Innate immune regulations and various siRNA modalities (original) (raw)
- 3466 Accesses
- 8 Citations
- 1 Altmetric
- Explore all metrics
Abstract
RNAi therapeutics are designed to produce the precise silencing effects against the gene-linked diseases which were known to be untreatable in the past. The highly immunostimulatory nature of siRNA enhances the off-target effects and easily get attacked by nucleases; hence, their modulation is essentially required for accurate alterations to be made in the structures to intensify the pharmacological attributes. The phosphonate modifications act as shield against undue phosphorylation effects, and the molecular changes in ribose sugar lowers the level of immunogenicity and increases the binding efficacy. When bases are substituted with virtual/or pseudo bases, they eventually reduce the off-target effects. These changes modulate the nucleic acid sensors and control the hyper-activation of innate immune response. Various modification designs based on STC (universal pattern), ESC, ESC + (advanced patterns) and disubstrate have been explored to silence the gene expression of various diseases e.g., hepatitis, HIV, influenza, RSV, CNV and acute kidney injury. This review describes the various innovative siRNA therapeutics and their implications on the developed immune regulations to silence the disease effects.
Graphical Abstract
siRNA causes the silencing effects through RISC processing. The innate immune signalling is induced by both TLR-dependent and TLR-independent pathways. Modification chemistries are utilized to modulate the immune response.

Similar content being viewed by others
Introduction
Interfering RNA (RNAi) technology is being employed to target the disease-associated gene to knockdown/or silence its debilitating effect by halting the activity of associated mRNA, therefore, repressing the translation process. Fire and Mello, in 1998, have demonstrated the RNAi concept for the first time, using dsRNA in Caenorhabditis elegans to observe the silencing effects. These discoveries on the RNA-mediated process of gene regulations have advanced our knowledge in this field [[1](/article/10.1007/s13346-023-01361-4#ref-CR1 "Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;19,391,6669:806–11. https://doi.org/10.1038/35888
.")\]. Later on, the work has been extended to watch the effects of siRNA molecular machines in plants. The final effects demonstrated the guide sequence-dependent endo-nucleolytic cleavage was linked to inhibit the translation by mRNA \[[2](/article/10.1007/s13346-023-01361-4#ref-CR2 "Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19(5):517–29.
https://doi.org/10.1101/gad.1284105
.")\]. By 2001, microRNA (miRNA) has also been explored and classified in small RNA regulators \[[3](/article/10.1007/s13346-023-01361-4#ref-CR3 "Bartel DP. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97.
https://doi.org/10.1016/s0092-8674(04)00045-5
.")\]. In the past, siRNA therapeutics development has faced many challenges regarding stability and specificity along with accurate drug delivery to the target tissues. But, the most recent advancements in the arena of chemistry, genetics and biotechnology have revolutionized the field of nucleic acid developments to the next level.In the past decade, incredible efforts have been made to study many chemical modification geometries and to analyse and evaluate their efficacy and biosafety in modified-siRNA drugs. siRNA/or mRNA used in higher concentration, without modification-treatments, can enhance the immunostimulatory effects of nucleic acid sensors initiating the strong innate immune response, by secreting excessive IFN-α. The nucleoside modifications are an effective tool to increase stability and reduce the immunogenicity of nucleic-acid therapeutics [[6](/article/10.1007/s13346-023-01361-4#ref-CR6 "Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Kariko K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39(21):9329–38. https://doi.org/10.1093/nar/gkr586
.")\]. Various siRNA modification chemistries are being used to control the innate immune stimulations in context with increasing potency and decreasing the involved toxicities. Off-target effects mediated by siRNAs, however, is one more parameter which considerably affect the precision and accuracy in the drug effectiveness. Chemical modifications of nucleotides can be placed at the phosphate backbone, ribose moiety and base. Phosphothioate (PS)-modified oligonucleotides are hydrophobic and more stable with an increased half-life of oligonucleotides and enhance their affinity to specific proteins. The protein binding capacity of oligonucleotides is advantageous to penetrate into the cells, but excessive bindings could lead the in-vivo toxicity with a consequence of eliminating of drug metabolites from the body. siRNA joined with PS will not make any changes in-vivo biodistribution and can accumulate in the liver, kidney, intestine, bone marrow, lymph nodes etc. \[[4](/article/10.1007/s13346-023-01361-4#ref-CR4 "Crooke ST, Wang S, Vickers TM, Shen W, Liang XH. Cellular uptake and trafficking of antisense oligonucleotides. Nat Biotechnol. 2017;35(3):230–7.")\]. 2′-Deoxy-2′-fluoro (2′-F) modification of adenosine reduces the cytokine production substantially while retaining knockdown activity of siRNA. Substitution with bases would provide the advantage of being resistant to the attack of nucleases, which is also a crucial point for nucleic acid-based drug development \[[5](/article/10.1007/s13346-023-01361-4#ref-CR5 "Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16(11):1833–40."), [6](/article/10.1007/s13346-023-01361-4#ref-CR6 "Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Kariko K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39(21):9329–38.
https://doi.org/10.1093/nar/gkr586
.")\].Additionally, various siRNA drug delivery platforms are being used and analysed in biodistribution studies such as, lipid nanoparticles (LNPs), drug phospholipid complex (DPC™), trimethylolpropane trimethacrylate (TRiM™), acetylgalactosamine (GalNAc-siRNA conjugates), local drug EluteR biodegradable (LODER™) polymers, exosomes and polypeptide nanoparticles (PNPs). The most popular platforms are, siRNA-LNPs have shown greater efficiency and GalNAc-siRNA also provided the better results through intravenous administration [7,8,9, 16].
RNA is constitutive of single-stranded oligonucleotides, which provides no apparent pathogenic and inflammatory response, despite being highly potent in nature [10, 11]. However, the long-double stranded dsRNA or RNA accumulated in higher concentration can induce the innate immune response, which could be TLR-mediated and non-TLR-mediated, in the cytoplasm to degrade the nucleic acid of the pathogens. Non-TLR-mediated immune response is mainly triggered by retinoic acid-inducible gene-1 (RIG-1), and its proteins bind to siRNA in the cytoplasm [12]. Toll-like receptors (TLR7) and myeloid differentiation primary response 88 (MyD88) adaptor recognize the influenza infection; however, TLR 7 also gets induced by molecules of non-viral origin and can sense the endosomal ssRNA to detect RNA viruses. dsRNA is known as the potent stimulator of innate response. GU-rich ssRNA oligonucleotides (RNA 40) derived from human immunodeficiency (HIV) virus activates the dendritic cells (DCs) to release interferon-α and proinflammatory cytokines. MyD88 is the central node linked to IL-1R receptor-associated kinases (IRAK), a family of kinases, including nuclear factor kappa B (NF-kB) [13,14,15].
The first RNAi-based therapeutic ONPATTRO® (patisiran, ALNTTRO2), for the treatment of hereditary transthyretin amyloidosis (hATTR amyloidosis) with polyneuropathy in adults, is approved by FDA and EC in 2018 for commercial manufacturing [16, 17]. The present review illustrates the various innovative siRNA therapeutic designs with modification chemistries and its subsequent impact on innate immune response to substantiate the silencing efficiency and lowering down the associated toxicities.
Mechanism of RNAi
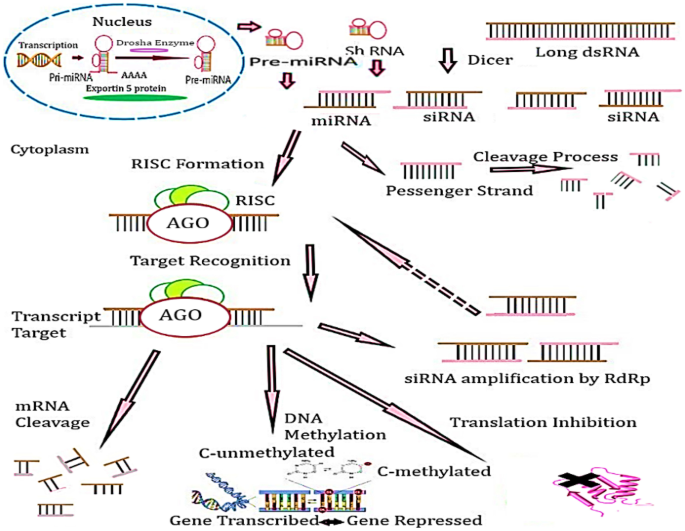
siRNA is ~ 21–22 bp long dsRNA molecules, which recognize the RNAi enzymatic machinery leading to a homology dependent degradation of target mRNA. Dicer is complexed with TAR (trans-activation response)-RNA binding protein (TRBP) presents siRNA to RNA-induced silencing complex (RISC). Protein argonaute-2 (Ago2) cleaves mRNA molecules between 10 and 11 basis relative to the 5′ end of antisense siRNA. The catalytic activity of RNA-induced silencing complex (RISC) cleaves the strand from loaded siRNA as ‘passenger strand’ released with a single-stranded guide RNA molecule that directs the specificity of the target molecule by intermolecular base pairing (Fig. 1) [18, 19]. siRNA unwinds the 5′ end, a less thermodynamically stable ends, of the guide strand which joins Ago-2. The complementary RNA molecules are recognized by guide RNA and cleaved by the catalytic activity of Ago-2. siRNA targets mRNA to destabilize the transcripts or repress the translation, if they bind to an endogenous substrate of RNAi as micro miRNA. The primary microRNA transcripts (pri-miRNA) are expressed within the nucleus. This is processed within nucleus into 60–70 bp hairpins processed by microprocessor complex-drosha enzyme (Drosha-DGCR8) (Fig. 1). The loop is escaped into the cytoplasm by RNase III Dicer. One of the two strands is loaded into the RISC. The mature miRNA shares partial complementarity with 3′UTR of the target mRNA. miRNA results in translation repression, which can be accompanied by degradation [20].
Fig. 1
RNAi gene silencing mechanism. Long ds RNA is cleaved by a dicer into siRNA/miRNAs. RNA-induced silencing complex assists in finding the complementary sequence in mRNA transcript, consequently deactivating the translation on that specific gene and increasing the cytosine methylation and mRNA cleavage. The pri-miRNA are expressed within the nucleus. This is processed within the nucleus into ‘Drosha-DGCR8’. The loop is escaped into the cytoplasm by RNase III Dicer. One of the two strands is loaded into the RISC
The siRNA duplexes are processed by their direct delivery to the target cells or by intracellular processing of the longer RNA hairpin transcripts which are mainly produced by DNA. Short hairpin RNA (shRNA) is transported to the cytoplasm and processed into siRNA by Dicer. The designed therapeutics work on the first strategy as the direct siRNA effectors are the consequence of potent gene silencing. But these require repeated administration of the drugs in a clinical setting; which is not a cost-effective process. However, DNA-based RNAi drugs have more potential of being stably introduced in a gene therapy setting, allowing a single treatment of viral vector delivered shRNA genes [21].
Modification chemistries for siRNA — a solution for increasing potency and reducing toxicities
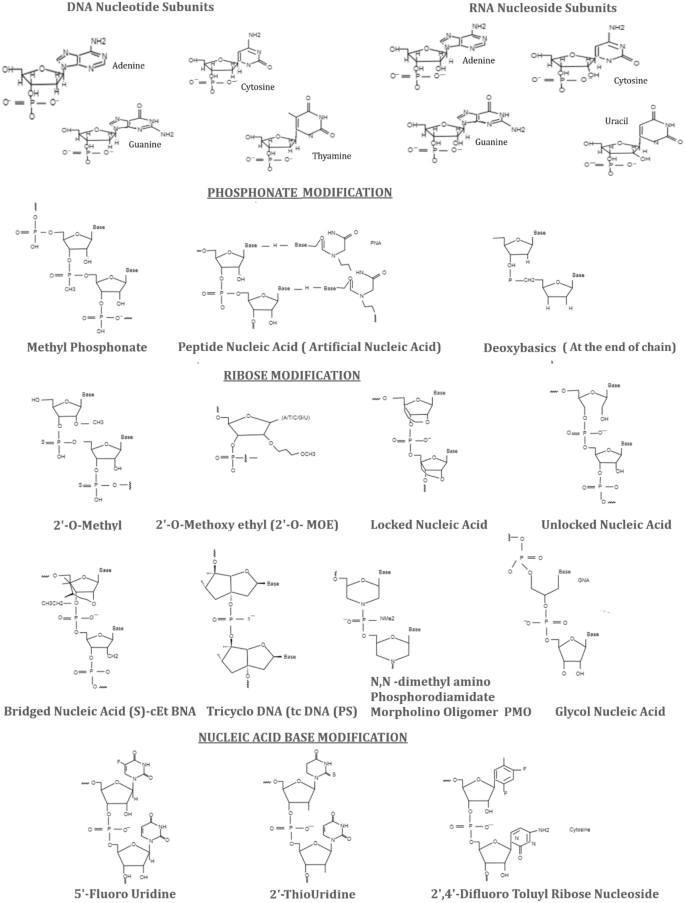
siRNAs and ASO can be modified structurally based on phosphonate; ribose and base analogs (Fig. 2). These changes are carried out on H/OH for RNA or DNA, ethyl bicyclic nucleic acid (S) (cEt-BNA(S)) and phosphorodiamidate morpholino oligomer (PMO). siRNA modifications by substituting 2′-OH with 2′-methoxyethyl (2′-OMe or 2′-MOE), with locked nucleic acid (LNA), or unlocked-nucleic acid (UNA) or glycol nucleic acid (GNA), have been competently observed to supress innate immunostimulatory response effectively driven by siRNA (Fig. 2). These various chemical geometries also offer enhanced potency, specificity and potentially abrogate the lethal effects [21, 22]. For instance, the combination of 2′-OMe and PS modifications facilitate the systemic administration of cholesterol-conjugated siRNA to achieve the efficient ApoB mRNA silencing in the liver and jejunum (in-vivo model) [23]. In addition, these combinations of 2′-OMe and 2′-F have also been used for ONPATTRO® [24, [25](/article/10.1007/s13346-023-01361-4#ref-CR25 "Adams D, Ganzalez-Duarte A, O’Riordan WD, Yang CC, Ueda M, Kristen AV, Tournev I, Schmidt HH, Coelho T, Berk JL, Lin KP, Vita G, Attarian S, Plante’-Bordeneuve V, Mezei MM, Campistol JM, Buades J, Brannagan TH, Kim BJ, Oh J, Parman Y, Sekijima Y, Hawkins PN, Solomon SD, Polydefkis M, Nochur SV, Sweetser M, Garg PP, Vaishnaw AK, Gollob JR and Suhr OB,. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med. 2018;379(1):11–21. https://doi.org/10.1056/NEJMoa1716153
.")\]. miRNA inhibitors (AntimiRs) or miRNA mimics down or upregulate the miRNAs. Miravirsen, an AntimiR-122, has been used in trials for hepatitis C virus infection.Fig. 2
Phosphonate, ribose and base modifications of siRNAs. The original forms of DNA nucleotide and RNA nucleoside subunits are also presented with all the bases [adenine, guanine, cytosine, thymine (DNA) and uracil (RNA)]. Phosphonate modifications secure the RNA from phosphorylation e.g. methyl phosphonate, peptide nucleic acid and deoxybasics. Ribose sugars often provide protection from nuclease attack and base affinity for stability, for example, 2′-O-methyl, 2-O-methoxy ethyl, locked nucleic acid, unlocked nucleic acid (UNA), (S)-cEt BNA bridged nucleic acid, tricyclo DNA tc DNA (PS), N, N-dimethyl amino phosphorodiamidate morpholino oligomer (PMO) and glycol nucleic acid (GNA). The base analogs reduce the excessive immune stimulations e.g. 5′-fluorouridine, 2′-thiouridine and 2,4-difluoro toluyl ribose nucleoside
In the beginning, unmodified and partially modified siRNA (21 nucleotide sequences or more) were tried to silence the genes CNV in-vivo on the local tissues such as, eyes. Modified and unmodified siRNA drugs e.g., Bevasiranib, AGN211745 and AGN21174526 were designed to inhibit the activity of VEGF, in the treatment of macular degeneration-AMD a leading cause of blindness and neovascular AMD. These drugs activate TLR3 and TRIF to induce IL-12 and IFN-γ could affect blood and lymphatic system [27, [28](/article/10.1007/s13346-023-01361-4#ref-CR28 "Garba AO, Mousa SA. Bevasiranib for the treatment of wet, age-related macular degeneration. Ophthalmol Eye Dis. 2010;2:75–83. https://doi.org/10.4137/OED.S4878
.")\].Phosphonate modification
ASO modification on the PS, where oxygen from phosphodiester is substituted by sulphur, is able to protect from nucleases. It also enables the rapid attachment to albumin assisting in cell penetration than naked siRNA. But the excessive binding could lead to in-vivo toxicity, consequently, accumulating the drug in liver, kidney, intestine, bone marrow, lymph nodes etc. with slow elimination of drug metabolites [4, 29, 30]. The neutral phosphodiester gp of phosphate backbone siRNA allows the easy delivery into cells, where thioesterase converts modified siRNA into its native form to attain the robust results [52]. PS-DNA oligomers bind to the active site of primers to inhibit the activity of reverse transcriptase (RT) of HIV-1 [31]. PS linkage stereoisomers Rp and Sp influence the performance of siRNA from 3′ to 5′ antisense strand and are the superior over RNase activities (Fig. 2) [9]. PS modifications increase the oligos; stability for their effective transportation.
The other PS modification chemistries offer special pharmacological properties. For instance, PS2 increase the affinity between RISC and siRNA; methyl phophonate (MP) and methoxy propyl phosphonate (MOP) reduce the ASO protein bindings, hence, decrease the toxicity especially MOP linkage at 2 and 3 position from 5′ end of DNA gap reduces the hepatotoxicity level of ASO. Peptide nucleic acid (PNA) and phosphotriesters increase the therapeutic capability of siRNA/ASO molecules and able to target certain nucleic acids to enhance its diagnostic skills [30,31,[32](#ref-CR32 "Ndeboko B, Ramamurthy N, Lemamy GJ, Jamard C, Nielsen PE, Cova L. Role of cell-penetrating peptidesin intracellular delivery of peptides nucleic acids targeting hepadnaviral replication. Mol Ther Nucleic Acids. 2017;9:162–9. https://doi.org/10.1016/j.omtn.2017.09.003
."),[33](/article/10.1007/s13346-023-01361-4#ref-CR33 "Singh RP, Oh BK, Choi JW. Application of peptide nucleic acid towards development of nanobiosensor arrays. Bioelectrochemistry. 2010;79:153–61.")\].For phosphonate modification, phosphate at 5′ of siRNA is introduced exogenously, modified by either phosphorylation mediated by cleavage and polyadenylation factor-I subunit-1 (Clp 1) or by chemical synthesis.
5′-Phosphate is rapidly dephosphorylated naturally; hence, it is imperative to select other analogs such as 5′-(E)-vinyl phosphonate (5′-(E)-VP), 5′-MP, (S)-5′-C-methyl with phosphate, 5′-PS etc. with similar conformations to protect from dephosphorylation and increase the activity, potency and stability. These analogues were evaluated for their effectiveness in-vivo and in-vitro. The substitute of oxygen and carbon with E-vinyl phosphonate moieties at 5′ end could improve the potency by 20 folds [34]. The intact stable oligonucleotides are effectively loaded on the RISC, help in achieving the appropriate pharmacokinetic properties. Therefore, this modification applied to the ds-siRNA improves the potency level in-vivo by its efficient accumulation in tissues [35].
Ribose and base modifications
Ribose sugar modifications at 2′ position protect siRNA from the attack of ribonucleases. 2′-OMe is a natural ribose sugar and used frequently in the course of ribose modification in the drug development process. 2′-OMe increases the stability, having a greater affinity to target its mRNA and reducing the immunogenicity [36]. Other analogs 2′MOE and 2′-F also help increase the binding affinity. 2′-O-benzyl or 2-methyl-4-Pyridine (six 2′-O-CH2Py (4)) are well tolerated on the guide strand and also help increase the activity, even these modifications have to be placed at the 8 and 15 positions on the guide strand [37, 38]. Other molecules e.g. UNAs, LNAs, GNAs, (S)-cEt-BNAs, tricyclo-DNA (tcDNA) and PMOs are able to increase the affinity of base pairing (Fig. 2). The conformational flexibility of nucleotides is decreased due to ribose and base modifications, which increase their binding affinity. For example, locked nucleic acid (LNA) is linked to 2′oxygen and 4′ carbon of ribose showed enhanced binding affinity. Its methylated analogue is also denoted as constraint (cEt) BNA. tcDNA is another constrained nucleotide having the attributes towards increasing the binding affinity; it is, however, a bit smaller than LNA (i.e. ΔTm ∼2 °C for tc DNA and ΔTm 4 to 8 °C per modification for LNA). Stable chimeric oligonucleotides may be prepared by various sugar modifications with higher affinity, which could help eliminate the negative effects caused by another modification [36, [39](/article/10.1007/s13346-023-01361-4#ref-CR39 "Watts JK. Locked nucleic acid: tighter is different. Chem Commun (Camb). 2013;49(50):5618–20. https://doi.org/10.1039/c3cc40340h
. Epub 2013 May 16.")\].Substitution with bases would provide the advantage of being resistant to nucleases in nucleic acid–based drug development. The base analogues e.g. pseudouridine, thiouridine, methyladenosine and methyl cytidine; their cytidine and uridine (Fig. 2) residues can help attenuate the innate immune response, which ensure that the designed ASO drugs are being more resistant to the attacks of nucleases. Pseudouridine was found to enhance the translational capacity and biological stability in mice models. These chemicals, indeed, are considered to play a significant role in research and development of molecular medicines. However, the main concerns for using these synthetic molecules, being added to the specific genome, are ought to be metabolized safely in the human body [5, [6](/article/10.1007/s13346-023-01361-4#ref-CR6 "Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Kariko K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39(21):9329–38. https://doi.org/10.1093/nar/gkr586
.")\].2′–5′ Oligoadenylate synthetase (2′–5′-OAS) and ribonuclease L (latent) RNase L, induced by interferons, are involved in the sensory and effector functions following the viral infections. OAS catalyses to produce 2′–5′-linked oligo adenylates (2–5A) that activates RNase L, resulting in breaking down the single-stranded self and non-self RNA. Therefore, modified nucleosides are present in cellular transcripts have been shown to suppress activation of several RNA sensors [[6](/article/10.1007/s13346-023-01361-4#ref-CR6 "Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Kariko K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39(21):9329–38. https://doi.org/10.1093/nar/gkr586
.")\].An adenosine analogue e.g. N-ethylpiperidine 7-EAA triazole (7-EAA, 7-ethynyl-8-aza-7-deazaadenosine), when added to the RNA strand pairs with uridine to form helix structure. This modified structure could attenuate the swift interactions between TLR8 and nucleotide of drug; therefore, it weakens the strong immunogenicity/immunostimulatory reaction to increase the safety of siRNA [40].
Phenylpyrrolocytosine (PhpC-6′) is a cytosine mimic, virtually identical to natural cytosine in siRNA, which could provide incredible base-pair fidelity, thermal stability and gene silencing activity. siRNA containing Php-C tends to accumulate in cytoplasm of HeLa cells, as revealed by real-time imaging for cellular trafficking [41]. Fucini et al. reported that adenosine is an important target for optimal modifications. 2′-F modification of adenosine results in a substantial decrease in cytokine production while retaining siRNA knockdown activity [37].
Passenger strand like guide strand of siRNA assembles on RISC causing off-target effects. 5-Nitroindole nucleotides (a universal base) which are incorporated at the 15th position of siRNA passenger strand eventually decrease the efficacy of the same strand, therefore, they help control the off-target-mediated effects [42]. 5-Fluoro-2′-deoxyuridine (FdU) substitution in siRNA can effectively suppress gene expression, induce the repair of damaged DNA, apoptosis and cell death [43]. More studies are being required to mitigate the off-target effects induced by undesired bindings of proteins to target mRNA [44].
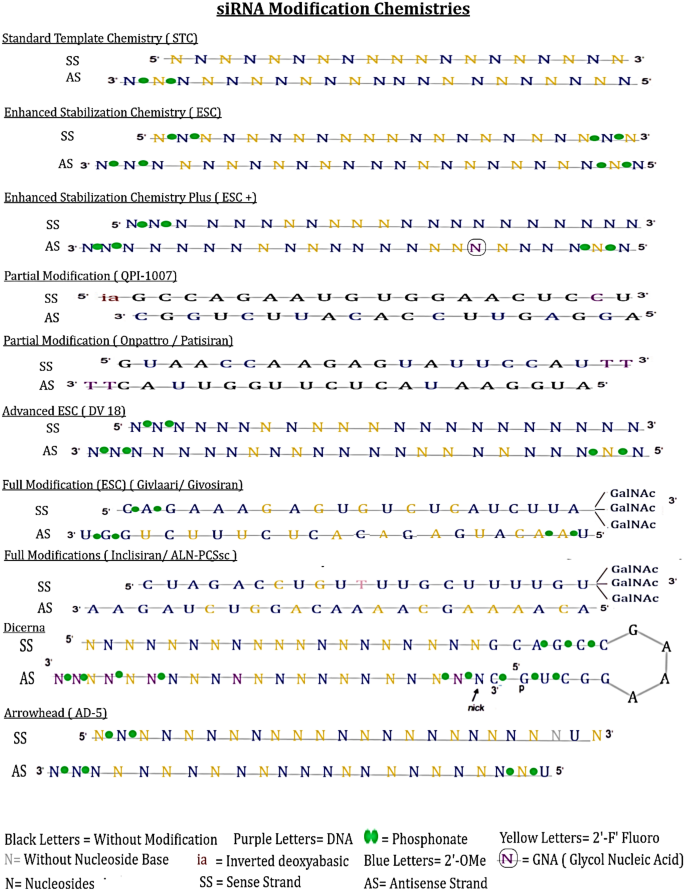
Modification designs of siRNA
The modification chemicals such as 2′-OMe, 2′-F and PS have been validated in many therapeutics in context with the elimination of immunogenic effects and hybridization-dependent off-target effects to improve the nuclear stability and potency [45, 46]. Dar et al. (2016) has prepared a specialized databank for chemically modified siRNAs to provide the understanding of effects through chemical modifications. This repository would furnish the essential information for developing stable and efficacious siRNA for future research [46]. In case of heavy modifications e.g. 2-OMe is placed in the antisense strand of QPI-1007, whereas L-DNA was placed in the sense strand could accelerate the siRNA stability/and biocompatibility. Pre-clinical and clinical validations of various modifications were described by Hu et al. [9]. The universal modification pattern, STC, was developed by Alnylam Pharmaceutical with improved stability and affinity for effective gene silencing. STC chemistry is known to compromise on toxicity point of view.
The new generation pattern, ESC, was proposed with extra 4 PS linkages at the 5′ antisense and 3′ sense strand with reduction in 2′-F substitutions in order to eliminate the associated toxicities essential for pharmacodynamic attributes that consequently reduce the dosing frequency (e.g. Cemdisiran-ALNCC5). The position of 2′-deoxy-2′-fluoro and 2′-O-methyl ribosugar modifications across both strands of the double-stranded siRNA duplex provide advantage with improved potency and sustainable stability without compromising intrinsic RNAi activity [9]. The N-acetyl glucosamine (NAG)-ligand for asialoglycoprotein (ASGPR) receptor is synthesized at the 5′ prime end of the sense strand and suitable for subcutaneous inoculations not intravenous. The NAG ligand directs to trigger the hepatocytes in the liver [47, 49]. This transformative approach can be applied for RNAi therapeutics including other investigational oligonucleotides for their accurate delivery to liver tissues. The nucleotides must remain stable against nucleolytic degradation especially by 5′ exonucleases. GILVAARI™, an approved siRNA therapeutic, is based on ESC design [47, 48].
The advanced ESC designs for DV18 comprised 6PS linkages at three strand terminals, with 2′F modifications at sites 7, 9, 10 and 11 in sense strand (SS) and at 2, 6, 8, 9, 14 and 16 in the antisense strand (AS). The 2′F modifications, in case of DV22, at sites 8 and 9 replaced by 2′-OMe, have provided effective gene silencing in non-human primates in preclinical trials. The conjugates of N-acetylgalactosamine-siRNA are attributed to the hepatotoxicity by escalating the off-target gene silencing mediated by miRNA-like recognition between siRNA and mistargeted RNA.
The technical design for ESC + has the glycol nucleic acid (GNA) substituted in the seed region; as compared to ESC that specifically mitigates the hepatotoxicity. Investigational siRNA therapeutics e.g. ALN-HBVO2, ALN-AATO2 and ALN-AGT of Alnylam therapeutics are based on ESC + design. Chemical modification of ribonucleotides is availed for enhancing stability and reducing the risk of innate immune stimulation, in case of ARC-AAT drug [9]. Arrowhead has added the inverted bases (e.g. deoxythymine-idT) at the strand terminus including unlocked nucleic acid (UNA) and X (without nucleoside base) for AD-5, flanking UAU or UAUAU motifs and siRNA conjugation with hydrophobic substrates, and this design is being used in clinical trials (Fig. 3).
Fig. 3
Representative designs of various siRNA modification patterns used in preclinical and clinical trials. STC — universal modification pattern. ESC — a new generation pattern proposed with extra 4PS linkages at the 5′ (antisense) and 3′ (sense) strand with reduction in 2′-F substitutions. Advanced ESC-designs comprised 6PS linkages at three strand terminals e.g. DV18 has 2′F modifications at sites 7, 9, 10 and 11 in sense strand (SS) and at 2, 6, 8, 9, 14 and 16 in the antisense strand (AS). ESC + design does provide a technical development by substituting GNA. Partial modification QPI-1007, partial modification ESC (Onpattro/Patisiran) and full modification ESC (Givlaari/Givosiran) are based on ESC design. Full modification inclisiran ALN PCSsc design. Disubstrate siRNA (DsiRNA) technology being recognized form Dicerna placing 3 consecutive 2′-F moieties at 9, 10 and 11 position at 5′-end and other site have alternative 2′-OMe and 2′-F moieties in the flank sequence. Arrowhead: at the end of terminus strand contain UNA and X without nucleoside base Hu et al. [9]
The technology on disubstrate siRNA (DsiRNA) has been recognized form Dicerna placing 3 consecutive 2′-F moieties at 9, 10 and 11 are positioned at 5′-end and other site have alternative 2′-OMe and 2′-F moieties in the flank sequence. A constant flank sequence ‘GCAGCCGAAAGGUGC’ contains inner complementary pairing motifs of ‘GCAGCC’ and ‘GGCUGC’, consecutive 2′-OMe moieties used to modify these motifs and consecutive DNA/RNA nucleosides can be positioned without additional modifications in ‘GAAA” bubble. DNA nucleosides can also be added at 2, 12, 16, 18, 20 and 21 positions in antisense strand at 5′-end. GalNAc moieties may be placed at unpaired GAAA nucleotides of DsiRNA. PS linkages are also required to be placed at specific positions in the antisense strand and flank sequences as well [9].
Innate immune signaling (TLR-dependent)
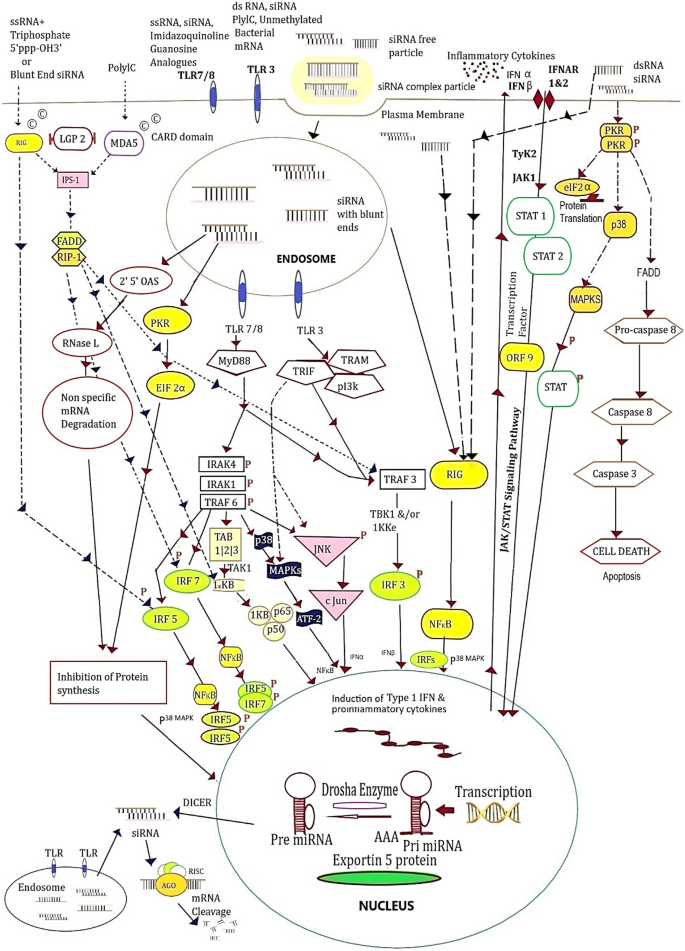
TLR-dependent innate immune signalling, with TLR 3 and other significant TLRs, occurs in the lung, aorta, dermis, choroidal and umbilical vein. Fibroblast cell lines also express TLR3 receptors. Exogenous siRNA/ and dsRNA can also activate TLR 3 and other RNA sensors to escalate the production of cytokines such as, IFNγ and IL-12 (in-vivo) triggering the inflammatory response [27]. siRNA can bind to the TLR 3 ectodomain, which triggers receptor dimerization [[50](/article/10.1007/s13346-023-01361-4#ref-CR50 "Pirher N, Pohar J, Manček-Keber M, Benčina M, Jerala R. Activation of cell membrane-localized Toll-like receptor 3 by siRNA. Immunol Lett. 2017;189. https://doi.org/10.1016/j.imlet.2017.03.019
.")\]. Notably, the use of naked and unmodified siRNA as therapeutics could activate the TLR3 localised on plasma membrane. The long dsRNA induces the endosomal TLRs producing type I IFN response \[[13](/article/10.1007/s13346-023-01361-4#ref-CR13 "Diebold SS, Kaisho T, Hemmi H, Akira S, Reise Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–31."), [51](/article/10.1007/s13346-023-01361-4#ref-CR51 "Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14.")\]. TLR response can also be generated by off-target sequences. It is hypothesized that both naked and conjugated siRNA are less likely to activate TLR3 in cytoplasm/ or endosome e.g. reduction in choroidal neovascularization in TLR3 mice is the cause of a similar effect.Most of the intracellular pathways are induced by activation of TLR 7/8 and TLR3 sensors. These pathways are depicted in Fig. 4. TLR 7/8 signal through myeloid differentiation primary response-88 (MyD88) pathways. TLR3 also signals through TIR domain containing adaptor inducing interferon-β (TRIF adaptor protein) in the cytoplasm. TRIF provides the signals for downstream production of IFN-β and IFN-α via TRAF family-associated NFkB activator binding kinase-1 (TANK-binding kinase-1/ or TBK1), which mediates through a transcription factor, interferon regulatory factor-3 (IRF3) [52, 53]. MyD88 and Toll/interleukin-1 receptors (TIR) are also associated through TLR7/8 to form a signalling complex of intermediate molecules like, IL-1 receptor associated kinase-1 (IRAK-1), IRAK-4 and TNFR-associated factor-6 (TRAF6). The subsequent signalling events and nuclear translocation of NFkB lead to the activation of transcription factors such as IRF5 and IRF7 that upregulate the expression of IFNα and inflammatory cytokines. This process occurs mostly in DC and B-cells. TLR7 and TLR8 ligands tend to follow the MyD88-IRF7 pathway (Fig. 4).
Fig. 4
TLR 3, TLR 7/8, RIG-1/MDA5 and PKR sense the siRNA and dsRNA. TLR7 and TLR8 generate the signals through their endosomal presence. MyD88, an adaptor protein, pathways through a complex IRAK-1, IRAK-4 and TRAF-6; and it also activates TRAF-3 pathway to regulate and translocate NF-κβ releasing cytokines, through IRF-5, IRF-7 and TAB1-3 via kinase 1KKB signaling with subunits p50 and p65. A stress stimulus for cytokines for autophagy/or apoptosis is produced via p38 (MAPK). IRAK-1, IRAK-4 and TRAF-6 also activate ATF2-c-Jun transcription factors to regulate the transcription of genes to express interferon IFNα. TLR-3 receptors from endosomes begin the sensing process via TRIF adaptor which activates IRF3 and IFNβ expression. NF-κβ and ATF2-c-Jun would also cause the release of inflammatory cytokines through TLR3 signaling. Triphosphate-ssRNA and blunt-ended dsRNA binds to the cell membrane followed by RIG-1 sensors through IPS-1 protein adaptor could generate IRF 5, IRF 7, TAK1-1KKB and TRAF-3 to IRF-3 to transcript and induce the formation of cytokines and IFN. The long dsRNA binding to PKR causes dimerization and transphosphorylation results in the formation of phosphorylated eIF2α and IκB; which cause the inhibition of translation and nuclear translocation, respectively. PKR phosphorylation also activates via p38 MAPK and STAT1 and 2 (through ORF-9) process the transcription through stimulation of IFN & ISG genes in the nucleus
Lysosomal TLRs are engaged to induce TNF-α, and inflammatory cytokine and IFN-α stimulation is associated with advanced signals. TIR family provides a platform for MyD88, which is the conserved central node for innate immune signaling. MyD88 DDs form a structure with IL-1 receptor-associated kinase (IRAK DD) denoted as ‘Myddosome’. TIR domain groups-TRIF, TRAF and p13k deem to recognize a plethora of RNAs from viral and bacterial pathogens, through binding and oligomerization of MyD88 adaptor. IFN-β and IFN-α upregulate the IRF7 and other IFN-inducible genes. This process allows the production of additional IFN-α, which amplifies the responses in a series of cycles. TLR3 gets activated through TRIF that activates receptor interacting protein-1 (RIP-1) and TRAF6, leading to the downstream production of NFkB, activating transcription factor (ATF) and c-Jun transcription factors finally inducing the inflammatory cytokines IL-6 and TNF-α (Fig. 4) [15, 54].
Innate immune signalling (TLR-independent)
The cytoplasmic sensors other than TLRs such as binding protein kinase-R (PKR) or retinoic acid inducible gene 1 (RIG-1) proteins perform via TLR-independent signaling to recognize the extrinsic siRNAs in viral infections [55, 56]. siRNA construct with blunt ends is responsible for IFN induction upregulated in an autocrine and paracrine manner.
RIG-1 contains two caspase recruitment domains (CARD) near its N-terminus that signals the activation of IRFs and NFkB. The dsRNA indulges into C-terminal of helicase domain eventually leading to the production of IFNβ and inflammatory mediators [57, 58]. RIG-1 adaptor proteins viz. IFN-β promotor stimulator 1(IPS-1), mitochondrial antiviral signaling (MAVS), virus-induced signaling adaptor (cardif or VISA), and fas-associated protein with death domain (FADD) adaptor produce the signals through RIG-1 for dsRNA. It performs the downstream activation of IPS-1 adaptor and the upstreaming of IRF7 for the production of type l IFN [59,60,61,62,63]. Protein kinase-R (PKR) pathway activates the signal transduction by proinflammatory stimuli, including bacterial lipopolysaccharide (LPS), TNF-α and IL-1. PKR is a component of the inhibitors of kappa B kinase complex that phosphorylate ELF-2α to play a catalytic role in its activation to inhibit translation. The stress-activated protein kinases p38 and c-Jun NH (2)-terminal kinase (JNK) are also regulated by PKR to induce the production of proinflammatory cytokines [64]. The higher sensitivity in kinase assays has revealed that siRNAs are able to mediate some level of PKR activation [65, 66]. The RNA recognition by cytoplasmic receptors RIG-1 and PKR are considered to be sequence-independent and RIG-1 is also involved in the induction of IFN through NFkB (Fig. 4).
The uncapped 5′-triphosphate groups of RNA bind to the activated RIG-1, consequently aggressive IFN response is developed in the cells expressing RIG-1 [67, 68]. These groups on RNA usually represent the pathogen-associated molecular pattern (PAMP), which is different from host RNA. siRNA constructs are synthesized using phage polymerase which usually activates RIG-1 to induce interferon, even in the absence of TLR7/8 expression e.g. engineered T7-siRNA help remove the initiation 5′-triphosphate synthesis, thereby alleviating interferon induction by this class [69]. The blunt-ended siRNA can also activate RIG-1. However, asymmetrical siRNA design having standard 3′ overhang at one end and 5′-antisense ends have become more widely used siRNAs to improve the potency of RNAi therapeutics [23, 70].
Immunostimulatory siRNA
The stimulation of TLR and non-TLR sensing pathways largely depend upon the RNA sequences presented in the process. The unmodified, blunt-ended siRNA activate TLR7/8 through endosomes and RIG into the cytoplasm to get the end product as type I IFN, which could excel the associated toxicities due to off-target effects [58, 65]. Cell lines of lung fibroblasts, MRC-5 or glioblastoma T98G express a strong level of RIG-1 response. Moreover, the innate signaling is mostly influenced by the used modification chemistries and delivery carriers.
The chemical modifications of siRNA using 2′-F, 2′-OMe or 2′-deoxyribonucleotides to substitute the purines and pyrimidines in a specific sequence manner could provide minimal immunostimulatory activity in-vivo (mice model) in lipid formulations. Incorporation of two 2′-OMe guanosine or uridine residues in the sense strand of highly immunostimulatory siRNA molecule is sufficient to minimise the IFN and inflammatory cytokines induction in-vitro (human PBMCs) and in-vivo (mice model) [71]. This largely inhibits TLR 7/8 pathway. The anti-inflammatory effects of RNA can be achieved by using either 2′OMe-uridine, -guanosine, or -adenosine residues in any combination; however, 2′OMe-cytidine combination have not shown much effect to lower down the immune stimulation [71, 72]. Nucleotide modifications using 2′OMe in antisense strand (with < 20% nucleotides) are generally well tolerated and could have an impact on gene silencing. The selective modification eliminates the subtle immune response, nevertheless, could lead to the production of antibodies in mice models [71, 73]. 2′OMe substitution at position 9 of sense strand reduces the efficiency to assemble on RISC, which might affect the RNAi mechanism. But this principle does not comply with all the RNA duplexes [74]. Therefore, it is critical to make changes on the antisense strand of siRNAs to minimize immunostimulatory activities.
Human TLRs get activated by the pathogen-derived RNAs. In addition, the modified nucleotides can occur naturally or by using chemicals and could reduce the innate response by antagonizing the TLR and RIG activities [72, 75]. However, DNA nucleotides incorporated at the blunt ends of siRNA still induce inflammatory response through TLR7/8-mediated immune stimulation. Locked nucleic acid contains ′O,4′-C methylene bridge in the sugar ring which is reported to display the partially reduced immunostimulatory activities with increased stability, but the siRNA containing inverted deoxy abasic end caps can retain the immunostimulatory activity (Table 1) [10]. Of note, 2′-fluoro and 2’-O methyl have unpredictable effects/or bystander effects on the immunostimulatory activities of modified siRNA.
Table 1 Typical features of immunostimulatory siRNAs to induce TLR and RIG immune signalling in the cytosol resulting in production of interferon and cytokines
The non-viral origin of ssRNA induces TLR dependent production of cytokines, which could also detect the RNA virus infections [13]. In addition, dsRNA can also lead to the direct stimulation of innate immune cells giving the indication of viral infection. Guanosine (G) and uridine (U)-rich ssRNA oligonucleotides derived from HIV-1 stimulate the DCs and macrophages to secrete IFNα, proinflammatory and regulatory cytokines. TLR-deficient mice showed that murine TLR7 and human TLR8 mediate the species-specific recognition of GU-rich ssRNA [14]. TLR and MyD88 are required to sense ssRNA viruses like vesicular stomatitis virus (VSV) and influenza and able to stimulate IFN-α in-vivo. Hence, TLR and MyD88 have an important role in receptor recognition of a wide range of pathogenic viruses to build immunity against ssRNA viruses [76]. AU-rich oligoribonucleotides (ORNs) mediate the human TLR8 activation, while GU-rich ORNs mediate the TLR7/8 activation. GU and AU rich ORNs stimulate the TLR-dependent innate and adaptive immune effects that would be beneficial against cancer, allergic and infectious diseases. Forsbach et al. identified the sequences stimulated the production of both cytokines and TNF-α, suggesting the existence of RNA motifs specifically for both or the single receptors. The motif analysis defined specific GU-rich 4-mer sequences such as, UUGU, GUUC, GUUU, UUUC, UGUU or UCUC activating the human TLR7/8 by inducing IFN, proinflammatory cytokines and chemokines from cells expressing only TLR7 or both TLR 7 and TLR8. On the other hand, AU-rich sequences such as AUGU, UAUA, AUAU, AUAC, UAUU, AAAU, CUAC, GUAC or UAUC were found to induce the strongest TNFα response, but not IFN stimulating monocytes and mDCs and not pDCs [77]. Host driving of CpG dinucleotide elimination at RNA level is a unique phenomenon in vertebrates. ssRNA with specific sequence motifs of AU- or GU-rich and CpG dinucleotides flanked by AU can significantly stimulate the antiviral immune response by secreting type I IFN (Table 1) [78].
It is critical to assess the valid immune response after siRNA treatment and to monitor IFN-α, cytokine IL-6 and TNF-α. However, IFNβ, IL-6, IL-8 and other chemokines in the supernatant of siRNA-treated cells have been used to monitor the activities through RIG-1/MDA5 and PKR pathways. Notably, the cytokines secreted through cell-lines may not indicate precise molecular pathways. The systemic administration of immunostimulatory siRNA formulations with a specific delivery system elevates the cytokines in 1–2 h [58, 65, 79]. Nevertheless, the negative cytokine interpretation always requires utmost care. For instance, lower level of IFN-α detection in the liver or spleen of treated mice does not manifest the systemic cytokine response. It suggests no immunotoxicity in mice, but is associated with off-target effects of gene expression via non-specific antiviral and antitumor activities [80].
Efficacy of siRNA could be identified by using green fluorescent protein (GFP), reporter systems and bacterial invasion. For instance, a chimeric luciferase-CCR5 gene by high-throughput assay is used for quantifying the expression, and level of luciferase would help determine the efficiency of the shRNA clones [[81](/article/10.1007/s13346-023-01361-4#ref-CR81 "Pang S, Pokomo L, Chen K, Kamata M, Mao SH, Zhang H, Razi E, An DS, Chen ISY. High- throughput screening of effective siRNAs using luciferase-linked chimeric mRNA. PLoS ONE. 2014. https://doi.org/10.1371/journal.pone.0096445
.")\]. The pharmacokinetic and biodistribution studies along with route of administration of siRNA drugs are the important parameters to be considered to control the magnitude of immune response and safety.Toxigenic effects and non-specific activities
The non-targeted siRNA (~ 21 nucleotides) suppressed the choroidal neovascularization (CNV) in mice-model in comparison to siRNA targeting vascular endothelial growth factor-A/VEGFA Receptor-1 (VEGFA/VEFAR1), without off-target RNAi and IFN αβ activation, reported by Kleinman et al. [27]. CNV suppression is performed via activation of TLR3 and TRIF inducing IFN and IL-12. Human choroidal endothelial cells expressing TLR3 coding variant 412FF may induce cytotoxicity, providing a direct clue to use this tool for personalized pharmacogenetic therapy. TLR 3 expressed in multiple human endothelial cells indicating that generic siRNA could treat 8% world’s population with CNV disorder, could also produce the immunostimulatory effects [27]. TLR-mediated response is the major cause for stimulating the immune cells and inflammatory cytokines. However, it can also activate the cytoplasmic RNA sensors to produce an effective response, especially in non-immune cells. The strong interactions of siRNA with nucleic acid sensors always results in inflammatory outcome; therefore, it is critical to abrogate this particular property in the candidate siRNA to develop safe and effective therapeutic [58]. Judge et al. reported that siRNA used with non-viral delivery vehicle can also act as potent stimulator for interferons and inflammatory cytokines production in-vivo (mice) and in-vitro (human blood culture). The achieved toxicity levels depend upon the used sequence. It is important to design siRNA with immunostimulatory motifs which could provide effective silencing through RNAi inducing minimal immune activation [11].
Off-target gene silencing has been noticed for increasing hepatotoxicity using GalNAc-siRNA (modified) conjugates. The gene silencing is mediated by miRNA in between the siRNA and mistargeted RNA. Furthermore, the disorganised nucleotides in the antisense strands without changing 2′OMe, 2′-F or PS/ or putting GNA at 7′ position could edge off both off-target effects and hepatotoxicity. These modifications affect the binding of siRNA with another target mRNA in a seed region-specific manner.
The immunostimulatory standards are to be prepared using unmodified siRNA with known immunostimulatory traits and considered for in-vivo quality test to analyse the siRNA integrity. Similarly, the modified siRNAs are also required to validate for precise efficacy with diminished lethal effects for both test and control and also to be compared with unmodified siRNA [7, 82]. The dsRNAs, irrespective of their GU contents, stimulate the I IFN induction in plasmocytoid dendritic cells (pDCs). Immunostimulatory motif in the sense strand exerts the immunostimulation and targets the silencing effect. Mice injected with immunostimulatory siRNA complexed with cationic liposomes are able to produce a response equal to TLR9 ligand CpG with IFN-α in T-cells and dendritic cells of spleen. On the other hand, the immunostimulatory effect was not noticed in TLR7-deficient mice. Therefore, TLR7-based immune recognition in a sequence specific manner could also be opted as an additional biological activity for the characterization of immunostimulatory siRNA [10].
The ssRNA induce TNF-α and IFN-α in human PBMCs. Activated macrophages to activate immunostimulatory TLR7, if treated with interferon γ could suppress the expression of TLR 7 by RNAi reduced the sensing of all immunostimulatory ssRNAs [88]. The bifunctional siRNA harbours both proinflammatory and specific silencing activities. miRNA with conserved uridine bulge design in human cells and can also produce the silencing efficiency. The increased cytokine production enhances the immunostimulatory activity protecting against Semliki Forest Virus infection (in-vitro); therefore, TLR8 and TLR7 get modified and become immunomodulatory in nature. The bifunctional D-siRNA strategy can be applied to any siRNA application, along with emerging CpG-siRNA delivery strategy which could modulate the immune cells to work against various viral infections and other cancer like diseases [90].
Systemic administration of synthetic siRNA duplex always ended up with high inflammatory innate response with IFN and cytokines; largely contribute to reducing down the overall efficacy [10, 11]. siRNA treatments have also been described to be efficacious in-vivo studies including influenza A [83]; herpes simplex virus [84]; respiratory syncytial virus and parainfluenza [85]; hepatitis B virus in mice and HepG cells [7, 89]; ebola in non-human primates [86] and SARS in monkeys and mice [87].
Conclusion
siRNA therapeutics have the biggest advantage over other small peptides and monoclonal therapeutics; as it executes base pairing with mRNA specifically to perform the required duty right on time. But monoclonal antibodies and other peptides need a spatial conformation of the target molecule to neutralize them. Therefore, the higher activity, specificity and affinity will not be identified. On the other hand, the gene of interest can be directly targeted by siRNA having the right nucleotide sequence. RNAi modalities confer a shorter time-period for research and development while eliminating the contamination issues observed by using animal products, in comparison with peptides and monoclonal antibodies. The diseases and other genetic disorders were left untreatable in the past due to lack of advancement in technology, but it is feasible now by using siRNA therapeutics [9, 91, 92].
RNAi opens new avenues for the therapeutic development industry, despite having its extensive clinical applications yet to be revealed. More studies are required to develop appropriate delivery carriers, erasing the associated toxicities, associated costs and other biological barriers for siRNA therapeutics [26]. The extensive strategies can be made to use RNAi applications for studying genes and their consecutive expressions e.g. producing animals encoding shRNAs (similar to siRNAs) or the use of viral vectors. However, the biggest advantage of RNAi applications is in the development of therapeutics. Different drug targets like p53, caspase 2 protein (CASP2), protein kinase N3 (PKN3), β2-adrenergic receptor, mutated kirsten rat sarcoma viral oncogene (KRAS) and microRNAs could be utilized, with various routes of administration such as ocular, intravenous, subcutaneous, intra-tumoural etc. Using appropriate siRNA modification portfolios with specific molecular geometries, delivery systems and optimization of effective dose (at zero toxicity level) with an increased half-life (from minutes to months to years) would help design the well-informed therapeutic interventions of siRNA for humans and animals against various genetic, cancer and infectious diseases in the foreseeable future.
Availability of data and materials
Not applicable.
Abbreviations
ASGPR:
Asialoglycoprotein
CARD:
Caspase recruitment domains
CNV:
Choroidal neovascularization
FADD:
Fas-associated protein with death domain
IRF:
Interferon regulatory factor
IRAK-1:
IL-1 receptor-associated kinase-1
IPS-1:
IFN-β promotor stimulator-1
JNK-c:
Jun NH (2)-terminal kinase
MAVS:
Mitochondrial antiviral signalling
MyD88:
Myeloid differentiation primary response 88
MDA5:
Melanoma differentiation-associated protein
MAPK:
Mitogen-activated protein kinase
NF-κβ:
Nuclear transcription factor
ORNs:
Oligoribonucleotides
OAS:
2′-5′-Oligoadenylate synthetase
PAMP:
Pathogen-associated molecular pattern (PAMP)
PhpC-6′ :
Phenylpyrrolocytosine
PKR:
Protein kinase-R
RIG-1:
Retinoic acid-inducible gene 1
RIP-1:
Receptor-interacting protein-1
RISC:
RNA-induced silencing complex
STAT:
Signal transducer and activators of transcription protein
TAK1:
TGF-β active kinase
TANK:
TRAF family member-associated NFkB activator
TBK1:
TANK-binding kinase-1
tcDNA:
Tricyclo DNA
TIR:
Toll/interleukin-1 receptors
TRAF6:
TNFR-associated factor 6
TRIF:
TIR domain-containing adaptor inducing interferon β
TRBP-TAR (trans activation response):
RNA binding protein
VEGF:
Vascular endothelial growth factor
VISA or CARDIF:
Virus-induced signalling adaptor
References
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;19,391,6669:806–11. https://doi.org/10.1038/35888.
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19(5):517–29. https://doi.org/10.1101/gad.1284105.
Article CAS PubMed Google Scholar - Bartel DP. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. https://doi.org/10.1016/s0092-8674(04)00045-5.
Article CAS PubMed Google Scholar - Crooke ST, Wang S, Vickers TM, Shen W, Liang XH. Cellular uptake and trafficking of antisense oligonucleotides. Nat Biotechnol. 2017;35(3):230–7.
Article CAS PubMed Google Scholar - Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16(11):1833–40.
Article CAS PubMed Google Scholar - Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Kariko K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39(21):9329–38. https://doi.org/10.1093/nar/gkr586.
Article CAS PubMed PubMed Central Google Scholar - Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, Vaish N, Zinnen S, Vargeese C, Bowman K, Shaffer CS, Jeffs LB, Judge A, Ian MacLachlan, Polisky B. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23(8):1002–7.
Article CAS PubMed Google Scholar - Maier MA, Jayaraman M, Matsuda S, Liu Ju, Barros S, Querbes W, Tam YK, Ansell SM, Kumar V, Qin J, Zhang X, Wang Q, Panesar S, Hutabarat R, Carioto M, Hettinger J, Kandasamy P, Butler D, Rajeev KG, pang B, Charisse K, Fitzgerald K, Mui BL, Du X, Cullis P, Madden TD, Hope MJ, Manoharan M, and Akinc A,. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol Ther. 2013;21(8):1570–8.
Article CAS PubMed PubMed Central Google Scholar - Hu B, Zhong L, Weng Y, Peng L, Huang Y, Zhao Y, Liang XJ. Therapeutic si RNA: state of the art. Signal Transduct Target Ther. 2020;5:101.
Article CAS PubMed PubMed Central Google Scholar - Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, Enders S, Hartmann G. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–70.
Article CAS PubMed Google Scholar - Judge AD, Sood V, Shaw JR, Fang D, Mcclintock K, Maclachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–62.
Article CAS PubMed Google Scholar - Zarember KA, Godowski PJ. Tissue expression of human Toll-like receptors and differential regulation of Toll-like receptor mRNAs in leukocytes in response to microbes, their products, and cytokines. J Immunol. 2002;168(2):554–61.
Article CAS PubMed Google Scholar - Diebold SS, Kaisho T, Hemmi H, Akira S, Reise Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–31.
Article CAS PubMed Google Scholar - Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–9.
Article CAS PubMed Google Scholar - Deguine J, Barton GM. MyD88: a central player in innate immune signaling. F1000Prime Rep. 2014;4, 6, 97.
- Weng Y, Xiao H, Zhang J, Liang XJ, Huang Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol Adv. 2019;37(5):801–25.
Article CAS PubMed Google Scholar - Huang YY. Approval of the first-ever RNAi therapeutics and its technological development history. Prog Biochem Biophys. 2019;46(3):313–22.
Google Scholar - Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(185–197):26.
Google Scholar - Liu J, Guo N, Gao C, Liu N, Zheng X, Tan Y, Lei J, Hao Y, Chen L, Zhang X. Effective gene silencing mediated by polypeptide nanoparticles LAH4-L1-siMDR1 in multi-drug resistant human breast cancer. J Biomed Nanotechnol. 2019;15(3):531–43.
Article CAS PubMed Google Scholar - Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27.
Article CAS PubMed PubMed Central Google Scholar - Sioud M, Furset G, Cekaite L. Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochem Biophys Res Commun. 2007;361(1):122–6.
Article CAS PubMed Google Scholar - Song X, Wang X, Ma Y, Liang Z, Zhenjun Y, Cao H. Site-specific modification using the 2′-methoxyethyl group improves the specificity and activity of siRNAs. Mol Ther Nucleic Acids. 2017;9:242–50.
Article CAS PubMed PubMed Central Google Scholar - Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–8.
Article CAS PubMed Google Scholar - Coelho T, Adam D, Silva A, Lozeron P, Hawkins PN, Mant T, Perez J, Chiesa J, Warrington S, Tranter E, Munisamy M, Falzone R, Harrop J, Cehelsky J, Bettencourt BR, Geissler M, Butler JS, Sehgal A, Meyers RE, Chen Q, Borland T, Hutabarat RM, Clausen VA, Alvarez R, Fitzgerald K, Gamba-Vitalo C, Nochur SV, Vaishnaw AK, Sah DWY, Gollob JA, Suhr OB. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med. 2013;369(9):819–29.
Article CAS PubMed Google Scholar - Adams D, Ganzalez-Duarte A, O’Riordan WD, Yang CC, Ueda M, Kristen AV, Tournev I, Schmidt HH, Coelho T, Berk JL, Lin KP, Vita G, Attarian S, Plante’-Bordeneuve V, Mezei MM, Campistol JM, Buades J, Brannagan TH, Kim BJ, Oh J, Parman Y, Sekijima Y, Hawkins PN, Solomon SD, Polydefkis M, Nochur SV, Sweetser M, Garg PP, Vaishnaw AK, Gollob JR and Suhr OB,. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med. 2018;379(1):11–21. https://doi.org/10.1056/NEJMoa1716153.
Article CAS PubMed Google Scholar - Titze-de-Almeida R, David C, Titze-de-Almeida SS. The race of 10 synthetic RNAi-based drugs to the pharmaceutical market. Pharm Res. 2017;34(7):1339–63.
Article CAS PubMed Google Scholar - Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, Albuquerque RJC, Yamasaki S, Itaya M, Pan Y, Appukuttan B, Gibbs D, Yang Z, Kariko K, Ambati BK, Wilgus TA, DiPietro LA, Sakurai E, Zhang K, Smith JR, Taylor EW, Ambati J. Sequence and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452(7187):591–7.
Article CAS PubMed PubMed Central Google Scholar - Garba AO, Mousa SA. Bevasiranib for the treatment of wet, age-related macular degeneration. Ophthalmol Eye Dis. 2010;2:75–83. https://doi.org/10.4137/OED.S4878.
Article CAS PubMed PubMed Central Google Scholar - Shen W, De Hoyos CL, Migawa MT, Vickers TA, Sun H, Low A, Bell TA, Rahdar M, Mukhopadhyay S, Hart CE, Bell M, Riney S, Murray SF, Greenlee S, Crooke RM, Liang Xh, Seth PP, Crooke ST. Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat Biotechnol. 2019;37:640–50.
Article CAS PubMed Google Scholar - Migawa MT, Shen W, Wan WB, Vasquez G, Oestergaard ME, Low A, De Hoyos CL, Gupta R, Murray S, Tanowitz M, Bell M, Nichols JG, Gaus H, Liang XH, Swayze EE, Crooke ST, Seth PP. Site-specific replacement of phosphorothioate with alkyl phosphonate linkages enhances the therapeutic profile of gapmer ASOs by modulating interactions with cellular proteins. Nucleic Acids Res. 2019;47(11):5465–79.
Article CAS PubMed PubMed Central Google Scholar - Marshall WS, Caruthers MH. Phosphorodithioate DNA as a potential therapeutic drug. Science. 1993;259:1564–70.
Article CAS PubMed Google Scholar - Ndeboko B, Ramamurthy N, Lemamy GJ, Jamard C, Nielsen PE, Cova L. Role of cell-penetrating peptidesin intracellular delivery of peptides nucleic acids targeting hepadnaviral replication. Mol Ther Nucleic Acids. 2017;9:162–9. https://doi.org/10.1016/j.omtn.2017.09.003.
Article CAS PubMed PubMed Central Google Scholar - Singh RP, Oh BK, Choi JW. Application of peptide nucleic acid towards development of nanobiosensor arrays. Bioelectrochemistry. 2010;79:153–61.
Article CAS PubMed Google Scholar - Parmar R, Willoughby JLS, Liu J, Foster DJ, Bringham B, Theile CS, Charisse K, Akinc A, Guidry E, Pei Y, Strapps W, Cancilla M, Stanton MG, Rajeev KG, Sepp-Lorenzino L, Manoharan M, Meyers R, Maier MA, Jadhav V. 5′-(E)-vinylphosphonate: a stable phosphate mimic can improve the RNAi activity of siRNA-GalNAc conjugates. ChemBioChem. 2016;17(11):985–9.
Article CAS PubMed Google Scholar - Haraszti RA, Roux L, Coles AH, Turanov AA, Alterman JF, Echeverria D, Godinho BMD, Aronin N, Khvorova A. 5-Vinylphosphonate improves tissue accumulation and efficacy of conjugated siRNAs in vivo. Nucleic Acids Res. 2017;45(13):7581–92.
Article CAS PubMed PubMed Central Google Scholar - Khvorova A, Watts JK. The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol. 2017;35:238–48.
Article CAS PubMed PubMed Central Google Scholar - Fucini RV, Haringsma HJ, Deng P, Flanagan WM, Willingham AT. Adenosine modification may be preferred for reducing siRNA immune stimulation. Nucleic Acid Ther. 2012;22(3):205–10.
Article CAS PubMed PubMed Central Google Scholar - Shen X, Corey DR. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018;46:1584–600.
Article CAS PubMed Google Scholar - Watts JK. Locked nucleic acid: tighter is different. Chem Commun (Camb). 2013;49(50):5618–20. https://doi.org/10.1039/c3cc40340h. Epub 2013 May 16.
Article CAS PubMed Google Scholar - Phelps KJ, Ibarra-Soza JM, Tran K, Fisher A, Beal PA. Click modification of RNA at adenosine: structure and reactivity of 7-ethynyl- and 7-triazolyl-8-aza-7-deazaadenosine in RNA. ACS Chem Biol. 2014;9(8):1780–7.
Article CAS PubMed PubMed Central Google Scholar - Wahba AS, Azizi F, Deleavey GF, Brown C, Robert F, Carrier M, Kalota A, Gewirtz AM, Pelletier J, Hudson RHE, Damha MJ. Phenylpyrrolocytosine as an unobtrusive base modification for monitoring activity and cellular trafficking of siRNA. ACS Chem Biol. 2011;6(9):912–9.
Article CAS PubMed Google Scholar - Zhang J, Zheng J, Lu C, Du Q, Liang Z, Xi Z. Modification of the siRNA passenger strand by 5-nitroindole dramatically reduces its off-target effects. ChemBioChem. 2012;13(13):1940–5.
Article CAS PubMed Google Scholar - Wu SY, Chen TM, Gmeiner WH, Chu E, Schmitz JC. Development of modified siRNA molecules incorporating 5-fluoro-2′-deoxyuridine residues to enhance cytotoxicity. Nucleic Acids Res. 2013;41(8):4650–9.
Article CAS PubMed PubMed Central Google Scholar - Peacock H, Kannan A, Beal PA, Burrows CJ. Chemical modification of siRNA bases to probe and enhance RNA interference. J Org Chem. 2011;76(18):7295–300.
Article CAS PubMed PubMed Central Google Scholar - Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: tools and applications. Drug Discov Today. 2008;13:842–55.
Article CAS PubMed Google Scholar - Dar SA, Thakur A, Qureshi A, Kumar M. siRNA mod: a database of experimentally validated chemically modified siRNAs. Sci Rep. 2016;6:20031.
Article CAS PubMed PubMed Central Google Scholar - Nair JK, Attarwala H, Sehgal A, Wang Q, Aluri K, Zhang X, Gao M, Liu J, Indrakanti R, Schofield S, Kretschmer P, Brown CR, Gupta S, Willoughby JLS, Boshar JA, Jadhav V, Charisse K, Zimmermann T, Fitzgerald K, Manoharan M, Rajeev KG, Akinc A, Hutabarat R, Maier MA. Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc-siRNA conjugates. Nucleic Acids Res. 2017;45(19):10969–77.
Article CAS PubMed PubMed Central Google Scholar - Foster DJ, Brown CR, Shaikh S, Trapp C, Schlegel MK, Qian K, Sehgal A, Rajeev KG, Jadhav V, Manoharan M, Kuchimanchi S, Maier MA, Milstein S. Advanced siRNA designs further improve in vivo performance of GalNAc-siRNA conjugates. Mol Ther. 2018;26(3):708–17.
Article CAS PubMed PubMed Central Google Scholar - Wooddell CI, Blomenkamp K, Peterson RM, Subbotin VM, Schwabe C, Hamilton J, Chu Q, Christianson DR, Hegge JO, Kolbe J, Hamilton HL, Branca-Afrazi MF, Given BD, Lewis DL, Gane E, Kanner SB, Teckman JH. Development of an RNAi therapeutic for alpha-1-antitrypsin liver disease. JCI Insight. 2020;5(12): e135348.
Article PubMed PubMed Central Google Scholar - Pirher N, Pohar J, Manček-Keber M, Benčina M, Jerala R. Activation of cell membrane-localized Toll-like receptor 3 by siRNA. Immunol Lett. 2017;189. https://doi.org/10.1016/j.imlet.2017.03.019.
- Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14.
Article CAS PubMed Google Scholar - Peters KL, Smith HL, Stark GR, Sen GC. IRF-3-dependent, NFkappa B- and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc Natl Acad Sci USA. 2002;99(9):6322–7.
Article CAS PubMed PubMed Central Google Scholar - Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K, Akira S. Cutting edge: a noveloll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor. J Immunol. 2002;169(12):6668–72.
Article CAS PubMed Google Scholar - Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–7.
Article CAS PubMed Google Scholar - Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23(1):19–28.
Article CAS PubMed Google Scholar - Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishil KJ, Yomaguchi O, Otsu K, Tsujimura T, Koh CS, eSousa CR, Matsura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–5.
Article CAS PubMed Google Scholar - Hiscott J, Lin R, Nakhaei P, Paz S. MasterCARD: a priceless link to innate immunity. Trends Mol Med. 2006;12:53–6.
Article CAS PubMed Google Scholar - Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19(2):89–101.
Article CAS PubMed Google Scholar - Kawai T, Takahashi K, sato s, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O and Akira S,. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–8.
Article CAS PubMed Google Scholar - Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–82.
Article CAS PubMed Google Scholar - Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager r and Tschopp J,. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–72.
Article CAS PubMed Google Scholar - Xu LG, Wang YY, Han KJ, LI LY, Zhai Z, and Shu HB,. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–40.
Article CAS PubMed Google Scholar - Balachandran S, Venkataraman T, Fisher PB, Barber GN. Fas-associated death domain-containing protein-mediated antiviral innate immune signaling involves the regulation of Irf7. J Immunol. 2007;178:2429–39.
Article CAS PubMed Google Scholar - Williams BR. Signal integration via PKR. Sci. STKE 2001. 2001;89:RE2.
- Marques JT, Devosse T, Wang D, Zamaniandaryoush M, Serbinowski P, Hartmann R, Fujita T, Devosse MA, Williams BR. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–65.
Article CAS PubMed Google Scholar - Puthenveetil S, Whitby L, Ren J, Kelnar K, Krebs JF, Beal PA. Controlling activation of the RNAdependent protein kinase by siRNAs using site-specific chemical modification. Nucleic Acids Res. 2006;34(17):4900–11.
Article CAS PubMed PubMed Central Google Scholar - Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis SC. RIGI-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314(5801):997–1001.
Article CAS PubMed Google Scholar - Ebert G, Poeck LJ, Baschuk N, Esser K, Esposito I, Hartmann G, Protzer U. 5′ Triphosphorylated small interfering RNAs control replication of hepatitis B virus and induce an interferon response in human liver cells and mice. Gastroenterology. 2011;141(2):696–706.
Article CAS PubMed Google Scholar - Kim DH, Longo M, Han Y, Lungberg P, Cantin E, Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol. 2004;22:321–5.
Article CAS PubMed Google Scholar - Kim DH, Behlke MA, Rose SD, Change MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–6.
Article CAS PubMed Google Scholar - Judge AD, Bola G, Lee AC, Maclachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505.
Article CAS PubMed Google Scholar - Kariko K, Buckstein M, NI H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–75.
Article CAS PubMed Google Scholar - Judge AD, Robbins M, Tavakoli I, Levi J, Hu L, Fronda A, Ambegia E, Mcclintock K, Maclachlan I. Confirming the RNAi-mediated mechanism of action of siRNAbased therapeutics in mice. J Clin Invest. 2009;119:661–73.
Article CAS PubMed PubMed Central Google Scholar - Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–20.
Article CAS PubMed PubMed Central Google Scholar - Zamanian-Dryoush M, Marques JT, Gantier MP, Behlke MA, John M, Rayman P, Finke J, Williams BR. Determinants of cytokine induction by small interfering RNA in human peripheral blood mononuclear cells. J Interferon Cytokine Res. 2008;28(4):221–33.
Article Google Scholar - Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA. 2004;101:5598–603.
Article CAS PubMed PubMed Central Google Scholar - Forsbach A, Nemorin JG, Montino C, Muller C, Samulowitz U, Vicari AP, Jurk M, Mutwiri GK, Kreig AM, Lipford GB, Vollmer J. Identifi cation of RNA sequence motifs stimulating sequence specific TLR8-dependent immune responses. J Immunol. 2008;180(6):3729–38.
Article CAS PubMed Google Scholar - Cheng X, Virk N, Chen W, Ji S, Ji S, Sun Y, Wu X. CpG usage in RNA viruses: data and hypotheses PLoS One. 2013;8(9): e74109.
CAS PubMed Google Scholar - Robbins M, Judge A, Liang L, Mcclintock k, Yaworski E, and Maclachlan I,. 2′-O-methylmodified RNAs Act as TLR7 Antagonists. Mol Ther. 2007;15(9):1663–9.
Article CAS PubMed Google Scholar - Geiss G, Jin G, Guo J, Bumgarner R, Katze MG, Sen GC. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J Biol Chem. 2001;276:30178–82.
Article CAS PubMed Google Scholar - Pang S, Pokomo L, Chen K, Kamata M, Mao SH, Zhang H, Razi E, An DS, Chen ISY. High- throughput screening of effective siRNAs using luciferase-linked chimeric mRNA. PLoS ONE. 2014. https://doi.org/10.1371/journal.pone.0096445.
Article PubMed PubMed Central Google Scholar - Robbins M, Judge A, Ambegia E, Choi C, Yaworski E, Plamer L, Maalintock L, Maclachlan I. Misinterpreting the therapeutic effects of siRNA caused by immune stimulation. Hum Gene Ther. 2008.
- Tompkins SM, Lo CY, Tumpey TM, Epstein SL. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc Natl Acad Sci USA. 2004;101(23):8682–6.
Article CAS PubMed PubMed Central Google Scholar - Palliser D, Chowdhury D, wang QY, Lee SJ, Bronson RT, Knipe DM, and Lieberman J,. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439(7072):89–94.
Article CAS PubMed Google Scholar - Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–5.
Article CAS PubMed Google Scholar - Geisbert TW, Lee ACH, Robbins M, Geisbert JB, Honko AN, Sood V, Johnson JC, de Jong S, Tavakoli I, Judge A, Hensley LE, Maclachlan I. Post exposure protection of non-human primates against a lethal Ebola virus challenged with RNA interference: a proof-of-concept study. Lancet. 2010;375(9729):1896–905.
Article CAS PubMed PubMed Central Google Scholar - Li BJ, Tang Q, Cheng D, Qin C, Xie FY, Wei Q, Xu J, Liu Y, Zheng BJ, Woodle MC, Zhang N, Lu PY. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat Med. 2005;11:944–51.
Article CAS PubMed PubMed Central Google Scholar - Gantier MP, Tong S, Behlke MA, Xu D, Phipps S, Foster PS, Williams BRG. TLR7 is involved in sequence specific sensing of single stranded RNAs in Human Macrophages. J Immunol. 2008;180(4):2117–24.
Article CAS PubMed Google Scholar - Chen X, Qianb Y, Yana F, Tu J, Yang X, Xing Y, Chen Z. 5′-Triphosphate-siRNA activates RIG-I-dependent type I interferon production and enhances inhibition of hepatitis B virus replication in HepG2.2.15 cells. Eur J Pharmocol. 2013;721(1–3):86–95.
- Gantier MP, Tong S, Behlke MA, Irving AT, Laapas M, Nilsson UW, Latz E, McMillan NAJ, Williams BRG. Rational design of immunostimulatory siRNA. Mol Ther. 2010;18(4):785–95.
Article CAS PubMed PubMed Central Google Scholar - Hu B, Weng Y, Xia XH, Liang XJ, Huang Y. Clinical advances of siRNA therapeutics. J Gene Med. 2019;21(7): e3097.
Article PubMed Google Scholar - Cho WG, Albuquerque RJC, Kleinman ME, Tarallo V, Greco A, Nozaki M, Green MG, Baffi JZ, Ambati BK, Falco MD, Alexander JS, Brunetti A, Falco SD, Ambati J. Small interfering RNA-induced TLR3 activation inhibits blood and lymphatic vessel growth. PNAS. 2009;106(17):7137–42.
Article CAS PubMed PubMed Central Google Scholar - Diebold SS, Massacrier C, Akira S, Paturel C, Morel Y, Reis SC. Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur J Immunol. 2006;36:3256–67.
Article CAS PubMed Google Scholar - Kabilova TO, Meschaninova MI, Venyaminova AG, Nikolin PV, Zenkova MA, Vlassov VV, Chernolovskaya EL. Short double stranded RNA with immunostimulatory activity: sequence dependence. Nucleic Acid Ther. 2012;22(3):196–204.
Article CAS PubMed Google Scholar - Han Q, Zhang C, Zhang J, Tian Z. Reversal of hepatitis B virus-induced immune tolerance by an immunostimulatory 3p-HBx-siRNAs in a retinoic acid inducible gene I-dependent manner. Hepatology. 2011;54(4):1179–89.
Article CAS PubMed Google Scholar
Acknowledgements
The author has conveyed her sincere gratitude to scientists and researchers who had worked in RNAi technology and contributed their valuable results in literature. The author expresses her sincere thanks to all the expert reviewers for their valuable time and suggestions for shaping up this manuscript.
Author information
Authors and Affiliations
- New Zealand Organization for Quality-Member, Auckland, New Zealand
Anju Kaushal
Authors
- Anju Kaushal
You can also search for this author inPubMed Google Scholar
Contributions
The author has contributed to the preparation of manuscript.
Corresponding author
Correspondence toAnju Kaushal.
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
The author has read and approved the final manuscript for publication.
Competing interests
The author declares no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kaushal, A. Innate immune regulations and various siRNA modalities.Drug Deliv. and Transl. Res. 13, 2704–2718 (2023). https://doi.org/10.1007/s13346-023-01361-4
- Accepted: 29 April 2023
- Published: 23 May 2023
- Issue Date: November 2023
- DOI: https://doi.org/10.1007/s13346-023-01361-4