Emerging colistin resistance in Salmonella enterica serovar Newport isolates from human infections - PubMed (original) (raw)
Emerging colistin resistance in Salmonella enterica serovar Newport isolates from human infections
Mohammed Elbediwi et al. Emerg Microbes Infect. 2020.
Abstract
Worldwide emergence of Salmonella enterica serovar Newport (S. Newport) infection in humans, in parallel with a significant increasing prevalence of antimicrobial resistance (AR), is a serious public health concern. However, the prevalence of S. Newport resistance in China remains largely unknown. A retrospective study of 287 S. Newport clinical isolates collected during 1997-2018 was undertaken for characterization of AR profiles using the micro-dilution assay. We found a recent emergence of colistin resistance in four Chinese clinical isolates, including _mcr_-1-positive isolates. Importantly, phylogenomic and microbiological investigations indicate multiple independent clonal transmission of colistin-resistant S. Newport isolates of different seafood origins. Our study highlights potential reservoirs for transmission of colistin resistance and suggests that the global food supply chain may facilitate this dissemination.
Keywords: Salmonella Newport; antimicrobial resistance; colistin resistance; mcr-1; seafood.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
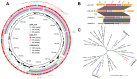
Figure 1.
Comparative sequence analysis of the _mcr_-carrying plasmids and phylogenomic features for all Chinese Salmonella Newport genomes available in the NCBI database in addition to four new colistin-resistant strains in this study. (a) Sequence comparison of two reconstructed _mcr_-1-positive plasmids from whole-genome sequence. The out layer circle refer to the CP01924.2 plasmid. (b) Genetic backbone flanking with _mcr_-1 sequence of SAL_ 276 and SAL_ 311 (CP019214.2 in E. coli isolated from sewage in China and CP033347.2 in S. Typhimurium isolated from pork in China). IS, insertion sequence; HP, hypothetical protein; PM, putative membrane; REC, recombinase. (c) Phylogenomic analysis of all available S. Newport Chinese isolates in this study, including 24 additional Chinese S. Newport isolates from different hosts retrieved from the NCBI database. The four isolates examined in this study in addition to their closely related seafood-origin isolates were clustered together in the three subclades (A, B and C). Tree scale represents the genetic distance between the isolates used to construct the tree. *These isolates were collected by US FDA, mentioning China as their place of origin.
Similar articles
- The characterization of mobile colistin resistance (mcr) genes among 33 000 Salmonella enterica genomes from routine public health surveillance in England.
Sia CM, Greig DR, Day M, Hartman H, Painset A, Doumith M, Meunier D, Jenkins C, Chattaway MA, Hopkins KL, Woodford N, Godbole G, Dallman TJ. Sia CM, et al. Microb Genom. 2020 Feb;6(2):e000331. doi: 10.1099/mgen.0.000331. Epub 2020 Jan 31. Microb Genom. 2020. PMID: 32003708 Free PMC article. - Acquisition of extended-spectrum cephalosporin- and colistin-resistant Salmonella enterica subsp. enterica serotype Newport by pilgrims during Hajj.
Olaitan AO, Dia NM, Gautret P, Benkouiten S, Belhouchat K, Drali T, Parola P, Brouqui P, Memish Z, Raoult D, Rolain JM. Olaitan AO, et al. Int J Antimicrob Agents. 2015 Jun;45(6):600-4. doi: 10.1016/j.ijantimicag.2015.01.010. Epub 2015 Feb 23. Int J Antimicrob Agents. 2015. PMID: 25769786 - Whole-Genome Sequencing of Drug-Resistant Salmonella enterica Isolates from Dairy Cattle and Humans in New York and Washington States Reveals Source and Geographic Associations.
Carroll LM, Wiedmann M, den Bakker H, Siler J, Warchocki S, Kent D, Lyalina S, Davis M, Sischo W, Besser T, Warnick LD, Pereira RV. Carroll LM, et al. Appl Environ Microbiol. 2017 May 31;83(12):e00140-17. doi: 10.1128/AEM.00140-17. Print 2017 Jun 15. Appl Environ Microbiol. 2017. PMID: 28389536 Free PMC article. - First report of mcr-1-harboring Salmonella enterica serovar Schwarzengrund isolated from poultry meat in Brazil.
Moreno LZ, Gomes VTM, Moreira J, de Oliveira CH, Peres BP, Silva APS, Thakur S, La Ragione RM, Moreno AM. Moreno LZ, et al. Diagn Microbiol Infect Dis. 2019 Apr;93(4):376-379. doi: 10.1016/j.diagmicrobio.2018.10.016. Epub 2018 Nov 1. Diagn Microbiol Infect Dis. 2019. PMID: 30477952 - Ceftriaxone-resistant Salmonella enterica serovar Newport: a case report from South India.
Sumana MN, Maheshwarappa YD, Rao MR, Deepashree R, Karthik MVSK, Shah NK. Sumana MN, et al. Front Public Health. 2024 Aug 8;12:1418221. doi: 10.3389/fpubh.2024.1418221. eCollection 2024. Front Public Health. 2024. PMID: 39175895 Free PMC article.
Cited by
- Identification of a broad-spectrum lytic Myoviridae bacteriophage using multidrug resistant Salmonella isolates from pig slaughterhouses as the indicator and its application in combating Salmonella infections.
Zhao M, Xie R, Wang S, Huang X, Yang H, Wu W, Lin L, Chen H, Fan J, Hua L, Liang W, Zhang J, Wang X, Chen H, Peng Z, Wu B. Zhao M, et al. BMC Vet Res. 2022 Jul 12;18(1):270. doi: 10.1186/s12917-022-03372-8. BMC Vet Res. 2022. PMID: 35821025 Free PMC article. - Prevalence and Genomic Investigation of Multidrug-Resistant Salmonella Isolates from Companion Animals in Hangzhou, China.
Teng L, Liao S, Zhou X, Jia C, Feng M, Pan H, Ma Z, Yue M. Teng L, et al. Antibiotics (Basel). 2022 May 5;11(5):625. doi: 10.3390/antibiotics11050625. Antibiotics (Basel). 2022. PMID: 35625269 Free PMC article. - Serotyping and Antimicrobial Resistance Profiling of Multidrug-Resistant Non-Typhoidal Salmonella from Farm Animals in Hunan, China.
Zhang Z, Li J, Zhou R, Xu Q, Qu S, Lin H, Wang Y, Li P, Zheng X. Zhang Z, et al. Antibiotics (Basel). 2023 Jul 12;12(7):1178. doi: 10.3390/antibiotics12071178. Antibiotics (Basel). 2023. PMID: 37508274 Free PMC article. - Changing Patterns of Salmonella enterica Serovar Rissen From Humans, Food Animals, and Animal-Derived Foods in China, 1995-2019.
Elbediwi M, Shi D, Biswas S, Xu X, Yue M. Elbediwi M, et al. Front Microbiol. 2021 Jul 29;12:702909. doi: 10.3389/fmicb.2021.702909. eCollection 2021. Front Microbiol. 2021. PMID: 34394048 Free PMC article. - A Burkholderia thailandensis DedA Family Membrane Protein Is Required for Proton Motive Force Dependent Lipid A Modification.
Panta PR, Doerrler WT. Panta PR, et al. Front Microbiol. 2021 Jan 12;11:618389. doi: 10.3389/fmicb.2020.618389. eCollection 2020. Front Microbiol. 2021. PMID: 33510730 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
This work was supported by the National Program on Key Research Project of China [grant number 2017YFC1600103, 2018YFD0500501, SQ2019YFE010999] as well as Horizon 2020 Framework Programme under Grant Agreement No. 861917 – SAFFI and Zhejiang Provincial Natural Science Foundation [grant number LR19C180001].
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials