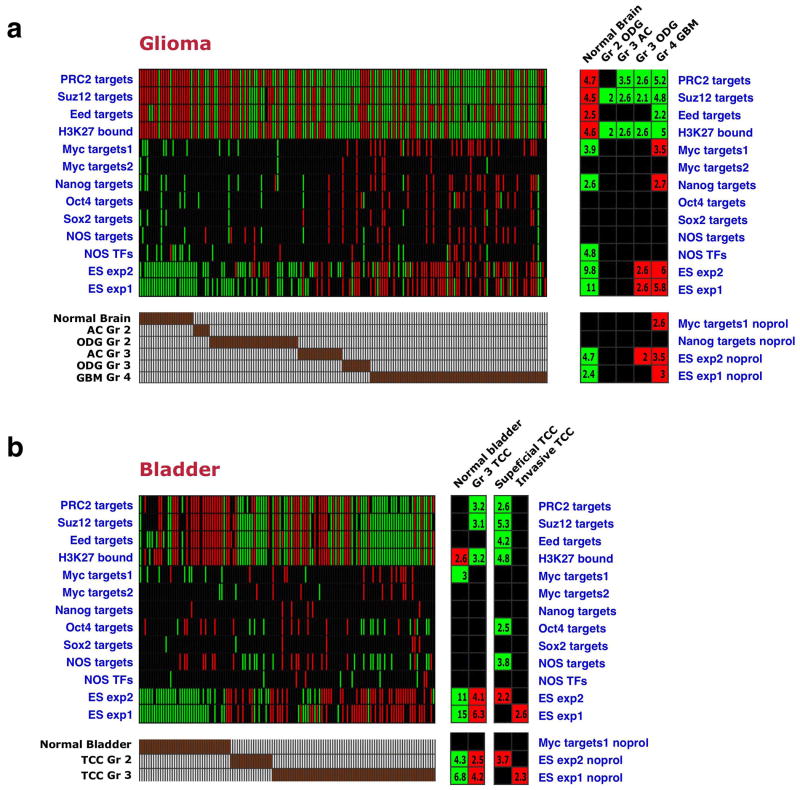
We examined whether the ES signature was present in tumors arising in tissues other than the breast. Analysis of the expression profiles of 157 gliomas41 revealed a striking correlation between tumor grade and the presence of the ES-like signature (Fig. 5a): grade 4 glioblastomas, which represent the most aggressive subtype of glioma, displayed significant gene-set enrichments corresponding to the ES signature, while the other glioma types included in this study (oligodendrogliomas and astrocytomas of grades 2 and 3) displayed lower enrichment levels for this signature in a manner that correlated with their lower grades. Normal brain tissue showed opposite enrichments (Fig. 5a), although a direct comparison is difficult in this case, due to the sampling of multiple distinct cell types.
Figure 5. ES signature in high-grade glioblastomas and bladder carcinomas.
(a) Gene set enrichment pattern across 157 normal brain and glioma samples41 of various subtypes (bottom). Gene sets subtracted for proliferation genes are indicated as noprol. ODG – oligodendroglioma, AC- astrocytoma, GBM – glioblastoma multiforme. (b) Gene set enrichment pattern across normal bladder samples and grade 2 and 3 transitional cell carcinomas (TCC)42. Invasive TCCs represent a tumor stage more advanced than superficial tumors.
We also analyzed an expression dataset of bladder carcinomas, which included normal urinary tract samples as well as grade 2 and 3 transitional cell carcinomas42. Here, as well, the high-grade tumors displayed an ES-like gene set enrichment pattern (Fig. 5b). Other parameters, such as superficial vs. invasive, did not show a strong correlation with the presence or absence of the ES signature (Fig. 5b). These results together indicate that an ES-like signature is present in poorly differentiated cancers arising in various tissues from distinct cells-of-origin.
The ES signature is not preferentially associated with the tumor-initiating fraction of breast cancer cells
Cancer stem cells, a subfraction of neoplastic cells possessing tumor-initiating capability, have recently been described in solid and hematopoietic tumors4. In breast cancers, a CD44high/CD24low tumor-initiating population has been identified, and the expression profiles of this population and of the non-tumor-initiating population (from 3 individual tumors) have recently been reported43. Since cancer stem cells have been suggested to possess stem cell-like traits3,4 we examined the behavior of the ES gene sets in these samples. Interestingly, the ES signature was not consistently associated with either the tumor-initiating or non-tumor-initiating population (Supplementary Fig. 3). This finding does not support a notion by which the tumor-initiating cells provide a high contribution to the ES signature observed in high-grade tumors. However, the small number of samples in this study does not allow definitive conclusions. We noted, however, that the CD24 gene is in fact highly expressed in ES cells22 and is also preferentially expressed in poorly-differentiated, ER-negative breast tumors (Supplementary Fig. 4c).
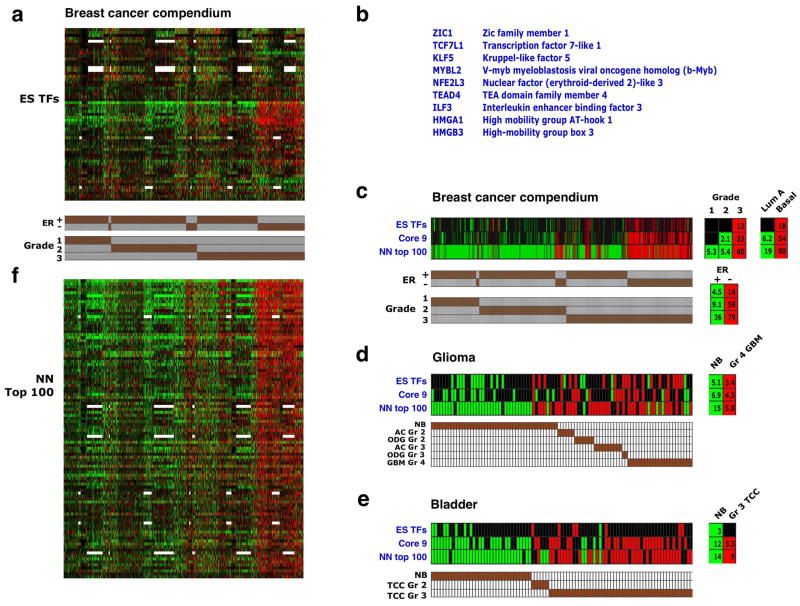
A set of ES cell-associated transcription regulators is expressed in poorly differentiated cancers
We hypothesized that among the genes expressed in ES cells, those encoding transcription regulators could provide an important contribution to tumor phenotypes. To identify such candidates, we extracted 68 genes encoding transcription regulators from the two ES Exp sets and the NOS TFs set (Supplementary Table 4). Hierarchical clustering across our breast cancer compendium revealed that a subset of 9 of these genes were preferentially and coordinately overexpressed in the high grade, ER-negative tumors (Fig. 6a,b, Supplementary Fig. 4a,b). Interestingly, several genes previously associated with adult stem/progenitor function, with ES and tumor cell proliferation, and/or with cancer progression were included in this “Core 9” subset. These included KLF5, which can replace KLF4 in somatic cell reprogramming44, TCF7L1, the ortholog of mouse Tcf3, which plays a pivotal role in skin stem cells45, and TEAD4, a co-factor of Yap1 in Hippo pathway signaling46.
Figure 6. A core set of ES-associated transcription factors is overexpressed in high-grade tumors.
(a) Expression pattern of 59 genes encoding ES-associated transcription regulators (rows) across the breast cancer compendium samples (columns), sorted by grade and ER status (indicated in bottom). 12 additional genes in the ES TFs set were not represented in most of the arrays used are therefore not shown. Red/green – two-fold or higher over- or underexpression, respectively. White – missing data. (b) Core set of 9 closely correlated ES transcription regulators, as determined by the pvclust method (Supplementary Figure 4b). (c) Gene set enrichment in the breast cancer compendium samples of the 68 ES-associated transcription regulators (ES TFs), the Core 9 gene subset (Core 9), and top ranking 100 genes in the nearest neighbor expression correlation analysis (NN top 100) – see panel f. Shown are enrichments in individual tumors and in tumors stratified by grade, ER status and intrinsic subtype. Only samples showing enrichment for at least one set are presented. (d) Analysis as in C in glioma samples. NB – normal brain, other annotations as in Figure 5a. (e) Analysis as in C in bladder carcinoma samples. NB – normal bladder, other annotations as in Figure 5b. (f) 1,700 transcription regulators in the human genome were ranked according to the similarity of their expression pattern in the breast cancer compendium to the expression of the Core 9 gene cluster (nearest neighbor analysis). Shown are the top-ranking 100 genes, in rank order (top down).
Examination of the enrichment pattern of the Core 9 genes, when defined as a distinct gene set, revealed that they are preferentially overexpressed not only in high grade, ER-negative and basal-like breast cancers (Fig. 6c), but also in high-grade glioblastomas and bladder carcinomas (Fig. 6d,e). These findings indicate that specific transcriptional regulators normally active in ES cells are often overexpressed in poorly differentiated tumors arising in distinct tissues. Notably, the known central regulators of ES identity – Nanog, Oct4 and Sox2, as well as Stat3 and Lin28 – are not broadly expressed in high-grade breast cancers (Supplementary Fig. 4a) and therefore are not included in the Core 9 set.
We next ranked all the genes encoding known and putative transcription regulators in the human genome by the degree of correlation of their expression with that of the Core 9 genes in the breast cancer compendium. The top ranked 100 genes (Supplementary Table 5) were generally enriched in high-grade cancers (Fig. 6c-f). These highly correlated genes included members of families encoding developmental regulators, such as Sox and Ets-domain proteins, as well as factors associated with proliferation, such as E2F1, c-Myc, and FoxM1. Importantly, the Polycomb complex components Ezh2 and Eed were also included in this list, indicating that high Polycomb activity goes hand-in-hand with expression of ES-associated transcription factors, and providing a possible mechanism for the increased repression of Polycomb targets observed in high-grade tumors. We suggest that the combined activity of these regulators contributes to generating a poorly differentiated state in tumors.
Discussion
The relationship between neoplastic cells and normal stem cells represents a question of great current interest. Some have postulated that the pathways conferring self-renewal capacity on normal stem cells may perform a similar function in cancer cells1. Examples of such appropriation of specific stem cell-associated regulators and signaling pathways by tumors have been described1,2. Here, we attempted to provide a broad view of the presence of molecular imprints of stemness in cancer, by examining the activity of gene sets associated with human ES cell identity in human tumors. Our analyses revealed an inverse relationship between the presence of an ES-like gene set enrichment signature in tumors and the degree of tumor differentiation. This finding is striking, since tumor differentiation/grade is defined by histopathological criteria, and it was unclear whether the absence of well-differentiated tissue traits would also entail a molecular similarity to an undifferentiated stem cell state. Viewed from the perspective of global gene expression patterns, our results indicate that this indeed may be the case.
We were struck by the association of the ES signature with high-grade tumors arising in distinct tissues. In the breast and brain, this signature was detected in tumor subtypes previously suggested to arise from oligopotent stem/progenitor cells37,47. Due to the currently limited characterization of most normal adult stem cells, we were unable to assess the similarity in gene expression profiles between such stem cells and ES cells. Moreover, we cannot definitively determine whether the ES signature is inherited from a stem cell-of-origin or, alternatively, is re-activated during the course of tumor progression.
Various parameters greatly affected our ability to associate gene-set enrichments with specific tumor subtypes within expression datasets; these included the number of samples profiled, tumor subtypes included, and clinical information available. The vast amount of data collected for breast cancers allowed us to derive the most definitive conclusions regarding this tumor type. Whether additional cancer types also possess the ES signature must be determined in future studies.
Proliferative capacity and the ability to self-renew are integral aspects of adult and embryonic stem cell identity. Distinct cell-cycle regulation mechanisms appear to control the high proliferation rate and truncated G1 phase typical of ES cells39. Accordingly, it is clear that genes associated with proliferation contribute to the ES signature described here and to its detection in specific tumors. The separation of proliferation from stemness is difficult, as exemplified by the dual roles of regulators such as Myc and β-catenin in both proliferation and differentiation. However, our results indicate that the combined expression patterns of the multiple genes within the analyzed gene sets reflect a complex ES-like phenotype, which goes beyond a general state of proliferation. Moreover, our findings may reflect a similarity in cell-cycle regulation between ES and cancer cells.
Breast cancer cells clearly differ from ES cells in multiple traits and do not possess their pluripotent ability; we could not infer in detail from our gene sets which specific properties are shared between ES and cancer cells. Among the activation targets of Nanog, Oct4 and Sox2, genes encoding transcription regulators were most consistently activated in high-grade breast tumors. Importantly, in germ cell tumors, which are considered closer to ES cells and often do express the NOS factors12,13,27, it is also the transcription factor-encoding NOS targets that are preferentially overexpressed. These genes may therefore represent the core of the regulatory network controlled by the NOS factors.
We identify a subgroup of transcription regulators that are highly (but not exclusively) expressed in ES cells and that are preferentially expressed in high-grade tumors. Interestingly, several of these genes are known to also function in adult stem/progenitor cells. We suggest that the degree of tumor differentiation is determined, at least in part, by the concerted activity of these factors, and in addition, that this activity contributes to aggressive tumor behavior. Further functional studies will be necessary to determine the roles of specific regulators in generating stem-like tumor phenotypes. In the longer term, detailed characterization of the stem-cell regulatory networks active in cancer is likely to yield powerful diagnostic and prognostic markers and, quite possibly, attractive targets for therapeutic intervention.
Methods
Gene set compilation
Gene sets were collected directly from indicated publications (Table 1). We included all genes for which we could convert the original gene identifiers into Entrez Gene IDs. Full gene-set lists are in Supplementary Table 1. We constructed 3 gene sets of proliferation-associated genes (Supplementary Table 3): the first including genes functionally associated with cell-cycle progression and cell division according to Gene Ontology (GO) annotations; the second, including cycling genes, was extracted from Whitfield et al.40; the third, including the tumor-based Proliferation Cluster was extracted from Hu et al.35. Where indicated, these genes were eliminated from our ES gene sets.
Expression data pre-processing
Expression data was imported from the referenced studies. Raw data was downloaded from the NCBI GEO website or from websites indicated in the original publications, and processed as described in Segal et al.18. Briefly, we first log2 transformed the expression values, and then calculated the mean expression level for each gene across all samples in a given dataset. These mean values were subtracted from all data points, such that expression was represented relative to each gene’s mean, negative values representing below-mean expression and vice versa. To construct the breast cancer compendium, we first normalized each of the 6 included studies independently, and then concatenated these sets. The compendium therefore does not represent a cross-comparison of expression levels between samples from different studies, but, rather, the over- or underexpression of each gene within the study in which it was performed. In cases where the same patients were included in more than one of the 6 studies comprising the compendium, such redundancy was eliminated so that each patient was included only once in the compendium. Specifically, patients of the Uppsala cohort analyzed in the Miller30 set were eliminated from the Sotiriou29 and Desmedt33 sets, and patients of the Oxford cohort present in the Sotiriou set were eliminated from the Desmedt set. The normalized expression data files as well as sample annotations can be found in http://jura.wi.mit.edu/bioc/benporath/.
Analysis of gene set enrichment patterns
To identify gene set enrichment patterns we used the methods described in Segal et al.18, as embedded in the Genomica software (http://genomica.weizmann.ac.il/). For each sample (array) we first scored the genes whose expression was at least 2-fold above or below the average expression level. We then assessed the fraction of over- or underexpressed genes that belong to each tested gene set, calculating a P value according to the hypergeometric distribution. This was repeated for every sample, using a threshold of P<0.05 for significant enrichment. To compare enrichment patterns across sample groups we included clinical annotations for each individual sample (e.g., grade, tumor size, ER status) derived from the original publications. For all samples showing enrichment for a particular gene set, we calculated the fraction of samples that possessed each annotation, and assigned a _P_-value according to the hypergeometric distribution. We used a more stringent threshold, P<0.01, for this calculation. In order to maintain consistent gene set enrichment significance results independently of the number of sample annotations and of the number of gene sets tested, we did not employ multiple hypothesis correction in these analyses. Heat maps showing gene set enrichments in individual samples include only those samples enriched for at least one set.
Classification of compendium samples to intrinsic subtypes
We employed the method published by Perou and colleagues to classify the 1,211 breast cancer samples in the compendium to the five intrinsic subtypes35,48. This method uses 306 classifying genes whose level of expression in each of the 5 subtypes is represented in centroids derived from a training set of breast cancers. A small number of the classifying genes were included in some of our gene sets; elimination of these genes from our gene sets did not substantially affect gene set enrichment patterns (data not shown).
Patient survival analysis
All patient survival data were extracted from the original publications. We defined individual tumors as possessing an ES signature for this purpose if they were enriched for both overexpression of the ES exp1 set and underexpression of the PRC2 targets set, and as possessing a non-ES signature if they displayed the opposite enrichments. P values were calculated using the log rank test and were calculated comparing the ES group to all other patients. Meta-analysis of the overall survival of patients from the five studies indicated was performed by combining the log hazard ratio estimated for each study, weighting by inverse estimator variance according to the random effects procedure of DerSimonian and Laird49. We eliminated patients of the Karolinska Institute cohort from the Desmedt33 patient set, to avoid overlap with other sets. Analyses were performed using R package version 0.8-2 (http://cran.r-project.org), Guido Schwarzer (2007) meta: Meta-Analysis.
Analysis of transcription factor expression
We compiled a set of 68 transcription factors and specific chromatin modifiers included in the NOS TFs, ES exp1 and ES exp2 gene sets (Supplementary Table 4). Hierarchical clustering was performed on the expression values of these genes in the breast cancer compendium data set using the R module pvclust50, with multiscale bootstrap resampling of 10,000 iterations to assess statistical significance, represented by a 1-100 score (Supplementary Fig. 4b). Clustering was performed using the average agglomeration method with correlation as the distance metric. This analysis identified a cluster of 9 genes showing coordinated expression associated with poor breast tumor differentiation. We searched for additional transcription regulators showing a similar expression pattern in the breast cancer compendium by calculating the Pearson correlation coefficient of each of 1,700 transcription regulators in the human genome to the mean expression levels of the original 9-gene cluster. Statistical significance was assessed as P<10-8 by reshuffling (106 iterations) of sample-gene associations.
Supplementary Material
Supplemenyary data
Acknowledgments
We thank Chris Fan and Charles Perou for assistance with the Intrinsic Subtype classification and the Proliferation Cluster, John Foekens for tumor data, Katherine Gurdziel and Joseph Rodriguez for bioinformatics assistance, and Yuval Dor, Thijn Brummelkamp, Wenjun Guo, Howard Cedar and Nir Friedman for reviewing of the manuscript and helpful discussions. IB is a Leukemia and Lymphoma Special Fellow, VJC was supported in part by NIH P41 HG 004059 (R. Gentleman, PI), and in part by the Whitehead Institute Bioinformatics Department (F. Lewitter, Director), AR is supported by the Burroughs Wellcome Career Award and the Scientific Interface, RAW is supported by grant NIH/NCI R01 CA078461, the Breast Cancer Research Foundation, and the Ludwig Cancer Center for Molecular Oncology at MIT.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 3.Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer. 2007;7:791–799. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- 4.Lobo NA, Shimono Y, Qian D, Clarke MF. The Biology of Cancer Stem Cells. Annu Rev Cell Dev Biol. 2007 doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 5.Andres AC, et al. Ha-ras and c-myc oncogene expression interferes with morphological and functional differentiation of mammary epithelial cells in single and double transgenic mice. Genes Dev. 1988;2:1486–1495. doi: 10.1101/gad.2.11.1486. [DOI] [PubMed] [Google Scholar]
- 6.Shachaf CM, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 10.Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- 12.Gidekel S, Pizov G, Bergman Y, Pikarsky E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell. 2003;4:361–370. doi: 10.1016/s1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 13.Santagata S, Ligon KL, Hornick JL. Embryonic stem cell transcription factor signatures in the diagnosis of primary and metastatic germ cell tumors. Am J Surg Pathol. 2007;31:836–845. doi: 10.1097/PAS.0b013e31802e708a. [DOI] [PubMed] [Google Scholar]
- 14.Li XL, et al. Expression of the SRY-related HMG box protein SOX2 in human gastric carcinoma. Int J Oncol. 2004;24:257–263. [PubMed] [Google Scholar]
- 15.Rodriguez-Pinilla SM, et al. Sox2: a possible driver of the basal-like phenotype in sporadic breast cancer. Mod Pathol. 2007;20:474–481. doi: 10.1038/modpathol.3800760. [DOI] [PubMed] [Google Scholar]
- 16.Rhodes DR, Chinnaiyan AM. Integrative analysis of the cancer transcriptome. Nat Genet. 2005;37(Suppl):S31–37. doi: 10.1038/ng1570. [DOI] [PubMed] [Google Scholar]
- 17.Segal E, Friedman N, Kaminski N, Regev A, Koller D. From signatures to models: understanding cancer using microarrays. Nat Genet. 2005;37(Suppl):S38–45. doi: 10.1038/ng1561. [DOI] [PubMed] [Google Scholar]
- 18.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 19.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 20.Wei CL, et al. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells. 2005;23:166–185. doi: 10.1634/stemcells.2004-0162. [DOI] [PubMed] [Google Scholar]
- 21.Odom DT, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Assou S, et al. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25:961–973. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez PC, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, et al. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci U S A. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sperger JM, et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci U S A. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 29.Sotiriou C, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 30.Miller LD, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chin K, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 33.Desmedt C, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 34.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Z, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen TO, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 37.Livasy CA, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 38.Pawitan Y, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 40.Whitfield ML, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun L, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 43.Liu R, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 46.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 48.Fan C, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 49.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemenyary data