Cerebellar c9RAN proteins associate with clinical and neuropathological characteristics of C9ORF72 repeat expansion carriers (original) (raw)
Abstract
Clinical and neuropathological characteristics associated with G4C2 repeat expansions in chromosome 9 open reading frame 72 (C9ORF72), the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia, are highly variable. To gain insight on the molecular basis for the heterogeneity among C9ORF72 mutation carriers, we evaluated associations between features of disease and levels of two abundantly expressed “c9RAN proteins” produced by repeat-associated non-ATG (RAN) translation of the expanded repeat. For these studies, we took a departure from traditional immunohistochemical approaches and instead employed immunoassays to quantitatively measure poly(GP) and poly(GA) levels in cerebellum, frontal cortex, motor cortex, and/or hippocampus from 55 C9ORF72 mutation carriers [12 patients with ALS, 24 with frontotemporal lobar degeneration (FTLD) and 19 with FTLD with motor neuron disease (FTLD-MND)]. We additionally investigated associations between levels of poly(GP) or poly(GA) and cognitive impairment in 15 C9ORF72 ALS patients for whom neuropsychological data were available. Among the neuroanatomical regions investigated, poly(GP) levels were highest in the cerebellum. In this same region, associations between poly(GP) and both neuropathological and clinical features were detected. Specifically, cerebellar poly(GP) levels were significantly lower in patients with ALS compared to patients with FTLD or FTLD-MND. Furthermore, cerebellar poly(GP) associated with cognitive score in our cohort of 15 patients. In the cerebellum, poly(GA) levels similarly trended lower in the ALS subgroup compared to FTLD or FTLD-MND subgroups, but no association between cerebellar poly(GA) and cognitive score was detected. Both cerebellar poly(GP) and poly(GA) associated with C9ORF72 variant 3 mRNA expression, but not variant 1 expression, repeat size, disease onset, or survival after onset. Overall, these data indicate that cerebellar abnormalities, as evidenced by poly(GP) accumulation, associate with neuropathological and clinical phenotypes, in particular cognitive impairment, of C9ORF72 mutation carriers.
Similar content being viewed by others
Introduction
Amyotrophic lateral sclerosis (ALS), the most common motor neuron disease (MND), is characterized by progressive upper and lower motor neuron weakness, typically resulting in death from ventilatory failure within three to five years of onset. In addition, up to 50 % of ALS patients develop cognitive impairments consistent with frontotemporal dementia (FTD) [15]. FTD, the clinical presentation of frontotemporal lobar degeneration (FTLD), encompasses a group of disorders characterized by changes in personality, behavior, and/or language. A proportion of FTD patients also develop ALS [33]. The clinical overlap between ALS and FTD, combined with neuropathological similarities—TDP-43 pathology is present in the majority of ALS cases and the most common pathological subtype of FTLD (FTLD-TDP)—suggest ALS and FTD are part of a disease spectrum. This concept was strengthened by the discovery that a G4C2 repeat expansion in chromosome 9 open reading frame 72 (C9ORF72) is the most common genetic cause of ALS and FTD [[10](/article/10.1007/s00401-015-1474-4#ref-CR10 "DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J et al (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72:245–256. doi: 10.1016/j.neuron.2011.09.011
"), [28](/article/10.1007/s00401-015-1474-4#ref-CR28 "Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L et al (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72:257–268. doi:
10.1016/j.neuron.2011.09.010
")\]. The molecular basis for the clinical variation among _C9ORF72_ patients is not known. In addition to TDP-43 pathology, patients with “c9FTD/ALS” develop cerebellar and hippocampal TDP-43-negative neuronal inclusions that are positive for p62, ubiquitin, and select ubiquitin-binding proteins \[[1](/article/10.1007/s00401-015-1474-4#ref-CR1 "Al-Sarraj S, King A, Troakes C, Smith B, Maekawa S, Bodi I, Rogelj B, Al-Chalabi A, Hortobagyi T, Shaw CE (2011) p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol 122:691–702. doi:
10.1007/s00401-011-0911-2
"), [5](/article/10.1007/s00401-015-1474-4#ref-CR5 "Bieniek KF, Murray ME, Rutherford NJ, Castanedes-Casey M, Dejesus-Hernandez M, Liesinger AM, Baker MC, Boylan KB, Rademakers R, Dickson DW (2013) Tau pathology in frontotemporal lobar degeneration with C9ORF72 hexanucleotide repeat expansion. Acta Neuropathol 125:289–302. doi:
10.1007/s00401-012-1048-7
"), [6](/article/10.1007/s00401-015-1474-4#ref-CR6 "Brettschneider J, Van Deerlin VM, Robinson JL, Kwong L, Lee EB, Ali YO, Safren N, Monteiro MJ, Toledo JB, Elman L et al (2012) Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol 123:825–839. doi:
10.1007/s00401-012-0970-z
"), [23](/article/10.1007/s00401-015-1474-4#ref-CR23 "Murray ME, DeJesus-Hernandez M, Rutherford NJ, Baker M, Duara R, Graff-Radford NR, Wszolek ZK, Ferman TJ, Josephs KA, Boylan KB et al (2011) Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol 122:673–690. doi:
10.1007/s00401-011-0907-y
"), [25](/article/10.1007/s00401-015-1474-4#ref-CR25 "Pikkarainen M, Hartikainen P, Alafuzoff I (2010) Ubiquitinated p62-positive, TDP-43-negative inclusions in cerebellum in frontotemporal lobar degeneration with TAR DNA binding protein 43. Neuropathology 30:197–199. doi:
10.1111/j.1440-1789.2009.01043.x
")\]. Other hallmark features of c9FTD/ALS result from the accumulation of sense G4C2 or antisense G2C4 repeat-containing RNA. These transcripts form nuclear RNA foci in cells throughout the central nervous system and are subject to repeat-associated non-ATG (RAN) translation \[[3](/article/10.1007/s00401-015-1474-4#ref-CR3 "Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R et al (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77:639–646. doi:
10.1016/j.neuron.2013.02.004
"), [10](/article/10.1007/s00401-015-1474-4#ref-CR10 "DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J et al (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72:245–256. doi:
10.1016/j.neuron.2011.09.011
"), [11](/article/10.1007/s00401-015-1474-4#ref-CR11 "Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J et al (2013) Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol 126:829–844. doi:
10.1007/s00401-013-1192-8
"), [21](/article/10.1007/s00401-015-1474-4#ref-CR21 "Mori K, Arzberger T, Grasser FA, Gijselinck I, May S, Rentzsch K, Weng SM, Schludi MH, van der Zee J, Cruts M et al (2013) Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol 126:881–893. doi:
10.1007/s00401-013-1189-3
"), [22](/article/10.1007/s00401-015-1474-4#ref-CR22 "Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C et al (2013) The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339:1335–1338. doi:
10.1126/science.1232927
"), [43](/article/10.1007/s00401-015-1474-4#ref-CR43 "Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH et al (2013) RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA 110:E4968–E4977. doi:
10.1073/pnas.1315438110
")\]. RAN translation of sense and antisense repeats, which occurs in all reading frames in the absence of an initiating ATG codon, yields five proteins of repeating dipeptides (GP, GA, GR, PR, PA), collectively referred to as c9RAN proteins. c9RAN proteins are a major component of TDP-43-negative, p62-positive inclusions \[[18](/article/10.1007/s00401-015-1474-4#ref-CR18 "Mann DM, Rollinson S, Robinson A, Bennion Callister J, Thompson JC, Snowden JS, Gendron T, Petrucelli L, Masuda-Suzukake M, Hasegawa M et al (2013) Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 1:68. doi:
10.1186/2051-5960-1-68
")\]. Indeed, c9RAN protein pathology, in particular inclusions of poly(GP) or poly(GA), is abundant in cerebellum, hippocampus, and neocortical regions \[[3](/article/10.1007/s00401-015-1474-4#ref-CR3 "Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R et al (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77:639–646. doi:
10.1016/j.neuron.2013.02.004
"), [16](/article/10.1007/s00401-015-1474-4#ref-CR16 "Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D et al (2013) Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol 126:859–879. doi:
10.1007/s00401-013-1181-y
"), [18](/article/10.1007/s00401-015-1474-4#ref-CR18 "Mann DM, Rollinson S, Robinson A, Bennion Callister J, Thompson JC, Snowden JS, Gendron T, Petrucelli L, Masuda-Suzukake M, Hasegawa M et al (2013) Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 1:68. doi:
10.1186/2051-5960-1-68
"), [21](/article/10.1007/s00401-015-1474-4#ref-CR21 "Mori K, Arzberger T, Grasser FA, Gijselinck I, May S, Rentzsch K, Weng SM, Schludi MH, van der Zee J, Cruts M et al (2013) Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol 126:881–893. doi:
10.1007/s00401-013-1189-3
"), [22](/article/10.1007/s00401-015-1474-4#ref-CR22 "Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C et al (2013) The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339:1335–1338. doi:
10.1126/science.1232927
"), [30](/article/10.1007/s00401-015-1474-4#ref-CR30 "Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, Arzberger T, Edbauer D (2015) Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. doi:
10.1007/s00401-015-1450-z
"), [43](/article/10.1007/s00401-015-1474-4#ref-CR43 "Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH et al (2013) RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA 110:E4968–E4977. doi:
10.1073/pnas.1315438110
")\].Several c9RAN proteins are toxic in a variety of models, thus implicating RAN translation as a mechanism of disease [[12](/article/10.1007/s00401-015-1474-4#ref-CR12 "Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW III, Sun S, Herdy JR, Bieri G et al (2015) Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18:1226–1231. doi: 10.1038/nn.4085
"), [14](/article/10.1007/s00401-015-1474-4#ref-CR14 "Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL (2014) Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345:1139–1145. doi:
10.1126/science.1254917
"), [19](/article/10.1007/s00401-015-1474-4#ref-CR19 "May S, Hornburg D, Schludi MH, Arzberger T, Rentzsch K, Schwenk BM, Grasser FA, Mori K, Kremmer E, Banzhaf-Strathmann J et al (2014) C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol 128:485–503. doi:
10.1007/s00401-014-1329-4
"), [20](/article/10.1007/s00401-015-1474-4#ref-CR20 "Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J et al (2014) C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345:1192–1194. doi:
10.1126/science.1256800
"), [35](/article/10.1007/s00401-015-1474-4#ref-CR35 "Tao Z, Wang H, Xia Q, Li K, Jiang X, Xu G, Wang G, Ying Z (2015) Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet 24:2426–2441. doi:
10.1093/hmg/ddv005
"), [37](/article/10.1007/s00401-015-1474-4#ref-CR37 "Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB et al (2014) Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84:1213–1225. doi:
10.1016/j.neuron.2014.12.010
"), [40](/article/10.1007/s00401-015-1474-4#ref-CR40 "Yamakawa M, Ito D, Honda T, Kubo K, Noda M, Nakajima K, Suzuki N (2015) Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum Mol Genet 24:1630–1645. doi:
10.1093/hmg/ddu576
")–[43](/article/10.1007/s00401-015-1474-4#ref-CR43 "Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH et al (2013) RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA 110:E4968–E4977. doi:
10.1073/pnas.1315438110
")\]. However, there have been conflicting reports on clinical and neuropathological correlations with c9RAN protein pathology in _C9ORF72_ mutation carriers \[[9](/article/10.1007/s00401-015-1474-4#ref-CR9 "Davidson YS, Barker H, Robinson AC, Thompson JC, Harris J, Troakes C, Smith B, Al-Saraj S, Shaw C, Rollinson S et al (2014) Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 2:70. doi:
10.1186/2051-5960-2-70
"), [16](/article/10.1007/s00401-015-1474-4#ref-CR16 "Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D et al (2013) Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol 126:859–879. doi:
10.1007/s00401-013-1181-y
"), [18](/article/10.1007/s00401-015-1474-4#ref-CR18 "Mann DM, Rollinson S, Robinson A, Bennion Callister J, Thompson JC, Snowden JS, Gendron T, Petrucelli L, Masuda-Suzukake M, Hasegawa M et al (2013) Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 1:68. doi:
10.1186/2051-5960-1-68
"), [30](/article/10.1007/s00401-015-1474-4#ref-CR30 "Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, Arzberger T, Edbauer D (2015) Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. doi:
10.1007/s00401-015-1450-z
")\]. For instance, among three studies, no associations between poly(GA) inclusion burden in various brain regions and either neurodegeneration, clinical phenotype, or neuropathological phenotype were detected \[[9](/article/10.1007/s00401-015-1474-4#ref-CR9 "Davidson YS, Barker H, Robinson AC, Thompson JC, Harris J, Troakes C, Smith B, Al-Saraj S, Shaw C, Rollinson S et al (2014) Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 2:70. doi:
10.1186/2051-5960-2-70
"), [16](/article/10.1007/s00401-015-1474-4#ref-CR16 "Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D et al (2013) Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol 126:859–879. doi:
10.1007/s00401-013-1181-y
"), [18](/article/10.1007/s00401-015-1474-4#ref-CR18 "Mann DM, Rollinson S, Robinson A, Bennion Callister J, Thompson JC, Snowden JS, Gendron T, Petrucelli L, Masuda-Suzukake M, Hasegawa M et al (2013) Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 1:68. doi:
10.1186/2051-5960-1-68
")\]. Conversely, it was recently reported that, in a cohort of 14 _C9ORF72_ patients, poly(GA) inclusions in cerebellar granule cells were significantly more abundant in patients with FTLD compared to patients with MND or FTLD-MND \[[30](/article/10.1007/s00401-015-1474-4#ref-CR30 "Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, Arzberger T, Edbauer D (2015) Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. doi:
10.1007/s00401-015-1450-z
")\]. These findings highlight the need for more thorough evaluations of the various c9RAN proteins in c9FTD/ALS, and that neuroanatomical regions not typically studied, such as the cerebellum, should be included in these investigations. Indeed, while often overlooked, neuroimaging and clinical evidence support the involvement of the cerebellum in cognitive and affective regulation \[[13](/article/10.1007/s00401-015-1474-4#ref-CR13 "Koziol LF, Budding D, Andreasen N, D’Arrigo S, Bulgheroni S, Imamizu H, Ito M, Manto M, Marvel C, Parker K et al (2014) Consensus paper: the cerebellum’s role in movement and cognition. Cerebellum 13:151–177. doi:
10.1007/s12311-013-0511-x
")\]. Combined with cerebellar atrophy emerging as a distinguishing feature of c9FTD/ALS compared to sporadic or other familial forms of these diseases \[[17](/article/10.1007/s00401-015-1474-4#ref-CR17 "Mahoney CJ, Beck J, Rohrer JD, Lashley T, Mok K, Shakespeare T, Yeatman T, Warrington EK, Schott JM, Fox NC et al (2012) Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: clinical, neuroanatomical and neuropathological features. Brain 135:736–750. doi:
10.1093/brain/awr361
"), [32](/article/10.1007/s00401-015-1474-4#ref-CR32 "Sha SJ, Takada LT, Rankin KP, Yokoyama JS, Rutherford NJ, Fong JC, Khan B, Karydas A, Baker MC, DeJesus-Hernandez M et al (2012) Frontotemporal dementia due to C9ORF72 mutations: clinical and imaging features. Neurology 79:1002–1011. doi:
10.1212/WNL.0b013e318268452e
"), [38](/article/10.1007/s00401-015-1474-4#ref-CR38 "Whitwell JL, Weigand SD, Boeve BF, Senjem ML, Gunter JL, Dejesus-Hernandez M, Rutherford NJ, Baker M, Knopman DS, Wszolek ZK et al (2012) Neuroimaging signatures of frontotemporal dementia genetics: C9ORF72, tau, progranulin and sporadics. Brain 135:794–806. doi:
10.1093/brain/aws001
")\], and our discovery of robust cerebellar transcriptome changes in c9ALS \[[27](/article/10.1007/s00401-015-1474-4#ref-CR27 "Prudencio M, Belzil V, Batra R, Ross CA, Gendron TF, Pregent L, Murray ME, Overstreet K, Piazza-Johnston A, Desaro P et al (2015) Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci 18:1175–1182. doi:
10.1038/nn.4065
")\], the question arises as to whether cerebellar pathology contributes to cognitive dysfunction in _C9ORF72_ mutation carriers. We thus evaluated whether poly(GP) and poly(GA), which are abundantly expressed in the cerebellum, associate with neuropathological and clinical characteristics of patients with a _C9ORF72_ repeat expansion.Materials and methods
Study design and participants
To evaluate associations between c9RAN protein levels and neuropathological diagnosis, we included all cases in the Mayo Clinic Jacksonville brain bank who were identified as C9ORF72 expansion carriers by repeat-primed PCR [[10](/article/10.1007/s00401-015-1474-4#ref-CR10 "DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J et al (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72:245–256. doi: 10.1016/j.neuron.2011.09.011
")\] and by immunostaining for poly(GP) pathology \[[3](/article/10.1007/s00401-015-1474-4#ref-CR3 "Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R et al (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77:639–646. doi:
10.1016/j.neuron.2013.02.004
")\], and for whom frozen tissue from the cerebellum, frontal cortex, motor cortex, and hippocampus was available (_n_ \= 55; Table [1](/article/10.1007/s00401-015-1474-4#Tab1)). Family history of disease was considered positive if patients were known to have a first or second degree relative with ALS, FTLD or mixed features of both. Age at onset was estimated based on the appearance of first disease symptoms.Table 1 Characteristics of C9ORF72 repeat expansion carriers
Of the 55 patients in our overall cohort, 15 composed our clinical cohort. An ALS-focused selection established a C9ORF72 patient cohort in which cognitive status ranged from normal to definitely abnormal. Study patients in the clinical cohort were ascertained from 2009 to 2014 on the basis of a diagnosis of ALS meeting El Escorial criteria, participation in the Mayo Clinic ALS Center deeded autopsy program, and genetically and pathologically confirmed to have a C9ORF72 repeat expansion (n = 15; Table 4; Supplementary Table 1). All patients in the clinical cohort were followed for multiple visits by the same neurologist (KBB) from initial encounter through advanced disease stage with severe, global disability. Assessments of cognitive function were derived from independent review of clinical records and test results by four evaluators, a neurologist (KBB) and three neuropsychologists (JAL, OP, and BKR), who all had access to the same clinical data. Clinical records for all patients included observations by clinical staff and family members involved in the patients’ care with regard to cognition and behavior, and for nine of the 15 patients, data from the ALS Cognitive Behavioral Screen, a validated neuropsychological screening instrument [[39](/article/10.1007/s00401-015-1474-4#ref-CR39 "Woolley SC, York MK, Moore DH, Strutt AM, Murphy J, Schulz PE, Katz JS (2010) Detecting frontotemporal dysfunction in ALS: utility of the ALS cognitive behavioral screen (ALS-CBS). Amyotroph Lateral Scler 11:303–311. doi: 10.3109/17482961003727954
")\], and/or a full neuropsychological examination (Supplementary Table 1). Examiners were aware of the patient’s diagnosis of c9ALS but were blinded to data on c9RAN protein levels. Owing to varied data available across the cohort on patients’ neuropsychological status, each patient with ALS was independently assessed by each of the evaluators on a 0–2 scale (0 = normal, 1 = mild or questionable impairment, or 2 = definitely abnormal) with respect to cognition. Scores were based on assessment of executive function, verbal fluency, and memory as supported by observational and quantitative data. Given our comparisons of cognitive status with autopsy findings, the assigned scores reflected the evaluator’s assessments of the patient’s cognitive function in late stage disease (i.e. near death with severe, global disability) based on available supporting data, which in all cases included caregiver information regarding the patient’s cognitive status at an advanced stage of disease. The rating of the 15 patients by the four evaluators resulted in 90 potential pairwise agreements; pairwise agreement for the cognitive score was 80 % (72/90). The final cognitive score for each patient represents the average of the scores of the four evaluators.Written informed consent was obtained from all participants or their legal next of kin if they were unable to give written consent, and biological samples were obtained with ethics committee approval.
Immunoassay analysis of c9RAN proteins in tissue homogenates
Brain homogenates were prepared as previously described [[2](/article/10.1007/s00401-015-1474-4#ref-CR2 "Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A et al (2013) Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol 126:385–399. doi: 10.1007/s00401-013-1149-y
")\]. In brief, approximately 50 mg of cerebellum, frontal cortex, motor cortex, or hippocampus tissue was homogenized in cold RIPA buffer (25 mM Tris–HCl pH 7.6, 150 mM NaCl, 1 % sodium deoxycholate, 1 % Nonidet P-40, 0.1 % sodium dodecyl sulfate, protease and phosphatase inhibitors) and sonicated on ice. Homogenates were cleared by centrifugation at 100,000_g_ for 30 min at 4 °C. The supernatant was collected and the concentration of RIPA-soluble proteins was determined by BCA assay. The resulting pellet was resuspended in RIPA buffer and, to prevent carry-over, re-sonicated and re-centrifuged. The RIPA-insoluble pellet was then extracted using 7 M urea buffer, sonicated and centrifuged at 100,000_g_ for 30 min at 22 °C. The protein concentration of the urea-soluble supernatant was determined by Bradford assay.Poly(GP) levels in lysates were measured using a previously described sandwich immunoassay that utilizes an affinity purified rabbit polyclonal poly(GP) antibody (Rb9259) and Meso Scale Discovery (MSD) electrochemiluminescence detection technology [[34](/article/10.1007/s00401-015-1474-4#ref-CR34 "Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P et al (2014) Discovery of a biomarker and lead small molecules to target r (GGGGCC)-associated defects in c9FTD/ALS. Neuron 83:1043–1050. doi: 10.1016/j.neuron.2014.07.041
")\]. This assay specifically detects poly(GP) and no other c9RAN protein \[[34](/article/10.1007/s00401-015-1474-4#ref-CR34 "Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P et al (2014) Discovery of a biomarker and lead small molecules to target r (GGGGCC)-associated defects in c9FTD/ALS. Neuron 83:1043–1050. doi:
10.1016/j.neuron.2014.07.041
")\]. Homogenates were diluted in Tris-buffered saline (TBS) and equal amounts of protein for all samples from the same neuroanatomical region and fraction were tested in duplicate wells. Serial dilutions of recombinant (GP)8 in TBS were used to prepare the standard curve. Response values corresponding to the intensity of emitted light upon electrochemical stimulation of the assay plate using the MSD QUICKPLEX SQ120 were acquired and background corrected using the average response from homogenates obtained from non-_C9ORF72_ repeat expansion carriers prior to interpolation of poly(GP) levels \[presented as ng of poly(GP) per mg protein\] using the standard curve. Poly(GA) levels in homogenates were similarly measured with an MSD-based sandwich immunoassay that utilizes a purified rabbit polyclonal poly(GA) antibody (Rb4333) provided by the Cleveland lab and a mouse monoclonal poly(GA) antibody (clone 5F2, \[[16](/article/10.1007/s00401-015-1474-4#ref-CR16 "Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D et al (2013) Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol 126:859–879. doi:
10.1007/s00401-013-1181-y
")\]) provided by the Edbauer lab. Serial dilutions of recombinant (GA)50 were used for the standard curve. This assay specifically detects poly(GA) and no other c9RAN protein (Supplementary Fig. 1).Immunohistochemistry and analysis of poly(GP) and phosphorylated TDP-43 pathology
To compare poly(GP) levels evaluated by immunoassay to poly(GP) pathology measured by immunohistochemistry, cerebellar tissue from 35 C9ORF72 repeat expansion carriers were immunostained for poly(GP). In addition, phosphorylated TDP-43 (pTDP-43) pathology was evaluated in frontal cortex and hippocampus tissues from the 15 patients in our clinical cohort to study associations of pTDP-43 with cognitive score or neuropathological diagnosis. Five micron thick slices were cut from formalin-fixed, paraffin-embedded tissue, and mounted on glass slides. After drying, slides were deparaffinized and rehydrated in xylene and alcohol washes before being steamed for 30 min in deionized water for antigen retrieval. All slides were processed on a Dako Autostainer with the Dako EnVision™+ system and 3′3-diaminobenzidine chromogen. Slides were stained with rabbit polyclonal poly(GP) antiserum (Rb5823, 1:5000) [[3](/article/10.1007/s00401-015-1474-4#ref-CR3 "Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R et al (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77:639–646. doi: 10.1016/j.neuron.2013.02.004
")\], rabbit polyclonal poly(GA) antiserum (Rb9880, 1:30,000) \[[42](/article/10.1007/s00401-015-1474-4#ref-CR42 "Zhang YJ, Jansen-West K, Xu YF, Gendron TF, Bieniek KF, Lin WL, Sasaguri H, Caulfield T, Hubbard J, Daughrity L et al (2014) Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol 128:505–524. doi:
10.1007/s00401-014-1336-5
")\], or a mouse monoclonal antibody against TDP-43 phosphorylated at S409/S410 (1:5000, Cosmo Bio Co., Tokyo, Japan), then counterstained with Lerner hematoxylin and coverslipped with Cytoseal permanent mounting media.To quantify poly(GP) pathology in the cerebellum, high resolution images of immunostained slides were captured using an Aperio ScanScope XT scanner and analyzed using ImageScope software (Leica Biosystems, Buffalo Grove, IL). For each case, twenty 1 mm2 annotated boxes were placed throughout the internal granule cell layer, analyzed using a color deconvolution algorithm tuned to strong immunopositivity per the 3′3-diaminobenzidine chromogen, and averaged among the 20 boxes. pTDP-43 pathology in the frontal cortex and hippocampus was similarly quantified from high resolution images using ImageScope software. In the hippocampus and frontal cortex, traced regions of interest included the dentate fascia, the dentate fascia with the hippocampus proper (CA4-subiculum), and the cross-sectional midfrontal gyrus (spanning crest to sulcal depth). All images were subsequently processed with a color deconvolution algorithm specifically tuned to pTDP-43 immunoreactivity.
Measurement of C9ORF72 transcript variant 1 or transcript variant 3 mRNA
Total RNA was extracted from frozen cerebellum and frontal cortex using the RNeasy Plus mini kit (Qiagen), following manufacturer’s protocols. RNA quality and quantity were determined with an Agilent 2100 Bioanalyzer using the RNA Nano Chip (Agilent Technologies). High quality RNA was obtained from the cerebellum and frontal cortex of 49 and 48 patients, respectively. We performed a reverse transcription reaction with 300 ng of RNA as template, using an equal ratio of Random Hexamers and Oligo dT primers and the SuperScript III Kit (Invitrogen). Quantitative real-time PCR was then done on an ABI7900 PCR system (Applied Biosystems) using TaqMan gene expression assays (Life Technologies) in accordance with the manufacturer’s recommendations. We assessed neuronal markers synaptophysin (SYP; Hs00300531_m1) and microtubule-associated protein 2 (MAP2; Hs00258900_m1) as well as C9ORF72 transcript variant 3 (NM_001256054.2; Hs00948764_m1). Using R Statistical Software (version 3.2.0; R Foundation for Statistical Computing), we calculated the median of replicates and took the geometric mean of neuronal markers, and subsequently determined expression levels relative to control subjects utilizing the ΔΔ Ct method. For transcript variant 1 (NM_145005.6), digital molecular barcoding (NanoString Technologies) was performed by the manufacturer using 100 ng of RNA as template. The NanoStringNorm R package and nSolver Analysis Software (NanoString Technologies, version 2.5) were used to account for technical assay variation using the geometric mean method. Maximum background subtraction was then performed, and data was normalized to the geometric mean of neuronal markers SYP and MAP2.
Measurement of C9ORF72 repeat size
For 53 patients for whom high quality DNA from the cerebellum and frontal cortex was available, the size of the G4C2 repeat expansion in these tissues was determined by Southern blotting as described elsewhere [[36](/article/10.1007/s00401-015-1474-4#ref-CR36 "van Blitterswijk M, Dejesus-Hernandez M, Niemantsverdriet E, Murray ME, Heckman MG, Diehl NN, Brown PH, Baker MC, Finch NA, Bauer PO et al (2013) Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol 12:978–988. doi: 10.1016/S1474-4422(13)70210-2
")\].Statistical analysis
In a cohort of 35 patients with the C9ORF72 repeat expansion, associations of cerebellar poly(GP) levels quantified by immunoassay with poly(GP) immunopositivity quantified from immunostained cerebellar sections were evaluated using a Spearman’s test of correlation; Spearman’s correlation coefficient r and 95 % confidence intervals (CI) were estimated.
In our cohort of 55 patients, levels of poly(GP) were compared among neuroanatomical regions (i.e. cerebellum, frontal cortex, motor cortex, and hippocampus) using a Friedman rank sum test. This was followed by a paired Wilcoxon signed rank test for pairwise comparisons when significant differences were encountered (p < 0.0083 considered significant after Bonferroni correction). Associations between soluble and insoluble poly(GP) or poly(GA) levels in a given neuroanatomical region were evaluated using a Spearman’s test of correlation. The difference between soluble and insoluble poly(GP) as a percentage of total poly(GP) levels was evaluated using a Wilcoxon signed rank test, and the same was done for poly(GA).
For a given neuroanatomical area, we assessed associations of poly(GP) or poly(GA) with six variables [i.e. disease subgroups (i.e. ALS, FTLD, FTLD-MND), C9ORF72 expansion size, C9ORF72 variant 1, C9ORF72 variant 3, age of onset, and survival after onset]. To adjust for multiple testing and control the family-wise error rate at 5 %, a Bonferroni correction was utilized separately for each outcome, and thus p < 0.0083 was considered significant. A Kruskal–Wallis rank sum test was performed to determine whether poly(GP) or poly(GA) levels differed significantly among disease subgroups, and when this test was significant, a Wilcoxon rank sum test was used for pairwise comparisons (p < 0.017 considered significant after Bonferroni correction). To assess associations between poly(GP) or poly(GA) levels and repeat size, C9ORF72 transcript variant 1 or 3 expression, and age of onset of the C9ORF72 expansion, a Spearman’s test of correlation was used. Associations between poly(GP) or poly(GA) levels and survival after disease onset were investigated using Cox proportional hazards regression models. Hazard ratios and 95 % CIs were estimated. Since all patients were deceased no censoring was necessary. Models were adjusted for age of onset and disease subgroup. Poly(GP) or poly(GA) levels were considered as dichotomous categorical variables using the median as the cut-off point.
In our cohort of 15 patients for whom cognitive status was evaluated, we assessed associations of seven outcomes [i.e. total poly(GP), soluble poly(GP), insoluble poly(GP), total poly(GA), soluble poly(GA), insoluble poly(GA), and pTDP-43 pathology] with two variables (i.e. cognitive score and neuropathological diagnosis). Associations of poly(GP), poly(GA) or pTDP-43 with cognitive score were assessed using Spearman’s test of correlation, and associations with neuropathological diagnosis were assessed using a Wilcoxon rank sum test (p < 0.025 considered significant after Bonferroni correction).
In a secondary analysis, associations between poly(GP) levels in all evaluated neuroanatomical regions and pTDP-43 pathology in the frontal cortex or hippocampus were analyzed using Spearman’s test of correlation (p < 0.025 considered significant after Bonferroni correction). Associations between cerebellar poly(GA) levels and pTDP-43 pathology in the frontal cortex or hippocampus were similarly assessed. A Wilcoxon rank sum test was used for pairwise comparisons between ALS and FTLD-MND subgroups in the clinical cohort with respect to sex, age of onset, survival after onset, education, and cognitive score (p < 0.01 considered significant after Bonferroni correction).
Results
Soluble and insoluble poly(GP) are detected in various neuroanatomical regions and are highest in the cerebellum
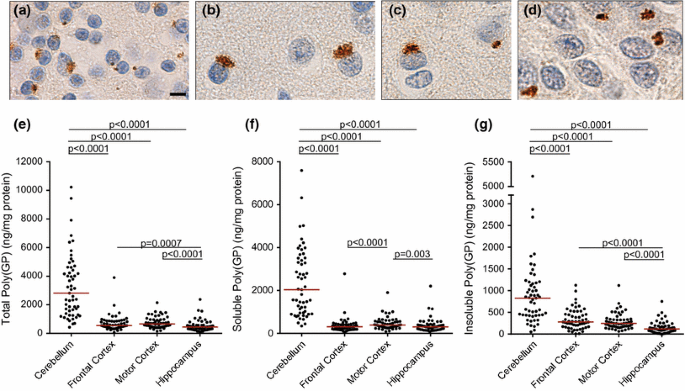
We previously reported, and show in Fig. 1a–d, that poly(GP) inclusions are detected in abundance in the cerebellum, neocortex, and hippocampus of C9ORF72 mutation carriers by immunohistochemistry [[3](/article/10.1007/s00401-015-1474-4#ref-CR3 "Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R et al (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77:639–646. doi: 10.1016/j.neuron.2013.02.004
")\]. In the present study, we utilized an immunoassay to measure levels of poly(GP) in various neuroanatomical regions. A comparison of cerebellar poly(GP) levels determined using this biochemical approach with poly(GP) immunopositivity in stained cerebellar sections from 35 _C9ORF72_ mutation carriers revealed a significant correlation between these two measures (Supplementary Table 2). One of the advantages of biochemical analyses of poly(GP) burden is the ability to measure different forms of the protein in a quantitative manner. Since soluble protein oligomers, like insoluble forms of aggregating proteins, may be pathogenic, we measured both soluble and insoluble poly(GP) levels in cerebellum, frontal cortex, motor cortex, and hippocampus homogenates sequentially extracted with RIPA buffer (soluble fraction) and urea buffer (insoluble fraction). As shown in Fig. [1](/article/10.1007/s00401-015-1474-4#Fig1)e–g, levels of soluble and insoluble poly(GP) were significantly higher in the cerebellum compared to the other neuroanatomical regions. Given that a significant correlation existed between these forms of poly(GP) in all regions (Supplementary Table 3), the analyses presented below are limited to total poly(GP) levels for simplicity. Data on soluble and insoluble poly(GP) can be found in the Electronic Supplementary Material (Supplementary Tables 4–8).Fig. 1
Comparison of poly(GP) levels in different neuroanatomical regions of C9ORF72 mutation carriers. a–d Poly(GP)-immunoreactive inclusions in the cerebellum (a), frontal cortex (b), motor cortex (c) and hippocampus (d) of a C9ORF72 repeat expansion carrier. Scale bar 5 µm. e–g Levels of total poly(GP) (e), soluble poly(GP) (f), or insoluble poly(GP) (g) in cerebellum, frontal cortex, motor cortex, and hippocampus of patients with C9ORF72 repeat expansions (n = 55). The median in a given group is denoted by a solid horizontal line
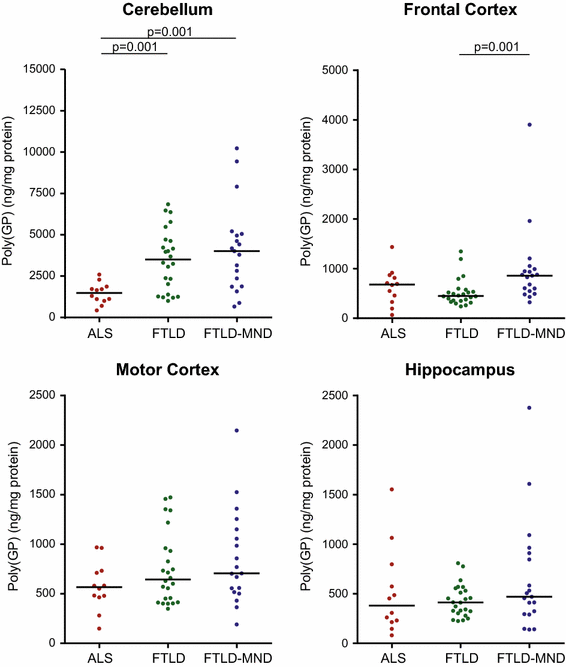
Cerebellar poly(GP) levels are significantly lower in C9ORF72 mutation carriers with ALS compared to patients with FTLD or FTLD-MND
We next evaluated whether poly(GP) levels in the cerebellum, frontal cortex, motor cortex, or hippocampus differ among patients neuropathologically diagnosed with ALS (n = 12), FTLD (n = 24), or FTLD-MND (n = 19). Of interest, poly(GP) levels in the cerebellum were significantly lower in the ALS subgroup (median 1481 ng/mg, IQR 1083–1758) compared to the FTLD (median 3501 ng/mg, IQR 1864–4652, p = 0.001) or FTLD-MND (median 4014 ng/mg, IQR 2119–5002, p = 0.001) subgroups (Table 2; Fig. 2). In the frontal cortex, poly(GP) levels also differed based on neuropathological subtype, although the expression profile varied from that observed in the cerebellum; patients with FTLD-MND (median 859 ng/mg, IQR 575–971) had significantly higher poly(GP) levels in the frontal cortex compared to patients with FTLD (median 452 ng/mg, IQR 358–546, p = 0.001) (Table 2; Fig. 2). No differences in poly(GP) levels among disease subgroups were detected in the motor cortex or hippocampus (Table 2; Fig. 2).
Table 2 Comparison of poly(GP) levels among disease subgroups
Fig. 2
Poly(GP) levels are significantly lower in the cerebellum of C9ORF72 mutation carriers with ALS compared to patients with FTLD or FTLD-MND. Shown are total levels of poly(GP) in the indicated neuroanatomical regions by disease subgroup [amyotrophic lateral sclerosis (ALS), n = 12; frontotemporal lobar degeneration (FTLD), n = 24; and FTLD with motor neuron disease (FTLD-MND); n = 19]. Poly(GP) levels in the cerebellum were significantly lower in patients with ALS compared to patients with FTLD or FTLD-MND. In the frontal cortex, poly(GP) levels were significantly higher in the FTLD-MND subgroup compared to the FTLD subgroup. The median in a given group is denoted by a solid horizontal line
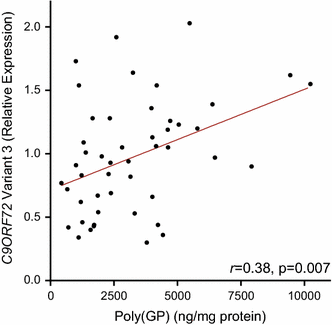
Cerebellar poly(GP) associates with C9ORF72 transcript variant 3 expression but not with expression of variant 1 or with repeat size
Because our studies uncovered associations between cerebellar or frontal cortical poly(GP) and the neuropathological diagnosis of C9ORF72 patients, we sought to identify factors that influence poly(GP) levels. We first investigated associations between poly(GP) and repeat size of the C9ORF72 expansion and found no association between these two variables in either the cerebellum or the frontal cortex (Table 3). We next evaluated associations between poly(GP) and transcript levels for C9ORF72 variant 1 or variant 3. The G4C2 repeat is located between exons 1a and 1b; if exon 1a is used, as is the case for variants 1 and 3, the repeat is located in intron 1 and is transcribed. If exon 1b is used, the repeat is located in the promoter region and is not transcribed (variant 2; NM_018325.4). In the cerebellum, poly(GP) levels associated with variant 3 (r 0.38, 95 % CI 0.10–0.62, p = 0.007; Fig. 3) but not with variant 1 (Table 3). In the frontal cortex, no association between poly(GP) levels and C9ORF72 transcript variant 1 or variant 3 was detected (Table 3).
Table 3 Associations of total poly(GP) levels with C9ORF72 expansion size, C9ORF72 variants 1 and 3, and age at onset
Fig. 3
Cerebellar poly(GP) associates with C9ORF72 transcript variant 3 expression. Associations of total poly(GP) levels in the cerebellum with expression of C9ORF72 transcript variant 3 (n = 49). The straight line is a linear regression line
Cerebellar poly(GP) associates with cognitive impairment
Despite the associations between poly(GP) in the cerebellum or frontal cortex and neuropathological subgroup, poly(GP) levels in these neuroanatomical regions did not associate with age of disease onset in our overall cohort of patients (Table 3). Furthermore, survival after onset, adjusted for age of onset and disease subgroup, was not associated with poly(GP) in these regions (Supplementary Table 6).
Since poly(GP) levels could nevertheless influence the clinical phenotype of patients, we examined associations between poly(GP) levels in different neuroanatomical regions and cognitive impairment. These studies were carried out on 15 c9ALS patients for whom neuropsychological data was available (Table 4; Supplementary Table 1). Ten of the 15 patients in this clinical cohort presented with ALS (67 %), four with ALS accompanied by cognitive impairment (27 %), and one with behavioral variant FTD (bvFTD-ALS, 7 %; Supplementary Table 1). At autopsy, nine patients were neuropathologically diagnosed with ALS (60 %) and six with FTLD-MND (40 %). No significant differences in sex, age of disease onset, survival after onset, education, or cognitive score were observed between the ALS and FTLD-MND subgroups (Table 4).
Table 4 Characteristics of patients in the clinical cohort
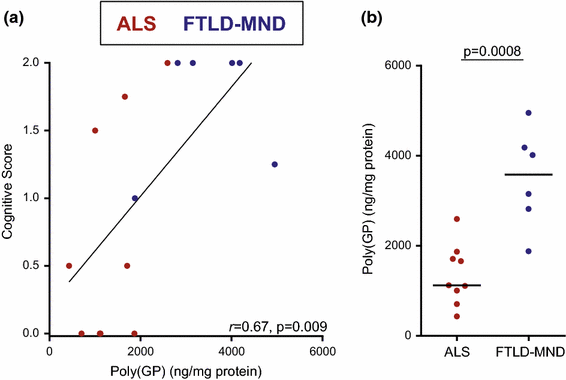
Notably, an association between poly(GP) levels in the cerebellum and cognitive score was detected (r = 0.67, 95 % CI 0.22–0.88, p = 0.009) (Table 5; Fig. 4a). In contrast, poly(GP) levels in frontal cortex, motor cortex, and hippocampus did not associate with cognitive score (Table 5). As anticipated, an association between pTDP-43 pathology in the frontal cortex and cognitive score was observed (r = 0.58, 95 % CI 0.09–0.85, p = 0.025) (Table 5).
Table 5 Associations of total poly(GP) levels or pTDP-43 pathology with cognitive score or neuropathological diagnosis in the clinical cohort
Fig. 4
Cerebellar poly(GP) associates with cognitive score and differs based on neuropathological subgroup in the clinical cohort of C9ORF72 mutation carriers. Four evaluators assessed cognitive function of patients in the clinical cohort (n = 15) from independent review of clinical records and test results, and assigned to each patient a score (0 = normal, 1 = mild or questionable impairment, or 2 = definitely abnormal). The final cognitive score for each patient is the average of the scores assigned by the four evaluators. Associations of total poly(GP) in the cerebellum with cognitive score (a) or neuropathological diagnosis (b) of patients are shown. In panel a, the straight line is a linear regression line. In panel b, the median in a given group is denoted by a solid horizontal line. Red circles represent patients neuropathologically diagnosed with amyotrophic lateral sclerosis (ALS), and blue circles represent patients with frontotemporal lobar degeneration with motor neuron disease (FTLD-MND)
Similarly to what was observed in the 55 C9ORF72 mutation carriers, cerebellar poly(GP) levels in patients with ALS (median 1122 ng/mg, IQR 1005–1708) were significantly lower compared to patients with FTLD-MND (median 3583 ng/mg, IQR 2901–4139, p = 0.0008) in the smaller clinical cohort (Table 5; Fig. 4b). In the other neuroanatomical regions, no difference in poly(GP) levels was detected between neuropathological subgroups (Table 5). As would be expected, pTDP-43 pathology in frontal cortex and hippocampus was significantly more abundant in the FTLD-MND subgroup compared to the ALS subgroup (Table 5).
We additionally investigated potential relationships between c9RAN protein expression and pTDP-43 pathology. No association between poly(GP) levels in the frontal cortex or hippocampus and pTDP-43 pathology in these same regions was detected. Likewise, poly(GP) levels in the motor cortex did not associate with frontal cortical or hippocampal pTDP-43 pathology. However, in line with our findings that poly(GP) levels in the cerebellum associated with neuropathological diagnosis, as did pTDP-43 pathology in the frontal cortex and hippocampus, we detected an association of cerebellar poly(GP) levels with pTDP-43 pathology in the frontal cortex (r = 0.69, 95 % CI 0.22–0.92, p = 0.005) or hippocampus (r = 0.69 95 % CI 0.25–0.89, p = 0.005) (Supplementary Table 8).
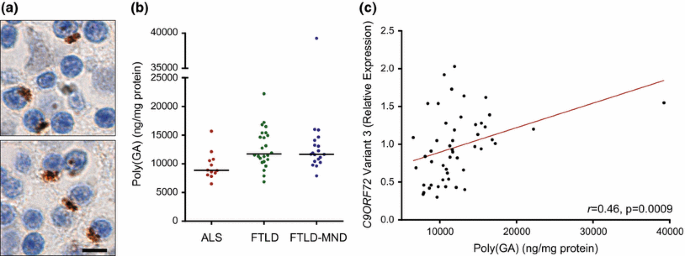
Associations of cerebellar poly(GA) with neuropathological and clinical features
Given our findings above and the fact that poly(GA) pathology is abundant in the cerebellum (Fig. 5a), and given a recent report showing that poly(GA) inclusion burden in cerebellar granule cells correlated with neuropathological subgroup [[30](/article/10.1007/s00401-015-1474-4#ref-CR30 "Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, Arzberger T, Edbauer D (2015) Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. doi: 10.1007/s00401-015-1450-z
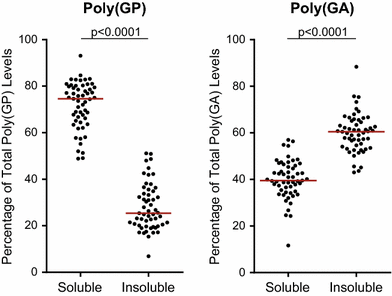
")\], we investigated whether cerebellar poly(GA) levels assessed by immunoassay differ based on neuropathological diagnosis or cognitive score in our case series. As observed for cerebellar poly(GP), there existed a significant correlation between soluble and insoluble poly(GA) levels in the cohort of 55 _C9ORF72_ mutation carriers (Supplementary Table 9). However, the solubility profile of poly(GA) differed from that of poly(GP); while soluble poly(GP) levels were significantly higher than insoluble poly(GP) levels in the cerebellum, the opposite was observed for poly(GA) (Fig. [6](/article/10.1007/s00401-015-1474-4#Fig6)).Fig. 5
Cerebellar poly(GA) levels in C9ORF72 mutation carriers with ALS, FTLD or FTLD-MND, and associations between poly(GA) and C9ORF72 transcript variant 3 expression. a Poly(GA)-immunoreactive inclusions in the cerebellum of a C9ORF72 repeat expansion carrier. Scale bar 5 µm. b Shown are total levels of cerebellar poly(GA) by disease subgroup [amyotrophic lateral sclerosis (ALS), n = 12; frontotemporal lobar degeneration (FTLD), n = 24; and FTLD with motor neuron disease (FTLD-MND), n = 19]. The median in a given group is denoted by a solid horizontal line. c Associations of poly(GA) levels in the cerebellum with expression of C9ORF72 transcript variant 3 (n = 49). The straight line is a linear regression line
Fig. 6
Solubility profiles of poly(GP) and poly(GA) in the cerebellum. Comparison of soluble and insoluble poly(GP) or poly(GA) in the cerebellum of patients with C9ORF72 repeat expansions (n = 55). Note that soluble poly(GP) levels were significantly higher than insoluble poly(GP) levels, whereas soluble poly(GA) levels were significantly lower than insoluble poly(GA) levels. The median in a given group is denoted by a solid horizontal line
Upon comparing cerebellar poly(GA) among neuropathological subgroups, differences in poly(GA) levels among subgroups neared statistical significance but the p value (p = 0.0088) did not meet the cut-off (p < 0.0083) established by Bonferroni correction. Nevertheless, total poly(GA) levels trended lower in the ALS subgroup (median 8877 ng/mg, 8327–10,616) in comparison to the FTLD (median 11,723 ng/mg, 10,666–15,054) or FTLD-MND (median 11,677 ng/mg, 10,627–13,673) subgroups (Fig. 5b; Supplementary Table 10). Compared to cerebellar poly(GP), the difference in poly(GA) levels among subgroups was less robust. Whereas poly(GP) levels in FTLD and FTLD-MND were approximately 2.5-fold higher than in ALS (Fig. 2), a 1.3-fold increase in poly(GA) levels was detected in FTLD and FTLD-MND compared to ALS (Fig. 5b).
Consistent with total poly(GP) in the cerebellum, total poly(GA) levels associated with C9ORF72 variant 3 (r 0.46, 95 % CI 0.22–0.66, p = 0.0009; Fig. 5c) but not with C9ORF72 variant 1, repeat size, age of onset, or survival after onset (Supplementary Tables 11 and 12). In contrast to cerebellar poly(GP), total poly(GA) levels did not associate with cognitive score or with neuropathological diagnosis (ALS vs. FTLD-MND) in our clinical cohort of 15 patients (Supplementary Table 13). An association between soluble poly(GA) and neuropathological diagnosis was nonetheless detected in this smaller cohort (Supplementary Table 13). Furthermore, poly(GA) levels in the cerebellum associated with pTDP-43 pathology in the frontal cortex or hippocampus (Supplementary Table 14), as did cerebellar poly(GP).
Discussion
Neuropathologic and clinical diversity characterize C9ORF72 repeat expansion carriers, but the molecular basis for this variation remains elusive. Our studies reveal intriguing findings involving c9RAN proteins in the cerebellum. We show that cerebellar poly(GP) levels were significantly lower in ALS patients compared to FTLD and FTLD-MND patients. This is not likely due to the shorter disease duration of ALS, which could afford less time for c9RAN proteins to accumulate, since poly(GP) levels in the frontal cortex, motor cortex, and hippocampus did not similarly differ between ALS and FTLD or FTLD-MND subgroups. Rather, the results suggest that expression of the C9ORF72 repeat expansion in cerebellum influences neuropathological phenotypes and this may consequently influence clinical features. Indeed, we found that cerebellar poly(GP) levels associated with cognitive impairment. Whether this occurs as a direct result of poly(GP) accumulation, or whether poly(GP) serves as a marker for aberrant events triggered by C9ORF72 repeat expansions, is not yet known. Similarly to our observations with poly(GP), cerebellar poly(GA) levels trended lower in patients with ALS compared to patients with FTLD or FTLD-MND, but the p value narrowly exceeded the threshold of significance after adjustment for multiple comparisons (p < 0.0083). This is likely due to the fact that differences in poly(GA) levels among disease subgroups were less robust than those observed for poly(GP), an observation that may also explain the lack of association between poly(GA) and cognitive impairment in our clinical cohort. A larger number of patients may be required to detect potential associations between poly(GA) levels and neuropathological diagnosis or cognitive score. Indeed, while findings in our retrospectively identified clinical cohort are of particular interest, further validation in larger cohorts of c9ALS and c9FTD patients in whom detailed cognitive and behavioral data are available is warranted in order to define contributions of c9RAN proteins to clinical phenotypes.
Our study supports the need for more sensitive and quantitative approaches when evaluating the relationship between putatively pathologic proteins and disease phenotypes. Previous studies that utilized semi-quantitative grading systems of c9RAN protein pathology, in particular for poly(GA), did not detect associations with clinical or neuropathological features [[9](/article/10.1007/s00401-015-1474-4#ref-CR9 "Davidson YS, Barker H, Robinson AC, Thompson JC, Harris J, Troakes C, Smith B, Al-Saraj S, Shaw C, Rollinson S et al (2014) Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 2:70. doi: 10.1186/2051-5960-2-70
"), [16](/article/10.1007/s00401-015-1474-4#ref-CR16 "Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D et al (2013) Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol 126:859–879. doi:
10.1007/s00401-013-1181-y
"), [18](/article/10.1007/s00401-015-1474-4#ref-CR18 "Mann DM, Rollinson S, Robinson A, Bennion Callister J, Thompson JC, Snowden JS, Gendron T, Petrucelli L, Masuda-Suzukake M, Hasegawa M et al (2013) Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 1:68. doi:
10.1186/2051-5960-1-68
")\]. It must be recognized, however, that semi-quantitative analyses may not be sufficiently sensitive to discern differences among patient subgroups, especially when cohorts are small. More recently, using a quantitative analysis of inclusion pathology, Schuldi et al. reported that inclusions of poly(GA) are less frequent in cerebellar granule cells of patients with ALS (_n_ \= 3) and FTLD-MND (_n_ \= 8) compared to patients with FTLD (_n_ \= 3) \[[30](/article/10.1007/s00401-015-1474-4#ref-CR30 "Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, Arzberger T, Edbauer D (2015) Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. doi:
10.1007/s00401-015-1450-z
")\]. This data differs from our observations that cerebellar poly(GA) trended lower in patients with ALS (_n_ \= 12) compared to patients with FTLD (_n_ \= 24) as well as patients with FTLD-MND (_n_ \= 19). Yet, it must be kept in mind that the two studies used different case series and different methods to quantify poly(GA) burden.The impetus for employing a biochemical approach to evaluate c9RAN proteins in this study was that it allows for non-biased, sensitive, and quantitative measures, and may permit the detection of a distinct pool of protein species not discernable by immunohistochemistry. With respect to the latter, we examined soluble and insoluble poly(GP) and poly(GA), finding a strong correlation between the soluble and insoluble forms of each of these c9RAN proteins. We additionally observed that poly(GP) and poly(GA) have different solubility profiles in brain tissue. A greater percentage of total poly(GP) was soluble, while a greater percentage of total poly(GA) was insoluble. Whether and how this influences the toxic potential of these c9RAN proteins is not yet known but worthy of investigation. Our data also suggest that poly(GA) levels are higher than levels of poly(GP); however, a direct comparison in the abundance of these two proteins should be made with some caution given the different biochemical properties of the proteins, differences in affinities of the poly(GP) and poly(GA) antibodies used for the immunoassays, and differences in the calibrators used for interpolating protein concentrations.
Little is known about factors that govern c9RAN protein expression. We detected an association between cerebellar poly(GP) or poly(GA) levels and C9ORF72 transcript variant 3, the pre-mRNA of which contains the expanded repeat that serves as a template for RAN translation. Surprisingly, the same association was not observed for transcript variant 1. Although pre-mRNA of variant 1 also contains the repeat expansion, one could speculate that the absence of an association may be a reflection of unique properties of each transcript variant (i.e. predisposition to RAN translation), which requires further study. There was no association between poly(GP) or poly(GA) levels in the cerebellum or frontal cortex with repeat size. This is congruent with our previous report that repeat size does not differ between disease subgroups [[36](/article/10.1007/s00401-015-1474-4#ref-CR36 "van Blitterswijk M, Dejesus-Hernandez M, Niemantsverdriet E, Murray ME, Heckman MG, Diehl NN, Brown PH, Baker MC, Finch NA, Bauer PO et al (2013) Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol 12:978–988. doi: 10.1016/S1474-4422(13)70210-2
")\], in contrast to what we show here for cerebellar poly(GP) levels. It is also worth noting that poly(GP) expression is higher in the cerebellum compared to the frontal cortex (Fig. [1](/article/10.1007/s00401-015-1474-4#Fig1)), whereas repeat lengths in the cerebellum are smaller than those in the frontal cortex \[[36](/article/10.1007/s00401-015-1474-4#ref-CR36 "van Blitterswijk M, Dejesus-Hernandez M, Niemantsverdriet E, Murray ME, Heckman MG, Diehl NN, Brown PH, Baker MC, Finch NA, Bauer PO et al (2013) Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol 12:978–988. doi:
10.1016/S1474-4422(13)70210-2
")\].Of importance, the findings herein add to a growing body of evidence supporting cerebellar involvement in c9FTD/ALS. For instance, we discovered far more transcriptome changes in the cerebellum than frontal cortex of c9ALS patients [[27](/article/10.1007/s00401-015-1474-4#ref-CR27 "Prudencio M, Belzil V, Batra R, Ross CA, Gendron TF, Pregent L, Murray ME, Overstreet K, Piazza-Johnston A, Desaro P et al (2015) Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci 18:1175–1182. doi: 10.1038/nn.4065
")\], and cerebellar atrophy is reported in _C9ORF72_ expansion carriers with neuroimaging methods \[[17](/article/10.1007/s00401-015-1474-4#ref-CR17 "Mahoney CJ, Beck J, Rohrer JD, Lashley T, Mok K, Shakespeare T, Yeatman T, Warrington EK, Schott JM, Fox NC et al (2012) Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: clinical, neuroanatomical and neuropathological features. Brain 135:736–750. doi:
10.1093/brain/awr361
"), [32](/article/10.1007/s00401-015-1474-4#ref-CR32 "Sha SJ, Takada LT, Rankin KP, Yokoyama JS, Rutherford NJ, Fong JC, Khan B, Karydas A, Baker MC, DeJesus-Hernandez M et al (2012) Frontotemporal dementia due to C9ORF72 mutations: clinical and imaging features. Neurology 79:1002–1011. doi:
10.1212/WNL.0b013e318268452e
"), [38](/article/10.1007/s00401-015-1474-4#ref-CR38 "Whitwell JL, Weigand SD, Boeve BF, Senjem ML, Gunter JL, Dejesus-Hernandez M, Rutherford NJ, Baker M, Knopman DS, Wszolek ZK et al (2012) Neuroimaging signatures of frontotemporal dementia genetics: C9ORF72, tau, progranulin and sporadics. Brain 135:794–806. doi:
10.1093/brain/aws001
")\]. Furthermore, with regards to the association of cerebellar poly(GP) with cognitive impairment, neuroanatomical studies have revealed reciprocal cerebellar connectivity with areas of the cerebral cortex involved in higher cognitive functioning. Output projections from the cerebellum first synapse on the dentate nucleus and then the thalamus before projecting to the cerebral cortex. Conversely, the cerebral cortex connects to the cerebellum via projections that synapse on the pons \[[7](/article/10.1007/s00401-015-1474-4#ref-CR7 "Buckner RL (2013) The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging. Neuron 80:807–815. doi:
10.1016/j.neuron.2013.10.044
")\]. It is also notable that cerebellar activation during cognitive tasks has been recorded by functional neuroimaging \[[24](/article/10.1007/s00401-015-1474-4#ref-CR24 "Petersen SE, Fox PT, Posner MI, Mintun M, Raichle ME (1989) Positron emission tomographic studies of the processing of singe words. J Cogn Neurosci 1:153–170. doi:
10.1162/jocn.1989.1.2.153
")\], and that neuropsychological studies on patients with cerebellar lesions have identified cognitive and affective disturbances, leading to the concept of “cerebellar cognitive affective syndrome” \[[31](/article/10.1007/s00401-015-1474-4#ref-CR31 "Schmahmann JD, Sherman JC (1998) The cerebellar cognitive affective syndrome. Brain 121(Pt 4):561–579")\]. A large spectrum of deficits has been described in patients with cerebellar damage, including executive dysfunction, mild language symptoms, blunting of affect, disinhibition, obsessive–compulsive traits, and psychosis. Patients with _C9ORF72_ repeat expansions present with many similar features \[[29](/article/10.1007/s00401-015-1474-4#ref-CR29 "Rohrer JD, Isaacs AM, Mizielinska S, Mead S, Lashley T, Wray S, Sidle K, Fratta P, Orrell RW, Hardy J et al (2015) C9orf72 expansions in frontotemporal dementia and amyotrophic lateral sclerosis. Lancet Neurol 14:291–301. doi:
10.1016/S1474-4422(14)70233-9
")\].Whereas TDP-43 pathology is reportedly consistent in FTLD and ALS patients with and without the C9ORF72 repeat expansion [[9](/article/10.1007/s00401-015-1474-4#ref-CR9 "Davidson YS, Barker H, Robinson AC, Thompson JC, Harris J, Troakes C, Smith B, Al-Saraj S, Shaw C, Rollinson S et al (2014) Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 2:70. doi: 10.1186/2051-5960-2-70
")\], neuropsychiatric symptoms, as well as cognitive and behavioral impairment, may be more common in FTD and ALS patients that carry the _C9ORF72_ mutation compared to non-carriers \[[8](/article/10.1007/s00401-015-1474-4#ref-CR8 "Byrne S, Elamin M, Bede P, Shatunov A, Walsh C, Corr B, Heverin M, Jordan N, Kenna K, Lynch C et al (2012) Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol 11:232–240. doi:
10.1016/S1474-4422(12)70014-5
"), [29](/article/10.1007/s00401-015-1474-4#ref-CR29 "Rohrer JD, Isaacs AM, Mizielinska S, Mead S, Lashley T, Wray S, Sidle K, Fratta P, Orrell RW, Hardy J et al (2015) C9orf72 expansions in frontotemporal dementia and amyotrophic lateral sclerosis. Lancet Neurol 14:291–301. doi:
10.1016/S1474-4422(14)70233-9
")\]. Features unique to the _C9ORF72_ repeat expansion, such as c9RAN proteins, may thus contribute to the increased frequency of certain phenotypes in c9FTD/ALS. Indeed, we noted that one ALS patient with definite cognitive impairment, who was neuropathologically diagnosed with ALS, had sparse extramotor TDP-43 pathology but relatively high cerebellar poly(GP) levels (Fig. [4](/article/10.1007/s00401-015-1474-4#Fig4)). In a similar manner, abundant poly(GA) pathology in cerebral cortical regions, hippocampus, and cerebellum, but only sparse TDP-43 pathology, have been reported in three _C9ORF72_ repeat expansion patients who developed fairly rapid cognitive decline, but died prematurely due to unrelated illnesses \[[4](/article/10.1007/s00401-015-1474-4#ref-CR4 "Baborie A, Griffiths TD, Jaros E, Perry R, McKeith IG, Burn DJ, Masuda-Suzukake M, Hasegawa M, Rollinson S, Pickering-Brown S et al (2015) Accumulation of dipeptide repeat proteins predates that of TDP-43 in Frontotemporal Lobar Degeneration associated with hexanucleotide repeat expansions in C9ORF72 gene. Neuropathol Appl Neurobiol 41:601–612. doi:
10.1111/nan.12178
")\]. While data from our group and others \[[4](/article/10.1007/s00401-015-1474-4#ref-CR4 "Baborie A, Griffiths TD, Jaros E, Perry R, McKeith IG, Burn DJ, Masuda-Suzukake M, Hasegawa M, Rollinson S, Pickering-Brown S et al (2015) Accumulation of dipeptide repeat proteins predates that of TDP-43 in Frontotemporal Lobar Degeneration associated with hexanucleotide repeat expansions in C9ORF72 gene. Neuropathol Appl Neurobiol 41:601–612. doi:
10.1111/nan.12178
"), [26](/article/10.1007/s00401-015-1474-4#ref-CR26 "Proudfoot M, Gutowski NJ, Edbauer D, Hilton DA, Stephens M, Rankin J, Mackenzie IR (2014) Early dipeptide repeat pathology in a frontotemporal dementia kindred with C9ORF72 mutation and intellectual disability. Acta Neuropathol 127:451–458. doi:
10.1007/s00401-014-1245-7
")\], indicate that clinical features can develop in _C9ORF72_ repeat expansion carriers with c9RAN protein pathology but limited TDP-43 pathology, we did nonetheless uncover a correlation between c9RAN proteins and pTDP-43\. While levels of poly(GP) and poly(GA) in the frontal cortex or hippocampus did not associate with pTDP-43 pathology in these same regions, there were significant associations of poly(GP) or poly(GA) levels in the cerebellum with pTDP-43 pathology in the frontal cortex or hippocampus. These findings could point toward a relationship whereby cerebellar c9RAN proteins influence TDP-43 in the frontal cortex and hippocampus, or simply reflect the fact that cerebellar c9RAN protein levels and pTDP-43 pathology in the frontal cortex and hippocampus independently associate with neuropathological diagnosis and consequently associate with each other. Nevertheless, the finding that poly(GA) levels in cerebellum associated with pTDP-43 pathology in the frontal cortex adds strength to the notion that, like poly(GP), cerebellar poly(GA) differs based on neuropathological subgroup.Overall, our findings implicate cerebellar abnormalities as a contributor to the neuropathological and clinical heterogeneity associated with the C9ORF72 repeat expansion, and support additional investigations on the role of this largely overlooked neuroanatomical region in c9FTD/ALS.
References
- Al-Sarraj S, King A, Troakes C, Smith B, Maekawa S, Bodi I, Rogelj B, Al-Chalabi A, Hortobagyi T, Shaw CE (2011) p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol 122:691–702. doi:10.1007/s00401-011-0911-2
Article CAS PubMed Google Scholar - Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A et al (2013) Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol 126:385–399. doi:10.1007/s00401-013-1149-y
Article PubMed Central CAS PubMed Google Scholar - Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R et al (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77:639–646. doi:10.1016/j.neuron.2013.02.004
Article PubMed Central CAS PubMed Google Scholar - Baborie A, Griffiths TD, Jaros E, Perry R, McKeith IG, Burn DJ, Masuda-Suzukake M, Hasegawa M, Rollinson S, Pickering-Brown S et al (2015) Accumulation of dipeptide repeat proteins predates that of TDP-43 in Frontotemporal Lobar Degeneration associated with hexanucleotide repeat expansions in C9ORF72 gene. Neuropathol Appl Neurobiol 41:601–612. doi:10.1111/nan.12178
Article CAS PubMed Google Scholar - Bieniek KF, Murray ME, Rutherford NJ, Castanedes-Casey M, Dejesus-Hernandez M, Liesinger AM, Baker MC, Boylan KB, Rademakers R, Dickson DW (2013) Tau pathology in frontotemporal lobar degeneration with C9ORF72 hexanucleotide repeat expansion. Acta Neuropathol 125:289–302. doi:10.1007/s00401-012-1048-7
Article PubMed Central CAS PubMed Google Scholar - Brettschneider J, Van Deerlin VM, Robinson JL, Kwong L, Lee EB, Ali YO, Safren N, Monteiro MJ, Toledo JB, Elman L et al (2012) Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol 123:825–839. doi:10.1007/s00401-012-0970-z
Article PubMed Central PubMed Google Scholar - Buckner RL (2013) The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging. Neuron 80:807–815. doi:10.1016/j.neuron.2013.10.044
Article CAS PubMed Google Scholar - Byrne S, Elamin M, Bede P, Shatunov A, Walsh C, Corr B, Heverin M, Jordan N, Kenna K, Lynch C et al (2012) Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol 11:232–240. doi:10.1016/S1474-4422(12)70014-5
Article PubMed Central CAS PubMed Google Scholar - Davidson YS, Barker H, Robinson AC, Thompson JC, Harris J, Troakes C, Smith B, Al-Saraj S, Shaw C, Rollinson S et al (2014) Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 2:70. doi:10.1186/2051-5960-2-70
Article PubMed Central PubMed Google Scholar - DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J et al (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72:245–256. doi:10.1016/j.neuron.2011.09.011
Article PubMed Central CAS PubMed Google Scholar - Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J et al (2013) Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol 126:829–844. doi:10.1007/s00401-013-1192-8
Article PubMed Central CAS PubMed Google Scholar - Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW III, Sun S, Herdy JR, Bieri G et al (2015) Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18:1226–1231. doi:10.1038/nn.4085
Article CAS PubMed Google Scholar - Koziol LF, Budding D, Andreasen N, D’Arrigo S, Bulgheroni S, Imamizu H, Ito M, Manto M, Marvel C, Parker K et al (2014) Consensus paper: the cerebellum’s role in movement and cognition. Cerebellum 13:151–177. doi:10.1007/s12311-013-0511-x
Article PubMed Central PubMed Google Scholar - Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL (2014) Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345:1139–1145. doi:10.1126/science.1254917
Article PubMed Central CAS PubMed Google Scholar - Lomen-Hoerth C, Murphy J, Langmore S, Kramer JH, Olney RK, Miller B (2003) Are amyotrophic lateral sclerosis patients cognitively normal? Neurology 60:1094–1097
Article CAS PubMed Google Scholar - Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D et al (2013) Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol 126:859–879. doi:10.1007/s00401-013-1181-y
Article CAS PubMed Google Scholar - Mahoney CJ, Beck J, Rohrer JD, Lashley T, Mok K, Shakespeare T, Yeatman T, Warrington EK, Schott JM, Fox NC et al (2012) Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: clinical, neuroanatomical and neuropathological features. Brain 135:736–750. doi:10.1093/brain/awr361
Article PubMed Central PubMed Google Scholar - Mann DM, Rollinson S, Robinson A, Bennion Callister J, Thompson JC, Snowden JS, Gendron T, Petrucelli L, Masuda-Suzukake M, Hasegawa M et al (2013) Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 1:68. doi:10.1186/2051-5960-1-68
Article PubMed Central PubMed Google Scholar - May S, Hornburg D, Schludi MH, Arzberger T, Rentzsch K, Schwenk BM, Grasser FA, Mori K, Kremmer E, Banzhaf-Strathmann J et al (2014) C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol 128:485–503. doi:10.1007/s00401-014-1329-4
Article PubMed Central CAS PubMed Google Scholar - Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J et al (2014) C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345:1192–1194. doi:10.1126/science.1256800
Article CAS PubMed Google Scholar - Mori K, Arzberger T, Grasser FA, Gijselinck I, May S, Rentzsch K, Weng SM, Schludi MH, van der Zee J, Cruts M et al (2013) Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol 126:881–893. doi:10.1007/s00401-013-1189-3
Article CAS PubMed Google Scholar - Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C et al (2013) The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339:1335–1338. doi:10.1126/science.1232927
Article CAS PubMed Google Scholar - Murray ME, DeJesus-Hernandez M, Rutherford NJ, Baker M, Duara R, Graff-Radford NR, Wszolek ZK, Ferman TJ, Josephs KA, Boylan KB et al (2011) Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol 122:673–690. doi:10.1007/s00401-011-0907-y
Article PubMed Central CAS PubMed Google Scholar - Petersen SE, Fox PT, Posner MI, Mintun M, Raichle ME (1989) Positron emission tomographic studies of the processing of singe words. J Cogn Neurosci 1:153–170. doi:10.1162/jocn.1989.1.2.153
Article CAS PubMed Google Scholar - Pikkarainen M, Hartikainen P, Alafuzoff I (2010) Ubiquitinated p62-positive, TDP-43-negative inclusions in cerebellum in frontotemporal lobar degeneration with TAR DNA binding protein 43. Neuropathology 30:197–199. doi:10.1111/j.1440-1789.2009.01043.x
Article PubMed Google Scholar - Proudfoot M, Gutowski NJ, Edbauer D, Hilton DA, Stephens M, Rankin J, Mackenzie IR (2014) Early dipeptide repeat pathology in a frontotemporal dementia kindred with C9ORF72 mutation and intellectual disability. Acta Neuropathol 127:451–458. doi:10.1007/s00401-014-1245-7
Article CAS PubMed Google Scholar - Prudencio M, Belzil V, Batra R, Ross CA, Gendron TF, Pregent L, Murray ME, Overstreet K, Piazza-Johnston A, Desaro P et al (2015) Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci 18:1175–1182. doi:10.1038/nn.4065
Article CAS PubMed Google Scholar - Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L et al (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72:257–268. doi:10.1016/j.neuron.2011.09.010
Article PubMed Central CAS PubMed Google Scholar - Rohrer JD, Isaacs AM, Mizielinska S, Mead S, Lashley T, Wray S, Sidle K, Fratta P, Orrell RW, Hardy J et al (2015) C9orf72 expansions in frontotemporal dementia and amyotrophic lateral sclerosis. Lancet Neurol 14:291–301. doi:10.1016/S1474-4422(14)70233-9
Article CAS PubMed Google Scholar - Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, Arzberger T, Edbauer D (2015) Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. doi:10.1007/s00401-015-1450-z
Google Scholar - Schmahmann JD, Sherman JC (1998) The cerebellar cognitive affective syndrome. Brain 121(Pt 4):561–579
Article PubMed Google Scholar - Sha SJ, Takada LT, Rankin KP, Yokoyama JS, Rutherford NJ, Fong JC, Khan B, Karydas A, Baker MC, DeJesus-Hernandez M et al (2012) Frontotemporal dementia due to C9ORF72 mutations: clinical and imaging features. Neurology 79:1002–1011. doi:10.1212/WNL.0b013e318268452e
Article PubMed Central PubMed Google Scholar - Snowden JS, Neary D, Mann DM (2002) Frontotemporal dementia. Br J Psychiatry 180:140–143
Article PubMed Google Scholar - Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P et al (2014) Discovery of a biomarker and lead small molecules to target r (GGGGCC)-associated defects in c9FTD/ALS. Neuron 83:1043–1050. doi:10.1016/j.neuron.2014.07.041
Article PubMed Central CAS PubMed Google Scholar - Tao Z, Wang H, Xia Q, Li K, Jiang X, Xu G, Wang G, Ying Z (2015) Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet 24:2426–2441. doi:10.1093/hmg/ddv005
Article CAS PubMed Google Scholar - van Blitterswijk M, Dejesus-Hernandez M, Niemantsverdriet E, Murray ME, Heckman MG, Diehl NN, Brown PH, Baker MC, Finch NA, Bauer PO et al (2013) Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol 12:978–988. doi:10.1016/S1474-4422(13)70210-2
Article PubMed Google Scholar - Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB et al (2014) Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84:1213–1225. doi:10.1016/j.neuron.2014.12.010
Article CAS PubMed Google Scholar - Whitwell JL, Weigand SD, Boeve BF, Senjem ML, Gunter JL, Dejesus-Hernandez M, Rutherford NJ, Baker M, Knopman DS, Wszolek ZK et al (2012) Neuroimaging signatures of frontotemporal dementia genetics: C9ORF72, tau, progranulin and sporadics. Brain 135:794–806. doi:10.1093/brain/aws001
Article PubMed Central PubMed Google Scholar - Woolley SC, York MK, Moore DH, Strutt AM, Murphy J, Schulz PE, Katz JS (2010) Detecting frontotemporal dysfunction in ALS: utility of the ALS cognitive behavioral screen (ALS-CBS). Amyotroph Lateral Scler 11:303–311. doi:10.3109/17482961003727954
Article PubMed Google Scholar - Yamakawa M, Ito D, Honda T, Kubo K, Noda M, Nakajima K, Suzuki N (2015) Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum Mol Genet 24:1630–1645. doi:10.1093/hmg/ddu576
Article CAS PubMed Google Scholar - Yang D, Abdallah A, Li Z, Lu Y, Almeida S, Gao FB (2015) FTD/ALS-associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions. Acta Neuropathol. doi:10.1007/s00401-015-1448-6
PubMed Google Scholar - Zhang YJ, Jansen-West K, Xu YF, Gendron TF, Bieniek KF, Lin WL, Sasaguri H, Caulfield T, Hubbard J, Daughrity L et al (2014) Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol 128:505–524. doi:10.1007/s00401-014-1336-5
Article PubMed Central CAS PubMed Google Scholar - Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH et al (2013) RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA 110:E4968–E4977. doi:10.1073/pnas.1315438110
Article PubMed Central CAS PubMed Google Scholar
Acknowledgments
This work was supported by the National Institutes of Health/National Institute of Neurological Disorders and Stroke [R21NS089979 (TFG, KBB), R21NS084528 (LP), R01NS088689 (LP), R01NS063964 (LP), R01NS077402 (LP), P01NS084974 (LP, DD, RR, KBB), R01NS080882 (RR), F32NS087842 (JJ)], Department of Defense [ALSRP AL130125 (LP)], Mayo Clinic Foundation (LP), ALS Association (TFG, KBB, LP, DWC), Target ALS (LP, CLT), Robert Packard Center for ALS Research at Johns Hopkins (LP), the ALS Therapy Alliance (RR), the Ludwig Institute for Cancer Research (DWC, CLT), and the European Union’s Seventh Framework Programme [FP7/2014-2019 grant 617198 (DE)]. Dr. Van Blitterswijk is supported by the Milton Safenowitz Post Doctoral Fellowship for ALS research from the ALS Association, and the Clinical Research in ALS and Related Disorders for Therapeutic Development (CReATe) Clinical Research Fellowship (U54NS092091).
Author information
Author notes
Authors and Affiliations
- Department of Neuroscience, Mayo Clinic, Jacksonville, FL, 32224, USA
Tania F. Gendron, Marka van Blitterswijk, Kevin F. Bieniek, Lillian M. Daughrity, Melissa E. Murray, Monica Castanedes-Casey, Linda Rousseau, Rosa Rademakers, Dennis W. Dickson & Leonard Petrucelli - Mayo Graduate School, Mayo Clinic, Rochester, MN, 55905, USA
Kevin F. Bieniek - Ludwig Institute, University of California at San Diego, La Jolla, CA, 92093, USA
Jie Jiang, Clotilde Lagier-Tourenne & Don W. Cleveland - Department of Psychiatry and Psychology, Mayo Clinic, Jacksonville, FL, 32224, USA
Beth K. Rush, Otto Pedraza & John A. Lucas - Department of Neurology, Mayo Clinic, Jacksonville, FL, 32224, USA
Pamela Desaro, Amelia Robertson, Karen Overstreet, Neill R. Graff-Radford & Kevin B. Boylan - Section of Biostatistics, Mayo Clinic, Jacksonville, FL, 32224, USA
Colleen S. Thomas & Julia E. Crook - Department of Neurology, Mayo Clinic, Rochester, MN, 55905, USA
Keith A. Josephs, Joseph E. Parisi, David S. Knopman, Ronald C. Petersen & Bradley F. Boeve - Department of Neurosciences, University of California at San Diego, La Jolla, CA, 92093, USA
Clotilde Lagier-Tourenne - German Center for Neurodegenerative Diseases (DZNE), Munich, Germany
Dieter Edbauer - Institute for Metabolic Biochemistry, Ludwig-Maximilians University Munich, Munich, Germany
Dieter Edbauer - Munich Cluster of Systems Neurology (SyNergy), Munich, Germany
Dieter Edbauer - Department of Cellular and Molecular Medicine, University of California at San Diego, La Jolla, CA, 92093, USA
Don W. Cleveland
Authors
- Tania F. Gendron
You can also search for this author inPubMed Google Scholar - Marka van Blitterswijk
You can also search for this author inPubMed Google Scholar - Kevin F. Bieniek
You can also search for this author inPubMed Google Scholar - Lillian M. Daughrity
You can also search for this author inPubMed Google Scholar - Jie Jiang
You can also search for this author inPubMed Google Scholar - Beth K. Rush
You can also search for this author inPubMed Google Scholar - Otto Pedraza
You can also search for this author inPubMed Google Scholar - John A. Lucas
You can also search for this author inPubMed Google Scholar - Melissa E. Murray
You can also search for this author inPubMed Google Scholar - Pamela Desaro
You can also search for this author inPubMed Google Scholar - Amelia Robertson
You can also search for this author inPubMed Google Scholar - Karen Overstreet
You can also search for this author inPubMed Google Scholar - Colleen S. Thomas
You can also search for this author inPubMed Google Scholar - Julia E. Crook
You can also search for this author inPubMed Google Scholar - Monica Castanedes-Casey
You can also search for this author inPubMed Google Scholar - Linda Rousseau
You can also search for this author inPubMed Google Scholar - Keith A. Josephs
You can also search for this author inPubMed Google Scholar - Joseph E. Parisi
You can also search for this author inPubMed Google Scholar - David S. Knopman
You can also search for this author inPubMed Google Scholar - Ronald C. Petersen
You can also search for this author inPubMed Google Scholar - Bradley F. Boeve
You can also search for this author inPubMed Google Scholar - Neill R. Graff-Radford
You can also search for this author inPubMed Google Scholar - Rosa Rademakers
You can also search for this author inPubMed Google Scholar - Clotilde Lagier-Tourenne
You can also search for this author inPubMed Google Scholar - Dieter Edbauer
You can also search for this author inPubMed Google Scholar - Don W. Cleveland
You can also search for this author inPubMed Google Scholar - Dennis W. Dickson
You can also search for this author inPubMed Google Scholar - Leonard Petrucelli
You can also search for this author inPubMed Google Scholar - Kevin B. Boylan
You can also search for this author inPubMed Google Scholar
Corresponding authors
Correspondence toLeonard Petrucelli or Kevin B. Boylan.
Additional information
T. F. Gendron and M. van Blitterswijk contributed equally to this work.
Electronic supplementary material
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Gendron, T.F., van Blitterswijk, M., Bieniek, K.F. et al. Cerebellar c9RAN proteins associate with clinical and neuropathological characteristics of C9ORF72 repeat expansion carriers.Acta Neuropathol 130, 559–573 (2015). https://doi.org/10.1007/s00401-015-1474-4
- Received: 20 July 2015
- Revised: 29 August 2015
- Accepted: 30 August 2015
- Published: 08 September 2015
- Issue Date: October 2015
- DOI: https://doi.org/10.1007/s00401-015-1474-4